Abstract
Purpose
Age-related macular degeneration (AMD) is one of the leading causes of blindness among the elderly, and the exact pathogenesis of the AMD remains unclear. The purpose of this review is to summarize potential metabolic biomarkers and pathways of AMD that might facilitate risk predictions and clinical diagnoses of AMD.
Methods
We obtained relevant publications of metabolomics studies of human beings by systematically searching the MEDLINE (PubMed) database before June 2020. Studies were included if they performed mass spectrometry–based or nuclear magnetic resonance–based metabolomics approach for humans. In addition, AMD was assessed from fundus photographs based on standardized protocols. The metabolic pathway analysis was performed using MetaboAnalyst 3.0.
Results
Thirteen studies were included in this review. Repeatedly identified metabolites including phenylalanine, adenosine, hypoxanthine, tyrosine, creatine, citrate, carnitine, proline, and maltose have the possibility of being biomarkers of AMD. Validation of the biomarker panels was observed in one study. Dysregulation of metabolic pathways involves lipid metabolism, carbohydrate metabolism, nucleotide metabolism, amino acid metabolism, and translation, which might play important roles in the development and progression of AMD.
Conclusions
This review summarizes the potential metabolic biomarkers and pathways related to AMD, providing opportunities for the construction of diagnostic or predictive models for AMD and the discovery of new therapeutic targets.
Keywords: age-related macular degeneration, metabolomics, biomarker, metabolic pathway
Age-related macular degeneration (AMD) is one of the leading causes of irreversible blindness in the elderly.1 The global elderly population is estimate to increase to 1.2 billion by the year 2025, and thus it is expected that visual impairment and blindness caused by AMD will become a global public concern.2 The early stage of AMD has no obvious clinical symptoms and is not easy to detect. The advanced stage of AMD develops into two subtypes with different characteristics, including geographical atrophy (GA) and wet age-related macular degeneration,3,4 leading to irreversible loss of vision. Studies have found that AMD has an obvious familial genetic predisposition,5 and the genetic variation located at the complement factor H and ARMS2/HTRA1 sites has a great risk effect.6 In addition, the development of AMD is also related to dietary and environmental factors,6 but the specific pathogenesis is still unclear. Although the vascular endothelial growth factor (VEGF) antibody has been proven to be effective in the treatment of wet AMD, this approach lacks a consistent administration regimen and is expensive.7,8 Macular atrophy usually develops after anti-VEGF therapy.9 Currently, there is no effective treatment for more than 80% of AMD patients in the early, middle, and atrophy stages.10 Therefore it is crucial to identify useful biomarkers that could effectively predict the occurrence of AMD and help with early diagnosis and prevention of the diseases.
Metabolomics, a method that combines high-throughput analysis with stoichiometric methods to analyze all the small molecules (<1000 Da) that make up a biological system as a whole.11 The successful application of metabolomics in diseases shows its great potential in the study of complex diseases such as drug discovery, cardiovascular diseases, and the response to drug therapy for AMD.12–14 The retina is a metabolically active tissue of the human body, and photoreceptors are the most numerous cells in the retina with the highest requirements for metabolism.15,16 Retinal pigment epithelial (RPE) provide metabolic and functional (VEGF) support for photoreceptors and choroid cells.17 Many small molecules, such as polyunsaturated fatty acids (PUFAs),18 amino acids, vitamin A analogues, phospholipids,19 and oxidative substances, are enriched in the outer layer of the retina, which are the key factors to maintain retinal homeostasis. 4,16 With age, metabolic exchange disorders between photoreceptors and RPE may cause retinal degeneration, particularly affecting the macular area, leading to AMD.20 The exchange of material between the retina, RPE, and choroid provides an important basis for the study of local metabolomics changes in AMD.21 The application of metabolomics in AMD research is expected to reveal the complex pathogenesis and identify potential biomarkers for disease prediction.
In this study, we systematically reviewed all the metabolomics studies of AMD on human beings and summarized metabolic biomarkers and pathways related to the occurrence and development of AMD. The findings would provide insights into potential pathogenic pathways for vision-threatening ocular disease and hold translational value in risk stratification from public health perspectives.
Methods
Search Strategy
We conducted a systematic review of metabolomics studies on AMD based on the Meta-analysis of Observational Studies in Epidemiology guidelines.22 We searched the electronic database of PubMed for relevant metabolomics studies on AMD published up to June 2020, with the following search terms: (“metabolomics” or “metabonomics” or “metabolome” or “metabolic profiling”) AND (“macular Degeneration” or “age-related maculopathy” or “age-related macular degeneration” ). Additional articles were identified through searching the reference lists from included studies.
Two authors (XWH and YW) searched articles independently, screened the included articles based on the title and abstract, and identified other articles by referring to the included articles. A senior author (CWP) made the final decision in case of disagreement.
Inclusion and Exclusion Criteria
We conducted a systematic review of all English-language studies if they performed mass spectrometry (MS)–based or nuclear magnetic resonance (NMR)–based metabolomics approach on humans. In addition, AMD was assessed from fundus photographs on the basis of standardized protocols. Studies were excluded if they were animal studies, in vivo studies, or drug therapy response reports, or if they did not provide specific metabolic information.
Quality Assessment
The study quality was assessed with the tool described by Lumbreras et al.23 QUADOMIC is an adaptation of QUADAS tool (a quality assessment tool used to systematically evaluate diagnostic accuracy studies), which takes into account the challenges presented by “-omics.” QUADOMICS incorporates four projects, covering sample characteristics, differential conditions in preanalytical, clinical, and physiological characteristics of research subjects, and overfitting during research and analysis. In addition, QUADOMICS divides the study into four clinical validation stages according to the study population, and this review does not involve the phase IV research.
