Abstract
BACKGROUND
Prognostic markers for meningioma are needed to risk-stratify patients and guide postoperative surveillance and adjuvant therapy.
OBJECTIVE
To identify a prognostic gene signature for meningioma recurrence and mortality after resection using targeted gene-expression analysis.
METHODS
Targeted gene-expression analysis was used to interrogate a discovery cohort of 96 meningiomas and an independent validation cohort of 56 meningiomas with comprehensive clinical follow-up data from separate institutions. Bioinformatic analysis was used to identify prognostic genes and generate a gene-signature risk score between 0 and 1 for local recurrence.
RESULTS
We identified a 36-gene signature of meningioma recurrence after resection that achieved an area under the curve of 0.86 in identifying tumors at risk for adverse clinical outcomes. The gene-signature risk score compared favorably to World Health Organization (WHO) grade in stratifying cases by local freedom from recurrence (LFFR, P < .001 vs .09, log-rank test), shorter time to failure (TTF, F-test, P < .0001), and overall survival (OS, P < .0001 vs .07) and was independently associated with worse LFFR (relative risk [RR] 1.56, 95% CI 1.30-1.90) and OS (RR 1.32, 95% CI 1.07-1.64), after adjusting for clinical covariates. When tested on an independent validation cohort, the gene-signature risk score remained associated with shorter TTF (F-test, P = .002), compared favorably to WHO grade in stratifying cases by OS (P = .003 vs P = .10), and was significantly associated with worse OS (RR 1.86, 95% CI 1.19-2.88) on multivariate analysis.
CONCLUSION
The prognostic meningioma gene-expression signature and risk score presented may be useful for identifying patients at risk for recurrence.
Keywords: Meningioma, Biomarker, Gene expression, Prognostic, WHO grade, Recurrence, Survival, Resection, Radiation, Expression, Gene
ABBREVIATIONS
- AUC
area under the curve
- CI
confidence interval
- CNV
copy number variation
- DNA
deoxyribonucleic acid
- GEO
Gene Expression Omnibus
- GTR
gross total resection
- IHC
immunohistochemistry
- LFFR
local freedom from recurrence
- OS
overall survival
- RR
relative risk
- TTF
time to failure
- WHO
World Health Organization
Meningiomas comprise 38% of all primary intracranial tumors diagnosed in the United States, and are the most common tumor of the central nervous system.1 Although many meningiomas are slow growing, a significant subset of meningiomas have high World Health Organization (WHO) histopathologic grade, including atypical meningiomas (WHO grade II, 10%-20%) and anaplastic meningiomas (WHO grade III, 3%-5%), and are prone to local recurrence despite optimal local control.1 Moreover, there are subsets of patients with WHO grade I meningiomas who develop paradoxical recurrences that could not be predicted from histopathologic or clinical features.2-5
Recent efforts to characterize the genetic, transcriptional, and epigenetic landscape of meningioma have identified mutually exclusive subgroups of meningiomas harboring recurrent mutations in TRAF4/KLF4, AKT1, and SMO, which almost exclusively occur in clinically indolent tumors.6-9 WHO grade II and III meningiomas tend to harbor loss of chromosome 22 or inactivating mutations in the tumor suppressor NF2 and, in some cases, less common alterations in TERT and other genes.10,11 Although high-grade meningiomas are also characterized by chromosomal instability with dramatic copy number variations (CNVs), the clinical and gene-expression significance of most CNVs in meningioma are not fully understood.12,13 Most recently, methylation-based classification of meningiomas has emerged as a robust prognostic assay, albeit clinically challenging to implement in most centers.6,13,14 Whole genome transcriptomic profiling has also identified gene-expression-based subgroups of meningiomas that appear to stratify according to location and clinical outcomes,13,15 but like deoxyribonucleic acid (DNA) methylation-based profiling, these methods remain challenging to implement clinically because of the financial, logistic, and quality assurance burden of these approaches.16,17
We and others have shown that high meningioma cell proliferation in resection specimens identifies tumors at risk for adverse clinical outcomes3,18-21 and that activation of the FOXM1 target genes drives meningioma cell proliferation across molecular subgroups and WHO grades.13 These data suggest that convergent gene-expression programs may underlie clinically aggressive meningiomas, which could be leveraged to develop prognostic biomarkers. Similar challenges in other cancer types have led to targeted gene-expression biomarkers now in widespread clinical use, particularly in breast cancer, in which a 21-gene-expression assay has been shown to be predictive of the need for adjuvant chemotherapy in a large randomized trial,22 and prostate cancer, in which similar assays are available to help risk-stratify patients and guide active surveillance.23,24
Objective
The aim of this study was to apply targeted gene-expression analysis to identify a prognostic gene-expression-based signature and risk score for meningioma recurrence after resection.
METHODS
Discovery and Validation Patient Cohorts
Our discovery cohort of patients with meningioma that were treated with resection was comprised of cases between 1990 and 2015 from the University of California San Francisco. Patients were retrospectively identified from an institutional clinical database and biorepository. Clinical data were collected as previously described (Text, Supplemental Digital Content 1).
In order to identify an independent validation dataset of patients with meningiomas treated with resection, a search was undertaken of the Gene Expression Omnibus (GEO) repository using the term “meningioma,” filtered for “expression profiling by array” and human samples (Table, Supplemental Digital Content 2); datasets were screened for availability of pre-specified clinical data (Text, Supplemental Digital Content 1). Only one dataset fit these criteria (GSE58037).25
Targeted Gene-Expression Analysis and Immunohistochemistry
As previously described,13 total RNA was extracted from tumor cores from formalin-fixed paraffin-embedded tissue blocks containing 75% or more tumor cells, as determined by hematoxylin and eosin staining. The GX Human Cancer Reference NanoString panel codeset, with 30 additional meningioma related genes (266 total gene probes; Table, Supplemental Digital Content 3), were synthesized by NanoString technologies (Seattle, Washington). RNA (200 ng per meningioma) was analyzed with the NanoString nCounter Analysis System at NanoString Technologies, according to the manufacturer's protocol.
