Abstract
Pyrroloquinoline quinone (PQQ) is a peptide-derived redox cofactor produced by prokaryotes that also plays beneficial roles in organisms from other kingdoms. We review recent developments on the pathway of PQQ biogenesis, focusing on the mechanisms of PqqE, PqqF/G, and PqqB. These advances may shed light on other, uncharacterized biosynthetic pathways.
Introduction
The 1960s and 1970s saw the emergence of bacterial glucose and alcohol dehydrogenases that act independently of nicotinamide or flavin-based cofactors, showing features of a novel quinone-like prosthetic group instead.[1–4] This “mystery cofactor” was later characterized by X-ray crystallography to be PQQ, also referred to as methoxatin (Figure 1A).[5,6]
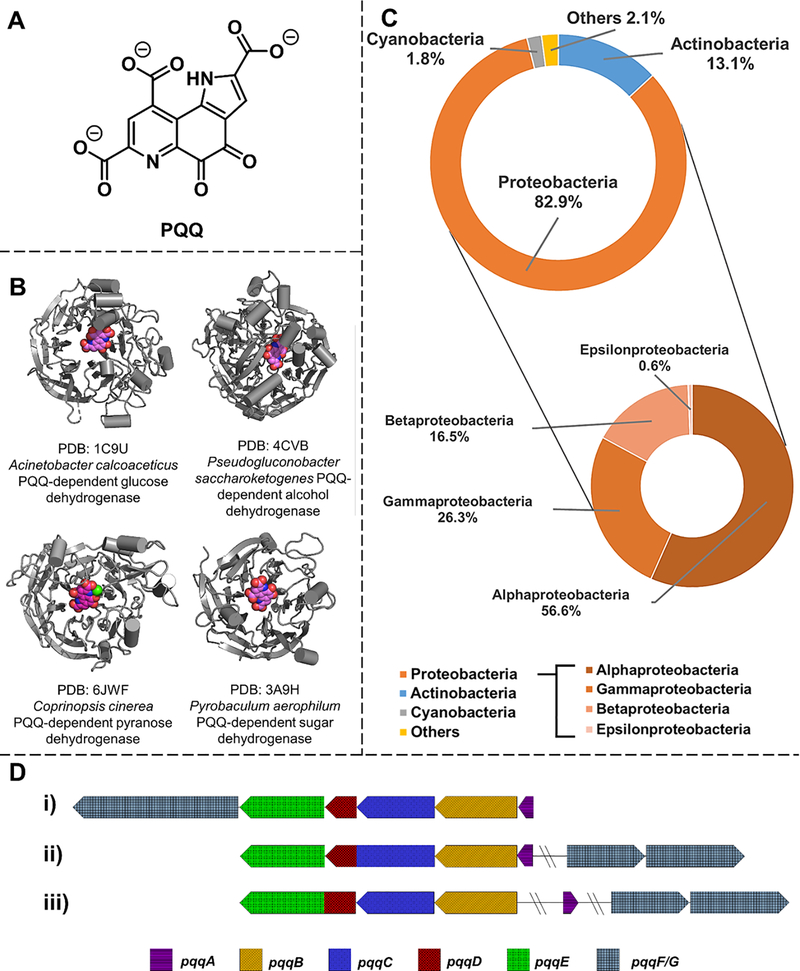
Figure 1. PQQ, PQQ-dependent enzymes and PQQ biogenesis in bacteria.
A. Structure of PQQ. B. Crystal structure of PQQ-dependent enzymes. PQQ-dependent dehydrogenases have been found in bacteria (e.g., Acinetobacter calcoaceticus and Pseudogluconobacter saccharoketogenes) as well as archaeon (e.g., Pyrobaculum aerophilum) and fungi (e.g., Coprinopsis cinerea). The bound-PQQ molecule is highlighted in magenta. C. Predicted PQQ producing bacteria are mostly γ-bacteria in the proteobacteria phylum. Doughnuts represent the distribution of species that are predicted to contain PQQ biosynthesis pathways. The initial pool of bacteria was selected from those that contain PqqA (PF08042)in the Pfam 32.0 database (https://pfam.xfam.org) since PqqA is absolutely required in PQQ biogenesis. D. Genes involved in PQQ biogenesis can be organized differently in various organisms: i) all six genes present in the pqq operon in Acinetobacter calcoaceticus; ii) pqqF/G located outside of the pqq operon containing pqqA-E in Methylobacterium extorquens; pqqD is fused with pqqC; iii) pqqA and pqqF/G are both distal from pqqB-E in Methylosinus trichosporium and pqqD is fused with pqqE.
The PQQ-dependent glucose and alcohol dehydrogenases (Figure 1B) participate in methylotrophic metabolism and ethanol/glucose utilization in prokaryotes.[7–9] More than 80% of PQQ-producing bacteria are proteobacteria, including a variety of opportunistic pathogens displaying antibiotic resistance (Figure 1C).[10–12] Bacteria can capture PQQ exogenously via a PQQ-binding protein, consistent with the proposal of inter-cellular PQQ trafficking.[13] There are recent reports of PQQ-dependent enzymes produced by archaea and fungi[14–16] implicating their symbiotic relationship with PQQ-producing microorganisms, although more biological studies are needed to explore this aspect further. The detection of PQQ in mammal fluid[17] and tissue[18] led to an early proposal that PQQ would be a eukaryotic vitamin.[19] This idea has been replaced by the concept of a “longevity vitamin,” whereby PQQ is not essential for humans but necessary for long-term health.[7,19–21] Echoing animal studies[22,23] that feeding PQQ improves mitochondria-related cellular functions is the finding that PQQ binds to human l-lactate dehydrogenase and regulates its activity, suggesting the active participation of PQQ within mammalian cells.[24] Given that many PQQ-producing bacteria, such as Acinetobacter and Pseudomonas, are associated with the human microbiome, the rising spotlight on the essential role of the human microbiota in health[25] introduces an intriguing new direction for the function of PQQ in eukaryotic organisms.
Unlike the pyridine nucleotide or flavin-based redox cofactors, our understanding of the biological origin of PQQ is a relatively recent event that began with the discovery of six conserved genes, pqqABCDEF/G, among PQQ-producing organisms (Figure 1D).[26–35] As the only known widespread redox cofactor that is derived from a ribosomally synthesized peptide, it is gratifying to see decades of research coalescing into a robust understanding of the molecular basis of PQQ production.
