Abstract
RNA interference (RNAi) is an antiviral pathway common to many eukaryotes that detects and cleaves foreign nucleic acids. In mammals, mitochondrially localized proteins such as mitochondrial antiviral signaling (MAVS), retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated protein 5 (MDA5) mediate antiviral responses. Here, we report that mitochondrial dysfunction in Caenorhabditis elegans activates RNAi-directed silencing via induction of a pathway homologous to the mammalian RIG-I helicase viral response pathway. The induction of RNAi also requires the conserved RNA decapping enzyme EOL-1/DXO. The transcriptional induction of eol-1 requires DRH-1 as well as the mitochondrial unfolded protein response (UPRmt). Upon mitochondrial dysfunction, EOL-1 is concentrated into foci that depend on the transcription of mitochondrial RNAs that may form double-stranded RNA (dsRNA), as has been observed in mammalian antiviral responses. Enhanced RNAi triggered by mitochondrial dysfunction is necessary for the increase in longevity that is induced by mitochondrial dysfunction.
Surveillance of mitochondrial dysfunction in the nematode Caenorhabditis elegans triggers the activation of an RNA interference pathway to mediate antiviral defense, in a manner homologous to the mammalian RIG-I helicase viral response pathway.
Introduction
Many RNA viruses carry an RNA-dependent RNA polymerase (RdRP) to replicate their RNA genome in the host, bypassing entirely information storage and replication with DNA. In this way, RNA viruses can replicate in nondividing cells with lower deoxyribonucleotide levels than those required for DNA viruses or retroviruses but with the substantial ribonucleotide levels needed for transcription of RNA and mRNAs in terminally differentiated cells. A double-stranded RNA (dsRNA) viral replication intermediate is a strong clue to the host cell that an RNA virus infection is underway. Recognition of the dsRNA replication intermediate is an initial step in antiviral immune responses. In mammals, the RNA helicases retinoic acid-inducible gene I (RIG-I) or melanoma differentiation-associated protein 5 (MDA5) recognize the RNA signatures of RNA virus replication [1]. RIG-I binds to dsRNA, as well as single-stranded RNA (ssRNA) with 5′-triphosphate that is a signature of the products from RdRPs that mediate viral RNA replication [2]. RIG-I or MDA5 associate with the mitochondrial antiviral signaling protein (MAVS) and elicit the downstream nuclear factor kappa B (NF-κB) and other interferon immune signaling that mediate systemic antiviral immune defenses [3].
The nematode Caenorhabditis elegans uses an RNA interference (RNAi) pathway to mediate antiviral defense instead of the interferon signaling of vertebrates and most invertebrates [4–7]. RNAi is a highly conserved mechanism for antiviral defense and broadly deployed by eukaryotes, including, fungi, nematodes, insects, plants, and vertebrates [8–10]. RNAi was initially discovered to mediate silencing triggered by engineered dsRNA triggers but soon discovered to produce natural small interfering RNAs (siRNAs) that target specific mRNAs for degradation and chromatin regions for epigenetic silencing. RNAi is mediated by 22 nt to 26 nt single-stranded short interfering or siRNAs that are produced and presented to target mRNAs by the Argonaute proteins and the Dicer dsRNA ribonuclease, conserved across many but not all eukaryotes [11].
Some of the genes in the vertebrate and insect NF-κB pathway are conserved between nematodes and other animals without RdRP genes. Like its mammalian orthologues RIG-I and MDA5, the C. elegans DRH-1 mediates antiviral RNAi and is essential for neutralization of an invading virus [12,13]. DRH-1 was initially identified as a binding partner with the Dicer protein DCR-1, the Argonaute protein RDE-1, and the RNA helicase RDE-4 [14]. In addition to drh-1, drh-2 is a pseudogene, and drh-3 encodes a component of an RdRP protein complex [15].
The DCR-1 ribonuclease is the first step in siRNA generation in most of the C. elegans RNAi pathways, including exogenous RNAi, endogenous RNAi, and antiviral RNAi [15,16]. siRNAs generated by these pathways engage the Mutator proteins that mediate siRNA-guided repression in both exogenous and endogenous RNAi [17,18]. Mutations in the Dicer, Argonaute, and accessory factors that generate and present siRNAs to target mRNAs are generally defective for RNAi or antiviral defense. But the Argonaute and RdRP gene families are expanded in C. elegans compared with most animals, and surprisingly, loss-of-function mutations in some of those genes causes an increase in the response to siRNAs: mutations in the RdRP RRF-3, the specialized Argonaute ERGO-1, the RNA helicase ERI-6/7, or the exoribonuclease ERI-1 enhance silencing by siRNAs [19,20]. Many of these enhanced RNAi (Eri) mutations desilence retroviral elements in the C. elegans genome so that antiviral response pathways are triggered to in turn induce the expression of RNAi-based antiviral defense [21].
A mutation in the C. elegans mitochondrial chaperone gene hsp-6 causes induction of a suite of drug detoxification and defense genes [22]. Here, we show that in addition to the induction of these defense pathways, RNAi pathways are also activated. Because RNAi is a key feature of C. elegans antiviral defense, and because of the association of mammalian viral defense pathways such as MAVS and RIG-I with the mitochondrion, we explored more fully how mitochondrial homeostatic pathways connect to RNAi and antiviral defense in C. elegans. We find that reduction of function mutations in a wide range of mitochondrial components robustly enhanced RNAi-mediated silencing of endogenous genes as well as a variety of reporters of RNAi. These antiviral responses to mitochondrial dysfunction are homologous to the RIG-I-based mitochondrial response in mammals because they depend on the RIG-I homologue, the DRH-1 RNA helicase. Comparing the C. elegans transcriptional response of a mitochondrial mutant and infection with the Orsay RNA virus, we found a striking overlap of expression of multiple members of C. elegans pals genes implicated in antiviral and anti-pathogen response pathways [23,24] and the eol-1/DXO RNA decapping enzyme gene. We found that eol-1 transcription is dramatically induced by mitochondrial dysfunction, and an eol-1 null mutation strongly suppresses the DRH-1-mediated antiviral RNAi response normally induced by mitochondrial dysfunction. We showed that the EOL-1 protein forms foci in the cytosol only if the mitochondrion is stressed, and the production of these foci is dependent on transcription of RNA from the mitochondrial genome. This is reminiscent of the central role that dsRNA released from the mitochondria plays in mammalian antiviral response pathways: During a viral infection, mitochondrial disruption by MAVS and other mitochondrial-associated viral immunity factors releases mitochondrial dsRNA and DNA to the cytosol, where dsRNAs trigger an MDA5-dependent interferon response [25,26], and DNA via cGAS and Sting activate NF-κB immune signaling [27].
Gene inactivation or mutations in a wide variety of nuclear-encoded mitochondrial genes cause one of the strongest increases in the life span in genome screens for life span extension [28]. We find that the uncoupling of the antiviral defense response via drh-1 or eol-1 mutations abrogates the increase in life span, suggesting that the antiviral axis of mitochondrial dysfunction is critical to the life span extension.
Results
C. elegans mitochondrial mutations enhance RNAi using the RNA helicase DRH-1
The hsp-6(mg585) allele is a reduction of function mutation (P386S) in the mitochondrial HSP70 chaperone in a region that is conserved between mammals and C. elegans (VQEIFGKVPSKAVNPDEAVA). hsp-6(mg585) causes induction of a suite of drug detoxification and defense genes that are also induced by a variety of mitochondrial mutations or toxins that disrupt mitochondrial function [22]. C. elegans HSP-6 and human mtHSP70 are orthologues of bacterial and archaeal dnaK; these mitochondrial chaperones were bacterial chaperones before the mitochondrial endosymbiosis event more than a billion years ago and the migration of these bacterial genes to the eukaryotic nuclear genome [22]. Null alleles of hsp-6 cause developmental arrest, presumably because of the defects in the folding or import of many mitochondrial client proteins [29,30]. But the viable hsp-6(mg585) allele allows mitochondrial roles in other pathways to be studied without the associated lethality. In addition to activating detoxification and immune responses, we found that the mitochondrial defects caused by hsp-6(mg585) also unexpectedly cause enhanced RNAi, which is an antiviral defense pathway.
RNAi in C. elegans can be induced by ingestion of approximately 1 kb of dsRNA corresponding to a particular gene. For feeding RNAi, Escherichia coli have been engineered to produce any of 18,000 dsRNAs corresponding to any C. elegans gene [31]. The specificity of RNAi in C. elegans is superior to the single siRNA approaches common in most animal systems, probably because, almost unique to animals, C. elegans uses RdRP genes to amplify the primary siRNAs produced by the Argonaute and Dicer proteins that nearly all eukaryotes also use for RNAi, and unlike in other animal systems, RNAi in C. elegans can be induced by 1-kb segments of dsRNA without the induction of interferon-related responses to dsRNA. Thus, thousands of siRNAs produced from feeding C. elegans with an E. coli expressing a 1-kb dsRNA sum for on-target effects on target mRNA inactivation and average for off-target mRNA inactivations. The specificity of dsRNA-induced RNAi was validated with genetic loci that had previously studied by genetics: Most dsRNAs corresponding to those genes with known phenotypes recapitulated the phenotypes predicted from genetics [32]. But a minority of genes expected to generate easily scored phenotypes by RNAi did not silence in wild type. These dsRNAs were used in genetic screens for C. elegans mutations that enhance RNAi, or eri- mutations [19]. For example, mutations in the conserved exonuclease eri-1 or the RdRP rrf-3 cause Eri [15,19]. Most mutants that enhance RNAi responses also cause silencing of multicopy transgenes, because the transgene is detected as a bearing some foreign signatures (lack of introns is a major signature of a viral origin) and silenced in the Eri state of these mutant strains. A set of dsRNA tester genes have been developed that cause strong phenotypes in Eri mutants, but no RNAi phenotype in wild type [33].
