Abstract
Purpose
MicroRNAs (miRNAs) are implicated in metabolic changes accompanying progression of obesity, insulin resistance (IR), and metabolic disorders in children. Identifying circulating miRNAs that uniquely associate with these disorders may be useful in early identification and prevention of obesity-related complications. We aimed to identify circulating miRNA signatures that distinguish adolescents with obesity and IR from those with obesity unaccompanied by IR.
Methods
Adolescents (aged 10–17 years) with obesity were recruited from a weight management clinic. Fasting serum samples were obtained from 33 participants. A total of 179 miRNAs were queried by a quantitative RT-PCR-based miRNA focus panel. Differentially expressed miRNAs were compared between groups using Student’s t-test or one-way ANOVA analysis, and the association between IR evaluated by homeostatic model assessment model (HOMA-IR > 4) and body mass index (BMI) status was assessed using Pearson’s correlation analysis.
Results
We found an expression pattern consisting of 12 elevated miRNAs linked to IR in obese adolescents. miR-30d, -221, and -122 were significantly correlated with clinical and biochemical markers of obesity and IR, suggestive of IR in adolescents at risk.
Conclusion
Specific signatures of circulating miRNAs reflected metabolic phenotypes and predicted the presence of IR in adolescents with obesity, suggesting that miRNA indicators may identify obesity-associated complications in childhood. Further studies will be needed to understand cause versus effect and the mechanisms by which IR status links to changes in blood miRNA profiles.
Keywords: serum miRNA, insulin resistance, adolescent, obesity
Introduction
Obesity is a complex and multifactorial disease characterized by excessive accumulation of body fat. Recent National Health and Nutrition Examination Survey revealed that the prevalence of childhood obesity in the United States was higher (20.6%) in adolescents aged 12–19 years than children aged 2–5 years (13.9%),1 with severe obesity accounting for 5.6% in the youth. Pediatric obesity is linked to metabolic disturbances, which also increases risk for further complications in adulthood, eg, insulin resistance (IR), non-alcoholic fatty liver disease (NAFLD), and type II diabetes (T2D).2,3 The degree of obesity classified by BMI percentile does not offer adequate information related to predicting or diagnosing obesity-related comorbidities,4 as not all children with obesity display metabolic disease. As a result, there is a critical need for early identification of the obesity-associated complications.
Obesity can induce various pathological changes in insulin-sensitive tissues (ie, adipose tissue, liver, skeletal muscle), and lead to localized inflammation and IR via autocrine and paracrine signaling.5 Obesity-associated chronic inflammation is a critical component in the pathogenesis of IR and development of metabolic disorders. Children with obesity and IR are more likely to develop metabolic diseases later in life.6 However, the molecular mechanisms linking obesity and IR in childhood are not entirely clear.
MicroRNAs (miRNAs) are a type of small non-coding RNA (about 19–24 nucleotides) that regulate metabolic pathways by translational repression or messenger RNA (mRNA) degradation at the post-transcriptional level.7 In the human genome, miRNAs regulate over 60% of protein-coding genes at the translation level8 and affect almost all metabolic pathways, where they act as a class of endocrine factors and molecular regulators involved in physiological and pathological processes.9 Dysfunction of miRNAs in tissues or systems can influence energy metabolism, insulin sensitivity, and glucose homeostasis, thus contributing to metabolic disorders and related diseases, eg, obesity, IR, NAFLD, T2D.10–12 Therefore, mechanistic investigations in animal and human studies have proposed miRNAs as potential biomarkers to aid in the prediction, diagnosis, and prognosis of obesity or metabolic diseases.13
Circulating miRNAs originate from cells or tissues and are released into the circulation where they can be found packaged in extracellular vesicles (EVs)14 or bound to lipoproteins or Argonaute-2 (AGO2).15 Via the circulation, they can reach and affect the metabolism of distant organs and are thought to contribute to the pathogenesis, development, and progression of obesity and its related chronic diseases. Select circulating miRNAs appear to be associated with obesity, eg, miR-140, -142, -125, and -423,16 or with obesity-related metabolic disturbances, eg, miR-122 and -34 with NAFLD and IR,17 and miR-103 and -107 with T2D and IR18 in children and adults. These miRNAs promote adipogenesis, adipocyte differentiation, and insulin resistance in insulin-sensitive tissues. In these clinical studies, the comparisons of differentially expressed miRNAs have always been with healthy weight or non-disease controls, in which there is a low miRNA abundance in biofluids. So, it is uncertain if these miRNA changes reflect the degree of pediatric obesity and its progression, and recapitulate risk factors for developing obesity-related complications.
Here, we determined if there are unique circulating miRNA signatures that indicate IR in a group of adolescents with obesity. Moreover, we also assessed the relationship between miRNAs and BMI values. The results may provide new insights into potential molecular mechanisms linking obesity and IR in adolescents, and highlight the potential of using miRNA-based biomarkers for prediction, diagnosis, and treatment of obesity-related metabolic disorders.
Materials and Methods
Study Design
This study was a secondary analysis of data collected to examine the role of Fibroblast Growth Factor 21 in the diagnosis of Non-Alcoholic Fatty Liver Disease (NAFLD) and prediction of changes in intrahepatic triglyceride percent in children with obesity.19 The study was approved by the Institutional Review Board of the University of Arkansas for Medical Sciences. Parental consent and participant assent from all participants < 18 years old were obtained.
