SUMMARY
The intestinal epithelium separates host tissue and gut-associated microbial communities. During inflammation, the host releases reactive oxygen and nitrogen species as an antimicrobial response. The impact of these radicals on gut microbes is incompletely understood. We discovered that the cryptic appBCX genes, predicted to encode a cytochrome bd-II oxidase, conferred a fitness advantage for E. coli in chemical and genetic models of non-infectious colitis. This fitness advantage was absent in mice that lacked epithelial NADPH oxidase 1 (NOX1) activity. In laboratory growth experiments, supplementation with exogenous hydrogen peroxide enhanced E. coli growth through AppBCX-mediated respiration in a catalase-dependent manner. We conclude that epithelial-derived reactive oxygen species are degraded in the gut lumen, which gives rise to molecular oxygen that supports aerobic respiration of E. coli. This work illustrates how epithelial host responses intersect with gut microbial metabolism in the context of gut inflammation.
eTOC blurb
During inflammation, the intestinal epithelium produces antimicrobial products to impede bacterial growth. Chanin et al. report that one of these antimicrobial products, reactive oxygen species, also promotes the outgrowth of E. coli. Detoxification of inflammatory reactive oxygen species through AppBCX allows E. coli to respire in an otherwise anaerobic environment.
Graphical Abstract
INTRODUCTION
The colonic epithelium creates a protective interface between gut-associated microbial communities and the host. The interaction between gut microbes and the epithelium is mutually beneficial. Bacterial products instruct the immune system, promote cellular differentiation, and support host metabolism (Pickard et al., 2017; Rakoff-Nahoum et al., 2004; Velazquez et al., 1997). Conversely, the metabolism of epithelial cells in the large intestine promotes an environment suitable for anaerobic bacteria to colonize in the lumen (Litvak et al., 2018).
During inflammation, reactive oxygen species (ROS) prevent microbes from entering the tissue (reviewed in (Aviello and Knaus, 2018)). One of the main sources of ROS during inflammation are NADPH oxidases, membrane bound protein complexes that produce superoxide from molecular oxygen. Superoxide dismutases catalyze the disproportionation of superoxide to hydrogen peroxide (H2O2). In turn, H2O2 is converted to hydroxyl radicals in the Fenton reaction, to hypochlorite by myeloperoxidase activity, or to molecular oxygen by catalases. NADPH oxidase 1 (NOX1) is primarily produced in the large intestine and smooth muscle (Panday et al., 2015; Suh et al., 1999), while phagocyte oxidase (PHOX, NOX2) and myeloperoxidase are predominantly expressed by professional phagocytic cells (Panday et al., 2015). Individuals lacking PHOX activity experience recurrent infection with fungal and bacterial pathogens and are at increased risk for atypical opportunistic infections (Curnutte et al., 1988; Dinauer et al., 1987). While the release of inflammatory ROS in the tissue is generally considered an antimicrobial host response, the impact of ROS on gut microbial communities is incompletely understood (Imlay, 2019).
The composition of the microbiota changes in inflammatory diseases (Arthur et al., 2012; Barman et al., 2008; Frank et al., 2007; Lupp et al., 2007; Normann et al., 2013; Stecher et al., 2007; Vujkovic-Cvijin et al., 2013). An increase in the relative abundance of facultative anaerobic bacteria, primarily members of the Enterobacteriaceae family (phylum Proteobacteria), characterizes inflammation-associated changes (dysbiosis) (Shin et al., 2015). In mouse models of non-infectious colitis, gut microbiota dysbiosis is not merely a bystander effect, but can worsen disease symptoms (Garrett et al., 2007; Zhu et al., 2018).
Oxygen availability impacts the composition and spatial organization of the gut microbiota (Albenberg et al., 2014; Byndloss et al., 2017). Increased oxygen availability during oral antibiotic treatment, enteric infection, and non-infectious colitis promotes the outgrowth of Enterobacteriaceae, including Escherichia coli (Byndloss et al., 2017; Hughes et al., 2017; Rivera-Chavez et al., 2016). The outgrowth of E. coli depends in part on the cytochrome bd-I oxidase, CydABX, a respiratory quinol:O2 oxidoreductase (Byndloss et al., 2017; Hughes et al., 2017). In contrast, the E. coli Cyo enzyme, a cytochrome bo3-type oxidase, requires highly oxygenated environments and does not function in the inflamed gut (Hughes et al., 2017).
The E. coli appBCX (cbd or cyx) genes encode an additional, putative cytochrome bd oxidase (Dassa et al., 1991; VanOrsdel et al., 2013) with approximately 60% amino acid sequence identity to CydABX (Dassa et al., 1991) (Fig. 1A). Biochemical analyses of the isolated AppBCX enzyme complex suggest it reduces oxygen while oxidizing quinol and thus functions as a terminal oxidase, akin to CydABX (Bekker et al., 2009; Sturr et al., 1996). Transcription of appBCX is regulated by low levels of oxygen, carbon, and phosphate starvation, as well as stationary phase growth (Atlung and Brondsted, 1994; Atlung et al., 1997; Brondsted and Atlung, 1996; Sousa et al., 2012). The physiological function of AppBCX is incompletely understood, likely because AppBCX expression is low when cultured under standard laboratory conditions. Mutants defective in AppBCX do not exhibit overt growth defects and phenotypes only manifest in the absence of other respiratory systems (Bekker et al., 2009; Dassa et al., 1991) (Fig. S1A). Here, we used chemical and genetic models of colitis to probe the physiological function of the putative cytochrome bd-II oxidase, AppBCX, in E. coli.