Data Synthesis and Analysis
For each study, information regarding the frequency of biological samples, detection and analysis platforms, sample size, study design methods, and repeated reports of biomarkers were extracted and summarized. The pathway analysis and topology results were performed on the basis of the potential disease-influencing metabolites reported from the all the included studies in the reviewed article, which was performed using the MetaboAnalyst software (version 3.0). Calculated P value was established on the basis of the pathway enrichment analysis whereas the pathway impact value was derived from the pathway topology analysis.24
Results
Study Characteristics
Thirteen articles met the inclusion criteria and were subsequently included in the review (Fig. 1). Three of the studies were by the same author on the same cohort, but there were differences in sample size, biological samples, test platform, and research grouping, so all of them were included in this review.25–27 A total of four types of sampled tissues were involved in the study, including serum (n = 3),4,10,28 plasma (n = 7),25,27,29–33 urine (n = 1),26 aqueous humor (n = 1), and both serum and plasma (n = 1).34,35 With regard to the analytical platforms used for metabolite detection, 10 studies used MS, and the other three used the NMR platform.27,26,34 The sample sizes ranged from 40 to 6533 individuals. The study with the largest sample size came from a recent study based on five cohorts from Europe. All studies compared metabolites in the case (AMD) and control groups (without AMD). Characteristics of the studies are presented in Table 1.
Figure 1.
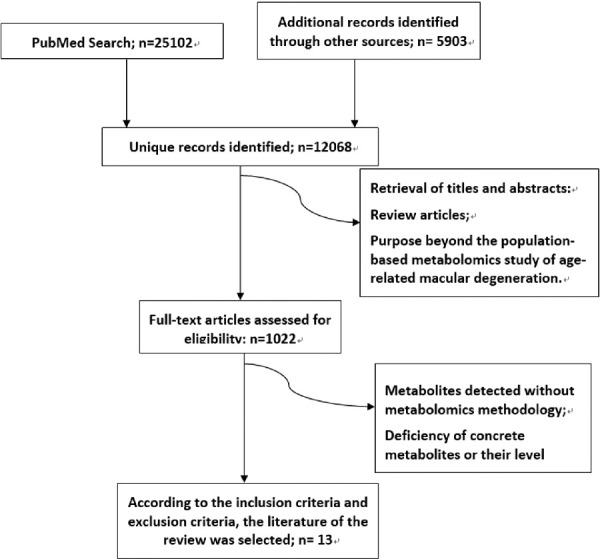
Flow diagram of literature search and study selection for metabolite markers of AMD.
Table 1.
Metabolomics Studies Analyzing Samples From Patients With AMD
| Biofluid | Source | Comparison | Targeted/Untargeted | Metabolomic Technique Used | Differential Metabolite Evaluation Method | References |
|---|---|---|---|---|---|---|
| Plasma | America | NVAMD patients (n = 100) and CP (n = 192) | Untargeted | LC-MS and LC-MS/MS | Nested feature selection | Mitchell et al.29 |
| Plasma | America | Early AMD, intermediate AMD, late AMD (n = 89) and CP (n = 30) | Targeted | UPLC-MS | Logistic regression analysis | Laíns et al.30 |
| Plasma | Two cohorts: the USA and Portugal | AMD patients (n = 391) and CP (n = 100) | Untargeted | UPLC-MS | Logistic regression models | Laíns et al.25 |
| Plasma | China | NVAMD patients (n = 20) and CP (n = 20) | Untargeted | UPLC-Q-TOF MS | PLS-DA, fold change analysis and t-test | Luo et al.32 |
| Serum | European | Patients with non-advanced AMD (n = 72) and CP (n = 72) | Targeted | HPLC-MS | sPLS-DA | Kersten et al.10 |
| Plasma | America | NVAMD patients (n = 26) and CP (n = 19) | Untargeted | LC-FTMS | Multiple testing | Osborn et al.31 |
| Plasma | Two cohorts: the USA and Portugal | AMD patients (n = 201) and CP (n = 42)(Coimbra); AMD patients (n = 113) and CP (n = 40) (Boston) | Untargeted | -NMR | PLS-DA | Laíns et al.27 |
| Serum | China | 21 PCV vs 19 CP | Untargeted | UPLC-MS | OPLS-DA | Li et al. 4 |
| Urine | America | AMD patients (n = 252) and CP (n = 53) (Coimbra); AMD patients (n = 147) and CP (n = 47) (Boston) | Untargeted | -NMR | PLS-DA | Laíns et al.26 |
| Plasma | France | Exudative-AMD patients (n = 40) and CP (n = 40) | Targeted | Mass spectrometry | Student's t test | Chao de la Barca et al.33 |
| Serum | China | AMD patients (n = 88), PCV patients (n = 102) and CP (n = 81) | Untargeted | GC-TOF/MS | OPLS-DA, Fold Change analysis and t test | Liu et al.28 |
| Aqueous humor | China | Wet AMD patients (n = 26) and CP (n = 20) | Untargeted | UHPLC-MS/MS | PLS-DA, OPLS-DA, and ANOVA | Han et al. 35 |
| S & P | European | 2267 AMD cases and 4266 CP | Targeted | High-throughput -NMR | Logistic regression | Acar et al.34 |
S & P, serum and plasma; CP, control patients; PCV, polypoidal choroidal vasculopathy; LC, liquid chromatogram; UPLC, ultra-performance liquid chromatography; FTMS, Fourier-transform mass spectrometry; GC, gas chromatography; TOF, time of flight; Q-TOF, quadrupole-time of flight; PLS-DA, partial least squares discriminant analysis; OPLS-DA, orthogonal partial least squares discriminant analysis; ANOVA, one-way analysis of variance.