Study Population
Children and adolescents aged 10−17 years were recruited from the Center for Obesity and its Consequences in Health (C.O.A.C.H.) Clinic, a clinical weight management program at the Arkansas Children’s Hospital, Little Rock, AR. Inclusion criteria were the presence of obesity as defined by age and sex-specific body mass index (BMI) ≥ 95th percentile per CDC growth charts, and attainment of puberty (minimum Tanner stage 2 as verified by a pediatric endocrinologist). Exclusion criteria were syndromic obesity, diabetes, liver disease including prior diagnosis of NAFLD, medications known to have a direct effect on hepatic lipid metabolism and glucose homeostasis (eg, metformin, statins, fibrates, steroids, thyroid hormones, growth hormones, hormonal contraceptives). Only those with available blood samples to measure miRNA profiles (N=33) were included in this secondary analysis.
Anthropometrics
The participants were characterized based on anthropometric measures, including BMI, BMI z-score (z-BMI), BMI percentile, and waist circumference (WC). Using a calibrated scale and stadiometer, weight and height were measured and rounded to the nearest tenth of a kilogram or a hundredth of a meter, respectively. Standard CDC growth charts were used to plot height and weight and calculate BMI, z-BMI, and BMI percentiles. WC (in centimeter) was measured using a non-elastic tape measure at the level of umbilicus while the subjects were standing.
Blood Analytes
Blood samples were obtained by venipuncture after 8- to 10-hour overnight fasting to measure serum concentrations of glucose, insulin, triglycerides (TG), high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein-cholesterol (LDL-C), and total cholesterol (TC) via a clinical analyzer (Siemens Atellica, Malvern, PA, USA) at the Arkansas Children’s Hospital Chemistry Laboratory. Serum adiponectin (Human Total Adiponectin/Acrp30 Quantikine) and leptin (Human Leptin Quantikine) concentrations were measured via ELISA per manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA) in the Metabolism and Bioenergetics Core at the Arkansas Children’s Research Institute (ACRI). Fasting free fatty acids (FFA) were measured via chemistry analyzer (Randox Daytona, Holliston, MA, USA) in the Metabolism and Bioenergetics Core at the ACRI.
Indices of Insulin Resistance
Whole-body and tissue-specific insulin actions were indirectly assessed by i) homeostatic model assessment of insulin resistance (HOMA-IR) calculated as fasting insulin (µIU/L) × fasting glucose (mmol/L)/22.5;20 ii) HOMA of adiponectin (HOMA-AD) calculated as fasting insulin (µIU/L) × glucose (mmol/L) divided by adiponectin (ng/mL);21 iii) triglycerides to high-density lipoprotein cholesterol (TG/HDL-C);22 iv) adiponectin to leptin ratio (adiponectin/leptin ratio), and v) adipose tissue IR (Adipo-IR) calculated as fasting free fatty acid (mmol/L) × fasting insulin (µIU/L).23 In the literature, IR assessed by HOMA and the euglycemic clamp, which is the standard reference test to quantitate IR, demonstrate a good correlation,24 In this study, IR was defined based on a HOMA-IR cut-off point of 4.0.25,26
Serum RNA Extraction and miRNA Profiling
miRNA was extracted from 200 µL serum using miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA, USA) per manufacturer’s instructions. RNA extraction was finally eluted in 14 µL. One microliter of RNA was reverse transcribed using the miRCURY LNA RT Kit (Qiagen). Samples were amplified utilizing locked nucleic acids (LNA) technology with 179 different primers on the Human Serum/Plasma miRCURY LNA miRNA Focus PCR panels (96-well format panel I& II, YAHS-106Y; Qiagen) and using miRCURY LNA SYBR Green PCR kit (Qiagen). C.elegans miRNA (cel-miR-39-3p), UniSp6, UniSp2, UniSp4, and Unisp5 as spike-in controls were added using the RNA Spike-in kit as well as UniSp6 predefined on plates for monitoring RNA isolation, cDNA synthesis, and PCR amplification. Sample quality and hemolysis were assessed using miScript PCR Controls (Qiagen). miRNA determination was performed with diluted samples using a quantitative RT-PCR (qRT-PCR) on a Fast 7500 Real-time PCR System, Applied Biosystems (Life Technologies, Foster City, CA, USA). Amplicons were analyzed for distinct melting curves, and the Tm was checked to the within known specifications for the assays. qRT-PCR data were analyzed using the ∆Ct method and normalized to a normalization factor calculated based on GeNorm methodology from the entire panel.27 All the detectable miRNAs were assessed for the least variance across all samples in groups. According to the GeNorm analysis, selecting miR-486-5p, -193-5p, -101-3p, and let-7a-5p as normalizers showed the least variance and was confirmed by comparison of the five suggested spike-in controls using Qiagen software. Inter-plate calibration (IPC) using UniSp3 controls was to correct for variance across plates. miRNA with Ct values > 35 in at least 65% of samples was excluded.