We found that the mitochondrial hsp-6(mg585) mutant showed the strong phenotype seen in Eri mutants on many of these Eri tester dsRNAs expressed from E. coli. For example, a dsRNA targeting the lir-1 (where lir is an abbreviation for lin-26-related) gene causes no phenotype in wild type, but causes lethal arrest in, for example, the eri-6(mg379)-Eri mutant, as well as on the mitochondrial hsp-6(mg585) mutant (Fig 1A). The enhanced lethality after exposure to lir-1 dsRNA in Eri mutants is due to siRNAs from this dsRNA targeting the duplicated and diverged genes with high regions of nucleotide homology on the same primary transcript of the lir-1, lir-2, and lin-26 operon [34]. One explanation for the enhanced response to lir-1 RNAi is that these genes have genetic interactions with hsp-6(mg585) that caused synthetic enhancement of certain phenotypes. Therefore, we tested whether other mitochondrial mutations enhance response to Eri tester dsRNAs. lir-1 RNAi also caused a lethal arrest on a variety of other nuclearly encoded mitochondrial protein point mutants, including nicotinamide adenine dinucleotide (NADH) dehydrogenase nuo-6/NDUFB4, ubiquinone biosynthesis clk-1/COQ7, and the iron–sulfur cluster isp-1/UQCRFS1 (Fig 1A). A distinct dsRNA that targets a histone 2B gene, his-44, causes larval arrest in Eri mutants but no lethality on wild type [35] and also caused larval arrest in the hsp-6/mtHSP70, nuo-6/NDUFB4, clk-1/COQ7, and isp-1/UQCRFS1 mitochondrial mutants (Fig 1A). his-44 maps to a cluster of histone genes including multiple histone 2B genes with nucleic acid homology; in the Eri mutants, the initial siRNAs produced from the his-44 dsRNA may spread to adjacent histone 2B genes. Thus, the enhanced response to lir-1 or his-44 RNAi was not due to strain background mutations in hsp-6(mg585) or the pleiotropy of a mitochondrial chaperone that may affect the function of many imported mitochondrial proteins. Three other mitochondrial point mutants that we tested (out of 6 tested), nduf-7(et19), mev-1(kn1), and gas-1(fc21) did not cause an Eri phenotype (S1A Fig). Many mutations in nuclearly encoded mitochondrial proteins are lethal, so that amino acid substitution reduction in function alleles that we tested constitutes most of the available viable mitochondrial mutations. NUO-6, GAS-1, and NDUF-7 are components of complex I, MEV-1 is from complex II, ISP-1 is a component of complex III, and CLK-1 produces the ubiquinone that also functions in electron transport. There was no obvious distinction between the types of mitochondrial mutations that caused Eri and those that did not, in terms of growth rate or severity of the phenotype. It is possible that only particular mitochondrial insults activate the RNAi pathway. The induction of Eri by multiple mitochondrial mutations favored the hypothesis that many but not all mitochondrial defects trigger an Eri phenotype.
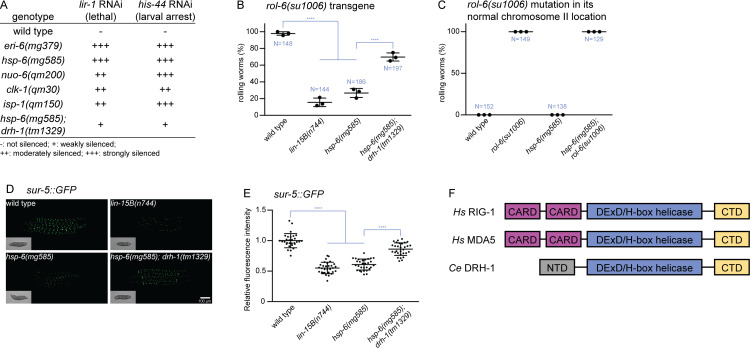
Fig 1. Mitochondrial mutants activate Eri through DRH-1.
(A) Eri response to lir-1 RNAi or his-44 RNAi in the mitochondrial mutants hsp-6, nuo-6, clk-1, or isp-1 or the eri-6 Eri mutant causes lethality/arrest but not on wild type. The Eri of the hsp-6 mitochondrial mutant is suppressed by a drh-1 mutation. (B) Transgene silencing test with rol-6(su1006) transgene. This multicopy transgene in wild type causes a rolling behavior due to the expression of an amino acid substitution in the rol-6 collagen gene. The expression of the rol-6 collagen mutation from the multicopy transgene is silenced by Eri in mitochondrial mutants, and this transgene silencing depends on drh-1 gene activity. Results of 3 replicate experiments are shown. N: total number animals tested; **** denotes p < 0.0001. (C) The mitochondrial mutations do not simply suppress the collagen defect of a rol-6 mutation: If the rol-6(su1006) mutation is chromosomally expressed, not from a transgene that is under RNAi control, the Rol phenotype is not suppressed by hsp-6(mg585) and an hsp-6(mg585) single mutation does not have any Rol phenotype. Results of 3 replicate experiments are shown. N: total number of animals tested. (D and E) Transgene silencing test with sur-5::GFP transgene. This transgene is ubiquitously expressed in all somatic cells. The expression of the sur-5::GFP from the transgene is silenced by Eri in mitochondrial mutants, and this transgene silencing depends on drh-1 gene activity. Animals were imaged in (D) and the fluorescence was quantified in (E). **** denotes p < 0.0001. (F) Diagrams of human RIG-I, MDA5, and C. elegans DRH-1. The helicase domain and CTD are conserved. The underlying numerical data can be found in S1 data. CARD, caspase recruitment domain; Ce, Caenorhabditis elegans; CTD, C-terminal domain; Eri, enhanced RNAi; Hs, Homo sapiens; NTD, N-terminal domain; RNAi, RNA interference.
A hallmark of an Eri phenotype is the silencing of transgenes, as increased RNAi detects the foreign genetic signatures (the fusion of non-C. elegans genes such as green fluorescent protein (GFP), the synthetic introns, and other engineered features) of transgenes [36]. We asked if the hsp-6(mg585) mutant displayed enhanced transgene silencing by introducing a transgene containing rol-6(su1006), a dominant mutation of a hypodermal collagen that causes an easily scored rolling movement phenotype [37]. lin-15B is a class B synthetic multivulva (synMuv B) gene, and loss of function of lin-15B significantly enhances transgene silencing [38]. A chromosomally integrated transgene carrying multiple copies of the rol-6(su1006) mutant collagen gene causes 100% of transgenic animals to roll (a Rol phenotype) in wild-type animals, but in strains with enhanced RNAi, this transgene is now silenced so that 16% of animals are Rol in the lin-15B(n744) mutant and 27% are Rol in the hsp-6(mg585) homozygous mutant (Fig 1B). The suppression of the Rol phenotype from the transgene carrying rol-6(su1006) was not due to a genetic interaction between hsp-6(mg585) and the rol-6(su1006) mutant collagen gene, because the hsp-6(mg585); rol-6(su1006) double mutant with the rol-6 collagen mutation located in its normal chromosomal location not subject to Eri silencing of a transgene, was still 100% Rol (Fig 1C). Rather, the Eri of the mitochondrial mutants, including hsp-6(mg585), causes a silencing of the rol-6(su1006) mutant collagen allele on the multicopy transgene to suppress the Rol phenotype.
To evaluate transgene silencing in other tissues, a ubiquitously expressed sur-5::GFP fusion gene was monitored in all somatic cells [39]. The bright GFP signal of sur-5::GFP was dramatically decreased in the Eri mutant lin-15B(n744) as well as in the hsp-6(mg585) mitochondrial mutant (Fig 1D and 1E). Thus, hsp-6(mg585) enhances exogenous RNAi and silences somatic transgenes. The somatic transgene silencing and Eri in the eri-1 mutant is associated with a failure to nuclearly localize the Argonaute transcriptional silencing factor NRDE-3 that acts downstream of siRNA generation [33,40]. The synMuvB Eri mutants cause a somatic misexpression of the normally germline P granules implicated in siRNA [40]. However, neither NRDE-3 nuclear delocalization nor the somatic expression of P granules occurred in hsp-6(mg585) mutant, suggesting that mitochondrial dysfunction does not induce the eri-1/rrf-3 or synMuvB classes of Eri.
