SUMMARY
Background.
IFNL4 genotype regulates the immune response by controlling the production of IFN-λ4, a type III interferon. We hypothesized that IFN-λ4 may play a role in infection clearance or alloreactivity in patients with acute leukemia who received myeloablative 10/10 HLA-matched hematopoietic cell transplant (HCT).
Methods.
We conducted a two-stage study using the Center for International Blood and Marrow Transplant Research repository and database. A discovery set of 404 patients (HCT: January 9, 2004 - December 26, 2008): IFNL4 polymorphisms (rs368234815, rs12979860, and rs117648444) were genotyped with TaqMan assays, and an independent validation of 1245 patients from the DISCOVeRY-BMT study (HCT: January 7, 2000 - December 26, 2008) with existing Illumina array genotype data. A combined analysis of 1591 was also performed.
Findings.
Donor, but not recipient IFNL4-positive genotype, was associated with increased risk of non-relapse mortality (NRM; HR-discovery=1.60, 95% CI=1.23–2.10, p=0.0005, HR-validation=1.22, 95% CI=1.05–1.40, p=0.007, and HR-combined =1.27, 95% CI=1.12–1.45, p=0.0001). The effect was driven by excess risk of deaths from graft-versus-host disease (HR-discovery= 2.0, 95% CI=1.05–3.84, p=0.03, and HR-validation=1.40, 95% CI=1.02–1.92, p=0.04, and HR-combined=1.46, 95% CI=1.11–1.94, p=0.008), and possibly infections (HR-discovery= 1.49, 95% CI=0.71–3.10, p=0.29, and HR-validation=1.54, 95% CI=1.13–2.10, p=0.006, and HR-combined=1.54, 95% CI=1.16–2.04, p=0.003). Associations with post-HCT overall survival (OS) were as follow: HR-discovery=1.24, 95% CI=1.02–1.51, p=0.03, and HR-validation=1.10, 95% CI=0.98–1.20, p=0.1, and HR-combined=1.11, 95% CI=1.02–1.22, p=0.02).
Interpretation.
Prioritizing HCT donors with IFNL4-Null genotype may decrease NRM and improve OS without significantly limiting the donor pool.
Funding.
The NCI Intramural Research Program. Full funding list is in the Acknowledgments.
INTRODUCTION
Allogeneic hematopoietic cell transplantation (HCT) is an effective therapy for patients with acute leukemia because of the anti-leukemia effect driven by the donor graft, known as the graft-versus-leukemia effect.1 The 3-year probabilities of matched unrelated donor post-HCT survival for adults with acute myeloid leukemia are 53%, 50% and 27% for patients in first, second remission and not in remission, respectively.2 Despite recent improvements in HCT modalities, the risks of transplant-related complications and non-relapse mortality (NRM) remain high, with deaths primarily caused by infections, graft-versus-host disease (GvHD), and organ failure. Together, these causes account for approximately 60% and 40% of deaths occurring within and beyond the first 100 days post-HCT, respectively.2
The interferon lambda 4 gene (IFNL4) encodes IFN-λ4, a recently discovered member of the type III interferon gene family.3 Protein expression of IFN-λ4 is controlled by a dinucleotide genetic variant (rs368234815-TT/dG) located in the first exon of IFNL4. The rs368234815-dG allele supports the production of IFN-λ4 protein, while the TT allele introduces a protein-abrogating frameshift. Thus, IFNL4 genotype can be categorized into IFNL4-positive (those producing 1 or 2 copies of IFN-λ4 protein), and IFNL4-Null (those not producing IFN-λ4 protein).3 Another IFNL4 polymorphism (rs12979860) is located within the first intron and can be used as a proxy when rs368234815 is not available. Individuals with IFNL4-positive genotype can be further genotyped for rs117648444 (a missense variant P70S in exon 2 of IFNL4) and categorized into those who can only produce functionally weak IFN-λ4-S70 protein and those who carry at least one haplotype for producing functionally strong IFN-λ4-P70 protein (reviewed in4; details are presented Appendix p10). IFNL4-Null individuals are more likely to clear hepatitis C virus (HCV) infection and less likely to reactivate cytomegalovirus (CMV) infection after solid organ transplantation.,5 IFN-λ4 activates the JAK/STAT pathway,3 and induces expression of negative regulators of cytokine and interferon signaling,6 which may have a broad impact on both the adaptive and innate immune responses, and explain diverse and generally negative clinical outcomes associated with its activity.7
We hypothesized that IFN-λ4 might be important for outcomes of HCT by affecting infection clearance and/or alloreactivity. In this study, we evaluated the association between the recipient and donor IFNL4 genetic polymorphisms with post-HCT survival outcomes in patients with acute leukemia.
METHODS
Study Design and Participants:
The Center for International Blood and Marrow Transplant Research (CIBMTR) maintains one of the world’s largest databases and biorepositories serving HCT and cellular therapy-related studies. CIBMTR collects detailed information on almost all allogeneic HCT performed in the United States. The research sample biorepository primarily includes samples collected in the context of unrelated donor transplants facilitated by the National Marrow Donor Program (NMDP/ Be The Match®). Data collected by the CIBMTR is thoroughly reviewed and audited to ensure high quality (details at https://www.cibmtr.org/Data/Available/Pages/index.aspx).
Eligibility criteria for this study included: patients with acute myeloid leukemia (AML) or acute lymphocytic leukemia (ALL), who received HCT at any age from an unrelated 10/10 HLA-matched donor, with myeloablative conditioning regimen, between January 1, 2000 - December 31, 2008, and had available pre-HCT recipient and/or donor blood sample.
