Abstract
Platinum(II) compounds are a critical class of chemotherapeutic agents. Recent studies have highlighted the ability of a subset of Pt(II) compounds, including oxaliplatin but not cisplatin, to induce cytotoxicity via nucleolar stress rather than a canonical DNA damage response. In this study, influential properties of Pt(II) compounds were investigated using redistribution of nucleophosmin (NPM1) as a marker of nucleolar stress. NPM1 assays were coupled to calculated and measured properties such as compound size and hydrophobicity. The oxalate leaving group of oxaliplatin is not required for NPM1 redistribution. Interestingly, although changes in diaminocyclohexane (DACH) ligand ring size and aromaticity can be tolerated, ring orientation appears important for stress induction. The specificity of ligand requirements provides insight into the striking ability of only certain Pt(II) compounds to activate nucleolar processes.
The chemotherapeutic agent cisplatin has inspired the synthesis and investigation of thousands of Pt(II) analogs.1 Of these, only two other compounds—carboplatin and oxaliplatin—have met FDA standards for medical use. Until recently, it was believed that the cytotoxicity of these compounds could be attributed solely to their DNA cross-linking abilities and subsequent induction of the DNA damage response (DDR), a known trigger of apoptotic pathways.2 As the body of research on Pt(II) reagents has grown, a more complex picture has emerged of the mechanisms of action behind these ubiquitous drugs.2 A striking recent discovery is that oxaliplatin, but not cisplatin or carboplatin, causes cytotoxicity via disruptions in ribosome biogenesis rather than DDR.3 Ribosome biogenesis occurs in the nucleolus, a conserved and highly structured membraneless organelle in eukaryotes. Disruptions of the nucleolus or ribosome biogenesis trigger the nucleolar stress response, which leads to cell death or senescence via activation of the tumor suppressor p53. Because its molecular mechanisms are not yet fully understood, and due to its potential role as a chemotherapeutic target, this fascinating stress process is an area of intense interest in the fields of molecular biology and medicine.4–7