Quality of the Included Studies
The results evaluated by the QUADOMICS Tool for the quality of the included studies are presented in Supplementary Table S1. All of the studies included in the systematic review were phase I studies and met the evaluation criteria of 60%, of which seven met the evaluation criteria of 80%. Among the 13 studies, two did not explicitly describe the inclusion criteria of the subjects,29,31 and four did not consider the overfitting of the analysis model.4,30,33,34
Metabolic Biomarkers Identified for AMD
Five studies aimed to identify metabolic differences between neovascular age-related macular degeneration (NVAMD) and control groups. The study by Mitchell et al.29 showed that multiple long-chain acylcarnitines that are part of the carnitine shuttle pathway in NVAMD patients were significantly increased. Using the UHPLC-QTOF MS platform, Luo et al.32 suggested that the different substances between NVAMD and controls were mainly amino acids, indicating that NVAMD is a disease associated with disorders of amino acid metabolism. Osborn et al.31 found that a panel of metabolites differed between NVAMD patients and controls, such as tyrosine metabolism, sulfur amino acid metabolism and amino acid. Chao de la Barca et al.33 revealed six new metabolites involved in the plasma metabolomic profile, suggesting mitochondrial energetic impairments and carnosine deficiency might play a role in NVAMD. Han et al.35 used aqueous humor samples and obtained 18 metabolites with significant differences. Li et al.4 investigated the difference between polypoidal choroidal vasculopathy (PCV) disease and controls, and identified 41 different substances. Laíns et al.25–27 conducted three metabolomics studies on the same cohort in the year 2017 to 2019 using plasma and urine samples separately. The results showed that AMD patients and controls had different plasma metabolomics characteristics,25,27 and the severity stages of AMD can be graded by NMR.26 Laíns et al.30 also observed that the most significant metabolites map related to AMD was the glycerophospholipid pathway. Kersten et al.10 conducted a targeted metabolomics study in advanced AMD and control group and identified four predictive metabolites. Liu et al.28 aimed to reveal a personalized metabolic pattern of macular neovascularization and extracted differential metabolites of AMD and PCV versus controls. Acar et al.34 identified metabolites associated with AMD from five European cohorts through the high-throughput NMR metabolomics platform. A total of 108 statistically significant metabolites were extracted from these studies. Table 2 summarizes potential metabolic biomarkers of AMD with a frequency of two or more reported in the literatures. Phenylalanine was totally reported three times, which was the most frequently reported biomarker.
Table 2.
Metabolic Biomarkers Related to AMD
| Metabolite Name | HMDB ID | Hits | Biological Samples to Be Analyzed |
|---|---|---|---|
| Adenosine | HMDB0000050 | 2 | Plasma25,30 |
| Hypoxanthine | HMDB0000157 | 2 | Serum28, Plasma25 |
| Tyrosine | HMDB0000158 | 2 | Serum34, Plasma32,34 |
| Phenylalanine | HMDB0000159 | 3 | Serum34, Plasma32,34, AH35 |
| Creatine | HMDB0000064 | 2 | Plasma27, AH35 |
| Citrate | HMDB0000094 | 2 | Urine26, Serum34, Plasma34 |
| Carnitine | HMDB0000062 | 2 | Plasma33, AH35 |
| Proline | HMDB0000162 | 2 | Plasma33, AH35 |
| Maltose | HMDB0000163 | 2 | Plasma25, Serum28 |
AH, aqueous humor.
Potential of Metabolite Marker Panels to Predict and Classify AMD
Several studies have assessed the potential of metabolic biomarker panels to distinguish AMD (Table 3). Mitchell et al.29 used 159 metabolic features to distinguish NVAMD from controls, which exhibited a balanced accuracy rate of 96.1% in the training set and 75.6% in the test set. Laíns et al.30 evaluated the predictive power of the first principal component of these 87 metabolites, yielding an area under curve (AUC) of 0.80 in discriminating patients with AMD from controls. Laíns et al.25 compared the predictive performance among the following four models: (1) Baseline model including only demographic covariates; (2) All-Met + EN model including baseline and the metabolites selected using elastic-net regression with all metabolites; (3) AMD/Control model including baseline and the metabolites identified in the logistic regression models; (4) Stage + 2 Eye model including baseline and the metabolites identified in the permutation-based cumulative logistic regression models, generating AUC values of 0.725, 0.745, 0.815, and 0.789, respectively. Kersten et al.10 used the selected predictors resulting from the sparse partial least squares discriminant analysis to predict nonadvanced AMD, achieving an AUC value of 0.71. Li et al.4 used the metabolites to predict PCV patients, with all achieving the AUC value of 0.8 or more.
Table 3.