Body Composition
Total body adiposity was assessed via bioimpedance technique using InBody® 570 body composition analyzer (InBody USA, Cerritos, CA. USA) according to the manufacturer’s protocols. In brief, tissue impedance was measured over 60 seconds when a low-intensity current travels between the bare feet and hands of the subjects. The total body fat (TBF) estimate was obtained from the equipment software. TBF estimate via InBody was previously shown to correlate well with the Dual-energy X-Ray Absorptiometry (DXA) scan.28
Statistical Analysis
Data are presented as mean ± standard deviation of the mean (SD) except where otherwise indicated. Categorical proportions (sex and race) were determined by Chi-square tests or Fisher’s exact test. For the multiple group comparison, one-way ANOVA was conducted, followed by Tukey or Dunn all-pairwise comparisons post hoc analysis to compare all groups to each other. Student’s t-test was used for comparison of two groups that were normally distributed. Group differences between obese with and without IR were assessed by a Mann–Whitney test for analytes that were not normally distributed (as defined by p < 0.05, determined by D’Agostino-Pearson normality test). Correlations between miRNA level (-log scale) and clinical and biological parameters (independent variables) were determined using Pearson’s correlation coefficients for normally distributed data or Spearman correlation coefficients for not normally distributed data. All statistical analyses were performed using GraphPad Prism7 (GraphPad Software, Inc., La Jolla, CA, USA). Significance was defined as p < 0.05.
Results
Characteristics of Participants with Insulin Resistance
The subjects’ age ranged from 10.4 to 17.3 years. Thirty HOMA-IR values were available from the adolescents, since three individuals were lacking the determination of insulin or glucose concentrations. We found that HOMA-IR values were tightly associated with fasting insulin levels (r > 0.99 and p < 0.01; Table S1), but not glucose levels (r < 0.50; Table S1). The participants were classified into obese groups with IR (OB with IR; HOMA-IR > 4) and without IR (OB; HOMA-IR ≤ 4), consisting of 21 and 9 subjects, respectively (Table 1). As expected, in the OB with IR group, various IR indices, such as HOMA-IR, HOMA-AD, Adipo-IR, TG, and TG/HDL-C significantly differed from OB without IR group (p < 0.05 or p < 0.01). Groups were comparable in regard to mean age, sex distribution, BMI, fasting glucose, leptin, adiponectin, adiponectin to leptin ratio, FFA, HDL-C, and systolic and diastolic blood pressures.
Table 1.
Characteristics of Participants Separated by Insulin Resistance
| Variables | OB | OB with IR | p-value |
|---|---|---|---|
| Number (n) | 9 | 21 | |
| Age (years) | 13.86 ± 1.28 | 14.61 ± 2.37 | 0.3793 |
| Sex, M/F (%) | 44.4/54.6 | 57.1/42.9 | 0.5229 + |
| BMI (kg/m2) | 35.07 ± 5.00 | 37.19 ± 4.61 | 0.2404 |
| Fasting glucose (mg/dL) | 88.33 ± 10.84 | 94.71 ± 9.1 | 0.1075 |
| Fasting insulin (µIU/mL) | 16.09 ± 4.46 | 56.98 ± 45.24 | 0.0124 |
| Fasting leptin (pg/mL) | 43.20 ± 25.40 | 51.41 ± 21.64 | 0.3778 |
| Fasting adiponectin (ng/mL) | 7.74 ± 3.80 | 7.49 ± 4.66 | 0.7637 |
| Adiponectin/leptin ratio | 0.23 ± 0.12 | 0.18 ± 0.13 | 0.3512 |
| HOMA-IR | 3.48 ± 0.70 | 10.90 ± 7.57 | 0.0071 |
| HOMA-AD | 11.83 ± 4.38 | 49.69 ± 77.90 | 0.0014 |
| FFA (mmol/L) | 0.16 ± 0.06 | 0.15 ± 0.06 | 0.9216 |
| Adipo-IR (mmol/L) | 2.43 ± 0.98 | 9.77 ± 10.07 | 0.0021 |
| TG (mg/dL) | 91.22 ± 53.96 | 152.62 ± 70.27 | 0.0183 |
| HDL-C (mg/dL) | 42.11 ± 6.57 | 38.67 ± 7.57 | 0.2458 |
| TG/HDL-C ratio | 2.36 ± 1.76 | 4.24 ± 1.76 | 0.0221 |
| Systolic BP (mmHg) | 127.78 ± 11.37 | 124.43 ± 12.07 | 0.8916 |
| Diastolic BP (mmHg) | 68.56 ± 7.70 | 69.76 ± 8.01 | 0.7051 |
Notes: Data are expressed as mean ± SD. Significant differences were determined by Student’s t-test or Mann–Whitney test. +Group comparison was examined using a Chi-square test. The p-value less than 0.05 is shown in bold and italic.
Abbreviations: TG, triglycerides; HDL-C, high-density lipoprotein-cholesterol; FFA, free fatty acids; OB, subjects with obesity; OB with IR, subjects with obesity and insulin resistance (IR).
miRNA Expressions Specific to Insulin Resistance
Of 179 miRNAs queried in the study, a total of 40 circulating miRNAs were identified across all the adolescents with obesity. Of these 40 identified, 21 miRNAs (Figure S1) have been reported in studies of obese populations, and their regulatory mechanisms or biological functions have been validated using Ingenuity Pathway Analysis (IPA, Qiagen, Valencia, CA, USA) or previously reported in human studies in the literature.11,13,16–18,29–32 The other 19 miRNAs have not previously been identified in the serum of children with obesity (Figure S2). Of these 40 miRNAs, miR-223-3p, -16-5p, -23a-3p, -25-3p, -150-5p, -30d-5p, -320a, and -21-5p were the 8 serum miRNAs with the highest miRNA levels (based on average of miRNA expression relative to normalizers) (Figures S1–S2).