The specificity of oxaliplatin as a nucleolar stress inducer is intriguing when considered alongside other data indicating a relationship between Pt(II) compounds and the nucleolus.8 Post-treatment fluorescent labeling of clickable Pt(II) drug analogs has shown localization of these compounds to the nucleolus,8,9 and there is significant evidence that Pt(II) compounds associate with ribosomes and ribosomal RNA.10–16 The structural determinants and molecular mechanisms by which only specific Pt(II) compounds may cause a nucleolar stress response are not understood. Here, we explore properties of oxaliplatin and other Pt(II) compounds and find that a narrow window of derivatives is able to induce nucleolar stress. The results define a set of constraints for Pt(II) compounds to induce this unique cell death pathway.
We selected Pt(II) compounds to test a variety of properties including steric bulk, hydrophobicity, cross-linking ability, and ligand orientation (Figure 1). The extent of nucleolar stress was measured by nucleophosmin (NPM1) imaging (Figure 2 and Figure S1). Translocation of NPM1 from the granular component (GC) of the nucleolus to the nucleoplasm is a hallmark of the nucleolar stress response.17,18 NPM1 translocation has been shown to be a necessary, but not sufficient, feature of p53-mediated cell death upon nucleolar stress18 and thus is a robust and appropriate marker. A549 cells were selected for this study as they are well-established to have a characteristic nucleolar stress response resulting in p53-mediated apoptosis.19,20
Figure 1.
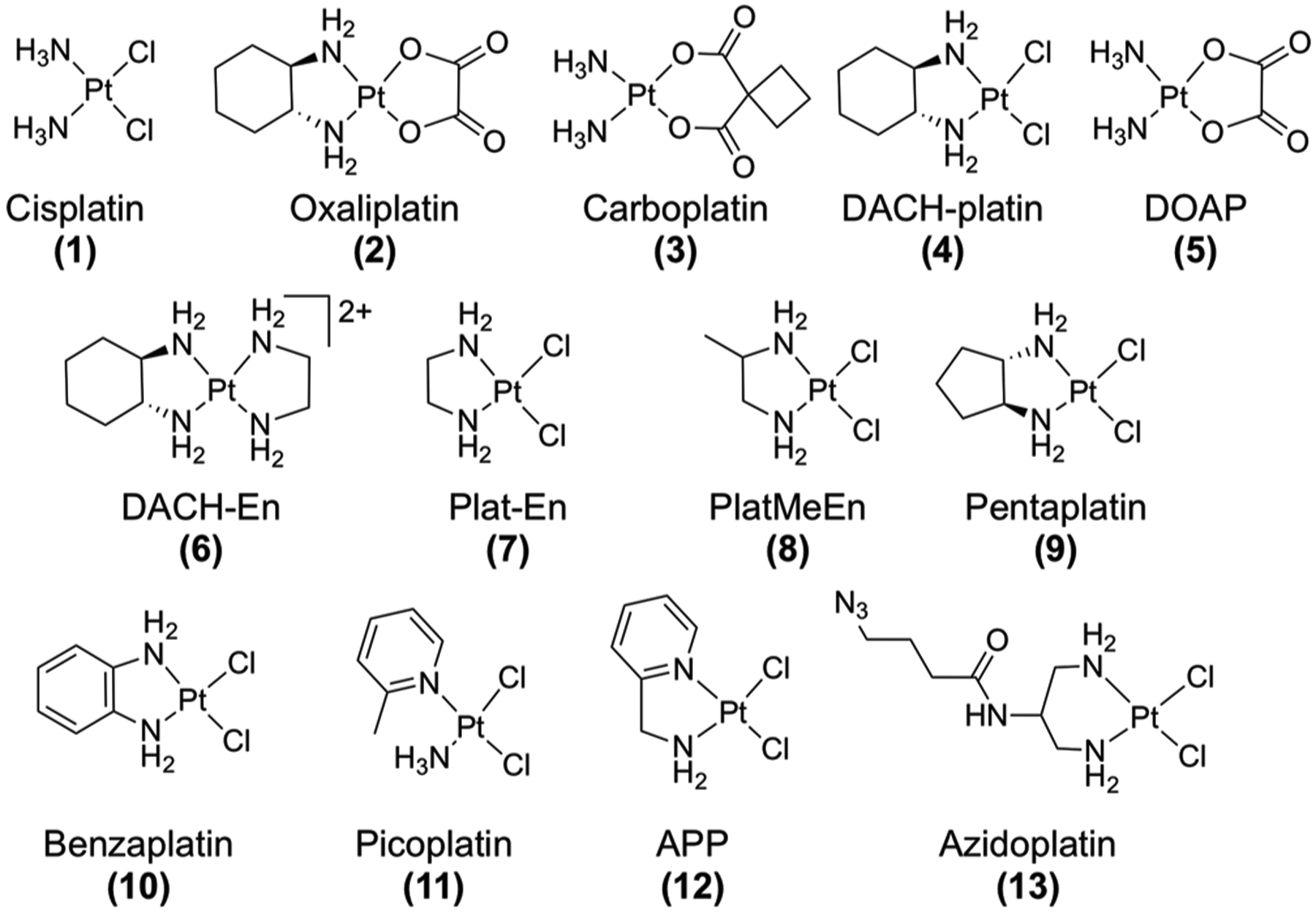
Compounds tested for inducing nucleolar stress via NPM1 relocalization in mammalian cells.
Figure 2.
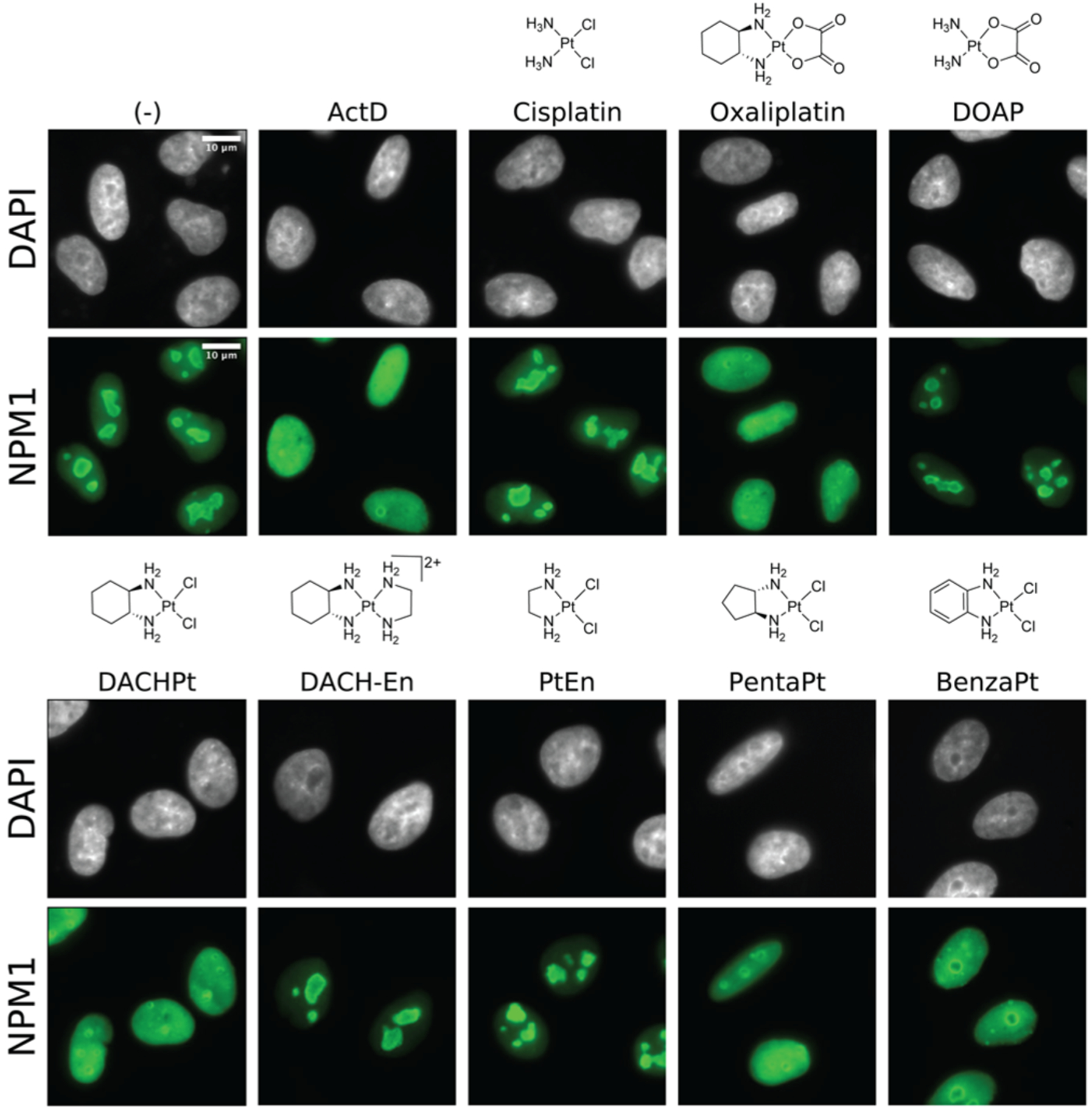
Nucleolar stress induced by Pt(II) compounds. NPM1 (green) relocalization following 24 h treatment in A549 cells. Treatment concentrations are 10 μM except for actinomycin D (5 nM). Scale bar = 10 μm.