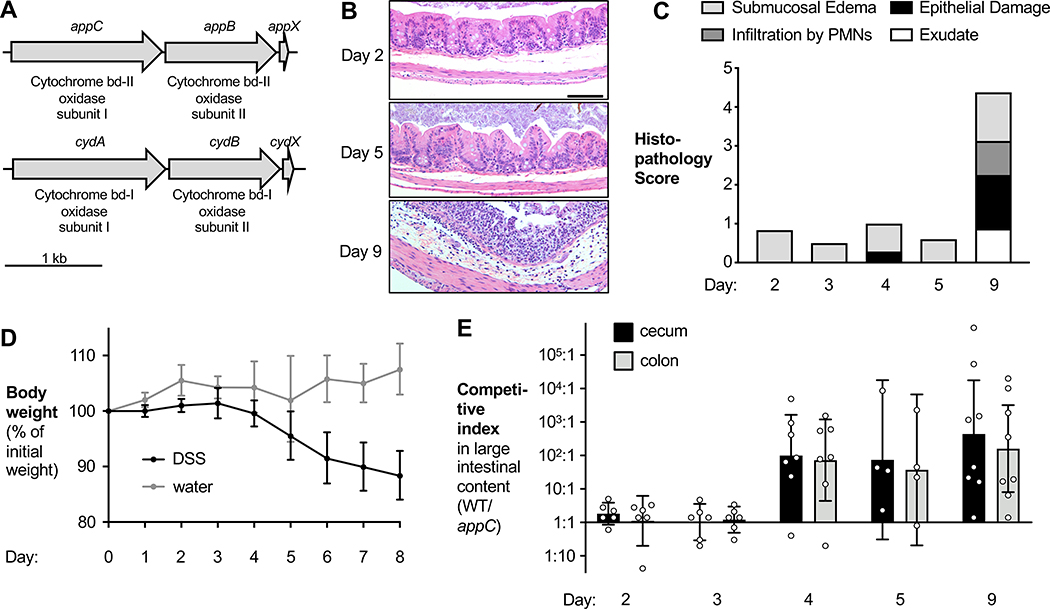
Figure 1. AppBCX provides a fitness advantage in the inflamed murine intestine.
See also Fig. S1.
(A) Schematic representation of the appBCX and cydABX gene loci in EcN. (B – E) Groups of wild-type C57BL/6 male and female mice were intragastrically inoculated with a 1:1 mixture of an EcN wild-type strain and an isogenic appC mutant. On the day of gavage, mice were given 3% DSS in the drinking water. On day 2, 3, 4, 5, and 9, samples were obtained. (B) Representative H+E stained sections of the cecum. Scale bar, 100 μm. (C) H+E stained sections were scored by a veterinary pathologist for submucosal edema (light gray bars), epithelial damage (black bars), infiltration by polymorphonuclear cells; PMN (dark gray bars), and exudate (white bars). Bars for each histopathology category are the average per group. (D) DSS-treated (black symbols) and mock-treated mice (gray symbols) were weighed daily and weight change was recorded. Symbols represent geometric means ± 95% confidence intervals. (E) The CI of the wild-type strain and the appC mutant in the cecal (black bars) and colonic (gray bars) content was determined.
Bars represent geometric means ± 95% confidence intervals.
RESULTS
The putative cytochrome oxidase AppBCX provides a fitness advantage in the inflamed gut
We surmised that more physiologically relevant conditions, such as the murine large intestine, may reveal the function of AppBCX. We therefore analyzed the function of AppBCX in a chemically-induced murine model of colitis (dextran sulfate sodium; DSS colitis) in which low levels of oxygen are available for respiration (Hughes et al., 2017). For our initial experiments, we used the human commensal strain E. coli Nissle 1917 (EcN). We colonized groups of male and female C57BL/6 mice with an equal ratio of EcN wild-type strain and an isogenic appC mutant. We then treated these animals with DSS in the drinking water and obtained samples at pre-determined time points. DSS elicited progressively worsening inflammatory responses (Fig. 1B – D; Fig. S1B). We plated cecal and colonic contents on selective agar and determined abundance of each strain (Fig. S1C and D). The competitive index (CI) was calculated by determining the ratio of wild-type to mutant bacteria, normalized by corresponding ratio in the inoculum (Fig. 1E). Initially, both strains were recovered in similar numbers. However, on day 4 and at later time points, the wild-type strain was recovered in higher numbers than the mutant. The phenotype of the appC mutant coincided with disease onset (Fig. 1B – D; Fig. S1B). The fitness advantage conferred by AppBCX was not unique to EcN; an appBC mutant in the murine commensal E. coli strain MP1 (Lasaro et al., 2014) was less efficient at colonizing the murine intestinal lumen in the DSS colitis model compared to the MP1 wild-type strain (Fig. S1E). These results suggest that AppBCX provides a fitness advantage during colitis, thus creating an opportunity to study the physiological role of AppBCX.
Epithelial-derived ROS provide a fitness advantage through AppBCX
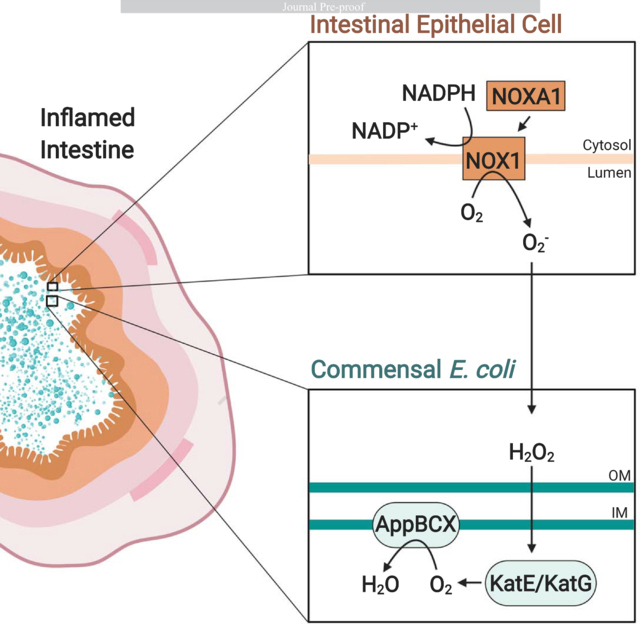
Next, we determined the origin of the AppBCX substrate in the murine gut. During inflammation, ROS and reactive nitrogen species change the metabolite landscape (Winter et al., 2010; Winter et al., 2013). We hypothesized that inflammatory ROS may degrade into oxygen in the gut lumen, thereby allowing for aerobic microbial respiration (Fig. 2A). One of the NADPH oxidases expressed in the intestinal tissue is the NOX1 complex (Suh et al., 1999). We therefore repeated the competitive colonization experiment in Nox1-deficient mice and littermate controls treated with DSS for 9 days (Fig. 2B and S2A). In the absence of Nox1, the fitness advantage supplied by AppBCX was ablated in the cecal content. We observed a similar trend in the colon content.