The discovery cohort (NCI set) used an available set (N=446) that was part of a previously published study to test this hypothesis.8 Patients in the original study were in first or second complete remission and had an available donor pre-HCT blood sample (since the original study focused on donor telomere length) in the CIBMTR biorepository. Of those, 410 recipient-donor pairs met study eligibility criteria; genotyping was completed for 404 donors and 319 recipients with available DNA.
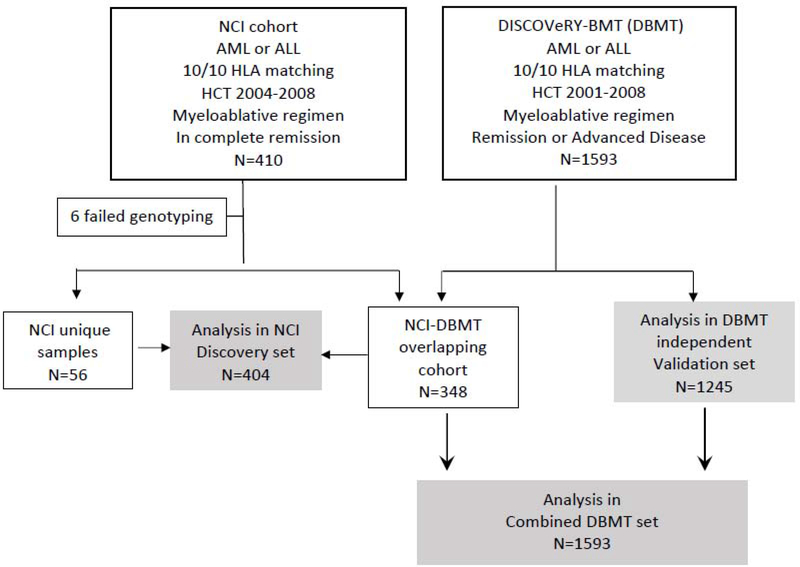
A validation cohort was sought after promising results were obtained in the first set. We used genomic and outcome data from the DISCOVeRY-BMT study (DBMT; a genome-wide association study aiming to determine the influence of susceptibility variants on one-year mortality after HCT).9 The DBMT included patients with AML, ALL, and myelodysplastic syndrome (MDS) selected from the CIBMTR database and biorepository. Of the 1593 HCT recipients meeting study eligibility criteria, we excluded 348 patients who were already included in the NCI set. This left 1245 unique patients for an independent validation analysis. We also performed a combined analysis using the full DBMT set of 1593 eligible patients (independent validation + overlapping patients in the NCI set) to improve statistical power. Figure 1 presents the study flow chart.
Figure 1:
Study flow chart.
The study was approved by the NMDP Institutional Review Board. All study participants provided written informed consent for participation in the CIBMTR repository and database.
Outcomes:
Non-relapse mortality (NRM) was defined as death during continuous complete remission. Overall survival (OS) was defined as time from date of HCT to death from any cause or last follow-up. Relapse definition was primarily based on not achieving complete hematologic recovery. Acute and chronic graft-versus-host disease (GvHD) were defined according to standard criteria.
Causes of death were ascertained at one year after HCT from adjudicated data generated as part of the DBMT study; detailed definitions are available elsewhere.9 Briefly, relapse mortality is death after leukemia relapse or disease progression. GvHD death is from severe acute or chronic GvHD under active treatment. Infection death included bacterial, viral, fungal, and/or protozoan infections causing end organ damage. Organ failure deaths were those not due to disease progression, GvHD or infection. The adjudicated causes of death data were available for 348 patients in the discovery set and all patients in the validation set.
Procedures:
In the NCI set, we genotyped IFNL4 polymorphisms rs368234815, rs117648444 and rs12979860 using TaqMan assays as was previously described.3 Blinded technical duplicates (10% of all samples) showed 100% genotype concordance. Polymorphisms rs368234815 and rs12979860 were in near-complete linkage disequilibrium (r2=0.99, Appendix p11), thus in the NCI set we focused on the functional rs368234815.
In the DBMT set, we used existing genotyping data from Illumina HumanOmniExpress-24 BeadChip available for the IFNL4-rs12979860, while rs117648444 was imputed using Haplotype Reference Consortium, hg19, via the Michigan Imputation server;10 only genotypes imputed with high confidence (>0.9) were used for analyses. Genotype concordance in the 348 overlapping samples between the NCI and the DBMT sets were 100% and 99% for rs12979860 and rs117648444, respectively.
Laboratory personnel were blinded to patient outcomes. Genotype and allele frequencies of all the markers in our study populations were similar to those from European populations in the 1000 Genomes Project (Appendix p. 2, 3). IFNL4 genotype, haplotype and protein groups were assigned to all individuals as outlined in Appendix (p 4, 5, 10).
We analyzed a publicly available single-cell RNA-sequencing (scRNA-seq) dataset 11 to evaluate the expression levels of all human interferons, including IFNL4. The analysis was done in the dataset11 of 32,333 bone marrow/progenitor CD34+ CD38- cells from 3 healthy (HIV, HCV and HBV-negative) bone marrow donors downloaded from Single Cell Expression Atlas (https://www.ebi.ac.uk/gxa/sc/experiments/E-HCAD-6/downloads). We used matrix.mtx, barcode and feature files for normalized scRNA-seq counts, and R packages “SingleCellExperiment” version 1.9.1 and “Scater” version 1.15.16 for the analysis. Expression values were computed as counts-per-million (CPM) and plotted using the R package “ggplot2”. Downloaded scRNA-seq BAM files were indexed using SAMtools and visualized with Integrative Genome Viewer against hg38 reference coordinates. IFNL4 genotypes were scored from sequencing reads for rs73930703 located in 3’UTR of IFNL4.