Cells were treated for 24 h with a given compound prior to fixation and secondary immunofluorescence to detect NPM1 (Figures 2 and S1). The extent of NPM1 redistribution was quantified using an image analysis pipeline (Figure S1) to calculate the coefficient of variation (CV) of NPM1 intensity in each cell (Figure 3). The uniform distribution of NPM1 in cells undergoing nucleolar stress yields a low CV, as seen in positive control samples treated with known stress-inducer actinomycin D (Figure 3). In addition to the observation of NPM1 redistribution, we noted a change in the shape of nucleoli from eccentrically shaped aggregates to round, sphere-like structures upon stress induction (Figure 2).
Figure 3.
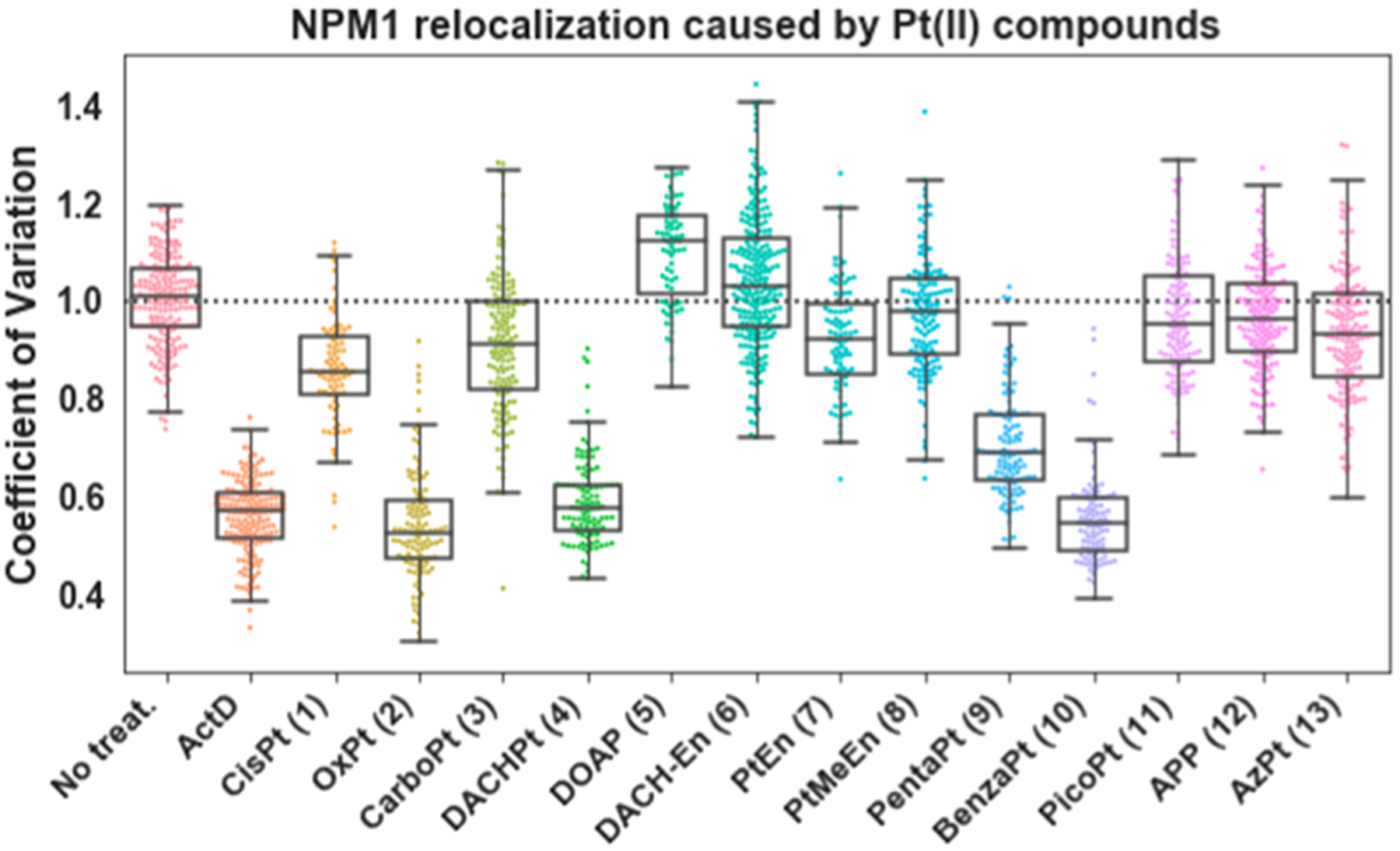
Quantification of NPM1 relocalization induced by Pt(II) compounds. Treatment conditions as in Figure 2; replicates, CV calculations, and boxplot presentation as described in the SI. For each treatment data set, boxes represent median, first, and third quartiles, and vertical lines are the range of data with outliers defined in the SI.