Figure 2. Epithelial-derived ROS induce an AppBCX-conferred fitness advantage in the inflamed murine intestine.
(A) Schematic of host-derived ROS detoxification in the intestinal lumen. (B) Groups of male Nox1-deficient mice and littermate controls (WT) were intragastrically inoculated with an equal mixture of the EcN wild-type strain and an isogenic appC mutant. On the day of gavage, mice received 3% DSS in the drinking water. Nine days later, the CI in the intestinal contents was determined. (C) Groups of male and female Noxa1ΔIEC mice and littermate controls (ctrl) were colonized with an equal mixture of EcN wild-type strain and an appC mutant and treated with DSS. After 5 days, the CI in the intestinal content was determined. (D) Groups of male and female C57BL/6 mice were given 1.5 % DSS in the drinking water for 9 days. On day 4, mice were intragastrically inoculated with an equal mixture of EcN wild-type strain and an appC mutant or a katE katG and a katE katG appC mutant. On day 9, the CI in the intestinal content was determined.
Bars represent geometric means ± 95% confidence intervals. *, p < .05; ***, p < 0.001.
NOX1 is expressed in colonocytes as well as other cell types (Konior et al., 2014). We therefore generated mice in which NOX1 function was defective in gut epithelial cells. NOX activator 1 (NOXA1) licenses NOX1 activity and is required for the generation of ROS (Banfi et al., 2003; Maehara et al., 2010). We then repeated the competitive colonization experiment in Noxa1ΔIEC (Noxa1fl/fl Tg(Vil-cre)+/−) mice and littermate controls (Noxa1fl/fl Tg(Vil-cre)−/−) (Fig. S3). The lack of NOXA1 in the epithelium had no discernable effect on the severity of inflammation in the DSS colitis model (Fig. S3A and B). Importantly, the fitness advantage conferred by AppBCX was significantly reduced in Noxa1ΔIEC mice, (Fig. S3C). Over the course of 9 days of DSS treatment, the integrity of the intestinal mucosa is compromised (Fig. 1B and C). To specifically interrogate the role of epithelial-derived ROS prior to epithelial damage, we decreased the length of DSS treatment to 5 days as this was sufficient to observe a fitness phenotype of the appC mutant (Fig. 1E). In wild-type littermate controls, appC provided a fitness advantage after 5 days of DSS treatment, while in Noxa1ΔIEC mice, this fitness advantage was ablated (Fig. 2C and S2B). These results indicate that epithelial NOX1 is required for E. coli to benefit from AppBCX function.
Reactive oxygen species and respiration confer an AppBCX-dependent growth advantage in vitro
We next sought to establish an in vitro culture system to study the physiological function of AppBCX. We hypothesized that ROS and oxygen availability may help mimic conditions found in the inflamed gut and induce AppBCX activity. We used mucin broth (Spiga et al., 2017), composed of hog mucin suspended in NCE minimal media, and added H2O2. H2O2 is a much longer-lived species than other forms of ROS and diffuses more freely through membranes. By modifying preincubation protocols in the anaerobic chamber, we were able to adjust initial oxygen levels between about 0.2 % and 18 % (ambient) oxygen (Fig. 3A and Fig. S4A).
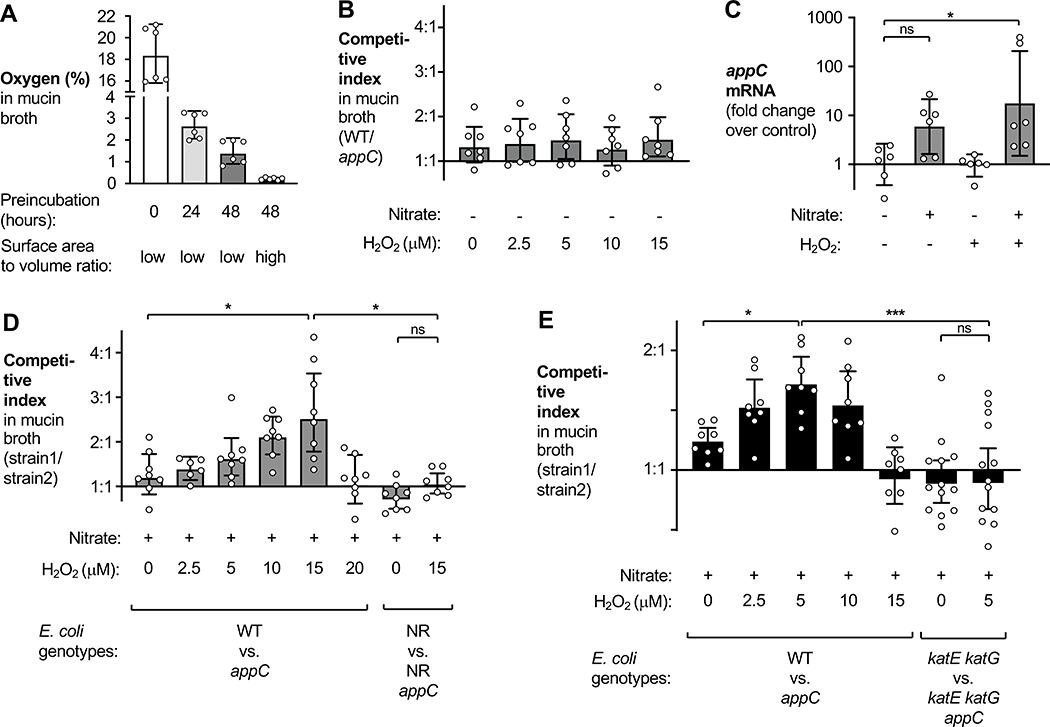
Figure 3. AppBCX provides a fitness advantage during anaerobic growth in the presence of low levels of H2O2.