Classification/Prediction Potential of Biomarker Panels
| References | Biomarker Panel | Discriminant Model | Discriminant Group; Precision |
|---|---|---|---|
| Mitchell et al.29 | 159 differential features | SVM | Training set; balanced accuracy rate = 96.1% test set; balanced accuracy rate = 75.6%, AUC = 0.83 |
| Laíns et al.30 | 87 differential features | Logistic regression | Differential material modeling (AUC = 0.8) Only contains age, gender, BMI and smoking status (AUC = 0.71) |
| Laíns et al.25 | * | Logistic regression | Baseline model (AUC = 0.725; 95% CI:0.671–0.779) All-Met + EN model (AUC = 0.745; 95% CI:0.692–0.797) Stage + 2Eye model (AUC = 0.815; 95% CI: 0.771–0.860) AMD/Control model (AUC = 0.789; 95% CI: 0.738–0.840) |
| Kersten et al.10 | Glutamine (Glu:Gln ratio, glutaminolysis, and PC.aa.C28.1) | sPLS-DA | Glutamine, Glu:Gln ratio, glutaminolysis, and PC.aa.C28.1 (AUC of 0.71, 95% CI: 0.62–0.79) glutamine (AUC of 0.66, 95% CI: 0.57–0.75) |
| Li et al.4 | 41 differential features | AUC | LPA (18:2), LPC (20:4), PC (20:1p/19:1), SM (d16:0/22:2), PAF (35:4), PC (16:0/22:5) and PC (18:1/20:4) are evaluated separately, AUC ≥ 0.8 |
SVM, support vector machine; sPLS-DA, sparse partial least squares discriminant analysis; CI, confidence interval; BMI, body mass index; Glu:Gln ratio, the ratio between glutamine and glutamate.
*Baseline: baseline model including only demographic covariates; All-Met + EN: all metabolites plus elastic net model including baseline + metabolites selected using elastic net regression with all metabolites; AMD/Control: AMD/Control model including baseline + metabolites identified in the logistic regression; Stage + 2Eye: stage + 2eye model including baseline + metabolites identified in the permutation-based cumulative logistic regression.
Pathway Analysis
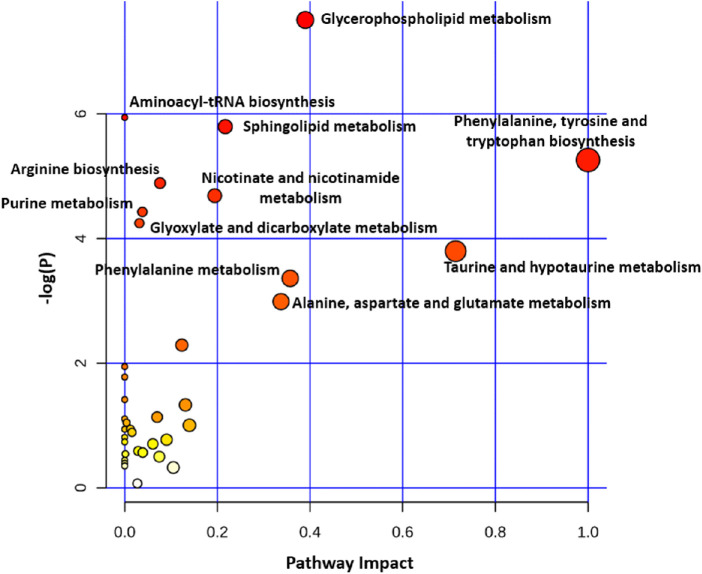
We extracted 108 potential candidate biomarkers from the included studies and performed the pathway enrichment analysis. As shown in Figure 2, phenylalanine, tyrosine, and tryptophan biosynthesis pathway had the largest Impact. Aminoacyl-tRNA biosynthesis was the most prominent pathway derived from the selected biomarkers. Other pathways were also enriched (P < 0.05), including alanine, aspartate, and glutamate metabolism; glyoxylate and dicarboxylate metabolism; arginine biosynthesis; glycerophospholipid metabolism; citrate cycle (tricarboxylic acid cycle [TCA] cycle); sphingolipid metabolism; lysine degradation; nicotinate and nicotinamide metabolism; and tyrosine metabolism. Detailed information regarding the enriched pathways are shown in Table 4.
Figure 2.
Results of the pathway analysis of metabolic biomarkers of AMD (circle color indicates significance level in the enrichment analysis, and circle size reflects pathway impact value from the topology analysis).
Table 4.
Results of the Pathway Analysis of Metabolic Biomarkers
| Pathway Name | Match Status | P Value | FDR | Impact |
|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 11/48 | 7.59E-06 | 6.37E-04 | 0.00000 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 3/4 | 4.04E-04 | 0.01698 | 1.00000 |
| Alanine, aspartate and glutamate metabolism | 6/28 | 0.00157 | 0.04406 | 0.33974 |
| Glyoxylate and dicarboxylate metabolism | 6/32 | 0.00325 | 0.05595 | 0.05556 |
| Arginine biosynthesis | 4/14 | 0.00333 | 0.05595 | 0.07614 |
| Glycerophospholipid metabolism | 6/36 | 0.00600 | 0.08396 | 0.38978 |
| Citrate cycle (TCA cycle) | 4/20 | 0.01296 | 0.15552 | 0.21656 |
| Sphingolipid metabolism | 4/21 | 0.01544 | 0.16212 | 0.21704 |
| Lysine degradation | 4/25 | 0.02825 | 0.26366 | 0.00469 |
| Nicotinate and nicotinamide metabolism | 3/15 | 0.03142 | 0.26391 | 0.19430 |
| Tyrosine metabolism | 5/42 | 0.04629 | 0.33754 | 0.24711 |
FDR, false discovery rate.