Statistical Analysis
We calculated hazard ratios (HRs) and 95% confidence intervals (CIs) for OS using multivariable Cox proportional hazard models and modeled cause-specific hazards for NRM and causes of death analyses. Competing events were relapse in NRM and cause of death other than that under study for cause-specific mortality. To select clinical factors predicting outcomes of interest to be included in the model, we used a stepwise forward-backward procedure in the NCI set with a p-value threshold of 0.05 for model entry and retention. All clinical factors in Table 1 were eligible to enter the model. For consistency, the same variables were used for the validation and combined analyses. Proportional hazards assumption was tested for all variables using a time-dependent approach. Factors violating the assumption were adjusted for through stratification. Final models were adjusted for donor and recipient age, GvHD prophylaxis, use of total body irradiation, and stratified by graft type. Follow-up started at the date of HCT and ended at the event of interest, death, or end of study (May 31, 2019). Associations with IFNL4 genotype were tested under an additive genetic model (based on 0, 1 or 2 copies of the risk alleles rs368234815-dG or rs12979860-T). The models calculated hazard ratios for NRM or OS per risk allele. For IFN-λ4 protein analysis, we compared the IFN-λ4-Weak and IFN-λ4-Strong with the IFNL4-Null group as the reference, adjusting for the same set of clinical factors as in the main models.
Table 1:
Characteristics of study participants in the discovery NCI and validation DBMT sets
| Discovery NCI Total, N=404 | Validation DBMT Total, N=1245 | P-value | |
|---|---|---|---|
| N (%) | |||
| Patient Age | 0.70 | ||
| <20 | 73 (18.07) | 254 (20.40) | |
| 20–<40 | 140 (34.65) | 420 (33.73) | |
| 40–50 | 90 (22.28) | 294 (23.61) | |
| 50+ | 101 (25.00) | 277 (22.25) | |
| Male Gender | 222 (54.95) | 689 (55.34) | 0.93 |
| Patient Race | 0.68 | ||
| Caucasian | 374 (92.57) | 1157 (92.93) | |
| African American | 12 (2.97) | 28 (2.25) | |
| Other | 18 (4.46) | 60 (4.82) | |
| Donor Age | 0.08 | ||
| 18–30 | 169 (41.94) | 463 (37.19) | |
| 31–<50 | 203 (50.37) | 705 (56.63) | |
| 50+ | 31 (7.69) | 77 (6.18) | |
| Donor Race | 0.33 | ||
| Caucasian | 361 (89.36) | 1084 (87.07) | |
| African American | 10 (2.47) | 28 (2.25) | |
| Other/unknown | 33 (8.17) | 133 (10.68) | |
| Disease Subtype | 0.65 | ||
| AML | 263 (65.10) | 793 (63.69) | |
| ALL | 141 (34.90) | 452 (36.31) | |
| Disease status at HCT | <0.001 | ||
| 1st complete remission | 253 (62.62) | 406 (32.61) | |
| 2nd complete remission | 151 (37.38) | 396 (31.81) | |
| Not in remission/advanced disease | 0 (0) | 463 (37.19) | |
| Recipient-donor CMV Match | 0.34 | ||
| Both negative | 131 (32.43) | 342 (27.47) | |
| Donor negative-recipient positive | 143 (35.40) | 426 (34.22) | |
| Donor positive-recipient negative | 55 (13.61) | 181 (14.54) | |
| Both positive | 75 (18.56) | 259 (20.80) | |
| Karnofsky Score | 0.001 | ||
| <90 | 88 (21.78) | 355 (28.51) | |
| 90–100 | 285 (70.54) | 755 (60.64) | |
| Missing | 31 (7.67) | 135 (10.84) | |
| Graft Type | <0.001 | ||
| Bone marrow | 129 (31.93) | 596 (47.87) | |
| Peripheral blood | 275 (68.07) | 649 (52.13) | |
| Use of Total Body Irradiation | 206 (50.99) | 761 (61.12) | <0.001 |
| GVHD Prophylaxis | <0.001 | ||
| Tacrolimus-based | 313 (77.48) | 772 (62.0) | |
| Cyclosporine-based | 91 (22.52) | 392 (31.49) | |
| Other | 0 (0) | 81 (6.51) | |
| Year of HCT | <0.001 | ||
| 2000–2003 | 0 (0) | 424 (34.06) | |
| 2004–2006 | 230 (56.93) | 532 (42.73) | |
| 2007–2008 | 174 (43.07) | 289 (23.21) | |
| Donor IFNL4 genotype | 0.88 | ||
| IFNL4-Null | 177 (43.81) | 556 (44.66) | |
| 1 copy IFNL4-positive | 179 (44.31) | 535 (42.97) | |
| 2 copies IFNL4-positive | 48 (11.88) | 154 (12.37) | |
| Recipient IFNL4 genotype* | 0.32 | ||
| IFNL4-Null | 156 (49.37) | 550 (44.90) | |
| 1 copy IFNL4-positive | 123 (38.92) | 531 (43.35) | |
| 2 copies IFNL4-positive | 37 (11.71) | 144 (11.76) | |
| Donor -rs117648444 genotype | 0.71 | ||
| G/G | 328 (81.38) | 1019 (82.11) | |
| G/A | 70 (17.36) | 212 (17.08) | |
| A/A | 5 (1.24) | 10 (0.81) | |
| Donor IFN-λ4 protein status | 0.91 | ||
| IFN-λ4-Null | 177 (43.92) | 556 (44.80) | |
| IFN-λ4-Weak | 56 (13.89) | 163 (13.13) | |
| IFN-λ4-Strong | 170 (42.18) | 522 (42.06) | |
based on non-overlapping 316 recipients from the discovery NCI set and 1225 recipients from the DBMT validation set with genotype information for rs368234815 in NCI set and rs12979860 in DBMT.
All statistical analyses were performed using SAS software version 9.4 (Cary, NC, USA).