PqqA/D/E: a radical beginning for the pathway
Early feeding experiments established the link between PQQ production and cellular tyrosine and glutamate levels.[36,37] Through transposon insertions into pqqA and point mutations that shift its reading frame, it was confirmed that pqqA expression is essential for PQQ production.[27,31,35] The pqqA is translated into a short peptide that contains the fully conserved residues, glutamate and tyrosine; site-direct mutagenesis identified these two residues on PqqA as the best candidate for the precursor to PQQ (Figure 2A).[31] No function could be proposed for PqqD or PqqE, even though their conservation in the pathway was well recognized via genetic knockouts experiments in various PQQ-producing bacteria.[26–35]
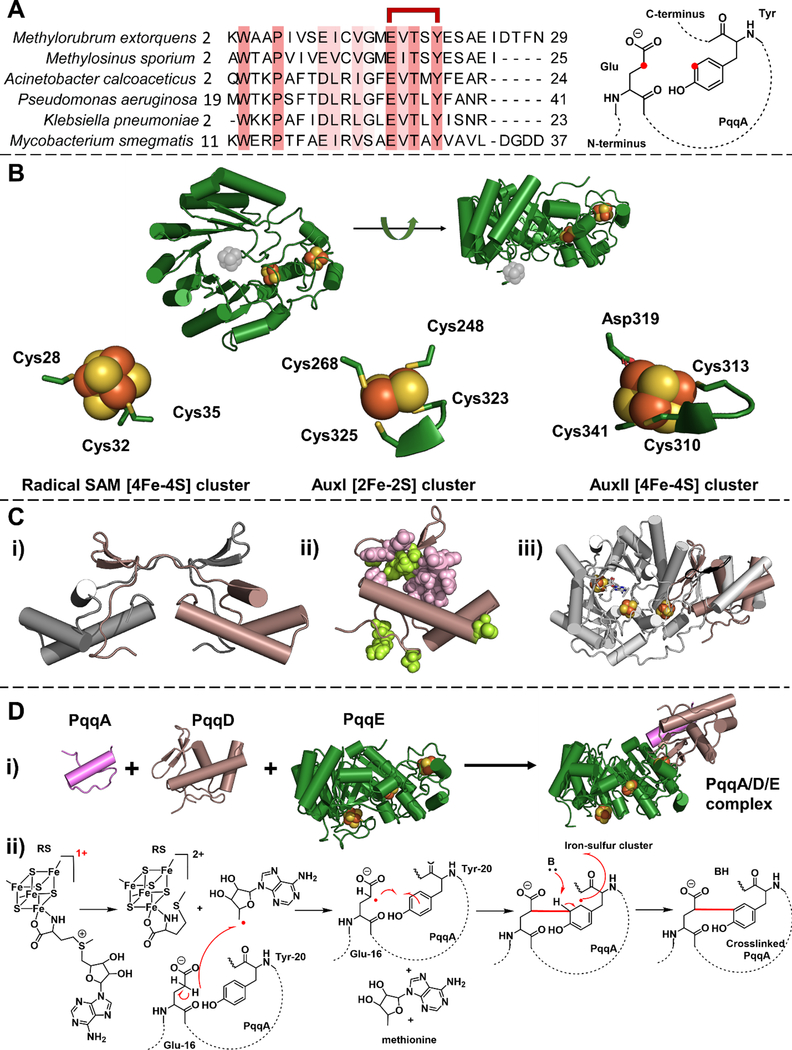
Figure 2. PqqA, PqqD and PqqE participate in the first step of the PQQ biogenesis.
A. PqqA is a short peptide. Left, the sequence alignment shows the conserved residues in PqqA (highlighted in salmon). The glutamate and tyrosine residues used to form PQQ are indicated as linked in red. Right, the representative structure of peptide PqqA. The to-be-crosslinked carbons on the glutamate and tyrosine residue are highlighted in red. B. X-ray crystal structure of Methylorubrum extorquens PqqE (PDB: 6C8V). The RS cluster in the N-terminal TIM barrel region was not well-resolved, possibly due to oxidative damage during the long delay times imposed by the protein crystallization process. The missing radical SAM [4Fe-4S] cluster is inserted (in gray). The ligands for the iron-sulfur cluster binding sites in PqqE are presented at the bottom. The ligand environment of the radical SAM [4Fe-4S] cluster is computationally modeled using CteB structure as a template by SWISS-MODEL.[62] C. Structures of PqqD. i) X-ray crystal structure of Xanthomonas campestris PqqD (PDB: 3G2B). A monomer of the PqqD dimer is shown in brown. ii) NMR structure of Methylorubrum extorquens PqqD (PDB: 3SXY) is shown in brown. The residues that interact with PqqA are highlighted in pink and the residues that interact with PqqE are highlighted in light green. iii) The PqqD NMR structure (brown) is superimposed with the peptide-binding domain in another radical SAM enzyme, CteB (gray) (PDB: 5WGG). D. The first step of PQQ biogenesis is catalyzed by a PqqA/D/E complex. i) A proposed structure for PqqA/PqqD/PqqE complex. The 3D structure of PqqA was predicted by PEP-FOLD3,[63] and the relative position of three components are modeled using CteB structure as a template by SWISS-MODEL.[62] ii) A proposed chemical mechanism of the reaction catalyzed in the PqqA/D/E complex. In the presence of PqqD and external reducing power, SAM is cleaved by PqqE, leading to the presumptive generation of 5’-deoxyadenosyl radical that abstracts a hydrogen atom from the γ-carbon of the conserved glutamate side chain of PqqA, to initiate C-C crosslinking to a conserved tyrosine.
Meanwhile, a group of enzymes within the radical S-adenosyl-l-methionine (SAM) superfamily[38] was linked to the biogenesis of important metabolites, such as biotin[39], lipoic acid[40], heme[41] and molybdopterin[42]. PqqE contains a conserved CxxxCxxC motif that coordinates a [4Fe-4S] cluster near the N-terminus, a common feature of radical SAM enzymes.[43] The ability of PqqE to conduct SAM cleavage was tested in the presence of the reducing agent sodium dithionite, confirming that PqqE is indeed a radical SAM enzyme that converts SAM to 5’-deoxyadenosine.[44] The iron content, electronic paramagnetic resonance (EPR)[44,45] and Mössbauer spectroscopy[46,47] of PqqE further suggested that the C-terminal domain, later classified as a SPASM domain[48], contains two more iron-sulfur clusters, the auxiliary site I (AuxI) and auxiliary site II (AuxII) (Figure 2B).