As predicted,21,22 oxaliplatin (2) induces robust redistribution of NPM1, similar to the positive control (Actinomycin D), while NPM1 distribution in cisplatin (1) and carboplatin (3) treated cells more closely resembles that of the no-treatment control (Figures 2 and 3).
We note that for cisplatin-treated cells, a small amount of NPM1 redistribution was observed at this treatment concentration. This is likely because the 24 h IC50 value of cisplatin (12.8 μM, Table S1) is close to the treatment concentration, which may result in a subset of cisplatin-treated cells experiencing abnormal NPM1 distribution downstream of other cell death pathways, such as those mediated by the DDR (see ref 23). Oxaliplatin, by contrast, shows robust NPM1 relocalization at treatment concentrations well below the 24 h IC50 value (81.5 μM, Table S1), suggesting that nucleolar stress significantly precedes cell death pathways.21 NPM1 relocalization at concentrations below IC50 values was observed with other stress-inducing compounds, some of which do not exhibit significant cell death until 48 h of treatment (Tables S1 and S2). Thus, observation of nucleolar stress at 24 h does not necessarily predict measured toxicity.
Oxaliplatin is distinct from cisplatin and carboplatin in both labile and nonlabile Pt(II) ligands. The labile, chelating oxalate ligand of oxaliplatin delays aquation and therefore biomolecule cross-linking27 in comparison with cisplatin. We exchanged the labile and nonlabile ligands of oxaliplatin and cisplatin with compounds 4 and 5. We found that compound 4, DACHPt, which has the nonlabile DACH ligand of oxaliplatin and labile chloride groups of cisplatin, induces robust nucleolar stress. By comparison, 5, or DOAP, which possesses the nonlabile ammine ligands of cisplatin and the labile oxalic acid ligand of oxaliplatin, does not induce stress (Figures 2 and 3). The oxalic acid ligand alone also had no influence on NPM1 redistribution, nor did the DACH ligand by itself (Figures S2 and S3). From this, we concluded that the nonlabile DACH ligand of oxaliplatin is responsible for the nucleolar stress response.
We next considered whether cross-linking of biomolecules by the Pt(II) compound is necessary for the induction of nucleolar stress. An alternate hypothesis is that the charged Pt(II) acts as a targeting agent to facilitate transport of the DACH moiety to the nucleolus where it disrupts nucleolar processes without forming a Pt(II)-DACH lesion on a biomolecule. Compound 6, DACH-En, retains the DACH ligand but is unable to form cross-links with biomolecules due to replacement of the oxalic acid with an ethylenediamine ligand (Figure 1). This positively charged compound did not induce stress (Figures 2 and 3), suggesting that cross-linking of Pt(II) to cellular targets is necessary to induce a nucleolar stress response.
To refine requirements of the Pt(II) ligands that cause nucleolar stress, we examined the effects of steric bulk by testing 7, 8, and 9. Pt-En (7) possesses a nonlabile ethylenediamine ligand. This small molecule did not induce stress in A549 cells (Figures 2 and 3), indicating that a chelating diamine ligand, a common feature between PtEn, DACHPt, and oxaliplatin, is not sufficient to induce stress. The addition of a methyl group to generate the bulkier PtMeEn (8) was also not sufficient to induce stress (Figures 3, S2, and S3). Compound 9, Pentaplatin, possesses a five-membered ring that places its volume between the non stress-inducing PtMeEn and the stress-inducing six-membered DACHPt. Pentaplatin was found to induce nucleolar stress (Figure 1), although with a slightly higher resultant CV than positive controls or oxaliplatin (Figure 3). These results suggest that bulk may be an important metric lending toward the ability of Pt(II) compounds to induce nucleolar stress. Using computed values for volume, we conclude that as a general trend Pt(II) compounds with more steric bulk are more likely to induce nucleolar stress (Figure 4A, Y axis). Compound length, or steric reach, also generally appears to correlate with stress induction (Figure 4A, X axis). Some exceptions to this trend are discussed below.
Figure 4.