In further comparison of miRNA profiles associated with IR, we found 12 miRNA species were significantly different in abundance in OB with IR compared to OB without IR, including increased levels of miR-223-3p, -23a-3p, -150-5p, -191-5p, -24-3p, -30d-5p, -122-5p, -30a-5p, -215-5p, -221-3p, -145-5p, and -342-3p (Figure 1 and Figure S3). Of these miRNAs, there was a positive correlation between miR-30d-5p levels with insulin level, TG/HDL-C ratio, HOMA-IR, and HOMA-AD (p < 0.05; Table 2). Similarly, miR-122-5p and miR-221-3p expressions were significantly correlated with insulin levels and IR indicators (Table 2). The miR-215-5p level was positively related with the TG/HDL-C ratio and insulin level (p < 0.05; Table 2).
Figure 1.
Relative abundance is of miRNA level specific to insulin resistance. Representative miRNA expressions (-log2 scale) are shown in (A–D). Bars indicate mean ± SD. Labeled (a, b) means differ for comparison of two groups, p < 0.05. OB: subjects with obesity; OB with IR: subjects with obesity and insulin resistance (IR).
Table 2.
miRNA Expressions Associated with Risk Factors and Predictive Indicators of IR
| Variables | OB with IR | |||||||
|---|---|---|---|---|---|---|---|---|
| miR-30d-5p | miR-122-5p | miR-221-3p | miR-215-5p | |||||
| r | p-value | r | p-value | r | p-value | r | p-value | |
| Insulin | 0.804 | <0.001 | 0.662 | 0.002 | 0.610 | 0.035 | 0.639 | 0.014 |
| HOMA-IR | 0.608 | 0.003 | 0.680 | <0.001 | 0.605 | 0.014 | 0.251 | 0.605 |
| HOMA-AD | 0.619 | 0.004 | 0.568 | <0.001 | 0.453 | 0.039 | 0.450 | 0.308 |
| Adipo-IR | 0.569 | 0.009 | 0.466 | 0.039 | 0.510 | 0.048 | 0.644 | 0.102 |
| TG/HDL-C | 0.544 | 0.011 | 0.596 | 0.004 | 0.524 | 0.002 | 0.656 | 0.019 |
Notes: Data on miRNA levels and biological analytes are transformed into a log scale (-log2). A significant difference was determined using Pearson’s correlation analysis. The p-value less than 0.05 is shown in bold and italic styles.
Abbreviation: OB with IR, subjects with obesity and insulin resistance.
Differential miRNA Expressions in Response to BMI Status
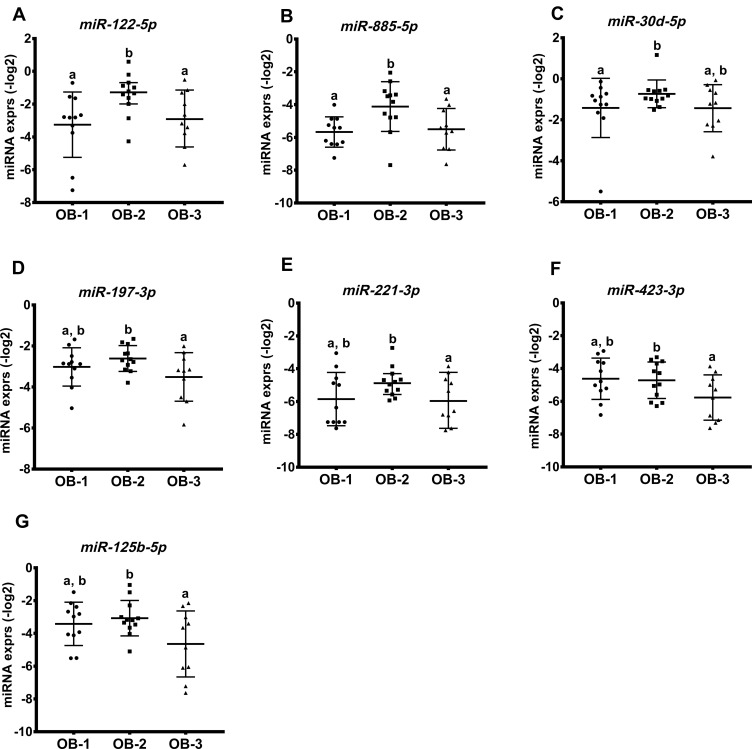
We further determined if miRNA expressions were associated with the status of obesity. In this cohort, BMI percentile did not discriminate participants in regards to clinical and biochemical phenotypes of obesity, but BMI values showed a positive correlation with body weight, WC, percent body fat, and leptin (r > 0.5 and p < 0.05; Table S1). Therefore, we determined miRNA species that differed by BMI values: OB-1, 30 kg/m2 < BMI < 35 kg/m2; OB-2, 35 kg/m2 ≤ BMI < 40 kg/m2; OB-3, BMI ≥ 40 kg/m2. All participants were well matched for sex, age, and ethnicities with no significant differences between BMI groups (p > 0.05). With respect to other weight parameters, z-BMI was significantly different between all three groups; but the BMI percentile did not differ between groups (Table S2). Weight and WC were higher in the OB-2 and OB-3 groups in comparison to OB-1 (p < 0.05), and percent body fat was significantly different between all three groups; there were no significant differences in height between groups.