Although members of the SPASM subfamily catalyze protein/peptide modification in the presence of substrate, SAM, and reductant, these components failed to produce any detectable chemical change in PqqA upon the addition of PqqE.[44] The missing factor has turned out to be PqqD, a small, cofactor-less protein that primarily exists as a free protein (and occasionally as a C-terminal fusion protein of PqqC or an N-terminal fusion of PqqE) (Figure 1D).[12] EPR and hydrogen-deuterium exchange analyses suggested that PqqD interacted directly with PqqE.[45] Further studies, using native mass spectrometry, surface plasmon resonance and isothermal titration calorimetry (ITC), supported a tight-binding (nanomolar) complex between PqqD and PqqA.[49] The confirmation of a “substrate chaperone” role for PqqD finally came from the in-vitro demonstration of Glu-Tyr cross-linking within PqqA when PqqD was incubated with PqqE.[50] While the X-ray structure of PqqD initially showed a “domain-swapped” dimer[51], solution NMR[52,53] and small-angle X-ray scattering[49] data have implicated a monomeric structure (Figure 2C). The PqqD monomer is highly similar to the peptide recognition domain in other peptide modification proteins, such as CteB[54] and SuiB[55], in support of a functional monomeric form of PqqD (Figure 2C). NMR studies also identified residues on PqqD that interact with PqqA and PqqE (Figure 2C).[53] In this manner, the interrelated functions of PqqA, PqqD, and PqqE in the first step of PQQ biogenesis were uncovered (Figure 2D).
When the first (and thus far only) crystal structure of PqqE was resolved,[56] a [2Fe-2S] cluster appeared in the AuxI site of the SPASM domain, distinct from other SPASM proteins that dominantly incorporate [4Fe-4S] at this site. This [2Fe-2S] cluster in PqqE was confirmed spectroscopically by EPR and Mössbauer spectroscopy.[47,56,57] Further EPR investigations on PqqE have also shown a low potential [4Fe-4S] cluster in AuxI that requires Ti(III)citrate for reduction.[57] Such a low redox potential contrasts with the majority of iron-sulfur clusters in other radical SAM enzymes that readily undergo reduction by sodium dithionite (with the exceptions being the low potential auxiliary [4Fe-4S] cluster in BtrN[58,59] and NeoN[60]). Native mass spectrometry and computational modeling studies of PqqE have alternatively supported the formation of either a [4Fe-4S] or [2Fe-2S] cluster within the AuxI site.[47,57,61] Site-specific mutagenesis, in which a cysteine ligand in the AuxI site is replaced by histidine, leads to an [4Fe-4S] only AuxI site, completely abrogating [2Fe-2S] cluster formation.[61] Of mechanistic significance, the Cys-to-His variant catalyzes reductive cleavage of SAM but loses all ability to cross-link the peptide substrate.[61] These findings set up a stage for uncovering the catalytically active form of PqqE at the AuxI site.[61]
PqqF/G: A labile element within the pqq operon
Following the modification of PqqA by PqqE in the presence of PqqD, it was clear that a peptidase would be required to generate PQQ. A protease-like protein encoded by pqqF in the pqq operon has been reported to be essential for PQQ biosynthesis in K. pneumoniae,[64] yet this gene was not detected in the A. calcoaceticus pqq operon nor did it seem essential for PQQ production in this organism.[26,27] In Serratia sp. Fs14, PqqF represents the type of protease that typically presents itself within the pqq operon. The first crystal structure of this protein uncovered an 84 kDa M16 metalloprotease containing a His-His-Glu-His motif for zinc-binding in the active site (Figure 3A).[65] In M. extorquens, the pqq operon does not contain a canonical pqqF.[8,32] Instead, two co-existing genes, pqqF and pqqG, that are located outside of the pqq operon, encode the protease associated with PQQ production.[8,32] A recent study reveals that M. extorquens PqqF and PqqG form a tight complex, catalyzing the hydrolysis of multiple peptide bonds in PqqA, with a preference for serine residues(Figure 3B).[65] It has been proposed that the heterodimeric PqqF/G first trims the cross-linked product from the PqqA/D/E reaction, with final hydrolysis to a cross-linked di-amino acid requiring the participation of one or more generic cellular proteases.[65]
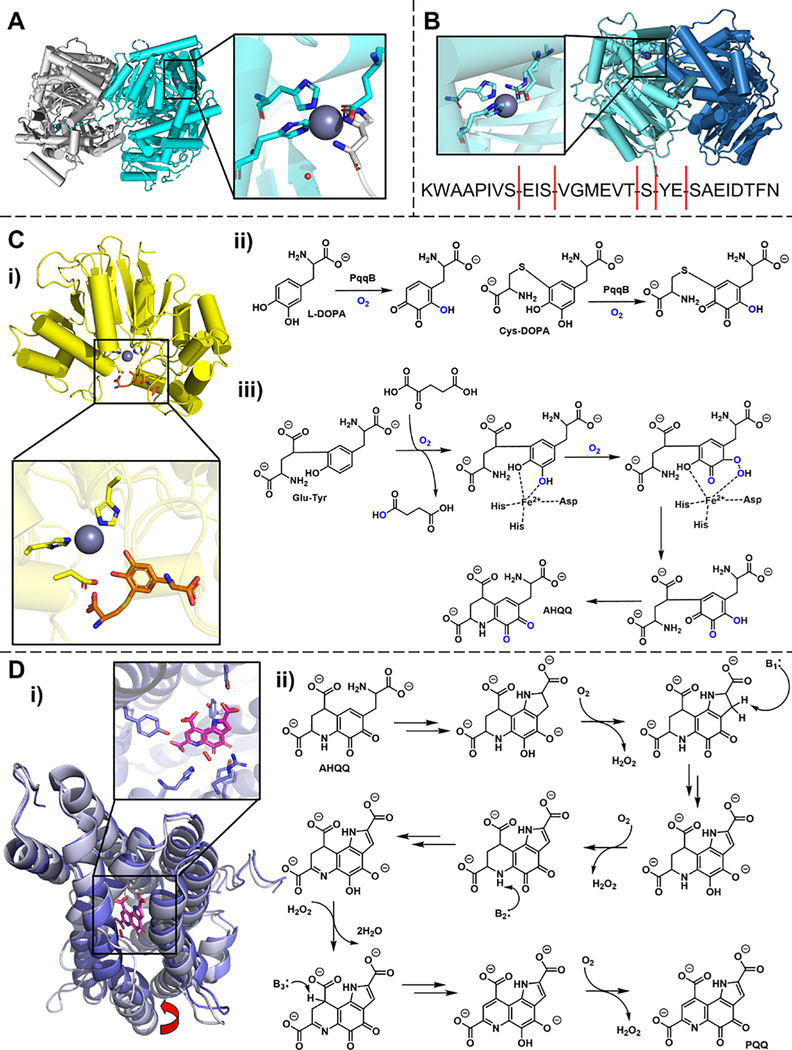
Figure 3. Properties of PqqF/G, PqqB and PqqC (proposed to catalyze the second, third and final steps in PQQ biogenesis).
