Abstract
A metabolic hallmark of many cancers is the increase in glucose consumption coupled to excessive lactate production. Mindful that L-lactate originates only from pyruvate, the question arises as to how can this be sustained in those tissues where pyruvate kinase activity is reduced due to dimerization of PKM2 isoform or inhibited by oxidative/nitrosative stress, posttranslational modifications or mutations, all widely reported findings in the very same cells. Hereby 17 pathways connecting glucose to lactate bypassing pyruvate kinase are reviewed, some of which transit through the mitochondrial matrix. An additional 69 converging pathways leading to pyruvate and lactate, but not commencing from glucose, are also examined. The minor production of pyruvate and lactate by glutaminolysis is scrutinized separately. The present review aims to highlight the ways through which L-lactate can still be produced from pyruvate using carbon atoms originating from glucose or other substrates in cells with kinetically impaired pyruvate kinase and underscore the importance of mitochondria in cancer metabolism irrespective of oxidative phosphorylation.
Keywords: cancer, glycolysis, mitochondria, metabolomics, Warburg effect, oncometabolism, lactate dehydrogenase
Glucose and Lactate in Cancer: Background
It is a well-known fact that most cancers exhibit increased rates in glucose consumption (Bose and Le, 2018). This is clinically exploited by following radionuclide-labeled glucose analogs for the purpose of tumor imaging in living human beings (Feng et al., 2019). The very same cancers are also known to be major lactate producers, which is important for their survival (de la Cruz-Lopez et al., 2019). The combination of an increased consumption of glucose with an increase in lactate output led to the assumption that cancers exhibit an increase in glycolysis; although this is true, serving the purpose of generating glycolytic metabolites which are diverted toward biosynthetic processes (DeBerardinis et al., 2008) and NADPH by the pentose phosphate pathway (Icard and Lincet, 2012), most tumors express a dimeric form of the M2 isoform of pyruvate kinase which has been reported to be much less active than that found in healthy cells; furthermore, numerous posttranslational modifications and mutations have been reported for this gene product, leading to a much reduced activity but still fueling cancer aggression (see section “Pyruvate Kinase”). Even more so, tumor cells with undetectable levels of pyruvate kinase still producing lactate can be found in vivo (Israelsen et al., 2013). On one hand, the decrease in pyruvate kinase activity is important for maintaining a metabolite “traffic jam,” forcing upstream metabolites toward biosynthetic pathways; on the other hand, it points to a metabolic conundrum because L-lactate may only originate from pyruvate, a metabolite arising from phosphoenolpyruvate (PEP) through pyruvate kinase in glycolysis (see Figure 1). The purpose of this review is to highlight the pathways that can lead to pyruvate and lactate—even commencing from glucose—bypassing pyruvate kinase. This is important because (i) carbon-labeled atoms in glucose may appear in lactate without net ATP production from glycolysis and (ii) hints on the possibility that other pathways leading to pyruvate/lactate could be crucial for cancer cell survival that are perhaps amenable to pharmacological and/or genetic manipulation. The list of pathways appearing below has been assembled by mining the following databases: Kyoto Encyclopedia of Genes and Genomes1 (Kanehisa and Goto, 2000), BRaunschweig ENzyme Database2 (Jeske et al., 2019), Metabolic Atlas3 (Robinson et al., 2020), Biochemical, Genetic, and Genomic knowledge base4 (King et al., 2016), MetaNetX5 (Moretti et al., 2016), Human Metabolome Database6 (Wishart et al., 2018), and Virtual Metabolic Human7 (Noronha et al., 2019).
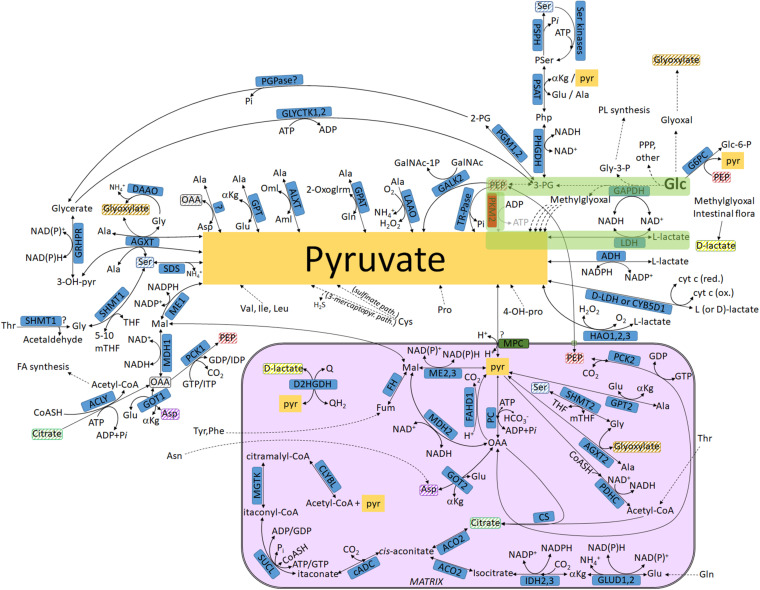
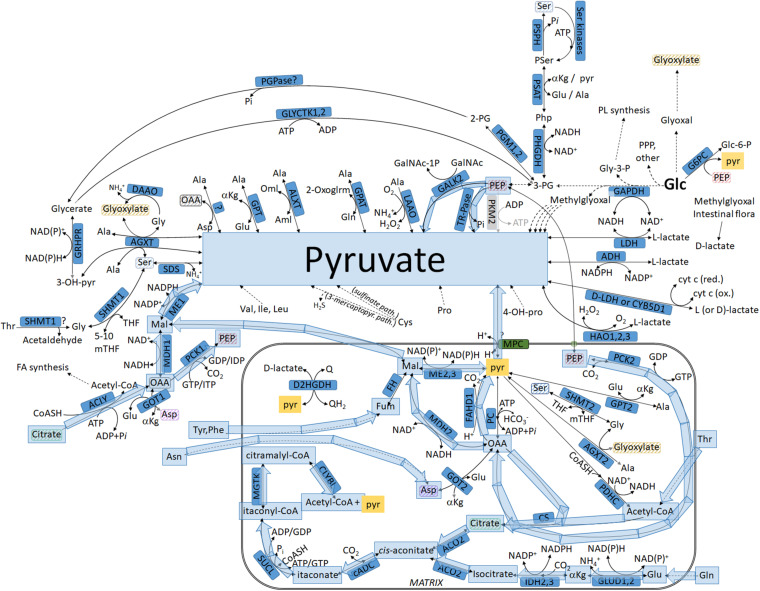
FIGURE 1.
Biochemical pathways connecting glucose or other metabolites to pyruvate and L- or D-lactate. The box in magenta represents a mitochondrion. Glycolysis is highlighted in green. Metabolites found both inside and outside the mitochondria that are not connected with an arrow are highlighted in matching striped colors (to avoid arrow clutter). For abbreviations, see Table 1.
Pyruvate Kinase
Pyruvate kinase generates ATP at the “substrate level” in the absence of oxygen by catalyzing the dephosphorylation of PEP to pyruvate (see Figure 1). There are four isoforms denoted as L, R, M1, and M2. For details regarding kinetic properties, tissue distribution, and regulation, the reader is referred to the review by Israelsen and Vander Heiden (2015). In the present review, the PKM2 isoform will be specifically examined; for a more thorough evaluation, the reader is referred to Li et al. (2014, 2018), Wong et al. (2015); Yang and Lu (2015), Dayton et al. (2016b), Hsu and Hung (2018), and Alquraishi et al. (2019). The non-enzymatic functions of PKM2 are examined elsewhere (Hoshino et al., 2007; Stetak et al., 2007; Luo et al., 2011; Yang et al., 2012; Yang and Lu, 2013).
Basically, PKM2 exhibits lower enzymatic activity compared to that by PKM1 (Yamada and Noguchi, 1999) and is allosterically regulated by fructose-1,6-bisphosphate (FBP); it exists either as a dimer with low affinity for PEP or as an FBP-bound tetramer with high affinity for PEP (Mazurek et al., 2005; Zhang et al., 2019). Although PKM2 has been branded as “the predominant isoform in cancer cells” (Altenberg and Greulich, 2004; Mazurek et al., 2005), further scrutiny in 25 human malignant cancers, six benign oncocytomas, tissue-matched controls, and several cell lines showed that “PKM2 dominance was not a result of a change in isoform expression, since PKM2 was also the predominant PKM isoform in matched control tissues.” Therefore, a switch from PKM1 to PKM2 isoform expression during malignant transformation may not be taking place, as previously postulated (Christofk et al., 2008). Mindful of the controversy surrounding the proposed functions of PKM2 (Hosios et al., 2015; Harris and Fenton, 2019), the group of Vander Heiden characterized the effects of cancer−associated PKM2 mutations on enzyme kinetics and allosteric regulation and reported that a decrease in PKM2 activity supports the rapid proliferation of cells (Liu V. M. et al., 2020). This is in line with earlier reports showing that a decrease in PKM2 activity due to posttranslational modifications (Lv et al., 2011) or inhibition by oxidative stress (Anastasiou et al., 2011) promotes tumor growth (Prakasam et al., 2018). Alternatively, exposure to small molecule PKM2 activators or expression of the constitutively active PKM1 thwarts cancer cell proliferation (Anastasiou et al., 2012). Finally, it has been also shown that PKM2 is not even required for the growth of many cancers (Cortes-Cros et al., 2013; Israelsen et al., 2013; Wang et al., 2014; Lunt et al., 2015; Dayton et al., 2016a, 2018; Lau et al., 2017; Tech et al., 2017; Hillis et al., 2018). In aggregate, the consensus seems to be that the lower the pyruvate kinase activity, the greater the stimulation of tumor growth. As discussed in the section below entitled “Evidence Showing That Pyruvate Kinase Inhibition Does Not Lead to a Proportional Decrease in Pyruvate/Lactate Formation,” even those cells exhibiting low—or even undetectable—pyruvate kinase activity still produce lactate, which begs the question: where does this lactate come from?
Evidence Showing That Pyruvate Kinase Inhibition Does Not Lead to a Proportional Decrease in Pyruvate/Lactate Formation
In Cortes-Cros et al. (2013), it was shown that knockdown of both PKM1 and PKM2 (PKM2 knockdown was on the order of > 95%) leading to an approximately fivefold decrease in overall pyruvate kinase activity yielded only a ∼50% decrease in the appearance of 13C originating from glucose to lactate.
In Chaneton et al. (2012), silencing of both PKM1 and PKM2 to an extent greater than 90% led to only a ∼30% decrease in pyruvate and lactate production, while PEP concentration increased by 100%.
In Vander Heiden et al. (2010), it was shown that cancer cell lysates expressing no pyruvate kinase activity produced 50% of pyruvate from PEP compared with the total cell lysates. Although in this work it was postulated that phosphate from PEP is transferred to the catalytic histidine on human PGAM1, this claim was subsequently rejected by the same authors, attributing their earlier findings to contaminating ATP−dependent protein kinases (Hosios et al., 2015).
In all of the abovementioned studies, it was assumed that, in view of severely diminished pyruvate kinase activity, pyruvate and lactate production is attributed to carbon sources other than glucose. Indeed Yu et al. (2019), determined that, in pancreatic ductal adenocarcinoma cells with PKM1 and PKM2 knockdown, cysteine catabolism generated ∼20% of intracellular pyruvate. The purpose of the present review is to not only outline these pathways but also show additional ways for obtaining 13C labeling in pyruvate or lactate originating from glucose; furthermore, since some of these pathways involve intermediates that transit through the matrix, the role of the mitochondria is emphasized, which is unrelated to the concept of oxidative phosphorylation.
Pathways Leading to Pyruvate Commencing From Glucose: Intermediates Not Transiting Through the Mitochondria
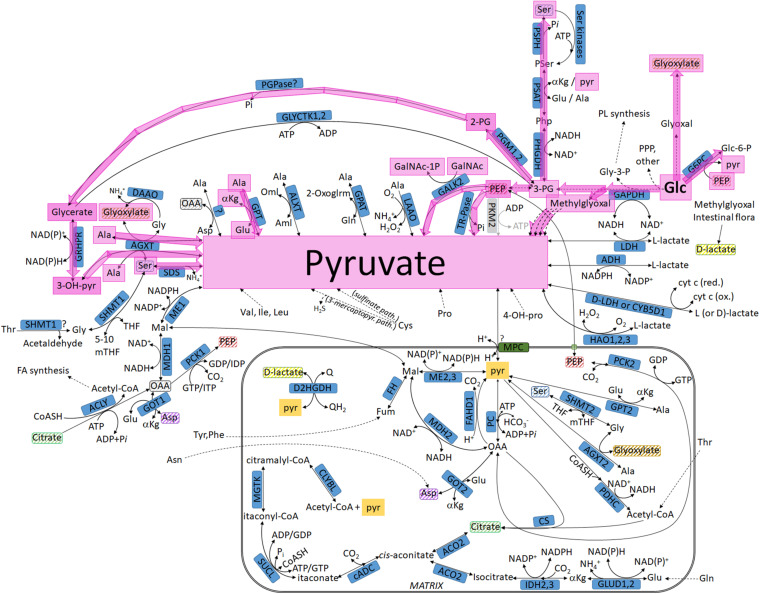
The pathways shown in this section refer to Figure 2 (lavender arrows). Multiple arrows imply multiple biochemical steps.
FIGURE 2.
Pathways leading to pyruvate commencing from glucose, highlighted in lavender: intermediates not transiting through the mitochondria. For abbreviations, see Table 1.
(1) Glc + PEP → Glc-6-P + pyruvate: This reaction is catalyzed by glucose-6-phosphatase (G6PC) (Nordlie, 1974; Colilla et al., 1975) (for abbreviations, see Table 1). In humans, G6PC expression was reported to be elevated in GBM when compared with normal brain (Abbadi et al., 2014), while in rodent hepatomas it was found to be decreased (Weber and Cantero, 1955).
TABLE 1.
