Abstract
Objective: To describe the methods used in a rapid review of the literature and to present the main epidemiological parameters that describe the transmission of SARS-Cov-2 and the illness caused by this virus, coronavirus disease 2019 (COVID-19).
Methods: This is a methodological protocol that enabled a rapid review of COVID-19 epidemiological parameters.
Findings: The protocol consisted of the following steps: definition of scope; eligibility criteria; information sources; search strategies; selection of studies; and data extraction. Four reviewers and three supervisors conducted this review in 40 days. Of the 1,266 studies found, 65 were included, mostly observational and descriptive in content, indicating relative homogeneity as to the quality of the evidence. The variation in the basic reproduction number, between 0.48 and 14.8; and the median of the hospitalization period, between 7.5 and 20.5 days stand out as key findings.
Conclusion: We identified and synthesized 10 epidemiological parameters that may support predictive models and other rapid reviews to inform modeling of this and other future public health emergencies.
Keywords: coronavirus infections, review, parameters, methods, models, statistical
Introduction
Public Health is confronted with the challenge of protecting poulations from emerging and reemerging diseases. Among the viruses capable of causing pandemics, special prominence is given to the family Coronaviridae (1–3). These viruses are responsible for three recent major epidemics: in 2009, the Severe Acute Respiratory Syndrome (SARS), caused by the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV); in 2012, the Middle East Respiratory Syndrome, caused by the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) (4); and, in 2019, the Corona Virus Disease−19 (COVID-19), caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (5). However, SARS-CoV-2 has peculiar clinical and epidemiological characteristics when compared with SARS-CoV, MERS-CoV, or others of the same family. These characteristics are reflected in the exponentially increasing numbers of COVID-19-related deaths (6).
The current epidemic goes back to December 31, 2019, when a pneumonia outbreak was reported in Wuhan, China, with 27 cases that were later identified as COVID-19 cases (7). In the following months, the epidemic evolved from a local problem to a pandemic with catastrophic consequences. As of August 13, ~20,5 million cases and 744,500 deaths had been reported to the World Health Organization (WHO), in all age ranges and nearly all continents—except Antarctica. The Americas are currently regarded as the epicenter of the pandemic, where 53.6% of the total recorded cases have been reported−54.7% of the cases recorded within the last 24 h in the world. The United States and Brazil are particularly affected. These countries have 5,094,500 cases (163,340 deaths) and 3,109,630 cases (103,026 deaths), respectively (8).
Understanding the parameters that influence the course of an epidemic is key for health-related decision-making and allows for planning of strategies to mitigate and control diseases, as well as provision of care to those infected and sick. The high transmissibility and virulence of SARS-CoV-2, lead to a significant rate of severe and critical cases requiring specialized care and intensive care beds, creates the need for predictive models capable of estimating health care demands and support decision-making (9–11).
Mathematical models are simplifications of complex processes involved in disease dynamics, which can lead to different results based on the method, assumptions and parameters adopted (12). To minimize uncertainties, parameters feeding the model must be valid, accurate, generalizable, and reliable, as well as adaptable in population-based terms. In an emergent situation these models may contain a series of uncertainties, due to the incipient availability of epidemiological characteristics (10, 11). This requires constant review of parameters as new information arises, as well as an ongoing literature review.
The COVID-19 emergency has prompted researchers to work toward describing different aspects of disease transmission and evolution. As a result, a significant number of scientific publications are being released daily, and the MEDLINE database alone already had 16,000 publications (keyword “COVID-19”) as of May 26, 2020—when this study was performed. Information from these publications can help decision-makers develop policies throughout the course of the emergency. However, due to the large number of studies available, identifying the relevant evidence in due time presents a great challenge and requires that the methods used in traditional literature reviews be adapted.
To support evidence-based decision-making using predictive models for the COVID-19 public health emergency, while the epidemic was establishing in Brazil, a rapid literature review method was proposed with the view to identify and describe clinical and epidemiological parameters relative to infection by SARS-CoV-2 and the illness caused by this virus, coronavirus disease 2019 (COVID-19). This article, therefore, aims to describe the methods employed in this rapid literature review and present the main epidemiological parameters describing SARS-CoV-2 transmission and the COVID-19 disease.
Materials and Methods
A methodological proposal for rapid review of epidemiological parameters and their application in the context of the current SARS-CoV-2 pandemic emergency.
Proposed Methodology
A rapid literature review, with the aim to identify clinical and epidemiological parameters to support mathematical models of COVID-19 transmission and disease. The proposed rapid review method developed by the authors includes the following steps: research scope definition; eligibility criteria; information sources; database search strategies; study selection; and data extraction. For method construction, we met with the group of modelers to identify the required parameters. The parameters defined for the search and their descriptions are provided in Table 1.
Table 1.
Description of identified epidemiological parameters of COVID-19.
| Parameters | Description |
|---|---|
| Basic reproduction number (R0) | The mean number of new infections arising from one infected person in a totally susceptible population (13). |
| Serial interval | Time between onset of symptoms in a primary case (infector) and onset of symptoms in a secondary case (infectee) (14). |
| Incubation period | Time between infection and onset of disease (15). |
| Transmissibility period | Time during which a person infected with SARS-CoV-2 transmits the virus to other people. |
| Proportion of detected cases | Proportion of cases identified as infected with SARS-CoV-2 among all cases tested. |
| Proportion of critical cases among hospitalized patients | Proportion of critical cases of COVID-19 among all hospitalized patients. |
| Proportion of deaths among critical cases | Proportion of deaths from COVID-19 among all critical cases of the disease. |
| Mean or median length of hospital stay | Time in days (mean or median) of hospital stay among COVID-19 cases. |
| Mean or median time between admission to hospital and onset of ARDSa | Time in days (mean or median) of hospital stay among COVID-19 cases before onset of ARDSa. |
| Length of hospital stay in wards before admission at ICUb | Time in days (mean or median) of hospital stay in wards among COVID-19 cases who required ICUb. |
Acute respiratory distress syndrome.
Intensive care unit.
Results
Methodological Protocol
During the preparation stage, the group developed a methodological protocol to guide construction of the methods employed in the rapid literature review. The protocol was composed of six stages, the respective descriptions of which are provided in Table 2.
Table 2.
Methodological proposal for quick literature review: identification of epidemiological parameters.
| Steps | Description |
|---|---|
| Search scope | It should be structured as follows: definition of the population to be studied; choice of epidemiological parameters; organizing groups of parameters according to similarity (e.g., types of studies that generate them). |
| Eligibility criteria | For a quick and reliable selection, only include studies published as from the date of the first outbreak of the disease in the world; presenting at least one of the parameters assessed in the abstract; original investigations, literature reviews; published in English or in other languages of the group domain; including studies published in other languages, but with the abstract in the languages of the domain that allow clear identification of any parameters of interest. It is suggested for reviewers to exclude: studies from preprint databases that analyzed primary data and have not been submitted to ethical evaluation; opinion articles; epidemiological bulletins with overlapping data of the same place, and studies that do not allow a reliable translation. |
| Sources of information | Literature search should be divided into two phases: the first should search at least two international databases, and the second should track the lists of references of studies identified in the first stage. |
| Search in the databases | The search syntax must represent the problem to be investigated, its primary endpoints and the date that best represents the beginning of the first outbreak in the world. For example: (name of the disease OR name of virus) AND (endpoint 1 OR endpoint 2) AND (start date AND final date). |
| Study selection | Study selection should comprise the following stages: selection of studies for complete assessing, from evaluation of titles and abstracts as per eligibility criteria; reading of full texts and new evaluation considering eligibility. Non-matching stages, but with the support of a more experienced researcher to clarify doubts and organize the process. |
| Data extraction | Data extraction should be guided by means of a structured tool, which allows the objective identification of parameters and a quick assessment of the quality of studies, in terms of validation and accuracy of data. Non-matching stage but overseen by a researcher with a trained in epidemiology. |
Operationalizing the Rapid Literature Review
The population of interest was composed of people living in high-risk areas of SARS-CoV-2 infection. The epidemiological parameters were divided into two groups, for better organization of the syntax and database search. The first group, referred to as Group 1, included the following parameters: basic reproduction number (R0); serial interval; incubation period; transmissibility period. The second, referred to as Group 2, included the following parameters: rate of detected cases; rate of critical cases among all hospitalized patients; rate of deaths among critical cases; mean or median length of hospital stay; mean or median time between hospital admission and ARDS (Acute Respiratory Distress Syndrome) onset; or mean or median length of hospital stay before ICU (Intensive Care Unit) admission.