A. Crystal structure of Serratia sp. PqqF (PDB: 5CIO). The monomer in the dimerized crystal structure is in cyan. The zoom-in picture shows the zinc-binding site in one of the monomers. Zn is shown in the purple sphere, a water molecule is in the red dot, the Zn-ligands from one monomer is shown in cyan stick, and the his-tag from another monomer bound to the active site Zn is shown in the gray stick. B. Structural model of Methylorubrum extorquens PqqF/G constructed via SWISS-MODEL using PDB: 5CIO as the template. PqqF is in cyan and PqqG is in blue. The zoom-in picture shows the modeled zinc-binding site in PqqF. The dominant protease cleavage sites on Methylorubrum extorquens PqqA by PqqF/G are highlighted by red lines at the bottom. C. Crystal structure of Pseudomonas putida PqqB (PDB: 6E13) and the chemical reaction catalyzed by PqqB. i) Cys-DOPA (orange), a substrate analog, can bind to the metal center of PqqB. The metal in the crystal structure is Zn (catalytically inactive) shown in the slate sphere. Metal-binding residues are shown in purple sticks. ii) PqqB is able to hydroxylate Cys-DOPA and L-DOPA in an O2-dependent manner. iii) A proposed mechanism for PqqB, based on the behavior of Cys-Tyr and Cys-DOPA. The mechanism is illustrated using the proposed native substrate, Glu-Tyr. The last step involves a spontaneous series of reactions to generate AHQQ, the substrate for PqqC. D. Crystal structures of PqqC and its catalytic mechanism. i) Crystal structure of Klebsiella pneumoniae PqqC in a complex with PQQ in a closed conformation (purple, PDB:1OTW) is superimposed with the open conformation (light purple, PDB: 1OTV). The bound PQQ in the closed conformation is in magenta, and the bound H2O2 is in red sticks. The red arrow at the bottom points to the large movement of the helix to accommodate the binding of PQQ. The zoom-in picture shows the ligands that interact with PQQ in the closed conformation. ii) Proposed reaction mechanism of PqqC.[74]
PqqB: The missing link and a new twist
Although pqqB is conserved in the pqq operon, conflicting results regarding the necessity of PqqB in PQQ production had been reported.[34,66,67] Sequence alignments, together with X-ray crystal structures, assigned PqqB to the metallo-β-lactamase (MBL) superfamily (Figure 3C).[68,69] Enzymes in this superfamily generally catalyze the hydrolysis of small molecules using metal centers that range from zinc to manganese, nickel, magnesium, and iron.[69,70] The latter led to the speculation that PqqB might serve as an oxygenase to modify the tyrosine sidechain within PqqA.[71] Metal reconstitutions of PqqB revealed fairly promiscuous and stable incorporation of copper, zinc, manganese, and magnesium, but not iron.[69] Chemical reactivity was pursued with metal reconstituted PqqB via the use of Cys-DOPA as an analog of the postulated Glu-Tyr di-amino acid substrate (Figure 3C). PqqB containing copper, zinc, manganese, or magnesium was, however, unable to catalyze any chemical conversion of Cys-DOPA. A surprising twist arose when PqqB was incubated with iron in the presence of Cys-Tyr or Cys-DOPA, leading not only to a significant increase in iron affinity but to two separate O2-dependent reactions: the decarboxylation of α-ketoglutatrate in the presence of Cys-Tyr and the aromatic ring hydroxylation of Cys-DOPA (Figure 3C).[72] Detailed kinetic analyses have confirmed that PqqB is a previously unknown iron-dependent hydroxylase, indicating a cross-over between the MBL superfamily and the non-heme iron hydroxylases. A mechanism has been proposed in which the Tyr of a Glu-Tyr di-amino acid is first hydroxylated to generate a 3,4-dihydoxy- intermediate that is subsequently oxidized again to a trihydroxy-derivative of Tyr (Figure 3C).[72] While each step consumes one equivalent of O2, neither produces hydrogen peroxide leading instead to water (Figure 3C). The organic product from the PqqB-catalyzed reaction with Cys-DOPA has further been shown to undergo a spontaneous cyclization in solution, leading to an analog of 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid (AHQQ) which is the substrate of PqqC in the final step of PQQ biogenesis.[73] These discoveries regarding the properties of PqqB have not only provided a critical step toward closing our understanding of the PQQ biosynthetic pathway, but also suggest an emerging presence of nonheme iron hydroxylating enzymes within the metallo-β-lactamase superfamily.[72]
PqqC: The last step but the first to be discovered
The function of PqqC was the first to be characterized among all the enzymes in the pathway. In pqqC knockout experiments, the accumulation of a “PQQ-like” intermediate, that disappeared upon PqqC complementation, led to the identification of the substrate for PqqC.[34,75,76] The chemical identity of this “PQQ-like” intermediate is AHQQ.[73] Through a combination of X-ray studies that yielded structures for PqqC with and without bound cofactor[77] and the detailed dissection of the four key oxidative steps[78] of PqqC, a catalytic mechanism has been described (Figure 3D). PqqC catalyzes an overall eight-electron and eight-proton oxidation on AHQQ in the absence of any organic/metal cofactor, consuming three equivalents of O2 in a single turnover manner. The stepwise utilization of O2 is accompanied by a progressive conformational change of PqqC, resulting in the reduction of bound product H2O2, rather than exogenous O2, during the 3rd step of the reaction.[78]
Overview of PQQ biogenesis
We can now propose the chemical steps that lead from PqqA to PQQ (Figure 4A). PqqA interacts with PqqD and guides the to-be-modified Glu and Tyr side chains of PqqA to the active side of PqqE. In the presence of reducing reagent and SAM, the [4Fe-4S] cluster in PqqE promotes the cleavage of SAM to 5’-deoxyadenosyl radical to initiate a series of steps culminating in a Glu-Tyr crosslink within PqqA. This product, PqqA’, is subsequently hydrolyzed by the protease PqqF (or a PqqF/ PqqG complex) that acts to trim away both N- and C-termini amino acids; the action of other (as yet unidentified) cellular proteases appears necessary in the generation of a Glu-Tyr di-amino acid substrate for PqqB. PqqB first hydroxylates the cross-linked Tyr to form Glu-DOPA, which is then further functionalized to a Glu-OH-DOPA quinone product. The latter is sufficiently reactive to undergo spontaneous cyclization, yielding AHQQ as the substrate for the final oxidative steps of the pathway catalyzed by PqqC.