To provide guidance for clinical decision-making, we calculated the absolute risks (AR) of NRM and OS, and Mean Risk Stratification (MRS) 12 in relation to donor IFNL4 genotype (online calculator is available at https://analysistools.cancer.gov/biomarkerTools/#meanRiskStratification). MRS is calculated based on the following equation (MRS=2t(1–t)×RD) where RD is absolute risk difference, and t is IFNL4 genotype frequency. As a measure of risk stratification, MRS is a more comprehensive metric for interpreting the potential value of a biomarker test than metrics of pure predictiveness, such as AUC (area under the curve).12 We calculated MRS using genotype frequency and risk estimates from the full DBMT set, under 2 possible clinical decision scenarios: 1) accepting only donors with IFNL4-Null (approximately 50% of our donor pool and rejecting the others), or 2) rejecting donors homozygous for IFNL4-positive genotype (accepting approximately 90% of our donor pool). See Supplemental Online Methods for detailed MRS explanation. To interpret the clinical significance of MRS in the context of HCT, we calculated the MRS for NRM and OS, based on 7/8 vs. 8/8 HLA-matching of donors after standardizing this calculation using NRM and OS probability estimates from our study and risk estimates from previous report.13 We used 8/8 vs. 7/8 HLA matching as an example of an MRS cutoff point that resulted in a successful actionable clinical decision change in HCT. See Online Methods for calculation details (Appendix p. 15–18).
Role of Funding Source:
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
RESULTS
Table 1 summarizes the patient characteristics by study sets. The discovery and validation sets (NCI; N=404 and DBMT; N=1245; all 10/10 HLA matched) were similar in the demographics of recipients and donors, disease frequency, and predominance of tacrolimus-based regimens for GvHD prophylaxis. Notable differences between the two sets included: graft source (bone marrow grafts in 129 patients (31.9%) of 404 vs. 596 (47.9%) of 1245, for the NCI and DBMT, respectively), and disease status at HCT (404 patients (100%) vs. 802 (64.2%) of 1245 were in remission, respectively). The distributions of recipient and donor IFNL4 genotypes were similar in both sets, such as 156 (49.4%) of 316 and 550 (44.9%) of 1225 of the NCI and DBMT recipients; and 177 (43.8%) of 404 and 556 (44.7%) of 1245 of the donors, respectively were IFNL4-Null (Table 1).
Results from both the discovery and validation sets showed no statistically significant association between recipient IFNL4 genotype and post-transplant NRM (HR=1.12, 95% CI=0.84–1.51, p=0.44, and HR=1.11, 95% CI=0.95–1.28, p=0.19, respectively) or OS (HR=1.01, 95% CI=0.81–1.26, p=0.96, and HR=1.07, 95 % CI=0.97–1.19, p=0.17). Analysis of the full DBMT set (combined set) showed similar results (Appendix p 6).
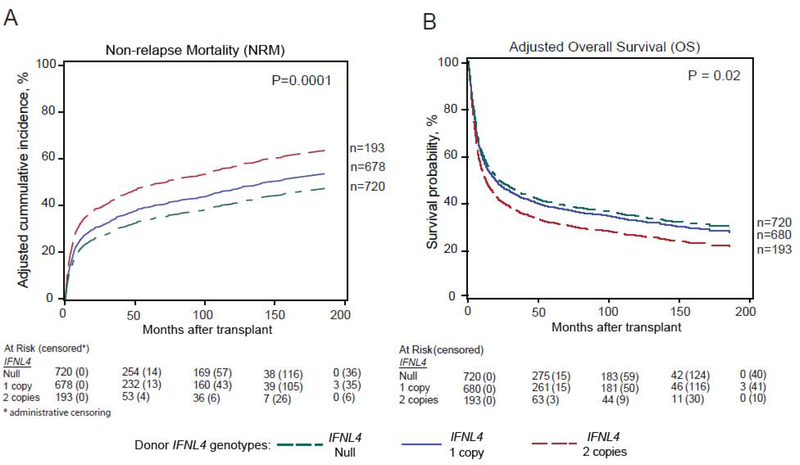
On the other hand, donor IFNL4-positive genotype was statistically significantly associated with an excess risk of NRM in both cohorts (HR=1.60, 95% CI=1.23–2.10, p=0.0005, and HR=1.22, 95% CI=1.05–1.41, p=0.007, in the discovery and validation sets, respectively) (Table 2). Similar result was noted in the combined set (HR =1.27, 95% CI=1.12–1.45, p=0.0001) (Table 2 & Figure 2A).