In Figure 2, miR-122-5p and miR-885-5p expression was higher in OB-2 when compared to OB-3 or OB-1; miR-30d-5p expression was highest in the OB-2 group, although this did not differ statistically when compared to the OB-3 group. Interestingly, expression levels of miR-197-3p, -221-3p, -423-4p, and -125b-5p were lower in the OB-3 compared to the OB-2 group. Additionally, we observed an inverse correlation of miR-197-3p expression with body weight in the OB-3 group (p < 0.001) and levels of miR-125b-5p and miR-221-3p were strongly and negatively correlated with percent body fat, but positively associated with muscle mass (only miR-221-3p; Table 3). The results showed that specific miRNAs were differentially expressed in response to overall adiposity.
Figure 2.
miRNA profiles associated with obesity in response to BMI status. Seven miRNAs were differentially expressed across the three groups shown in (A–G). miRNA levels are presented on log-transformed data (-log2). Bars show mean ± SD. Labeled (a or b) means difference for comparison of two groups following the ANOVA analysis, p < 0.05.
Table 3.
Correlation Analysis Between Anthropometric Measures and miRNA Levels in Obesity Groups
| Variables | OB-2 | OB-3 | ||||||
|---|---|---|---|---|---|---|---|---|
| miR-30d-5p | miR-197-3p | miR-125b-5p | miR-221-3p | |||||
| r | p-value | r | p-value | r | p-value | r | p-value | |
| WC (cm) | 0.605 | 0.037 | 0.134 | 0.622 | 0.410 | 0.635 | 0.239 | 0.614 |
| Weight (kg) | 0.389 | 0.325 | 0.770 | 0.009 | 0.205 | 0.814 | 0.251 | 0.605 |
| Body fat (%) | 0.458 | 0.245 | 0.268 | 0.703 | 0.724 | 0.018 | 0.772 | 0.009 |
| Muscle mass (kg) | 0.221 | 0.478 | 0.166 | 0.496 | 0.210 | 0.521 | 0.759 | 0.011 |
Notes: Data on miRNA levels are transformed into a log scale (-log2). A significant difference was determined using Pearson’s correlation analysis. The p-value less than 0.05 is shown in bold and italic. OB-2: 35 kg/m2 ≤ BMI < 40 kg/m2; OB-3: BMI ≥ 40 kg/m2.
Abbreviation: WC, waist circumference.
Associations Between miR-122, -221, -30d and IR and BMI
Among the identified miRNAs, we found that adolescents in the OB-2 group (35 kg/m2 ≤ BMI < 40 kg/m2) had increased miR-122-5p, -221-3p, and -30d-5p that were also observed in the OB-IR group. These three miRNAs accounted for 42.9% of miRNA species differentially expressed in obesity and 25% in IR, respectively (Figure S4). The individuals in the OB-2 had a 90.9% of IR presence and the highest insulin level of 51.68 µU/mL, compared with the OB-1 (BMI < 35) or OB-3 (BMI ≥ 40) (21.89 µIU/mL in the OB-1 and 37.48 µIU/mL in the OB-3), which was consistent with findings that were seen in individuals with IR (Table 1).
Discussion
Adolescent obesity is a global public health problem and a major risk factor for obesity and associated metabolic diseases in adults. Conventional serum biomarkers, eg, leptin,33 adiponectin,34 are not always associated with obesity-related complications such as IR. In the literature, circulating miRNA expressions targeting IR and obesity have been profiled via comparison with healthy and normal weight controls; whereas subjects with different degrees of obesity are grouped into one group.17,31 These miRNA signatures may discriminate individuals with a predisposition to IR, but do not distinguish disease progression within an obese population. In the present study, we found a panel of altered circulating miRNAs that characterized obese adolescents with IR from those without IR. We also identified differential expressions of miRNAs depending on degree of obesity status. These miRNA signatures may provide insights into underlying mechanisms associated with obesity progression and its complications. As an example, we found obese adolescents have increased miRNAs (eg, miR-122, -221 or -30d), indicating developed IR, although no changes were observed in fasting glucose, adiponectin, and leptin. To our knowledge, this is the first report showing that circulating miRNAs can serve as independent markers to identify adolescents who have IR (ie, HOMA-IR > 4) within an obese cohort (35 kg/m2 ≤ BMI < 40 kg/m2). This finding indicates that circulating miRNA signatures might improve IR screening in obese adolescents. These findings will require validation in larger cohorts of children in a longitudinal study.