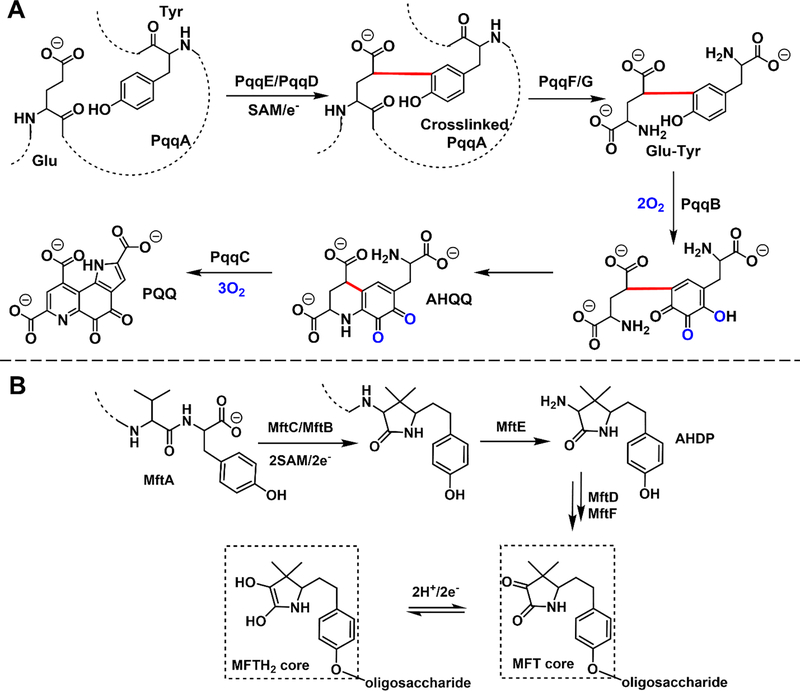
Figure 4. Key features of the proposed PQQ biosynthetic pathway and the newly proposed mycofactocin pathway.
A. The production of one molecule of PQQ requires the participation of at least six proteins, five equivalents of O2, one equivalent of SAM and the participation of an electron donor system to initiate the reductive SAM cleavage. B. The newly proposed redox cofactor mycofactocin (MFT) biosynthetic pathway shares high similarity to PQQ biogenesis. [82,84] In the proposed pathway, MftA, a short peptide, is modified by radical SAM enzyme, MftC, in the presence of a chaperone, MftB. The modified MftA is hydrolyzed by protease, MftE, followed by oxidative deamination and glycosylation. The mature MFT contains a presumed redox-active MFT core that can be reduced to MFTH2.[84]
Outlook
Beyond what is shown here, many intriguing questions remain in PQQ biogenesis. Perhaps the most compelling is the question of how the individual steps of the pathway coordinate with each other such that anaerobic reaction and oxygen-dependent reactions can operate sequentially, and whether this is related to the highly conserved order of open reading frame within the pqq operon. While the formation of a functional ternary complex of PqqA/D/E is established, it is intriguing to speculate that additional transient protein-protein interactions may function to segregate the aerobic and anaerobic portions of the pathway. The deeper understanding of the PQQ biosynthesis pathway will encourage further studies into antibiotic design targeting PQQ biogenesis selective against opportunistic pathogens. The utilization of PQQ by non-PQQ producing organisms, especially in eukaryotic cells, is also an intriguing aspect for future research in microbiology and systems biology.
Looking to the future, as new organisms and new biosynthetic pathways are discovered, much exciting chemistry is anticipated. The combination of genetic knockout/editing technologies, genome sequencing, bioinformatics tools (e.g., RODEO[79], antiSMASH[80]), and machine learning[81] is enabling rapid progress. Mycofactocin (MFT) biosynthesis [82–84] is an example of the ability of experimental and bioinformatics methods to drive new peptide-derived cofactor discovery.[82](Figure 4B) Even though the molecular function of MFT remains to be confirmed,[83] its structure and properties strongly suggest its role as a second, peptide-derived, prokaryotic redox cofactor.[84]
Acknowledgments
We thank the National Institute of Health for funding support (GM118117 to J.P.K.)
Footnotes
Conflict of interest statement
Nothing declared.
References
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hauge J Kinetics and specificity of glucose dehydrogenase from Bacterium anitratum Biochim Biophys Acta, 45 (1960), pp. 263–269 [DOI] [PubMed] [Google Scholar]
- 2.Hauge J Glucose dehydrogenase of Bacterium anitratum: an enzyme with a novel prosthetic group J Biological Chem, 239 (1964), pp. 3630–9•• This study presented the first experimental characterization of PQQ as a prosthetic group in PQQ-dependent glucose dehydrogenase in situ and in isolation.
- 3.Anthony C, Zatman L The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group Biochem J, 104 (1967), pp. 960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westerling J, Frank J, Duine J The prosthetic group of methanol dehydrogenase from Hyphomicrobium X: Electron spin resonance evidence for a quinone structure Biochem Bioph Res Co, 87 (1979), pp. 719–724 [DOI] [PubMed] [Google Scholar]
- 5.Salisbury S, Forrest H, Cruse W, Kennard O A novel coenzyme from bacterial primary alcohol dehydrogenases Nature, 280 (1979), pp. 843–844•• This study was the first structural characterization of PQQ molecule using X-ray crystallography.
- 6.Duine JA, Frank J The prosthetic group of methanol dehydrogenase: purification and some of its properties Biochem J, 187 (1980), pp. 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duine JA The PQQ story J Biosci Bioeng, 88 (1999), pp. 231–236• This article provided a historical view of the milestones in the discovery of PQQ, including the initial identification, chemical structure assignment, and development in the quinoproteins as well as the PQQ-dependent enzymes.
- 8.Chistoserdova L, Chen S-W, Lapidus A, Lidstrom ME Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view J Bacteriol,185 (2003), pp. 2980–2987•• This is a review on the metabolic pathways in methylotrophs and an overview of the essential genes involved in C1 metabolism in model organism Methylobacterium extorquens AM1
- 9.Matsutani M, Yakushi T Pyrroloquinoline quinone-dependent dehydrogenases of acetic acid bacteria Appl Microbiol Biot, 102 (2018), pp. 9531–9540 [DOI] [PubMed] [Google Scholar]
- 10.Fernández M, Conde S, de la Torre J, Molina-Santiago C, Ramos JL, Duque E Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440 Antimicrob Agents Ch, 56 (2011), pp. 1001–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin P The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes Adv Microb Physiol, 40 (1998), pp. 1–80 [DOI] [PubMed] [Google Scholar]
- 12.Shen YQ, Bonnot F, Imsand EM, RoseFigura JM, Sjölander K, Klinman JP Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone Biochemistry, 51 (2012), pp. 2265–2275•• This study showed a wide distribution of PQQ biosynthetic pathway among bacteria via bioinformatic analysis. It also demonstrates the strict order of pqqB, pqqC, pqqD, and pqqE in the open reading frames in various bacteria.
- 13.Ho J, Cotruvo J A periplasmic binding protein for pyrroloquinoline quinone Biochemistry, 58 (2019), pp. 2665–2669• PqqT was identified as a periplasmic protein that binds PQQ in a 1:1 ratio, suggesting a role in the intercellular trafficking of PQQ.