The Eri of hsp-6(mg585) most resembled the eri-6/7 RNA helicase and ergo-1 Argonaute Eri phenotypes, associated with desilencing of recently acquired viral genes and induction of viral immunity, without nuclear NRDE-3 or somatic P granule expression [21,36,41]. In mammalian cells, the RNA helicase MDA5 mediates an interferon antiviral immune response that is strongly enhanced by a mitochondrial RNA degradation mutation that enhances production of mitochondrial dsRNAs [25]. Intriguingly, mutations in the C. elegans homologue of MDA5, drh-1, suppress the synthetic lethality of Eri and dsRNA editing double mutants [21,42]. C. elegans DRH-1 contains 3 domains, including conserved helicase domain and C-terminal domain (CTD) (Fig 1F) and an N-terminal domain (NTD) that is only conserved in nematodes (S1A Fig). The caspase activation and recruitment domains (CARD) of RIG-I and MDA5 associate with the mitochondrial protein MAVS, which is unique to mammals [3]. Conversely, the NTD of DRH-1 is nematode-specific and essential for the inhibition of viral replication [13]. To test if DRH-1 is required for hsp-6(mg585) induced transgene silencing, 2 transgenes rol-6(su1006) and sur-5::GFP were tested for silencing in an hsp-6; drh-1 double mutant. The drh-1(tm1329) allele disrupts the NTD and renders the strain susceptible to viral infection [12]. drh-1(tm1329) suppressed the transgene silencing induced by hsp-6(mg585): hsp-6(mg585) animals carrying the rol-6(su1006) transgene were 27% Rol, but the hsp-6(mg585); drh-1(tm1329) double mutant was 70% Rol (Fig 1B), showing that the transgene silencing induced by hsp-6(mg585) was suppressed by drh-1(tm1329). Similarly, the GFP fluorescence of sur-5::GFP was significantly brighter in the hsp-6(mg585), drh-1(tm1329) double mutant compared with hsp-6(mg585) (Fig 1D and 1E). The lir-1 and his-44 Eri lethal or larval arrest phenotypes of hsp-6(mg585) were also suppressed by drh-1(tm1329) (Fig 1A). drh-1 mutant animals carrying a distinct mutant allele from a wild strain of C. elegans with a C-terminal truncation are competent for RNAi but show defects in antiviral RNAi [16]. Therefore, hsp-6(mg585) enhancement of transgene silencing and his-44 and lir-1 RNAi responses require DRH-1 gene activity.
EOL-1 acts downstream of DRH-1 for somatic silencing
Upon virus infection, human RIG-I and MDA5 mediate the up-regulation of interferon genes. Although the interferon signaling pathway is not conserved in C. elegans, we suspected DRH-1, like its mammalian orthologues, might promote the transcriptional activation of downstream response genes. Comparison of the expression profile of hsp-6(mg585) [22] and wild-type animals infected with the Orsay RNA virus [43] showed that of the 126 genes most up-regulated by Orsay virus infection, 45 genes were also the top up-regulated genes in the hsp-6(mg585) mutant (Fig 2A and S1 Table). Of these 45 genes, 12 are members of the pals-1 to pals-40 genes, which located in a few clusters, and remarkably, a null allele in pals-22 causes an Eri phenotype, multicopy transgene silencing [23,24]. Thus, the increased expression of pals- genes in virus-infected and mitochondrial defective animals is not just associated with enhanced RNAi, but a mutation in one pals-22 gene can cause increased RNAi and transgene silencing.
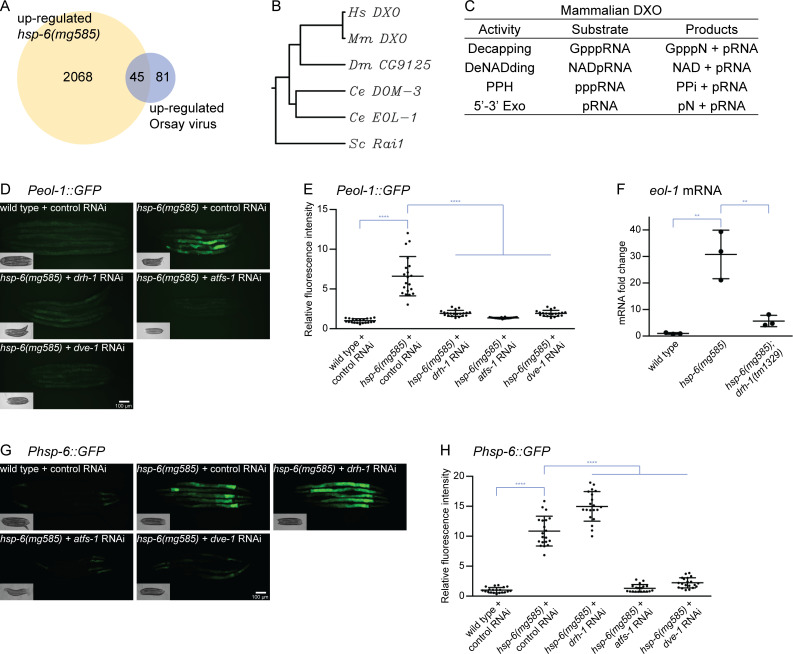
Fig 2. Increased eol-1 expression depends on DRH-1.
(A) Venn diagram of genes up-regulated in hsp-6(mg585) and Orsay virus infection. (B) Phylogenetic tree of EOL-1/DXO. EOL-1 is conserved from yeast to mammals. (C) The enzyme activities of human DXO. Mammalian DXO modifies the 5′ end of mRNAs: decapping, deNADing, pyrophosphohydrolase and 5′-3′ exonuclease. (D and E) The induction of Peol-1::GFP transcriptional fusion reporter requires drh-1 and UPRmt. The Peol-1::GFP is strongly induced by hsp-6(mg585) mitochondrial mutant, and this induction is abrogated by drh-1 RNAi as well as RNAi of genes involved in UPRmt (atfs-1 and dve-1). Animals were imaged in (D) and the fluorescence was quantified in (E). **** denotes p < 0.0001. (F) The hsp-6(mg585) mutant causes increased mRNA level of eol-1 in a drh-1-dependent manner. RT-qPCR assays showed the mRNA level of eol-1 was induced by hsp-6(mg585) mutant, and this induction was abolished in hsp-6(mg585); drh-1(tm1329) double mutant. ** denotes p < 0.01. (G and H) DRH-1 does not contribute to UPRmt. The benchmark reporter of UPRmt Phsp-6::GFP is induced by the hsp-6(mg585) mutant. And the induction is suppressed by RNAi of atfs-1 or dev-1, but not drh-1. Animals were imaged in (G) and the fluorescence was quantified in (H). **** denotes p < 0.0001. The underlying numerical data can be found in S1 data. Ce: Caenorhabditis elegans; Dm: Drosophila melanogaster; Hs: Homo sapiens; Mm: Mus musculus; RT-qPCR, quantitative reverse transcription PCR; Sc: Saccharomyces cerevisiae. UPRmt, mitochondrial unfolded protein response.
The eol-1 RNA decapping gene was also strongly induced in virally infected and mitochondrial mutant C. elegans (S1 Table). We selected eol-1 for detailed analysis because (1) it is conserved from yeast to mammals (Fig 2B); (2) eol-1 expression is induced by multiple mitochondrial mutations as well as in a eri-6; adr-1/2 Eri mutant that desilences endogenous retroviruses and retrotransposons [21,44]; (3) the induction of eol-1 by Orsay virus infection is dependent on drh-1 [45]; and (4) the human orthologue of EOL-1, DXO, represses hepatitis C virus replication [46]. C. elegans eol-1 was initially identified as a mutant that enhanced olfactory learning after Pseudomonas aeruginosa infection [47]. eol-1 encodes a decapping exoribonulease orthologous to yeast Rai1 and mammalian DXO (Fig 2B). Mammalian DXO processes the 5′ end of mRNA [48], including a decapping activity that removes the unmethylated guanosine cap and the first nucleotide (GpppN), the deNADing activity that removes the nicotinamide adenine dinucleotide (NAD) cap, and the pyrophosphohydrolase (PPH) activity that releases pyrophosphate (PPi) from 5′ triphosphorylated RNA, and 5′-3′ exonuclease (5′-3′ Exo) to degrade the entire RNA (Fig 2C). The removal of the N7-methyl guanosine (m7G) cap and subsequent degradation of mammalian mRNAs are directed by the decapping exoribonuclease DXO. Expression of Mus musculus DXO rescued the enhanced olfactory learning phenotype in C. elegans eol-1 mutant indicating not only the amino acid sequence but also the function of EOL-1 is conserved [47]. eol-1 is one of 10 C. elegans DXO homologues. Interestingly, 6 of these genes (M01G12.7, M01G12.9, M01G12.14, Y47H10A.3, Y47H10A.4, and Y47H10A.5) are clustered as tandemly duplicated genes, adjacent to rrf-2, one of 4 C. elegans RdRPs, and one (C37H5.14) is adjacent to hsp-6.
To verify the transcriptional up-regulation of eol-1 in hsp-6(mg585) mutant and test if the induction requires DRH-1, a transcriptional fusion reporter Peol-1::GFP containing the eol-1 promoter, GFP, and eol-1 3’UTR was constructed. In wild-type animals treated with control RNAi, Peol-1::GFP was barely detectable (Fig 2D and 2E); in the hsp-6(mg585) mitochondrial mutant, Peol-1::GFP was strongly induced (Fig 2D and 2E). The induction of Peol-1::GFP was abrogated in hsp-6(mg585) treated with drh-1 RNAi (Fig 2D and 2E). Since the Peol-1::GFP transcriptional fusion reporter is a transgene that is subject to Eri transgene silencing by hsp-6(mg585) and suppressed by drh-1, it might not reflect the accurate expression level of eol-1 in these mutants. To evaluate the actual mRNA level of chromosomal eol-1, quantitative reverse transcription PCR (RT-qPCR) was performed. The mRNA level of chromosomal eol-1 was 30-fold increased in the hsp-6(mg585) single mutant, and this induction was almost completely abolished in the hsp-6(mg585); drh-1(tm1329) double mutant (Fig 2F). Thus, mitochondrial dysfunction of hsp-6(mg585) triggers the transcriptional activation of eol-1 in a DRH-1-dependent manner.