Bioinformatic and Statistical Analyses
Cases were dichotomized based on time to local recurrence (Text, Supplemental Digital Content 1). NanoString data were pre-processed according to manufacturer guidelines (Text, Supplemental Digital Content 1). Prediction analysis for microarrays (PAM), an extension of the nearest centroid classifier,26 was used to identify a subset of genes from the discovery cohort that were associated with poor outcomes (pamr: Pam: PAM, R package version 1.56.1; R Foundation for Statistical Computing).27 K-fold cross-validation was performed using the pamr.cv function to determine the optimal shrinkage threshold, resulting in a subset of genes minimizing the classification error (Text, Supplemental Digital Content 1).
In order to generate a generalizable risk score based on the genes of interest identified by PAM, Z- and log2-transformed counts of genes of interest were further scaled and constrained using the softmax transformation.28
Next, an elastic net regression classifier was trained utilizing K-fold cross-validation, and using the above transformed values as input and the probability of classification as poor-outcome as output. The probability of poor-outcome between 0 and 1 was defined as the meningioma gene-signature risk score. Elastic net regression was performed using the ElasticNetCV function of the Scikit-learn package in Python.29
Microarray data from the validation cohort were pre-processed as described previously.30 An identical set of transformations was applied to the data, and the elastic net classifier from above was applied to the validation cohort to obtain gene-signature risk scores.
CNV data were also obtained from the validation cohort, as previously described.30 Gene set enrichment analysis was performed using ConsensusPathDB,31 and protein-protein interaction analysis, clustering, and visualization was performed with the STRING database.32 All other statistical analyses including Cox proportional hazards regression, Kaplan-Meier survival analysis and log-rank tests, and other standard statistical tests were performed in JMP (JMP®, Version 14.0. SAS Institute Inc, Cary, North Carolina, 1989-2019).
RESULTS
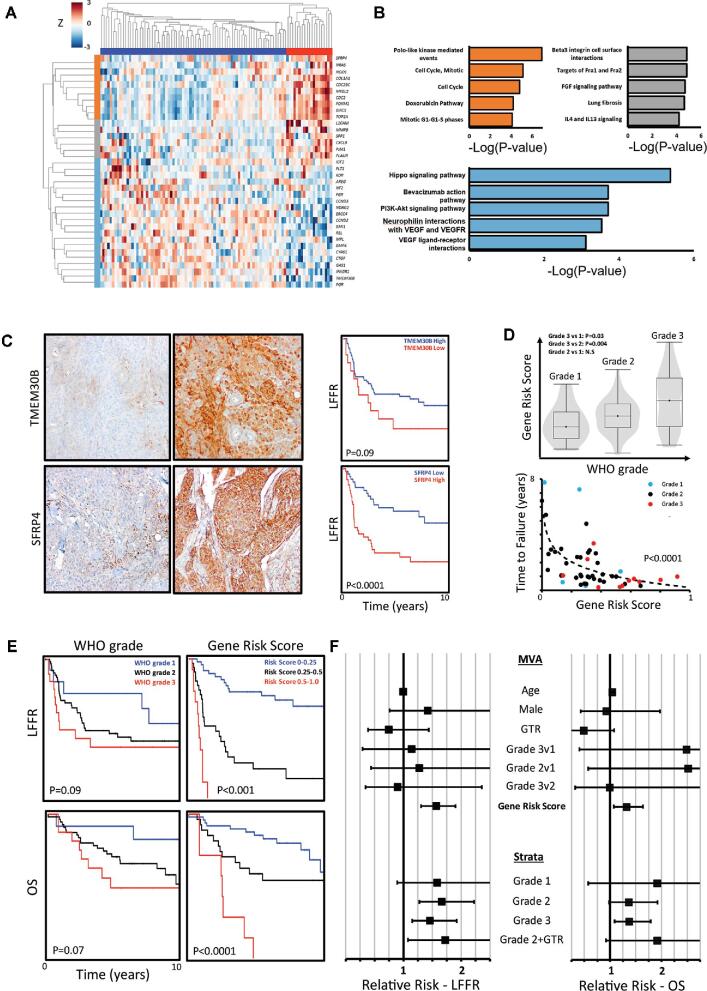
The characteristics of the discovery and validation cohorts are summarized in Table. After dichotomizing the discovery cohort into poor-outcome (N = 25, median local freedom from recurrence [LFFR] 0.70 yr, median overall survival (OS) 2.5 yr) and baseline-outcome cases (N = 71, median LFFR not reached, median OS 11.9 yr), the method of shrunken centroids identified a set of 36 genes that distinguished between outcome subgroups (Figure 1A; Table, Supplemental Digital Content 4). In order to confirm the prognostic significance of these genes, unsupervised hierarchical clustering was performed on the discovery cohort (Figure 1A), which demonstrated robust clustering of cases into 2 subgroups with significant differences in LFFR (median 0.92 vs 7.8 yr, P < .0001, log-rank test) and OS (4.0 vs 14.4 yr, P = .0003, log-rank test). The subgroup of meningioma cases with the worst outcomes showed increased expression of genes associated with cell-cycle regulation and mitosis (Figure 1B), including FOXM1,33BIRC5,34TOP2A,35CDC2,36SFRP4,37 and MYBL2,38 as well as concomitant decreased expression of BMP4, a signaling molecule involved in embryonic development, differentiation, and bone and cartilage morphogenesis39; CTGF, which is important for wound healing and fibrosis40; GAS1, a tumor suppressor41; progesterone receptor, which has been implicated in low grade meningiomas42; TMEM30B, a transmembrane gene product with unknown function.43,44 Immunohistochemistry (IHC) for representative enriched or suppressed gene products from each cluster confirmed that cases with high SFRP4 staining had worse LFFR (Figure 1C; P < .0001), whereas cases with low or absent TMEM30B staining showed a trend towards worse LFFR (Figure 1C; P = .09). In further support of the prognostic value of these genes, we have previously shown increased IHC staining of FOXM1 to be strongly associated with worse LFFR and OS.13 Closer interrogation of the gene signature revealed that multiple prognostic genes were contained at chromosomal loci frequently affected by CNVs in high-grade meningiomas,12 including 1p, 1q, 6q, 17q, and 20q (Figure, Supplemental Digital Content 5A). Consistently, the expression of 4 genes that were enriched in meningiomas with poor outcomes, FOXM1, TOP2A, BIRC5, and CDC25C, was positively associated with the number of CNVs in cases from the validation cohort (Figure, Supplemental Digital Content 5; Table, Supplemental Digital Content 6). These data suggest our gene-expression signature may capture genes that are recurrently altered through CNVs associated with clinically aggressive meningiomas.12,13
TABLE.