See also Fig. S4.
(A) Mucin broth, in microcentrifuge tubes (low surface area to volume ratio) or in glass flasks (high surface area to volume ratio), was incubated anaerobically for the indicated period of time. The oxygen concentration in the media was determined using a Clark-type oxygen sensor. (B) Mucin broth in microcentrifuge tubes (low surface area to volume ratio) was preincubated anaerobically for 48 h and then inoculated with a 1:1 mixture of an EcN wild-type strain and an isogenic appC mutant. H2O2 was added at the indicated concentrations. (C) EcN wild-type bacteria were grown in mucin broth, prepared as in (B), supplemented with either water or nitrate (0.4 mM). After four hours of growth, H2O2 (12.5 μM) or water was added. Thirty minutes later, RNA was isolated and samples analyzed via RT-qPCR. appC mRNA levels were normalized to gmk mRNA. (D) Mucin broth was prepared as in (B) and supplemented with 0.4 mM nitrate and various concentrations of H2O2. This media was inoculated with a 1:1 mixture of the indicated strains and after 6 h, the CI was determined. (E) Mucin broth was preincubated anaerobically in a flask (high surface area to volume ratio) for 48 h. Nitrate (0.4 mM) and various concentrations of H2O2 were added and the media inoculated with a 1:1 mixture of the indicated strains. After 6 h, the CI was determined.
Bars represent geometric means ± 95% confidence intervals. *, p < .05; ***, p < 0.001; ns, not statistically significant)
We first determined the competitive fitness of the EcN wild-type strain and the appC mutant in mucin broth, with an initial concentration of approximately 1.5 % oxygen (Fig. 3B). We added various concentrations of H2O2to the media at the beginning of the experiment. After 6 h, we plated cultures on selective agar and the CI was determined. Regardless of the H2O2 concentration, AppBCX provided no significant growth advantage as the wild-type EcN and the appC mutant were recovered in similar numbers (Fig. 3B). Under these conditions, H2O2 concentrations exceeding 15 μM impeded growth of E. coli.
The ArcAB two-component system regulates transcription of appBCX (Park et al., 2013). ArcAB senses the redox state of membrane-bound quinones in the electron transport chain. We hypothesized that active electron transport, sensed by ArcAB, may be required to observe AppBCX activity. We therefore quantified appC mRNA levels in mucin broth supplemented with the alternative electron acceptor nitrate and H2O2 (Fig. 3C). Addition of both nitrate and H2O2 significantly increased the transcription of appC (Fig. 3C), suggesting that optimal AppBCX induction might require both cues. In contrast, neither nitrate or H2O2 alone or together increased transcription of cydA (Fig. S4B). We next repeated the competitive growth experiment and added varying concentrations of H2O2 to the media (Fig. 3D). In the presence of nitrate, the EcN wild-type strain outcompeted the appC mutant upon the addition of 15 μM H2O2. Consistent with the idea that nitrate induces appBCX transcription via the respiration-sensing ArcAB system, the fitness advantage conferred by AppBCX was ablated in a mutant lacking all three nitrate reductases (NarZYWV, NarGHJI, and NapABC; NR mutant) (Fig. 3D).
We hypothesized that the substrate for AppBCX is derived from H2O2. However, under these experimental conditions, a small amount of oxygen (1.5%) was present (Fig. 3A) and it is possible that residual oxygen serves as the terminal electron acceptor under these conditions. We reduced background levels of oxygen to about 0.2 % by increasing the surface area-to-volume ratio of the culture vessel (Fig. 3A and E). Akin to our previous results, the EcN wild-type strain exhibited a significant fitness advantage over the appC mutant in nitrate-containing media upon the addition of H2O2 in a dose-dependent manner (Fig. 3E), supporting the idea that the substrate for AppBCX is indeed derived from H2O2. Under these culture conditions, the other cytochrome bd oxidase CydABX does not function (Fig. S4B and S4C).
We next determined whether H2O2 or its breakdown product oxygen is the substrate for AppBCX. The catalases KatE and KatG could convert H2O2 to oxygen, which in return could serve as the substrate for AppBCX. Alternatively, cytochrome bd-type oxidases exhibit catalase activity (Borisov et al., 2013) in vitro. To test the former idea, we analyzed the fitness advantage conferred by AppBCX in the absence of KatE and KatG (katE katG mutant vs. katE katG appC mutant) (Fig. 3E). At a concentration of 5 μM H2O2, the wild-type strain outcompeted the appC mutant, while the katE katG mutant and the katE katG appC mutant were recovered in similar numbers under identical culture conditions (Fig. 3E). At this H2O2 concentration, growth of the catalase deficient mutants was not significantly inhibited (data not shown). We also performed an analogous experiment in the DSS colitis model (Fig. 2D and S2C). Catalase-deficient mutants are highly susceptible to oxidative stress and we thus decreased the DSS concentration. Wild-type mice were treated with 1.5% DSS in the drinking water for 9 days. On day 4, mice were colonized with a mixture of the EcN wild-type strain and the appC mutant or with a mixture of the katE katG and the katE katG appC mutant. The magnitude of the fitness advantage conferred by appC in this modified DSS model (Fig. 2D) was somewhat reduced compared to our previous observations, yet catalase-deficient mutants were proficient for gut colonization under these conditions (Fig. S2C). Most importantly, the competitive growth advantage conferred by appC was abolished in the absence of catalase activity. Taken together, these experiments suggest that oxygen, generated by catalase-mediated degradation of ROS, serves as the terminal electron acceptor for AppBCX.