Abbreviations.
| 2-Oxoglrm | 2-oxoglutaramate (a-ketoglutaramate) |
| 2-PG | 2-Phosphoglycerate |
| 3-OH-pyr | 3-hydroxypyruvate |
| 3-PG | 3-Phosphoglycerate |
| 4-OH-proline | 4-hydroxyproline |
| 5-10 mTHF | 5-10 methylene-Tetrahydrofolate |
| ACLY | ATP Citrate Lyase |
| ACO | Aconitase |
| ADH | Alcohol Dehydrogenase |
| AGXT | Alanine-glyoxylate Aminotransferase |
| aKG | a-ketoglutarate |
| Ala | Alanine |
| ALXT | Alanine-Ketomalonate Transaminase |
| Aml | Aminomalonate |
| Asn | Asparagine |
| Asp | Aspartate |
| cADC | cis-Aconitate Decarboxylase |
| CLYBL | Citramalyl-CoA Lyase |
| CS | Citrate Synthase |
| CYB5D1 | Cytochrome B5 Domain-Containing Protein 1 |
| Cys | Cysteine |
| D2HGDH | D-2-Hydroxyglutarate Dehydrogenase |
| DAAO | D-amino acid Oxidase |
| D-LDH | D-Lactate Dehydrogenase |
| FAHD | Acylpyruvase |
| FH | Fumarate Hydratase |
| Fum | Fumarate |
| G6PC | Glucose 6 phosphatase |
| GALK | N-acetylgalactosamine Kinase |
| GalNAc | N-Acetylgalactosamine |
| GalNAc-1-P | N-Acetylgalactosamine-1-Phosphate |
| GAPDH | Glyceraldehyde 3 Phosphate Dehydrogenase |
| Glc | Glucose |
| Glc-6-P | Glucose-6-phosphate |
| Gln | Glutamine |
| Glu | Glutamate |
| GLUD | Glutamate Dehydrogenase |
| Gly | Glycine |
| Gly-3-P | Glyceraldehyde-3-Phosphate |
| GLYCTK | Glycerate Kinase |
| GOT | Aspartate Aminotransferase |
| GPAT | Glutamine-Pyruvate Transaminase |
| GPT | Alanine Aminotransferase |
| GRHPR | Glyoxylate Reductase |
| HAO | Hydroxyacid Oxidase |
| IDH | Isocitrate Dehydrogenase |
| Ile | Isoleucine |
| KGDHC | a-Ketoglutarate Dehydrogenase Complex |
| LAAO | L-amino-acid Oxidase |
| LDH | Lactate Dehydrogenase |
| Leu | Leucine |
| Mal | Malate |
| MDH | Malate Dehydrogenase |
| ME | Malic Enzyme |
| MGTK | Methylglutaconase |
| MPC | Mitochondrial Pyruvate Carrier |
| mTHF | methyl-Tetrahydrofolate |
| OAA | Oxaloacetate |
| Oml | Oxomalonate |
| PCK | Phosphoenolpyruvate Carboxykinase |
| PCK | Pyruvate Carboxylase |
| PDHC | Pyruvate Dehydrogenase Complex |
| PEP | Phosphoenolpyruvate |
| PGM | Phosphoglucomutase |
| PGPase | 2-phosphoglyceric acid Phosphatase |
| Phe | Phenylalanine |
| PHGDH | Phosphoglycerate Dehydrogenase |
| Php | Phosphohydroxypyruvate |
| PKM2 | Pyruvate Kinase isoform M2 |
| PL | Phospholipids |
| PPP | Pentose Phosphate Pathway |
| PSAT | Phosphoserine Aminotransferase |
| Pser | Phosphoserine |
| PSPH | Phosphoserine Phosphatase |
| pyr | Pyruvate |
| Q | Quinone |
| QH2 | Quinol |
| SDH | Succinate Dehydrogenase |
| SDS | Serine Dehydratase |
| Ser | Serine |
| SHMT | Serine Hydroxymethyltransferase |
| SUCL | Succinate-CoA Ligase |
| THF | Tetrahydrofolate |
| Thr | Threonine |
| TR-Pase | Tartrate-resistant acid Phosphatase |
| Tyr | Tyrosine |
| Val | Valine |
(2) Glc →→→ methylglyoxal →→→ pyruvate: This may occur through four different routes involving aldehyde dehydrogenase 9, zinc binding alcohol dehydrogenase domain containing two [more recently renamed to prostaglandin reductase 3 (Yu et al., 2013)] and at least two oxoaldehyde dehydrogenases; for details, see Vander Jagt and Hunsaker (2003). Methylglyoxal has been reported to trigger metastasis in breast, anaplastic thyroid, and colorectal cancer (Chiavarina et al., 2017; Antognelli et al., 2019; Nokin et al., 2019).
(3) Glc →→→ PEP → pyruvate: the terminal reaction is catalyzed by tartrate-resistant acid phosphatases (TRAP), the molecular identity of which remained unknown well after their biochemical characterization (Helwig et al., 1978; Chen and Chen, 1988; Hayman et al., 1989); they are most likely substantiated by a metalloprotein enzyme with the ability to catalyze the hydrolysis of orthophosphate monoesters under acidic conditions (Bull et al., 2002). The expression of this enzyme (TRAP) is a marker of bone disease in cancer patients (Nguyen et al., 1991; Koizumi and Ogata, 2002; Mose et al., 2003; Terpos et al., 2003; Chao et al., 2005).
(4) Glc →→→ PEP; PEP + GalNAc → GalNAc-1P + pyruvate: Terminal reaction catalyzed by N-acetylgalactosamine kinase isoforms 1 or 2 (Pastuszak et al., 1996). These enzymes are implicated in many signaling pathways inherent to carcinogenesis (Zeidan and Hart, 2010).
(5) Glc →→→ 3-PG → 2-PG (by phosphoglucomutase 1 or 2) → glycerate [probably through 2-phosphoglyceric acid phosphatase (Baranowski et al., 1968)] → 3-OH-pyr [by glyoxylate reductase (Mdluli et al., 2005)]; 3-OH-pyr + Ala (or glyoxylate) → Gly + pyruvate (or Ser): the terminal reaction is catalyzed by alanine-glyoxylate aminotransferase (Danpure et al., 2003). The mitochondrial isoform of the latter enzyme (alanine-glyoxylate aminotransferase isoform 2, AGXT2) has been reported to form glycine and pyruvate from alanine and glyoxylate; this reaction has been confirmed in normal tissues (Holmes and Assimos, 1998) and HepG2 cancer cells (Baker et al., 2004). The same reaction has been reported to take place in peroxisomes (Poore et al., 1997). On the other hand, loss of alanine-glyoxylate aminotransferase (AGXT) expression has been reported to accelerate the progression of hepatocellular carcinoma (Sun et al., 2019). A “futile cycle” may exist between 3-PG and glycerate through 2-phosphoglyceric acid phosphatase and glycerate kinase 1 and 2; glycerate kinase 2 is also found in the mitochondria (Guo et al., 2006).
(6) Glc →→→ 3-PG → phosphohydroxypyruvate (Php), catalyzed by phosphoglycerate dehydrogenase; Php + Ala → phosphoserine (Pser) + pyruvate, catalyzed by phosphoserine aminotransferase (PSAT) (Hirsch and Greenberg, 1967): PSAT overexpression is associated with increased tumorigenicity in human esophageal squamous cell carcinoma (Liu et al., 2016) and colon carcinomas (Yoon et al., 2015) and a poor outcome on tamoxifen therapy in recurrent breast cancer (De Marchi et al., 2017); conversely, its selective loss suppresses migration, invasion, and experimental metastasis in triple negative breast cancer (Metcalf et al., 2020).
(7) Glc →→→ 3-PG →Php (catalyzed by phosphoglycerate dehydrogenase); Php + Ala (or Glu) → Pser + pyruvate (or →Kg); the latter reaction is catalyzed by phosphoserine aminotransferase; Pser → Ser → pyruvate, catalyzed by serine dehydratase (Ogawa et al., 2006) or serine dehydratase-like (SDSL) (Ogawa et al., 2006). Notably, SDS was reported to be absent from human colon carcinomas (Snell et al., 1988).
(8) Glc →→→ Glyoxal →→→ glyoxylate (Lange et al., 2012); glyoxylate + 3-OH-pyr (or Ala) → Gly + pyruvate (or Ser): the terminal reaction is catalyzed by AGXT (for considerations related to cancer, see pathway no. 5).
(9) Glc →→→ 3-PG →Php (catalyzed by phosphoglycerate dehydrogenase); Php + Glu → Pser + →Kg; latter reaction catalyzed by phosphoserine aminotransferase; →Kg + Ala → Glu + pyruvate, catalyzed by alanine aminotransferase (GPT; for considerations related to cancer, see pathway no. 6).
Pathways Leading to Pyruvate Commencing From Glucose: Intermediates Transiting Through the Mitochondria
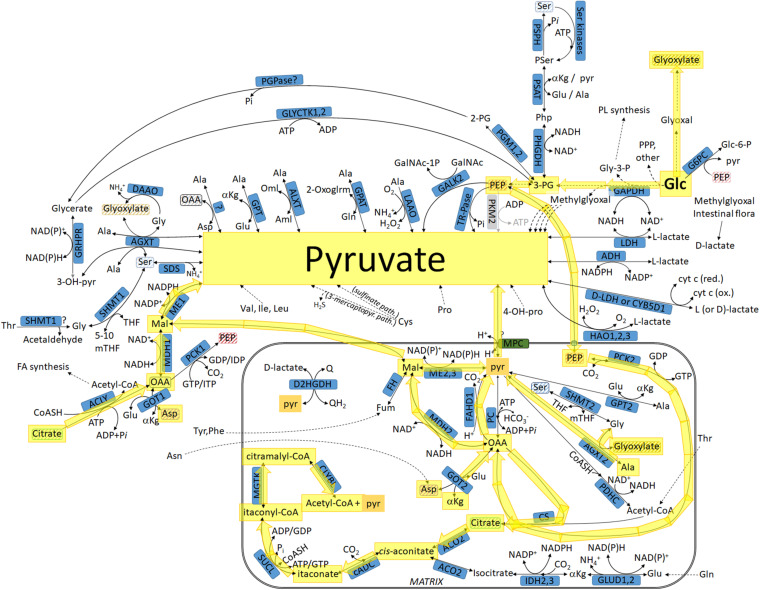
These pathways depend on one or more of three critical parameters: (1) glyoxylate entry into the mitochondria, (2) reversibility of the matrix phosphoenolpyruvate carboxykinase (PCK2), and (3) reversibility of the mitochondrial pyruvate carrier (MPC). Regarding glyoxylate, I was unable to find information on its transport across the inner mitochondrial membrane; however, it is known that it can be processed by the matrix-localized AGXT2 (Kakimoto et al., 1969). PCK2 expression and activity level are critical for many cancer types: in tumor-initiating enriched prostate cancer cell clones, PCK2 was overexpressed, and this correlated with more aggressive tumors and lower survival rates (Zhao et al., 2017); in lung cancer cell lines and in non-small cell lung cancer samples, PCK2 expression and activity were enhanced under low-glucose conditions (Leithner et al., 2015); finally, it was reported that PCK2 is required for glucose-independent cancer cell proliferation and tumor growth in vivo (Vincent et al., 2015). Regarding PCK2 reversibility, the enzyme has been shown to operate in the reaction toward OAA synthesis in mitochondria from rabbit liver (Carlsen et al., 1988), pigeon and rat liver (Wiese et al., 1996), guinea pig liver (Garber and Ballard, 1970; Garber and Salganicoff, 1973), rabbit enterocytes (Wuensch and Ray, 1997), chicken liver (Hebda and Nowak, 1982; Makinen and Nowak, 1983; Wilson et al., 1983; Erecinska and Wilson, 1984), and bullfrog liver (Goto et al., 1980). However, in Vincent et al. (2015), it was shown that a fraction of pyruvate originated from glutamine from PEP through PCK2. With respect to the reversibility of the MPC, this is a working hypothesis because there are no data showing pyruvate release from normally polarized mitochondria. Nevertheless, this is not a far-fetched hypothesis: succinate and other metabolites are effluxed from the mitochondria for non-metabolic roles against a hyperpolarized membrane potential (Mills et al., 2016), demonstrating that this is possible under appropriate conditions. It may be also relevant that pyruvate catabolism through the pyruvate dehydrogenase complex is associated with suppression of tumor growth in vitro and in vivo (Michelakis et al., 2008); relevant to this, genes coding for both the pyruvate dehydrogenase complex and pyruvate carboxylase in certain cancers are usually downregulated (Yuen et al., 2016); furthermore, pyruvate is found in blood plasma, urine, and cerebrospinal fluid, and its presence there is not associated with damage of plasma membranes. Of course, this does not mean that extracellular pyruvate originated from the mitochondria, but it indicates that it can cross the plasma membrane through monocarboxylate transporters, some of which are distributed both in plasma and in the inner mitochondrial membrane (Hussien and Brooks, 2011); indeed monocarboxylate transporter 1, which is one of the four known pyruvate transport mechanisms, was recently shown to export pyruvate from the cell (Hong et al., 2016); however, mitochondrial pyruvate export remains hypothetical especially in view of the fact that its exit is influenced by the membrane potential and →pH. It was also recently reported that loss of an MPC isoform prior to a tumorigenic stimulus doubled the frequency of adenoma formation and produced higher-grade tumors, and this was associated with a glycolytic metabolic phenotype and increased expression of stem cell markers (Bensard et al., 2020). Mindful of the above, these pathways are as shown in Figure 3 (yellow arrows).
FIGURE 3.
Pathways leading to pyruvate commencing from glucose, highlighted in yellow: intermediates transiting through the mitochondria. For abbreviations, see Table 1.
(10) Glc →→→ glyoxal →→→ glyoxylate: Glyoxylate enters the mitochondria; glyoxylate + Ala → Gly + pyruvate through AGXT2. Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathway no. 5).
(11) Glc →→→ PEP which enters the mitochondria; PEP transport across the inner membrane of mammalian mitochondria has been demonstrated to occur by the tricarboxylate carrier by Robinson (1971) and the group of Soling et al. (1971) and Kleineke et al. (1973) and to a lesser extent by the adenine nucleotide carrier, shown by the Shug and Shrago (1973); Sul et al. (1976) and in Drahota et al. (1983) and reviewed in Passarella et al. (2003). The possibility of a PEP/pyruvate transporter has also been put forward (Satrustegui et al.,2007). More recently, PEP cycling via mitochondrial PEPCK evoking PEP transport across the inner mitochondrial membrane has also been demonstrated by the group of Kibbey (Stark et al., 2009); PEP → OAA by PCK2; OAA → pyruvate by reverse operation of PC. However, this is expected to be a very minor path. Pyruvate may exit the mitochondria through the MPC.
(12) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11. PEP → OAA by PCK2; OAA → pyruvate by FAHD1 (Pircher et al., 2011, 2015). FAHD1 also converts 3-acylpyruvate, acetylpyruvate, and fumarylpyruvate to pyruvate (Pircher et al., 2011). It is not known where acetylpyruvate comes from, but its existence is known since Krebs reported it (Krebs and Johnson, 1937). Pyruvate may exit the mitochondria through the MPC. FAHD1 depletion has been shown to induce premature senescence in human endothelial cells by inhibiting mitochondrial metabolism (Petit et al., 2017); however, this might be a double-edged sword since OXPHOS capacity has been inversely correlated with malignancy in several cell types (Zhou et al., 2003; Matoba et al., 2006; Hu et al., 2012; Hall et al., 2013; Bartesaghi et al., 2015; Nicolay et al., 2015; Capala et al., 2016; Smith et al., 2020).
(13) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11; PEP → OAA by PCK2; OAA → Mal by MDH2; Mal → pyruvate by ME2,3 (Zelewski and Swierczynski, 1991). Pyruvate may exit the mitochondria through the MPC. ME2 knockdown suppresses tumor growth in lung cancer (Ren et al., 2014), while ME2,3 deletions confer lethality in pancreatic cancer (Dey et al., 2017).
(14) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11; PEP → OAA by PCK2; OAA → Mal by MDH2; Mal exits the mitochondria; Mal → pyruvate by ME1 (Zelewski and Swierczynski, 1991; Loeber et al., 1994). ME1 knockdown inhibits the growth of colon cancer cells (Murai et al., 2017), and its overexpression is associated with larger breast tumor size, higher incidence of lymph node metastasis, and higher incidence of lymph–vascular invasion (Liu C. et al., 2020). In the same line, ME1 is associated with tumor budding—a phenomenon representing epithelial to mesenchymal transition—in oral squamous cell carcinomas (Nakashima et al., 2020).
(15) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11; PEP → OAA by PCK2; OAA + acetyl-CoA → citrate by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY (Chypre et al., 2012); OAA → Mal by MDH1; Mal → pyruvate by ME1 (for considerations related to cancer, see pathway no. 14).
