Abstract
Solid-state nanopore (SSN)-based analytical methods have found abundant use in genomics and proteomics with fledgling contributions to virology – a clinically critical field with emphasis on both infectious and designer-drug carriers. Here we demonstrate the ability of SSN to successfully discriminate adeno-associated viruses (AAVs) based on their genetic cargo [double-stranded DNA (AAVds-DNA), single-stranded DNA (AAVss-DNA) or none (AAVEmpty)], devoid of digestion steps, through nanopore-induced electro-deformation (characterized by relative current change; ΔI/I0). The deformation order was found to be AAVEmpty > AAVssDNA > AAVdsDNA. A deep learning algorithm was developed by integrating support vector machine with an existing neural network, which successfully classified AAVs from SSN resistive-pulses (characteristic of genetic cargo) with >95% accuracy – a potential tool for clinical and biomedical applications. Subsequently, the presence of AAVEmpty in spiked AAVds-DNA was flagged using the ΔI/I0 distribution characteristics of the two types for mixtures composed of ~75:25 and ~40:60 (in concentration) AAVEmpty: AAVds-DNA.
Keywords: Adeno-associated virus, solid-state nanopores, deep learning, support vector machine, resistive pulse sensing
Graphical Abstract
Solid-state nanopore based electro-deformation coupled with deep learning to distinguish AAV particles based on their cargo content
INTRODUCTION
With over 200 clinical studies globally and the recent FDA approval of Luxturna – the first approved gene therapy in the United States to treat hereditary blindness1, 2 – adeno-associated virus (AAV) vectors are gaining substantial traction in viral gene therapy. One considerable challenge in the translation of AAV vectors, once produced, is the difficulty of characterizing the vectors based on their transgene packaging. Key characterization metrics of AAV include titer (capsid and genome titers), exact genomic content [single-stranded (ssDNA) versus self-complementary double-stranded (dsDNA) and overall genome length], and heterogeneity of a vector preparation (empty versus full capsids). To obtain these metrics, a combination of multiple assays has to be performed, including quantitative polymerase chain reaction (qPCR) or droplet digital PCR (ddPCR) for genomic titer,3 enzyme-linked immunosorbent assay (ELISA) for capsid titer, and analytical ultracentrifugation for vector prep composition.4 Alarmingly, the variability associated with vector characterization assays was revealed through a blind study in which AAV samples were sent to several groups for vector tittering using qPCR and ELISA. The mean and standard deviation (SD) for genomic and capsid titer were 3.82×1010±2.97×1010 and 9.43×1011±3.19×1011, respectively.5 These are highly concerning outcomes especially for dose-dependent therapeutics where overdosing, for example through underestimation of the empty capsids, could trigger unexpected immune responses. The genomic content could also be analyzed through alkaline gel assay or a Southern blot which, however, requires overnight runs and are semi-quantitative at best. In this study, we demonstrate the use of solid-state nanopores (SSNs) – low-cost, ostensibly simple, low-sample requiring, high sensing throughput, label-free single-molecule sensor class – to characterize each vector type based on electro-deformation, discriminate between the vectors, and flag the presence of empty capsid from a mixture (a critical development step towards quality assurance of AAV preps).
A solitary nano-scale aperture that spans an impervious membrane (biological or solid-state) separating two electrolyte reservoirs – a nanopore – has been used to characterize a myriad of biomolecules6–10 and particles11–14, nanoparticles15, 16 and synthetic polymers17 using a multitude of molecular level features6, 18, 19 and membrane mechanical properties (i.e., stiffness, deformability).20, 21 However, compared to the plethora of DNA and protein studies, the virus footprint in the nanopore community is surprisingly meager perhaps because nanopores were mostly recognized for small molecule analysis (driven by potential commercial interests) while studies on filamentous,22 rod-like,23, 24 and spherical viruses12, 25 have redrawn the boundaries of nanopore technology. With the emergence of new viral threats challenging the very fabric of human existence, the importance of developing low-cost, high throughput, portable technologies for diagnostic purposes has gained substantial focus with the dawn of 2020. While biological nanopores have been used to sequence the genome of viruses such as Zaire Ebola26 – emphasizing the clinical importance of this technology – our proposed method using SSN would analyze the virus particles, devoid of digestions steps, and would eventually pave the way for rapid assessment of the genetic cargo and the purity of an AAV prep. In this work, we designed and tested a silicon nitride (SixNy) based SSN device to characterize three AAV vector types – empty (AAVEmpty), AAV with ssDNA (AAVss-DNA) and AAV with dsDNA (AAVds-DNA) – using the demonstrated ability of SSN to estimate the electro-deformation of soft nanoparticles in response to an electric field11, 12 and numerical predictions to quantify the deformation. In addition, we used a deep convolutional neural network to classify AAVs based on their cargo from the resistive pulse data. Machine learning approaches have been shown to distinguish biomolecules using ionic current-time waveforms.27–29 The deep neural network used here was developed by modifying an existing residual neural network (ResNet50) with a support vector machine. The electro-deformation was apparent through the voltage trend of the relative-current change originating from particle transit (ΔI/I0) – a departure from the linear (Ohmic) nature was observed. The extent to which a given particle deforms is a function of its spring constant for which both membrane mechanical properties and intra-particle properties such as transgene packaging are paramount. For example, an AAVEmpty is expected to deform more than a cargo-carrying counterpart of the same serotype. Therefore, the deformation characteristics of the three AAV types are expected to be fundamentally different and we intend to use this property to discriminate each type. The expected deformation order is AAVEmpty > AAVssDNA > AAVdsDNA, which as will be shown later, agrees well with our obtained results. Such discrimination would be useful to flag the presence of AAVEmpty in a sample consisting of genetic cargo-carrying AAV capsids. This is important as previous studies demonstrated that a high dose of AAV vector can cause severe toxicity which may be triggered by the high capsid dose or cargo expression.30, 31 Therefore, it is especially crucial in dose-dependent studies to know not only the AAV concentration but also the composition of the vector distribution to reduce the delivery of excessive AAV capsids.