AppBCX provides a fitness advantage in a genetic model of colitis
At later time points, DSS treatment results in profound epithelial erosion. Therefore, we investigated the role of AppBCX in Il10−/− mice. These animals develop colitis spontaneously over their lifetime, a process that can be accelerated by providing piroxicam in the diet (Hale et al., 2005). We fed groups of Il10−/− mice or wild-type BALB/c mice a piroxicam-fortified or control diet (Fig. 4). To facilitate engraftment in this model, we used the mouse-adapted commensal E. coli strain MP1. We colonized mice with a 1:1 ratio of wild-type MP1 and an isogenic appBC mutant. After 14 days, piroxicam administration resulted in increased mRNA levels of proinflammatory markers (Fig. 4A and B) and the development of inflammation in the cecal tissue (Fig. 4C and D). The MP1 wild-type strain outcompeted the appBC mutant in piroxicam-fed Il10−/− mice, while we observed no fitness advantage in mice on the standard diet (Fig. 4E and S4D). These data suggest that AppBCX provides a fitness advantage in a murine model of inflammatory bowel disease.
Figure 4. AppBCX provides a fitness advantage in a piroxicam-accelerated Il10−/− model of colitis.
See also Fig. S4.
Groups of male and female wild-type BALB/c mice or Il10−/− BALB/c were intragastrically inoculated with a 1:1 ratio of the MP1 wild-type strain and an isogenic appBC mutant. Beginning with the day of the gavage, one group of BALB/c Il10−/− were fed a piroxicam fortified diet. After 14 days, mice were euthanized and samples analyzed. (A – B) mRNA levels of Tnfa (A) and Nos2 (B) in the cecal tissue. (C) Samples were scored by a veterinary pathologist for submucosal edema (light gray bars), epithelial damage (black bars), infiltration by polymorphonuclear cells; PMN (dark gray bars), and exudate (white bars). Bars for each histopathology score category are the average per treatment group. (D) Representative images of H+E stained sections of the cecum. Scale bar, 500 μm. (E) CI of the MP1 wild-type strain and the appBC mutant in the cecum content.
Bars represent geometric means ± 95% confidence intervals. *, p < 0.05; **, p < 0.01; ns, not statistically significant.
DISCUSSION
The mechanisms driving the bloom of Enterobacteriaceae family members in IBD patients or in models of non-infectious inflammatory diarrhea are not completely understood. During antibiotic- and pathogen-induced dysbiosis, changes in colonocyte metabolism allow oxygen to diffuse into the gut lumen, supporting growth of facultative anaerobic bacteria (Byndloss et al., 2017; Rivera-Chavez et al., 2016). Furthermore, in mouse models of colitis, genes encoding components of the bacteria respiratory chain are a signature of a dysbiotic microbiota (Hughes et al., 2017), suggesting that oxygen respiration may occur during non-infectious colitis. At the same time, recruitment of inflammatory cells, especially neutrophils, influences local oxygen levels. The oxidative burst mounted by neutrophils consumes oxygen, creating a hypoxic microenvironment termed inflammatory hypoxia (reviewed in (Campbell et al., 2016; Taylor and Colgan, 2017)). Our study provides an explanation for these seemingly contradictory observations on oxygen availability and bacterial respiratory processes during non-infectious colitis. The picture emerging from our work is that ROS generated by NOX1 at the epithelial interface serve as a local source of oxygen to support AppBCX-mediated respiration by E. coli and possibly other Enterobacteriaceae family members. Curiosuly, E. coli utilizes sublethal amounts of ROS as a substrate for in vitro growth through a cytochrome c peroxidase (Khademian and Imlay, 2017). It is conceivable that the E. coli cytochrome c peroxidase provides a fitness advantage by direct degradation of hydrogen peroxide in the inflamed gut.
Very early onset IBD patients, those diagnosed before the age of 6, often have genetic mutations in genes related to the generation of host-derived ROS (reviewed in (Moran, 2017)). This finding suggests a complex role of host-derived ROS in the development of disease. Many of the ROS damage DNA, proteins, and lipids, and are bactericidal in high concentrations. Lack of this immune response may allow closer microbial contact, from both commensal and pathogenic bacteria, initiating an inflammatory event. Work in a mouse model of NADPH oxidase deficiency suggests that a lack of host-generated ROS in the gut can be partially rescued by production of H2O2 by Lactobacilli populations (Pircalabioru et al., 2016). It is conceivable that lack of NOX1 activity in the Nox1−/− and Noxa1ΔIEC mice could alter microbial community composition and metabolism in our colitis model, however, this did not compensate for the lack of host-produced ROS since the fitness defect of the appC mutant was abolished in both Nox1-deficient and Noxa1ΔIEC mice.
In addition to the antimicrobial properties of ROS, many ROS play important roles in host signaling during health and disease. While further research is needed to fully elucidate the role of ROS in host response, the concentration, subcellular localization, and route of signaling (autocrine/paracrine/microbiota-derived) appear to be important factors (Schmidt et al., 1995; Voltan et al., 2008). In particular, both NOX1- and DUOX2-derived ROS contribute to wound repair responses in the epithelium, pathogen recognition, and immune cell recruitment (Coant et al., 2010; Kawahara et al., 2004; Leoni et al., 2013; Thiagarajah et al., 2017). While we cannot formally rule out the possibility that the signaling function of NOX1-derived ROS contributes to the appC phenotype, we did not observe any overt changes in the overall inflammatory responses in Noxa1ΔIEC animals in our model.