Meningioma Gene Signature Discovery and Validation Cohort Characteristics
| Discovery cohort | Validation cohort | |
|---|---|---|
| Clinical characteristic by patient | ||
| Number of unique patients | 84 | 56 |
| Median patient age | 60.4y (IQR 52.6-67.6) | 64.0 (53.0-76.0) |
| Number of male patients | 33 (39%) | 23 (41%) |
| Number of patients with recurrent meningioma at presentation | 22 (26%) | 11 (20%) |
| Number of patients with prior meningioma resections | 15 (18%) | Unknown |
| Number of patients with prior meningioma radiotherapy | 18 (21%) | 5 (9%) |
| Number of gross total resections | 39 (46%) | 47 (84%) |
| WHO grade of first resection | ||
| I | 12 (14%) | 35 (63%) |
| II | 64 (76%) | 16 (29%) |
| III | 7 (8%) | 5 (9%) |
| Median clinical follow up (yr) | 6.4 (IQR 3.6-9.1) | 5.4 (2.9-7.0) |
| Number of patients who died | 35 (42%) | 10 (18%) |
| Number of patients who died of meningioma progression | 17 (20%) | Unknown |
| Clinical characteristics by meningioma | ||
| Number of meningiomas | 96 | 56 |
| Number of meningiomas recurrent at presentation | 33 (34%) | 11 (20%) |
| Number of meningiomas with prior surgery | 19 (20%) | Unknown |
| Number of meningiomas with prior radiotherapy | 29 (30%) | 5 (9%) |
| Number of gross total resections | 45 (47%) | 47 (84%) |
| WHO grade | ||
| I | 13 (14%) | 35 (63%) |
| II | 64 (67%) | 16 (29%) |
| III | 19 (20%) | 5 (9%) |
| Number recurred | 49 (58%) | 11 (20%) |
FIGURE 1.
Targeted gene-expression analysis of clinically aggressive meningiomas identifies a prognostic gene signature. A, Unsupervised hierarchical clustering of prognostic genes identified using PAM confirms the ability of the gene set to stratify meningioma patients into high-risk (red cluster) and lower-risk categories (blue cluster, log-rank test, P < .0001). Gene expression is normalized by row. B, Gene enrichment analysis of prognostic gene clusters from A identifies a tightly correlated set of genes involved in cell-cycle processes (orange cluster), and clusters of genes involved in cellular signaling and extracellular matrix interactions (light blue and grey clusters). C, Representative IHC images demonstrating high TMEM30B staining on the top right (20x magnification) and low/absent TMEM30B staining on the top left. Similarly, representative IHC images demonstrating low SFRP4 staining (20x magnification) on the bottom left and high SFRP4 staining on the bottom right are shown. Low TMEM30B staining (15 of 96 meningiomas, 16%) is associated with a trend towards worse LFFR, and high SFRP4 staining (46 of 94 meningiomas, 49%) is significantly associated with worse LFFR. D, Elastic net regression was used to generate a gene-signature risk score between 0 and 1 per tumor sample (accuracy 0.80, AUC 0.86). Gene risk score correlates with tumor grade and is correlated with a faster time to failure (TTF) (TTF vs log(gene risk), P < .0001, F-test). Meningiomas with a gene risk score of greater than 0.5 uniformly recur within 2 yr of resection. Meningiomas which did not recur are not plotted. E, The gene-signature risk score outperforms WHO grade in stratifying LFFR (P < .001 vs P = .09, log-rank test) and OS (P < .0001 vs P = .07, log-rank test). F, After adjusting for age, sex, extent of resection, and grade using multivariate Cox regression, the gene-signature risk score is independently associated with recurrence (RR 1.56 per 0.1 risk score increase, 95% CI 1.30-1.90) and mortality (RR 1.32 per 0.1 increase, 95% CI 1.07-1.64). After stratifying patients by grade, the gene-signature risk score remains significantly prognostic for meningioma recurrence and mortality on univariate Cox regression. Further, among gross totally resected grade 2 tumors (Grade 2 + gross total resection (GTR)), the gene risk score is significantly prognostic of recurrence.