(16) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11; PEP → OAA by PCK2; OAA + Glu →→Kg + Asp by GOT2; Asp exits the mitochondria; Asp + →Kg → Glu + OAA by GOT1; OAA → Mal by MDH1; Mal → pyruvate by ME1 (for considerations related to cancer, see pathway no. 14).
(17) Glc →→→ PEP; PEP enters the mitochondria through the means outlined in pathway 11; PEP → OAA by PCK2; OAA + acetyl-CoA → citrate by CS; citrate → cis-aconitate, intermediate of ACO2 reaction; cis-aconitate → itaconate by cADC; itaconate + CoASH + ATP (or GTP) → itaconyl-CoA + Pi + ADP (or GDP) by SUCL (Nemeth et al., 2016); itaconyl-CoA → citramalyl-CoA by methylglutaconase (MGTK); citramalyl-coA → acetyl-CoA + pyruvate by CLYBL (Shen et al., 2017). Pyruvate may exit the mitochondria through the MPC. CLYBL has been reported to be associated with colorectal cancer metastasis (Li and Peng, 2013). Furthermore, CLYBL was reported to be overexpressed in 465 out of 38,258 tumor samples in the COSMIC database8.
Pathways Leading to Pyruvate But Not Commencing From Glucose: Intermediates Not Transiting Through the Mitochondria
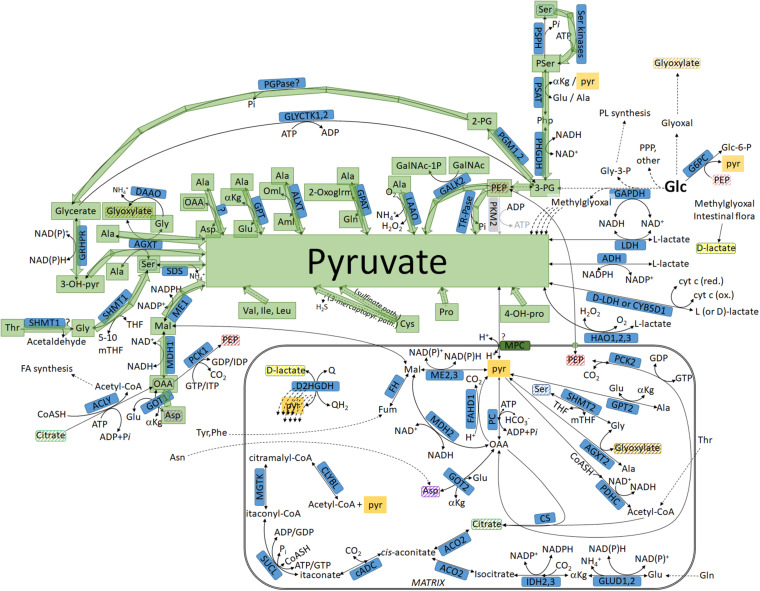
These pathways are shown in Figure 4 (green arrows).
FIGURE 4.
Pathways leading to pyruvate but not commencing from glucose, highlighted in green: intermediates not transiting through the mitochondria. For abbreviations, see Table 1.
(18) Ser → pyruvate, catalyzed by SDS or SDSL (for considerations related to cancer, see pathway no. 6).
(19) Ser →→→ PEP; PEP → pyruvate; terminal reaction catalyzed by tartrate-resistant acid phosphatase (TR-Pases; for considerations related to cancer, see pathway no. 3).
(20) Ser →→→ PEP; PEP + GalNAc → GalNAc-1P + pyruvate. The terminal reaction is catalyzed by N-acetylgalactosamine kinase isoforms 1 or 2 (for considerations related to cancer, see pathway no. 4).
(21) Ala → pyruvate, catalyzed by L-amino-acid oxidases (LAAO) (Nakano et al., 1967): Several mammalian LAAOs have been described, of which the enzyme “interleukin-4 induced gene 1” (IL4I1) is the best characterized (Castellano and Molinier-Frenkel, 2017); IL4I1 expression was reported to be associated with poor prognosis in human breast cancers (Finak et al., 2008).
(22) Ala + 2-oxoglrm → Gln + pyruvate, catalyzed by glutamine-pyruvate transaminase (GPAT) (Cooper and Meister, 1972; Cooper and Kuhara, 2014). GPAT is upregulated in many cancers in a MYC-dependent manner (Dong et al., 2020).
(23) Ala + 2-Oml → Aml + pyruvate, catalyzed by alanine-ketomalonate transaminase (ALXT) (Nagayama et al., 1958). I was unable to find relevant literature on ALXT expression or aminomalonate levels and cancer.
(24) Ala + αKg → Glu + pyruvate, catalyzed by GPT: GPT—similar to GPAT—is upregulated in many cancers in a MYC-dependent manner (Dong et al., 2020).
(25) Ala + OAA → Asp + pyruvate; enzyme unknown (Rowsell, 1956).
(26) Ala + Glyoxylate → Gly + pyruvate, catalyzed by alanine-glyoxylate aminotransferase (for considerations related to cancer, see pathway no. 5).
(27) Ala + 3-OH-pyr → Ser + pyruvate, catalyzed by alanine-glyoxylate aminotransferase (for considerations related to cancer, see pathway no. 5).
(28) Thr → Gly + acetaldehyde, catalyzed by SHMT1 (Garrow et al., 1993; Pinthong et al., 2014); Gly + 5,10 mTHF → THF + Ser, catalyzed by serine hydroxymethyltransferase 1; Ser → pyruvate, catalyzed by SDS or SDSL. SHMT1 knockdown induces apoptosis in lung cancer cells (Paone et al., 2014), and SHMT inhibitors block the growth of many human cancer cells (Ducker et al., 2017). Patients with high SHMT2 expression exhibit a shorter overall survival rate compared with patients with low expression (Koseki et al., 2018; for further considerations related to SDS or SDSL and cancer, see pathway no. 6).
(29) Asp + αKg → Glu + OAA, catalyzed by GOT1; OAA → Mal by MDH1; Mal → pyruvate by ME1 (for considerations related to cancer, see pathway no. 14).
(30) 4-OH-proline →→→ pyruvate, through glyoxylate formation (see pathway no. 26).
(31) Cys →→→ pyruvate through the sulfinate pathway (Stipanuk, 1979, 2020). Notably, in pancreatic cancer cells exhibiting PKM1/2 knockdown, 20% of intracellular pyruvate originated from cysteine (Yu et al., 2019). The contribution of cysteine catabolism to cancer has been extensively reviewed by Serpa (2020).
(32) Cys → 3-sulfino-L-alanine catalyzed by aspartate 4-decarboxylase (Liu et al., 2012); 3-sulfino-L-alanine is transaminated to 3-sulfinopyruvate by either aspartate aminotransferase or deaminated to the same product by cysteine sulfinic acid deaminase; 3-sulfinopyruvate is non-enzymatically converted to sulfite and pyruvate (Stipanuk, 2020; for considerations related to cancer, see pathway no. 31).
(33) Cys →→→ H2S + pyruvate through the 3-mercaptopyruvate pathway (Nagahara and Sawada, 2006). Cys can also transaminate with →-ketoglutarate to form glutamate and 3-mercaptopyruvate though GOT1, exhibiting cysteine transaminase activity. The catabolism of 3-mercaptopyruvate toward pyruvate is outlined in the reactions below (pathway no. 34; for considerations related to cancer, see pathway no. 31).
(34) L-cysteine is isomerized to D-cysteine by cysteine racemase (2-amino-3-mercaptopropionic acid racemase) (Soda and Osumi, 1969); D-Cys is converted to 3-mercaptopyruvate by D-amino acid oxidase and, in turn, to pyruvate and H2S by 3-mercaptopyruvate sulfurtransferase (3MST) (Shibuya et al., 2013) or thiosulfate sulfurtransferase (TST) (Pallini et al., 1991). The possibility of conversion of D-Cys to pyruvate by D-cysteine desulfhydrase (Nagasawa et al., 1985) in mammalian cells is yet to be reported. 3-Mercaptopyruvate can also react with hydrogen cyanide, forming pyruvate and thiocyanate in a reaction catalyzed by 3MST or TST; obviously, this is only a very minor route of pyruvate production due to cyanide toxicity (Bhandari et al., 2014; for further considerations related to cancer, see pathway no. 31).
(35) Ser → dehydroalanine (2-aminoacrylate) by serine dehydratase (SDS), serine dehydratase-like protein (SDSL), or serine racemase (SRR): Dehydroalanine can further hydrolyze to NH3 and pyruvate through SDS, SDSL, or SRR (Kashii et al., 2005); sometimes this reaction is referred to as hydrolysis by “2-aminoacrylate aminohydrolase.” Dehydroalanine can also spontaneously hydrolyze to NH3 and pyruvate through the intermediate 2-iminopropanoate; the latter later part of this spontaneous hydrolysis can be accelerated by 2-iminopropanoate deaminase (Lambrecht et al., 2012). Dehydroalanine can also be derived from 2 3,5-diiodo-L-tyrosine or 3,5-diiodo-L-tyrosine by thyroid peroxidase in the process of forming thyroxine and triiodothyronine, respectively (Gavaret et al., 1980). The crucial importance of serine metabolism for the growth and survival of proliferating cells is extensively reviewed in Yang and Vousden (2016) and Newman and Maddocks (2017).
(36) Se-methyl-L-selenocysteine (SeMSC, Se-methylselenocysteine, methyl selenocysteine) can be deaminated to methaneselenol, NH3, and pyruvate by selenocysteine lyase (Esaki et al., 1982). SeMSC can be found in many edible plants, including garlic, onions, and broccoli, as well as in dietary supplements (Yang and Jia, 2014). SeMSC was shown to exhibit anticarcinogenic properties (Ip et al., 1991; Medina et al., 2001) and even potentiate the antitumor activity of anticancer drugs (Cao et al., 2014).
(37) Val →→→ 2-methyl-3-oxopropanoate; 2-methyl-3-oxopropanoate can get transaminated with alanine by AGXT2 to D-3-amino-isobutanoate + pyruvate (Kakimoto et al., 1969). The overexpression of enzymes participating in valine catabolism is associated with poor prognosis in prostate cancer (Mayers et al., 2016) and tumors of the colon (Shan et al., 2019). The role of valine in cancer has been extensively reviewed in Ananieva and Wilkinson (2018) and Lieu et al. (2020).
(38) Leu →→→ 3-methylbutanoyl-CoA; the latter compound is converted to isobutyryl-CoA through branched-chain fatty acid metabolism (many steps); isobutyryl-CoA →→→ 2-methyl-3-oxopropanoate; 2-methyl-3-oxopropanoate can get transaminated with alanine by AGXT2 to D-3-amino-isobutanoate + pyruvate (Kakimoto et al., 1969). Because leucine catabolism shares many steps with that of valine, for considerations related to cancer, see pathway no. 37.
(39) Ile →→→ 2-methylbutanoyl-CoA; the latter compound is converted to isobutyryl-CoA through branched-chain fatty acid metabolism (many steps); isobutyryl-CoA →→→ 2-methyl-3-oxopropanoate; 2-methyl-3-oxopropanoate can get transaminated with alanine by AGXT2 to D-3-amino-isobutanoate + pyruvate (Kakimoto et al., 1969). Because isoleucine catabolism shares many steps with that for valine, for considerations related to cancer, see pathway no. 37.
(40) Pro + αKg + O2 → CO2 + succinate + trans-4-hydroxy-L-proline, catalyzed by prolyl 4-hydroxylase subunit alpha (isoforms 1, 2, or 3); trans-4-hydroxy-L-proline is then converted to L-1-pyrroline-3-hydroxy-5-carboxylate, also yielding NAD(P)H, by either pyrroline-5-carboxylate reductase (isoforms 1, 2, or 3) or left–right determination factor 1 (LEFTY1), a member of the TGF-→ family of proteins; L-1-pyrroline-3-hydroxy-5-carboxylate can be converted to L-erythro-4-hydroxyglutamate, also yielding NAD(P)H, by aldehyde dehydrogenase 4 family member A1; in turn, L-erythro-4-hydroxyglutamate is transaminated with either OAA by GOT2, yielding 4-hydroxy-2-oxoglutarate + aspartate, or →Kg by GOT1 or GOT2, yielding 4-hydroxy-2-oxoglutarate + glutamate; finally, 4-hydroxy-2-oxoglutarate is converted to glyoxylate and pyruvate by 4-hydroxy-2-oxoglutarate glyoxylate-lyase. It is relevant that increased proline catabolism has been recently reported to support metastasis (Elia et al., 2017). Arg, through either interconversion to metabolites as for proline catabolism or through citrulline/ornithine and the fumarate nucleotide cycle will also lead to pyruvate formation; however, this probably requires inter-organ communication and, thus, may not be found within a single cell. The crucial role of proline catabolism in tumor growth and metastatic progression is extensively reviewed in Phang (2019) and D’Aniello et al. (2020).
Pathways Leading to Pyruvate But Not Commencing From Glucose: Intermediates Transiting Through the Mitochondria
These pathways are shown in Figure 5 (blue arrows).
FIGURE 5.
Pathways leading to pyruvate but not commencing from glucose, highlighted in blue: intermediates transiting through the mitochondria. For abbreviations, see Table 1.
(41) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → Acetyl-coA + ADP + Pi + OAA by ACLY (Chypre et al., 2012); OAA → Mal by MDH1; Mal → pyruvate by ME1. The potential role of threonine catabolism in cancer is reviewed in Tsun and Possemato (2015) and Lieu et al. (2020) (for further considerations regarding ME1 and cancer, see pathway no. 14).
(42) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate → cis-aconitate, intermediate of ACO2 reaction; cis-aconitate → itaconate by cADC; itaconate + CoASH + ATP (or GTP) → itaconyl-CoA + Pi + ADP (or GDP) by SUCL; itaconyl-CoA → citramalyl-CoA by MGTK; citramalyl-coA → acetyl-CoA + pyruvate by CLYBL. Pyruvate may exit the mitochondria through the MPC (regarding threonine and cancer, see pathway no. 41; regarding CLYBL and cancer, see pathway no. 17).
(43) Asn →→→ Asp; Asp + αKg → Glu + OAA by GOT2; OAA by PCK2; OAA → pyruvate by reverse operation of PC. However, this is expected to be a path of a very minor flux. Pyruvate may exit the mitochondria through the MPC. The crucial role of asparagine availability in cancer is explored in Panosyan et al. (2014); Krall et al. (2016), and Knott et al. (2018). However, more emphasis on asparagine availability for anabolic, rather than catabolic, purposes is given.
(44) Asn →→→ Asp; Asp + αKg → Glu + OAA by GOT2; OAA → pyruvate by acylpyruvase (FAHD1). Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathways no. 12 and 37).
(45) Asn →→→ Asp; Asp + αKg → Glu + OAA by GOT2; OAA → Mal by MDH2; Mal → pyruvate by ME2,3. Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathways no. 13 and 37).
(46) Asn →→→ Asp; Asp + αKg → Glu + OAA by GOT2; OAA → Mal by MDH2; Mal exits the mitochondria; Mal → pyruvate by ME1 (for considerations related to cancer, see pathways no. 14 and 37).
(47) Tyr, Phe →→→ Fum; Fum → Mal by FH; Mal → pyruvate by ME2,3 (for considerations related to cancer, see pathway no. 13).
(48) Tyr, Phe →→→ Fum; Fum → Mal by FH; Mal exits the mitochondria; Mal → pyruvate by ME1 (for considerations related to cancer, see pathway no. 14).