- 14.Matsumura H, Umezawa K, Takeda K, Sugimoto N, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N Discovery of a eukaryotic pyrroloquinoline quinone-dependent oxidoreductase belonging to a new auxiliary activity family in the database of carbohydrate-active enzymes Plos One, 9 (2014), Article e104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda K, Ishida T, Yoshida M, Samejima M, Ohno H, Igarashi K, Nakamura N Crystal structure of the catalytic and cytochrome b domains in a eukaryotic pyrroloquinoline quinone-dependent dehydrogenase Appl Environ Microb, 85 (2019), Article e01692–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakuraba H, Yokono K, Yoneda K, Watanabe A, Asada Y, Satomura T, Yabutani T, Motonaka J, Ohshima T Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum Arch Biochem Biophys, 502 (2010), pp. 81–88organization diversity of the pqq operon in various organisms.
- 17.Mitchell AE, Jones AD, Mercer RS, Rucker RB Characterization of pyrroloquinoline quinone amino acid derivatives by electrospray ionization mass spectrometry and detection in human milk Anal Biochem, 269 (1999), pp. 317–325 [DOI] [PubMed] [Google Scholar]
- 18.Kumazawa T, Seno H, Urakami T, Matsumoto T, Suzuki O Trace levels of pyrroloquinoline quinone in human and rat samples detected by gas chromatography/mass spectrometry Biochimica Et Biophysica Acta Bba - Gen Subj, 1156 (1992), pp. 62–66 [DOI] [PubMed] [Google Scholar]
- 19.Kasahara T, Kato T A new redox-cofactor vitamin for mammals Nature, 422 (2003), pp. 832–832 [DOI] [PubMed] [Google Scholar]
- 20.Klinman J Quinoenzymes in biology Annu Rev Biochem, 63 (1994), pp. 299–344•• This comprehensive review summarized the peptide/protein-derived cofactors and their functions in biology.
- 21.Ames B Prolonging healthy aging: Longevity vitamins and proteins Proc National Acad Sci, 115 (2018), pp. 10836–10844• In this perspective, the role of PQQ in human health is modified to be a longevity vitamin.
- 22.Stites T, Storms D, Bauerly K, Mah J, Harris C, Fascetti A, Tchaparian QE, Satre M, Rucker R Pyrroloquinoline quinone modulates mitochondrial quantity and function in mice J Nutrition, 136 (2006), pp. 390–396 [DOI] [PubMed] [Google Scholar]
- 23.Chowanadisai W, Bauerly KA, Tchaparian E, Wong A, Cortopassi GA, Rucker RB Pyrroloquinoline quinone stimulates mitochondrial biogenesis through cAMP response element-binding protein phosphorylation and increased PGC-1α expression J Biol Chem, 285 (2010), pp. 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akagawa M, Minematsu K, Shibata T, Kondo T, Ishii T, Uchida K Identification of lactate dehydrogenase as a mammalian pyrroloquinoline quinone (PQQ)-binding protein Sci Rep, 6 (2016), Article srep26723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg ED, Sonnenburg JL The ancestral and industrialized gut microbiota and implications for human health Nat Rev Microbiol, 17 (2019), pp. 383–390. [DOI] [PubMed] [Google Scholar]
- 26.Horsman H, Huinen R, Putte P Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrroloquinoline-quinone: nucleotide sequence and expression in Escherichia coli K-12 J Bacteriol, 171 (1989), pp. 447–455• This is one of the initial studies showing the key biosynthesis genes for PQQ production in A. calcoaceticus.
- 27.Goosen N, Vermaas DA, van de Putte P Cloning of the genes involved in synthesis of coenzyme pyrrolo-quinoline-quinone from Acinetobacter calcoaceticus J Bacteriol, 169 (1987), pp. 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meulenberg J, Sellink E, Loenen W, Riegman N, Kleef M, Postma P Cloning of Klebsiella pneumoniae pqq genes and PQQ biosynthesis in Escherichia coli FEMS Microbiol Lett, 71 (1990), pp. 337–343• In this study, K. pneumoniae pqq genes were expressed in the non-PQQ producer, E. coli, leading to the production of PQQ. The importance of each gene was tested using this hetero-expression method as well.
- 29.Biville F, Turlin E, Gasser F Cloning and genetic analysis of six pyrroloquinoline quinone biosynthesis genes in Methylobacterium organophilum DSM 760 Microbiology, 135 (1989), pp. 2917–2929• This is one of the first studies that identified the pqq genes in methylotrophic bacteria.
- 30.Biville F, Mazodier P, Turlin E, Gasser F Mutants of Methylobacterium organophilum unable to synthesize PQQ Antonie Van Leeuwenhoek, 56 (1989), pp. 103–107 [DOI] [PubMed] [Google Scholar]
- 31.Goosen N, Huinen RG, van de Putte P A 24-amino-acid polypeptide is essential for the biosynthesis of the coenzyme pyrrolo-quinoline-quinone J Bacteriol, 174 (1992), pp. 1426–1427•• This study used site specific mutagenesis to demonstrate that one glutamate and one tyrosine residue in PqqA are essential for PQQ biogenesis.
- 32.Morris CJ, Biville F, Turlin E, Lee E, Ellermann K, Fan WH, Ramamoorthi R, Springer AL, Lidstrom ME Isolation, phenotypic characterization, and complementation analysis of mutants of Methylobacterium extorquens AM1 unable to synthesize pyrroloquinoline quinone and sequences of pqqD, pqqG, and pqqC J Bacteriol, 176 (1994), pp. 1746–1755• This work established the importance of the six pqq genes in PQQ production in M. extorquens AM1. It is worth noting that the initial nomenclature in early studies of the PQQ pathway in M. extorquens AM1 is different from later studies. The pqqD, pqqG and pqqC in this article correspond to pqqA, pqqB and pqqC in the later literature.
- 33.Ramamoorthi R, Lidstrom ME Transcriptional analysis of pqqD and study of the regulation of pyrroloquinoline quinone biosynthesis in Methylobacterium extorquens AM1 J Bacteriol, 177 (1995), pp. 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velterop JS, Sellink E, Meulenberg JJ, David S, Bulder I, Postma PW Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway J Bacteriol, 177 (1995), pp. 5088–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goosen N, Horsman HPA, Huinen RGM, van de Putte P Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrrolo-quinoline-quinone: nucleotide sequence and expression in Escherichia coli K-12. J Bacteriol 171 (1989), pp. 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Kleef MA, Duine JA L-tyrosine is the precursor of PQQ biosynthesis in Hyphomicrobium X FEBS Lett, 237 (1988), pp. 91–97• This is one of the first studies that showed that PQQ is derived from tyrosine.