About half of all genes encoded by E. coli are not well characterized (Riley et al., 2006). By using conditions found in the inflamed gut, such as nitrate (Winter et al., 2013) and H2O2, we were able to deduce cues needed for expression of genes that were considered cryptic. In vitro experiments have identified additional layers of regulation of AppBCX (Atlung and Brondsted, 1994; Atlung et al., 1997; Brondsted and Atlung, 1996), some of which are critical for gut colonization (Lasaro et al., 2014). In some bacteria, oxidative and nitrosative stress induce expression of bd oxidases and bd oxidase mutants are more susceptible to H2O2 (Lindqvist et al., 2000; Wall et al., 1992). E. coli has two well described regulators that detect H2O2stress (OxyR) and redox-active compounds (SoxS and SoxR). In previous research, AppBCX has not been found to be a part of the SoxS or OxyR regulon. Similarly, we found no effect of sublethal amounts of H2O2 on appC transcription. The exact mechanisms that govern AppBCX expression remain to be determined.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to Sebastian E. Winter (Sebastian.Winter@UTSouthwestern.edu).
Materials Availability
All plasmids and bacterial strains generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate any datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
The E. coli strains used in this study are listed in the Key Resource Table. Strains were grown aerobically in LB broth (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl) or on LB agar plates (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl, 15 g/l agar) at 37°C. When appropriate, carbenicillin (Carb), chloramphenicol (Cm), and kanamycin (Kan) were added to the media at a final concentration of 100 μg/ml, 20 μg/ml, and 50 μg/ml, respectively. Strains deficient for cydA, katE, and katG were generated and cultured anaerobically (90 % N2, 5 % CO2, 5 % H2; Sheldon Manufacturing) using preincubated media.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| zxx::RP4 2-(Tetr::Mu) (Kanr::Tn7) λpir | (Simon, 1983) | S17–1 λpir |
| F−endA1 hsdR17 (r−m+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U189 φ80lacZΔM15 λpir | (Pal et al., 2005) | DH5α λpir |
| E. coli Nissle 1917 (O6:K5:H1) | (Grozdanov et al., 2004) | E. coli Nissle 1917 |
| E. coli Nissle 1917 ΔcydA | (Hughes et al., 2017) | SW1362 |
| E. coli Nissle 1917 ΔappC | This study | MW139 |
| E. coli Nissle 1917 ΔkatE | This study | RC32 |
| E. coli Nissle 1917 ΔkatE ΔkatG | This study | RC76 |
| E. coli Nissle 1917 ΔkatE ΔkatG ΔappC | This study | RC115 |
| E. coli Nissle 1917 ΔnarG ΔnapA ΔnarZ | (Winter et al., 2013) | SW930 |
| E. coli Nissle 1917 ΔnarG ΔnapA ΔnarZ ΔappC | This study | RC147 |
| E. coli MP1 | (Lasaro et al., 2014) | MP1 |
| E. coli MP1 ΔappBC | This study | RC75 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SYBR Green qPCR Master Mix | Life Technologies | Cat# 4309155 |
| Mucin from porcine stomach, Type II | Sigma | Cat# M2378 |
| Sodium Nitrate | Sigma | Cat# S5506 |
| Dextran sulfate sodium | Alfa Aesar | Cat# J63606 (lots U03C023, T11D032, M06B017, N15F027) |
| TRI Reagent | Molecular Research | Cat# TR118 |
| Piroxicam diet (100ppm) | Envigo | Custom diet |
| Hydrogen Peroxide (3%) | Henry Schein | Cat# 112–7069 |
| LB Broth, Miller (Luria Bertani) | Becton Dickinson | Cat# 244520 |
| LB Agar, Miller (Luria Bertani) | Becton Dickinson | Cat# 244620 |
| Lysozyme from chicken egg white | Sigma | Cat# L4919 |
| Kanamycin Sulfate | Fisher | Cat# BP906 |
| Chloramphenicol | Fisher | Cat# BP904 |
| Carbenicillin, Disodium Salt | VWR | Cat# J358 |
| Critical Commercial Assays | ||
| Gibson Assembly Cloning Kit | New England Biolabs | Cat# E2611 |
| Q5 Hot Start 2x Master Mix | New England Biolabs | Cat# M0494L |
| TURBO DNA-free Kit | Invitrogen | Cat# AM1907 |
| QIAfilter Plasmid Midi Kit | Qiagen | Cat# 12245 |
| QIAEX II Gel Extraction Kit | Qiagen | Cat# 20021 |
| Aurum Total RNA Mini Kit | Bio-Rad | Cat# 7326820 |
| Dynabeads mRNA Direct Kit | Life Technologies | Cat# 61012 |
| TaqMan Reverse Transcription Reagents | Applied Biosystems | Cat# N8080234 |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT: C57BL/6J | The Jackson Laboratory | Cat# 000664 |
| Mouse: Nox1: B6.12X1-Nox1tm1Kkr/J | The Jackson Laboratory | Cat# 018787 |
| Mouse: Noxa1ΔIEC: Noxa1fl/fl Tg(Vil-cre)+/− | This study | |
| Mouse: BALB/c: BALB/cJ | The Jackson Laboratory | Cat# 000651 |
| Mouse: Il10−/−: C.129P2(B6)-Il10tm1Cgn/J | The Jackson Laboratory | Cat# 004333 |
| Mouse: B6.Cg-Tg(Vil1-cre)997Gum/J | The Jackson Laboratory | Cat# 004586 |
| Oligonucleotides | ||
| Information regarding oligonucleotides used in this study is listed in Table S1 | ||
| Recombinant DNA | ||
| ori(R101) repA101ts Carbr | (Winter et al., 2013) | pSW172 |
| ori(pSC101) Carbr | (Wang and Kushner, 1991) | pWSK29 |
| ori(pSC101) Kanr | (Wang and Kushner, 1991) | pWSK129 |
| ori(R6K) mobRP4 sacRB Cmr Tetr | (Kingsley et al., 1999) | pRDH10 |
| ori(R6K) mobRP4 sacRB Kanr | (Gillis et al., 2018) | pGP706 |
| Upstream and downstream regions of the MP1 appBC genes in pGP706 | This study | pRC11 |
| Upstream and downstream regions of the Nissle appC gene in pRDH10 | This study | pSW296 |
| Upstream and downstream regions of the Nissle katG gene in pGP706 | This study | pRC8 |
| Upstream and downstream regions of the Nissle katE gene in pGP706 | This study | pRC6 |
| Software and Algorithms | ||
| Excel for Mac 16.16 | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel#pivot-forPersonal |
| Prism V8.0 | Graph Pad | https://www.graphpad.com/scientific-software/prism/ |
| MacVector V13.5.2 | Mac Vector | https://macvector.com/ |
| QuantStudio6 | ThermoFisher | https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-7-flex-real-time-pcr-system.html |
| Photoshop Elements 15 | Adobe | https://www.adobe.com/products/photoshop-elements.html |
| Other | ||
| Oxygen Probe | Unisense | OX-N-14839 |
| Anaerobic Chamber | Sheldon Manufacturing | Bactron300 |
Animal experimentation
All mouse experiments were reviewed and approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center. Mice were kept in a specific pathogen free environment with standard 12 h light/dark cycles. Animals had ad libitum access to feed and water.