Discussion
In this review, thirteen metabolomics studies on AMD were comprehensively reviewed and analyzed to identify valuable metabolic biomarkers and pathways involved in AMD. Among these studies, four articles included are from the same authors, and some of them analyze the same (or similar) cohorts. However, these studies were performed on different technology platforms so that different metabolite ranges were detected. In addition, some used different biological samples (plasma and urine), so we assume that these studies provide complementary information. Finally, nine metabolites, repeatedly recognized twice or more, were regarded as potential biomarkers for AMD, including phenylalanine, adenosine, hypoxanthine, tyrosine, creatine, citrate, carnitine, proline and maltose. The pathway analysis revealed a series of metabolic disorders related to AMD.
The Potential Advantages of Metabolomics in AMD
Eyes have blood-water and blood-retina barriers, which give the eye unique metabolic environment. Macromolecules cannot be detected through the barriers, but some small metabolites can be.36 This makes it possible to use metabolomics to study metabolites in the blood of AMD patients to reveal the mechanism of the disease. As opposed to genomics, metabolic methods for eye diseases may be applicable to patients with any mutation as long as the same pathogenic pathway is involved.
Amino Acid Metabolites and Metabolism Pathways
Phenylalanine, tyrosine, and tryptophan biosynthesis had the highest impact in the pathway analysis. In the ketogenic pathway, phenylalanine and tyrosine metabolize to catecholamines and melanin. Catecholamine, such as dopamine, is an important neurotransmitter that could influence cell-to-cell coupling in the retina.37 Functionally, oxidative degradation of melanin was proposed to be associated with promoting reactive oxygen species generation and formation of metabolic byproducts, such as melanolipofuscin, which was regarded as a possible pathogenesis of AMD.38 In the glycogenic pathway, they are transformed to fumaric acid and acetoacetic acid under the action of enzymes, in which way they enter cellular energy metabolism. In addition, tyrosine may undergo phosphorylation, sulfation, or nitration and so influence protein function.39–41 However, whether the increase or decrease in phenylalanine and tyrosine is the risk factor for AMD is still unclear. The eyes, especially the maculae, are susceptible to oxidative stress because of their high metabolic activity and the high content of PUFAs. Proline is another amino acid that has been reported to regulate the development of AMD. Oxidative stress mainly affects RPE cells in AMD.42 It was reported that proline was the main nutrient for RPE cells43 and could mediate metabolic communication between RPE cells and the retina.44 Proline serves as an alternative source of energy during stress or hypoxia and an antioxidant for maintaining redox balance by controlling mitochondrial function.43
Significant “amino-acyl-tRNA biosynthesis” pathways dysregulation in AMD was observed in our study, but it showed no impact. Changes in this pathway are not AMD specific, which involves disturbances between extensive cellular translation events and cellular energy requirements.45 This may be a bias caused by the change of the concentration amino acid, which is often identified by MetaboAnalyst algorithm.
Energy Metabolism
Decreased citrate level in AMD patients has been detected by Acar et al.34 and Laíns et al.26, which reflected an enhancement in energy requirement in this disease. Citrate is a central integral of the TCA, participating in energy metabolism. Previous research on mice showed that replenishing TCA cycle metabolites via oral supplementation was useful for maintaining retinal function and protecting nerves on the photoreceptor cells and inner retinal network.46 The depletion of maltose identified in two studies25,28 could be another indication of increasing energy requirement. Creatine, a substance synthesized by three amino acids (arginine, glycine and methionine) is also the important compound for energy storage and utilization. A recent study by Kanow et al.47 emphasized the importance of maintaining a gradient of aerobic glycolysis activity between the retina and RPE. When the glycolysis activity in RPE is abnormally high, the glucose available for consumption in the retina will decrease, which will also cause the death of photoreceptors.48 With the growth of human age, the glucose consumed by the retinal photoreceptor is becoming less and less, which leads to dysfunction of mitochondria and DNA damage of RPE, leading to AMD.47
Purine Nucleotide Metabolites and Metabolism Pathways
The metabolism of adenosine and hypoxanthine mainly affect purine nucleotide cycle. Purines not only participate in intracellular energy metabolism, but also activate intercellular communication through receptors, leading to apoptosis of photoreceptors and RPE cells, all of which are the pathogenesis of AMD.49,50 Prior work has proposed that purine metabolism might act as alternative pathway to overcome inadequate glucose supply and energy crisis in neurodegeneration.51 Laíns et al.25 and Liu et al.28 observed the opposite alteration of hypoxanthine level between AMD patients and the controls, which might be because of the different biofluids they analyzed. Uric acid is a terminal metabolite of purine metabolism. Increasing evidences have showed the positive effect of uric acid on antioxidant stress.52 Increased levels of plasma adenosine have been observed in patients with AMD relative to controls.30,25 Adenosine is the raw material for adenosine triphosphate (ATP) synthesis, so its accumulation may suggest energy metabolism dysfunction in the pathology of AMD. ATP could induce photoreceptor death and retinal remodeling,53 and blockade of microglial adenosine A2A receptor could impact inflammatory mechanisms and prevent photoreceptor loss.54 Adenosine was also reported to be a key regulator of sustained inflammasome activation.55 Persistent inflammation has been proven to underlie many diseases including AMD.42 Overall, purine nucleotide metabolism has been shown to be crucial for the development and progression of AMD and thus possesses potential as a target of interventions for AMD.
Lipids Metabolites and Metabolism Pathways
Abnormal lipid metabolism is one of the main pathways of AMD.56 Carnitine and its metabolites are mainly involved in fatty acid metabolism. Some people believe that plasma carnitine level is an indicator of energy metabolism in cellular diet.57 Decreased carnitine level will lead to energy metabolism disorder.58,59 However, Mitchell et al.29 found the increasing of multiple long-chain acyl carnitine that are part of the carnitine shuttle pathway in NVAMD patients. The different conclusions on the association between carnitine alteration and metabolic dysregulation could be explained by carnitine's diphasic function of β-oxidation and anti-oxidation.35,60 Because of the limited number of studies, other metabolites related to fatty acid metabolism have not been repeatedly verified. Therefore more researches should be carried out to describe the characteristic fatty acid metabolism profile of AMD.