Next, we utilized our 36-gene signature of poor meningioma outcomes to generate a tumor specific gene-signature risk score between 0 and 1 based on an elastic net regression classifier that achieved a cross-validation accuracy of 0.80 and area under the curve (AUC) of 0.86 in distinguishing poor- and baseline-outcome cases in the discovery cohort. The meningioma gene-signature risk score based on this classifier achieved a concordance index (c-index) of 0.75 ± 0.03 (P < .0001, Wald test) for LFFR, and 0.72 ± 0.04 for OS (P < .0001, Wald test), within the discovery cohort. The risk score was correlated with WHO grade (Figure 1D) and was strongly correlated with faster time to failure (F-test, P < .0001; Figure 1D) and compared favorably to WHO grade in stratifying cases by LFFR and OS (Figure 1E). In order to investigate the clinical utility of the meningioma gene-signature risk score, we constructed a multivariate Cox model of LFFR and OS, incorporating age, sex, extent of resection, WHO grade, and meningioma gene-signature risk score (Figure 1D). After adjusting for these clinical covariates, a higher meningioma gene-signature risk score remained significantly associated with worse LFFR (Figure 1F; relative risk [RR] 1.56 per 0.1 risk score increase, 95% CI 1.30-1.90, P < .0001) and OS (RR 1.32 per 0.1 increase, 95% CI 1.07-1.64, P = .01). Similarly, after stratifying cases in the discovery cohort by WHO grade, the meningioma gene-signature risk score remained significantly associated with worse LFFR among WHO grade II (RR 1.67 per 0.1 increase, 95% CI 1.27-2.22, P = .0003) and III (RR 1.45 per 0.1 increase, 95% CI 1.15-1.92, P = .003) tumors on univariate analysis, and trended towards significance among WHO grade I tumors (P = .10), likely owing to the small sample size of grade I tumors in the discovery cohort. The meningioma gene-signature risk score was similarly associated with worse LFFR among the subgroup of atypical WHO grade II meningiomas status post-gross total resection (GTR) (N = 26, RR 1.72 per 0.1 increase, 95% CI 1.08-2.86, P = .03), and remained significantly associated with worse LFFR among primary meningiomas without prior radiation (N = 60, RR 2.0 per 0.1 increase, 95% CI 1.44-2.81, P < .0001) with a trend towards worse OS (RR 1.50 per 0.1 increase, 95% CI 0.98-2.35, P = .06) in this subgroup (Figure 1F).
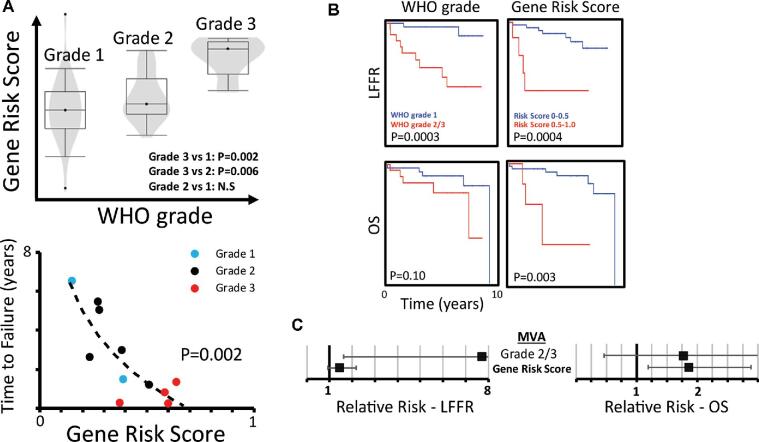
Finally, we sought to validate the prognostic utility of our meningioma gene-signature risk score in an independent cohort of meningiomas status post resection at an independent institution. The validation cohort we identified was more representative of a general population of patients with meningiomas, with fewer events of local recurrence (20% vs 58%; Table) or mortality (18 vs 42%). Nevertheless, the meningioma gene-signature risk score was again associated with WHO grade and strongly correlated with faster time to failure (F-test, P = .002; Figure 2A). Moreover, our meningioma gene-signature risk score was able to accurately stratify cases by LFFR (Figure 2B, P = .0004, log-rank test; Figure, Supplemental Digital Content 7), and compared favorably to WHO grade in stratifying cases by OS (P = .003 vs P = .10, log-rank test), achieving a c-index of 0.76 ± 0.07 (P = .01, Wald test) for LFFR, and 0.76 ± 0.11 for OS (P = .002, Wald test). Finally, after adjusting for WHO grade, a higher meningioma gene-signature risk score remained significantly associated with worse OS (RR 1.86 per 0.1 increase, 95% CI 1.19-2.88, P = .005) (Figure 2C).
FIGURE 2.
Prognostic gene-signature risk score validation in an independent dataset. A, Meningioma gene-signature risk scores were calculated on an independent validation dataset from an outside institution. The gene-signature risk score remains correlated with WHO grade and with faster TTF among patients who recurred (TTF vs log(GS risk), P = .002, F-test). Meningiomas which did not recur are not plotted. B, The gene risk score remains significantly associated with worse LFFR (P = .0004, log-rank test) and compares favorably to WHO grade in stratifying patients by OS (P = .003 vs P = .10, log-rank test). C, The gene-signature risk score remains significantly prognostic for mortality (RR 1.86 per 0.1 increase, 95% CI 1.19-2.88) after adjusting for WHO grade on Cox regression.