(49) Tyr, Phe →→→ Fum; Fum → Mal by FH; Mal → OAA by MDH2; OAA → pyruvate by acylpyruvase (FAHD1). Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathway no. 12).
(50) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP enters the mitochondria; PEP → OAA by PCK2; OAA → pyruvate by acylpyruvase (FAHD1). Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathway no. 12).
(51) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP enters mitochondria; PEP → OAA by PCK2; OAA → Mal by MDH2; Mal → pyruvate by ME2,3. Pyruvate may exit the mitochondria through the MPC (for considerations related to cancer, see pathway no. 13).
(52) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → acetyl-CoA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP enters the mitochondria; PEP → OAA by PCK2; OAA → Mal by MDH2; Mal exits the mitochondria; Mal → pyruvate by ME1 (for considerations related to cancer, see pathway no. 14).
(53) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → Acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP + GalNAc → GalNAc-1P + pyruvate. Terminal reaction catalyzed by N-acetylgalactosamine kinase isoforms 1 or 2 (for considerations related to cancer, see pathway no. 4).
(54) Thr →→→ acetyl-CoA; acetyl-CoA + OAA → citrate, catalyzed by CS; citrate exits the mitochondria through the dicarboxylate carrier; citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP → pyruvate; the terminal reaction is catalyzed by tartrate-resistant acid phosphatases (for considerations related to cancer, see pathway no. 3).
Incompletely Characterized Reactions Forming Pyruvate
In the literature, some reactions have been described to produce pyruvate but are incompletely characterized. These are collectively listed below:
(55) O-carbamoyl-L-serine + H2O → pyruvate + 2 NH3, catalyzed by carbamoyl-serine ammonia lyase (Copper and Meister, 1973). O-Carbamoyl-L-serine is a weak inhibitor of a phosphate-dependent glutaminase (Shapiro et al., 1979); mindful of the crucial importance of glutamine catabolism through glutaminases in many cancer types, this route of pyruvate provision is probably minor.
(56) L-Cysteine-S-conjugate + H2O → a thiol + NH3 + pyruvate, catalyzed by cysteine S-conjugate →-lyases (Cooper and Pinto, 2006). The possibility of cysteine S-conjugate β-lyases metabolizing anticancer agents is reviewed in Cooper et al. (2011).
(57) cystathionine + H2O → L-homocysteine + pyruvate + NH3 or cysteine + H2O → sulfide + NH3 + pyruvate or cystine → thiocysteine + pyruvate + NH3, all catalyzed by cystathionine gamma-lyase (Stipanuk et al., 2006; Chiku et al., 2009). Cystathionine gamma-lyase was reported to be upregulated in bone−metastatic PC3 cells, and its knockdown suppressed tumor growth and metastasis (Wang et al., 2019). In the same line, this enzyme was shown to be upregulated and played a crucial role in the proliferation and migration of breast cancer cells (You et al., 2017).
(58) L-Serine O-sulfate + H2O → pyruvate + NH3 + sulfate catalyzed by serine-sulfate ammonia-lyase (Tudball and Thomas, 1972). I was unable to find relevant literature on serine-sulfate ammonia-lyase expression or L-serine O-sulfate levels and cancer.
(59) N-Acetylneuraminate → N-acetyl-D-mannosamine + pyruvate catalyzed by N-acetylneuraminate lyase (Brunetti et al., 1962); relevant to this, treatment of HL-60 cells by phorbol esters leads to a marked increase in the activity of this enzyme (Warren, 1986).
(60) D-Alanine + H2O + O2 → pyruvate + NH3 + H2O2 catalyzed by DAAO (Nagata et al., 1992; Abe et al., 2005; Fuchs et al., 2005; Smith et al., 2009). The interaction of D-alanine (and other D-amino acids) with tumors is reviewed in Bastings et al. (2019).
(61) L-Alanine → pyruvate + NH3 catalyzed by glutamate dehydrogenase; this reaction exhibits a weak activity (Silverstein, 1974). The role of glutamate dehydrogenase in cancer cells has been extensively reviewed in Moreno-Sanchez et al. (2020).
(62) 2-Oxosuccinamic acid + Ala → Asn + pyruvate, catalyzed by asparagine aminotransferase (Cooper, 1977; Maul and Schuster, 1986). The origin of 2-oxosuccinamic acid is not known (Cooper et al., 1987). I was unable to find relevant literature on 2-oxosuccinamic acid levels and cancer.
(63) Pyruvate oxime + acetone → pyruvate + acetone oxime, catalyzed by oximinotransferase (Omura et al., 1956). Due to acetone volatility, this is probably a very minor pathway for pyruvate production.
(64) Methylmalonyl-CoA + pyruvate → propionyl-CoA + oxaloacetate catalyzed by methylmalonyl-CoA carboxytransferase (Swick and Wood, 1960). This reaction is reversible and thus may yield pyruvate. I was unable to find relevant literature on methylmalonyl-CoA carboxytransferase and cancer.
(65) L-Alanine + 3-oxopropanoate → pyruvate + →-alanine, catalyzed by either →-alanine-pyruvate transaminase (Ito et al., 2001) or alanine-glyoxylate aminotransferase isoform 2 (Lee et al., 1995) (for considerations related to cancer, see pathway no. 5).
(66) Phenylpyruvate + L-alanine → L-phenylalanine + pyruvate catalyzed by phenylalanine (histidine) transaminase (Minatogawa et al., 1977). Phenylpyruvate has been reported to inhibit pyruvate kinase activity in human brain (Weber, 1969), thus enhancing PK-bypassing pathways. Phenylpyruvate levels were also found to be increased in ovarian cancers (Fong et al., 2011).
(67) 2-Oxoisohexanoate + L-alanine → L-leucine + pyruvate, catalyzed by the mitochondrial branched-chain L-amino acid aminotransferase (Schadewaldt et al., 1995). The role of branched-chain L-amino acid aminotransferase in cancer has been reviewed in Ananieva and Wilkinson (2018).
(68) PCK1, ME1, and ME2,3 may also convert OAA to CO2 and pyruvate (Sauer, 1973; Carlson et al., 1978; Bukato et al., 1995; Lee et al., 1995) (for considerations related to cancer, see pathway nos. 13 and 14).
(69) Salsolinol can be converted to salsolinol-1-carboxylate by salsolinol synthetase which can then be catabolized to dopamine and pyruvate (by an unknown enzyme); salsolinol is an endogenous catechol isoquinoline detected in humans derived from dopamine metabolism (Sandler et al., 1973; Collins et al., 1979). Salsolinol has been implicated in the initiation and promotion of alcohol-related breast carcinogenesis (Murata et al., 2016).
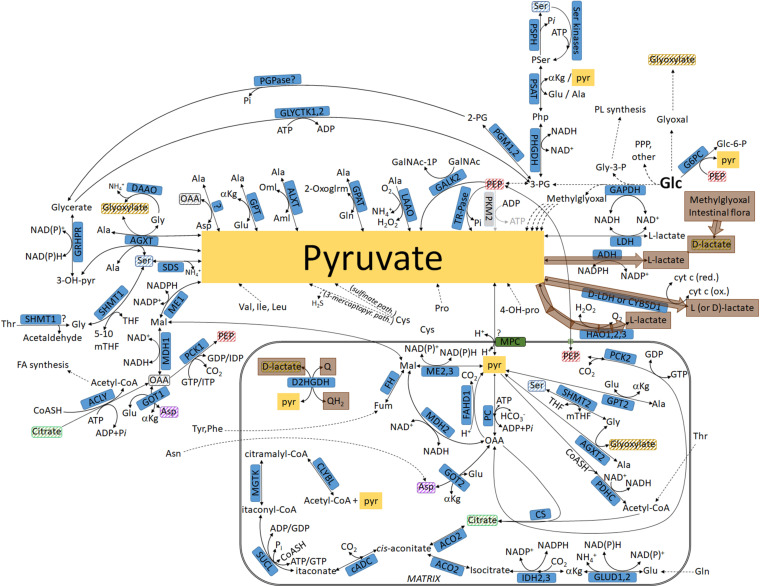
Pathways Leading to L-Lactate and D-Lactate Including Those Not Going Through Lactate Dehydrogenase
These pathways are shown in Figure 6 (brown arrows).
FIGURE 6.
Pathways leading to L- and D-lactate, including those not going through lactate dehydrogenase, highlighted in brown. For abbreviations, see Table 1.
Lactate—unlike pyruvate—exhibits chirality; thus, it exists in L- or D- configuration. In humans, a putative D-lactate dehydrogenase is known to exist (Flick and Konieczny, 2002; Ewaschuk et al., 2005; Chen et al., 2015). In metabolomics experiments, it is uncommon to distinguish between L- and D-lactate even although it is possible by using special columns. In this section, D- and L-lactate-forming pathways are outlined, including those not going through LDH:
(70) D-lactate formation by methylglyoxal and intestinal flora (Chen et al., 2015) (for considerations related to cancer, see pathway no. 2).
(71) Pyruvate + QH2 → D-lactate + Q, catalyzed by D2HGDH in the mitochondrial matrix (Cammack, 1969, 1970). Mutations in D2HGDH have been reported to be involved in multiple types of cancers but render the enzyme hypoactive or inert (Ye et al., 2018); thus, it is unlikely for this route to be important regarding pyruvate production.
(72) D- (or L-) Lactate + 2 ferricytochrome → 2 ferrocytochrome C + 2 H+ + pyruvate, catalyzed by D-lactate dehydrogenase; this reaction is mentioned in several databases, but no reference is given.
(73) D- (or L-) Lactate + 2 ferricytochrome → 2 ferrocytochrome C + 2 H+ + pyruvate, catalyzed by cytochrome B5 domain-containing protein 1; this reaction is mentioned in several databases, but no reference is given.
(74) Pyruvate + NADPH → NADP+ + L-lactate, catalyzed by ADH (Bosron and Prairie, 1972). The many roles of ADH in malignant neoplasms have been extensively reviewed in Orywal and Szmitkowski (2017).
(75) Pyruvate + H2O2 → L-lactate + O2, catalyzed by hydroxyacid oxidases (HAO1,2,3) (Fry and Richardson, 1979; Vignaud et al., 2007). However, in Jones et al. (2000), no HAO activity was reported. In primary pancreatic tumors, HAO3 is strongly downregulated (Thakur et al., 2008). HAO2 was reported to inhibit the malignancy of clear cell renal cell carcinoma cells. Overall, it is unlikely for this to be a substantial pathway in yielding pyruvate in cancer.
(76) Protein deglycase (E.C. 3.5.1.124) may form D-lactate from proteins (Richarme et al., 2015; Richarme and Dairou, 2017). Relevant to this, the deglycase DJ-1/Park7 is important for cancer cell survival (Vasseur et al., 2009).
(77) Methylglyoxal spontaneously forms a hemithioacetal adduct with GSH; subsequently, glyoxalase I (lactoylglutathione lyase; EC 4.4.1.5) produces S-D-lactoylglutathione from this adduct (Thornalley, 1990), and glyoxalase II (hydroxyacylglutathione hydrolase; EC 3.1.2.6), in turn, hydrolyzes S-D-lactoylglutathione to D-lactate + GSH (Cordell et al., 2004) (for considerations related to cancer, see pathway no. 2).
Finally, it is worth mentioning that LDH may process substrates other than pyruvate and lactate, interconverting glyoxylate + NAD+ to oxalate + NADH or α-ketobutyrate to →-hydroxybutyrate or L-glycerate to hydroxypyruvate (Dawkins and Dickens, 1965; Kim and Whitesides, 1988).
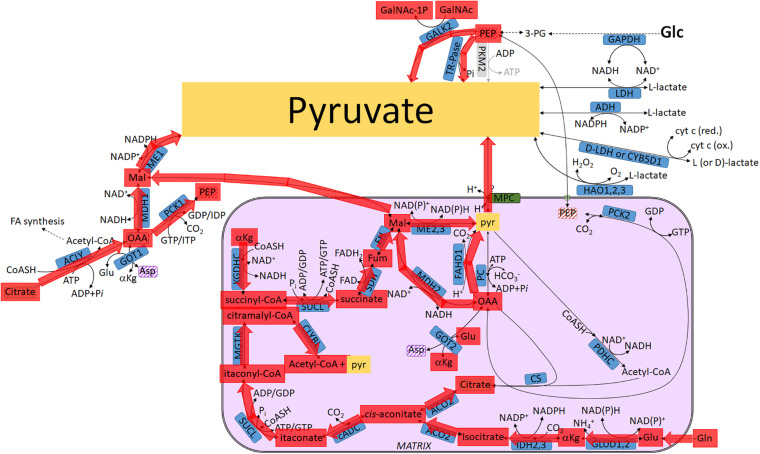
Pathways Leading to Pyruvate Commencing From Glutamine (Glutaminolysis)
It is a well-known fact that most cancer cells grow much better when feeding media contain glutamine; this spurred from the pioneering studies of Eagle et al. (1956), showing the dependence of cancer cells growing in monolayer cultures on glutamine. The many critical roles of glutamine in tumor metabolism is reviewed in Altman et al. (2016). From the energetic point of view it were Reitzer et al. (1979) who first showed that glutamine, not sugars, is the main energy source in cultured HeLa cells and that carbon atoms from glutamine incorporate into lactate, but not more than 13%. Zielke et al. (1980), likewise reported that human diploid fibroblasts metabolize up to 13% of media glutamine to lactate. In the same line of thought, Scott et al. (2011), showed that, in human melanoma cell lines, glutamine did not significantly label lactate, in agreement with the data of Ta and Seyfried (2015) reporting that, in a murine glioblastoma cell line, minimal amounts of lactate derived from glutamine were detected. Le et al. (2012), as well as Son et al. (2013) likewise showed that 13C-labeled atoms in glutamine appear in lactate also to a minimal extent. However, in a study published by DeBerardinis et al. (2007), ∼60% of the glutamine metabolized by SF188 cells was claimed to be converted to lactate, although they seemed to combine this percentage with that of alanine production. The pathway of converting glutamine to pyruvate (and lactate), referred to by McKeehan (1982) as “glutaminolysis,” has been considered a hallmark of tumor metabolism; however, this is a misconception: in normal tissues, ∼18% of glutamine carbons appear in lactate (Windmueller and Spaeth, 1974), as opposed to ∼10–13% (or less) in tumor cells (see the references above). Thus, if anything, cancer cells exhibit a decrease in glutamine-to-lactate conversion exactly as anticipated, mindful that glutamine provides both energy and building blocks for several biosynthetic processes of cancer. Although glutaminolysis was originally attributed to the pathway Gln → Glu → aKg → succinyl-CoA → succinate → fumarate → malate (exiting the mitochondria) → pyruvate (through malic enzyme), several other routes may also contribute (outlined below; see Figure 7).
FIGURE 7.
Pathways leading to pyruvate commencing from glutamine (glutaminolysis), highlighted in red. For abbreviations, see Table 1.
(78) (For the sake of completion, the glutaminolysis pathway proposed by McKeehan (1982) is repeated in the present entry) Gln → Glu → aKg → succinyl-CoA → succinate → fumarate → malate; malate exits the mitochondria → pyruvate; this last step is catalyzed by cytosolic malic enzyme (ME1).
(79) Gln → Glu → aKg → isocitrate → cis-aconitate → itaconate by cADC; itaconate + CoASH + ATP (or GTP) → itaconyl-CoA + Pi + ADP (or GDP) by SUCL (Nemeth et al., 2016); itaconyl-CoA → citramalyl-CoA by methylglutaconase (MGTK); citramalyl-coA → acetyl-CoA + pyruvate by CLYBL (Shen et al., 2017). Pyruvate may exit the mitochondria through the MPC.