In C. elegans, the mitochondrial unfolded protein response (UPRmt) is a transcriptional program responding to mitochondrial dysfunction and essential for mitochondrial recovery, immunity, detoxification, and aging [49]. RNAi of the genes atfs-1 or dev-1, which encode transcription factors that mediate the expression of UPRmt genes [50,51], suppresses the induction of Peol-1::GFP (Fig 2D and 2E). We then tested if DRH-1 contributes to the activation of UPRmt by observing the induction of Phsp-6::GFP, the canonical reporter of UPRmt. The Phsp-6::GFP was strongly induced in the hsp-6(mg585) mutant and suppressed by RNAi of atfs-1 or dev-1 as expected (Fig 2G and 2H). RNAi of drh-1 in the hsp-6(mg585); Phsp-6::GFP strain did not disrupt induction of Phsp-6::GFP; in fact, induction of Phsp-6::GFP by hsp-6(mg585) after drh-1(RNAi) was more than in hsp-6(mg585) alone, perhaps because drh-1 inactivation inhibited hsp-6(mg585)-induced Eri and transgene silencing. Thus, the transcriptional activation of eol-1 requires UPRmt signaling, whereas DRH-1 is not part of the general UPRmt.
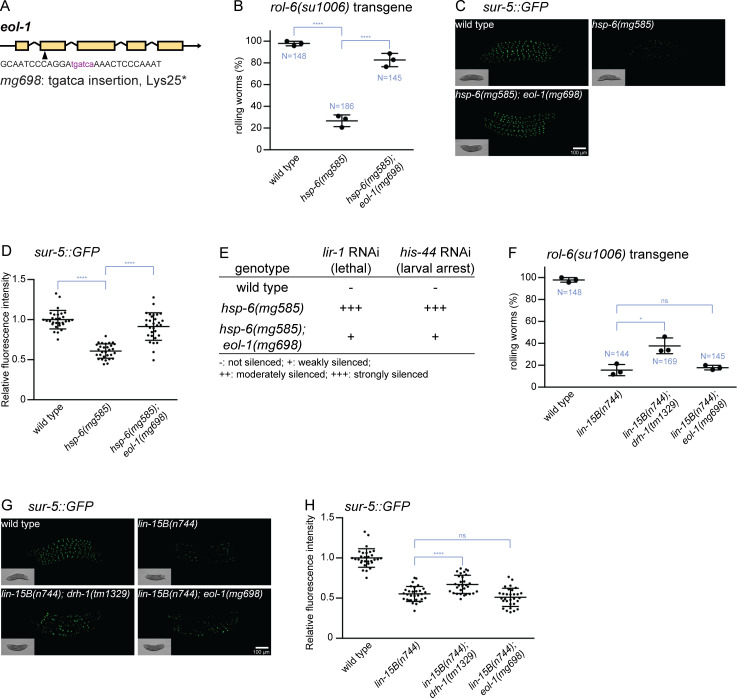
The DRH-1-dependent up-regulation of eol-1 implies that EOL-1 might act in the enhanced RNAi pathway induction of the hsp-6(mg585) mutant. To test this possibility, a loss-of-function mutant eol-1(mg698) was generated by CRISPR-Cas9 [52]. In eol-1(mg698), 6 nucleotides “TGATCA,” which contains a stop codon and a Bcl I endonuclease recognition sequence to facilitate genotyping, was inserted into the eol-1 locus and resulted in a “Lys25 to stop” nonsense mutation (Fig 3A). The rol-6(su1006) and sur-5::GFP transgenes were tested for silencing in the hsp-6; eol-1 double mutant. For the rol-6(su1006) transgene, the hsp-6(mg585); eol-1(mg698) double mutant, showed 82% rolling compared with 27% in the hsp-6(mg585) single mutant (Fig 3B). For the sur-5::GFP transgene, the GFP signal in the hsp-6(mg585); eol-1(mg698) double mutant was substantially increased relative to the hsp-6(mg585) single mutant (Fig 3C and 3D). Moreover, the Eri phenotype of hsp-6(mg585) as assessed using the lethality of the lir-1 or his-44 dsRNAs was also suppressed by eol-1(mg698) (Fig 3E).
Fig 3. EOL-1 gene activity is required for Eri.
(A) The eol-1(mg698) mutant allele generated by CRISPR-Cas9. (B) Transgene silencing test with the rol-6(su1006) multicopy transgene. The expression of the rol-6 collagen mutation from the transgene is silenced by the Eri in the hsp-6(mg585) mutant, and this transgene silencing depends on eol-1 gene activity. Results of 3 replicate experiments are shown. N: total number of animals tested; **** denotes p < 0.0001. (C and D) Transgene silencing test with sur-5::GFP transgene. This transgene was ubiquitously expressed in all somatic cells. The expression of the sur-5::GFP from the transgene is silenced by Eri in the hsp-6(mg585) mutant, and this transgene silencing requires eol-1. Animals were imaged in (C), and the fluorescence was quantified in (D). **** denotes p < 0.0001. (E) Eri response to lir-1 RNAi or his-44 RNAi in hsp-6(mg585) mutant requires eol-1. RNAi of lir-1 or his-44 causes lethality/arrest on hsp-6(mg585) mutant but not wild type. The Eri is suppressed by the eol-1(mg698) mutation. (F) Silencing of rol-6(su1006) transgene caused by synMuvB Eri mutations does not depend on eol-1. The rol-6(su1006) transgene is silenced by the synMuvB lin-15(n744) mutation, and this transgene silencing was slightly suppressed by drh-1(tm1329), but not by eol-1(mg698). Results of 3 replicate experiments are shown. N: total number of animals tested; * denotes p < 0.05; ns denotes p > 0.05. (G and H) Silencing of sur-5::GFP transgene caused by synMuvB Eri mutations does not require eol-1. The sur-5::GFP transgene is silenced by the synMuvB lin-15(n744) mutation, and this transgene silencing was slightly suppressed by drh-1(tm1329), but not eol-1(mg698). Animals were imaged in (G) and the fluorescence was quantified in (H). **** denotes p < 0.0001; ns denotes p > 0.05. The underlying numerical data can be found in S1 data. Eri, enhanced RNAi; ns, not significant; RNAi, RNA interference.
Because mammalian DXO, a homologue of C. elegans EOL-1, is involved in mRNA decay, we tested whether EOL-1 mediates the degradation of mRNAs from transgenes. In such a model, eol-1 might also be required for the somatic transgene silencing caused by other Eri mutants, for example, by the large synMuv B class of Eri mutants. Although drh-1(tm1329) slightly suppressed the transgene silencing of a lin-15B null mutant, eol-1 did not contribute to transgene silencing by the synMuvB mutants (Fig 3F–3H). Thus, EOL-1 does not act in the synMuvB RNAi pathway that silences transgenes and is specifically required for mitochondrial dysfunction-induced transgene silencing.
Formation of EOL-1 foci requires biogenesis of mitochondrial RNAs
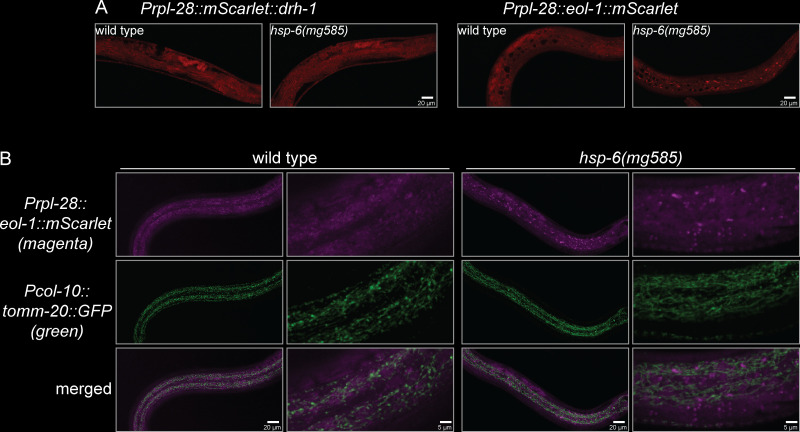
In order to understand the function of DRH-1 and EOL-1 in silencing of transgenes, we monitored their subcellular localization by fusing mScarlet at the N-terminus of DRH-1 and C-terminus of EOL-1, respectively, under the control of rpl-28 promoter in a miniMOS vector [53]. The miniMOS-generated single copy transgene is able to avoid multicopy transgene silencing by the RNAi pathway, and the ribosome promoter Prpl-28 is a constitutive promoter with universal expression in all tissues. In wild-type animals, mScarlet::DRH-1 and EOL-1::mScarlet were localized diffusely in the cytosol without any notable pattern (Fig 4A). In the hsp-6(mg585) mitochondrial mutant, strikingly, EOL-1::mScarlet formed massive puncta in many cell types (Fig 4A). mScarlet::DRH-1 remained diffusely localized in hsp-6(mg585) (Fig 4A), just as no change was observed in DRH-1::GFP subcellular distribution upon Orsay virus infection [16]. The formation of EOL-1::mScarlet foci in the hsp-6 mitochondrial mutant, and the specificity of EOL-1 in the suppression of hsp-6(mg585) induced silencing, and the mammalian finding that MDA5 recognizes mitochondrial dsRNAs generated a hypothesis that the target of EOL-1 decapping are RNAs derived from the mitochondrial genome. First, we examined whether the cytoplasmic EOL-1 foci localized at the mitochondria surface. Because loss-of-function eol-1(mg698) suppresses the mitochondrial defect-induced rol-6(su1006) transgene silencing of in the hypodermis, the hypodermal-specific col-10 promoter-driven TOMM-20::GFP miniMOS single copy transgene was used to monitor mitochondria in hypodermal cells. However, EOL-1::mScarlet foci were not specifically associated with the mitochondria (Fig 4B). Therefore, EOL-1 accumulates as foci in response to mitochondrial dysfunction, but these foci as not mitochondrially associated and may function in the cytosol.