DISCUSSION
Key Findings
More than 15%-20% of meningiomas are high grade, and in clinical practice a subset of patients with meningiomas of all grades experience a clinically aggressive course associated with significant morbidity and mortality.45-48 Thus, there is an urgent need for better prognostic markers to help delineate clinically aggressive meningiomas. To that end, here, we perform targeted gene-expression analysis on a discovery cohort of meningioma cases that are enriched for clinical endpoints of local recurrence and disease-specific mortality. We identify a 36-gene signature of clinically aggressive meningioma and derive a meningioma gene-signature risk score between 0 and 1 that compares favorably to WHO grade in stratifying cases by risk of recurrence and survival. Moreover, we demonstrate the utility of this gene signature in risk stratifying meningioma patients from an independent validation cohort that is more representative of typical meningioma patients.
Clinical Significance
Longitudinal studies of meningioma patients with long-term follow-up indicate that the 10-yr recurrence rates after primary resection of benign, WHO grade I tumors are upwards of 20%-30%,45-47 and 40%-50% for WHO grade II tumors.48-53 These recurrences and subsequent therapies in the form of repeat craniotomy and ionizing radiation are causes of significant morbidity and, in many cases, mortality.46,54,55 Younger patients, in particular, may stand to gain most from appropriate adjuvant management in preventing morbidity and mortality associated with local recurrence, yet may also be more likely to experience long-term toxicities of aggressive therapy, which can include cognitive or neurological effects because of radiation or repeat surgery,56,57 radiation necrosis, and risk of secondary malignancies or malignant transformation because of radiation.56,58 The gene signature and risk score identified here could be used to identify high-risk patients who may benefit from aggressive adjuvant management and, conversely, to spare low-risk patients the potential toxicities of more aggressive interventions.
A Gene-Expression Signature of Clinically Aggressive Meningioma
The meningioma gene signature we report consists of enriched genes involved in cell-cycle regulation and proliferation, and suppressed genes involved in stem cell differentiation, wound healing, and tumor suppressor functions.38-48 As an added marker of external validity, many prognostic genes we identify have previously been implicated in aggressive meningiomas, including FOXM1,13,59-61TOP2A,13,62BIRC5,62MYBL2,15 and CDC2.62 Prior work from our group demonstrated that elevated expression of FOXM1 and FOXM1 target genes, including TOP2A, was associated with poorer outcome.13BIRC5, whose gene product is also known as survivin, is co-expressed with FOXM1 in breast cancer patients with poor outcomes and drug-resistance.63 Similarly, FOXM1 and MYBL2 are associated with meningiomas identified by gene-expression clustering to have poorer outcomes.15 Thus, these components of our meningioma gene signature and risk score may be representative of a common or convergent set of genes associated with meningioma cell proliferation and mitosis.
A prior study by Olar et al64 utilized a similar gene-expression approach to identify an 18-gene marker prognostic for meningioma recurrence; our work builds on this study and goes further by utilizing a novel discovery cohort enriched for clinical events (49 recurrences vs 18), and by also examining mortality as a clinical endpoint. Notably, prognostic genes in both our studies appear to be involved in proliferation, angiogenesis, and invasion.
Strengths and Limitations
The strengths of the present study that distinguish it from previous investigations include (i) the use of a discovery cohort significantly enriched for adverse clinical endpoints, including mortality, the majority of which were documented to be secondary to disease progression; (ii) validation of our meningioma gene-signature risk score using an independent cohort of meningiomas that were representative of the general population of meningioma patients; and (iii) integration of multiple genes whose altered expression have previously been described to be prognostic in meningioma into a unified prognostic model.
Generalizability
This study has several limitations. First, our study is retrospective and thus limited by the inherent biases of retrospective investigations. We attempted to mitigate these by utilizing multiple sources for collection of clinical endpoints and by careful re-review of meningioma pathology and radiology. Second, both our discovery and validation cohorts represent cases from two academic institutions. Although the validation cohort is more representative of a general population of meningioma patients, it nevertheless may not be representative of the larger clinical population encountered outside of academic institutions. Our validation cohort was also limited by a small sample size with relatively few clinical events, precluding the performance of robust multivariate analysis, and tissue was unavailable to us for immunohistochemistry. Ultimately, further validation and calibration of our risk score in a larger cohort will be needed prior to clinical use.
CONCLUSION
A gene signature and prognostic risk score based on targeted gene-expression analysis of meningiomas compared favorably to WHO grade in stratifying cases by LFFR and OS, and may be useful for guiding surveillance or adjuvant therapy after surgery.
Funding
This work was supported by grants from the NIH (K08CA212279-01), University of California San Francisco Wolfe Meningioma Program Project, and the University of California San Francisco Physician Scientist Scholar Program to Dr Raleigh.
Disclosures
The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
Supplementary Material
Contributor Information
William C Chen, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Harish N Vasudevan, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Abrar Choudhury, Department of Radiation Oncology, University of California San Francisco, San Francisco, California; Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
Melike Pekmezci, Department of Pathology, University of California San Francisco, San Francisco, California.
Calixto-Hope G Lucas, Department of Pathology, University of California San Francisco, San Francisco, California.
Joanna Phillips, Department of Pathology, University of California San Francisco, San Francisco, California.
Stephen T Magill, Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
Matthew S Susko, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Steve E Braunstein, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Nancy Ann Oberheim Bush, Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
Lauren Boreta, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Jean L Nakamura, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Javier E Villanueva-Meyer, Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, California.
Penny K Sneed, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
Arie Perry, Department of Pathology, University of California San Francisco, San Francisco, California.
Michael W McDermott, Department of Radiation Oncology, University of California San Francisco, San Francisco, California.