(80) Gln → Glu → aKg → isocitrate → cis-aconitate → citrate, exiting the mitochondria → citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY (Chypre et al., 2012); OAA → Mal by MDH1; Mal → pyruvate by ME1.
(81) Gln → Glu → aKg → isocitrate → cis-aconitate → citrate, exiting the mitochondria → citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP + GalNAc → GalNAc-1P + pyruvate. The terminal reaction is catalyzed by N-acetylgalactosamine kinase isoforms 1 or 2.
(82) Gln → Glu → aKg → isocitrate → cis-aconitate → citrate, exiting the mitochondria → citrate + ATP + CoASH → acetyl-coA + ADP + Pi + OAA by ACLY; OAA → PEP by PCK1; PEP → pyruvate; the terminal reaction is catalyzed by tartrate-resistant acid phosphatases.
(83) Gln → Glu → aKg → succinyl-CoA → succinate → fumarate → malate → pyruvate by ME2,3; pyruvate may exit the mitochondria through the MPC.
(84) Gln → Glu → aKg; aKg transaminates with Asp forming Glu and OAA, by GOT2; OAA → pyruvate by FAHD1 (Pircher et al., 2011, 2015); pyruvate may exit the mitochondria through the MPC.
(85) Gln → Glu → aKg; aKg transaminates with Asp forming Glu and OAA, by GOT2; OAA → Mal by MDH2; Mal exits the mitochondria; Mal → pyruvate by ME1 (Zelewski and Swierczynski, 1991; Loeber et al., 1994).
(86) Gln → Glu → aKg; aKg transaminates with Asp forming Glu and OAA, by GOT2; OAA → Mal by MDH2; malate → pyruvate by ME2,3; pyruvate may exit the mitochondria through the MPC.
Energetics of Glycolysis With Kinetically Inactive PK
Glycolysis yields a net of two ATP molecules per glucose molecule; however, in view of an inactive PK while pyruvate is made through PK-bypass pathways, net ATP production from glycolysis is expected to be zero. Although the importance of high-energy phosphate generation has been downplayed in cancer tissues (Vander Heiden et al., 2009), it cannot be ignored that—according to the BRENDA database—among the 336 enzymatic reactions requiring ATP in a cell (without even considering quantitatively important, non-enzymatic mechanisms such as Na+/K+ ATPase), 125 of them occur in the cytosol. Clearly, while it is imperative to prevent phosphofructokinase and hexokinase from ATP-dependent feedback inhibition and allow a high flux of glycolysis for the sake of generating intermediates shuttled toward other pathways, ATP is still needed for many other reactions. Crunching the numbers regarding cytosolic energetics is a daunting task, but what is definite is that a cell with nearly zero ATP production from glycolysis may not harbor ATP-consuming mitochondria, for whatever reason (hypoxia, mtDNA mutations, etc.). This can be solved by maintaining the adenine nucleotide translocase in “forward” mode, i.e., providing ATP to the cytosol which is made by SUCL supported by glutaminolysis (Chinopoulos et al., 2010). Production of pyruvate and, therefore lactate is still maintained by the PK-bypassing pathways so as to thwart a reductive stress as pyruvate-to-lactate by LDH maintains a low NADH/NAD+ ratio. Finally, it is important to emphasize that this lack of ATP generation by glycolysis due to PK inhibition does not only occur in neoplastic tissues, but it seems to be a more general pathophysiological mechanism also present in tissue ischemia: it was recently reported that during acute kidney injury, PK was inhibited by oxidative/nitrosative stress for the purpose of diverting glycolytic intermediates toward the pentose phosphate pathway which, in turn, yielded reducing equivalents and mounted a better response during the reperfusion phase where ROS are formed, thus increasing the chances for organ survival (Zhou et al., 2019).
Conclusion
The above considerations aim to (i) highlight that L-lactate can still be produced from pyruvate using carbon atoms originating from glucose or other substrates in cells with kinetically impaired pyruvate kinase and (ii) show that the mitochondria may contribute to cancer metabolism irrespective of oxidative phosphorylation by providing means of contributing to pyruvate production. Having said that, it is important to emphasize that none of the aforementioned reactions take into account the potential regulatory effects of metabolites on other reactions such as those occurring on PK by amino acids (Chaneton et al., 2012; Yuan et al., 2018). In addition, each enzyme probably exhibits different kinetic and thermodynamic constraints which control the overall flux, which also means that many of these pathways may not operate simultaneously. Such exponentially increasing complexity of a system precludes the possibility of predictions and modeling, though I would be happy to be proven wrong.
Author Contributions
CC wrote and edited the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I am indebted to Dr. Dora Ravasz, Dr. David Bui, Jeonghyoun Lee, and Seung Won Jeong for assistance with database mining and to Prof. Thomas N. Seyfried for helpful discussions.
Funding. This work was supported by grants from NKFIH (FIKP-61822-64888-EATV, VEKOP 2.3.3-15-2016-00012, 2017-2.3.4-TET-RU-2017-00003, KH129567, and K135027) to CC.
References
- Abbadi S., Rodarte J. J., Abutaleb A., Lavell E., Smith C. L., Ruff W., et al. (2014). Glucose-6-phosphatase is a key metabolic regulator of glioblastoma invasion. Mol. Cancer Res. 12 1547–1559. 10.1158/1541-7786.mcr-14-0106-t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yoshikawa N., Sarower M. G., Okada S. (2005). Physiological function and metabolism of free D-alanine in aquatic animals. Biol. Pharm. Bull. 28 1571–1577. 10.1248/bpb.28.1571 [DOI] [PubMed] [Google Scholar]
- Alquraishi M., Puckett D. L., Alani D. S., Humidat A. S., Frankel V. D., Donohoe D. R., et al. (2019). Pyruvate kinase M2: a simple molecule with complex functions. Free Radic. Biol. Med. 143 176–192. 10.1016/j.freeradbiomed.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenberg B., Greulich K. O. (2004). Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84 1014–1020. 10.1016/j.ygeno.2004.08.010 [DOI] [PubMed] [Google Scholar]
- Altman B. J., Stine Z. E., Dang C. V. (2016). From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16 619–634. 10.1038/nrc.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva E. A., Wilkinson A. C. (2018). Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 21 64–70. 10.1097/mco.0000000000000430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., et al. (2011). Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334 1278–1283. 10.1126/science.1211485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Yu Y., Israelsen W. J., Jiang J. K., Boxer M. B., Hong B. S., et al. (2012). Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 8 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antognelli C., Moretti S., Frosini R., Puxeddu E., Sidoni A., Talesa V. N. (2019). Methylglyoxal Acts as a Tumor-Promoting Factor in Anaplastic Thyroid Cancer. Cells 8 547. 10.3390/cells8060547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. R., Cramer S. D., Kennedy M., Assimos D. G., Holmes R. P. (2004). Glycolate and glyoxylate metabolism in HepG2 cells. Am. J. Physiol. Cell Physiol. 287 C1359–C1365. [DOI] [PubMed] [Google Scholar]
- Baranowski T., Wolna E., Morawiecki A. (1968). Purification and properties of crystalline 2-phospho-D-glycerate hydro-lyase from human muscle. Eur. J. Biochem. 5 119–123. 10.1111/j.1432-1033.1968.tb00345.x [DOI] [PubMed] [Google Scholar]
- Bartesaghi S., Graziano V., Galavotti S., Henriquez N. V., Betts J., Saxena J., et al. (2015). Inhibition of oxidative metabolism leads to p53 genetic inactivation and transformation in neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 112 1059–1064. 10.1073/pnas.1413165112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastings J., van Eijk H. M., Olde Damink S. W., Rensen S. S. (2019). d-amino acids in health and disease: a focus on cancer. Nutrients 11:2205. 10.3390/nu11092205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensard C. L., Wisidagama D. R., Olson K. A., Berg J. A., Krah N. M., Schell J. C., et al. (2020). Regulation of tumor initiation by the mitochondrial Pyruvate Carrier. Cell Metab. 31 284–300.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R. K., Oda R. P., Petrikovics I., Thompson D. E., Brenner M., Mahon S. B., et al. (2014). Cyanide toxicokinetics: the behavior of cyanide, thiocyanate and 2-amino-2-thiazoline-4-carboxylic acid in multiple animal models. J. Anal. Toxicol. 38 218–225. 10.1093/jat/bku020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S., Le A. (2018). Glucose metabolism in cancer. Adv. Exp. Med. Biol. 1063 3–12. [DOI] [PubMed] [Google Scholar]
- Bosron W. F., Prairie R. L. (1972). Triphosphopyridine nucleotide-linked aldehyde reductase. I. Purification and properties of the enzyme from pig kidney cortex. J. Biol. Chem. 247 4480–4485. [PubMed] [Google Scholar]
- Brunetti P., Jourdian G. W., Roseman S. (1962). The sialic acids. III. Distribution and properties of animal N-acetylneuraminic aldolase. J. Biol. Chem. 237 2447–2453. [PubMed] [Google Scholar]
- Bukato G., Kochan Z., Swierczynski J. (1995). Purification and properties of cytosolic and mitochondrial malic enzyme isolated from human brain. Int. J. Biochem. Cell Biol. 27 47–54. 10.1016/1357-2725(94)00057-3 [DOI] [PubMed] [Google Scholar]
- Bull H., Murray P. G., Thomas D., Fraser A. M., Nelson P. N. (2002). Acid phosphatases. Mol. Pathol. 55 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack R. (1969). Assay, purification and properties of mammalian D-2-hydroxy acid dehydrogenase. Biochem. J. 115 55–64. 10.1042/bj1150055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack R. (1970). Mammalian D-2-hydroxy acid dehydrogenase. Effect of inhibitors and reaction sequence. Biochem. J. 118 405–408. 10.1042/bj1180405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Durrani F. A., Toth K., Rustum Y. M. (2014). Se-methylselenocysteine offers selective protection against toxicity and potentiates the antitumour activity of anticancer drugs in preclinical animal models. Br. J. Cancer 110 1733–1743. 10.1038/bjc.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capala M. E., Pruis M., Vellenga E., Schuringa J. J. (2016). Depletion of SAM50 Specifically Targets BCR-ABL-expressing leukemic stem and progenitor cells by interfering with mitochondrial functions. Stem Cells Dev. 25 427–437. 10.1089/scd.2015.0151 [DOI] [PubMed] [Google Scholar]
- Carlsen B. D., Lambeth D. O., Ray P. D. (1988). Synthesis of malate from phosphoenolpyruvate by rabbit liver mitochondria: implications for lipogenesis. Biochim. Biophys. Acta 965 1–8. 10.1016/0304-4165(88)90143-2 [DOI] [PubMed] [Google Scholar]
- Carlson G. M., Colombo G., Lardy H. A. (1978). A vicinal dithiol containing an essential cysteine in phosphoenolpyruvate carboxykinase (guanosine triphosphate) from cytosol of rat liver. Biochemistry 17 5329–5338. 10.1021/bi00618a002 [DOI] [PubMed] [Google Scholar]
- Castellano F., Molinier-Frenkel V. (2017). An overview of l-amino acid oxidase functions from bacteria to mammals: focus on the immunoregulatory phenylalanine oxidase IL4I1. Molecules 22:2151. 10.3390/molecules22122151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B., Hillmann P., Zheng L., Martin A. C. L., Maddocks O. D. K., Chokkathukalam A., et al. (2012). Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491 458–462. 10.1038/nature11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T. Y., Yu J. C., Ku C. H., Chen M. M., Lee S. H., Janckila A. J., et al. (2005). Tartrate-resistant acid phosphatase 5b is a useful serum marker for extensive bone metastasis in breast cancer patients. Clin. Cancer Res. 11 544–550. [PubMed] [Google Scholar]
- Chen C. H., Chen S. C. (1988). Evidence of acid phosphatase in the cytoplasm as a distinct entity. Arch. Biochem. Biophys. 262 427–438. 10.1016/0003-9861(88)90394-3 [DOI] [PubMed] [Google Scholar]
- Chen C. M., Chen S. M., Chien P. J., Yu H. Y. (2015). Development of an enzymatic assay system of D-lactate using D-lactate dehydrogenase and a UV-LED fluorescent spectrometer. J. Pharm. Biomed. Anal. 116 150–155. 10.1016/j.jpba.2015.07.018 [DOI] [PubMed] [Google Scholar]
- Chiavarina B., Nokin M. J., Bellier J., Durieux F., Bletard N., Sherer F., et al. (2017). Methylglyoxal-mediated stress correlates with high metabolic activity and promotes tumor growth in colorectal cancer. Int. J. Mol. Sci. 18:213. 10.3390/ijms18010213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiku T., Padovani D., Zhu W., Singh S., Vitvitsky V., Banerjee R. (2009). H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 284 11601–11612. 10.1074/jbc.m808026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinopoulos C., Gerencser A. A., Mandi M., Mathe K., Torocsik B., Doczi J., et al. (2010). Forward operation of adenine nucleotide translocase during F0F1-ATPase reversal: critical role of matrix substrate-level phosphorylation. FASEB J. 24 2405–2416. 10.1096/fj.09-149898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., et al. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452 230–233. 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
- Chypre M., Zaidi N., Smans K. (2012). ATP-citrate lyase: a mini-review. Biochem. Biophys. Res. Commun. 422 1–4. 10.1016/j.bbrc.2012.04.144 [DOI] [PubMed] [Google Scholar]
- Colilla W., Jorgenson R. A., Nordlie R. C. (1975). Mammalian carbamyl phosphate : glucose phosphotransferase and glucose-6-phosphate phosphohydrolase: extended tissue distribution. Biochim. Biophys. Acta 377 117–125. 10.1016/0005-2744(75)90292-2 [DOI] [PubMed] [Google Scholar]
- Collins M. A., Nijm W. P., Borge G. F., Teas G., Goldfarb C. (1979). Dopamine-related tetrahydroisoquinolines: significant urinary excretion by alcoholics after alcohol consumption. Science 206 1184–1186. 10.1126/science.505002 [DOI] [PubMed] [Google Scholar]
- Cooper A. J. (1977). Asparagine transaminase from rat liver. J. Biol. Chem. 252 2032–2038. [PubMed] [Google Scholar]
- Cooper A. J., Krasnikov B. F., Niatsetskaya Z. V., Pinto J. T., Callery P. S., Villar M. T., et al. (2011). Cysteine S-conjugate beta-lyases: important roles in the metabolism of naturally occurring sulfur and selenium-containing compounds, xenobiotics and anticancer agents. Amino Acids 41 7–27. 10.1007/s00726-010-0552-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. J., Kuhara T. (2014). alpha-Ketoglutaramate: an overlooked metabolite of glutamine and a biomarker for hepatic encephalopathy and inborn errors of the urea cycle. Metab. Brain Dis. 29 991–1006. 10.1007/s11011-013-9444-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. J., Pinto J. T. (2006). Cysteine S-conjugate beta-lyases. Amino Acids 30 1–15. 10.1007/s00726-005-0243-4 [DOI] [PubMed] [Google Scholar]
- Cooper A. J., Raps S. P., Meister A. (1987). Fluorometric determination of alpha-ketosuccinamic acid in rat tissues. Anal. Biochem. 167 312–320. 10.1016/0003-2697(87)90170-9 [DOI] [PubMed] [Google Scholar]
- Cooper J. L., Meister A. (1972). Isolation and properties of highly purified glutamine transaminase. Biochemistry 11 661–671. 10.1021/bi00755a001 [DOI] [PubMed] [Google Scholar]