Table 2:
Risk of non-relapse mortality and overall survival in relation to donor IFNL4 genotype
| Non-relapse Mortality (NRM) | Overall Survival (OS) | |||
|---|---|---|---|---|
| Na events/total | HRb (95% CI) P-value | N events/total | HRb (95% CI) P-value | |
| Discovery NCI setc (N=404) | ||||
| IFNL4 genotype (per risk allele)d | 115/402 | 1.60 (1.23–2.10) 0.0005 | 223/404 | 1.24 (1.02–1.51) 0.03 |
| IFN-λ4 protein status | ||||
| IFN-λ4-Null | 38/177 | reference | 91/177 | reference |
| IFN-λ4-Weak | 14/55 | 1.32 (0.71–2.48) 0.38 | 30/56 | 1.06 (0.69–1.62) 0.79 |
| IFN-λ4-Strong | 62/169 | 2.18 (1.43–3.30) 0.0002 | 101/170 | 1.39 (1.04–1.86) 0.03 |
| DBMT validation setc (N=1245) | ||||
| IFNL4 genotype (per risk allele)d | 407/1244 | 1.22 (1.05–1.41) 0.007 | 875/1245 | 1.10 (0.98–1.20) 0.10 |
| IFN-λ4 protein status | ||||
| IFN-λ4-Null | 165/556 | reference | 390/556 | reference |
| IFN-λ4-Weak | 50/163 | 1.07 (0.77–1.47) 0.6 | 112/163 | 1.01 (0.82–1.2) 0.86 |
| IFN-λ4-Strong | 191/521 | 1.29 (1.05–1.60) 0.01 | 371/522 | 1.08 (0.93–1.24) 0.28 |
| DBMT full set (overlapping + validation; N=1593) | ||||
| IFNL4 genotype (per risk allele)d | 518/1591 | 1.27 (1.12–1.45) 0.0001 | 1078/1593 | 1.11 (1.02–1.22) 0.02 |
| IFN-λ4 protein status | ||||
| IFN-λ4-Null | 211/720 | reference | 482/720 | reference |
| IFN-λ4-Weak | 65/212 | 1.11 (0.84–1.47) 0.44 | 140/212 | 1.03 (0.85–1.25) 0.70 |
| IFN-λ4-Strong | 241/655 | 1.37 (1.14–1.65) 0.0007 | 454/657 | 1.13 (0.99–1.28) 0.06 |
Number of events and at risk by genotype is available in Supplemental Appendix p 9
Additive genetic models (HR presented per risk allele), unless specified. All models were adjusted for: donor and recipient age, GvHD prophylaxis, use of total body irradiation, and stratified by graft type
348/404 samples were also genotyped in the DBMT (overlapping set)
IFNL4 genotype risk allele is defined as the presence of rs368234815-dG allele in NCI set and rs12979860-T allele in DBMT
Figure 2:
IFNL4 genotype and hematopoietic cell transplant: Donor IFNL4 genotype and the probability of post-transplant non-relapse mortality (A); Overall survival (B) in the combined DBMT set.
The observed higher risk of NRM in patients receiving HCT from donors with IFNL4-positive genotype resulted in statistically significant inferior OS in the discovery and combined sets (HR=1.24, 95% CI=1.02–1.51, p=0.03, and HR =1.11, 95% CI=1.02–1.22, p=0.02, respectively) (Table 2 & Figure 2B). The association did not reach statistical significance in the validation cohort (HR=1.10, 95% CI=0.98–1.20, p=0.10) (Table 2).
The associations between donor IFNL4 genotypes and one-year post-HCT causes of death are summarized in Table 3. The data showed statistically significant associations between donor IFNL4-positive genotype and risk of death from GvHD in all study sets. A significant relationship with infection-related death was only noted in the validation, and combined sets. No associations were noted with death from organ failure (HR-combined=1.05, 95% CI=0.78–1.40, p=0.74) or leukemia relapse (HR-combined=0.96, 95% CI=0.82–1.12, p=0.60).
Table 3:
Post-HCT one-year cause-specific mortality in relation to donor IFNL4 genotype in the DBMT set
| Primary cause of death | NCI validation seta N=348 | DBMT independent set N=1245 | Combined DBMT sets N=1953 | ||
|---|---|---|---|---|---|
| Nb events | HRb (95% CI) | Nb events | HRc (95% CI) p-value | HR (95% CI) p-value | |
| GvHD | 20 | 2.0 (1.05–3.84) P=0.03 | 78 | 1.40 (1.02–1.92) P=0.04 | 1.46 (1.11–1.94) 0.008 |
| Infections | 14 | 1.49 (0.71–3.10) 0.29 | 80 | 1.54 (1.13–2.1) 0.006 | 1.54 (1.16–2.04) P=0.003 |
| Organ failure | 13 | 1.80 (0.83–3.91) P=0.14 | 86 | 0.91 (0.65–1.26) 0.57 | 1.05 (0.78–1.40) P=0.74 |
| Relapse | 52 | 0.89 (0.58–1.37) P=0.60 | 313 | 0.97 (0.82–1.14) P=0.68 | 0.96 (0.82–1.12) P=0.60 |
Patients with adjudicated causes of death data generated as part of the DBMT study were included in this analysis
Number of events and at risk by genotype is available in Supplemental Appendix p 9
Cause-specific hazards accounting for competing mortality of causes other than those under study. HRs are calculated based on additive genetic models in relation to IFNL4 genotype (rs12979860-T allele; risk allele) and adjusted for donor and recipient age, GvHD prophylaxis, use of total body irradiation, and stratified by graft type.
We then used the combined set and complete the following secondary analyses: 1) restricted to patients who received HCT while in remission, this showed similar results to that observed in all patients (HR for NRM =1.27, p=0.001, and for OS=1.12, p=0.04); 2) stratified on graft source, the HR for NRM=1.36, p=0.0003; and HR=1.18, p=0.11, for peripheral blood vs. bone marrow stem cell transplants respectively, p-interaction=0.56); 3) stratified on donor-recipient pre-HCT CMV serostatus as a proxy for risk of post-HCT CMV viremia or use of anti-viral prophylaxis, and found no risk differences in NRM or OS in relation to donor IFNL4 genotype by donor-recipient CMV matching or donor seropositivity (Appendix p.8).
In further analysis, compared with IFNL4-Null donors, a higher risk of NRM was observed in patients who received HCT from donors with IFN-λ4-Strong, but not from donors with IFN-λ4-Weak protein; similar results were noted in all study sets. Results for OS followed the same trend, but did not reach statistical significance in the validation cohort or the combined analysis (Table 2 & Appendix p.12).