RESULTS AND DISCUSSION
Basic operation principle of a nanopore is outlined in Figure 1a where the analyte (AAV in this case) is added to the cis side and a voltage (negative for AAV) is applied to the trans side to drive the analyte across the nanopore from the cis to the trans side. This perturbs the open-pore ionic current stamping particle specific information. All experiments were conducted with <10 nM AAV – this minimal sample usage complements the tedious AAV preparation methods. Given the AAV size (~25 nm in diameter), SSN is an obvious requirement since the narrowest constriction of ubiquitous biological nanopores such as α-hemolysin and MspA are not wide enough for such a particle to transit. A rich blend of fabrication techniques are accessible to us, such as controlled dielectric breakdown (CDB)32, 33, focused ion beam (FIB)34, 35 and transmission electron microscope (TEM)36. Since it is difficult to fabricate larger diameter nanopores using CDB due to non-opening failure among other factors37, and preliminary studies with FIB fabricated pores produced poor event frequencies, we ultimately fabricated nanopores of ~100 nm in diameter through ~12 nm thin free-standing SixNy on silicon using TEM (Figure S1a). Any pore showing significant current rectification was discarded and only those with rectification ~1 was used (Figure S1b). Although the nanopore devices are low-cost and high throughput sensors (in general), the fabrication method (both the membrane and the pore) would, to a large extent, govern the overall cost associated with the device. Since, TEM-based nanopore fabrication methods are not as high throughput as other methods such as controlled dielectric breakdown, we note, the workflow could be limited by the pore fabrication step. However, if this limitation could be overcome, the overall throughput (combination of the fabrication and sensing time scales) and cost of the nanopore device could be improved significantly. The voltage polarity used for AAV translocations herein (−20 mV to −175 mV; Figure 1b) has an added advantage of being immune to any DNA contaminations during AAV preparation (i.e., any DNA that did not get encapsidated) since DNA would only respond to a positive voltage bias at this operational electrolyte chemistry (2M LiCl buffered at pH~7). All experiments were in triplicate (unless otherwise noted) with unique nanopores of comparable size and each nanopore was discarded after running a given virus type to avoid any cross-contamination. On average, a minimum of 500 resistive pulses were recorded at −20 mV whereas a minimum of 1000 resistive pulses were recorded at subsequent voltages.
Figure 1:
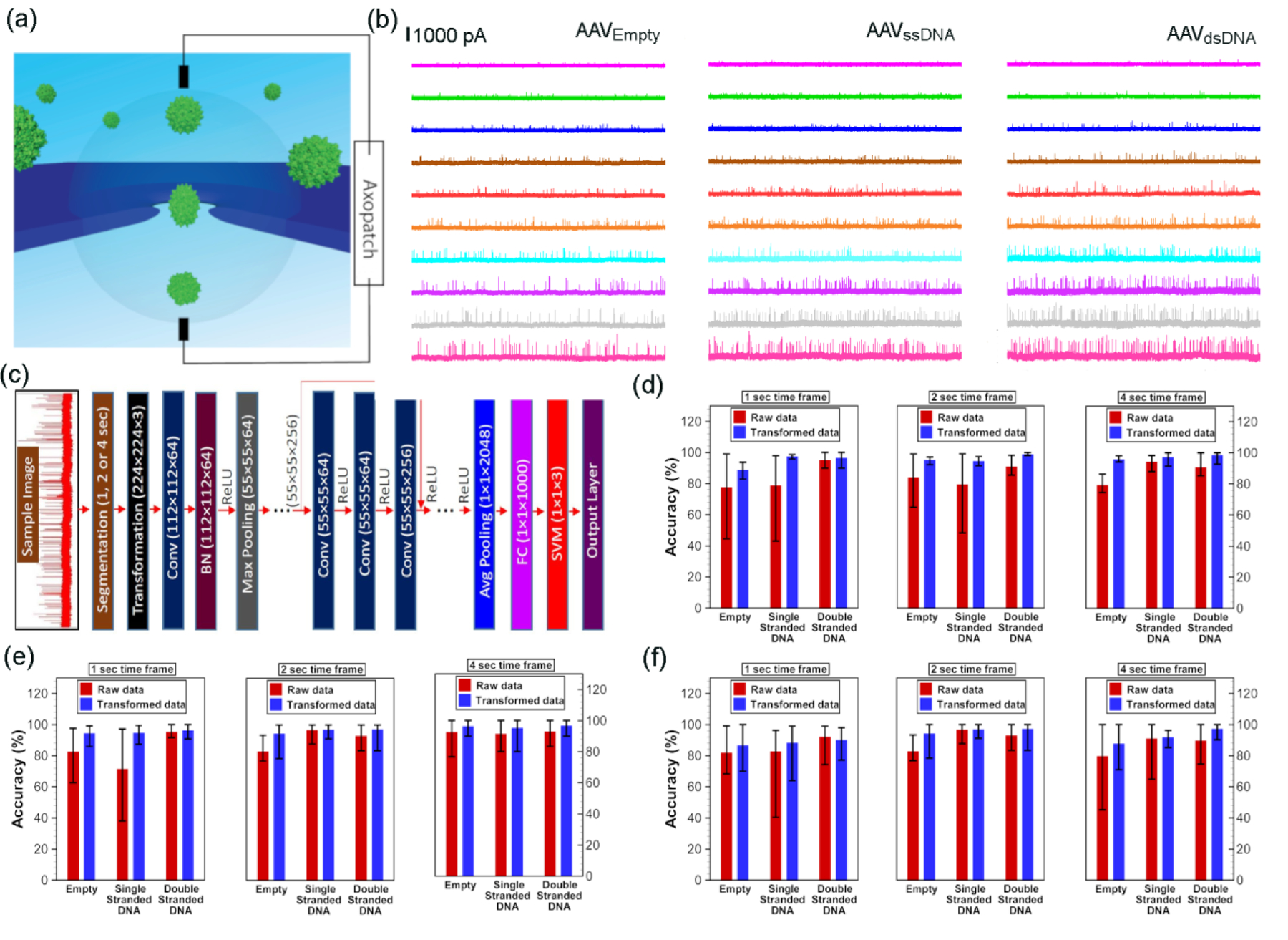
(a) A schematic representation of a typical nanopore setup where the analyte is driven across the nanopore in response to an applied electric field stamping analyte-specific resistive-pulses as shown in (b). (b) Representative 10 second current traces corresponding to the translocation of “AAVEmpty (left column)”, “AAVssDNA (middle column)” and, “AAVdsDNA (right column)” at −20 mV (magenta), −30 mV (green), −40 mV (blue), −50 mV (brown), −60 mV (red), −75 mV (orange), −100 mV (cyan), −125 mV (purple), −150 mV (gray) and −175 mV (pink). (c) Architecture of the deep convolutional neural network used for the classification of AAVs based on the genetic cargo. All current signals (images) obtained from solid-state nanopore experiments were segmented based on time frames (1, 2, or 4 sec) and resized to have the dimensions of 224×224×3 to comply with ResNet50 requirements before those images are inputted into the network. Both raw data as well as transformed data was used for the classification, where the Conv: convolutional block; BN: batch normalization; ReLU: rectified linear activation unit; Max Pooling: maximum pooling; FC: fully connected layer; SVM: support vector machine; Avg Pooling: average pooling. Efficacy of deep convolutional neural networks in classification of AAVs from nanopore experimental data was obtained for the applied bias voltages of (d) −175 mV, (e) −150 mV and (f) −100 mV using 1 sec, 2 sec and 4 sec frames. For a particular class, 80% of the data were randomly selected from all images of that class for training the network, while the rest of the images of that class were used for validation. Mean and error bars were obtained from training and validation of the model by randomly selecting the training and validation data set for 10 times.