Fig 4. EOL-1 protein forms cytoplasmic foci if mitochondria are dysfunctional.
(A) EOL-1 protein, but not DRH-1 protein form puncta in the hsp-6(mg585) mitochondrial mutant but not in wild type. Mitochondrial dysfunction caused by hsp-6(mg585) mutation triggered the formation of EOL-1::mScarlet foci, while mScarlet::DRH-1 remained diffusely localized. (B) EOL-1 foci are not associated with the mitochondria. In the hsp-6(mg585) mutant, the EOL-1::mScarlet foci do not colocalize with mitochondria that were visualized by the mitochondrial outer membrane protein TOMM-20::GFP.
We explored whether C. elegans mitochondria release RNA into the cytosol, as has been observed in the MDA5 mitochondrial surveillance pathway in human and D. melanogaster [25,26]. The human mtDNA transcribes 37 genes, including 13 mRNAs encoding subunits of electron transport chain (ETC), 2 ribosomal RNAs (rRNA), and 22 transfer RNAs (tRNA), residing on both the heavy and light strands of the mitochondrial DNA [54]. Transcription on both strands generated 2 genome size polycistronic transcripts that are processed into individual genes. During this processing, the complementary noncoding RNAs in mammals are degraded by the mitochondrial degradosome formed by RNA helicase SUV3 and polynucleotide phosphorylase PNPase [55]. Loss of SUV3 or PNPase stabilizes the noncoding RNAs to form dsRNAs with the genes derived from the opposite strand [25]. The C. elegans mitochondrial genome encodes 36 genes with one less ETC subunit [56]. However, these 36 genes are encoded exclusively on the heavy strand of C. elegans mtDNA, and no transcription of the light strand has been detected [57], which disfavors the mitochondrial dsRNA model.
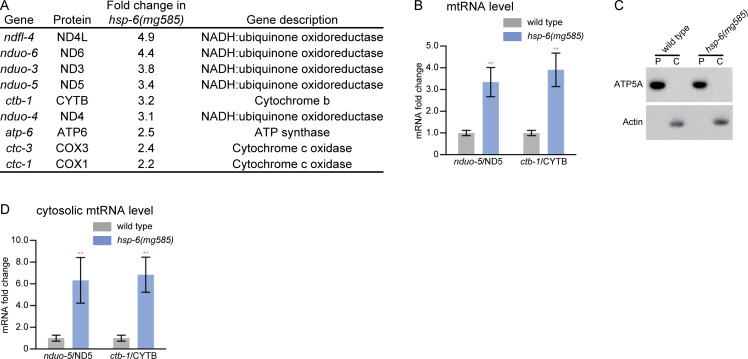
mRNA-seq analysis of hsp-6(mg585) revealed that 9 of 12 mitochondrial mRNA encoding ETC subunits were up-regulated (Fig 5A). The increased level of mitochondrial mRNA was further verified by RT-qPCR analysis of nduo-5/ND5 and ctb-1/CYTB (Fig 5B). In order to examine if the increased abundance of mitochondrial mRNAs within the mitochondrial matrix caused their release into the cytosol, the cytosolic fraction (actin antibody was used as a control protein for this fractionation) was separated by centrifugation from the pellet fraction containing mitochondria (ATP5A antibody used as a control for this fractionation) (Fig 5C). RT-qPCR analyses showed that the mRNA levels of nduo-5/ND5 and ctb-1/CYTB were substantially increased in the cytosol of hsp-6(mg585) compared with wild type (Fig 5D). Analysis of siRNA abundance in C. elegans supports the surveillance of mitochondrial mRNAs by the RNAi pathway. Mitochondrially translated mRNAs from the mitochondrial genome generate some of the most abundant siRNAs (S2 Table). For example, the mitochondrial genes ctc-1, nduo-2, and nduo-5, all mitochondrial genome-encoded electron transport chain genes, generate siRNA levels at thousands of reads per million, rivaling those generated by transposable elements in C. elegans (the retr-1 transposable element at rank 24, ctc-1 at rank 29, nduo-2 is at rank 104, and nduo-5 at rank 119).
Fig 5. Release of mitochondrial RNA into the cytosol.
(A) Electron transport chain genes encoded in the mitochondrial genome are induced by the hsp-6(mg585) mitochondrial chaperone gene mutation. Of the 12 genes in the mitochondrial genome that encode subunits of electron transport chain, 9 genes were up-regulated in hsp-6(mg585) mutation. (B) The mRNA level of nduo-5/ND5 or ctb-1/CYTB was induced by hsp-6(mg585) mitochondrial mutation. Both nduo-5/ND5 (NADH:ubiquinone oxidoreductase) and ctb-1/CYTB (cytochrome b) are transcribed from the mitochondrial genome, and their expression level was evaluated by RT-qPCR assays. **p < 0.01. (C and D) The mRNA level of nduo-5/ND5 or ctb-1/CYTB in the cytosol was increased in hsp-6(mg585) mitochondrial mutation. The pellet and cytosolic fractions were separated by centrifugation and probed with ATP5A and actin antibodies, respectively, to assess purification in (C). The mRNA level of nduo-5/ND5 or ctb-1/CYTB in the mitochondria-free cytosolic fraction was evaluated by RT-qPCR in (D). The underlying numerical data can be found in S1 data. The original blot images can be found in S1 raw images. C, cytosolic fraction; P, pellet fraction; RT-qPCR, quantitative reverse transcription PCR.
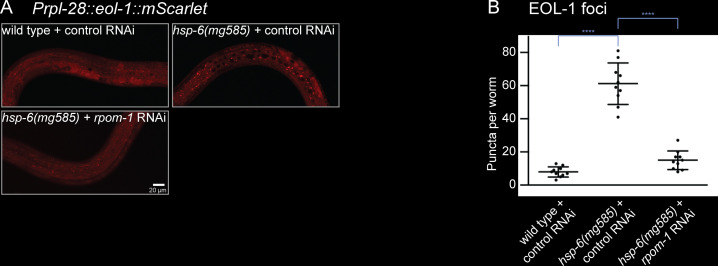
The strategy of single strand transcription from the mitochondrial genome is not just parochial to C. elegans; it is a strategy used by many members of the genus Caenorhabditis [58]. If the unique strand transcriptional architecture of the Caenorhabditis mitochondrial genome is linked with production of dsRNA from the mitochondria, and if the release of mitochondrial RNA into the cytosol in mitochondrial mutants is the trigger for the formation of EOL-1 foci, we would expect a suppression of EOL-1 focus formation by shutting down mitochondrial RNA biogenesis. The human gene POLRMT encodes a mitochondrial DNA-directed RNA polymerase that catalyzes the transcription of mitochondrial DNA [54]. RNAi of rpom-1, the C. elegans orthologue of human POLRMT, potently reduced the number of EOL-1 foci in hsp-6(mg585) mutant (Fig 6A and 6B). Therefore, the formation EOL-1 foci required the biogenesis of mitochondrial RNA. Mitochondrial RNA polymerase is central to all gene expression in the mitochondrion, including the 12 to 13 protein coding genes, the tRNA genes, and the rRNA genes. So a defect in rpom-1 is expected to be pleiotropic and thus difficult to ascribe to production of dsRNA from mitochondrial transcripts. But paradoxically, with 36 client genes in the mitochondrion, rprom-1 has a rather circumscribed sphere of genes it controls, so its function upstream of EOL-1 foci could very well be via dsRNAs produced from mitochondria but released at even higher levels in mitochondrial mutant strains.
Fig 6. Formation of EOL-1 foci requires the biogenesis of mitochondrial RNA.
(A and B) Formation of EOL-1 cytoplasmic foci requires the transcription of mitochondrial RNA from mitochondrial genome by RPOM-1 RNA polymerase. The EOL-1::mScarlet foci formed in the hsp-6(mg585) mutant were inhibited by RNAi of mitochondrial RNA polymerase rpom-1/ POLRMT. Animals were imaged in (A), and the number of foci was quantified in (B). **** denotes p < 0.0001. The underlying numerical data can be found in S1 data.