Circulating miRNA signatures associated with IR have been identified in adults with obesity and obesity-related metabolic disorders.13,17,30 miRNAs linking to causation have been suggested as promising biomarkers for early diagnosis, progression, and treatment of IR related diseases, eg, T2D, NAFLD.11 In the present study, the miRNA signature integrated several individual miRNAs presented in adolescents with IR. It displayed an increased expression pattern, suggestive of miRNA features of obesity-mediated IR in adolescents different from those reported in adults.29,30 Increased concentrations of miR-342, -223, -30d, -215, -221, and -122 were found to be associated with insulin levels. These findings are consistent with previous reports on obesity and its related metabolic diseases.17,32,35–40 miRNAs have been demonstrated to be involved in multiple metabolic pathways, including insulin signaling, adipokine expression, adipogenesis, and lipid metabolism.17 For example, adipose tissue-derived miR-342 alters insulin sensitivity due to the introduction of transcription factor CEBPA (CCAT/enhancer-binding protein, alpha),39 which regulates fatty acid synthesis and glucose metabolism. Adipose tissue-derived miR-30d upregulates the production of secreted frizzled-related protein 4 (SFRP4), which plays a critical role in the pathogenesis of obesity and T2D;38 miR-30d overexpression also increased glucose-stimulated insulin gene transcription in MIN6 cells.41 Liver-specific miR-122 directly targets PTP1B (protein-tyrosine phosphatase 1B)42 and regulates the insulin/IGF (insulin-like growth factor) signaling pathway. The increased miR-221 selectively expressed in the livers of ob/ob mice regulates adiponectin43 and leptin expressions.37 Therefore, we speculate that the co-regulation of increased miR-122, -221, and -30d in the setting of IR may play a role in metabolic dysfunction in adipose tissue and liver. Altered miRNAs may impair cellular insulin signaling in insulin-sensitive tissues and lipid metabolism and cause imbalance of leptin-adiponectin regulation. Therefore, it is possible that altered miRNAs may be mechanistic indicators of obesity-mediated IR in adolescents. The investigation of circulating miRNAs associated with IR in insulin-sensitive tissues will aid in a better understanding of systemic IR in the body.
Interestingly, we also identified miRNA expression patterns that were associated with weight status in obese adolescents. Specifically, in those classified as morbidly obese (BMI ≥ 40 kg/m2) miR-125b, -423, and -221 negatively correlated with BMI, body fat, and WC, which is consistent with recent reports in adults with severe obesity in comparison to healthy weight or normal BMI individuals.29,44,45 These miRNA species have been identified to be either up- or down-regulated during obesity progression and adipogenesis. For example, decreased miR-221 regulates adipocyte differentiation and promotes inflammation of the adipose tissue in the development of obesity, which may be implicated in tumor necrosis factor-α (TNF-α)-mediated chronic inflammation.37,46 Other miRNA species, eg, miR-122, -150, and -483-5p, have been previously demonstrated to correlate with BMI in children or adults with obesity.29,47,48 Taken together, these findings suggest these distinct miRNA signatures may offer insight into how adipogenesis pathways are regulated with increased BMI.
We acknowledge the difference between adults and adolescents may result from the hormone regulation of miRNA expressions.49 Other factors, eg, genetic predisposition, or age, may also influence our results. There are other limitations to our study. We used HOMA-IR as a surrogate for IR assessment instead of the euglycemic clamp, albeit previous studies have shown that HOMA model can closely mirror the findings in the glucose clamp technique in assessing IR.24 Although the HOMA value to define the presence of IR is not well characterized, and puberty-associated physiological IR, independent of sex and BMI, brings extra challenges in identifying the optimum HOMA-IR level to define IR in adolescents, a cut-point of 4 is highly conservative and is very likely to identify subjects with true IR. The HOMA-AD model incorporates adiponectin in the glucose–insulin interaction and could be a superior tool to assess “insulin sensitivity” even in subjects with diabetes; however, it is comparable to the HOMA model in assessing IR.50 Given the additional costs of calculating HOMA-AD, it has not been widely used when primary outcome measure is IR as in this study. Similarly, Adipo-IR as a marker to assess insulin action in adipose tissue51 gives similar information about IR compared to HOMA-IR, of the glucose and lipid metabolisms in adipocytes, and is not as widely used as HOMA-IR. The size of the cohort was relatively small; thus, more studies are needed for validations. Additionally, changes in the level of circulating miRNAs as indicators do not necessarily reflect dysfunction of miRNA specific to tissues; therefore, further studies are needed to determine miRNA regulation in different tissues including intracellular and extracellular miRNAs and tissue-specific miRNAs to illustrate how miRNAs are implicated in obesity progression and metabolic disorders. Such studies will aid in a better understanding of mechanisms of adolescent obesity and its related metabolic diseases in target tissues or via cell-to-cell communication and metabolic organ crosstalk.
In conclusion, specific circulating miRNA signatures displayed increased expression in adolescents with IR and obesity, revealing associations between aberrant miRNAs and IR in children. We also identified a relationship between miRNA expressions and BMI in morbidly obese adolescents. To our knowledge, this is the first report that serum miRNA signatures are linked to the severity of obesity and IR in children, highlighting the unique miRNA expressions in obese adolescents not found in obese adults. Overall, circulating miRNA signatures can reflect IR and obesity status in children. This might have clinical implications in establishing practical strategies for weight management, monitoring the prognosis of obesity-related complications, and evaluating the prevention of metabolic diseases.
Acknowledgments
We would like to express our gratitude to Matthew Cotter and Oleksandra Pavliv in the Metabolism and Bioenergetics Core and the staff at the Center for Childhood Obesity Prevention at the Arkansas Children’s Research Institute for valuable help in clinical data generation and technical assistance. We also thank Dr. Sean Adams for the critical review of the manuscript.
Funding Statement
Research reported in this publication was partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 5P20GM109096. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. HL, EB, and KM were also supported by the United States Department of Agriculture/Agricultural Research Service (USDA-ARS Project 6026-51000-012-06S).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors have no conflicts of interest to disclose.