- 37.Houck D, Hanners J, Unkefer C, van Kleef MA, Duine JA PQQ: biosynthetic studies in Methylobacterium AM1 and Hyphomicrobium X using specific 13C labeling and NMR Antonie Van Leeuwenhoek 56 (1989), pp. 93–101• Authors in this paper discovered that PQQ produced by two methylotrophs was derived from glutamate and tyrosine via feeding experiments and NMR.
- 38.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods Nucleic Acids Res, 29 (2001), pp. 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begley T, Xi J, Kinsland C, Taylor S, McLafferty F The enzymology of sulfur activation during thiamin and biotin biosynthesis Curr Opin Chem Biol, 3 (1999), pp. 623–629 [DOI] [PubMed] [Google Scholar]
- 40.Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE, Marletta MA Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein Biochemistry, 39 (2000), pp. 15166–15178 [DOI] [PubMed] [Google Scholar]
- 41.Layer G, Grage K, Teschner T, Schünemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein A, Jahn D Radical s-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-l-methionines J Biol Chem, 280 (2005), pp. 29038–29046 [DOI] [PubMed] [Google Scholar]
- 42.Hanzelmann P, Schindelin H Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans Proc National Acad Sci, 101 (2004), pp. 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broderick JB, Duffus BR, Duschene KS, Shepard EM Radical s-adenosylmethionine enzymes Chem Rev, 114 (2014), pp. 4229–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu S, King D, Britt DR, Klinman JP Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-l-methionine enzyme Biochemistry, 48 (2009), pp. 10151–10161•• This study is the first to demonstrate that PqqE is a radical SAM enzyme exhibiting the SAM cleavage reactivity in the presence of reducing reagent and SAM.
- 45.Wecksler SR, Stoll S, Iavarone AT, Imsand EM, Tran H, Britt RD, Klinman JP Interaction of PqqE and PqqD in the pyrroloquinoline quinone (PQQ) biosynthetic pathway links PqqD to the radical SAM superfamily Chem Comm, 46 (2010), pp. 7031–7033• This study provided the first evidence that PqqD interacts with PqqE via an EPR signal shift upon the binding of PqqD.
- 46.Saichana N, Tanizawa K, Pechoušek J, Novák P, Yakushi T, Toyama H, Frébortová J PqqE from Methylobacterium extorquens AM1: a radical S-adenosyl-l-methionine enzyme with an unusual tolerance to oxygen J Biochem, 159 (2015), pp. 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saichana N, Tanizawa K, Ueno H, Pechoušek J, Novák P, Frébortová J Characterization of auxiliary iron–sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1 FEBS Open Bio, 7 (2017), pp. 1864–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grell TAJ, Goldman PJ, Drennan CL SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes J Biol Chem, 290 (2014), pp. 3964–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latham JA, Iavarone AT, Barr I, Juthani PV, Klinman JP PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway J Biol Chem, 290 (2015), pp. 12908–12918•• This study proposed the functional role of PqqD as a chaperone in the PQQ biogenesis and determined the dissociation constants between PqqA, PqqD and PqqE.
- 50.Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, Klinman JP Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD J Biol Chem, 291 (2016), pp. 8877–8884•• This is the first observation of the peptide modification reactivity of PqqE on PqqA in the presence of PqqD.
- 51.Tsai T-Y, Yang C-Y, Shih H-L, Wang AH-J, Chou S-H Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier Proteins Struct Funct Bioinform, 76 (2009), pp. 1042–1048 [DOI] [PubMed] [Google Scholar]
- 52.Evans RL, Latham JA, Klinman JP, Wilmot CM, Xia Y 1H, 13C, and 15N resonance assignments and secondary structure information for Methylobacterium extorquens PqqD and the complex of PqqD with PqqA Biomol NMR Assign, 10 (2016), pp. 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans RL, Latham JA, Xia Y, Klinman JP, Wilmot CM Nuclear magnetic resonance structure and binding studies of PqqD, a chaperone required in the biosynthesis of the bacterial dehydrogenase cofactor pyrroloquinoline quinone Biochemistry. 56 (2017), pp. 2735–2746•• This study not only provided the NMR structure of the PqqD monomer in solution, but also probe the key residues on PqqD that are in contact with PqqA and PqqE in the PqqA/D/E complex.
- 54.Grove TL, Himes P, Hwang S, Yumerefendi H, Bonanno JB, Kuhlman B, Almo SC, Bowers AA Structural insights into thioether bond formation in the biosynthesis of sactipeptides J Am Chem Soc, 139 (2017), pp. 11734–11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis KM, Schramma KR, Hansen WA, Bacik JP, Khare SD, Seyedsayamdost MR, Ando N Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition Proc Natl Acad Sci, 114 (2017), pp. 10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barr I, Stich TA, Gizzi AS, Grove TL, Bonanno JB, Latham JA, Chung T, Wilmot CM, Britt RD, Almo SC, et al. X-ray and EPR characterization of the auxiliary Fe-S clusters in the radical SAM enzyme PqqE Biochemistry, 57 (2018), pp. 1306–1315•• This study provided the first and, so far, the only crystal structure of PqqE. It illustrated the molecular basis of the distinct iron-sulfur cluster binding environments at the SPASM domain comparing to other radical SAM proteins in the subfamily.
- 57.Tao L, Zhu W, Klinman JP, Britt DR Electron paramagnetic resonance spectroscopic identification of the Fe-S clusters in the SPASM domain-containing radical SAM enzyme PqqE Biochemistry, 58 (2019), pp. 5173–5187• This study characterized the electronic properties of all the iron-sulfur clusters in PqqE using multiple EPR techniques. It proposed the existence of the [4Fe-4S] cluster in the AuxI site of PqqE, which was not observed in the X-ray crystal structure previously.
- 58.Maiocco SJ, Grove TL, Booker SJ, Elliott SJ Electrochemical resolution of the [4Fe-4S] centers of the AdoMet radical enzyme BtrN: evidence of proton coupling and an unusual, low-potential auxiliary cluster J Am Chem Soc, 137 (2015), pp. 8664–8667 [DOI] [PubMed] [Google Scholar]
- 59.Grove TL, Ahlum JH, Sharma P, Krebs C, Booker SJ A consensus mechanism for radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters Biochemistry, 49 (2010), pp. 3783–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kudo F, Hoshi S, Kawashima T, Kamachi T, Eguchi T Characterization of a radical S-adenosyl-l-methionine epimerase, NeoN, in the last step of neomycin B biosynthesis J Am Chem Soc, 136 (2014), pp. 13909–13915 [DOI] [PubMed] [Google Scholar]
- 61.Zhu W, Walker LM, Tao L, Iavarone AT, Wei X, Britt RD, Elliot SJ, Klinman JP The structural properties and catalytic implications of the SPASM domain iron-sulfur clusters in Methylorubrum extorquens PqqE Manuscript under review [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer F, de Beer T, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modeling of protein structures and complexes Nucleic Acids Res, 46 (2018), pp. W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex Nucleic Acids Res, 44 (2016), pp. W449–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meulenberg J, Sellink E, Riegman N, Postma P Nucleotide sequence and structure of the Klebsiella pneumoniae pqq operon Mol Gen Genetics MGG, 232 (1992), pp. 284–294• This study is one of the early studies of the PQQ biosynthetic genes in K. pneumoniae.