To identify Group 1 and Group 2 parameters, we selected studies indexed in databases: Medical Literature Analysis and Retrieval System Online (MEDLINE) and Excerpta Medica dataBASE (EMBASE). For each group of parameters, we organized search syntaxes on MEDLINE, via PubMed and on EMBASE, based, respectively, on MeSH (Medical Subject Headings) and Emtree (Embase Subject Headings) terms. Searches were performed in two stages, one on March 27, 2020 and the second on April 13, 2020. Additional studies were obtained from mannualy searches in the references of the selected articles and reviews.
We organized four search syntaxes based on the group of parameters and the database. Table 3 shows the search syntaxes used to identify studies on MEDLINE via PubMED, which were adapted for EMBASE. Duplicates were removed with the help of reference management software programs Mendeley Desktop version 1.19.4 and Covidence.
Table 3.
Search syntax: MEDLINE.
| MEDLINE/GROUP 1: ((“coronavirus”[MeSH Terms] OR “severe acute respiratory syndrome coronavirus 2”[Supplementary Concept] OR “severe acute respiratory syndrome coronavirus 2”[All Fields]) OR “COVID-19”[All Fields] OR “COVID-19”[Supplementary Concept] OR “Novel Coronavirus” [All Fields]) AND ((((“basic reproduction number” [MeSH Terms] OR (“basic” [All Fields] AND “reproduction”[All Fields] AND “number”[All Fields]) OR “basic reproduction number”[All Fields]) OR R0[All Fields] OR “basic reproductive number”[All Fields] OR (basic[All Fields] AND (“reproduction”[MeSH Terms] OR “reproduction”[All Fields] OR “reproductive”[All Fields]) AND number[All Fields])) OR (“infectious disease incubation period”[MeSH Terms] OR (“infectious”[All Fields] AND “disease”[All Fields] AND “incubation”[All Fields] AND “period”[All Fields]) OR “infectious disease incubation period”[All Fields])) OR ((“disease transmission, infectious”[MeSH Terms] OR (“disease”[All Fields] AND “transmission”[All Fields] AND “infectious”[All Fields]) OR “infectious disease transmission”[All Fields] OR (“disease”[All Fields] AND “transmission”[All Fields] AND “infectious”[All Fields]) OR “disease transmission, infectious”[All Fields]) OR (communicable[All Fields] AND period[All Fields]))) AND (“2020/01/01”[PDAT] : “2020/04/13”[PDAT]) |
| MEDLINE/GROUP 2: ((“coronavirus”[MeSH Terms] OR “severe acute respiratory syndrome coronavirus 2”[Supplementary Concept] OR “severe acute respiratory syndrome coronavirus 2”[All Fields]) OR “COVID-19”[All Fields] OR “COVID-19”[Supplementary Concept] OR “Novel Coronavirus”[All Fields]) AND ((((((((“Hospitalization”[Mesh] OR “Critical Care”[Mesh]) OR “Intensive Care Units”[Mesh]) OR “Critical Illness”[Mesh]) OR “Mortality”[Mesh]) OR “Hospital Mortality”[Mesh]) OR “Pneumonia, Ventilator-Associated”[Mesh]) OR “Disease Attributes”[Mesh] OR “hospitalized patient*”[All Fields] OR (epidemiological characteristic[All Fields] OR epidemiological characteristics[All Fields]) OR (clinical characteristic[All Fields] OR clinical characteristics[All Fields]) OR (clinical outcome[All Fields] OR clinical outcomes[All Fields] OR clinical outcomes,[All Fields]) OR “hospitalization rate”[All Fields] OR (undocumented[All Fields] AND (“infections”[MeSH Terms] OR “infections”[All Fields] OR “infection”[All Fields])) OR “documented infection”[All Fields]) OR (((((((“Epidemiologic Studies”[Mesh] OR “Clinical Studies as Topic”[Mesh]) OR “Epidemiologic Study Characteristics”[Mesh]) OR “Decision Support Techniques”[Mesh]) OR “Case Reports”[Publication Type]) OR “Observational Study”[Publication Type]) OR “Cohort Studies”[Mesh]) OR ((“IEEE Int Conf Automation Sci Eng (CASE)”[Journal] OR “CASE (Phila)”[Journal] OR “case”[All Fields]) AND (“SERIEs (Berl)”[Journal] OR “series”[All Fields])))) AND (“2020/01/01”[PDAT] : “2020/12/31”[PDAT]) AND (“2020/01/01”[PDAT] : “2020/04/13”[PDAT]) |
The eligibility criteria included studies published as of January 1, 2020. We included original research studies, epidemiological bulletins and literature reviews addressing any of the parameters of interest, published in English, Spanish or Portuguese. Studies in other languages were included only when any of the parameters of interest could be identified in the Abstract published in English, Spanish or Portuguese. The list of elegibility criteria is presented in Table 2.
For study selection, the titles and abstracts identified were classified as per the inclusion and exclusion criteria. Studies that met the inclusion criteria and none of the exclusion criteria were selected for full reading and reassessed for eligibility. Data were extracted based on three spreadsheets specifically developed for the parameters addressed in the review.
Data search, inclusion, reading, and extraction were not conducted in a paired fashion, and each study was reviewed by an investigator under supervision by a second, more experienced investigator with an epidemiology background. The supervisor supported every stage of the review, providing guidance and answering questions, and that data extraction was entirely verified by two supervisors.
Epidemiological Parameters
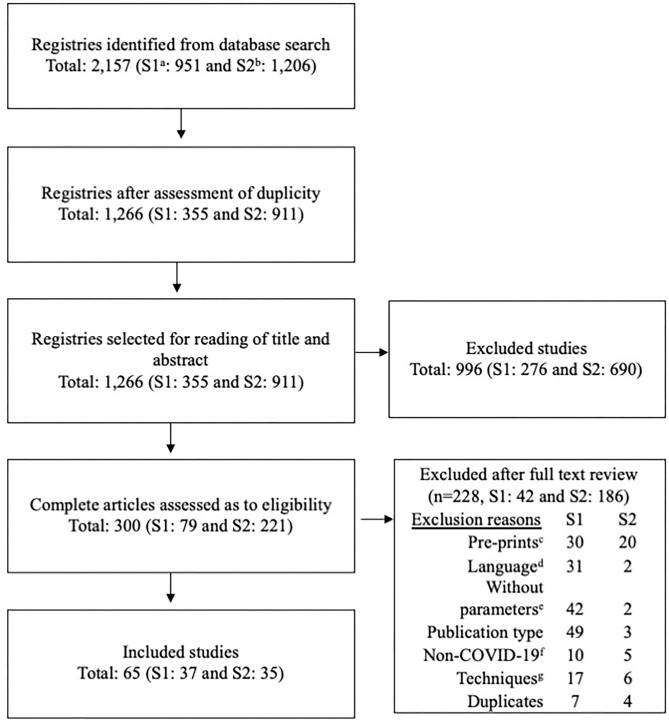
Figure 1 describes the flow of information at different stages of the review. At first, we found 951 studies using the strategies set up to identify parameters in Group 1 and 1,206 studies using the strategies to retrieve parameters in Group 2. After assessing for duplicates, we were left with 1,266 studies (Group 1: 355 and Group 2: 911), of which 65 were included (Group 1: 37 and Group 2: 35).
Figure 1.
Flowchart of the selection process of evidence of clinical and epidemiological parameters of COVID-19. a, First group of parameters (syntax group 1); b, Second group of parameters (syntax group 2); c, articles published as pre-prints; d, article in non-English, Spanish, or Portuguese and the parameter data was not included in the abstract; e, it was not possible to extract the parameters of interest; f, did not provide data on COVID-19; g, Laboratory studies or other techniques.
Epidemiological parameters were divided into 3 datasets according to the groups searched.