Enhanced RNAi mediates life span extension
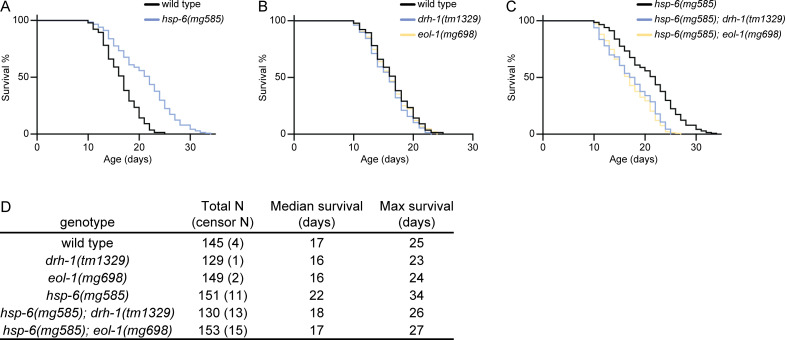
While the majority of mitochondrial components are essential for eukaryotes and null alleles of most mitochondrial genes are lethal, C. elegans reduction-of-function mitochondrial mutants, including hsp-6(mg585) (Fig 7A–7D and S2A–S2D and S2G Fig) increase longevity, in some cases dramatically [28,59,60]. But other mitochondrial mutants, such as mev-1(kn1) and gas-1(fc21), shorten the life span (S2E–S2G Fig). We explored whether the Eri output of the hsp-6(mg585) mitochondrial mutant contributes to its longevity. drh-1(tm1329) or eol-1(mg698) single mutants did not shift the survival curve compared with wild type (Fig 7B–7D), showing that these mutants are not short-lived or sickly in some way. But drh-1(tm1329) or eol-1(mg698) mutations strongly suppressed the life span extension of hsp-6(mg585), showing that drh-1 or eol-1 gene activities are required for the extension of life span by hsp-6(mg585) (Fig 7C and 7D). Because drh-1 and eol-1 mediate the enhanced RNAi, an antiviral response, of mitochondrial mutants, and because these outputs also mediate in the increased longevity of mitochondrial mutants, but not the normal longevity of wild type, these data support the model that enhanced antiviral defense as a key antiaging output from mitochondrial mutations.
Fig 7. Decoupling of the Eri response to mitochondrial dysfunction also abrogates the life span enhancement of mitochondrial mutations, but not wild-type life span.
(A) The hsp-6(mg585) mitochondrial mutation extends life span. (B) drh-1(tm1329) or eol-1(mg698) single mutants do not extend or shorten wild life span. (C) drh-1(tm1329) or eol-1(mg698) suppresses the extension of life span caused by the hsp-6(mg585) mutation. (D) Median and maximal survival and number of animals surveyed and censored due to losses during the month-long assay. The underlying numerical data can be found in S1 data.
Discussion
The central role of mitochondria in antiviral immune response
We found that mutations in 4 nuclearly encoded mitochondrial genes hsp-6, nuo-6, clk-1, and isp-1 strongly enhance RNAi. These mitochondrial mutations enhanced response to dsRNAs introduced by feeding (Fig 1A) and caused the silencing of multicopy transgenes (Fig 1B–1E), a capacity that is strongly associated with enhanced surveillance of foreign genes [36]. NUO-6, GAS-1, and NDUF-7 are components of complex I, MEV-1 is from complex II, ISP-1 is a component of complex III, and CLK-1 produces ubiquinone that transports electrons between complexes I or II and complex III. Three other mitochondrial point mutants, nduf-7(et19), mev-1(kn1), and gas-1(fc21) did not enhance RNAi (S1A Fig). Like most mitochondrial mutations, nduf-7(et19) lengthens life span (S2D Fig); however, mev-1(kn1) or gas-1(fc21) shorten life span (S2E and S2F Fig). Mitochondrial components have been identified in previous genome wide screens for Eri and RNAi defective gene inactivations: coq-1, a coenzyme Q biosynthetic gene; aco-2, aconitase; idha-1, mitochondrial isocitrate dehydrogenase; asb-1 and asg-1, subunits of the mitochondrial F0 ATP synthase; ucr-2.3, ubiquinol cytochrome c reductase; tufm-1, mitochondrial translation elongation factor; tin-44 mitochondrial inner membrane translocase; and phb-2 prohibitin, a chaperone for electron transport chain components, constitute a large fraction of the 27 gene inactivations that desilenced transposons in the germline; like viruses, transposons are silenced by RNAi [31,61]. sdha-2, which encodes a subunit of complex II, emerged from an Eri screen [36]. There is precedent from other animals for mitochondrial engagement of small RNA pathways, most especially for the piRNAs of Drosophila, which depend on the zucchini mitochondrial phosphatase [62].
Consistent with mitochondrial disruption causing induction of the RNAi pathway, many of the gene inductions in the hsp-6(mg585) mitochondrial mutant [22] overlap with those up-regulated by Orsay virus infection (Fig 2A and S1 Table) [43], many of which are the pals-1 to pals-40 genes. Mutations in pal-22 cause an Eri response, suggesting that induction of the pals- genes may mediate enhanced siRNA-based viral immunity [23,24].
In a viral infection, mitochondria release RNA into the cytosol, as has been observed in the MDA5 mitochondrial surveillance pathway in human and D. melanogaster [25,26]. The C. elegans mitochondrial genome encodes 36 genes [56] transcribed from one strand [57]. Like mammalian MDA5, we showed that the mRNA levels of nduo-5/ND5 and ctb-1/CYTB were substantially increased in the cytosol of hsp-6(mg585) compared with wild type (Fig 5D). Consistent with the model that dsRNA produced from mitochondrially transcribed and translated mRNAs engage the RNAi machinery in the C. elegans cytoplasm, the mitochondrial genes ctc-1, nduo-2, and nduo-5, mitochondrial electron transport chain genes, generate some of the most abundant levels of siRNAs in the genome, similar in abundance to those generated by transposable elements (S2 Table).
Mitochondrial dysfunction may be a key pathogenic feature of viral infection, and systems have evolved to trigger antiviral defense if mitochondria dysfunction is detected. The mitochondria have also been implicated in the Drosophila and mammalian antiviral piRNA pathway: In Drosophila, retroviruses are recognized by the dedicated piRNA pathway that generates piRNAs from integrated flamenco as well as other retroviral elements [63] to target newly encountered viruses that are related to these integrated viruses. A variety of mitochondrial outer membrane proteins couple piRNAs to target loci [64,65]. Similarly, the Sting interferon pathway also uses the MAVS and RIG-I interaction with mitochondria to signal antiviral responses.
Human mitochondrial mutations also trigger antiviral immunity [25,27]. The human RIG-I RNA helicase recognizes viral dsRNA or ssRNA 5′-triphosphate [1]. We showed that mutations in the RIG-1 orthologue drh-1(tm1329) suppressed the Eri induced by hsp-6(mg585) (Fig 1). The eol-1 RNA decapping gene was also strongly induced in virally infected and mitochondrial mutant C. elegans in a DRH-1-dependent manner (S1 Table and Fig 2F). The Eri and transgene silencing phenotypes of hsp-6(mg585) was suppressed by a eol-1(mg698) null mutation (Fig 3E). The localization of the EOL-1 protein was regulated by mitochondrial function: In wild type, EOL-1::mScarlet was localized diffusely in the cytosol (Fig 4A), whereas in the hsp-6(mg585) mitochondrial mutant, EOL-1::mScarlet formed massive puncta in many cell types (Fig 4A).
The EOL-1/DXO exonuclease may remove the 5′ cap of mitochondrial RNA, such as a 5′-NAD modification [66], to facilitate the recognition by DRH-1. The 5′ ends of the majority of mRNAs are chemically modified or capped to protect from RNA nucleases [67]. Prokaryotic and eukaryotic cells have different strategies to modify their mRNA with di- or triphosphate in bacteria and m7G cap in eukaryotes. Thus, the eukaryotic 5′ cap is a conspicuous sign to identify self, rather than the non-self-uncapped RNA from virus or bacteria.
The RNAi pathway drives gene silencing of mRNAs that have signatures of being foreign. For example, many of the Eri mutants mediate the silencing of integrated viruses in the C. elegans genome that are recently acquired [21]. These viral genes are recognized as foreign by their small number of introns, for example, and by poor splice sites [68]. The antiviral defense of C. elegans is more closely related to that of fungi and plants than most animals, for example, Drosophila and vertebrates. Nematodes, plants, and most fungi use the common pathway genes Dicer, specialized Argonaute/PIWI proteins, and most distinctively, RdRPs to produce primary siRNAs and secondary siRNAs against invading viruses. Because these pathways are common across many (but not all) eukaryotes, it is likely that the common ancestor of animals, plants, fungi, and some protists carried the Dicer, Argonaute, and RdRP genes that mediate RNAi defense.
The loss of RdRP genes in most animal species may be associated with the evolution of the Sting and NF-κB interferon RNA virus defense pathway of most insects and vertebrates, a pathway that is not present in nematodes or plants or fungi [69]. In addition to the interferon pathway, the piRNA pathway has specialized in insects and vertebrates to mediate antiviral defense [70]. The piRNA pathway is not present in fungi or plants and is not as central to viral defense in C. elegans, perhaps because it has continued to depend on its RdRP [11].
Viral defense in C. elegans is strongly induced in the many mutants that cause enhanced RNAi, eri-1 to eri-10, the many synMuvB mutations, and the mitochondrial mutants we characterized here. These mutations may induce antiviral defense because viral infections may compromise these genetic pathways or because these mutations genetically trigger an antiviral state that is normally under physiological regulation. A surveillance system to monitor the states of these pathways may be an early detection system for viruses to induce expression and activities of Dicer, Argonaute proteins, and RdRPs in siRNA production, targeting, and amplification [71,72]. Regardless of whether antiviral defense is mediated by RdRPs in nematodes or by MAVS and interferon signaling in vertebrates and most insects, the mitochondria are woven into the pathways. For Flock House virus and other nodaviruses, the mitochondrion is a center of viral replication; mutations that compromise the mitochondrion in yeast, for example, cause defects in antiviral responses [73–75].