To explain the association between IFNL4 genotype and outcomes of HCT, we hypothesized that the transplanted graft is the source of IFNL4-expressing cells. We analyzed mRNA expression of all human interferons - IFNL4 and 21 other transcripts in bone marrow samples of three healthy donors. We showed that the IFNL4 was the main interferon transcript expressed in bone marrow of all three individuals tested, with expression detected in 0.67% of all sequenced cells (range of expression of other interferons=0–0.07%), and in 83.1% of all interferon-expressing cells (range for other interferons=0–8.8%). Based on the IFNL4 proxy variant rs73930703 within the transcribed region, all three individuals are likely to be IFNL4-Null (Appendix p 7&13).
Using risk stratification tools to guide donor selection strategy based on IFNL4 genotype testing, we found that selecting IFNL4-Null donors was associated with an absolute risk reduction of post-HCT NRM of 6.4% (95% CI=1.2–11.6%) versus 11.0% (95% CI=2.7–19.4%) if rejecting donors homozygous for the IFNL4-positive genotype. The Mean Risk Stratification (MRS) provided by selecting HCT donors with the common IFNL4-Null status (~50% frequency) changed the average risk of NRM by MRS=3.2% (95% CI=0.67% to 5.74%). Our estimate for the corresponding MRS for 8/8 vs. 7/8 HLA-matching is 3.5% (95% CI=1.83%−5.23%). In contrast, the MRS of the rare homozygous IFNL4-positive genotype (frequency=12.1%) was only 2.4% (95% CI=0.56% to 4.15%), despite contributing the highest AR difference (11.0%). These results suggest that the strongest impact on population-average risk of NRM would be achieved by attempting to use donors with IFNL4-Null status rather than merely avoiding homozygous IFNL4-positive donors. For OS, the MRS for selecting donors with the IFNL4-Null status was 1.4% (95% CI= −1.3% to 4.1%), which is less than that for HLA matching (2.1%; 95% CI= 0.53% to 3.7%). See Supplemental Results for MRS and AUC comparison. (Appendix p. 19–24)
DISCUSSION
In this study of acute leukemia HCT recipients, we showed that donor IFNL4 genotype is associated with patient risk of NRM. The observed risk reduction when selecting IFNL4-Null donors was beyond that already provided by the HLA matching since all patients in this study received an HCT from a 10/10 HLA-matched donor. Our findings suggest potential clinical utility for prioritizing 10/10 HLA-matched donors with IFNL4-Null genotype to further reduce patient risk of NRM and improve OS.
Donor selection is key for HCT success, with HLA matching being the most important factor. A CIBMTR study of 3,857 patients who received myeloablative regimens for malignant conditions showed an associated mortality risk of 1.25 with every single HLA mismatch.13 Growing evidence suggests a role for additional donor genetic factors in post-HCT survival. For example, longer donor telomere length was associated with an improved OS after unrelated donor HCT in patients with SAA.14–16 Yet, results for the possible post-HCT benefit of longer donor telomere length in acute leukemia are still inconclusive.17 Other examples include polymorphisms in Vitamin D receptor gene, cytokine genes, and HLA-DP permissiveness and expression.18–20 None of these markers made it to the donor selection clinical practice with the exception of considering HLA-DP.
Consistent with the unfavorable effect of IFNL4-positive genotype detected in our study, a large study of 840 solid organ transplant recipients showed a high incidence of CMV reactivation in patients with the rs368234815-dG/dG genotype (HR=1.46, p=0.05), while no association was noted in patients receiving antiviral prophylaxis.5 Two small studies evaluated the possible effect of IFNL4 genotype on CMV viremia after HCT with inconclusive results.21,22 The use of infection prophylaxis after HCT is a common practice. In 2009, a guideline for preventing infection-related complications after HCT was published in an effort to mainstream recommendations and practices.23 In recent years, new and more effective drugs to prevent and treat infections- particularly CMV and invasive fungal disease- became available,24,25 leading to decrease in post-HCT infections and improving patient survival. In our current analysis, donor IFNL4 genotype was associated with increased risk of death from infections, but detailed information on infections or infection prophylaxis was not available to further explore this question. We found no risk differences in NRM or OS in relation to donor IFNL4 genotype by donor-recipient CMV matching or donor seropositivity, as a proxy for CMV risk of infection or need for prophylaxis. The use of actual detailed data is necessary to answer the question since strategies of infection prophylaxis may differ between transplant centers.
The impact of donor IFNL4 genotype on HCT survival was less pronounced in the validation cohort where bone marrow grafts were more frequent than in the discovery set. Our evaluation suggested a possible higher risk of NRM associated with donor IFNL4-positive genotype in patients receiving peripheral blood than bone marrow stem cell source, although the difference was not statistically significant. This needs further evaluation since unrelated donor bone marrow transplants are associated with lower risk of NRM and improved OS, and may explain the observed difference in the impact of IFLN4 genotype between study sets.
The donor IFNL4 genotype was also associated with a higher risk of death due to GvHD (HR=1.54, p=0.003). This association may reflect a higher risk of developing severe forms of GvHD or a lower response to GvHD immunosuppressive therapies. We evaluated the risk of severe acute GvHD (grade 3–4) and chronic GvHD in relation to donor IFNL4 genotype in the combined DBMT set. No statistically significant association was noted with acute GvHD grade 3–4 (HR=1.11, p=0.16), but a suggestive association with the risk of chronic GvHD existed (HR=1.12, p=0.03). Type III interferons, including IFN-λ4, signal through a distinct receptor complex of IFNLR1 and IL-10R2, to activate the expression of IFN-stimulated genes (ISG) via the JAK/STAT signaling pathway (reviewed in26). Donor IL10R2 genotype and JAK/STAT signaling have been associated with risk of GvHD,27 which was reduced by JAK inhibitors,28 thus providing a plausible link between GvHD and type III interferon signaling.