References
- 1.Hales C, Carroll M, Fryar C, Ogden C. Prevalence of obesity among adults and youth: United States, 2015–2016. NCHS Data Brief. 2017. [PubMed] [Google Scholar]
- 2.Poirier P, Giles TD, Bray GA, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on Obesity and Heart Disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113(6):898–918. doi: 10.1161/CIRCULATIONAHA.106.171016 [DOI] [PubMed] [Google Scholar]
- 3.Weiss R, Dziura J, Burgert TS, et al. Obesity and the metabolic syndrome in children and adolescents. N Engl J Med. 2004;350(23):2362–2374. doi: 10.1056/NEJMoa031049 [DOI] [PubMed] [Google Scholar]
- 4.Skinner AC, Skelton JA. Prevalence and trends in obesity and severe obesity among children in the United States, 1999-2012. JAMA Pediatrics. 2014;168(6):561–566. doi: 10.1001/jamapediatrics.2014.21 [DOI] [PubMed] [Google Scholar]
- 5.de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582(1):97–105. doi: 10.1016/j.febslet.2007.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy-Marchal C, Arslanian S, Cutfield W, et al. Insulin resistance in children: consensus, perspective, and future directions. J Clin Endocrinol Metab. 2010;95(12):5189–5198. doi: 10.1210/jc.2010-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 10.Dumortier O, Hinault C, Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18(3):312–324. doi: 10.1016/j.cmet.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 11.Ji C, Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol. 2019;15(12):731–743. doi: 10.1038/s41574-019-0260-0 [DOI] [PubMed] [Google Scholar]
- 12.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landrier J-F, Derghal A, Mounien L. MicroRNAs in obesity and related metabolic disorders. Cells. 2019;8(8):8. doi: 10.3390/cells8080859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallo A, Tandon M, Alevizos I, Illei GG, Afarinkia K. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. doi: 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23(2):91–97. doi: 10.1097/MOL.0b013e328350a425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prats-Puig A, Ortega FJ, Mercader JM, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab. 2013;98(10):E1655–60. doi: 10.1210/jc.2013-1496 [DOI] [PubMed] [Google Scholar]
- 17.Oses M, Margareto Sanchez J, Portillo MP, Aguilera CM, Labayen I. Circulating miRNAs as biomarkers of obesity and obesity-associated comorbidities in children and adolescents: a systematic review. Nutrients. 2019;11(12):12. doi: 10.3390/nu11122890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112 [DOI] [PubMed] [Google Scholar]
- 19.Tas E, Bai S, Ou X, et al. Fibroblast growth factor-21 to adiponectin ratio: a potential biomarker to monitor liver fat in children with obesity. Front Endocrinol. 2020;11. doi: 10.3389/fendo.2020.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keskin M, Kurtoglu S, Kendirci M, Atabek ME, Yazici C. Homeostasis model assessment is more reliable than the fasting glucose/insulin ratio and quantitative insulin sensitivity check index for assessing insulin resistance among obese children and adolescents. Pediatrics. 2005;115(4):e500–3. doi: 10.1542/peds.2004-1921 [DOI] [PubMed] [Google Scholar]
- 21.Matsuhisa M, Yamasaki Y, Emoto M, et al. A novel index of insulin resistance determined from the homeostasis model assessment index and adiponectin levels in Japanese subjects. Diabetes Res Clin Pract. 2007;77(1):151–154. doi: 10.1016/j.diabres.2006.10.005 [DOI] [PubMed] [Google Scholar]
- 22.Giannini C, Santoro N, Caprio S, et al. The triglyceride-to-HDL cholesterol ratio: association with insulin resistance in obese youths of different ethnic backgrounds. Diabetes Care. 2011;34(8):1869–1874. doi: 10.2337/dc10-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams-Huet B, Devaraj S, Siegel D, Jialal I. Increased adipose tissue insulin resistance in metabolic syndrome: relationship to circulating adipokines. Metab Syndr Relat Disord. 2014;12(10):503–507. doi: 10.1089/met.2014.0092 [DOI] [PubMed] [Google Scholar]
- 24.Bonora E, Targher G, Alberiche M, et al. Homeostasis model assessment closely mirrors the glucose clamp technique in the assessment of insulin sensitivity: studies in subjects with various degrees of glucose tolerance and insulin sensitivity. Diabetes Care. 2000;23(1):57–63. doi: 10.2337/diacare.23.1.57 [DOI] [PubMed] [Google Scholar]
- 25.Reinehr T. Changes in the atherogenic risk factor profile according to degree of weight loss. Arch Dis Child. 2004;89(5):419–422. doi: 10.1136/adc.2003.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valerio G, Licenziati MR, Iannuzzi A, et al. Insulin resistance and impaired glucose tolerance in obese children and adolescents from southern Italy. Nutr Metab Cardiovasc Dis. 2006;16(4):279–284. doi: 10.1016/j.numecd.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 27.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller RM, Chambers TL, Burns SP. Validating InBody®570 multi-frequency bioelectrical impedance analyzer versus DXA for body fat percentage analysis. JEPonline. 2016;19(5):8. doi: 10.1249/01.mss.0000487979.68551.d7 [DOI] [Google Scholar]
- 29.Ortega FJ, Mercader JM, Catalan V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59(5):781–792. doi: 10.1373/clinchem.2012.195776 [DOI] [PubMed] [Google Scholar]