David A Solomon, Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
Philip V Theodosopoulos, Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
David R Raleigh, Department of Radiation Oncology, University of California San Francisco, San Francisco, California; Department of Neurological Surgery, University of California San Francisco, San Francisco, California.
Supplemental Digital Content 1. Text. Additional detailed methods.
Supplemental Digital Content 2. Table. List of candidate GEO accession numbers. GEO accession numbers for publicly available meningioma gene-expression datasets.
Supplemental Digital Content 3. Table. List of targeted gene-expression panel genes. List of all 266 genes comprising the NanoString targeted gene-expression panel, with corresponding chromosome locations (between Chrloc and Chrlocend, mapped to Genome Reference Consortium Human Build 38, GRCh38, accessed 3/13/2018). In addition, univariate Cox regression P-values, Bonferroni corrected p-values, and beta-coefficients are displayed for each gene.
Supplemental Digital Content 4. Table. List of gene names and descriptions for the 36-gene signature. List of all 36 genes included in the prognostic gene signature, including gene symbol, name, and description taken from Entrez.
Supplemental Digital Content 5. Figure. Correlation of prognostic gene-signature genes with chromosome location and CNV. A, All 266 genes from the discovery dataset are displayed by chromosome location. A moving average of neighboring gene-gene correlation (ρ, window size 4 genes) identified chromosome regions with highly co-expressed genes corresponding to areas of known frequent CNVs in meningioma, including 1p, 1q, 3p, 6q, 7q, 11q, 14q, 17q, 20q, and 22q. Coefficients of univariate Cox regression between gene expression and local recurrence are displayed (β, color-scale -3 to 3), as well as P-values (color-scale 0.05-0). Areas of negative β, shown in blue, correspond to areas in which presumed CNV deletions are associated with worse outcome, and areas of positive β, shown in red, correspond to areas in which presumed CNV amplifications are associated with worse outcome. Multiple genes from the prognostic gene signature appear to cluster in the 1p, 1q, 6q, 17q, and 20q regions, although most prognostic genes exist in areas of low neighboring gene correlation, which may represent conserved areas infrequently affected by CNV. B, Analysis of the total number of CNVs and gene expression in the validation microarray cohort identified 397 genes significantly correlated with CNV number (FDR q-value < .05). 4 gene-signature genes were among these: FOXM1, CDC25C, TOP2A, and BIRC5, which form a tightly co-expressed gene network highly correlated with CNV number (P < .0001, F-test). STRING protein-protein interaction analysis and clustering of prognostic genes (confidence level threshold 0.7, MCF clustering, inflation parameter = 3) yielded a cluster of proliferative genes (red) containing these CNV-correlated genes: FOXM1, CDC25C, TOP2A, and CDC25C, and a cluster of mesenchymal genes involved in osteoblast differentiation and collagen development (yellow).
Supplemental Digital Content 6. Table. Genes correlated with increased CNV in the validation cohort. Univariate linear regression was performed using the Z-score, log-transformed gene-expression values from all probes in the validation dataset as the independent variable, and number of CNVs as the dependent variable. The resulting P-value was Bonferroni corrected, and all genes with corrected p-value less than 0.05 are shown, with genes included in the gene signature highlighted in yellow.
Supplemental Digital Content 7. Figure. Grade and gene risk score and expanded validation Kaplan-Meier curves. A, Proportion of WHO grades within gene-signature risk score strata in the discovery and validation cohorts. N = 50, 32, and 14, in the 0-0.25, 0.25-0.50, 0.50-1.0 groups, respectively, in the discovery cohort, and N = 18, 30, and 8 in the same groups, respectively, in the validation cohort. B, Expanded Kaplan-Meier curves for LFFR and OS in the validation cohort, broken down by risk score strata. C, Expanded Kaplan-Meier curves for LFFR and OS in the validation cohort using the gene-signature risk score, linearly re-scaled within the validation cohort between 0 and 1, demonstrating further stratification into low, intermediate, and higher risk groupings. D, Expanded Kaplan-Meier curves for LFFR and OS in the validation cohort, broken down by grade.
COMMENT
This is a well-written paper that adds to the literature on the use of gene expression signatures as a means of better predicting the risk of meningioma recurrence following resection. The authors utilized a discovery cohort enriched for clinical events where the 36-gene signature was constructed from enriched genes involved in cell cycle and proliferation and suppressed genes involved in stem cell differentiation, wound healing and tumor suppressor functions and performed a validation with an independent cohort. This is similar to the work done by Olar et al1 where a 18-gene expression signature was used. The results of both studies help to identify patients at higher risk for tumor recurrence that may benefit from more aggressive adjuvant management.