It is intriguing that death from both immune complications of HCT – inefficient ability to clear infection and overreaction in GvHD - are associated with the genetically determined ability to produce IFN-λ4. It is possible that this observation is attributed to ISG expression; furthermore, IFN-λ4 expression was associated with enhanced negative regulator of IFN signaling upon exposure to viral infection,6 and interferon-related DNA damage resistance signature in prostate cancer.29 Notably, IFNL4 was the predominant interferon transcript detected by scRNA-seq in the bone marrow of healthy blood donors. It is possible that IFN-λ4 produced in bone marrow in individuals with IFNL4-positive genotype can reprogram progenitor cells that later contribute to multiple negative phenotypes, especially when placed into an immunosuppressed environment of transplant recipients. The results suggest a novel functional role of IFN-λ4 in hematopoietic stem cells in normal and disease conditions; our scRNA-seq analysis was limited by the small sample size warranting future investigation.
IFN-λ4-P70S status only affects IFN-λ4 and not other IFN-λs encoded in the same genomic region,4 and therefore can provide evidence of a direct relationship with IFNL4. Patients receiving HCT from donors with IFN-λ4-Weak protein had NRM risk comparable to those with IFNL4-Null donors, while high NRM risk was observed in patients who received HCT from donors with IFN-λ4-Strong protein. This is in line with increased interferon gene signature and impaired HCV clearance in patients with the IFN-λ4-Strong compared to patients with IFN-λ4-Weak protein and IFNL4-Null genotype. 30
Our calculated absolute risk difference (RD) and mean risk stratification (MRS) for NRM showed a potential benefit for selecting donors based on their IFNL4 genotype in the current era of optimal HLA recipient-donor matching. Our data showed an additional 6.4% AR reduction in NRM in donors with IFNL4-Null genotype and 10/10 HLA matching. The effect of selecting donors based on their IFNL4 genotype on the reduction of NRM risk was comparable to that of selecting 8/8 vs. 7/8 HLA matched donors (3.2% vs. 3.5%, respectively), but a lower MRS for risk of OS was noted (1.4% vs. 2.1%, respectively). Thus, our findings reinforce the primary importance of optimal HLA matching, while suggesting that using donors with IFNL4-Null genotype may further reduce NRM risk. A comprehensive study is needed to definitively decide which of the two strategies (merely avoiding donors homozygous for IFNL4-positive vs. actively selecting those with IFNL4-Null genotype) is more effective for clinical practice.
The strengths of this study include the availability of comprehensive clinical and outcome data for the study population, together with pre-HCT samples from both recipients and donors for a relatively large number of patients. Yet, the study was limited by the unavailability of information on infection prophylaxis regimens or episodes of post-HCT infections. Our results may not be generalizable beyond the target study population and need to be studied further for other HCT indications.
In conclusion, our study showed that donor IFNL4-positive genotype is associated with an increased risk of NRM and inferior OS in patients with acute leukemia who received a 10/10 HLA-matched unrelated donor myeloablative HCT. If confirmed, prioritizing donors with the IFNL4-Null genotype may improve HCT survival.
Supplementary Material
RESEARCH IN CONTEXT.
Evidence before this study
We searched PubMed from English articles published between January 1, 2007 and June 28, 2020 evaluating the association between IFNL4 genotype and outcomes after hematopoietic cell transplant (HCT). We used the following search terms: (IFNL4) or (IFNL3) or (IL28B) and (allogeneic cell transplant). Two reports were identified focusing on cytomegalovirus (CMV) viremia with inconclusive results, mostly because of the small sample size.
Added value of this study
Allogeneic hematopoietic cell transplantation (HCT) is an effective therapy for patients with acute leukemia, but patients still suffer from high morbidity and mortality. The discovery of new molecular and genetic markers that can guide donor selection and/or provide risk stratification tools are essential to improve patient survival. We explored the possible association between IFNL4 genotype, a common genetic variant with a strong functional role in host immune modulation and viral clearance, and survival outcomes after 10/10 HLA-matched unrelated donor HCT in patients with acute leukemia. Our results showed that donor IFNL4-positive genotype was associated with excess risk of post-HCT non-relapse mortality (NRM) driven by increased deaths from infections and graft-versus-host disease.
Implications of all available evidence
Our findings suggest potential clinical utility for IFNL4 genotype in donor selection.
ACKNOWLEDGMENTS:
The study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI); a Public Health Service grant (5U24CA076518) from NCI, the National Heart Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a grant/cooperative agreement 4U10HL069294 from NHLBI and NCI; a contract from the Health Resources and Services Administration (HHSH234200637015C); grants N00014-17-1-2388, N00014-16-1-2020 and N0014-17-1-2850 from the Office of Naval Research; and R01 HL102278 from NHLBI.
Footnotes
COMPETING INTERESTS: L.P.-O. is a co-inventor on an IFN-λ4-related patent issued to NCI/NIH, and receives royalties for antibodies for IFN-λ4 detection. Other authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shahinaz M. Gadalla, Clinical Genetics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD
Youjin Wang, Clinical Genetics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD.
Tao Wang, Division of Biostatistics, Medical College of Wisconsin, Milwaukee, WI; Center for International Blood and Marrow Transplant Research, Medical College of Wisconsin, Milwaukee, WI.
Olusegun O. Onabajo, Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD
A. Rouf Banday, Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD.
Adeola Obajemu, Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD.
Ezgi Karaesman, College of Pharmacy, Ohio State University Columbus, OH.
Lara Sucheston-Campbell, College of Pharmacy, Ohio State University Columbus, OH.
Theresa Hahn, Roswell Park Comprehensive Cancer Center, Buffalo, NY.
Jennifer A. Sees, Center for International Blood and Marrow Transplant Research, Minneapolis, MN
Stephen R. Spellman, Center for International Blood and Marrow Transplant Research, Minneapolis, MN
Stephanie J. Lee, Center for International Blood and Marrow Transplant Research, Medical College of Wisconsin, Milwaukee, WI Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA.