- Copper A. J., Meister A. (1973). Enzymatic conversion of O-carbamyl-L-serine to pyruvate and ammonia. Biochem. Biophys. Res. Commun. 55 780–787. 10.1016/0006-291x(73)91212-6 [DOI] [PubMed] [Google Scholar]
- Cordell P. A., Futers T. S., Grant P. J., Pease R. J. (2004). The Human hydroxyacylglutathione hydrolase (HAGH) gene encodes both cytosolic and mitochondrial forms of glyoxalase II. J. Biol. Chem. 279 28653–28661. 10.1074/jbc.m403470200 [DOI] [PubMed] [Google Scholar]
- Cortes-Cros M., Hemmerlin C., Ferretti S., Zhang J., Gounarides J. S., Yin H., et al. (2013). M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc. Natl. Acad. Sci. U.S.A. 110 489–494. 10.1073/pnas.1212780110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello C., Patriarca E. J., Phang J. M., Minchiotti G. (2020). Proline metabolism in tumor growth and metastatic progression. Front. Oncol. 10:776. 10.3389/fonc.2020.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danpure C. J., Lumb M. J., Birdsey G. M., Zhang X. (2003). Alanine:glyoxylate aminotransferase peroxisome-to-mitochondrion mistargeting in human hereditary kidney stone disease. Biochim. Biophys. Acta 1647 70–75. 10.1016/s1570-9639(03)00055-4 [DOI] [PubMed] [Google Scholar]
- Dawkins P. D., Dickens F. (1965). The Oxidation of D- and L-Glycerate by Rat Liver. Biochem. J. 94 353–367. 10.1042/bj0940353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton T. L., Gocheva V., Miller K. M., Bhutkar A., Lewis C. A., Bronson R. T., et al. (2018). Isoform-specific deletion of PKM2 constrains tumor initiation in a mouse model of soft tissue sarcoma. Cancer Metab. 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton T. L., Gocheva V., Miller K. M., Israelsen W. J., Bhutkar A., Clish C. B., et al. (2016a). Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 30 1020–1033. 10.1101/gad.278549.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton T. L., Jacks T., Vander Heiden M. G. (2016b). PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17 1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz-Lopez K. G., Castro-Munoz L. J., Reyes-Hernandez D. O., Garcia-Carranca A., Manzo-Merino J. (2019). Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front. Oncol. 9:1143. 10.3389/fonc.2019.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi T., Timmermans M. A., Sieuwerts A. M., Smid M., Look M. P., Grebenchtchikov N., et al. (2017). Phosphoserine aminotransferase 1 is associated to poor outcome on tamoxifen therapy in recurrent breast cancer. Sci. Rep. 7:2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7 11–20. 10.1016/j.cmet.2007.10.002 [DOI] [PubMed] [Google Scholar]
- DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., et al. (2007). Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104 19345–19350. 10.1073/pnas.0709747104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P., Baddour J., Muller F., Wu C. C., Wang H., Liao W. T., et al. (2017). Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542 119–123. 10.1038/nature21052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Tu R., Liu H., Qing G. (2020). Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 5:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahota Z., Rauchova H., Mikova M., Kaul P., Bass A. (1983). Phosphoenolpyruvate shuttle–transport of energy from mitochondria to cytosol. FEBS Lett. 157 347–349. 10.1016/0014-5793(83)80573-0 [DOI] [PubMed] [Google Scholar]
- Ducker G. S., Ghergurovich J. M., Mainolfi N., Suri V., Jeong S. K., Hsin-Jung Li S., et al. (2017). Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 114 11404–11409. 10.1073/pnas.1706617114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle H., Oyama V. I., Levy M., Horton C. L., Fleischman R. (1956). The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J. Biol. Chem. 218 607–616. [PubMed] [Google Scholar]
- Elia I., Broekaert D., Christen S., Boon R., Radaelli E., Orth M. F., et al. (2017). Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 8:15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erecinska M., Wilson D. F. (1984). Relationship of the intra- and extramitochondrial adenine nucleotide ratios during synthesis of phosphoenolpyruvate using extramitochondrial ATP. J. Biol. Chem. 259 10904–10906. [PubMed] [Google Scholar]
- Esaki N., Nakamura T., Tanaka H., Soda K. (1982). Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian distribution and purification and properties of pig liver enzyme. J. Biol. Chem. 257 4386–4391. [PubMed] [Google Scholar]
- Ewaschuk J. B., Naylor J. M., Zello G. A. (2005). D-lactate in human and ruminant metabolism. J. Nutr. 135 1619–1625. 10.1093/jn/135.7.1619 [DOI] [PubMed] [Google Scholar]
- Feng H., Wang X., Chen J., Cui J., Gao T., Gao Y., et al. (2019). Nuclear imaging of glucose metabolism: beyond (18)F-FDG. Contrast Media Mol. Imaging 2019:7954854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., Bertos N., Pepin F., Sadekova S., Souleimanova M., Zhao H., et al. (2008). Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14 518–527. [DOI] [PubMed] [Google Scholar]
- Flick M. J., Konieczny S. F. (2002). Identification of putative mammalian D-lactate dehydrogenase enzymes. Biochem. Biophys. Res. Commun. 295 910–916. 10.1016/s0006-291x(02)00768-4 [DOI] [PubMed] [Google Scholar]
- Fong M. Y., McDunn J., Kakar S. S. (2011). Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One 6:e19963. 10.1371/journal.pone.0019963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D. W., Richardson K. E. (1979). Isolation and characterization of glycolic acid oxidase from human liver. Biochim. Biophys. Acta 568 135–144. 10.1016/0005-2744(79)90281-x [DOI] [PubMed] [Google Scholar]
- Fuchs S. A., Berger R., Klomp L. W., de Koning T. J. (2005). D-amino acids in the central nervous system in health and disease. Mol. Genet. Metab. 85 168–180. 10.1016/j.ymgme.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Garber A. J., Ballard F. J. (1970). Regulation of phosphoenolpyruvate metabolism in mitochondria from guinea pig liver. J. Biol. Chem. 245 2229–2240. [PubMed] [Google Scholar]
- Garber A. J., Salganicoff L. (1973). Regulation of oxalacetate metabolism in liver mitochondria. Evidence for nicotinamide adenine dinucleotide-malate dehydrogenase equilibrium and the role of phosphoenolpyruvate carboxykinase in the control of oxalacetate metabolism in intact guinea pig and rat liver mitochondria. J. Biol. Chem. 248 1520–1529. [PubMed] [Google Scholar]
- Garrow T. A., Brenner A. A., Whitehead V. M., Chen X. N., Duncan R. G., Korenberg J. R., et al. (1993). Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J. Biol. Chem. 268 11910–11916. [PubMed] [Google Scholar]
- Gavaret J. M., Nunez J., Cahnmann H. J. (1980). Formation of dehydroalanine residues during thyroid hormone synthesis in thyroglobulin. J. Biol. Chem. 255 5281–5285. [PubMed] [Google Scholar]
- Goto Y., Shimizu J., Shukuya R. (1980). Purification and molecular characteristics of mitochondrial phosphoenolpyruvate carboxykinase from bullfrog (Rana catesbeiana) liver. J. Biochem. 88 1239–1249. 10.1093/oxfordjournals.jbchem.a133092 [DOI] [PubMed] [Google Scholar]
- Guo J. H., Hexige S., Chen L., Zhou G. J., Wang X., Jiang J. M., et al. (2006). Isolation and characterization of the human D-glyceric acidemia related glycerate kinase gene GLYCTK1 and its alternatively splicing variant GLYCTK2. DNA Seq. 17 1–7. 10.1080/10425170500476665 [DOI] [PubMed] [Google Scholar]
- Hall A., Meyle K. D., Lange M. K., Klima M., Sanderhoff M., Dahl C., et al. (2013). Dysfunctional oxidative phosphorylation makes malignant melanoma cells addicted to glycolysis driven by the (V600E)BRAF Oncogene. Oncotarget 4 584–599. 10.18632/oncotarget.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. A., Fenton A. W. (2019). A critical review of the role of M2PYK in the Warburg effect. Biochim. Biophys. Acta Rev. Cancer 1871 225–239. 10.1016/j.bbcan.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman A. R., Warburton M. J., Pringle J. A., Coles B., Chambers T. J. (1989). Purification and characterization of a tartrate-resistant acid phosphatase from human osteoclastomas. Biochem. J. 261 601–609. 10.1042/bj2610601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebda C. A., Nowak T. (1982). The purification, characterization, and activation of phosphoenolpyruvate carboxykinase from chicken liver mitochondria. J. Biol. Chem. 257 5503–5514. [PubMed] [Google Scholar]
- Helwig J. J., Farooqui A. A., Bollack C., Mandel P. (1978). Purification and some properties of tartrate-sensitive acid phosphatase from rabbit kidney cortex. Biochem. J. 175 321–329. 10.1042/bj1750321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis A. L., Lau A. N., Devoe C. X., Dayton T. L., Danai L. V., Di Vizio D., et al. (2018). PKM2 is not required for pancreatic ductal adenocarcinoma. Cancer Metab. 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H., Greenberg D. M. (1967). Studies on phosphoserine aminotransferase of sheep brain. J. Biol. Chem. 242 2283–2287. [PubMed] [Google Scholar]
- Holmes R. P., Assimos D. G. (1998). Glyoxylate synthesis, and its modulation and influence on oxalate synthesis. J. Urol. 160 1617–1624. 10.1097/00005392-199811000-00003 [DOI] [PubMed] [Google Scholar]
- Hong C. S., Graham N. A., Gu W., Espindola Camacho C., Mah V., Maresh E. L., et al. (2016). MCT1 Modulates Cancer Cell Pyruvate Export and Growth of Tumors that Co-express MCT1 and MCT4. Cell Rep. 14 1590–1601. 10.1016/j.celrep.2016.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Hirst J. A., Fujii H. (2007). Regulation of cell proliferation by interleu-induced nuclear translocation of pyruvate kinase. J. Biol. Chem. 282 17706–17711. 10.1074/jbc.m700094200 [DOI] [PubMed] [Google Scholar]
- Hosios A. M., Fiske B. P., Gui D. Y., Vander Heiden M. G. (2015). Lack of Evidence for PKM2 Protein Kinase Activity. Mol. Cell 59 850–857. 10.1016/j.molcel.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. C., Hung W. C. (2018). Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer 17:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lu W., Chen G., Wang P., Chen Z., Zhou Y., et al. (2012). K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 22 399–412. 10.1038/cr.2011.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussien R., Brooks G. A. (2011). Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol. Genomics 43 255–264. 10.1152/physiolgenomics.00177.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icard P., Lincet H. (2012). A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim. Biophys. Acta 1826 423–433. 10.1016/j.bbcan.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Ip C., Hayes C., Budnick R. M., Ganther H. E. (1991). Chemical form of selenium, critical metabolites, and cancer prevention. Cancer Res. 51 595–600. [PubMed] [Google Scholar]
- Israelsen W. J., Dayton T. L., Davidson S. M., Fiske B. P., Hosios A. M., Bellinger G., et al. (2013). PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155 397–409. 10.1016/j.cell.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen W. J., Vander Heiden M. G. (2015). Pyruvate kinase: function, regulation and role in cancer. Semin. Cell Dev. Biol. 43 43–51. 10.1016/j.semcdb.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Ohyama T., Kontani Y., Matslida K., Sakata S. F., Tamaki N. (2001). Influence of dietary protein levels on beta-alanine aminotransferase expression and activity in rats. J. Nutr. Sci. Vitaminol. 47 275–282. 10.3177/jnsv.47.275 [DOI] [PubMed] [Google Scholar]
- Jeske L., Placzek S., Schomburg I., Chang A., Schomburg D. (2019). BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 47 D542–D549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. M., Morrell J. C., Gould S. J. (2000). Identification and characterization of HAOX1, HAOX2, and HAOX3, three human peroxisomal 2-hydroxy acid oxidases. J. Biol. Chem. 275 12590–12597. 10.1074/jbc.275.17.12590 [DOI] [PubMed] [Google Scholar]
- Kakimoto Y., Taniguchi K., Sano I. (1969). D-beta-aminoisobutyrate:pyruvate aminotransferase in mammalian liver and excretion of beta-aminoisobutyrate by man. J. Biol. Chem. 244 335–340. [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashii T., Gomi T., Oya T., Ishii Y., Oda H., Maruyama M., et al. (2005). Some biochemical and histochemical properties of human liver serine dehydratase. Int. J. Biochem. Cell Biol. 37 574–589. 10.1016/j.biocel.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Kim M. J., Whitesides G. M. (1988). L-Lactate dehydrogenase: substrate specificity and use as a catalyst in the synthesis of homochiral 2-hydroxy acids. J. Am. Chem. Soc. 110 2959–2964. 10.1021/ja00217a044 [DOI] [PubMed] [Google Scholar]
- King Z. A., Lu J., Drager A., Miller P., Federowicz S., Lerman J. A., et al. (2016). BiGG Models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44 D515–D522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleineke J., Sauer H., Soling H. D. (1973). On the specificity of the tricarboxylate carrier system in rat liver mitochondria. FEBS Lett. 29 82–86. 10.1016/0014-5793(73)80531-9 [DOI] [PubMed] [Google Scholar]
- Knott S. R. V., Wagenblast E., Khan S., Kim S. Y., Soto M., Wagner M., et al. (2018). Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554 378–381. 10.1038/nature25465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Ogata E. (2002). Bone metabolic markers as gauges of metastasis to bone: a review. Ann. Nucl. Med. 16 161–168. 10.1007/bf02996296 [DOI] [PubMed] [Google Scholar]
- Koseki J., Konno M., Asai A., Colvin H., Kawamoto K., Nishida N., et al. (2018). Enzymes of the one-carbon folate metabolism as anticancer targets predicted by survival rate analysis. Sci. Rep. 8:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall A. S., Xu S., Graeber T. G., Braas D., Christofk H. R. (2016). Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 7:11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H. A., Johnson W. A. (1937). Acetopyruvic acid (alphagamma-diketovaleric acid) as an intermediate metabolite in animal tissues. Biochem. J. 31 772–779. 10.1042/bj0310772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht J. A., Flynn J. M., Downs D. M. (2012). Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5’-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 287 3454–3461. 10.1074/jbc.m111.304477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J. N., Wood K. D., Knight J., Assimos D. G., Holmes R. P. (2012). Glyoxal formation and its role in endogenous oxalate synthesis. Adv. Urol. 2012:819202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. N., Israelsen W. J., Roper J., Sinnamon M. J., Georgeon L., Dayton T. L., et al. (2017). PKM2 is not required for colon cancer initiated by APC loss. Cancer Metab. 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., et al. (2012). Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15 110–121. 10.1016/j.cmet.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. S., Muragaki Y., Ideguchi T., Hase T., Tsuji M., Ooshima A., et al. (1995). Molecular cloning and sequencing of a cDNA encoding alanine-glyoxylate aminotransferase 2 from rat kidney. J. Biochem. 117 856–862. 10.1093/oxfordjournals.jbchem.a124787 [DOI] [PubMed] [Google Scholar]