Table 4 shows search results by basic reproduction number (R0) and time-varying reproduction number (Rt)—when present—in the 19 studies identified (16–18, 20–35). Analyses relied mostly on data from China, followed by Japan and South Korea, and were performed between December 2019 and March 2020. The highest R0 identified was 14.8, estimated for the Diamond Princess cruise-ship during its quarantine in Japan (22), and the lowest was 0.48 - in South Korea (30). Decreases in SARS-CoV-2 reproduction numbers have been seen after restrictive measures were implemented.
Table 4.
Presentation of parameters: basic reproduction number (R0).
| Author, year [references] | Country | Date of study | Method: | R0a | Others |
|---|---|---|---|---|---|
| Chen et al., 2020 (16) | China | Between December 2019 and January 2020 | Reservoir-people transmission network modelc | 3,58. | – |
| Chen et al., 2020 (16) | Japan (Diamond Princess cruise ship) | February 2020 | Package “earlyR” | 2.28 (95%CI: 2.06–2.52) | – |
| Kuniya, 2020 (17) | Japan | Between January and February 2020 | SEIR Modeld | 2.6 (95%CI: 2.4–2.8) | – |
| Fang et al., 2020 (18) | China | Between January and February 2020 | SEIR Modeld | – | R according to date: Day 0: 2.4; day 10: 3.2; day 20: 2.98, etc. |
| Zhao et al., 2020 (19) | China | January 2020 | Statistical exponential growth modele | 2.24 (95%CI: 1.96—2.55) | – |
| Kucharski et al., 2020 (20) | China | Between January and February 2020 | Stochastic transmission model | – | Rtb(before and after restriction measures): 2.35 (95%CI: 1.15–4.77) to 1.05 (95%CI: 0.41–2.39) |
| Wang et al., 2020 (21) | China | Between December 2019 and February 2020 | SEIR Modeld | 3.1 | Rtb measured in distinct phases after restriction measures: 2.6; 1.9 and 0.5 |
| Rocklöv et al., 2020 (22) | Japan (Diamond Princess cruise ship) | January 2020 | SEIR Modeld | 14.8 | Rtb after restriction measures: 1.74 |
| Mizumoto and Chowell, 2020 (23) | Japan (Diamond Princess cruise ship) | Between January and February 2020 | Next generation matrix (NGM)f in a totally susceptible population | Maximum Rtb: 11.2 (95%CI: 7.5—16.2) and median Rt: 5.8 (95%CI: 0.6–11.0) | |
| Tang et al., 2020 (24) | China | January 2020 | SEIR Modeld | 6.47 (95%CI: 5.71—7.23) | – |
| Wu et al., 2020 (25) | China | Between December 2019 and January 2020 | SEIR Modeld | 2.68 (95%CI: 2.47–2.86) | – |
| Li et al., 2020 (26) | China | January 2020 | Statistical exponential growth modele | 2.2 (95%CI: 1.4–3.9). | – |
| Zhao et al., 2020 (27) | China | January 2020 | Exponential growth model by Poisson in a completely susceptible population | 2.56 (95%CI: 2.49–2.63) | – |
| Wang et al., 2020 (28) | China | Between January and February 2020 | Statistical exponential growth modele | 3.49 (95%CI: 3.42–3.58) | After containment measures Rtb: 2.95 (95%CI: 2.86–3.03) |
| Zhou et al., 2020 (29) | China | January 2020 | SEIR Modeld | 2.8–3.3 | – |
| Ki and Task Force for 2019-nCoV, 2020 (30) | South Korea | Between January and February 2020 | Not informed | 0.48 (95%CI: 0.25–0.84) | – |
| Du et al., 2020 (31) | China | Between January and February 2020 | Exponential growth model based on public data from Wuhan | 1.32 (95%CI: 1.16–1.48) | – |
| Anastassopoulou et al., 2020 (32) | China | Between January and February 2020 | SIDR modelg | 2–2.6 | Other R0a estimates based on linear regression varied between 3.2 (95%CI: 2.4–4) and 5.14 (95%CI: 4.25–6.03). |
| Song et al., 2020 (33)* | China | January 2020 | Weibull distribution methods, Gamma Lognormal, |
Weibull distribution methods: 3.74 (95%CI: 3.63–3.87); Gamma: 3.16 (95%CI: 2.90–3.43); Lognormal: 3.91 (95%CI: 3.71–4.11) |
– |
| Pan et al., 2020 (34) | China | Between December 2019 and March 2020 | Used a method proposed by Cori et al. (2013)h in R version 3.6.2. | – | Maximum Rtb — 03/01/2020: 3.94 (95%CI: 3.32—-4.63); Minimum Rtb: 08/03/2020: 0.10 (95%CI: 0.08—-0.13) |
R0, basic reproduction number.
Rt, effective basic reproduction number (variable in time).
Reservoir–People transmission network.
SEIR: Susceptible – Exposed – Infected – Recovered.
Statistical exponential growth model.
NGM: next–generation matrix.
SIDR: Susceptible – Infected – Recovered – Death.
Cori A et al. (2013). doi: 10.1093/aje/kwt133.
Article in Chinese, only the abstract in English was assessed.
Data extracted from studies on incubation and transmissibility periods and serial interval can be seen in Table 5. We identified 22 studies (14, 15, 26, 30, 31, 33, 36–51) of which 19 (14, 15, 26, 30, 33, 36–41, 43, 44, 46–51) addressed the incubation period, with means ranging from 3.6 days (48) to 6.7 days (39); one addressed the transmissibility period [Median: 9.5 days, interquartile range (IQR) 3.5–13.0] (42); and five (26, 30, 31, 36, 45) addressed the serial interval, with means ranging from 3.96 (31) to 7.5 days (26). The parameters presented in the chart were predominantly estimated based on descriptive observational studies from China, South Korea, and Singapore.
Table 5.
Presentation of serial interval parameters, incubation period, and transmissibility period.