Franco DeMonte
Houston, Texas
REFERENCE
- 1. Olar A, Goodman LD, Wani KM, et al.A gene-expression signature predicts recurrence-free survival in meningioma. Oncotarget. 2018;9(22):16087-16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united states in 2011-2015. Neuro Oncol. 2018;20(Suppl_4):iv1-iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aizer AA, Bi WL, Kandola MS et al. Extent of resection and overall survival for patients with atypical and malignant meningioma. Cancer. 2015;121(24):4376-4378. [DOI] [PubMed] [Google Scholar]
- 3. Chen WC, Magill ST, Wu A et al. Histopathologic features predictive of local control of atypical meningioma after surgery and adjuvant radiotherapy. J Neurosurg. 2018;130(2):443-450. [DOI] [PubMed] [Google Scholar]
- 4. Condra KS, Buatti JM, Mendenhall WM, Friedman WA, Marcus RB, Rhoton AL. Benign meningiomas: primary treatment selection affects survival. Int J Radiat Oncol Biol Phys. 1997;39(2):427-36. [DOI] [PubMed] [Google Scholar]
- 5. Cahill KS, Claus EB.. Treatment and survival of patients with nonmalignant intracranial meningioma: results from the surveillance, epidemiology, and end results program of the national cancer institute. Clinical article. J Neurosurg. 2011;115(2):259-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahm F, Schrimpf D, Stichel D et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;2045(17)30155-30159. [DOI] [PubMed] [Google Scholar]
- 7. Preusser M, Brastianos PK, Mawrin C. Advances in meningioma genetics: novel therapeutic opportunities. Nat Rev Neurol. 2018;14(2):106-115. [DOI] [PubMed] [Google Scholar]
- 8. Clark VE, Erson-Omay EZ, Serin A et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark VE, Harmancl AS, Bai H et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahm F, Schrimpf D, Olar A et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016;108(5): djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shankar GM, Santagata S. BAP1 mutations in high-grade meningioma: implications for patient care. Neuro Oncol. 2017;19(11):1447-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bi WL, Abedalthagafi M, Horowitz P et al. Genomic landscape of intracranial meningiomas. J Neurosurg. 2016;125(3):525-535. [DOI] [PubMed] [Google Scholar]
- 13. Vasudevan HN, Braunstein SE, Phillips JJ et al. Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep. 2018;22(13):3672-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olar A, Wani KM, Wilson CD et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017;133(3):431-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patel AJ, Wan YW, Al-Ouran R et al. Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc Natl Acad Sci USA. 2019;116(43):21715-21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horak P, Fröhling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1(5):e000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Locke WJ, Guanzon D, Ma C et al. DNA methylation cancer biomarkers: translation to the clinic. Front Genet. 2019;10:1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun SQ, Cai C, Murphy RKJ et al. Radiation therapy for residual or recurrent atypical meningioma: the effects of modality, timing, and tumor pathology on long-term outcomes. Neurosurgery. 2016;79(1):23-32. [DOI] [PubMed] [Google Scholar]
- 19. Olar A, Wani KM, Sulman EP et al. Mitotic index is an independent predictor of recurrence-free survival in meningioma. Brain Pathol. 2015;25(3):266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bruna J, Brell M, Ferrer I, Gimenez-Bonafe P, Tortosa A. Ki-67 proliferative index predicts clinical outcome in patients with atypical or anaplastic meningioma. Neuropathology. 2007;27(2):114-120. [DOI] [PubMed] [Google Scholar]
- 21. Perry A, Stafford SL, Scheithauer BW, Suman VJ, Lohse CM. The prognostic significance of MIB-1, p53, and DNA flow cytometry in completely resected primary meningiomas. Cancer. 1998;82(11):2262-2269. [PubMed] [Google Scholar]
- 22. Sparano JA, Gray RJ, Makower DF et al. Adjuvant chemotherapy guided by a 21-gene-expression assay in breast cancer. N Engl J Med. 2018;379(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knezevic D, Goddard AD, Natraj N et al. Analytical validation of the oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cuzick J, Berney DM, Fisher G et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106(6):1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee Y, Liu J, Patel S et al. Genomic landscape of meningiomas. Brain Pathol. 2010;20(4):751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene-expression. Proc Natl Acad Sci USA. 2002;99(10):6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hastie T, Tibshirani R, Narasimhan B, Chu G. pamr: Pam: Prediction Analysis for Microarrays. 2019. https://cran.r-project.org/package=pamr. Accessed March 23, 2020. [Google Scholar]
- 28. Kishan K, Rui L, Feng C, Qi Y, Haake AR. GNE: a deep learning framework for gene network inference by aggregating biological information. BMC Syst Biol. 2019;13(Suppl 2):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedrogosa F, Varoquaux G, Gramfort A, Al E. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12(Oct):2825-2830. [Google Scholar]
- 30. Lee Y, Liu J, Patel S et al. Genomic landscape of meningiomas. Brain Pathol. 2010;20(4):751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB - A database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(Database issue):D623-D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szklarczyk D, Gable AL, Lyon D et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bin LG, XZ Li, Zeng S et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Obexer P, Hagenbuchner J, Unterkircher T et al. Repression of BIRC5/Survivin by FOXO3/FKHRL1 sensitizes human neuroblastoma cells to DNA damage-induced apoptosis. Mol Biol Cell. 2009;20(7):2041-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9(5):338-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan J, Yan R, Krämer A et al. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23(34):5843-5852. [DOI] [PubMed] [Google Scholar]