Figure 1: OriginLab 2018b, TecPlot 9.0, GIMP 2.10.8
The question that intrigued us was, can the resistive pulses (each resistive pulse corresponds to a single AAV particle translocating through the nanopore) shown in Figure 1b be used to characterize and distinguish each AAV class? For this, we used a deep neural network (DNN) framework initially which is based on an existing deep residual network (ResNet50 – an award-winning platform developed Microsoft for ImageNet38) as shown in Figure 1c. Since ResNet50 is not trained for virus detection, we had to make necessary changes to our network by modifying the last few steps from the ResNet50. Therefore, the output features of fully connected layer (1000 1×1) of the ResNet50 are fed into a support vector machine (SVM) as shown in Figure 1c. Thus, based on the extracted features of the fully connected layer (fc1000) of ResNet50, we trained a multiclass SVM using the one-versus-one method39 for three different classes (see Deep Neural Network for Classification of Virus Cargoes from Electrical Signal Section under Methods for more details). Classification results obtained from our deep learning model are shown in Figures 1d–f for AAVs translocation data at voltage −175 mV, −150 mV and −100 mV (see SI Table S1 for the number of images used in each class for different time frames). Our results show that accuracy (Fig. 1d–f) can be improved by transforming (see Methods for transformation of data) the experimental data. Although maximum accuracy is sometimes higher for raw data than transformed data, the mean accuracy for any class (or any time frame) is always lower for raw data than that of transformed data (Figures 1d–f). The transformed data has always outperformed the raw data because distinct features of signals are more preserved in transformed data. Unlike classical machine learning problem, we used a few experimental data for training (for a given applied electric field), but segmentation of each experimental signal helped us to attain our desired goal in the data-driven classification. The accuracy of the prediction is improved significantly for 4 second time frame data, even though the number of images used to train the support vector machine is much smaller for 4 second case compared to 1 or 2 second cases. Further increase in the time frame window would probably help us to get a better prediction, but one must be mindful of the reduction in the training data with an increase in the time frame. Our model results show that as long as our deep learning algorithm is trained with the appropriate data, we can get accurate prediction despite the fact that data-based techniques such as machine learning is never 100% accurate. The reason for no false positive (or negative) in our proposed method is due to flexibility of testing multiple frames from a single experiment by segmenting resistive-pulse data into hundreds of smaller time frames. This comprehensive analysis indicates that our approach to identify AAVs based on electric current signal is robust, and this algorithm can be used to detect viruses quickly from SSN experiments.
After successfully identifying each AAV class using our DNN model, we then investigated the possibility of using relative current change (ΔI/I0) to discriminate each AAV type as this metric is dependent on the membrane rigidity.21, 40 The scatter plots of ΔI/I0 versus translocation time and histograms corresponding to ΔI/I0 are shown in Figures 2a–c. Each of the histograms were fitted with a single Gaussian function (see SI Section 1 for the histogram and fitting details). The behaviour of ΔI/I0 with voltage for each AAV class as shown in Figure 2d, is indicative of electro-deformation due to deviation from the Ohmic linear scaling (see the associated discussion of SI Section 5 for more details). A single AAV can only house a single DNA molecule.41, 42 Iodixanol ultracentrifugation can effectively separate the empty capsids formed during production,43, 44 thus reducing the complexity arising from the number of DNA copies inside an AAV– either a given particle will have a single copy of the intended genome package or it will not. The deformation profiles (ΔI/I0 versus applied voltage) as seen in Figure 2d indicate that electro-deformation follow an inverse relationship with cargo content: lesser the void within the AAV (i.e., higher the volume occupied by the cargo material), lesser would be the deformation (AAVEmpty > AAVssDNA > AAVdsDNA). At higher voltages, we see that the differences in the deformation profiles become less, and almost within error at ~−175 mV. If the trends of the deformation profiles seen in Figure 2d continue, one would pragmatically not expect to see any