| Author, year [references] | Country | Study date | Characteristics of the sample (sample, age, and sex) | Incubation period | Transmissibility period | Serial interval |
|---|---|---|---|---|---|---|
| Linton et al., 2020 (14) | China | January 2020 | N = 158 cases; predominance of age group 30–39 years; Female sex: 42% | Mean: 5.6. (95%CI: 5.0–6.3 days). |
– | – |
| Song et al., 2020 (33)* | China | January 2020 | No information | Mean: 5.01. (95%CI: 4.31–5.69 days). |
– | – |
| Pung et al., 2020 (36) | Singapore | February 2020 | N = 36 cases; 17 positive cases; Age [median (IQR)]: 40 (36–51) years; Female sex: 59% | Median: 4 days (IQR: 3–6 days). |
– | Range: 3–8 days |
| Chan et al., 2020 (37) | China | January 2020 | N = 6 cases; 5 adults and one child, Age (mean): 45 years; Female sex: 50% | Range: 3–6 days | – | – |
| Lauer et al., 2020 (38) | 50 places in and outside China | January and February 2020 | N = 181 cases; Age [median (IQR)]: 44.5 (34–55,5) years; Female sex: 38% | Median: 5.1 days (95%CI: 4.5–5.8 days). |
– | – |
| Fan et al., 2020 (39) | China | January and February 2020 | N = 54 cases; Age [mean (lower and upper limits)]: 38 (1, 67–94) years; Female sex: 33.3%; 19 cases subsidized the estimated incubation period | Mean: 6.7 days | – | – |
| Yang et al., 2020 (40)* | China | January and February 2020 | N = 325 cases | Median: 7 days; Mode: 4 days Range: 1–20 days |
– | – |
| Backer et al., 2020 (41) | China | January 2020 | N = 88 cases; Age (lower and upper limits): 2–72 years; Female sex: 35.2% | Mean: 6.4 days (95%CI: 5.6–7.7 days). |
– | – |
| Hu et al., 2020 (42) | China | January and February 2020 | N = 24 cases; Age [median (IQR)]: 32.5 (19.0–57.0) years; Female sex: 77.7% | – | Median: 9.5 days (IQR 3.5–13.0 days) |
|
| Li et al., 2020 (26) | China | January 2020 | N = 425 cases; Age [median (lower and upper limits)]: 59 (15–89) years; Female sex: 49%; 10 cases subsidized the estimated incubation period, and 6 pairs subsidized the serial interval calculation | Mean: 5.2 days (95%CI: 4.1–7.0 days) |
– | Mean (SD): 7.5(±3.4) days (95%CI: 5.3–19 days) |
| Wu et al., 2020 (43)* | China | January 2020 | N = 40 cases; 35% of the aged over 60 years | Median: 6 days | – | – |
| Zhang et al., 2020 (44)* | China | January 2020 | N = 17 cases; Age [median (lower and upper limits)]: 55 (19–79) years; Female sex: 23.5% | Median: 4 days Range: 0–12 days |
– | – |
| Ki and Task Force for 2019-nCoV, 2020 (30) | South Korea | January and February 2020 | N = 28 cases; Age [median (lower and upper limits)]: 42 (21–73) years; Female sex: 46.4% | Mean: 3.9 days Median: 3 days |
– | Mean: 6.6 days; Median: 4 days |
| Nishiura et al., 2020 (45) | China | February 2020 | Data from articles and reports: 28 pairs. | – | – | Median: 4.6 days (95%CI: 3.5–5.9 days) |
| Lian et al., 2020 (46) | China | Between January 17 and February 12, 2020 | N = 778 cases, divided into elderly group, with 136 cases and mean age of 68.28 years; and non-elderly adult group with 652 cases and mean age of 41.15 years. | Median: 5 days (IQR: 2–9 days) |
– | – |
| Sun et al., 2020 (15) | China | January 2020 | N = 33 cases | Median: 4.5 days (IQR: 0–5.5 days) Maximum: 9.5 days |
– | – |
| Jia et al., 2020 (47) | China | Between January and February 2020 | N = 44 cases; Age [median (lower and upper limits)]: 46 (1 and 90 years); Female sex: 65.9% | Mean: 6.28 days Range: 1–14 days |
– | – |
| Du et al., 2020 (31) | China | Between January and February 2020 | N = 468 cases | – | – | Mean: 3.96 (95%CI: 3.53–4.39 days) SD ± 4.75 days (95%CI: 4.46–5.07 days) |
| Guan et al., 2020 (48) | China | Between December 2019 and January 2020 | N = 1,590 hospitalized patients; Mean age: 48.9 (SD ± 16.3) years; Female sex: 42.7% | Mean: 3.6 days (SD ± 4.2 days) |
– | – |
| Jin et al., 2020 (49) | China | Between January and February 2020 | N = 651, divided into group with gastrointestinal symptoms (GI) (N = 21), Age [mean (SD)]: 46.14 ± 14.19 years; Female sex: 50%; and asymptomatic group GI (N = 577), Age [mean (SD)]: 45.09 ± 14.45 years; Female sex: 49.05% | With GI symptoms: median: 4 days (IQR: 3–7 days). With no GI symptoms: median: 5 days (IQR: 3–8 days) |
– | – |
| Sun et al., 2020 (50) | China | Between January and February 2020 | N = 8 children; Age (lower and upper limits): 2 months to 15 years; Female sex: 25% | Range: 5–10 days | – | – |
| Guan et al., 2020 (51) | China | Up to January 2020 | N = 1,099; Age (median): 47 (IQR 35–58) years; Female sex: 41.9% | Median: 4 days (IQR: 2–7 days) |
– | – |
N: total number of participants in the study.
GI: gastrointestinal symptoms.
95%CI: 95% confidence interval.
IQR: Interquartile range.
SD: Standard deviation.
Article in Chinese, only the abstract in English was assessed.
Table 6 shows results for Group 2 and the rate of detected cases; rate of critical cases among all hospitalized patients; rate of deaths among critical cases; mean or median length of hospital stay; mean or median time between hospital admission and ARDS onset; or mean or median length of hospital stay before ICU admission. These parameters were mostly extracted from studies in China, predominantly with adult and elderly subjects aged between 41 and 68 years, and males. Of the 35 studies reviewed (34, 46, 49–79), only three (52, 53, 79) showed the proportion of cases identified as infected with SARS-CoV-2 among all cases tested (detected cases), and numbers ranged between 4.45% (79) and 61.8% (53). In the studies, there was variability in the criteria used to define cases as critical, and data showed that the proportion of critical COVID-19 cases among all hospitalized patients ranged between 0.06% (51) and 86.9% (56). Not all studies reported the number of deaths among critical cases, and when they did (49, 51, 56, 57, 60–62, 65–67, 70, 73, 75, 77, 78), the rate identified was 1.35% (49) to 78% (60). The median length of total stay among COVID-19 cases ranged from a minimum of 7.5 days (cases with a death outcome) (60) and a maximum of 20.5 days (77). The median length of outpatient stays prior to ARDS onset ranged between two (66) and 14 (62) days. The median lenght of outpatients stays prior to ICU admission was reported in one study as one day (57) and the mean as 8 days (64).
Table 6.
Presentation of parameters: detected cases, critical cases among hospitalized patients, deaths among the critical cases, hospitalization period, and hospitalization period before ARDS or ICU.
| Author, year [references] | Country | Study date | Characteristics of the sample (sample, age, and sex) | Cases detected N/D (%) | Critical cases among hospitalized patients N/D (%) | Deaths among critical cases | Total length of stay (N)—mean/median | Period between hospitalization before ARDS OR ICU (N)—mean/median |
|---|---|---|---|---|---|---|---|---|
| R. Li et al., 2020 (52) | China | March 2020 | No information | ?/? (14%; 95%CI: 10–18%)a | – | - | – | – |
| Novel Coronavirus Pneumonia Emergency Response Epidemiology Team, 2020 (53)* | China | February 2020 | N = 672 cases; 86.6% aged between 30 and 79 years | 44,672 /72,314 (61.8%) | – | - | – | - |
| Zheng et al., 2020 (54) | China | February 2020 | N = 25 cases; Age [median (IQR; range)]: 3 years (2–9 years; 3 months−14 years); Female sex: 44% | – | 2b/25 (8%) | – | – | – |
| Peng et al., 2020 (55)* | China | Between January and February 2020 | N = 112 cases; adults with cardiovascular disease, age (mean): 62 years; Female sex: 52.67% | – | 16c/ 112 (14.28%) | – | – | – |
| Liu et al., 2020 (56) | China | Between December 2019 and January 2020 | N = 137 hospitalized patients; Age [median (lower and upper limits)]: 57 (20–83) years: Female sex: 55.47% | – | 119b/137 (86.9%) | 16/137 (11.7%) | – | (137)—Median of 7 days, ranging from 1 to 20 days – ARDSg |