- 37. Liang CJ, Wang ZW, Chang YW, Lee KC, Lin WH, Lee JL. SFRPs are biphasic modulators of wnt-signaling-elicited cancer stem cell properties beyond extracellular control. Cell Rep. 2019;28(6):1511-1525.e5. [DOI] [PubMed] [Google Scholar]
- 38. Musa J, Aynaud MM, Mirabeau O, Delattre O, Grünewald TG. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8(6):e2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kallioniemi A. Bone morphogenetic protein 4-a fascinating regulator of cancer cell behavior. Cancer Genet. 2012;205(6):267-277. [DOI] [PubMed] [Google Scholar]
- 40. Braig S, Wallner S, Junglas B, Fuchshofer R, Bosserhoff AK. CTGF is overexpressed in malignant melanoma and promotes cell invasion and migration. Br J Cancer. 2011;105(2):231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Del Sal G, Ruaro ME, Philipson L, Schneider C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell. 1992;70(4):595-607. [DOI] [PubMed] [Google Scholar]
- 42. Wolfsberger S, Doostkam S, Boecher-Schwarz HG et al. Progesterone-receptor index in meningiomas: correlation with clinico-pathological parameters and review of the literature. Neurosurg Rev. 2004;27(4):238-245. [DOI] [PubMed] [Google Scholar]
- 43. Pérez-Magán E, De Lope ÁR R T et al. Differential expression profiling analyses identifies downregulation of 1p, 6q, and 14q genes and overexpression of 6p histone cluster 1 genes as markers of recurrence in meningiomas. Neuro Oncol. 2010;12(12):1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pérez-Magán E, Campos-Martín Y, Mur P et al. Genetic alterations associated with progression and recurrence in meningiomas. J Neuropathol Exp Neurol. 2012;71(10):882-893. [DOI] [PubMed] [Google Scholar]
- 45. Kotecha RS, Pascoe EM, Rushing EJ et al. Meningiomas in children and adolescents: a meta-analysis of individual patient data. Lancet Oncol. 2011;12(13):1229-1239. [DOI] [PubMed] [Google Scholar]
- 46. Kotecha RS, Jacoby P, Cole CH, Gottardo NG. Morbidity in survivors of child and adolescent meningioma. Cancer. 2013;119(24):4350-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Alkemade H, De Leau M, Dieleman EMT et al. Impaired survival and long-term neurological problems in benign meningioma. Neuro Oncol. 2012;14(5):658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen WC, Magill ST, Wu A et al. Histopathological features predictive of local control of atypical meningioma after surgery and adjuvant radiotherapy. J Neurosurg. 2018;130(2):443-450. [DOI] [PubMed] [Google Scholar]
- 49. Aizer AA, Arvold ND, Catalano P et al. Adjuvant radiation therapy, local recurrence, and the need for salvage therapy in atypical meningioma. Neuro Oncol. 2014;16(11):1547-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adeberg S, Hartmann C, Welzel T et al. Long-Term outcome after radiotherapy in patients with atypical and malignant meningiomas—clinical results in 85 patients treated in a single institution leading to optimized guidelines for early radiation therapy. Int J Radiat Oncol. 2012;83(3):859-864. [DOI] [PubMed] [Google Scholar]
- 51. Bagshaw HP, Burt LM, Jensen RL et al. Adjuvant radiotherapy for atypical meningiomas. J Neurosurg. 2016;126(6):1822-1828. [DOI] [PubMed] [Google Scholar]
- 52. Hammouche S, Clark S, Wong AHL, Eldridge P, Farah JO. Long-term survival analysis of atypical meningiomas: survival rates, prognostic factors, operative and radiotherapy treatment. Acta Neurochir (Wien). 2014;156(8):1475-1481. [DOI] [PubMed] [Google Scholar]
- 53. Komotar RJ, Iorgulescu JB, Raper DMS et al. The role of radiotherapy following gross-total resection of atypical meningiomas. J Neurosurg. 2012;117(4):679-686. [DOI] [PubMed] [Google Scholar]
- 54. Chen WC, Hara J, Magill ST et al. Salvage therapy outcomes for atypical meningioma. J Neurooncol. 2018;138(2):425-433. [DOI] [PubMed] [Google Scholar]
- 55. Magill ST, Lee DS, Yen AJ et al. Surgical outcomes after reopertaion for recurrent non-skull base meningiomas. J Neurosurg. 2019;131:1179-1187. [DOI] [PubMed] [Google Scholar]
- 56. Kaur G, Sayegh ET, Larson A et al. Adjuvant radiotherapy for atypical and malignant meningiomas: a systematic review. Neuro Oncol. 2014;16(5):628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Najafabadi A, Van Der Meer P, Boele F et al. The long-term disease burden of meningioma patients: results on health-related quality of life, cognitive function, anxiety and depression. Neuro Oncol. 2018;20(Suppl_6):vi154-vi155. [Google Scholar]
- 58. Pollock BE, Link MJ, Stafford SL, Parney IF, Garces YI, Foote RL. The risk of radiation-induced tumors or malignant transformation after single-fraction intracranial radiosurgery: results based on a 25-Year experience. Int J Radiat Oncol Biol Phys. 2017;97(5):919-923. [DOI] [PubMed] [Google Scholar]
- 59. Laurendeau I, Ferrer M, Garrido D et al. Gene-expression profiling of the hedgehog signaling pathway in human meningiomas. Mol Med. 2010;16(7-8):262-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim H, Park KJ, Ryu BK et al. Forkhead box M1 (FOXM1) transcription factor is a key oncogenic driver of aggressive human meningioma progression. Neuropathol Appl Neurobiol. 2019;46(2):125-141. [DOI] [PubMed] [Google Scholar]
- 61. Kang SH, Kim H, Park K et al. Altered FOXM1 expression contributes to the meningioma malignancy and can be a critical target for the tumor progression. Neuro Oncol. 2018;20(Suppl 3):iii271-iii272. [Google Scholar]
- 62. Stuart JE, Lusis EA, Scheck AC et al. Identification of gene markers associated with aggressive meningioma by filtering across multiple sets of gene-expression arrays. J Neuropathol Exp Neurol. 2011;70(1):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Moraes GN, Delbue D, Silva KL et al. FOXM1 targets XIAP and survivin to modulate breast cancer survival and chemoresistance. Cell Signal. 2015;27(12):2496-2505. [DOI] [PubMed] [Google Scholar]
- 64. Olar A, Goodman LD, Wani KM et al. A gene-expression signature predicts recurrence-free survival in meningioma. Oncotarget. 2018;9(22):16087-16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.