- Leithner K., Hrzenjak A., Trotzmuller M., Moustafa T., Kofeler H. C., Wohlkoenig C., et al. (2015). PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene 34 1044–1050. 10.1038/onc.2014.47 [DOI] [PubMed] [Google Scholar]
- Li X., Peng S. (2013). Identification of metastasis-associated genes in colorectal cancer through an integrated genomic and transcriptomic analysis. Chin. J. Cancer Res. 25 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Li X. F., Liu J. T., Wang H., Fan L. L., Li J., et al. (2018). PKM2, a potential target for regulating cancer. Gene 668 48–53. 10.1016/j.gene.2018.05.038 [DOI] [PubMed] [Google Scholar]
- Li Z., Yang P., Li Z. (2014). The multifaceted regulation and functions of PKM2 in tumor progression. Biochim. Biophys. Acta 1846 285–296. 10.1016/j.bbcan.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Lieu E. L., Nguyen T., Rhyne S., Kim J. (2020). Amino acids in cancer. Exp. Mol. Med. 52 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Jia Y., Cao Y., Wu S., Jiang H., Sun X., et al. (2016). Overexpression of Phosphoserine Aminotransferase 1 (PSAT1) Predicts Poor Prognosis and Associates with Tumor Progression in Human Esophageal Squamous Cell Carcinoma. Cell. Physiol. Biochem. 39 395–406. 10.1159/000445633 [DOI] [PubMed] [Google Scholar]
- Liu C., Cao J., Lin S., Zhao Y., Zhu M., Tao Z., et al. (2020). Malic enzyme 1 indicates worse prognosis in breast cancer and promotes metastasis by manipulating reactive oxygen species. Onco Targets Ther. 13 8735–8747. 10.2147/ott.s256970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Ge X., Ding H., Jiang H., Christensen B. M., Li J. (2012). Role of glutamate decarboxylase-like protein 1 (GADL1) in taurine biosynthesis. J. Biol. Chem. 287 40898–40906. 10.1074/jbc.m112.393728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu V. M., Howell A. J., Hosios A. M., Li Z., Israelsen W. J., Vander Heiden M. G. (2020). Cancer-associated mutations in human pyruvate kinase M2 impair enzyme activity. FEBS Lett. 594 646–664. 10.1002/1873-3468.13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber G., Dworkin M. B., Infante A., Ahorn H. (1994). Characterization of cytosolic malic enzyme in human tumor cells. FEBS Lett. 344 181–186. 10.1016/0014-5793(94)00386-6 [DOI] [PubMed] [Google Scholar]
- Lunt S. Y., Muralidhar V., Hosios A. M., Israelsen W. J., Gui D. Y., Newhouse L., et al. (2015). Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57 95–107. 10.1016/j.molcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Hu H., Chang R., Zhong J., Knabel M., O’Meally R., et al. (2011). Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145 732–744. 10.1016/j.cell.2011.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., et al. (2011). Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42 719–730. 10.1016/j.molcel.2011.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen A. L., Nowak T. (1983). 3-Mercaptopicolinate. A reversible active site inhibitor of avian liver phosphoenolpyruvate carboxykinase. J. Biol. Chem. 258 11654–11662. [PubMed] [Google Scholar]
- Matoba S., Kang J. G., Patino W. D., Wragg A., Boehm M., Gavrilova O., et al. (2006). p53 regulates mitochondrial respiration. Science 312 1650–1653. 10.1126/science.1126863 [DOI] [PubMed] [Google Scholar]
- Maul D. M., Schuster S. M. (1986). Kinetic properties and characteristics of mouse liver mitochondrial asparagine aminotransferase. Arch. Biochem. Biophys. 251 585–593. 10.1016/0003-9861(86)90367-x [DOI] [PubMed] [Google Scholar]
- Mayers J. R., Torrence M. E., Danai L. V., Papagiannakopoulos T., Davidson S. M., Bauer M. R., et al. (2016). Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353 1161–1165. 10.1126/science.aaf5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S., Boschek C. B., Hugo F., Eigenbrodt E. (2005). Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 15 300–308. 10.1016/j.semcancer.2005.04.009 [DOI] [PubMed] [Google Scholar]
- McKeehan W. L. (1982). Glycolysis, glutaminolysis and cell proliferation. Cell Biol. Int. Rep. 6 635–650. 10.1016/0309-1651(82)90125-4 [DOI] [PubMed] [Google Scholar]
- Mdluli K., Booth M. P., Brady R. L., Rumsby G. (2005). A preliminary account of the properties of recombinant human Glyoxylate reductase (GRHPR), LDHA and LDHB with glyoxylate, and their potential roles in its metabolism. Biochim. Biophys. Acta 1753 209–216. 10.1016/j.bbapap.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Medina D., Thompson H., Ganther H., Ip C. (2001). Se-methylselenocysteine: a new compound for chemoprevention of breast cancer. Nutr. Cancer 40 12–17. 10.4324/9781410608000-4 [DOI] [PubMed] [Google Scholar]
- Metcalf S., Dougherty S., Kruer T., Hasan N., Biyik-Sit R., Reynolds L., et al. (2020). Selective loss of phosphoserine aminotransferase 1 (PSAT1) suppresses migration, invasion, and experimental metastasis in triple negative breast cancer. Clin. Exp. Metastasis 37 187–197. 10.1007/s10585-019-10000-7 [DOI] [PubMed] [Google Scholar]
- Michelakis E. D., Webster L., Mackey J. R. (2008). Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br. J. Cancer 99 989–994. 10.1038/sj.bjc.6604554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E. L., Kelly B., Logan A., Costa A. S. H., Varma M., Bryant C. E., et al. (2016). Succinate Dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167 457–470.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minatogawa Y., Noguchi T., Kido R. (1977). Species distribution and properties of hepatic phenylalanine (histidine):pyruvate aminotransferase. Hoppe Seylers Z. Physiol. Chem. 358 59–67. 10.1515/bchm2.1977.358.1.59 [DOI] [PubMed] [Google Scholar]
- Moreno-Sanchez R., Marin-Hernandez A., Gallardo-Perez J. C., Pacheco-Velazquez S. C., Robledo-Cadena D. X., Padilla-Flores J. A., et al. (2020). Physiological Role of Glutamate Dehydrogenase in Cancer Cells. Front. Oncol. 10:429. 10.3389/fonc.2020.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S., Martin O., Van Du Tran T., Bridge A., Morgat A., Pagni M. (2016). MetaNetX/MNXref–reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 44 D523–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mose S., Menzel C., Kurth A. A., Obert K., Breidert I., Borowsky K., et al. (2003). Tartrate-resistant acid phosphatase 5b as serum marker of bone metabolism in cancer patients. Anticancer Res. 23 2783–2788. [PubMed] [Google Scholar]
- Murai S., Ando A., Ebara S., Hirayama M., Satomi Y., Hara T. (2017). Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis 6:e329. 10.1038/oncsis.2017.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Midorikawa K., Kawanishi S. (2016). “Chapter 25 - Molecular Link Between Alcohol and Breast Cancer: the Role of Salsolinol,” in Molecular Aspects of Alcohol and Nutrition, ed. Patel V. B. (San Diego, CA: Academic Press; ), 315–324. 10.1016/b978-0-12-800773-0.00025-2 [DOI] [Google Scholar]
- Nagahara N., Sawada N. (2006). The mercaptopyruvate pathway in cysteine catabolism: a physiologic role and related disease of the multifunctional 3-mercaptopyruvate sulfurtransferase. Curr. Med. Chem. 13 1219–1230. 10.2174/092986706776360914 [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Ishii T., Kumagai H., Yamada H. (1985). D-Cysteine desulfhydrase of Escherichia coli. Purification and characterization. Eur. J. Biochem. 153 541–551. 10.1111/j.1432-1033.1985.tb09335.x [DOI] [PubMed] [Google Scholar]
- Nagata Y., Masui R., Akino T. (1992). The presence of free D-serine, D-alanine and D-proline in human plasma. Experientia 48 986–988. [DOI] [PubMed] [Google Scholar]
- Nagayama H., Muramatsu M., Shimura K. (1958). Enzymatic formation of aminomalonic acid from ketomalonic acid. Nature 181 417–418. 10.1038/181417a0 [DOI] [PubMed] [Google Scholar]
- Nakano M., Tsutsumi Y., Danowski T. S. (1967). Crystalline L-amino-acid oxidase from the soluble fraction of rat-kidney cells. Biochim. Biophys. Acta 139 40–48. 10.1016/0005-2744(67)90111-8 [DOI] [PubMed] [Google Scholar]
- Nakashima C., Kirita T., Yamamoto K., Mori S., Luo Y., Sasaki T., et al. (2020). Malic enzyme 1 is associated with tumor budding in oral squamous cell carcinomas. Int. J. Mol. Sci. 21:7149. 10.3390/ijms21197149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth B., Doczi J., Csete D., Kacso G., Ravasz D., Adams D., et al. (2016). Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. FASEB J. 30 286–300. 10.1096/fj.15-279398 [DOI] [PubMed] [Google Scholar]
- Newman A. C., Maddocks O. D. K. (2017). Serine and functional metabolites in cancer. Trends Cell Biol. 27 645–657. 10.1016/j.tcb.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Nguyen M., Bonneterre J., Hecquet B., Desoize B., Demaille A. (1991). Plasma acid and alkaline phosphatase in patients with breast cancer. Anticancer Res. 11 831–833. [PubMed] [Google Scholar]
- Nicolay B. N., Danielian P. S., Kottakis F., Lapek J. D., Jr., Sanidas I., Miles W. O., et al. (2015). Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev. 29 1875–1889. 10.1101/gad.264127.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokin M. J., Bellier J., Durieux F., Peulen O., Rademaker G., Gabriel M., et al. (2019). Methylglyoxal, a glycolysis metabolite, triggers metastasis through MEK/ERK/SMAD1 pathway activation in breast cancer. Breast Cancer Res. 21:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlie R. C. (1974). Metabolic regulation by multifunctional glucose-6-phosphatase. Curr. Top. Cell. Regul. 8 33–117. 10.1016/b978-0-12-152808-9.50009-2 [DOI] [PubMed] [Google Scholar]
- Noronha A., Modamio J., Jarosz Y., Guerard E., Sompairac N., Preciat G., et al. (2019). The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 47 D614–D624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Gomi T., Nishizawa M., Hayakawa Y., Endo S., Hayashi K., et al. (2006). Enzymatic and biochemical properties of a novel human serine dehydratase isoform. Biochim. Biophys. Acta 1764 961–971. 10.1016/j.bbapap.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Omura H., Shimamura M., Yamafuji K. (1956). Measurement of transoximase action. Enzymologia 17 359–362. [PubMed] [Google Scholar]
- Orywal K., Szmitkowski M. (2017). Alcohol dehydrogenase and aldehyde dehydrogenase in malignant neoplasms. Clin. Exp. Med. 17 131–139. 10.1007/s10238-016-0408-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallini R., Guazzi G. C., Cannella C., Cacace M. G. (1991). Cloning and sequence analysis of the human liver rhodanese: comparison with the bovine and chicken enzymes. Biochem. Biophys. Res. Commun. 180 887–893. 10.1016/s0006-291x(05)81148-9 [DOI] [PubMed] [Google Scholar]
- Panosyan E. H., Wang Y., Xia P., Lee W. N., Pak Y., Laks D. R., et al. (2014). Asparagine depletion potentiates the cytotoxic effect of chemotherapy against brain tumors. Mol. Cancer Res. 12 694–702. 10.1158/1541-7786.mcr-13-0576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone A., Marani M., Fiascarelli A., Rinaldo S., Giardina G., Contestabile R., et al. (2014). SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 5:e1525. 10.1038/cddis.2014.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarella S., Atlante A., Valenti D., de Bari L. (2003). The role of mitochondrial transport in energy metabolism. Mitochondrion 2 319–343. 10.1016/s1567-7249(03)00008-4 [DOI] [PubMed] [Google Scholar]
- Pastuszak I., Drake R., Elbein A. D. (1996). Kidney N-acetylgalactosamine (GalNAc)-1-phosphate kinase, a new pathway of GalNAc activation. J. Biol. Chem. 271 20776–20782. 10.1074/jbc.271.34.20776 [DOI] [PubMed] [Google Scholar]
- Petit M., Koziel R., Etemad S., Pircher H., Jansen-Durr P. (2017). Depletion of oxaloacetate decarboxylase FAHD1 inhibits mitochondrial electron transport and induces cellular senescence in human endothelial cells. Exp. Gerontol. 92 7–12. 10.1016/j.exger.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Phang J. M. (2019). Proline metabolism in cell regulation and cancer biology: recent advances and hypotheses. Antioxid. Redox Signal. 30 635–649. 10.1089/ars.2017.7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinthong C., Maenpuen S., Amornwatcharapong W., Yuthavong Y., Leartsakulpanich U., Chaiyen P. (2014). Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: implications for selective inhibition. FEBS J. 281 2570–2583. 10.1111/febs.12803 [DOI] [PubMed] [Google Scholar]
- Pircher H., Straganz G. D., Ehehalt D., Morrow G., Tanguay R. M., Jansen-Durr P. (2011). Identification of human fumarylacetoacetate hydrolase domain-containing protein 1 (FAHD1) as a novel mitochondrial acylpyruvase. J. Biol. Chem. 286 36500–36508. 10.1074/jbc.m111.264770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H., von Grafenstein S., Diener T., Metzger C., Albertini E., Taferner A., et al. (2015). Identification of FAH domain-containing protein 1 (FAHD1) as oxaloacetate decarboxylase. J. Biol. Chem. 290 6755–6762. 10.1074/jbc.m114.609305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore R. E., Hurst C. H., Assimos D. G., Holmes R. P. (1997). Pathways of hepatic oxalate synthesis and their regulation. Am. J. Physiol. 272 C289–C294. [DOI] [PubMed] [Google Scholar]
- Prakasam G., Iqbal M. A., Bamezai R. N. K., Mazurek S. (2018). Posttranslational Modifications of Pyruvate Kinase M2: tweaks that benefit cancer. Front. Oncol. 8:22. 10.3389/fonc.2018.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Wice B. M., Kennell D. (1979). Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 254 2669–2676. [PubMed] [Google Scholar]
- Ren J. G., Seth P., Clish C. B., Lorkiewicz P. K., Higashi R. M., Lane A. N., et al. (2014). Knockdown of malic enzyme 2 suppresses lung tumor growth, induces differentiation and impacts PI3K/AKT signaling. Sci. Rep. 4:5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richarme G., Dairou J. (2017). Parkinsonism-associated protein DJ-1 is a bona fide deglycase. Biochem. Biophys. Res. Commun. 483 387–391. 10.1016/j.bbrc.2016.12.134 [DOI] [PubMed] [Google Scholar]
- Richarme G., Mihoub M., Dairou J., Bui L. C., Leger T., Lamouri A. (2015). Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 290 1885–1897. 10.1074/jbc.m114.597815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. H. (1971). Transport of phosphoenolpyruvate by the tricarboxylate transporting system in mammalian mitochondria. FEBS Lett. 14 309–312. 10.1016/0014-5793(71)80287-9 [DOI] [PubMed] [Google Scholar]
- Robinson J. L., Kocabas P., Wang H., Cholley P. E., Cook D., Nilsson A., et al. (2020). An atlas of human metabolism. Sci. Signal. 13:eaaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowsell E. V. (1956). Transaminations with pyruvate and other alpha-keto acids. Biochem. J. 64 246–252. 10.1042/bj0640246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler M., Carter S. B., Hunter K. R., Stern G. M. (1973). Tetrahydroisoquinoline alkaloids: in vivo metabolites of L-dopa in man. Nature 241 439–443. 10.1038/241439a0 [DOI] [PubMed] [Google Scholar]
- Satrustegui J., Pardo B., Del Arco A. (2007). Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol. Rev. 87 29–67. 10.1152/physrev.00005.2006 [DOI] [PubMed] [Google Scholar]