| Wang et al., 2020 (57) | China | Between January and February 2020 | N = 138 hospitalized patients; Age [median (IQR; lower and upper limits)]: 56 (42–68; 22–92) years: Female sex: 45, 7% | – | 36c/138 (26.1%) | 6/36 (16.66%) | (47)—Median (among those who were discharged): 10 days (IQR 7–14 days) | (138)—Median: 5 days (IQR 1-10 days)g
median of 1 day (IQR 0 – 3 days)c |
| Cheng et al., 2020 (58)* | China | Up to February 2020 | N = 1,079 cases; Age [mean (±SD)]: 46 (24) years: Female sex: 46, 8% | – | 72d/ 1265 (5.7%) | – | – | – |
| Guan et al., 2020 (51) | China | Up to January 2020. | N = 1,099 cases; Age (median): 47 (IQR 35–58) years; Female sex: 41.9% | – | 67c/1,099 (0.06%) | 15/173 (8.67%) | Median of 13 (IQR 11.5–17.0 days) | Median of 5 days (IQR 2–7 days) between onset of symptoms and onset of pneumonia |
| Mo et al., 2020 (59) | China | Between January and February 2020 | N = 155 hospitalized patients; Age [median (IQR)]: 54 (42–66) years; Female sex: 44.50% | – | 37e/ 155 (23.9%) | – | (22)—Median of fatal cases: 10.5 (IQR: 8–16 days); 133-−10 recovered cases (IQR 7–15 days) | - |
| Zhou et al., 2020 (60) | China | Up to January 2020 | N = 191 hospitalized patients; Age [median (IQR)]: 56 (46–67) years; Female sex: 38% | – | 53e/191 (28%) | 42/53 (78%) | (191)—median: 11 days (IQR: 7–14 days)k; (54)—Fatal cases: 7.5 (IQR: 5–11 days); (137)—Survivors: 12 (IQR: 9–15 days) |
(191)—median: 12 days (IQR: 8-−15 days)g |
| Wan et al., 2020 (61) | China | Between January and February 2020 | N = 137 hospitalized patients; Age [median (IQR)]: 47 (36–55) years; Female sex: 46.70% | – | 40e/135 (29.6%) | 1/40 (2.5%) 1g/21 (4.8%) |
– | – |
| Huang et al., 2020 (62) | China | Between December 2019 and January 2020 | N = 41 hospitalized patients; Age [median (IQR)]: 49 (41–58) years; Female sex: 27% | – | 13c/41 (31.7%) | 5/13c (38%) | – | (12) Median between 8 and 14 daysg |
| Wang et al., 2020 (63) | China | Between January and February 2020 | N = 55 asymptomatic cases; Age [median (lower and upper limits)]: 49 (2–69) years; Female sex: 60% | – | 2f/55 (3.6%) | – | - | – |
| Young et al., 2020 (64) | Singapore | Between January and February 2020 | N = 18 cases in isolation at hospital; Age [median (lower and upper limits)]: 47 (3–73) years; Female sex: 50% | – | 2e/18 (11%) | – | – | (18) Mean – 8 daysc |
| Chen et al., 2020 (65) | China | January 2020 | N = 99 cases; Age [mean (±SD)]: 55 (13.1) years; Female sex: 32% | – | 17g/99 (17%) | 11/17 (64.7%) | – | – |
| Wu et al., 2020 (66) | China | Between December 2019 and January 2020 | N = 201 hospitalized patients; Age [median (IQR)]: 51 (43–60) years; Female sex: 36.30% | – | 53c/201 (26.4%) | 44/84 (52.4%) | (201)—median of 13 days (IQR: 10–16 days) | (201)—Median: 2 days (IQR−1–4 days)g |
| He et al., 2020 (67)* | China | February 2020 | N = 54 hospitalized patients (severe and critical); Age (mean): 68 years; Female sex: 37.0% | – | – | 26/54 (48.1%) | – | – |
| Wu et al., 2020 (68) | China | Between January and February 2020 | N = 80 cases; Age [median (IQR)]: 46.1 (30.7–61.5) years; Female sex: 51.25% | – | – | – | (21)—mean of patients who were discharged after 8 days | - |
| Sun et al., 2020 (50) | China | Between January and February 2020 | N = 8 cases (children); Age (lower and upper limits): 2 months to 15 years; Female sex: 25% | – | 3e/8c (37.5%) | – | (5 severe/critical patients): mean of 18.2 days (SD: 4.02 days) | – |
| Liu et al., 2020 (69) | China | Between January and February 2020 | N = 15 cases; Age [mean (±SD) (lower and upper limits)]: 32 (5; 23–40) years; Female sex: 100% (all pregnant women) | – | (2)—mean: 16 days | – | ||
| Cao et al., 2020 (70) | China | Between January and February 2020 | N = 199 hospitalized patients; Age [median (IQR)]: 58 (49–68) years; Female sex: 39.70% | – | 61b/199 (30.65%) | – | (199)—Median: 15 days (IQR: 12–17 days)—(with pneumonia) Median: 10 days (IQR: 5–14 days) at ICU |
- |
| Pan et al., 2020 (34) | China | Between December 2019 and March 2020 | N = 88 cases; Age [median (IQR)]: 56.7 (43.4–66.8) years; Female sex: 51.60% | – | 970e/32,325 (3%) | – | – | – |
| Liu et al., 2020 (71) | China | Between January and February 2020 | N = 73 hospitalized patients; Age [mean (±SD)]: 41.6 (14.5) years; Female sex: 43.84% | – | 3e/73 (4%) | – | – | – |
| Cao et al., 2020 (72) | China | Between January and February 2020 | N = 135 cases; Age [mean (±SD)]: 48.87 (17.12) years; Female sex: 48.06% | – | 2e/135 (1.48%) | – | – | – |
| Simonnet et al., 2020 (73) | France | April 2020 | N = 124 hospitalized patients at ICU; Age [median (IQR)]: 60 (51–70) years; Female sex: 27% | – | 85h/ 124c (68.6%) | 18/78c (23%) | Time to mechanical ventilation (n = 85): 62 cases upon admission; 13 cases at day 1; 4 cases at day 2; and 6 cases within 7 daysc. | |
| Lian et al., 2020 (46) | China | Between January and February 2020 | N = 778 cases, divided into elderly group, with 136 cases and mean age of 68.28 years; and non-elderly adult group with 652 cases and mean age of 41.15 years. | – | Group < 60 years: 9c/652 (1.38%); 35g/652 (5.37%); 1i/652 (0.15%); 72j/652 (11.04%). Group ≥ 60: 13c/136 (9.56%); 23g/136 (16.91%);1j/136 (0.74%);10j/ 136 (7.35%). |
– | – | – |
| Liu et al., 2020 (74) | China | Between January and February 2020 | N = 56 hospitalized patient, divided into group ≥60 years (N = 18), Age [median (IQR)]: 68 (65.25–69.75) years, female sex: 50%; and group < 60 years (N = 38), age [median (IQR)]: 47 (35.75–51.25) years; female sex: 33.337% (group < 60 years) | – | 4k/18 (22.22%) (group ≥ 60) and 2k/38 (5.26%) (group < 60 years) | – | – | – |
| Guan et al., 2020 (75) | China | Between January and February 2020 | N = 575 hospitalized patients; Age [mean (±SD)]: 48.9 (16.3) years; Female sex: 42.70% | – | 50h/1590 (3.1%); 99c/1590 (6.2%) | 50/1590 (6.2%) | – | – |
| Petrie, 2020 (76) | Australia | Between January and February 2020 | N = 6,606 cases; Age [median (IQR)]: 60.5 (42–72) years; Female sex: 50.00% | – | 39b/810 (4.8%); 141c/810 (17%) | – | – | – |
| Cai et al., 2020 (77) | China | Between December 2019 and January 31, 2020 | N = 298 cases; Age [median (IQR)]: 47 (33-61) years; Female sex: 51.34% | – | 30c, h/298(10.1%) | 3/58d (5.2%) | (298)—Median: 20.5 days (IQR: 15–26 days) | – |
| Bhatraju et al., 2020 (78) | USA | Between February and March 2020 | N = 24 critical cases; Age [mean (±SD)]: 64 (18) years; Female sex: 38% | – | – | 12/24c (50%) | (24)—Median: 12 days (IQR: 8–12 days) at hospital; at ICU: 9 days (IQR: 4–14 days) Among survivors: (12) 17 days (IQR: 16–23 days) at hospital; at ICU (survivors): 14 days (IQR: 4–17 days). |
– |
| – | – | Duration of mechanical ventilation: general 10 days (IQR: 7–12 days); among those who were extubated (n = 6/18, 33%) 11 days (IQR: 4–17 days) | – | |||||
| Jin et al., 2020 (49) | China | Between January and February 2020 | N = 651, divided into group with gastrointestinal symptoms (GI) (N = 21), Age [mean (SD)]: 46.14 ± 14.19 years; Female sex: 50%; and asymptomatic group GI (N = 577), Age [mean (SD)]: 45.09 ± 14.45 years; Female sex: 49.05% | – | With GI symptoms: 5g/74c (6.76%); 1i/74c (1.35%); 13j/74c (17.57%) asymptomatic GI: 12g/577c (2.08%); 1i/477c (0.17%); 51j/577c (8.84%) | 1/74c (1.35%) in the group with GI symptoms | – | – |
| Republic of Korea, 2020 (79) | Korea | January and March 2020 | N = 94,635; Sex female: 62% | 4,212/94,635 (4.45%) | – | – | – | – |
The proportion of registry of infections at the first moment was estimated at 0.65 (95%CI: 0.60–0.69), that is, 65% of infections were registered/detected, in period 1. This proportion dropped to 14% before travel restrictions and was kept as such throughout the period 2.
Cases who required mechanical ventilation.
Cases who required admission to intensive care unit (ICU).
Cases reported as critical, with no mentioning of case definition.