discernible differences between each virus type (based on the genetic cargo) at voltages >−175 mV. We then plotted the percentage difference of the ΔI/I0 of each AAV type referenced to AAVempty (, see SI Section 2 for the definition). The trends of Figure 2e show a sharp drop in at voltages above −125 mV. The AAVds-DNA and AAVss-DNA showcased an averaged of ~19.9±2.7% and ~7.8±2.1%, respectively up to −125 mV. It is not surprising to see AAVds-DNA having the greatest difference with respect to AAVempty (i.e., highest ) as it is expected to deform the least. It typically takes ~2 hours in total to acquire the minimum event count for all voltages noted previously (at least 500 for −20 mV and 1000 for the rest). Among these voltages, all AAV types showed >1 resistive pulses/s at ≥−50 mV. Taking the useful voltage regime for electro-deformation-based discrimination (≤−125 mV) and appreciable event frequency (≥−50 mV), one can bracket −50mV to −125 mV as the optimized voltage range for this study. Consequently, it merely takes ~30 minutes in total to acquire ~1000 events for all voltages for this optimized voltage range – a testament to the sensing throughput of the nanopore platform.
Figure 2:
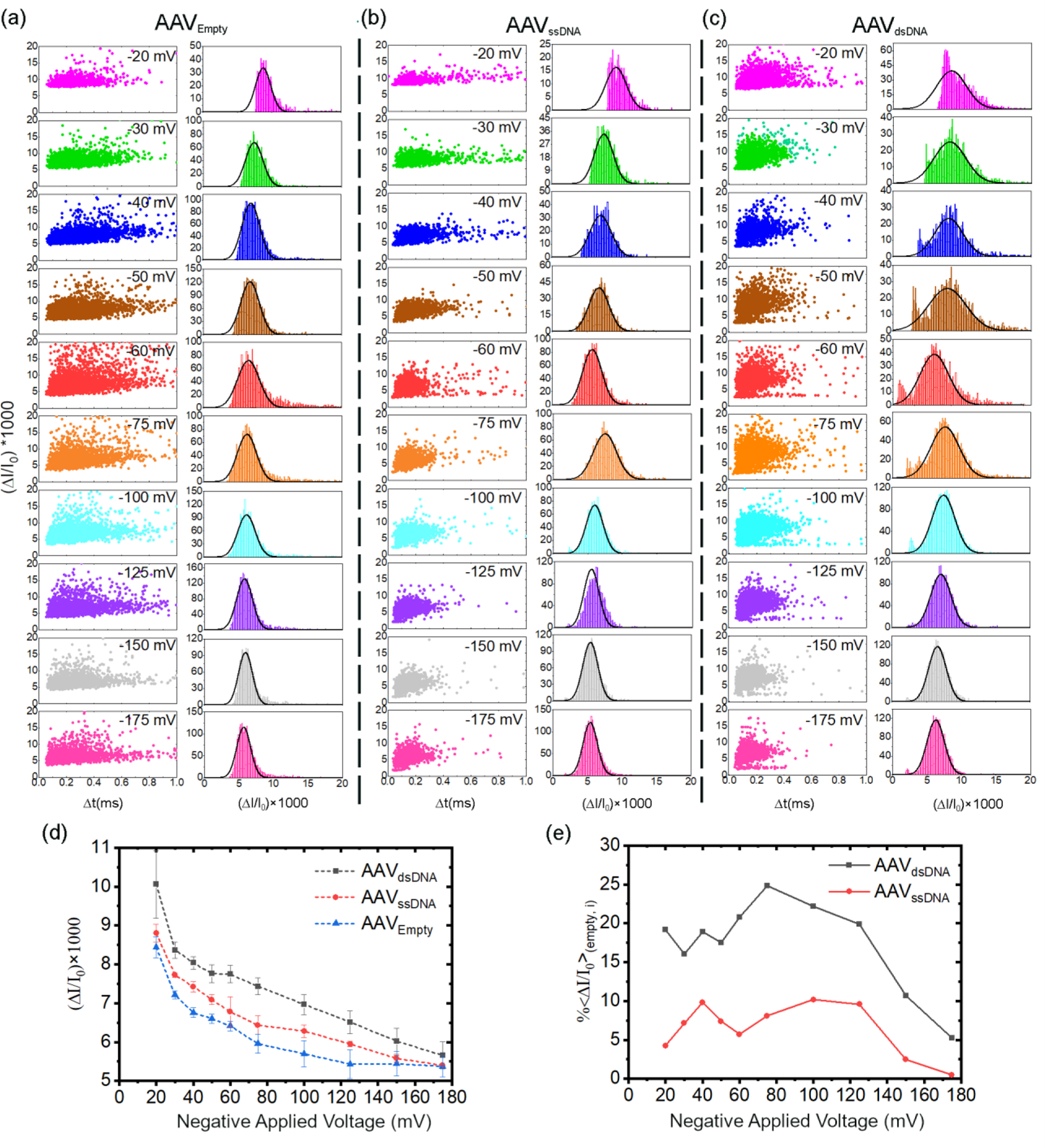
Scatter plots of change in relative current change (ΔI/I0) versus translocation time and, corresponding histograms to the ΔI/I0 (right column) of (a) AAVEmpty, (b) AAVssDNA, (c) AAVdsDNA at −20 mV (magenta), −30 mV (green), −40 mV (blue), −50 mV (brown), −60 mV (red), −75 mV (orange), −100 mV (cyan), −125 mV (purple), −150 mV (gray) and −175 mV (pink). The histograms were fitted with a single Gaussian function (see SI Section 1 for the histogram and fitting details). (d) vs voltage corresponding to AAVds-DNA (black), AAVss-DNA (red) and AAVempty (blue). (e) Percentage difference of AAV encapsulating dsDNA (black) and ssDNA (red) with respect to AAVempty (, calculated using Equation S2). All experiments were in triplicate with unique nanopores and performed using ~98 ± 5 nm diameter nanopores in 2M LiCl (buffered at pH~7 using 10 mM Tris buffer).
Figure 2: OriginLab 2018b, GIMP 2.10.8
To numerically model the deformation of each AAV type, we used an immersed interface method (IIM), which has been developed and validated earlier.45–47 The IIM can estimate the electric potential distribution inside the nanopore geometry, which is used to calculate electric current at any particular section using , where σ is the local conductivity, A is the cross-sectional area, and is the direction vector normal to the particular section. To quantify the extent of deformation, we defined aspect ratio (α) of the virus as the ratio of its equatorial (along the electric field) length over the polar (perpendicular to the electric field) length. For a circular shape, α = 1.0, and it is greater than unity when the virus is deformed in the direction of the applied field46. As shown in Figures 3a–c, when the viruses are allowed to deform with an increasing electric field, we observe consistent nonlinear behavior in the ΔI/I0 due to competing electrostatic and electrophoretic forces on the virus capsid.40, 47 For a properly chosen set of conductivity ratios, the numerical predictions of ΔI/I0 (green circles) fall within the experimental bounds (blue limits) and reveal an interesting power-law like behavior (green dashed line, Figures 3a–c) in all three cases. The corresponding change in the virus shape with increasing potential is presented in terms of the aspect ratio (red diamonds), which shows a linearly increasing behavior with increasing electric field. The slope of this aspect ratio vs. applied voltage plot was found to be decreasing with increasing conductivity ratio (λ) (red dashed line with slopes of 0.0358, 0.0276, and 0.0216 mV−1 for AAVempty, AAVss-DNA, and AAVds-DNA, respectively). The decrease in the slope also corresponds to a higher degree of deformation of the AAV samples. Hence, one can use the slope of the aspect ratio vs. applied voltage plot as a characteristic identifier of each virus type with its signature inner conductivity and deformation attributes.
Figure 3:
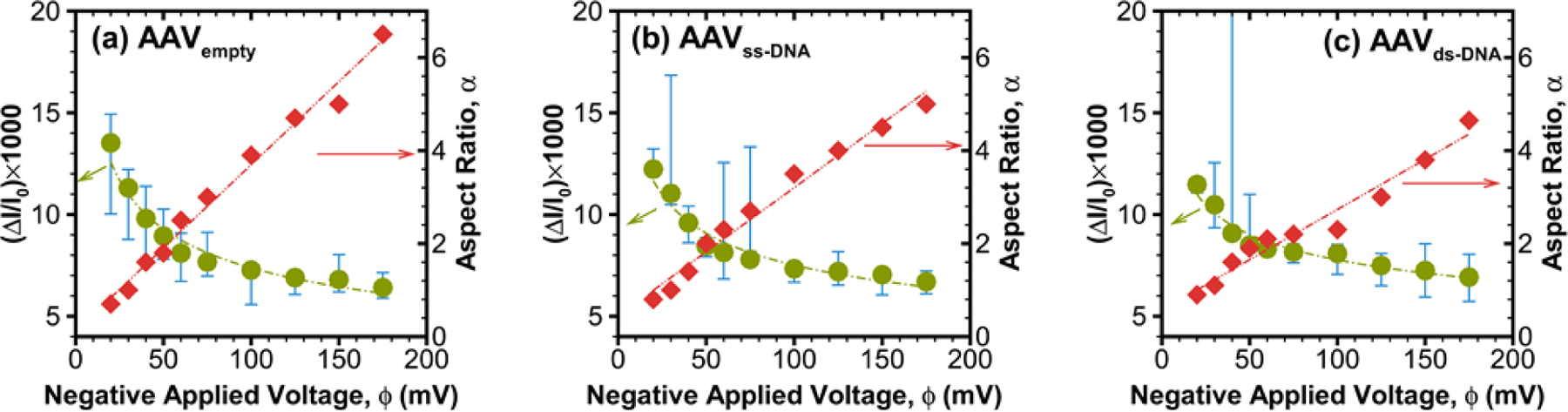
Nondimensional current drop ratio (ΔI/I0) and corresponding aspect ratio (α) with negative applied voltage (ϕ) corresponding to (a) AAVempty, (b) AAVss-DNA and (c) AAVds-DNA. The numerically calculated relative current drop data (green circle) are fitted with a power-law curve drawn in green lines while the estimated aspect ratio data (red diamonds) are fitted with a straight line (red dashed lines). Three sets of independent experimental data are used to calculate the bounding upper and lower values (blue bounding bars) for ΔI/I0.