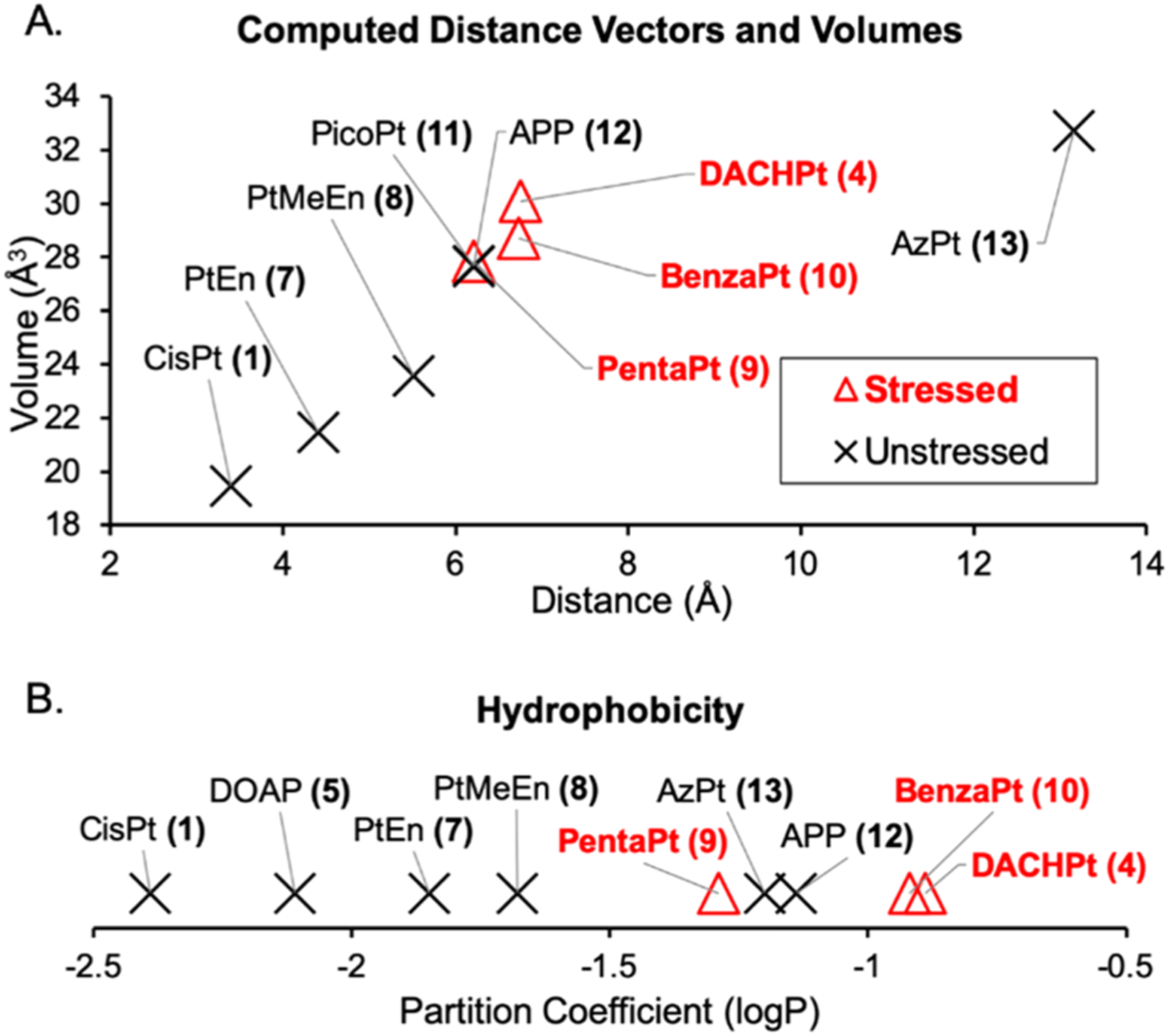
Size and hydrophobicity correlate with stress induction, with some exceptions. Compounds with a higher partition coefficient (measured as log P) are more hydrophobic than those with a lower logP. LogP measurements and calculations of compound volume and Pt-edge distance are described in the SI.
The chair confirmation of the DACH ligand is not essential for stress induction. BenzaPt (10), in which the DACH cyclohexane is replaced with a planar aromatic ring (Figure 5B), also induces robust NPM1 redistribution (Figures 2 and 3). Like DACHPt, BenzaPt is more hydrophobic than the simpler diam(m)ine compounds. To estimate the relative hydrophobicity of our compounds of interest, we measured their water/octanol partition coefficients (Supporting Information and Methods). All of the stress-inducing compounds were found to be relatively hydrophobic (Figure 4B), leading to the conclusion that hydrophobicity, like steric bulk, positively correlates with stress induction. Similarly to steric bulk, however, exceptions to this trend were observed.
Figure 5.