A decrement in mitochondrial function is one of the most potent mechanisms to increase longevity in a variety of species [28]. drh-1(tm1329) or eol-1(mg698) mutations strongly suppressed the life span extension of hsp-6(mg585), showing that drh-1 or eol-1 gene activities are required for the extension of life span by hsp-6(mg585) (Fig 7C and 7D). Because drh-1 and eol-1 mediate the enhanced RNAi, an antiviral response, of mitochondrial mutants, and because these outputs also mediate in the increased longevity of mitochondrial mutants, but not the normal longevity of wild type, these data support the model that enhanced antiviral defense as a key antiaging output from mitochondrial mutations.
It is possible that the dramatic increase in frailty in old age could reflect viral vulnerability. In fact, one of the most dramatic hallmarks of the recent Coronavirus Disease 2019 (COVID-19) epidemic is that the virus is far more lethal to the elderly. Conversely, a decrement in antiviral capacity may accelerate aging: In mammals, a reduction in Dicer expression in heart, adipose, and brain is a hallmark of aging [76,77], and a loss-of-function mutant of DCR-1/Dicer shortens C. elegans life span [78].
Materials and methods
C. elegans maintenance and strains used in this study
C. elegans strains were cultured at 20°C. Strains used in this study are listed in S3 Table.
Generation of transgenic animals
For Peol-1::GFP, the plasmid harboring [Peol-1::GFP::eol-1 3′UTR + cbr-unc-119(+)] was injected into unc-119(ed3). For single-copy transgenes Prpl-28::mScarlet::drh-1, Prpl-28::eol-1::mScarlet, and Pcol-10::tomm-20::GFP, the plasmid was injected into unc-119(ed3) following the miniMOS protocol [53]. For CRISPR of eol-1(mg698), we chose dpy-10(cn64) as the co-CRISPR marker [52] and pJW1285 (Addgene) to express both of guide-RNA (gRNA) and Cas9 enzyme [79].
Microscopy
The fluorescent signals of sur-5::GFP, Peol-1::GFP, and Phsp-6::GFP transgenic animals were photographed by Zeiss AX10 Zoom.V16 microscope (Carl Zeiss AG, Oberkochen, Germany). The subcellular pattern of Prpl-28::mScarlet::drh-1, Prpl-28::eol-1::mScarlet, and Pcol-10::tomm-20::GFP were carried out on the Leica TCS SP8 confocal microscope (Leica Camera AG, Wetzlar, Germany). Photographs were analyzed by Fiji-ImageJ (https://fiji.sc/; https://imagej.net/Welcome).
Sample collection and RNA isolation
For estimation of the mRNA level of eol-1, nduo-5, and ctb-1, around 200 L4 larvae were handpicked from mixed population and frozen by liquid nitrogen. The total RNA was isolated by TRIzol extraction (Thermo Fisher, Waltham, Massachusetts, United States of America, #15596026). For analyses of cytosolic mRNA level of nduo-5 and ctb-1, approximately 2,000 worms were synchronized by bleach preparation of eggs, hatching progeny eggs to L1 larvae, grown until L4 stage, and frozen by liquid nitrogen. Worm lysates were generated by TissueLyser with steel beads (Qiagen, Venlo, the Netherlands, #69989), and resuspended in 500 μl of the lysis buffer containing 0.8-M sucrose, 10-mM Tris-HCl, 1-mM EDTA, 1× protease inhibitor cocktail (Roche, Basel, Switzerland, 11873580001), and 1-U/μl murine RNase inhibitor (New England BioLabs, Ipswich, Massachusetts, USA, #M0314L). The lysate was centrifuged at 2,500 × g for 10 min at 4°C to remove the pellet debris. And the supernatant was centrifuged at 20,000 × g for 10 min at 4°C. Collect the pellet for western blot, and the supernatant was centrifuged at 20,000 × g for 10 min at 4°C. Take 100 μl of the supernatant for western blot and 300 μl for RNA isolation with TRIzol.
RT-qPCR
The cDNA was generated by ProtoScript II First Strand cDNA Synthesis Kit (New England BioLabs, #E6560L). qPCR was performed toward eol-1, nduo-5, and ctb-1 with act-1 as control by iQ SYBR Green Supermix (BIO-RAD, Hercules, California, USA, #1708880).
Western blot
The pellet or supernatant from the previous centrifugation were mix NuPAGE LDS sample buffer (Thermo Fisher #NP0007) and heated at 70°C for 10 min. Samples were loaded onto NuPAGE 4% to 12% Bis-Tris Protein Gels (Thermo Fisher #NP0323BOX) and run with NuPAGE MES SDS running buffer (Thermo Fisher #NP0002). After semi-dry transfer, PVDF membrane (Millipore, Burlington, Massachusetts, USA, #IPVH00010) was blocked with 5% nonfat milk, and probed with anti-ATP5A (Abcam, Cambridge, United Kingdom, ab14748) or anti-actin (Abcam #ab3280) primary antibodies. The membrane was developed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher #34096) and visualized by Amersham Hyperfilm (GE Healthcare, Chicago, Illinois, USA, #28906845).
Life span analysis
Animals were synchronized by egg laying and grown until the L4 stage as day 0. Adults were separated from their progeny by manual transfer to new plates. Survival was examined on a daily basis, and the survival curve was generated by GraphPad Prism (Graphpad Software, San Diego, California, USA).
Supporting information
(A) Enhanced RNAi response to lir-1 RNAi or his-44 RNAi in the eri-6 control enhanced RNAi mutant or any of the mitochondrial mutants (nduf-7, mev-1, or gas-1), causes lethality/arrest on the mitochondrial mutants but not wild type. (B) Protein alignment of DRH-1 NTD in nematode species. Ac: Ancylostoma ceylanicum; Ce: Caenorhabditis elegans; Cl: Caenorhabditis latens; Cr: Caenorhabditis remanei; Nb: Nippostrongylus brasiliensis; NTD, N-terminal domain; RNAi, RNA interference.
(TIF)
(A–D) Mitochondrial mutants (nuo-6, clk-1, isp-1, or nduf-7) extend life span. (E and F) Mitochondrial mutants (mev-1 or gas-1) shorten life span. (G) Median and maximal survival and number of animals surveyed and censored due to losses during the month-long assay. The underlying numerical data can be found in S1 data.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Acknowledgments
We thank Caenorhabditis Genetics Center and National BioRescource Project (Tokyo, Japan) for providing strains.
Abbreviations
- CARD
caspase activation and recruitment domains
- CTD
C-terminal domain
- dsRNA
double-stranded RNA
- Eri
enhanced RNAi
- ETC
electron transport chain
- gRNA
guide-RNA
- lir
lin-26-related
- MAVS
mitochondrial antiviral signaling
- MDA5
melanoma differentiation-associated protein 5
- m7G
N7-methyl guanosine
- NAD
nicotinamide adenine dinucleotide
- NTD
N-terminal domain
- PPH
pyrophosphohydrolase
- PPi
pyrophosphate
- RdRP
RNA-dependent RNA polymerase
- RIG-I
retinoic acid-inducible gene I
- RNAi
RNA interference
- rRNA
ribosomal RNAs
- RT-qPCR
quantitative reverse transcription PCR
- siRNA
small interfering RNA
- ssRNA
single-stranded RNA
- tRNA
transfer RNAs
- UPRmt
unfolded protein response
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
K.M. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2213-15). This work is supported by a grant from the National Institute of Health awarded to G.R. (NIH GM044619 and AG16636). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chow KT, Gale M Jr., Loo YM. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol. 2018;36:667–94. 10.1146/annurev-immunol-042617-053309 . [DOI] [PubMed] [Google Scholar]
- 2.Lassig C, Hopfner KP. Discrimination of cytosolic self and non-self RNA by RIG-I-like receptors. J Biol Chem. 2017;292(22):9000–9. 10.1074/jbc.R117.788398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zevini A, Olagnier D, Hiscott J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017;38(3):194–205. 10.1016/j.it.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–3. 10.1038/nature03870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9(1):e1000586 10.1371/journal.pbio.1000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 2005;436(7053):1044–7. 10.1038/nature03957 . [DOI] [PubMed] [Google Scholar]
- 7.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102(51):18420–4. 10.1073/pnas.0507123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gammon DB, Mello CC. RNA interference-mediated antiviral defense in insects. Curr Opin Insect Sci. 2015;8:111–20. 10.1016/j.cois.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szittya G, Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol 2013;371:153–81. 10.1007/978-3-642-37765-5_6 . [DOI] [PubMed] [Google Scholar]
- 10.Ding SW, Han Q, Wang J, Li WX. Antiviral RNA interference in mammals. Curr Opin Immunol. 2018;54:109–14. 10.1016/j.coi.2018.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493(7434):694–8. 10.1038/nature11779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammon DB, Ishidate T, Li L, Gu W, Silverman N, Mello CC. The Antiviral RNA Interference Response Provides Resistance to Lethal Arbovirus Infection and Vertical Transmission in Caenorhabditis elegans. Curr Biol. 2017;27(6):795–806. 10.1016/j.cub.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X, Zhang R, Wang J, Ding SW, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A. 2013;110(40):16085–90. 10.1073/pnas.1307453110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109(7):861–71. 10.1016/s0092-8674(02)00793-6 . [DOI] [PubMed] [Google Scholar]
- 15.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr., Pang K, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124(2):343–54. 10.1016/j.cell.2005.11.036 . [DOI] [PubMed] [Google Scholar]
- 16.Ashe A, Belicard T, Le Pen J, Sarkies P, Frezal L, Lehrbach NJ, et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. elife. 2013;2:e00994 10.7554/eLife.00994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips CM, Montgomery TA, Breen PC, Ruvkun G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26(13):1433–44. 10.1101/gad.193904.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, et al. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108(4):1201–8. 10.1073/pnas.1018695108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 2004;427(6975):645–9. 10.1038/nature02302 . [DOI] [PubMed] [Google Scholar]