Figure 3: TecPlot 9.0
We then ventured to mimic a sample of AAVds-DNA contaminated with AAVempty by spiking an AAVds-DNA aliquot with a significant amount of AAVempty (~75% AAVempty and ~25% AAVds-DNA; AAV75:25%). Identification of AAVempty in vector batches is especially important for clinical studies to minimize adverse immune responses in patients. Since, in a real-world sample-scenario, the operator would not have pre-knowledge of such contamination, to stay true to such a situation, we fitted each profile with a single Gaussian rather than two or more Gaussians. Unlike the histograms of AAVds-DNA (Figure 2c), spiked mixtures (Figure 4b and 4c) cannot be well fitted with a single Gaussian – a clear population outside the Gaussian fit exist at higher . The existence of an apparent outlying population compared to the one residing within the Gaussian fit may also serve as a visual and qualitative metric to qualify the presence of a significant AAVempty population in the sample. Thus, one can use the ratio of the population higher and lower than the mean of the Gaussian fit as a metric to flag the presence of AAVempty in each sample: a perfect fit would have a value of 1 for . Looking at Figure 2a, it is evident that AAVempty has a broader distribution along the ΔI/I0 axis. The tail along the ΔI/I0 may indicate the presence of a secondary population, although not as prominent as the lower (ΔI/I0) and denser population (i.e., the ΔI/I0 distribution can be well fitted with a single Gaussian function). However, such a tail along the ΔI/I0 axis is absent in its cargo-carrying counterparts. It could mean, the deformation is more restrictive in the presence of a cargo whereas it is more diverse in the absence of a cargo. Thus, it is not surprising to see AAVds-DNA having a value closer to one (Figure 4d and 4e) whereas AAVEmpty deviating somewhat away from the ideal value. The mixture significantly deviated from the ideal value which could be inextricably linked to the presence of populations corresponding to both AAVds-DNA and AAVempty. It is evident from Figure 4d, the profiles corresponding to AAVds-DNA and AAV75:25% are well separated and indicative of a departure from a AAVds-DNA sample (i.e., presence of a contaminant). We were also able to flag the presence of AAVempty in the mixture using deformation profiles ( vs voltage) as evident by Figure S4 (see SI Section 6 for more details). Using the metric, we were able to flag the presence of AAVempty in a mixture of ~40% AAVempty and ~60% AAVds-DNA (Figure 4e) through the visual separation of the mixture similar to above. The second mixture was deliberately limited to three voltages (−50 to −75 mV) as these three yielded the greatest separation of in the ~75:25% mixture evident from Figure 4d. It is worthwhile noting, the error associated with the profile of AAVds-DNA is much lower compared to the rest, which could also serve as a visual clue to the purity of the sample under investigation. One could potentially expand this study to cover a range of AAVds-DNA:AAVempty ratios and develop a correlation between the AAVempty percentage and as a function of voltage.
Figure 4:
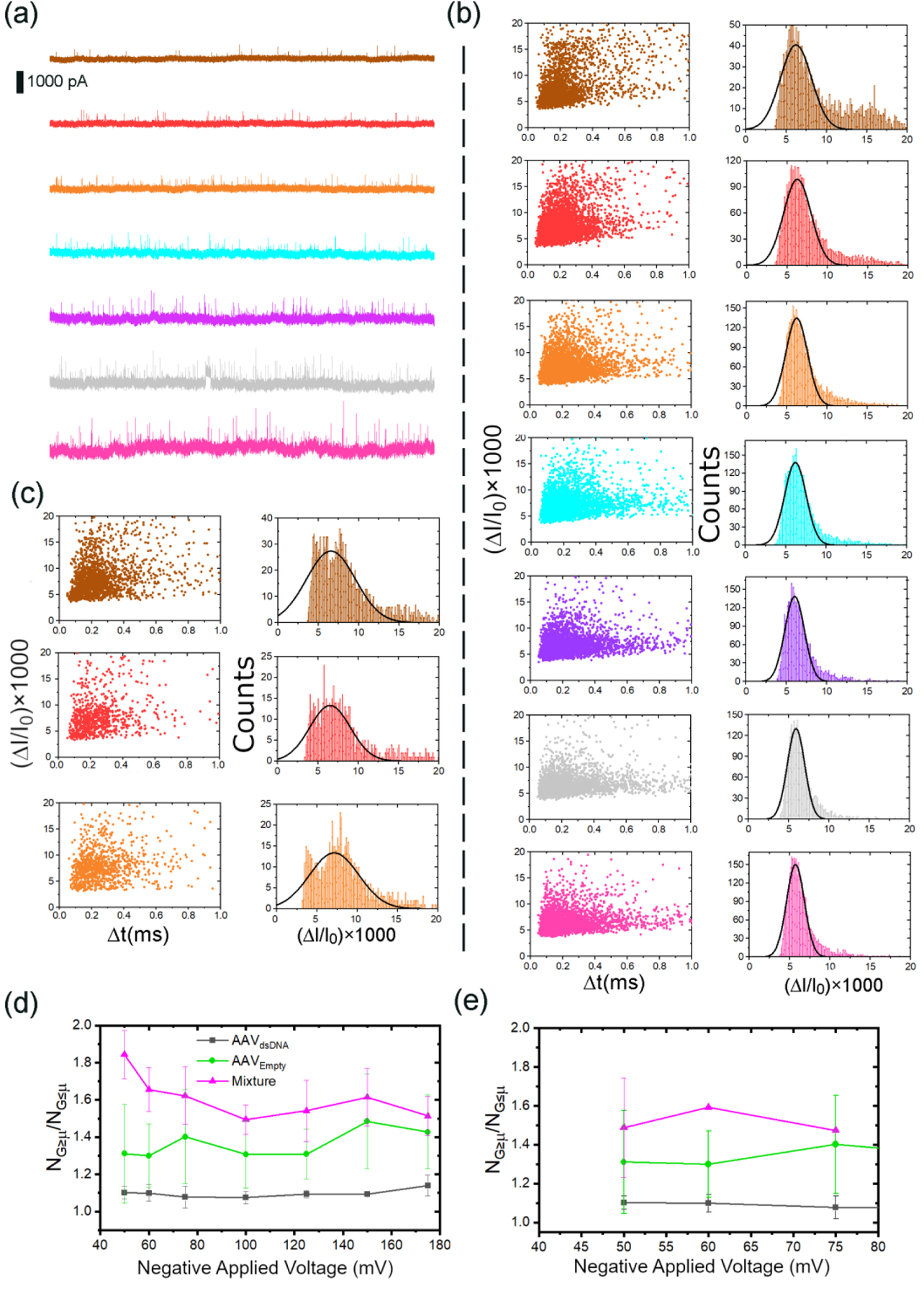
(a) 5-second representative current traces corresponding to a 75:25% molar mixture of AAVEmpty and AAVds-DNA in response to −50 mV (brown), −60 mV (red), −75 mV (orange), −100 mV (cyan), −125 mV (purple), −150 mV (gray) and −175 mV (pink). (b) The corresponding scattered plots and histograms. (c) The scattered plots and histograms of 40:60% molar mixture of AAVEmpty and AAVds-DNA in response to −50 mV, −60 mV, −75 mV. (d)-(e) The ratio of the population above and below the mean of each Gaussian fit corresponding of histograms at each applied voltage corresponding to AAVds-DNA (black), AAVempty (green) and mixtures (magenta) of (d) 75:25% and (e) 40:60% (in molar concentration) of AAVEmpty and AAVds-DNA. Each mixture was measured in duplicate using unique nanopores.