Limitations and Future Directions
Our study lays a foundation for the discovery of AMD biomarkers and providing new ideas for the research of AMD mechanism. However, the summary of current AMD studies shows few reproductively verified conclusions, limiting the extrapolation of the results. First, the existing research sample size is small, the subjects come from different ethnic groups, and the standardized operating procedures are still unavailable to metabolomics of AMD.61,62 Second, the eye is a relatively independent organ of metabolism, and eye tissue is more closely related to ocular lesions. However, because of its fine ocular structure and low tissue availability, the eye tissue is mainly derived from surgical resection or postmortem sampling, which limits its application in metabolomics research.61,63
The approach based on pathway analysis is to combine a series of metabolic feature changes shown by statistical analysis in metabolomics studies with prior biological knowledge to simplify the interpretability of data analysis.24 In a disease state, the original metabolic state in the body will be changed, cells and biological fluids that provide metabolic support for cells will enter a new metabolic balance state, and abnormal disturbance effects will be reflected in all aspects of the body.64 Using metabolic pathways to study changes in metabolic states of diseases is to study diseases from a relatively holistic perspective. Therefore, in this study, all the different metabolites in different tissues were included in the pathway analysis to obtain as many descriptions of the metabolic state of the disease as possible under the condition of limited research. This has been done in many other reviews on other disorders such as the systematic review of pancreatic cancer,65 ischemic stroke,66 and diabetes.67 Still, we acknowledge that the study has a bias because of the uneven distribution of metabolites in tissues (the systemic and local distribution of metabolites remains unclear), but this opens the possibility for a comprehensive description of disease metabolic changes. In future studies, more attention can be paid to metabolic differences of different biological tissues of the same disease, as well as joint studies of multiple omics, so as to gradually draw a complete blueprint for the disease mechanism of AMD.
Conclusions
In summary, this systematic review showed that there were some metabolic differences between AMD patients and controls. Phenylalanine, adenosine, hypoxanthine, tyrosine, creatine, citrate, carnitine, proline and maltose were repeatedly reported as AMD-related metabolites that affect nucleotide metabolism. Metabolic pathways, such as phenylalanine, tyrosine and tryptophan biosynthesis, and alanine, aspartate, and glutamate metabolism, are closely related to the development of AMD. In the future, these potential metabolite markers might be combined with clinical parameters to construct AMD risk prediction models on the basis of changes in metabolic pathways.
Supplementary Material
Acknowledgments
Supported by the National Natural Science Foundation of China (no. 81973061) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure: X.-W. Hou, None; Y. Wang, None; C.-W. Pan, None
References
- 1. Solomon SD, Lindsley K, Vedula SS, Krzystolik MG, Hawkins BS. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2019; 8(8): Cd005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajan SI, Sarma PS, Mishra US. Demography of Indian aging, 2001-2051. J Aging Soc Policy. 2003; 15(2-3): 11–30. [DOI] [PubMed] [Google Scholar]
- 3. Brown CN, Green BD, Thompson RB, den Hollander AI, Lengyel I. Metabolomics and age-related macular degeneration. Metabolites. 2018; 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li M, Zhang X, Liao N, et al.. Analysis of the serum lipid profile in polypoidal choroidal vasculopathy. Sci Rep. 2016; 6: 38342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kersten E, Paun CC, Schellevis RL, et al.. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv Ophthalmol. 2018; 63(1): 9–39. [DOI] [PubMed] [Google Scholar]
- 6. Fritsche LG, Igl W, Bailey JN. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016; 48(2): 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayordomo-Febrer A, López-Murcia M, Morales-Tatay JM, Monleón-Salvado D, Pinazo-Durán MD. Metabolomics of the aqueous humor in the rat glaucoma model induced by a series of intracamerular sodium hyaluronate injection. Exp Eye Res. 2015; 131: 84–92. [DOI] [PubMed] [Google Scholar]
- 8. Eandi CM, Alovisi C, De Sanctis U, Grignolo FM. Treatment for neovascular age related macular degeneration: the state of the art. Eur J Pharmacol. 2016; 787: 78–83. [DOI] [PubMed] [Google Scholar]
- 9. Sahel JA, Bennett J.. Depicting brighter possibilities for treating blindness. Sci Transl Med. 2019; 11(494): eaax2324. [DOI] [PubMed] [Google Scholar]
- 10. Kersten E, Dammeier S, Ajana S, et al.. Metabolomics in serum of patients with non-advanced age-related macular degeneration reveals aberrations in the glutamine pathway. PloS One. 2019; 14: e0218457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bino RJ, Hall RD, Fiehn O, et al.. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004; 9(9): 418–425. [DOI] [PubMed] [Google Scholar]
- 12. Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016; 15(7): 473–484. [DOI] [PubMed] [Google Scholar]