Hormuzd A. Katki, Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD
Ludmila Prokunina-Olsson, Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD.
REFERENCES
- 1.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood 1990; 75(3): 555–62. [PubMed] [Google Scholar]
- 2.D’Souza A, Fretham C, Lee SJ, et al. Current Use of and Trends in Hematopoietic Cell Transplantation in the United States. Biol Blood Marrow Transplant 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature genetics 2013; 45(2): 164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prokunina-Olsson L Genetics of the Human Interferon Lambda Region. J Interferon Cytokine Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manuel O, Wojtowicz A, Bibert S, et al. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis 2015; 211(6): 906–14. [DOI] [PubMed] [Google Scholar]
- 6.Obajemu AA, Rao N, Dilley KA, et al. IFN-lambda4 Attenuates Antiviral Responses by Enhancing Negative Regulation of IFN Signaling. Journal of immunology 2017; 199(11): 380–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onabajo OO, Muchmore B, Prokunina-Olsson L. The IFN-lambda4 Conundrum: When a Good Interferon Goes Bad. J Interferon Cytokine Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadalla SM, Wang T, Loftus D, et al. No association between donor telomere length and outcomes after allogeneic unrelated hematopoietic cell transplant in patients with acute leukemia. Bone marrow transplantation 2018; 53(4): 383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn T, Sucheston-Campbell LE, Preus L, et al. Establishment of Definitions and Review Process for Consistent Adjudication of Cause-specific Mortality after Allogeneic Unrelated-donor Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant 2015; 21(9): 1679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karaesmen E, Rizvi AA, Preus LM, et al. Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood 2017; 130(13): 1585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe’er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol 2019; 37(4): 451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katki HA, Schiffman M. A novel metric that quantifies risk stratification for evaluating diagnostic tests: The example of evaluating cervical-cancer screening tests across populations. Prev Med 2018; 110: 100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007; 110(13): 4576–83. [DOI] [PubMed] [Google Scholar]
- 14.Gadalla SM, Wang T, Dagnall C, et al. Effect of Recipient Age and Stem Cell Source on the Association between Donor Telomere Length and Survival after Allogeneic Unrelated Hematopoietic Cell Transplantation for Severe Aplastic Anemia. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadalla SM, Wang T, Haagenson M, et al. Association between donor leukocyte telomere length and survival after unrelated allogeneic hematopoietic cell transplantation for severe aplastic anemia. JAMA : the journal of the American Medical Association 2015; 313(6): 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadalla S, Aubert G, Wang T, et al. Donor telomere length and causes of death after unrelated hematopoietic cell transplantation in patients with marrow failure. Blood 2018; 131(21): 2393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadalla SM, Wang T, Loftus D, et al. Donor Telomere Length and Outcomes after Allogeneic Unrelated Hematopoietic Cell Transplant in Patients with Acute Leukemia. Bone marrow transplantation 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Malkki M, O’HUigin C, et al. High HLA-DP Expression and Graft-versus-Host Disease. N Engl J Med 2015; 373(7): 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickinson AM. Non-HLA genetics and predicting outcome in HSCT. Int J Immunogenet 2008; 35(4–5): 375–80. [DOI] [PubMed] [Google Scholar]
- 20.Dehn J, Spellman S, Hurley CK, et al. Selection of unrelated donors and cord blood units for hematopoietic cell transplantation: guidelines from the NMDP/CIBMTR. Blood 2019; 134(12): 924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bravo D, Solano C, Gimenez E, et al. Effect of the IL28B Rs12979860 C/T polymorphism on the incidence and features of active cytomegalovirus infection in allogeneic stem cell transplant patients. Journal of medical virology 2014; 86(5): 838–44. [DOI] [PubMed] [Google Scholar]
- 22.Corrales I, Solano C, Amat P, et al. IL28B genetic variation and cytomegalovirus-specific T-cell immunity in allogeneic stem cell transplant recipients. Journal of medical virology 2017; 89(4): 685–95. [DOI] [PubMed] [Google Scholar]
- 23.Center for International B, Marrow Transplant R, National Marrow Donor P, et al. Guidelines for preventing infectious complications among hematopoietic cell transplant recipients: a global perspective. Bone Marrow Transplant 2009; 44(8): 453–558. [PubMed] [Google Scholar]
- 24.Marty FM, Ljungman P, Chemaly RF, et al. Letermovir Prophylaxis for Cytomegalovirus in Hematopoietic-Cell Transplantation. N Engl J Med 2017; 377(25): 2433–44. [DOI] [PubMed] [Google Scholar]
- 25.Tissot F, Agrawal S, Pagano L, et al. ECIL-6 guidelines for the treatment of invasive candidiasis, aspergillosis and mucormycosis in leukemia and hematopoietic stem cell transplant patients. Haematologica 2017; 102(3): 433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-lambda4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res 2014; 34(11): 829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin MT, Storer B, Martin PJ, et al. Genetic variation in the IL-10 pathway modulates severity of acute graft-versus-host disease following hematopoietic cell transplantation: synergism between IL-10 genotype of patient and IL-10 receptor beta genotype of donor. Blood 2005; 106(12): 3995–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi J, Cooper ML, Alahmari B, et al. Pharmacologic blockade of JAK1/JAK2 reduces GvHD and preserves the graft-versus-leukemia effect. PLoS One 2014; 9(10): e109799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang W, Wallace TA, Yi M, et al. IFNL4-DeltaG Allele Is Associated with an Interferon Signature in Tumors and Survival of African-American Men with Prostate Cancer. Clin Cancer Res 2018; 24(21): 5471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terczynska-Dyla E, Bibert S, Duong FH, et al. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun 2014; 5: 5699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.