Figure 4: OriginLab 2018b, GIMP 2.10.8
CONCLUSIONS
We have demonstrated the ability of solid-state nanopores of ~100 nm diameter, fabricated using TEM through nominally ~12 nm SixNy membranes, to discriminate AAV based on their genetic cargo (i.e., single-stranded DNA, self-complementary DNA or none). All experiments were conducted using negative voltages and translocations were recorded from ~−20 mV to ~−175 mV in sufficiently small voltage increments. A deep neural network platform, developed based on ResNet50 with appropriate modifications by support vector machine, were used to identify the current profiles of each AAV type. The accuracy of the machine learning prediction can be improved significantly by segmenting each experimental resistive-pulse signal into hundreds of data and running the model for tens of data sets from each experiment. More importantly, the prediction accuracy increases with the length of the time frame (1 sec versus 2 sec versus 4 sec) of experimental data. For transformed data, the mean accuracy of the network was always 90% or higher for any class regardless of the voltage bias or time frame. The electro-deformation was numerically modelled using an immersed interface approach. The model results indicated a power-law behavior for the nondimensional current drop (ΔI/I0) with applied potential for all three cases. Interestingly, The ΔI/I0 profiles with voltage clearly showed distinct deformation patterns for each AAV type with deformation being more prominent as the internal cavity of AAV is less occupied by its cargo: . The average percentage ΔI/I0 with respect to was, as expected, higher for than with the two having an averaged value of ~19.9±2.7% and ~7.8±2.1% respectively, up to −125 mV. Since displayed the highest difference, we ventured to see if could be flagged from a mixture of and . Other than the difference associated with another significant difference in the distribution of ΔI/I0 is being more Gaussian than with the latter having a tail along higher ΔI/I0 values. This feature was used to successfully flag the presence of in mixtures of ~75% AAVempty and ~25% AAVds-DNA and ~40% AAVempty and ~60% AAVds-DNA. Taken together, SSN platforms along with their advantages such as low cost and sample requirement, rapid analysis, user friendliness with minimal training requirement (as seen with other nanopore technologies) could potentially transform the method discussed herein to a widely accessible tool to profile and discriminate each AAV class and to flag the presence of AAVempty which could be crucial for minimizing safety issues with human gene therapy.
METHODS
AAV Production
AAV particles were produced using HEK293T cells (ATCC) using 25 kDa linear polyethylenimine (PEI, Thermo) mediated triple transfection48. Briefly, HEK293T cells were cultured to 70% confluency on 15cm poly-L-lysine coated cell-culture plates using DMEM (LONZA) with 10% FBS (Atlanta Biologics) and 1% penicillin-streptomycin (Gibco). Adenovirus helper genes (pXX6–80), AAV9 rep-cap (pAAV2/9), and a transgene cassette plasmid (self-complement or single-stranded GFP) were mixed in a 1:1:1 molar ratio with the PEI transfection mix and allowed to incubate at room temperature for 30 minutes before adding to cells. The cell pellet was harvested 48 hours after transfection and underwent three cycles of freeze-thaw followed by benzonase treatment before purification using iodixanol (OptiPrep) step gradient (15%, 25%, 40%, 54%) ultracentrifugation. The 40% fraction was extracted, followed by concentration and buffer exchange using Amicon 150 kDa MWCO filtration unit (Millipore-Sigma) into GB-buffer (50mM Tris, pH 7.6, 150mM NaCl, 10mM MgCl2). Concentration of virus particles was established using qPCR using primers against cytomegalovirus (CMV) promoter (forward: TCACGGGGATTTCCAAGTCTC, reverse: AATGGGGCGGAGTTGTTACGAC) on the transgene cassette. The empty capsids were collected from the layer between the 25% and 40% fraction of the iodixanol column, and concentration was measured using western blot with B1 antibodies against a standard of AAV9 particles.
Nanopore Electrical Measurements
All electrical measurements were conducted using Ag/AgCl electrodes connected to an Axopatch 200B (Molecular Devices LLC, USA). The data were acquired at 250 kHz (except I-V measurements which were done at 10 kHz), filtered using the inbuilt 4-pole Bessel low-pass filter at 10 kHz setting and digitized either using a BNC 2110 connector block (National Instruments, USA) or 1440A Digitizer (Molecular Devices LLC, USA). For pore diameter measurements the former was used and for other temporal acquisitions, the latter was used. When the BNC 2110 was used, the instrument control was done using custom-coded LabVIEW scripts and pClamp (version 10.6, Molecular Devices LLC., USA) was used otherwise. Before each measurement, the pipette offset setting of the Axopatch 200B was used to nullify the zero-voltage current. The electrodes were prepared in the following manner: a ~2-inch-long Ag wire was sanded to remove any oxide residuals and contaminants on the surface. Then it was dipped in a bleach solution (425044, Sigma Aldrich) for at least one hour (preferably overnight) until the electrode turns black. It is then soldered to a TE connectivity contact gold pin and connected to the head stage of the Axopatch 200B system. The electrodes were checked after each experiment to see whether it has retained its color or whether it has turned white. The latter indicates that the electrode needs to be sanded down and put in the bleach solution for it to function as a reversible electrode.
Nanopore Fabrication
Nanopores were fabricated through as supplied silicon nitride chips (NBPX5001Z-HR, Norcada, Canada) that are nominally ~12 nm thick using TEM (JEM-2100F, JEOL, Japan) at 200 keV as described previously (see Figure S1 for a representative TEM image of a pore and its current-voltage curve).49 The size was initially validated through TEM as shown in Figure S1 and subsequently crosschecked with Equation 1.
Nanopore Characterization
The fabricated nanopore chips were mounted between two Teflon half cells using PDMS gaskets to be watertight. Each chamber can hold ~450 µL of electrolyte. The schematic diagram of the cell is shown in Figure S2. The chambers were initially filled with ethanol (A4094, Fisher Scientific), placed in a vacuum desiccator and connected to a mechanical pump to remove the air bubbles along the channel connecting the chip and the electrolyte reservoir. Upon the appearance of bubbles from both the channels, the pump was disconnected, and the system was brought to atmospheric pressure gently to avoid re-entry of air bubbles. The content was then thoroughly exchanged with ultra-pure water followed by 1 M KCl (P9333, Sigma-Aldrich, USA) buffered at pH~7 (phosphate buffer saline, P5493, Sigma-Aldrich, USA). A voltage ramp of +200 mV to −200 mV is then applied to acquire a current-voltage (I-V) curve. The I-V curve was then linearly fitted and the slope (G) was used to estimate the nanopore size using,
| (1) |
where , , and are the ionic conductance, electrolyte conductivity, nanopore length, and diameter, respectively. If the pore is not properly wet, the I-V curve would either showcase a significantly less than the expected G value. Thus, all pores, before usage were subjected to a 2-second +8 V pulse to ensure proper-wetting.