- 13. Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012; 126: 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao Y, Teo YCK, Beuerman RW, Wong TY, Cheung CMG. A serum metabolomics study of patients with nAMD in response to anti-VEGF therapy. Sci Rep. 2020; 10(1): 1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Country MW. Retinal metabolism: a comparative look at energetics in the retina. Brain Res. 2017; 1672: 50–57. [DOI] [PubMed] [Google Scholar]
- 16. Wang J, Westenskow PD, Fang M, Friedlander M, Siuzdak G. Quantitative metabolomics of photoreceptor degeneration and the effects of stem cell-derived retinal pigment epithelium transplantation. Philos Trans A Math Phys Eng Sci. 2016; 374(2079): 20150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rowan S, Jiang S, Korem T, et al.. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc Natl Acad Sci USA. 2017; 114(22): E4472–E4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Merle BM, Benlian P, Puche N, Bassols A, Delcourt C, Souied EH. Circulating omega-3 Fatty acids and neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2014; 55(3): 2010–2019. [DOI] [PubMed] [Google Scholar]
- 19. Shen JJ, Matern D, Millington DS, et al.. Acylcarnitines in fibroblasts of patients with long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency and other fatty acid oxidation disorders. J Inherit Metab Dis. 2000; 23(1): 27–44. [DOI] [PubMed] [Google Scholar]
- 20. Thompson RB, Reffatto V, Bundy JG, et al.. Identification of hydroxyapatite spherules provides new insight into subretinal pigment epithelial deposit formation in the aging eye. Proc Natl Acad Sci USA. 2015; 112(5): 1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartlett H, Eperjesi F. Age-related macular degeneration and nutritional supplementation: a review of randomised controlled trials. Ophthalmic Physiol Opt. 2003; 23(5): 383–399. [DOI] [PubMed] [Google Scholar]
- 22. Stroup DF, Berlin JA, Morton SC, et al.. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000; 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 23. Lumbreras B, Porta M, Márquez S, Pollán M, Parker LA, Hernández-Aguado I. QUADOMICS: an adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the evaluation of the methodological quality of studies on the diagnostic accuracy of '-omics'-based technologies. Clin Biochem. 2008; 41(16-17): 1316–1325. [DOI] [PubMed] [Google Scholar]
- 24. Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016; 55: 14.10.11–14.10.91. [DOI] [PubMed] [Google Scholar]
- 25. Laíns I, Chung W, Kelly RS, et al.. Human plasma metabolomics in age-related macular degeneration: meta-analysis of two cohorts. Metabolites. 2019; 9: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laíns I, Duarte D, Barros AS, et al.. Urine nuclear magnetic resonance (NMR) metabolomics in age-related macular degeneration. J Proteome Res. 2019; 18(3): 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laíns I, Duarte D, Barros AS, et al.. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. PloS One. 2017; 12: e0177749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu K, Fang J, Jin J, et al.. Serum metabolomics reveals personalized metabolic patterns for macular neovascular disease patient stratification. J Proteome Res. 2020; 19: 699–707. [DOI] [PubMed] [Google Scholar]
- 29. Mitchell SL, Uppal K, Williamson SM, et al.. The carnitine shuttle pathway is altered in patients with neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2018; 59(12): 4978–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laíns I, Kelly RS, Miller JB, et al.. Human plasma metabolomics study across all stages of age-related macular degeneration identifies potential lipid biomarkers. Ophthalmology. 2018; 125: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osborn MP, Park Y, Parks MB, et al.. Metabolome-wide association study of neovascular age-related macular degeneration. PloS One. 2013; 8: e72737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo D, Deng T, Yuan W, Deng H, Jin M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017; 17: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chao de la Barca JM, Rondet-Courbis B, Ferré M, et al.. A plasma metabolomic profiling of exudative age-related macular degeneration showing carnosine and mitochondrial deficiencies. J Clin Med. 2020; 9(3): 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Acar İE, Lores-Motta L, Colijn JM, et al.. Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration: the EYE-RISK Consortium. Ophthalmology. 2020; 127(12): 1693–1709. [DOI] [PubMed] [Google Scholar]
- 35. Han G, Wei P, He M, Teng H, Chu Y. Metabolomic profiling of the aqueous humor in patients with wet age-related macular degeneration using UHPLC-MS/MS. J Proteome Res. 2020; 19: 2358–2366. [DOI] [PubMed] [Google Scholar]
- 36. Greenwood J, Pryce G, Devine L, et al.. SV40 large T immortalised cell lines of the rat blood-brain and blood-retinal barriers retain their phenotypic and immunological characteristics. J Neuroimmunol. 1996; 71: 51–63. [DOI] [PubMed] [Google Scholar]
- 37. Witkovsky P. Dopamine and retinal function. Doc Ophthalmol. 2004; 108: 17–40. [DOI] [PubMed] [Google Scholar]
- 38. Yacout SM, McIlwain KL, Mirza SP, Gaillard ER. Characterization of retinal pigment epithelial melanin and degraded synthetic melanin using mass spectrometry and in vitro biochemical diagnostics. Photochem Photobiol. 2019; 95(1): 183–191. [DOI] [PubMed] [Google Scholar]
- 39. Murdaugh LS, Wang Z, Del Priore LV, Dillon J, Gaillard ER. Age-related accumulation of 3-nitrotyrosine and nitro-A2E in human Bruch's membrane. Exp Eye Res. 2010; 90: 564–571. [DOI] [PubMed] [Google Scholar]