DSS model of colitis
We used male and female 7–12-week-old wild-type C57BL/6 WT and C57BL/6 Noxa1ΔIEC mice. For C57BL/6 Nox1tm1Kkr/J, we used 7–12-week-old male mice. Colitis was induced by administering dextran sulfate sodium (DSS; Alfa Aesar) in the drinking water. For all experiments, 3 % DSS was used, except for the experiments shown in Fig. 2D and S2C, in which 1.5 % was used. Mice were randomly assigned into cages or separated by genotype 3 days before the experiment. In the competitive colonization experiments, animals were inoculated with 5 × 108 CFU of each E. coli strain at the indicated time point. For experiments in the DSS colitis model that used 3% DSS and lasted for a total of 9 days, DSS-supplemented drinking water was switched to filter-sterilized water one day prior to the end of the experiment. After euthanization, cecal and colonic content were harvested in sterile PBS and stored on ice. The E. coli burden in the luminal contents was determined by plating serial 10-fold dilutions on LB plates supplemented with the appropriate antibiotics.
Piroxicam-accelerated Il10−/− model of colitis
7–12-week-old male and female Il10−/− BALB/c were randomly assigned into cages before oral inoculation with 5 × 108 CFU of the E. coli MP1 wild-type strain and 5 × 108 CFU of the appBC mutant. Regular mouse chow was replaced with Piroxicam-fortified diet (100 ppm; Teklad custom research diets, Envigo) and changed daily. After 14 days, mice were euthanized and the samples were collected as described above.
METHOD DETAILS
Plasmids
All plasmids used in this study are listed in the Key Resource Table. EcN and MP1 strains were differentially marked with the low-copy number plasmids pWSK29 and pWSK129 (Wang and Kushner, 1991) to facilitate recovery and enumeration of strains (Winter et al., 2013). All plasmids were created using the Gibson Assembly Cloning kit (New England Biolabs) and following standard molecular cloning techniques. For plasmids created in this study (pRC6, pRC8, pRC11, pSW296), DNA regions immediately upstream and downstream of katE, katG, appBC and appC were PCR amplified using Q5 Hot Start High Fidelity DNA Polymerase (New England Biolabs) from EcN or MP1 using the primers specified in the Table S1. DNA fragments were ligated using the Gibson method into the SphI-digested plasmid pGP706 or pRDH10 as noted. Plasmid inserts were verified by Sanger DNA sequencing.
Construction of bacterial mutants by allelic exchange
The routine host for suicide plasmids was DH5α λpir. For conjugations, plasmids were transferred to S17–1 λpir hosts to serve as the donor strain. To allow for recovery of exconjugants, EcN and MP1 recipient strains contained the heat-sensitive plasmid pSW172. Exconjugants, in which the suicide plasmid had integrated into the chromosome by single crossover, were selected for using LB plates containing Kan or Cm, respectively. All conjugation experiments with EcN and MP1 were carried out at 30 °C to enable stable replication of the heat sensitive plasmid pSW172. Integration of the plasmids in the chromosome was confirmed by PCR when appropriate. Subsequently, second crossover events were selected for using sucrose plates (5 % sucrose, 15 g/L agar, 8 g/L nutrient broth base). This second event leads to an unmarked deletion, which was confirmed by PCR. pSW172 was cured by growing the bacteria overnight at 37 °C. MW139 and RC147 were generated by introducing pSW296 into EcN and SW930, respectively. RC75 and RC32 were generated by introducing pRC11 and pRC6 into EcN and MP1, respectively. The EcN katE katG appC mutant (RC115) was created similarly by introducing the katG mutation into RC32 (katE) and by subsequently creating a mutation of appC in RC76 (katE katG). Mutations were confirmed by PCR.
Histopathology
Fomalin-fixed (10% buffered formalin phosphate; Thermo Fischer) tissue was embedded in paraffin and stained with hematoxylin and eosin. The samples were blinded and scored by a veterinary pathologist according to criteria described in (Winter et al., 2013). Representative images were uniformly linearly adjusted using Adobe Photoshop Elements 15.
Intestinal mRNA analysis
The relative transcription levels of Nos2 and Tnf genes was determined by RT-qPCR. Cecal tissue was homogenized in a Mini beadbeater (Biospec Products, Bartlesville) and RNA was extracted using the TRI reagent method (Molecular Research Center, Cincinnati). For samples from DSS-treated mice, residual DSS contaminants were further purified using the Dynabeads mRNA Direct Kit (Life Technologies) per the manufacturer`s instructions. cDNA was generated by TaqMan reverse transcription reagents (Life Technologies). Real-time PCR was performed using SYBR Green (Life Technologies). Data was processed in a QuantStudio 6 Flex instrument (Life Technologies) and analyzed using the comparative CT method. The primers listed in the Table S1 were added at a final concentration of 250 nM. Target gene transcription of each sample was normalized to Eef2 mRNA (Eissa et al., 2016).