Cases who presented shock or who required mechanical ventilation or admission to intensive care unit (ICU).
Cases of severe respiratory failure, but with no need for monitoring at intensive care unit (ICU).
Cases who developed acute respiratory distress syndrome.
Critical cases were defined according to WHO guidelines.
Cases who presented shock.
Cases who had liver injury.
Cases with pneumonia severity index (PSI) 4 and 5.
ARDS, Acute respiratory distress syndrome; ICU, Intensive care unit; GI, gastrointestinal; 95%CI, 95% confidence interval; IQR, Interquartile range; SD, Standard deviation.
Article in Chinese, only the abstract in English was assessed.
Discussion
This study presented a proposal of a rapid literature review method, which identified a set of epidemiological parameters aiming to support construction of predictive models and evidence-based decision-making in view of the COVID-19 pandemic. The syntaxes developed and the rapid review method proposed allowed for identification and synthesizing of all epidemiological parameters of interest in only 40 days. This required the joint effort of researchers and adjustments to the method usually recommended for systematic literature reviews.
Although complex, it is imperative to select good parameters to support mathematical and epidemiological models that predict diseases dynamics in different territories, especially emergent and reemergent epidemics. For COVID-19, the models presented to date are mostly based on local parameters of early stages of the epidemic, as well as the viral behavior of other coronaviruses, such as those causing SARS and MERS outbreaks. Our results show that it is possible to overcome said difficulties by rapidly and systematically gathering evidence produced with different methodologies and in different settings, facilitating identification of parameters that are more suitable to the context and the purpose of the predictive model, improving quality and accuracy of results, and potentially helping territories enhance their COVID-19 preparedness and emergency response.
For all parameters assessed, we found a higher frequency of studies from China. We believe this was due to fact that COVID-19-related cases first emerged in China, which favors a higher number of studies coming from there. Only a few studies were from Europe, the Americas and regions other than Asia, however, with the spread of COVID-19 throughout the world, studies from these regions will be increasingly frequent in the literature, allowing for a more in-depth analysis of other contexts.
One of the parameters most affected by the local context is the reproduction number (R). Cultural habits, control measures in place—such as contact tracing, lockdown or border closures—and the stage of the disease in the territory will directly impact the value and evolution of R (80). Also, limitations concerning data quality and the number of observations have been reported in many studies and may impact estimates. In this sense, we found three outliers in this review. The one with the lowest R (0.48) was developed in South Korea (30) using massive testing, contact tracing and quarantine strategies, in addition to case isolation (81). One of the highest R values (more than 14) was from data on a cruise-ship [i.e., an enclosed population for which, although some restrictive measures were put in place, social distancing was not possible (22, 23)].
Therefore, for construction of predictive models, in order to use the most appropriate R value, it is imperative to understand health systems and their surveillance strategies, as well as consider the social, economic, demographic and cultural contexts of the population for which the estimates are made. It is also worth mentioning that some studies (18, 20, 21, 28, 34) showed a lower R value after restrictive measures were implemented.
The incubation period, infectious period and serial interval are also crucial for understanding the evolution of epidemics. In this regard, there was no wide variation in the incubation period and serial interval among the selected studies, which may contribute to the accuracy of predictive models, however, these results must be consistently confirmed outside of Asia. The scarcity of studies on the transmissible period is another important aspect, and there is a need for new studies estimating this parameter for different populations.
The parameters were mostly extracted for adult, male subjects. Studies suggest that children develop mild symptoms or remain asymptomatic, which hinders case identification, however they play a crucial role in the disease transmission cycle (82). Also, the predominance of males can be explained due to the larger proportion of males in the Chinese population (83). Work conditions of males may also put them at higher risk of exposure to the pathogen, and some health conditions may increase the risk of severe disease (84).
The parameters pertaining to the rate of critical cases among all COVID-19 cases are extremely relevant for managers to anticipate and put in place the logistics and technologies required for critical patient care. Due to the different criteria adopted to define critical cases, it was difficult to establish a homogeneous classification. However, we identified different situations that led to cases being classified as critical, allowing for application of the parameter in predictive models based on the local context or demand. As for the proportion of deaths among critical cases, we also found heterogeneity in the studies. We believe that the criteria used to classify cases as critical may have influenced the way the fatality rate was presented in this clinical classification, leading to inconsistent results.
Variability in case classification is a difficulty in several diseases (85). This heterogeneity is an obstacle in literature reviews and other epidemiological studies, since it precludes head-to-head comparison of research studies. In that sense, we recommend that researchers use a standard classification, based on a protocol such as that of the WHO (86), to standardize case presentation and facilitate data use by other groups. We highlight that in this review, we presented the different classifications of critical cases, allowing modelers and decision-makers to identify parameters according to the context.
The length of hospital stays identified in the studies ranged from one to nearly 3 weeks, and the length of outpatient stay until ARDS onset or ICU admission ranged from immediate up to 2 weeks. This information is relevant so that mathematical models can anticipate the demand for hospital beds, estimated costs and even potential complications arising from long stays, supporting decision-making by managers.
Although the usual method employed in systematic literature reviews is the gold standard (87), particularly due to its minimizing of the risk of bias and ensuring critical and adequate data review, it is time-consuming (88) and usually takes between 6 months and 2 years for completion (89), which limits its use in the current emergency context. By simplifying or omitting components usually included in systematic reviews, rapid literature reviews can be produced faster, although with a higher risk of bias (90).
Thus, this protocol was considered a rapid review because, among the limitations, we highlight the inclusion of only two databases, the language restriction, the non-paired data selection and extraction processes, as well as the absence of a careful evidence quality assessment (90). However, to reduce these limitations, we used sensitive syntaxes in comprehensive databases; all review stages were supervised by experienced researchers with an epidemiology background; meetings were held to standardize concepts and organize the execution of all steps. Also, most parameters were extracted from descriptive observational studies, including cohort studies and case series, using similar methods, leading to relative homogeneity in respect to evidence quality. Furthermore, in terms of limitations, we included studies with different populations—groups restricted to enclosed spaces such as cruise-ships, hospitalized patients and specific professionals, for example—and reviewed data collected using primary and secondary instruments. However, study characteristics are presented in all extraction charts, to make for easier reading.
It should also be noted that some parameters for monitoring the disease progress were not included. These parameters, such as 7-days, or 14-days averages of cases and deaths can be important for health authorities that are using the mathematical models to make decisions regarding the reopening of various societal sectors. However, this rapid review explored the parameters requested by the group of Brazilian mathematical modelers to determine assistance measures, and these parameters, at that time, were not demanded. When replicating this method, the syntax can be easily adapted to obtain these and other parameters, as needed.
Due to the difficulties to define good parameters, we recommend that, when using the data presented in this article, researchers pay attention to disease transmission chains; the contribution of different age ranges to infection strength; the stage of implementation of control measures; and the current and projected health situation in each territory. Modelers must also consider the accuracy of results, assess the number of studies selected, and test uncertainties. We recommend the use of the syntaxes developed and presented in this article when performing new searches to update parameters, contemplating studies conducted in other contexts of time, place, and people, when needed. Also, we believe that these syntaxes can be adapted according to the types of models that are being constructed (e.g., microsimulations, agent-based modeling, systems dynamic modeling, causal inference analysis, economic analysis and other epidemiological and mathematical models) and how impact outcomes are being looked at/predicted.
Knowing the parameters that help understand the dynamics of the SARS-CoV-2 pandemic, such as those presented in this study, allows for modeling of the impact of surveillance and control measures on virus transmission. Mathematical models of transmission estimate the number of infections over time and their consequences, allow for sizing of the resources needed for patient care, and assessment of the impact of non-pharmaceutical interventions (91), supporting decision-making and public policy management.
The rapid literature review methodology used in this study was developed and operationalized in slightly more than 1 month, and showed that it is feasible to rapidly identify and summarize a set of epidemiological parameters in the context of public health emergencies, where an expressive and increasing number of publications can be found. The epidemiological parameters presented here describe information from different scenarios of COVID-19 transmission, disease and deaths and may be used to support predictive models used to estimate the societal impact of the disease, helping decision-makers develop evidence-based preventive measures and ensure preparedness of health systems.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.