- 40. Nag TC, Kathpalia P, Gorla S, Wadhwa S. Localization of nitro-tyrosine immunoreactivity in human retina. Ann Anat. 2019; 223: 8–18. [DOI] [PubMed] [Google Scholar]
- 41. Thao MT, Karumanchi DK, Yacout SM, Gaillard ER. Nitrite ion modifies tyrosine and lysine residues of extracellular matrix proteins. Nitric Oxide. 2018; 79: 51–56. [DOI] [PubMed] [Google Scholar]
- 42. Datta S, Cano M, Ebrahimi K, Wang L, Handa JT. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017; 60: 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010; 15: 89–97. [DOI] [PubMed] [Google Scholar]
- 44. Yam M, Engel AL, Wang Y, et al.. Proline mediates metabolic communication between retinal pigment epithelial cells and the retina. J Biol Chem. 2019; 294: 10278–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sissler M, González-Serrano LE, Westhof E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol Med. 2017; 23(8): 693–708. [DOI] [PubMed] [Google Scholar]
- 46. Wert KJ, Velez G, Kanchustambham VL, et al.. Metabolite therapy guided by liquid biopsy proteomics delays retinal neurodegeneration. EBioMedicine. 2020; 52: 102636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanow MA, Giarmarco MM, Jankowski CS, et al.. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017; 6: e28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du J, Rountree A, Cleghorn WM, et al.. Phototransduction influences metabolic flux and nucleotide metabolism in mouse retina. J Biol Chem. 2016; 291: 4698–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu W, Meng YF, Xing Q, Tao JJ, Lu J, Wu Y. Identification of lncRNAs involved in biological regulation in early age-related macular degeneration. Int J Nanomedicine. 2017; 12: 7589–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reichenbach A, Bringmann A.. Purinergic signaling in retinal degeneration and regeneration. Neuropharmacology. 2016; 104: 194–211. [DOI] [PubMed] [Google Scholar]
- 51. Kori M, Aydin B, Unal S, Arga KY, Kazan D. Metabolic biomarkers and neurodegeneration: a pathway enrichment analysis of Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis. OMICS. 2016; 20: 645–661. [DOI] [PubMed] [Google Scholar]
- 52. Glantzounis GK, Tsimoyiannis EC, Kappas AM, Galaris DA. Uric acid and oxidative stress. Curr Pharm Des. 2005; 11: 4145–4151. [DOI] [PubMed] [Google Scholar]
- 53. Vessey KA, Greferath U, Aplin FP, et al.. Adenosine triphosphate-induced photoreceptor death and retinal remodeling in rats. J Comp Neurol. 2014; 522: 2928–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Madeira MH, Rashid K, Ambrosio AF, Santiago AR, Langmann T. Blockade of microglial adenosine A2A receptor impacts inflammatory mechanisms, reduces ARPE-19 cell dysfunction and prevents photoreceptor loss in vitro. Sci Rep. 2018; 8: 2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ouyang X, Ghani A, Malik A, et al.. Adenosine is required for sustained inflammasome activation via the A(2)A receptor and the HIF-1alpha pathway. Nat Commun. 2013; 4: 2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brantley MA Jr, Osborn MP, Sanders BJ, et al.. Plasma biomarkers of oxidative stress and genetic variants in age-related macular degeneration. Am J Ophthalmol. 2012; 153: 460–467.e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kendler BS. Carnitine: an overview of its role in preventive medicine. Prev Med. 1986; 15: 373–390. [DOI] [PubMed] [Google Scholar]
- 58. Winter SC, Simon M, Zorn EM, et al.. Relative carnitine insufficiency in children with type I diabetes mellitus. Am J Dis Child. 1989; 143: 1337–1339. [DOI] [PubMed] [Google Scholar]
- 59. Stumpf DA, Parker WD Jr., Angelini C. Carnitine deficiency, organic acidemias, and Reye's syndrome. Neurology. 1985; 35: 1041–1045. [DOI] [PubMed] [Google Scholar]
- 60. Ohno Y, Otsuka Y, Nohara M, et al.. Characterization of an L-Carnitine Transport System in Murine Photoreceptor Cell Line. Biol Pharm Bull. 2017; 40: 2110–2116. [DOI] [PubMed] [Google Scholar]
- 61. Yanshole VV, Yanshole LV, Snytnikova OA, Tsentalovich YP. Quantitative metabolomic analysis of changes in the lens and aqueous humor under development of age-related nuclear cataract. Metabolomics. 2019; 15: 29. [DOI] [PubMed] [Google Scholar]
- 62. Heazell AE, Brown M, Worton SA, Dunn WB. Review: The effects of oxygen on normal and pre-eclamptic placental tissue–insights from metabolomics. Placenta. 2011; 32(Suppl 2): S119–S124. [DOI] [PubMed] [Google Scholar]
- 63. Ji Y, Rao J, Rong X, Lou S, Zheng Z, Lu Y. Metabolic characterization of human aqueous humor in relation to high myopia. Exp Eye Res. 2017; 159: 147–155. [DOI] [PubMed] [Google Scholar]
- 64. Lindon JC, Holmes E, Nicholson JK. So what's the deal with metabonomics? Anal Chem. 2003; 75: 384A–391A. [DOI] [PubMed] [Google Scholar]
- 65. Long NP, Yoon SJ, Anh NH, et al.. A systematic review on metabolomics-based diagnostic biomarker discovery and validation in pancreatic cancer. Metabolomics. 2018; 14: 109. [DOI] [PubMed] [Google Scholar]
- 66. Ke C, Pan CW, Zhang Y, Zhu X, Zhang Y. Metabolomics facilitates the discovery of metabolic biomarkers and pathways for ischemic stroke: a systematic review. Metabolomics. 2019; 15: 152. [DOI] [PubMed] [Google Scholar]
- 67. Sun Y, Gao HY, Fan ZY, He Y, Yan YX. Metabolomics Signatures in Type 2 Diabetes: A Systematic Review and Integrative Analysis. J Clin Endocrinol Metab. 2020; 105(4): dgz240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.