- Sauer L. A. (1973). An NAD- and NADP-dependent malic enzyme with regulatory properties in rat liver and adrenal cortex mitochondrial fractions. Biochem. Biophys. Res. Commun. 50 524–531. 10.1016/0006-291x(73)90871-1 [DOI] [PubMed] [Google Scholar]
- Schadewaldt P., Wendel U., Hammen H. W. (1995). Human branched-chain L-amino acid aminotransferase: activity and subcellular localization in cultured skin fibroblasts. Amino Acids 9 147–160. [DOI] [PubMed] [Google Scholar]
- Scott D. A., Richardson A. D., Filipp F. V., Knutzen C. A., Chiang G. G., Ronai Z. A., et al. (2011). Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J. Biol. Chem. 286 42626–42634. 10.1074/jbc.m111.282046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpa J. (2020). Cysteine as a Carbon Source, a Hot Spot in Cancer Cells Survival. Front. Oncol. 10:947. 10.3389/fonc.2020.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Gao Y., Jin W., Fan M., Wang Y., Gu Y., et al. (2019). Targeting HIBCH to reprogram valine metabolism for the treatment of colorectal cancer. Cell Death Dis. 10 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. A., Clark V. M., Curthoys N. P. (1979). Inactivation of rat renal phosphate-dependent glutaminase with 6-diazo-5-oxo-L-norleucine. Evidence for interaction at the glutamine binding site. J. Biol. Chem. 254 2835–2838. [PubMed] [Google Scholar]
- Shen H., Campanello G. C., Flicker D., Grabarek Z., Hu J., Luo C., et al. (2017). The human knockout gene CLYBL connects Itaconate to Vitamin B12. Cell 171 771–782.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N., Koike S., Tanaka M., Ishigami-Yuasa M., Kimura Y., Ogasawara Y., et al. (2013). A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat. Commun. 4:1366. [DOI] [PubMed] [Google Scholar]
- Shug A. L., Shrago E. (1973). Inhibition of phosphoenolpyruvate transport via the tricarboxylate and adenine nucleotide carrier systems of rat liver mitochondria. Biochem. Biophys. Res. Commun. 53 659–665. 10.1016/0006-291x(73)90712-2 [DOI] [PubMed] [Google Scholar]
- Silverstein E. (1974). Equilibrium kinetic study of bovine liver glutamate dehydrogenase at high pH. Biochemistry 13 3750–3754. 10.1021/bi00715a021 [DOI] [PubMed] [Google Scholar]
- Smith A. L., Whitehall J. C., Bradshaw C., Gay D., Robertson F., Blain A. P., et al. (2020). Age-associated mitochondrial DNA mutations cause metabolic remodelling that contributes to accelerated intestinal tumorigenesis. Nat. Cancer 1 976–989. 10.1038/s43018-020-00112-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M., Uslaner J. M., Yao L., Mullins C. M., Surles N. O., Huszar S. L., et al. (2009). The behavioral and neurochemical effects of a novel D-amino acid oxidase inhibitor compound 8 [4H-thieno [3,2-b]pyrrole-5-carboxylic acid] and D-serine. J. Pharmacol. Exp. Ther. 328 921–930. 10.1124/jpet.108.147884 [DOI] [PubMed] [Google Scholar]
- Snell K., Natsumeda Y., Eble J. N., Glover J. L., Weber G. (1988). Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br. J. Cancer 57 87–90. 10.1038/bjc.1988.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda K., Osumi T. (1969). Crystalline amino acid racemase with low substrate specificity. Biochem. Biophys. Res. Commun. 35 363–368. 10.1016/0006-291x(69)90507-5 [DOI] [PubMed] [Google Scholar]
- Soling H. D., Walter U., Sauer H., Kleineke J. (1971). Effects of synthetic analogues of phosphoenolpyruvate on muscle and liver pyruvate kinase, muscle enolase, liver phosphoenolpyruvate carboxykinase and on the intra-/extra-mitochondrial tricarboxylic acid carrier transport system. FEBS Lett. 19 139–143. 10.1016/0014-5793(71)80498-2 [DOI] [PubMed] [Google Scholar]
- Son J., Lyssiotis C. A., Ying H., Wang X., Hua S., Ligorio M., et al. (2013). Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496 101–105. 10.1038/nature12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Pasquel F., Turcu A., Pongratz R. L., Roden M., Cline G. W., et al. (2009). Phosphoenolpyruvate cycling via mitochondrial phosphoenolpyruvate carboxykinase links anaplerosis and mitochondrial GTP with insulin secretion. J. Biol. Chem. 284 26578–26590. 10.1074/jbc.m109.011775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A., Veress R., Ovadi J., Csermely P., Keri G., Ullrich A. (2007). Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 67 1602–1608. 10.1158/0008-5472.can-06-2870 [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H. (1979). Effect of excess dietary methionine on the catabolism of cysteine in rats. J. Nutr. 109 2126–2139. 10.1093/jn/109.12.2126 [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H. (2020). Metabolism of Sulfur-Containing Amino Acids: how the Body Copes with Excess Methionine, Cysteine, and Sulfide. J. Nutr. 150(Suppl. 1), 2494S–2505S. [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H., Dominy J. E., Jr., Lee J. I., Coloso R. M. (2006). Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 136(6 Suppl.), 1652S–1659S. [DOI] [PubMed] [Google Scholar]
- Sul H. S., Shrago E., Shug A. L. (1976). Relationship of phosphoenolpyruvate transport, acyl coenzyme A inhibition of adenine nucleotide translocase and calcium ion efflux in guinea pig heart mitochondria. Arch. Biochem. Biophys. 172 230–237. 10.1016/0003-9861(76)90071-0 [DOI] [PubMed] [Google Scholar]
- Sun Y., Li W., Shen S., Yang X., Lu B., Zhang X., et al. (2019). Loss of alanine-glyoxylate and serine-pyruvate aminotransferase expression accelerated the progression of hepatocellular carcinoma and predicted poor prognosis. J. Transl. Med. 17:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swick R. W., Wood H. G. (1960). The Role of Transcarboxylation in Propionic Acid Fermentation. Proc. Natl. Acad. Sci. U.S.A. 46 28–41. 10.1073/pnas.46.1.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta N. L., Seyfried T. N. (2015). Influence of Serum and Hypoxia on Incorporation of [(14)C]-D-Glucose or [(14)C]-L-Glutamine into Lipids and Lactate in Murine Glioblastoma Cells. Lipids 50 1167–1184. 10.1007/s11745-015-4075-z [DOI] [PubMed] [Google Scholar]
- Tech K., Tikunov A. P., Farooq H., Morrissy A. S., Meidinger J., Fish T., et al. (2017). Pyruvate Kinase Inhibits Proliferation during Postnatal Cerebellar Neurogenesis and Suppresses Medulloblastoma Formation. Cancer Res. 77 3217–3230. 10.1158/0008-5472.can-16-3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., de la Fuente J., Szydlo R., Hatjiharissi E., Viniou N., Meletis J., et al. (2003). Tartrate-resistant acid phosphatase isoform 5b: a novel serum marker for monitoring bone disease in multiple myeloma. Int. J. Cancer 106 455–457. 10.1002/ijc.11247 [DOI] [PubMed] [Google Scholar]
- Thakur A., Bollig A., Wu J., Liao D. J. (2008). Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol. Cancer 7:11. 10.1186/1476-4598-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269 1–11. 10.1042/bj2690001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun Z. Y., Possemato R. (2015). Amino acid management in cancer. Semin. Cell Dev. Biol. 43 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudball N., Thomas P. (1972). The enzymic degradation of L-serine O-sulphate, Mechanism of the reaction. Biochem. J. 128 41–46. 10.1042/bj1280041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324 1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., Locasale J. W., Swanson K. D., Sharfi H., Heffron G. J., Amador-Noguez D., et al. (2010). Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329 1492–1499. 10.1126/science.1188015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Jagt D. L., Hunsaker L. A. (2003). Methylglyoxal metabolism and diabetic complications: roles of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase and 2-oxoaldehyde dehydrogenase. Chem. Biol. Interact. 14 341–351. 10.1016/s0009-2797(02)00212-0 [DOI] [PubMed] [Google Scholar]
- Vasseur S., Afzal S., Tardivel-Lacombe J., Park D. S., Iovanna J. L., Mak T. W. (2009). DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc. Natl. Acad. Sci. U.S.A. 106 1111–1116. 10.1073/pnas.0812745106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud C., Pietrancosta N., Williams E. L., Rumsby G., Lederer F. (2007). Purification and characterization of recombinant human liver glycolate oxidase. Arch. Biochem. Biophys. 465 410–416. 10.1016/j.abb.2007.06.021 [DOI] [PubMed] [Google Scholar]
- Vincent E. E., Sergushichev A., Griss T., Gingras M. C., Samborska B., Ntimbane T., et al. (2015). Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60 195–207. 10.1016/j.molcel.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Huang J. T., Chen W. L., Wang R. H., Kao M. C., Pan Y. R., et al. (2019). Dysregulation of cystathionine gamma-lyase promotes prostate cancer progression and metastasis. EMBO Rep. 20:e45986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Israelsen W. J., Lee D., Yu V. W. C., Jeanson N. T., Clish C. B., et al. (2014). Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158 1309–1323. 10.1016/j.cell.2014.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L. (1986). Sialic acid lyase in human promyelocytic leukemic cells (HL-60) during phorbol-ester-induced differentiation. Biochim. Biophys. Acta 888 278–281. 10.1016/0167-4889(86)90226-0 [DOI] [PubMed] [Google Scholar]
- Weber G. (1969). Inhibition of human brain pyruvate kinase and hexokinase by phenylalanine and phenylpyruvate: possible relevance to phenylketonuric brain damage. Proc. Natl. Acad. Sci. U.S.A. 63 1365–1369. 10.1073/pnas.63.4.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., Cantero A. (1955). Glucose-6-phosphatase activity in normal, pre-cancerous, and neoplastic tissues. Cancer Res. 15 105–108. [PubMed] [Google Scholar]
- Wiese T. J., Wuensch S. A., Ray P. D. (1996). Synthesis of citrate from phosphoenolpyruvate and acetylcarnitine by mitochondria from rabbit, pigeon and rat liver: implications for lipogenesis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 114 417–422. 10.1016/0305-0491(96)00035-1 [DOI] [PubMed] [Google Scholar]
- Wilson D. F., Erecinska M., Schramm V. L. (1983). Evaluation of the relationship between the intra- and extramitochondrial [ATP]/[ADP] ratios using phosphoenolpyruvate carboxykinase. J. Biol. Chem. 258 10464–10473. [PubMed] [Google Scholar]
- Windmueller H. G., Spaeth A. E. (1974). Uptake and metabolism of plasma glutamine by the small intestine. J. Biol. Chem. 249 5070–5079. [PubMed] [Google Scholar]
- Wishart D. S., Feunang Y. D., Marcu A., Guo A. C., Liang K., Vazquez-Fresno R., et al. (2018). HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 46 D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N., Ojo D., Yan J., Tang D. (2015). PKM2 contributes to cancer metabolism. Cancer Lett. 356 184–191. 10.1016/j.canlet.2014.01.031 [DOI] [PubMed] [Google Scholar]
- Wuensch S. A., Ray P. D. (1997). Synthesis of citrate from phosphoenolpyruvate and acetylcarnitine by mitochondria from rabbit enterocytes: implications for lipogenesis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 118 599–605. 10.1016/s0305-0491(97)00242-3 [DOI] [PubMed] [Google Scholar]
- Yamada K., Noguchi T. (1999). Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem. J. 337(Pt 1), 1–11. 10.1042/0264-6021:3370001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jia X. (2014). Safety evaluation of Se-methylselenocysteine as nutritional selenium supplement: acute toxicity, genotoxicity and subchronic toxicity. Regul. Toxicol. Pharmacol. 70 720–727. 10.1016/j.yrtph.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Yang M., Vousden K. H. (2016). Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16 650–662. 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- Yang W., Lu Z. (2013). Nuclear PKM2 regulates the Warburg effect. Cell Cycle 12 3154–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Lu Z. (2015). Pyruvate kinase M2 at a glance. J. Cell Sci. 128 1655–1660. 10.1242/jcs.166629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., et al. (2012). ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 14 1295–1304. 10.1038/ncb2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D., Guan K. L., Xiong Y. (2018). Metabolism, Activity, and Targeting of D- and L-2-Hydroxyglutarates. Trends Cancer 4 151–165. 10.1016/j.trecan.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Kim J. G., Seo A. N., Park S. Y., Kim H. J., Park J. S., et al. (2015). Clinical implication of serine metabolism-associated enzymes in colon cancer. Oncology 89 351–359. 10.1159/000439571 [DOI] [PubMed] [Google Scholar]
- You J., Shi X., Liang H., Ye J., Wang L., Han H., et al. (2017). Cystathionine- gamma-lyase promotes process of breast cancer in association with STAT3 signaling pathway. Oncotarget 8 65677–65686. 10.18632/oncotarget.20057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Teoh S. T., Ensink E., Ogrodzinski M. P., Yang C., Vazquez A. I., et al. (2019). Cysteine catabolism and the serine biosynthesis pathway support pyruvate production during pyruvate kinase knockdown in pancreatic cancer cells. Cancer Metab. 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. H., Chang Y. C., Su T. H., Nong J. Y., Li C. C., Chuang L. M. (2013). Prostaglandin reductase-3 negatively modulates adipogenesis through regulation of PPARgamma activity. J. Lipid Res. 54 2391–2399. 10.1194/jlr.m037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., McNae I. W., Chen Y., Blackburn E. A., Wear M. A., Michels P. A. M., et al. (2018). An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor. Biochem. J. 475 1821–1837. 10.1042/bcj20180171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C. A., Asuthkar S., Guda M. R., Tsung A. J., Velpula K. K. (2016). Cancer stem cell molecular reprogramming of the Warburg effect in glioblastomas: a new target gleaned from an old concept. CNS Oncol. 5 101–108. 10.2217/cns-2015-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan Q., Hart G. W. (2010). The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123 13–22. 10.1242/jcs.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelewski M., Swierczynski J. (1991). Malic enzyme in human liver. Intracellular distribution, purification and properties of cytosolic isozyme. Eur. J. Biochem. 201 339–345. 10.1111/j.1432-1033.1991.tb16291.x [DOI] [PubMed] [Google Scholar]
- Zhang Z., Deng X., Liu Y., Liu Y., Sun L., Chen F. (2019). PKM2, function and expression and regulation. Cell Biosci. 9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li J., Fan T. W. M., Hou S. X. (2017). Glycolytic reprogramming through PCK2 regulates tumor initiation of prostate cancer cells. Oncotarget 8 83602–83618. 10.18632/oncotarget.18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. L., Zhang R., Anand P., Stomberski C. T., Qian Z., Hausladen A., et al. (2019). Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 565 96–100. 10.1038/s41586-018-0749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Kachhap S., Singh K. K. (2003). Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis 18 287–292. 10.1093/mutage/18.3.287 [DOI] [PubMed] [Google Scholar]
- Zielke H. R., Sumbilla C. M., Sevdalian D. A., Hawkins R. L., Ozand P. T. (1980). Lactate: a major product of glutamine metabolism by human diploid fibroblasts. J. Cell. Physiol. 104 433–441. 10.1002/jcp.1041040316 [DOI] [PubMed] [Google Scholar]