- 65.Martins AM, Latham JA, Martel PJ, Barr I, Iavarone AT, Klinman JP A two-component protease in Methylorubrum extorquens with high activity toward the peptide precursor of the redox cofactor pyrroloquinoline quinone J Biol Chem, 294 (2019), pp. 15025–15036•• This is the first experimental evidence to demonstrate that the complex of PqqF and PqqG exhibits peptidase activity.
- 66.Puehringer S, Metlitzky M, Schwarzenbacher R The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach BMC Biochem, 9 (2008), pp. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan H, Xia Y, Li J, Kang Z, Zhou J Identification of transporter proteins for PQQ-secretion pathways by transcriptomics and proteomics analysis in Gluconobacter oxydans WSH-003 Front Chem Sci Eng, 11 (2017), pp. 72–88. [Google Scholar]
- 68.Metlitzky M, Puehringer S, Fisher SJ Crystal structure of PqqB from Pseudomonas putida at 2.2 Å resolution J Biophysical Chem, 3 (2012), pp. 206–210 [Google Scholar]
- 69.Tu X, Latham JA, Klema VJ, Evans RL, Li C, Klinman JP, Wilmot CM Crystal structures reveal metal-binding plasticity at the metallo-β-lactamase active site of PqqB from Pseudomonas putida J Biol Inorg Chem, 22 (2017), pp. 1089–1097• In this study, multiple crystal structures of PqqB were resolved with a range of metal ions incorporated in the active site.
- 70.Bebrone C Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily Biochem Pharmacol 74 (2007), pp. 1686–1701 [DOI] [PubMed] [Google Scholar]
- 71.Klinman JP, Bonnot F Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ Chem Rev 114 (2013), pp. 4343–4365• This is a comprehensive review of the biosynthesis of quinocofactors and features of quinoproteins.
- 72.Koehn EM, Latham JA, Armand T, Evans RL, Tu X, Wilmot CM, Iavarone AT, Klinman JP Discovery of hydroxylase activity for PqqB provides a missing link in the pyrroloquinoline quinone biosynthetic pathway J Am Chem Soc, 141 (2019), pp. 4398–4405•• This study is the first functional characterization of PqqB using a combination of kinetics analysis, X-ray crystallography, isotopic labeling in substrate analog, product analysis mass spectrometry and ITC measurements.
- 73.Magnusson OT, Toyama H, Saeki M, Schwarzenbacher R, Klinman JP The structure of a biosynthetic intermediate of pyrroloquinoline quinone (PQQ) and elucidation of the final step of PQQ biosynthesis J Am Chem Soc, 126 (2004), pp. 5342–5343•• This study isolated and characterized the chemical structure of the key intermediate AHQQ in the PQQ biogenesis.
- 74.Bonnot F, Iavarone AT, Klinman JP Multistep, eight-electron oxidation catalyzed by the cofactorless oxidase, PqqC: identification of chemical intermediates and their dependence on molecular oxygen Biochemistry, 52 (2013), pp. 4667–4675.• This detailed mechanistic study revealed the catalytic mechanism of PqqC as well as the identity of intermediates during the reaction.
- 75.van Kleef MA, Duine JA A search for intermediates in the bacterial biosynthesis of PQQ Biofactors Oxf Engl, 1 (1988), pp. 297–302 [PubMed] [Google Scholar]
- 76.Toyama H, Fukumoto H, Saeki M, Matsushita K, Adachi O, Lidstrom ME PqqC/D, which converts a biosynthetic intermediate to pyrroloquinoline quinone Biochem Bioph Res Co, 299 (2002), pp. 268–272. [DOI] [PubMed] [Google Scholar]
- 77.Magnusson OT, Toyama H, Saeki M, Rojas A, Reed JC, Liddington RC, Klinman JP, Schwarzenbacher R Quinone biogenesis: structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone Proc Natl Acad Sci, 101 (2004), pp. 7913–7918•• This study, for the first time, provided the molecular basis of PqqC using X-ray crystallography and the kinetics analysis of PqqC catalysis. The structures of apo- and the PQQ-bound PqqC shed light on how PqqC activates oxygen without the presence of other cofactors.
- 78.Magnusson O, RoseFigura J, Toyama H, Schwarzenbacher R, Klinman JP Pyrroloquinoline quinone biogenesis: characterization of PqqC and its H84N and H84A active site variants Biochemistry, 46 (2007), pp. 7174–7186• The kinetic study on the PqqC variants in this study confirms that the active site residue His-84 functions as a proton donor to the oxyanion of the quinoid species.
- 79.Tietz JI, Schwalen CJ, Patel PS, Blair T, Tai H-C, Zakai UI, Mitchell DA A new genome-mining tool redefines the lasso peptide biosynthetic landscape Nat Chem Biol, 13 (2017), pp. 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T,antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res, 47 (2019) pp. W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tallorin L, Wang J, Kim WE, Sahu S, Kosa NM, Yang P, Thompson M, Gilson MK, Frazier PI, Burkart MD, et al. Discovering de novo peptide substrates for enzymes using machine learning Nat Commun, 9 (2018), pp. 5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ayikpoe R, Govindarajan V, Latham JA Occurrence, function, and biosynthesis of mycofactocin Appl Microbiol Biot, 103 (2019), pp. 2903–2912•• This review summarized the current knowledge on mycofactocin biosynthesis and proposed that it could be a new redox cofactor that is crucial for mycobacterium tuberculosis.
- 83.Haft DH Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners BMC Genomics, 12 (2011), pp. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peña-Ortiz L, Graça AP, Guo H, Braga D, Köllner TG, Regestein L, Beemelmanns C and Lackner G, Structure elucidation of the redox cofactor mycofactocin reveals oligo-glycosylation by MftF Chem Sci, (2020), Advance Article. DOI: 10.1039/d0sc01172j•• This research article determined the chemical structure of glycosylated mycofactocin (MFT) and uncovered the functional role of MftF in MFT biosynthesis pathway in Mycolicibacterium smegmatis.