- 30.Iacomino G, Siani A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017;12:23. doi: 10.1186/s12263-017-0577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson MD, Cismowski MJ, Serpico M, Pusateri A, Brigstock DR. Elevation of circulating microRNA levels in obese children compared to healthy controls. Clin Obes. 2017;7(4):216–221. doi: 10.1111/cob.12192 [DOI] [PubMed] [Google Scholar]
- 32.Iacomino G, Russo P, Marena P, et al. Circulating microRNAs are associated with early childhood obesity: results of the I.Family Study.. Genes Nutr. 2019;14(1):2. doi: 10.1186/s12263-018-0622-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers MG, Leibel RL, Seeley RJ, Schwartz MW. Obesity and leptin resistance: distinguishing cause from effect. Trends Endocrinol Metab. 2010;21(11):643–651. doi: 10.1016/j.tem.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nigro E, Scudiero O, Monaco ML, et al. New insight into adiponectin role in obesity and obesity-related diseases. Biomed Res Int. 2014;2014:658913. doi: 10.1155/2014/658913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masotti A, Baldassarre A, Fabrizi M, et al. Oral glucose tolerance test unravels circulating miRNAs associated with insulin resistance in obese preschoolers. Pediatr Obes. 2017;12(3):229–238. doi: 10.1111/ijpo.12133 [DOI] [PubMed] [Google Scholar]
- 36.Chuang T-Y, Wu H-L, Chen -C-C, et al. MicroRNA-223 expression is upregulated in insulin resistant human adipose tissue. J Diabetes Res. 2015;2015:943659. doi: 10.1155/2015/943659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia. 2013;56(9):1971–1979. doi: 10.1007/s00125-013-2950-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nunez Lopez YO, Garufi G, Pasarica M, Seyhan AA. Elevated and correlated expressions of miR-24, miR-30d, miR-146a, and SFRP-4 in human abdominal adipose tissue play a role in adiposity and insulin resistance. Int J Endocrinol. 2018;2018:7351902. doi: 10.1155/2018/7351902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matulewicz N, Stefanowicz M, Nikolajuk A, Karczewska-Kupczewska M. Markers of adipogenesis, but not inflammation, in adipose tissue are independently related to insulin sensitivity. J Clin Endocrinol Metab. 2017;102(8):3040–3049. doi: 10.1210/jc.2017-00597 [DOI] [PubMed] [Google Scholar]
- 40.Li WD, Xia JR, Lian YS. Hepatic miR215 target Rictor and modulation of hepatic insulin signalling in rats. Mol Med Rep. 2019;19(5):3723–3731. doi: 10.3892/mmr.2019.10031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou T, Meng X, Che H, et al. Regulation of insulin resistance by multiple MiRNAs via targeting the GLUT4 signalling pathway. Cell Physiol Biochem. 2016;38(5):2063–2078. doi: 10.1159/000445565 [DOI] [PubMed] [Google Scholar]
- 42.Nigi L, Grieco GE, Ventriglia G, et al. MicroRNAs as regulators of insulin signaling: research updates and potential therapeutic perspectives in type 2 diabetes. Int J Mol Sci. 2018;19(12):12. doi: 10.3390/ijms19123705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lustig Y, Barhod E, Ashwal-Fluss R, et al. RNA-binding protein PTB and microRNA-221 coregulate AdipoR1 translation and adiponectin signaling. Diabetes. 2014;63(2):433–445. doi: 10.2337/db13-1032 [DOI] [PubMed] [Google Scholar]
- 44.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes. 2016;40(1):88–101. doi: 10.1038/ijo.2015.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ameling S, Kacprowski T, Chilukoti RK, et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med Genomics. 2015;8(1):61. doi: 10.1186/s12920-015-0136-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou -W-W, Wang Y-T, Liao Y-C, Chuang S-C, Wang S-N, Juo S-HH. Decreased MicroRNA-221 is associated with high levels of TNF-a in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell Physiol Biochem. 2013;32(1):127–137. doi: 10.1159/000350131 [DOI] [PubMed] [Google Scholar]
- 47.Hijmans JG, Diehl KJ, Bammert TD, et al. Influence of overweight and obesity on circulating inflammation-related microRNA. Microrna. 2018;7(2):148–154. doi: 10.2174/2211536607666180402120806 [DOI] [PubMed] [Google Scholar]
- 48.Gallo W, Esguerra JLS, Eliasson L, Melander O. miR-483-5p associates with obesity and insulin resistance and independently associates with new onset diabetes mellitus and cardiovascular disease. PLoS One. 2018;13(11):e0206974. doi: 10.1371/journal.pone.0206974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reza A, Choi YJ, Han SG, et al. Roles of microRNAs in mammalian reproduction: from the commitment of germ cells to peri-implantation embryos. Biol Rev Camb Philos Soc. 2019;94(2):415–438. doi: 10.1111/brv.12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vilela BS, Vasques AC, Cassani RS, et al. The HOMA-Adiponectin (HOMA-AD) closely mirrors the HOMA-IR Index in the screening of insulin resistance in the Brazilian Metabolic Syndrome Study (BRAMS). PLoS One. 2016;11(8):e0158751. doi: 10.1371/journal.pone.0158751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryden M, Andersson DP, Arner P. Usefulness of surrogate markers to determine insulin action in fat cells. Int J Obes. 2020. [DOI] [PubMed] [Google Scholar]