Author Contributions
LG, WA, MO, and HP outlined the review. LG, MO, and HP coordinated the review. HP developed the syntaxes. LG conducted literature searches, imported the publications, and removed duplicates. AO, AA, LS, and YM performed study selection and data extraction. MO and HP oversaw data extraction and resolved conflicts. LG, AO, MO, and HP wrote the first version of the manuscript. FS and WA contributed with data analysis and interpretation. MA contributed with data interpretation and manuscript translation. All authors critically reviewed, read, and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our gratitude to all health care professionals who have been working to mitigate the impacts of this pandemic.
References
- 1.Cheng VCC, Lau SKP, Woo PCY, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. (2007) 20:660–94. 10.1128/CMR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao GF. From “A” IV to “Z” IKV: attacks from emerging and re-emerging pathogens. Cell. (2018) 172:1157–9. 10.1016/j.cell.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. (2011) 81:85–164. 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Director-General's Opening Remarks at the Media Briefing on COVID-19–11 March 2020. Geneve: World Health Organization; (2020) Available online at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-−11-march-2020 (accessed March 17, 2020). [Google Scholar]
- 6.Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D, Jin H-J, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. (2020) 7:11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Pneumonia of Unknown Cause—China. Disease Outbreak News: 5 January 2020. Geneve: World Health Organization; (2020). Available online at: https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ (accessed March 17, 2020). [Google Scholar]
- 8.WHO Coronavirus Disease (COVID-19) Situation Report–206. Geneve: World Health Organization; (2020). Available online at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200813-covid-19-sitrep-206.pdf?sfvrsn=bf38f66b_6 (accessed August 13, 2020). [Google Scholar]
- 9.Verma V, Vishwakarma RK, Verma A, Nath DC, Khan HTA. Time-to-death approach in revealing chronicity and severity of COVID-19 across the World. PLoS ONE. (2020) 15:e0233074. 10.1371/journal.pone.0233074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. (2020) 20:669–77. 10.1016/S1473-3099(20)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam D. Special report: the simulations driving the world's response to COVID-19. Nature. (2020) 580:316–8. 10.1038/d41586-020-01003-6 [DOI] [PubMed] [Google Scholar]
- 12.Panayidou K, Gsteiger S, Egger M, Kilcher G, Carreras M, Efthimiou O, et al. GetReal in mathematical modelling: a review of studies predicting drug effectiveness in the real world. Res Synth Methods. (2016) 7:264–77. 10.1002/jrsm.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul EM. The interval between successive cases of an infectious disease. Am J Epidemiol. (2003) 158:1039–47. 10.1093/aje/kwg251 [DOI] [PubMed] [Google Scholar]
- 14.Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung S, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. (2020) 9:538. 10.3390/jcm9020538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun K, Chen J, Viboud C. Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: a population-level observational study. Lancet Digit Heal. (2020) 2:e201–8. 10.1016/S2589-7500(20)30026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T-M, Rui J, Wang Q-P, Zhao Z-Y, Cui J-A, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty. (2020) 9:24. 10.1186/s40249-020-00640-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuniya T. Prediction of the epidemic peak of coronavirus disease in Japan, 2020. J Clin Med. (2020) 9:789. 10.3390/jcm9030789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, Nie Y, Penny M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: a data-driven analysis. J Med Virol. (2020) 92:645–59. 10.1002/jmv.25750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Qianyin L, Jinjun R, Musa SS, Yang G, Wang W, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-NCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Inf Dis. (2020) 92:214–17. 10.1016/j.ijid.2020.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. (2020) 20:553–8. 10.1101/2020.01.31.20019901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Wang Z, Dong Y, Chang R, Xu C, Yu X, et al. Phase-adjusted estimation of the number of Coronavirus Disease 2019 cases in Wuhan, China. Cell Discov. (2020) 6:10. 10.1038/s41421-020-0148-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocklöv J, Sjödin H, Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J Travel Med. (2020) 27:taaa030. 10.1093/jtm/taaa030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizumoto K, Chowell G. Transmission potential of the novel coronavirus (COVID-19) onboard the diamond Princess Cruises Ship, 2020. Infect Dis Model. (2020) 5:264–70. 10.1101/2020.02.24.20027649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang B, Wang X, Li Q, Bragazzi NL, Tang S, Xiao Y, et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med. (2020) 9:462. 10.2139/ssrn.3525558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. (2020) 395:689–97. 10.1016/S0140-6736(20)30260-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. (2020) 382:1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, Musa SS, Lin Q, Ran J, Yang G, Wang W, et al. Estimating the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of January 2020: a data-driven modelling analysis of the early outbreak. J Clin Med. (2020) 9:388. 10.3390/jcm9020388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, You XY, Wang YJ, Peng LP, Du ZC, Gilmour S, et al. Estimating the basic reproduction number of COVID-19 in Wuhan, China. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:476–9. 10.3760/cma.j.cn112338-20200210-00086 [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Liu Q, Yang Z, Liao J, Yang K, Bai W, et al. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Based Med. (2020) 13:3–7. 10.1111/jebm.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ki M. Task Force for 2019-nCoV. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol Health. (2020) 42:e2020007. 10.4178/epih.e2020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis. (2020) 26:25452. 10.1101/2020.02.19.20025452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anastassopoulou C, Russo L, Tsakris A, Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS ONE. (2020) 15:e0230405. 10.1371/journal.pone.0230405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song QQ, Zhao H, Fang LQ, Liu W, Zheng C, Zhang Y. Study on assessing early epidemiological parameters of coronavirus disease epidemic in China. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:461–5. 10.3760/cma.j.cn112338-20200205-00069 [DOI] [PubMed] [Google Scholar]
- 34.Pan A, Liu L, Wang C, Guo H, Hao X, Wang Q, et al. Association of public health interventions with the epidemiology of the COVID-19 outbreak in Wuhan, China. JAMA. (2020) 323:1915–23. 10.1001/jama.2020.6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: a data-driven analysis. Int J Infect Dis. (2020) 93:201–4. 10.1016/j.ijid.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pung R, Chiew CJ, Young BE, Chin S, Chen MI-C, Clapham HE, et al. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. (2020) 395:1039–46. 10.1016/S0140-6736(20)30528-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. (2020) 395:514–23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The Incubation period of coronavirus disease 2019 (covid-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. (2020) 172:577–82. 10.7326/M20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J, Liu X, Pan W, Douglas MW, Bao S. Epidemiology of 2019 novel coronavirus disease-19 in Gansu Province, China, 2020. Emerg Infect Dis. (2020) 26:251 10.3201/eid2606.200251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang HY, Xu J, Li Y, Liang X, Jin YF, Chen SY, et al. The preliminary analysis on the characteristics of the cluster for the Corona Virus Disease. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:623–8. 10.3760/cma.j.cn112338-20200223-00153 [DOI] [PubMed] [Google Scholar]
- 41.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance. (2020) 25:62. 10.2807/1560-7917.ES.2020.25.5.2000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. (2020) 63:706–11. 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu WS, Li YG, Wei ZF, Zhou PH, Lyu LK, Zhang GP, et al. Investigation and analysis on characteristics of a cluster of COVID-19 associated with exposure in a department store in Tianjin. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:489–93. 10.3760/cma.j.cn112338-20200221-00139 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Su X, Chen W, Fei CN, Guo LR, Wu XL, et al. Epidemiological investigation on a cluster epidemic of COVID-19 in a collective workplace in Tianjin. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:649–53. 10.3760/cma.j.cn112338-20200219-00121 [DOI] [PubMed] [Google Scholar]