Nanopore Electrolyte Preparation
All electrolytes including LiCl (L4408, Sigma-Aldrich, USA) and KCl were dissolved in ultra-pure water (ARS-102 Aries high purity water systems) with ~18 MΩ∙cm resistivity. Each solution contains 10 mM of either tris buffer (J61036, Fisher Scientific, USA) or phosphate buffer saline (P5493, Sigma-Aldrich, USA). The former was used for translocation experiments whereas the latter was used to acquire current-voltage (I-V) curves for pore-diameter estimation. The solutions were then filtered using a filtration system with a Polyethersulfone membrane (S2VPU02RE, Fisher Scientific). Caution: dissolving LiCl in water is an exothermic process. After the electrolyte solution reached the room temperature, the pH was adjusted by adding HCl (H1758, Sigma-Aldrich, USA) or KOH (306568, Sigma-Aldrich, USA) dropwise while gently stirring the electrolyte solution continuously. Caution: these are concentrated solutions and should only be open inside a properly functioning fume hood. Both pH and conductivity of the electrolyte solutions were measured and typically, a 2M LiCl solution at pH ~7 would have a conductivity of ~12 S/m whereas a 1 M KCl solution at pH~7 would have a conductivity of ~11 S/m.
Event Characterization
A custom MATLAB (version 9.4, USA) script was used, where events were characterized as perturbations at least 5 times the standard deviation of the open-pore current. In brief, the code scans through the open-pore current using custom moving windows. This ensures any subtle variations in the open-pore current of a given window is independent of the rest. The window size is typically set as 1/10th the acquisition frequency (100 ms long window). Although larger windows can be used, we have observed the translocation times are mostly <1 ms, thus justifying the moving window size. This is also evident from the scatter plots shown in Figure 2. The average of the data points in the window is then used to calculate a preliminary baseline, and any perturbation that is 5 times the standard deviation of the baseline is flagged and assigned temporarily the value of the baseline. Then using the new values, a secondary baseline is calculated and used as the open-pore baseline of that window. After detecting an event, its duration (Δt), maximum depth (ΔI) and the local baseline (I0) to perform analysis shown in the manuscript.
Image Preparation for Deep Neural Network:
Due to the unavailability of large training data (from experiments), we have segmented the electrical (resistive-pulse) signals of each experiment into 4N, 2N, and N number of images (graphs) depending on the time frames (1, 2, or 4 sec). While it is possible to maintain the x-axis length constant in each image for a particular time frame, keeping the same scale range (the difference between the upper and lower bound of current) was challenging for the y-axis (Fig. 1b) during the auto plotting of graphs. Thus, we have trained and validated the model with two sets of images. The first set of images are plotted (aka raw data) automatically, while the y-axis of the second set was transformed as
| (2) |
where and are upper bound and lower bound of current values, respectively and is the current change. In other words, the second set of graphs (termed as transformed data) are plotted by considering a fixed current change (ΔI) in the vertical axis for all three classes.
Deep Neural Network for Classification of Virus Cargoes from Electrical Signal
We have developed a deep neural network algorithm by modifying last couple of layers of the ResNet50 – a residual deep neural network developed by Microsoft research team. The ResNet50 has been trained for 1000 different classes with 13,000,000 natural images, and it requires a 224×224×3 color image as input for proper identification within its database. However, for our classification problem, we have only three classes based on the cargo inside AAVs: empty, single stranded DNA, and double stranded DNA. Thus, from the extracted features of the fully connected layer (fc1000) of ResNet50, we have trained a multiclass support vector machine (SVM) using the one-versus-one method39 for three different classes. For the three class scenario, the one-versus-one method yields three binary classifiers where each one is trained on data from two classes. For example, to train data from the and the classes, we solved an optimization problem as
| (3) |
where , and are the weight, bias, slack variable, and the penalty parameter, respectively. Eq. (3) is subjected to the following constraints
| (4) |
where is the training data and is the class of . The function maps the training data to a higher dimensional space. In Eq. (3), the penalty (second) term is used to reduce the number of training errors in case the data are not linearly separable, while optimization of the regularization (first) term provides the maximum margin between two classes of data. Thus, the basic concept behind SVM is to find a balance between the regularization term and the training errors.
Based on the optimized weight and bias, scores are calculated for each class from an unseen test/validation image, and the highest score is used for classification of that image. If is the classifier to distinguish a pair of classes (positive examples) and (negative examples), the classification criteria for a new image
| (5) |
Supplementary Material
ACKNOWLEDGEMENTS
This material is based upon work supported by a National Science Foundation Fellowship to W.C. (2018253392). We acknowledge the University of North Carolina at Chapel Hill Gene Therapy Center Vector Core for providing us with pXX6–80 and scAAV2-CMV-GFP, and the University of Pennsylvania Vector Core for providing us with pAAV2/9. The work was partly funded by NSF CMMI 1712069, NIH R03EB022759, and NIH R21GM134544. We would like to thank Prof. Moon J. Kim and Mr. Qingxiao Wang at the University of Texas at Dallas and Mr. Jung Soo Lee at Southern Methodist University for fabricating solid-state nanopores.
Footnotes
Software’s used for Machine Learning
MATLAB 2018b
DECLARATION OF INTERESTS
J.S. is an employee of Biogen as of August 2019.
W.T.C. is an employee of Biogen as of March 2020.
B.I.K is an employee of the Australian National University as of March 2020.
Y.M.N.D.Y.B is affiliated with the Australian National University as of September 2020.
An invention disclosure is filled for the work discussed herein.