- 20.Fischer SE, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455(7212):491–6. 10.1038/nature07274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer SEJ, Ruvkun G. Caenorhabditis elegans ADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons. Proc Natl Acad Sci U S A 2020. 10.1073/pnas.1919028117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao K, Ji F, Breen P, Sewell A, Han M, Sadreyev R, et al. Mitochondrial Dysfunction in C. elegans Activates Mitochondrial Relocalization and Nuclear Hormone Receptor-Dependent dDetoxification gGenes. Cell Metab. 2019. 10.1016/j.cmet.2019.01.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy KC, Dror T, Sowa JN, Panek J, Chen K, Lim ES, et al. An Intracellular Pathogen Response Pathway Promotes Proteostasis in C. elegans. Curr Biol. 2017;27(22):3544–53 e5. 10.1016/j.cub.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyva-Diaz E, Stefanakis N, Carrera I, Glenwinkel L, Wang G, Driscoll M, et al. Silencing of Repetitive DNA Is Controlled by a Member of an Unusual Caenorhabditis elegans Gene Family. Genetics. 2017;207(2):529–45. 10.1534/genetics.117.300134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560(7717):238–42. 10.1038/s41586-018-0363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pajak A, Laine I, Clemente P, El-Fissi N, Schober FA, Maffezzini C, et al. Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo. PLoS Genet. 2019;15(7):e1008240 10.1371/journal.pgen.1008240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–7. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33(1):40–8. 10.1038/ng1056 . [DOI] [PubMed] [Google Scholar]
- 29.Wiedemann N, Pfanner N. Mitochondrial mMachineries for pProtein iImport and aAssembly. Annu Rev Biochem. 2017;86:685–714. 10.1146/annurev-biochem-060815-014352 . [DOI] [PubMed] [Google Scholar]
- 30.Kim HE, Grant AR, Simic MS, Kohnz RA, Nomura DK, Durieux J, et al. Lipid Biosynthesis Coordinates a Mitochondrial-to-Cytosolic Stress Response. Cell. 2016;166(6):1539–52 e16. 10.1016/j.cell.2016.08.027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308(5725):1164–7. 10.1126/science.1109267 . [DOI] [PubMed] [Google Scholar]
- 32.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2(1):RESEARCH0002 Epub 2001/02/24. 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321(5888):537–41. 10.1126/science.1157647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 2009;183(4):1283–95. 10.1534/genetics.109.108134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Ruvkun G. Regulation of Caenorhabditis elegans RNA interference by the daf-2 insulin stress and longevity signaling pathway. Cold Spring Harb Symp Quant Biol. 2004;69:429–31. 10.1101/sqb.2004.69.429 . [DOI] [PubMed] [Google Scholar]
- 36.Fischer SE, Pan Q, Breen PC, Qi Y, Shi Z, Zhang C, et al. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev. 2013;27(24):2678–95. 10.1101/gad.233254.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer JM, Johnson JJ. Analysis of mutations in the sqt-1 and rol-6 collagen genes of Caenorhabditis elegans. Genetics. 1993;135(4):1035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Kennedy S, Conte D Jr., Kim JK, Gabel HW, Kamath RS, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436(7050):593–7. 10.1038/nature04010 . [DOI] [PubMed] [Google Scholar]
- 39.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149(3):1323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Shi Z, Cui M, Han M, Ruvkun G. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 2012;8(3):e1002542 10.1371/journal.pgen.1002542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, et al. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011;7(11):e1002369 10.1371/journal.pgen.1002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich DP, Tyc KM, Bass BL. C. elegans ADARs antagonize silencing of cellular dsRNAs by the antiviral RNAi pathway. Genes Dev. 2018;32(3–4):271–82. 10.1101/gad.310672.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen K, Franz CJ, Jiang H, Jiang Y, Wang D. An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes. BMC Genomics. 2017;18(1):303 10.1186/s12864-017-3689-3 PubMed Central PMCID: PMC5392922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senchuk MM, Dues DJ, Schaar CE, Johnson BK, Madaj ZB, Bowman MJ, et al. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet 2018;14(3):e1007268 10.1371/journal.pgen.1007268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowa JN, Jiang H, Somasundaram L, Tecle E, Xu G, Wang D, et al. The Caenorhabditis elegans RIG-I Homolog DRH-1 Mediates the Intracellular Pathogen Response upon Viral Infection. J Virol. 2020;94(2). 10.1128/JVI.01173-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amador-Canizares Y, Bernier A, Wilson JA, Sagan SM. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5' end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018;46(10):5139–58. 10.1093/nar/gky273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y, Zhang J, Calarco JA, Zhang Y. EOL-1, the homolog of the mammalian Dom3Z, regulates olfactory learning in C. elegans. J Neurosci. 2014;34(40):13364–70. 10.1523/JNEUROSCI.0230-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kramer S, McLennan AG. The complex enzymology of mRNA decapping: Enzymes of four classes cleave pyrophosphate bonds. Wiley Interdiscip Rev RNA. 2019;10(1):e1511 10.1002/wrna.1511 . [DOI] [PubMed] [Google Scholar]
- 49.Lin YF, Haynes CM. Metabolism and the UPR(mt). Mol Cell. 2016;61(5):677–82. 10.1016/j.molcel.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–90. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, et al. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell. 2016;165(5):1197–208. 10.1016/j.cell.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198(3):837–46. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frokjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, et al. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods 2014;11(5):529–34. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hallberg BM, Larsson NG. Making proteins in the powerhouse. Cell Metab. 2014;20(2):226–40. 10.1016/j.cmet.2014.07.001 . [DOI] [PubMed] [Google Scholar]
- 55.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res 2013;41(2):1223–40. 10.1093/nar/gks1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics. 1992;130(3):471–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blumberg A, Rice EJ, Kundaje A, Danko CG, Mishmar D. Initiation of mtDNA transcription is followed by pausing, and diverges across human cell types and during evolution. Genome Res. 2017;27(3):362–73. 10.1101/gr.209924.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li R, Ren X, Bi Y, Ding Q, Ho VWS, Zhao Z. Comparative mitochondrial genomics reveals a possible role of a recent duplication of NADH dehydrogenase subunit 5 in gene regulation. DNA Res. 2018;25(6):577–86. 10.1093/dnares/dsy026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer- Graiwer J, Hsin H, Fraser AG, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–401. 10.1126/science.1077780 . [DOI] [PubMed] [Google Scholar]
- 60.Zhang Q, Wu X, Chen P, Liu L, Xin N, Tian Y, et al. The Mitochondrial Unfolded Protein Response Is Mediated Cell-Non-autonomously by Retromer-Dependent Wnt Signaling. Cell. 2018;174(4):870–83 e17. Epub 2018/07/31. 10.1016/j.cell.2018.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442(7105):882 Epub 2006/08/25. 10.1038/442882a . [DOI] [PubMed] [Google Scholar]
- 62.Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, Zamore PD. The RNA-Binding ATPase, Armitage, Couples piRNA Amplification in Nuage to Phased piRNA Production on Mitochondria. Mol Cell. 2019;74:982–995. 10.1016/j.molcel.2019.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–35. 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20(3):376–87. 10.1016/j.devcel.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munafo M, Manelli V, Falconio FA, Sawle A, Kneuss E, Eastwood EL, et al. Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery. Genes Dev. 2019;33(13–14):844–56. 10.1101/gad.325662.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bird JG, Basu U, Kuster D, Ramachandran A, Grudzien-Nogalska E, Towheed A, et al. Highly efficient 5' capping of mitochondrial RNA with NAD(+) and NADH by yeast and human mitochondrial RNA polymerase. elife. 2018;7 10.7554/eLife.42179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grudzien-Nogalska E, Kiledjian M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip Rev RNA. 2017;8(1). 10.1002/wrna.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newman MA, Ji F, Fischer SEJ, Anselmo A, Sadreyev RI, Ruvkun G. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Genes Dev. 2018;32(9–10):670–81. 10.1101/gad.311514.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aravind L, Anantharaman V, Iyer LM. Evolutionary connections between bacterial and eukaryotic signaling systems: a genomic perspective. Curr Opin Microbiol. 2003;6(5):490–7. Epub 2003/10/24. 10.1016/j.mib.2003.09.003 . [DOI] [PubMed] [Google Scholar]
- 70.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–4. 10.1126/science.1146484 . [DOI] [PubMed] [Google Scholar]
- 71.Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149(2):452–66. 10.1016/j.cell.2012.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Govindan JA, Jayamani E, Zhang X, Breen P, Larkins-Ford J, Mylonakis E, et al. Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans. Nat Cell Biol. 2015;17(10):1294–303. 10.1038/ncb3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller DJ, Ahlquist P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J Virol 2002;76(19):9856–67. 10.1128/jvi.76.19.9856-9867.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 2001;75(23):11664–76. 10.1128/JVI.75.23.11664-11676.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao L, Lindenbach B, Wang X, Dye B, Kushner D, He Q, et al. Genome-wide analysis of host factors in nodavirus RNA replication. PLoS One. 2014;9(4):e95799 10.1371/journal.pone.0095799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149(4):847–59. 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–30. 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16(3):336–47. 10.1016/j.cmet.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ward JD. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics. 2015;199(2):363–77. 10.1534/genetics.114.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]