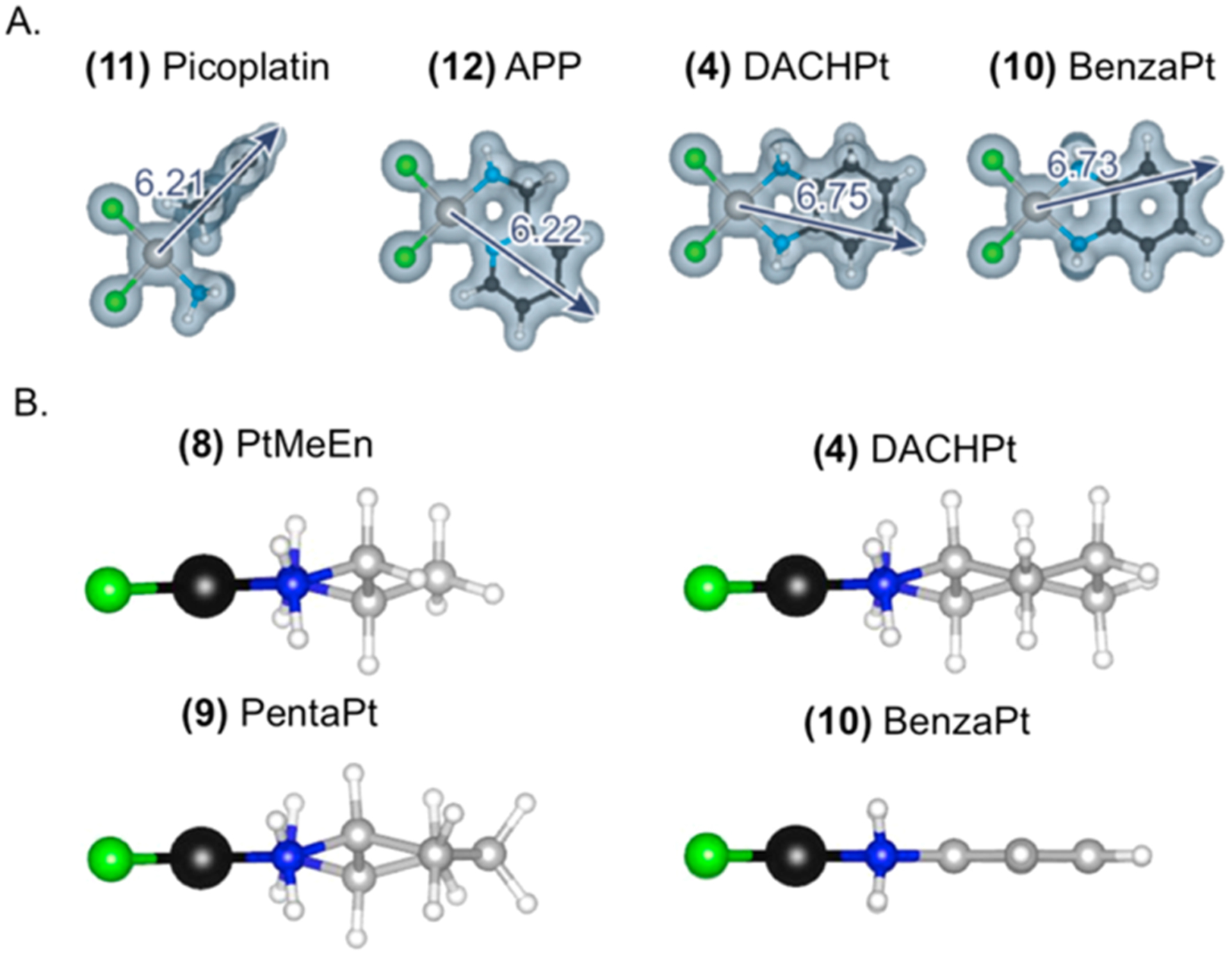
(A) Computed distance measurements and volume representations for non stress-inducing compounds 11 and 12 alongside stress-inducing compounds 4 and 10. (B) Ball and stick drawings of non stress-inducing compound 8 alongside stress-inducing 4, 9, and 10.
Compounds 11, 12, and 13 do not cause NPM1 relocalization despite being similar or higher in terms of size and hydrophobicity to compounds that do cause nucleolar stress (Figure 4). These exceptions may provide insight into the elements responsible for causing stress.
One particularly interesting comparison is between APP (12)28,29 and BenzaPt (10) (Figure 5A). These two Pt(II) compounds both have an aromatic ring, but differ in the orientation of the ring relative to the Pt(II) and, by extension, ring orientation relative to a biomolecule to which the compound is bound. While BenzaPt causes nucleolar stress, APP does not. Similarly, picoplatin (11) does not cause nucleolar stress despite having volume and reach similar to other compounds (Figures 4 and 5). These results demonstrate a critical role for ring orientation in the ability of Pt(II) compounds to induce nucleolar stress.
The observation that azidoplatin (13) does not cause stress is of interest as this compound has extended volume and has previously been shown to localize to the nucleolus.9 Thus, nucleolar localization, even when combined with relatively high hydrophobicity and larger bulk and length, is not sufficient to induce nucleolar stress.
Taken together, the results described provide significant insight into the structural determinants of nucleolar stress induction among Pt(II) compounds. We conclude that there is an important role for ligand orientation and a general correlation between steric bulk and stress induction (Figure 5).
The differential responses induced by these compounds have clinical implications as the three currently FDA-approved Pt(II) chemotherapeutics are known to have different treatment and side effect profiles. Other important differences between these compounds have been observed in the literature. For example, oxaliplatin is noted to cause immunogenic cell death (ICD), while cisplatin does not.30–32 Although this contrast is also observed in nucleolar stress, connections between ICD and nucleolar stress are not well-studied. Oxaliplatin has also been shown to cause changes in the size of neuronal nucleoli correlating with peripheral neuropathy,33 a common side effect associated with oxaliplatin chemotherapy regimens. The relationship between nucleolar stress and platinum-induced neurotoxicity has not been explored. Additionally, there is some evidence that p53 mutations in colon cancer cell lines result in resistance to oxaliplatin-mediated cell death.34 This may be of interest given oxaliplatin’s use in colon cancer treatments and p53’s role in nucleolar stress-induced cell death.