- 45.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. (2020) 93:284–6. 10.1101/2020.02.03.20019497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lian J, Jin X, Hao S, Cai H, Zhang S, Zheng L, et al. Analysis of epidemiological and clinical features in older patients with coronavirus disease 2019 (COVID-19) outside Wuhan.Clin Infect Dis. (2020) 71:740–7. 10.1093/cid/ciaa242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia J, Hu X, Yang F, Song X, Dong L, Zhang J, et al. Epidemiological characteristics on the clustering nature of COVID-19 in Qingdao city, 2020: a descriptive analysis. Disaster Med Public Health Prep. (2020) 1–5. 10.1017/dmp.2020.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan W, Liang W, Zhao Y, Liang H, Chen Z, Li Y, et al. Comorbidity and its impact on 1,590 patients with Covid-19 in China: a nationwide analysis. Eur Respir J. (2020) 55:2000547. 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin X, Lian J-S, Hu J-H, Gao J, Zheng L, Zhang Y-M, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. (2020) 69:1002–9. 10.1136/gutjnl-2020-320926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Li H, Lu X-X, Xiao H, Ren J, Zhang F-R, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center's observational study. World J Pediatr. (2020) 16:251-9. 10.1007/s12519-020-00354-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. (2020) 382:1708–20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science. (2020) 368:489–93. 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team . The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:145–51. 10.3760/cma.j.issn.0254-6450.2020.02.003 [DOI] [PubMed] [Google Scholar]
- 54.Zheng F, Liao C, Fan Q-H, Chen H, Zhao X-G, Xie Z-G, et al. Clinical characteristics of children with coronavirus disease 2019 in Hubei, China. Curr Med Sci. (2020) 40:275–80. 10.1007/s11596-020-2172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng YD, Meng K, Guan HQ, Leng L, Zhu RR, Wang BY, et al. Clinical characteristics and outcomes of 112 cardiovascular disease patients infected by 2019-nCoV. Zhonghua Xin Xue Guan Bing Za Zhi. (2020) 48:450–5. 10.3760/cma.j.cn112148-20200220-00105 [DOI] [PubMed] [Google Scholar]
- 56.Liu K, Fang Y-Y, Deng Y, Liu W, Wang M-F, Ma J-P, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J. (2020) 133:1025–31. 10.1097/CM9.0000000000000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng JL, Huang C, Zhang GJ, Liu DW, Li P, Lu CY, et al. Epidemiological characteristics of novel coronavirus pneumonia in Henan. Zhonghua Jie He He Hu Xi Za Zhi. (2020) 43:327–31. 10.3760/cma.j.cn112147-20200222-00148 [DOI] [PubMed] [Google Scholar]
- 59.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. (2020) ciaa270. 10.1093/cid/ciaa270. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y, et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J Med Virol. (2020) 92:797–806. 10.1002/jmv.25783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Liu Y, Liu L, Wang X, Luo N, Li L. Clinical outcomes in 55 patients with severe acute respiratory syndrome coronavirus 2 who were asymptomatic at hospital admission in Shenzhen, China. J Infect Dis. (2020) 221:1770–4. 10.1093/infdis/jiaa119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. (2020) 323:1488. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. (2020) 395:507–13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. (2020) 180:934–43. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He XW, Lai JS, Cheng J, Wang MW, Liu YJ, Xiao ZC, et al. Impact of complicated myocardial injury on the clinical outcome of severe or critically ill COVID-19 patients. Zhonghua Xin Xue Guan Bing Za Zhi. (2020) 48:E011 10.3760/cma.j.cn112148-20200228-00137 [DOI] [PubMed] [Google Scholar]
- 68.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, et al. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu province: a multicenter descriptive study. Clin Infect Dis. (2020) 71:706–12. 10.1093/cid/ciaa199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu D, Li L, Wu X, Zheng D, Wang J, Yang L, et al. Pregnancy and perinatal outcomes of women with coronavirus disease (COVID-19) pneumonia: a preliminary analysis. AJR Am J Roentgenol. (2020) 215:127–32. 10.2139/ssrn.3548758 [DOI] [PubMed] [Google Scholar]
- 70.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. (2020) 382:1787–99. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu K-C, Xu P, Lv W-F, Qiu X-H, Yao J-L, Gu J-F, et al. CT manifestations of coronavirus disease-2019: a retrospective analysis of 73 cases by disease severity. Eur J Radiol. (2020) 126:108941. 10.1016/j.ejrad.2020.108941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao C, Li Y, Liu S, Fan H, Hao L. Epidemiologic features of 135 patients with coronavirus disease (COVID-19) in Tianjin, China. Disaster Med Public Health Prep. (2020) 1–5. 10.1017/dmp.2020.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simonnet A, Chetboun M, Poissy J, Raverdy V, Noulette J, Duhamel A, et al. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity. (2020) 28:1195–9. 10.1002/oby.22831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu K, Chen Y, Lin R, Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. (2020) 80:e14–8. 10.1016/j.jinf.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guan W, Liang W, Zhao Y, Liang H, Chen Z, Li Y, et al. Comorbidity and its impact on 1,590 patients with Covid-19 in China: a nationwide analysis. Eur Respir J. (2020) 55:2000547. 10.1183/13993003.01227-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petrie S. COVID-19, Australia: epidemiology report 12: reporting week ending 23:59 AEST 19 April 2020. Commun Dis Intell. (2020) 44:36. 10.33321/cdi.2020.44.36 [DOI] [PubMed] [Google Scholar]
- 77.Cai Q, Huang D, Ou P, Yu H, Zhu Z, Xia Z, et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. (2020) 75:1742–52. 10.1101/2020.02.17.20024018 [DOI] [PubMed] [Google Scholar]
- 78.Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, et al. Covid-19 in critically ill patients in the Seattle region—case series. N Engl J Med. (2020) 382:2012–22. 10.1056/NEJMoa2004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Republic of Korea . Report on the epidemiological features of coronavirus disease 2019 (COVID-19) outbreak in the Republic of Korea from January 19 to March 2, 2020. J Korean Med Sci. (2020) 35:e112. 10.3346/jkms.2020.35.e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maier BF, Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. (2020) 368:eabb4557. 10.1101/2020.02.18.20024414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pietrewicz O. Preparedness and surveillance: South Korea's response to COVID-19. Polski Instytut Spraw Miedzynarodowych. (2020) 66:1–2. Available online at: https://pism.pl/publications/Preparedness_and_Surveillance__South_Koreas_Response_to_COVID19 [Google Scholar]
- 82.Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang F, et al. Epidemiology of COVID-19 among children in China. Pediatrics. (2020) 145:e20200702. 10.1542/peds.2020-0702 [DOI] [PubMed] [Google Scholar]
- 83.The World Bank Population, female (% of total population)—China. World Bank staff estimates based on age/sex distributions of United Nations Population Division's World Population Prospects: 2019 Revision. (2020) Available online at: https://data.worldbank.org/indicator/SP.POP.TOTL.FE.ZS?locations=CN (accessed May 26, 2020).
- 84.Eurofound and International Labour Organization Working Conditions in a Global Perspective. Luxembourg; Geneva: Publications Office of the European Union; International Labour Organization; (2019). [Google Scholar]
- 85.Ward A, Arrighi HM, Michels S, Cedarbaum JM. Mild cognitive impairment: disparity of incidence and prevalence estimates. Alzheimer's Dement. (2012) 8:14–21. 10.1016/j.jalz.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 86.WHO Coronavirus Disease (COVID-10) Technical Guidance: Patient Management. Technical Guidance, Case Management. Geneve: World Health Organization; (2020). Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/patient-management (accessed May 27, 2020). [Google Scholar]
- 87.Snyder H. Literature review as a research methodology: an overview and guidelines. J Bus Res. (2019) 104:333–9. 10.1016/j.jbusres.2019.07.039 [DOI] [Google Scholar]
- 88.McKenzie JE, Clarke MJ, Chandler J. Why do we need Evidence-Based Methods in Cochrane? In: Tovey D. editor. Cochrane Database of Systematic Reviews. Chichester: John Wiley & Sons, Ltd. (2015). p. ED000102. 10.1002/14651858.ED000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khangura S, Konnyu K, Cushman R, Grimshaw J, Moher D. Evidence summaries: the evolution of a rapid review approach. Syst Rev. (2012) 1:10. 10.1186/2046-4053-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tricco AC, Antony J, Zarin W, Strifler L, Ghassemi M, Ivory J, et al. A scoping review of rapid review methods. BMC Med. (2015) 13:224. 10.1186/s12916-015-0465-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flaxman S, Mishra S, Gandy A, Unwin HJT, Coupland H, Mellan TA, et al. Estimating the number of infections and the impact of non-pharmaceutical interventions on COVID-19 in European countries: technical description update. arXiv. (2020) 1–7. 10.1038/s41586-020-2405-7 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.