REFERENCES
- 1.Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA and Maguire AM, Lancet, 2017, 390, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ledford H, Nature, 2017, 550, 314. [DOI] [PubMed] [Google Scholar]
- 3.Lock M, Alvira MR, Chen S-J and Wilson JM, Human Gene Therapy Methods, 2013, 25, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnham B, Nass S, Kong E, Mattingly M, Woodcock D, Song A, Wadsworth S, Cheng SH, Scaria A and O’Riordan CR, Human Gene Therapy Methods, 2015, 26, 228–242. [DOI] [PubMed] [Google Scholar]
- 5.Lock M, McGorray S, Auricchio A, Ayuso E, Beecham EJ, Blouin-Tavel V, Bosch F, Bose M, Byrne BJ, Cation T, Chiorini JA, Chrarto A, Clark KR, Conlon T, Darmon C, Doria M, Douar A, Flotte TR, Francis JD, Francois A, Giacca M, Korn MT, Korytov I, Leon X, Leuchs B, Lux G, Melas C, Mizukami H, Moullier P, Müller M, Ozawa K, Philipsberg T, Poulard K, Raupp C, Rivière C, Roosendaal SD, Samulski RJ, Soltys SM, Surosky R, Tenenbaum L, Thomas DL, van Montfort B, Veres G, Wright JF, Xu Y, Zelenaia O, Zentilin L and Snyder RO, Human Gene Therapy, 2010, 21, 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saharia J, Bandara YND, Goyal G, Lee JS, Karawdeniya BI and Kim MJ, ACS Nano, 2019, 13, 4246–4254. [DOI] [PubMed] [Google Scholar]
- 7.Karawdeniya BI, Bandara YMNDY, Nichols JW, Chevalier RB and Dwyer JR, Nature Communications, 2018, 9, 3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman KJ, Jürgens M, Prabhu A, Ahn CW, Jemth P, Edel JB and Kim MJ, Analytical Chemistry, 2011, 83, 5137–5144. [DOI] [PubMed] [Google Scholar]
- 9.Plesa C, Verschueren D, Pud S, van der Torre J, Ruitenberg JW, Witteveen MJ, Jonsson MP, Grosberg AY, Rabin Y and Dekker C, Nature Nanotechnology, 2016, 11, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 10.Hagan JT, Sheetz BS, Bandara YND, Karawdeniya BI, Morris MA, Chevalier RB and Dwyer JR, Analytical and Bioanalytical Chemistry, 6, 10. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Saharia J, Bandara YND, Karawdeniya BI, Goyal G, Darvish A, Wang Q, Kim MJ and Kim MJ, Electrophoresis, 2019. [DOI] [PubMed]
- 12.Darvish A, Lee JS, Saharia J, Sundaram RVK, Goyal G, Bandara N, Ahn CW, Kim J, Dutta P and Chaiken I, Electrophoresis, 2018. [DOI] [PMC free article] [PubMed]
- 13.Tsutsui M, Yoshida T, Yokota K, Yasaki H, Yasui T, Arima A, Tonomura W, Nagashima K, Yanagida T, Kaji N, Taniguchi M, Washio T, Baba Y and Kawai T, Scientific Reports, 2017, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsui M, Tanaka M, Marui T, Yokota K, Yoshida T, Arima A, Tonomura W, Taniguchi M, Washio T, Okochi M and Kawai T, Analytical Chemistry, 2018, 90, 1511–1515. [DOI] [PubMed] [Google Scholar]
- 15.Lee JS, Peng B, Sabuncu AC, Nam S, Ahn C, Kim MJ and Kim M, Electrophoresis, 2018, 39, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darvish A, Goyal G, Aneja R, Sundaram RV, Lee K, Ahn CW, Kim K-B, Vlahovska PM and Kim MJ, Nanoscale, 2016, 8, 14420–14431. [DOI] [PubMed] [Google Scholar]
- 17.Robertson JW, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV and Kasianowicz JJ, Proceedings of the National Academy of Sciences, 2007, 104, 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandara YMNDY, Tang J, Saharia J, Rogowski LW, Ahn CW and Kim MJ, Analytical Chemistry, 2019, 91, 13665–13674. [DOI] [PubMed] [Google Scholar]
- 19.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A and Akeson M, Nature Methods, 2007, 4, 315. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Saharia J, Bandara YND, Karawdeniya BI, Goyal G, Darvish A, Wang Q, Kim MJ and Kim MJ, Electrophoresis, 2019, 40, 1337–1344. [DOI] [PubMed] [Google Scholar]
- 21.Darvish A, Lee JS, Peng B, Saharia J, VenkatKalyana Sundaram R, Goyal G, Bandara N, Ahn CW, Kim J and Dutta P, Electrophoresis, 2019, 40, 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMullen A, De Haan HW, Tang JX and Stein D, Nature Communications, 2014, 5, 4171. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Chen Y, Zhou Q, Wang R, Xia B, Ma D, Luo K and Liu Q, Analytical Chemistry, 2016, 88, 2502–2510. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Wu H, Kong J and Liu Q, Science of Advanced Materials, 2013, 5, 2039–2047. [Google Scholar]
- 25.Zhou K, Li L, Tan Z, Zlotnick A and Jacobson SC, Journal of the American Chemical Society, 2011, 133, 1618–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoenen T, Groseth A, Rosenke K, Fischer RJ, Hoenen A, Judson SD, Martellaro C, Falzarano D, Marzi A and Squires RB, Emerging Infectious Diseases, 2016, 22, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi M, ACS Omega, 2020, 5, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arima A, Tsutsui M, Harlisa IH, Yoshida T, Tanaka M, Yokota K, Tonomura W, Taniguchi M, Okochi M and Washio T, Scientific Reports, 2018, 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arima A, Harlisa IH, Yoshida T, Tsutsui M, Tanaka M, Yokota K, Tonomura W, Yasuda J, Taniguchi M, Washio T, Okochi M and Kawai T, Journal of the American Chemical Society, 2018, 140, 16834–16841. [DOI] [PubMed] [Google Scholar]
- 30.Hinderer C, Katz N, Buza EL, Dyer C, Goode T, Bell P, Richman LK and Wilson JM, Human Gene Therapy, 2018, 29, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hordeaux J, Wang Q, Katz N, Buza EL, Bell P and Wilson JM, Molecular Therapy, 2018, 26, 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bandara YMNDY, Saharia J, Karawdeniya BI, Hagan JT, Dwyer JR and Kim MJ, Nanotechnology, 2020, 31, 335707. [DOI] [PubMed] [Google Scholar]
- 33.Kwok H, Briggs K and Tabard-Cossa V, PLoS One, 2014, 9, e92880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhu AS, Jubery TZN, Freedman KJ, Mulero R, Dutta P and Kim MJ, Journal of Physics: Condensed Matter, 2010, 22, 454107. [DOI] [PubMed] [Google Scholar]
- 35.Prabhu AS, Freedman KJ, Robertson JW, Nikolov Z, Kasianowicz JJ and Kim MJ, Nanotechnology, 2011, 22, 425302. [DOI] [PubMed] [Google Scholar]
- 36.Kim MJ, Wanunu M, Bell DC and Meller A, Advanced Materials, 2006, 18, 3149–3153. [Google Scholar]
- 37.Yanagi I, Akahori R and Takeda K.-i., Scientific Reports, 2019, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He KM, Zhang XY, Ren SQ, Sun J and Ieee, Seattle, WA, 2016. [Google Scholar]
- 39.Hsu CW and Lin CJ, Ieee Transactions on Neural Networks, 2002, 13, 415–425. [DOI] [PubMed] [Google Scholar]
- 40.Morshed A, Karawdeniya BI, Bandara Y, Kim MJ and Dutta P, Electrophoresis, 2020, 41, 449–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weitzman MD and Linden RM, in Adeno-Associated Virus, Springer, 2012, pp. 1–23. [DOI] [PubMed] [Google Scholar]
- 42.Horowitz ED, Rahman KS, Bower BD, Dismuke DJ, Falvo MR, Griffith JD, Harvey SC and Asokan A, Journal of Virology, 2013, 87, 2994–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura T, Ferran B, Tsukahara Y, Shang Q, Desai S, Fedoce A, Pimentel DR, Luptak I, Adachi T, Ido Y, Matsui R and Bachschmid MM, Scientific Reports, 2019, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z and Wilson JM, Human Gene Therapy, 2010, 21, 1259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hossan MR, Dillon R and Dutta P, Journal of Computational Physics, 2014, 270, 640–659. [Google Scholar]
- 46.Morshed A, Dutta P, Hossan MR and Dillon R, Physical Review Fluids, 2018, 3:103702, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morshed A, Dutta P and Kim MJ, Electrophoresis, 2019, 40, 2584–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Judd J, Wei F, Nguyen PQ, Tartaglia LJ, Agbandje-McKenna M, Silberg JJ and Suh J, Molecular Therapy-Nucleic Acids, 2012, 1, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saharia J, Bandara YND, Lee JS, Wang Q, Kim MJ and Kim MJ, Electrophoresis, 2020, 41, 630–637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