Further study is warranted to provide clarification on the molecular mechanisms by which these compounds induce such different responses in the cell. For example, the stress-inducers may be interfering with progression of ribosome biogenesis,17,19 disrupting an intermolecular interaction of NPM1 that sequesters it in the nucleolus,18 altering biophysical properties of nucleic acids,35,36 or globally perturbing the biomolecular interactions that maintain nucleolar integrity. More work is needed to understand this fascinating biological stress process and to define the specific properties of Pt(II) compounds that cause it.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation [CHE1710721 to V.J.D.], the National Institutes of Health [T32 GM007759-29 to E.C.S.] and used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation [ACI-1548562]. Computations were also performed on the PICS Coeus high performance computer, which is supported by the National Science Foundation [1624776]. This work is also supported by the Department of Chemistry and Biochemistry, Department of Biology, Institute of Molecular Biology and the Material Science Institute at the University of Oregon.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b10319.
Supplementary figures, tables, and materials and methods (PDF)
Supplementary NMR spectra (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Kelland L The Resurgence of Platinum-Based Cancer Chemotherapy. Nat. Rev. Cancer 2007, 7 (8), 573–584. [DOI] [PubMed] [Google Scholar]
- (2).Wexselblatt E; Yavin E; Gibson D Cellular Interactions of Platinum Drugs. Inorg. Chim. Acta 2012, 393, 75–83. [Google Scholar]
- (3).Bruno PM; Liu Y; Park GY; Murai J; Koch CE; Eisen TJ; Pritchard JR; Pommier Y; Lippard SJ; Hemann MT A Subset of Platinum-Containing Chemotherapeutic Agents Kills Cells by Inducing Ribosome Biogenesis Stress. Nat. Med 2017, 23 (4), 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tsai RYL; Pederson T Connecting the Nucleolus to the Cell Cycle and Human Disease. FASEB J 2014, 28 (8), 3290–3296. [DOI] [PubMed] [Google Scholar]
- (5).Woods SJ; Hannan KM; Pearson RB; Hannan RD The Nucleolus as a Fundamental Regulator of the P53 Response and a New Target for Cancer Therapy. Biochim. Biophys. Acta, Gene Regul. Mech 2015, 1849 (7), 821–829. [DOI] [PubMed] [Google Scholar]
- (6).Boulon S; Westman BJ; Hutten S; Boisvert F-MM; Lamond AI The Nucleolus under Stress. Mol. Cell 2010, 40 (2), 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Farley-Barnes KI; McCann KL; Ogawa LM; Merkel J; Surovtseva YV; Baserga SJ Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number. Cell Rep 2018, 22 (7), 1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pickard AJ; Bierbach U The Cell’s Nucleolus: An Emerging Target for Chemotherapeutic Intervention. ChemMedChem 2013, 8 (9), 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wirth R; White JD; Moghaddam AD; Ginzburg AL; Zakharov LN; Haley MM; DeRose VJ Azide vs. Alkyne Functionalization in Pt(II) Complexes for Post-Treatment Click Modification: Solid State Structure, Fluorescent Labeling, and Cellular Fate. J. Am. Chem. Soc 2015, 137 (48), 15169–15175. [DOI] [PubMed] [Google Scholar]
- (10).Plakos K; DeRose VJ Mapping Platinum Adducts on Yeast Ribosomal RNA Using High-Throughput Sequencing. Chem. Commun 2017, 53 (95), 12746–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hostetter AA; Osborn MF; DeRose VJ Characterization of RNA-Pt Adducts Formed from Cisplatin Treatment of Saccharomyces cerevisiae. ACS Chem. Biol 2012, 7 (1), 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Osborn MF; White JD; Haley MM; DeRose VJ Platinum-RNA Modifications Following Drug Treatment in S. cerevisiae Identified by Click Chemistry and Enzymatic Mapping. ACS Chem. Biol 2014, 9 (10), 2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Melnikov SV; Soll D; Steitz TA; Polikanov YS Insights into RNA Binding by the Anticancer Drug Cisplatin from the Crystal Structure of Cisplatin-Modified Ribosome. Nucleic Acids Res 2016, 44 (10), 4978–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Rijal K; Chow CS A New Role for Cisplatin: Probing Ribosomal RNA Structure. Chem. Commun 2008, 0 (1), 107–109. [DOI] [PubMed] [Google Scholar]
- (15).Saunders AM; DeRose VJ Beyond Mg2+: Functional Interactions between RNA and Transition Metals. Curr. Opin. Chem. Biol 2016, 31, 153–159. [DOI] [PubMed] [Google Scholar]
- (16).Sutton EC; McDevitt CE; Yglesias MV; Cunningham RM; DeRose VJ Tracking the Cellular Targets of Platinum Anticancer Drugs: Current Tools and Emergent Methods. Inorg. Chim. Acta 2019, 498, 118984. [Google Scholar]
- (17).Rubbi CP; Milner J Disruption of the Nucleolus Mediates Stabilization of p53 in Response to DNA Damage and Other Stresses. EMBO J. 2003, 22 (22), 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yang K; Wang M; Zhao Y; Sun X; Yang Y; Li X; Zhou A; Chu H; Zhou H; Xu J; Wu M; Yang J; Yi J A Redox Mechanism Underlying Nucleolar Stress Sensing by Nucleophosmin. Nat. Commun 2016, 7, 13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bursac S; Brdovcak MC; Donati G; Volarevic S Activation of the Tumor Suppressor P53 upon Impairment of Ribosome Biogenesis. Biochim. Biophys. Acta, Mol. Basis Dis 2014, 1842 (6), 817–830. [DOI] [PubMed] [Google Scholar]
- (20).Nicolas E; Parisot P; Pinto-Monteiro C; de Walque R; De Vleeschouwer C; Lafontaine DLJ Involvement of Human Ribosomal Proteins in Nucleolar Structure and P53-Dependent Nucleolar Stress. Nat. Commun 2016, 7, 11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bruno PM; Liu Y; Park GY; Murai J; Koch CE; Eisen TJ; Pritchard JR; Pommier Y; Lippard SJ; Hemann MT A Subset of Platinum-Containing Chemotherapeutic Agents Kills Cells by Inducing Ribosome Biogenesis Stress. Nat. Med 2017, 23 (4), 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).McDevitt CE; Yglesias MV; Mroz AM; Sutton EC; Yang MC; Hendon CH; DeRose VJ Monofunctional Platinum(II) Compounds and Nucleolar Stress: Is Phenanthriplatin Unique? J. Biol. Inorg. Chem 2019, 24 (6), 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).This model is supported by previously published data demonstrating that cisplatin causes significantly more DNA damage than oxaliplatin24–26 and that DDR-mediated cell death occurs upon cisplatin treatment, but not oxaliplatin treatment.3 Additionally, data from our lab shows that DNA damage occurs at early time points in cisplatin-treated cells, prior to any putative NPM1 relocation, whereas no such damage occurs prior to observed NPM1 distribution in oxaliplatin-treated cells (unpublished).
- (24).Chaney SG; Campbell SL; Bassett E; Wu Y Recognition and Processing of Cisplatin- and Oxaliplatin-DNA Adducts. Crit. Rev. Oncol. Hematol 2005, 53 (1), 3–11. [DOI] [PubMed] [Google Scholar]
- (25).Faivre S; Chan D; Salinas R; Woynarowska B; Woynarowski JM DNA Strand Breaks and Apoptosis Induced by Oxaliplatin in Cancer Cells. Biochem. Pharmacol 2003, 66 (2), 225–237. [DOI] [PubMed] [Google Scholar]
- (26).Woynarowski JM; Faivre S; Herzig MC; Arnett B; Chapman WG; Trevino AV; Raymond E; Chaney SG; Vaisman A; Varchenko M; Juniewicz PE Oxaliplatin-Induced Damage of Cellular DNA. Mol. Pharmacol 2000, 58 (5), 920–927. [DOI] [PubMed] [Google Scholar]
- (27).Jerremalm E; Videhult P; Alvelius G; Griffiths WJ; Bergman T; Eksborg S; Ehrsson H Alkaline Hydrolysis of Oxaliplatin—Isolation and Identification of the Oxalato Monodentate Intermediate. J. Pharm. Sci 2002, 91 (10), 2116–2121. [DOI] [PubMed] [Google Scholar]
- (28).Zacharioudakis E; Agarwal P; Bartoli A; Abell N; Kunalingam L; Bergoglio V; Xhemalce B; Miller KM; Rodriguez R Chromatin Regulates Genome Targeting with Cisplatin. Angew. Chem., Int. Ed 2017, 56 (23), 6483–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Brunner H; Schellerer K-M New Porphyrin Platinum Conjugates for the Cytostatic and Photodynamic Tumor Therapy. Inorg. Chim. Acta 2003, 350, 39–48. [Google Scholar]
- (30).Siew Y-Y; Neo S-Y; Yew H-C; Lim S-W; Ng Y-C; Lew S-M; Seetoh W-G; Seow S-V; Koh H-L Oxaliplatin Regulates Expression of Stress Ligands in Ovarian Cancer Cells and Modulates Their Susceptibility to Natural Killer Cell-Mediated Cytotoxicity. Int. Immunol 2015, 27 (12), 621–632. [DOI] [PubMed] [Google Scholar]
- (31).Englinger B; Pirker C; Heffeter P; Terenzi A; Kowol CR; Keppler BK; Berger W Metal Drugs and the Anticancer Immune Response. Chem. Rev 2019, 119 (2), 1519–1624. [DOI] [PubMed] [Google Scholar]
- (32).Terenzi A; Pirker C; Keppler BK; Berger W Anticancer Metal Drugs and Immunogenic Cell Death. J. Inorg. Biochem 2016, 165, 71–79. [DOI] [PubMed] [Google Scholar]
- (33).McKeage MJ; Hsu T; Screnci D; Haddad G; Baguley BC Nucleolar Damage Correlates with Neurotoxicity Induced by Different Platinum Drugs. Br. J. Cancer 2001, 85 (8), 1219–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Toscano F; Parmentier B; Fajoui ZE; Estornes Y; Chayvialle J-A; Saurin J-C; Abello J P53 Dependent and Independent Sensitivity to Oxaliplatin of Colon Cancer Cells. Biochem. Pharmacol 2007, 74 (3), 392–406. [DOI] [PubMed] [Google Scholar]
- (35).Keck MV; Lippard SJ Unwinding of Supercoiled DNA by Platinum-Ethidium and Related Complexes. J. Am. Chem. Soc 1992, 114 (9), 3386–3390. [Google Scholar]
- (36).Malina J; Novakova O; Vojtiskova M; Natile G; Brabec V Conformation of DNA GG Intrastrand Cross-Link of Antitumor Oxaliplatin and Its Enantiomeric Analog. Biophys. J 2007, 93 (11), 3950–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