In vitro growth assays
To limit background levels of oxygen in the culture media, all assays were conducted in an anaerobic chamber and bacterial strains were inoculated, grown, and prepared in the anaerobic chamber. Competitive growth assays were performed in LB broth or mucin broth, as indicated in the figure legend. Mucin broth contained 0.5% (w/v) hog mucin (Sigma-Aldrich, St. Louis) in no-carbon E medium (3.9 g/L KH2PO4, 5.0 g/L anhydrous K2HPO4, and 3.5 g/L NaNH4HPO4 ∗ 4 H2O) (Berkowitz et al., 1968) and was supplemented with 1 mM MgSO4. Hog mucin was sterilized by dissolving 100 mg in 70% Ethanol, heating to 65 °C for 2 h, and incubating at 25 °C for 18 h prior to dehydration (Vacufuge plus, Eppendorf). Sodium nitrate (Sigma Aldrich, St. Louis) and H2O2 (Henry Schein) were added at the indicated final concentrations.
For competitive growth assays, 1 mL of mucin broth was inoculated with the indicated strains at a final concentration of 1 × 103 CFU/mL per strain in a 1.5 mL microcentrifuge tube. After addition of the indicated concentration of H2O2 or water, tubes were closed and incubated for 6 h at 37 °C in the anaerobic chamber. In some experiments, all media and reagents were preincubated in 200 mL flasks for 48 h; the high surface area to volume ratio facilitates gas exchange and the removal of oxygen.
For comparison of aerobic and anaerobic growth of various E. coli mutants, 100 mL of LB broth was inoculated with 1 mL of an overnight culture. Aerobically grown samples were placed in a shaker at 37 °C. Cultures grown under anaerobic conditions were kept in an anaerobic chamber and preincubated media was used (Bactron EZ Anaerobic Chamber; 5% hydrogen, 5% CO2, 90% nitrogen; Sheldon Manufacturing). The OD600 of each sample was taken every 45 min. Generation time was calculated using logarithmic growth rate. Experiments were done in triplicate.
In vitro mRNA analysis
Mucin broth was preincubated in the anaerobic chamber for 48 h prior to experimental start. Five mL of medium was inoculated with 100 μl of overnight EcN in a 15 mL sealable conical tube. Nitrate (0.4mM) was added as indicated. After 4 h of growth in the anaerobic chamber, H2O2 (12.5 μM final concentration) or water was added to the cultures. Cultures were allowed to grow for an additional 30 minutes. RNA was extracted using the Aurum total RNA minikit (Bio-Rad). Double the indicated volumes of lysozyme, lysis buffer, and isopropanol were used. The RNA was then DNase treated (Invitrogen) twice according to the manufacturer’s instructions. cDNA was generated by TaqMan reverse transcription reagents (Life Technologies). A reaction with no reverse transcriptase was performed to quantify contamination with DNA. Real-time PCR was performed as above. The primers listed in the Table S1 were added at a final concentration of 250 nM. Target gene transcription of each sample was normalized to gmk mRNA.
Oxygen measurement
A Clark-type oxygen sensor (Unisense) was used to quantify the amount of oxygen in pre-incubated media. 750 μL aliquots of mucin broth were removed from the anaerobic chamber with a 1 mL layer of pre-incubated mineral oil placed on top. The probe was polarized and calibrated according to manufacturer’s recommendation. Signal (S) was converted from partial pressure to concentration (C) using the following equation, where atmospheric level solubility (a) was estimated to be 230.2, (S0) is calibration for zero oxygen, and (Sat) is the calibration for atmospheric reading: C = a x (S - S0) / (Sat - S0).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed and graphs created using Prism and Microsoft Excel. p values of less than 0.05 were considered statistically significant. In some instances, the p value is shown in the graph. Unless otherwise stated, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not statistically significant. The definition of bar height and error bars is listed in each figure legend.
In vitro experiments
The number of dots shown in each graph corresponds to samples obtained from independent repeats of the experiments. Bacterial growth experiments performed under laboratory conditions were analyzed using the Kruskal-Wallis test with Dunn’s post hoc multiple analyses test.
Mouse experiments
The number of dots shown in each graph refers to the number of animals from which samples were taken. Sample sizes, such as the number of animals per group, were not estimated a priori since effect sizes in our system are difficult to predict. Animals that had no detectable colonization by experimentally introduced bacterial strains were removed from the analysis. We determined colonization to be greater than 1 colony per 100 μL of resuspended intestinal content. In total, 18 mice were excluded from our study. The majority of these excluded mice (16 of the 18) were from non-inflamed control groups or mice treated with DSS for 5 days or less. Mice that had to be euthanized for humane reasons prior to the predetermined time point were also not further analyzed. All competitive indices in murine models were analyzed using the Mann-Whitney U-test. All bacterial colonization abundances were analyzed using the Wilcoxon signed rank test (for paired comparisons).
DATA AND CODE AVAILABILITY
No large datasets or code were generated as part of this study.
Supplementary Material
Highlights.
AppBCX promotes the outgrowth of E. coli in murine models of gut inflammation.
AppBCX respiration requires reactive oxygen species produced by epithelial NOX1.
Hydrogen peroxide, detoxified to oxygen, supports AppBCX respiration in vitro.
ACKNOWLEDGEMENTS
We thank Drs. Jessica Moreland, Vanessa Sperandio, and Julie Pfeiffer (UT Southwestern Medical Center) for helpful discussions. Work in S. E.W.’s lab was funded by the The Welch Foundation (I-1969-20180324), NIH (AI118807, AI128151), the Burroughs Wellcome Fund (1017880), and a Research Scholar Grant (RSG-17-048-01-MPC) from the American Cancer Society. R.C. was in part supported by T32 training grant AI007520 and W.Z. was supported by a Research Fellows Award from the Crohn’s and Colitis Foundation (454921). Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies. The funders had no role in study design, data collection, interpretation, or decision to submit the work for publication.
Footnotes
DECLARATION OF INTEREST
The corresponding author (SEW) is listed as an inventor on patent application WO2014200929A1, which describes a treatment to prevent the inflammation-associated expansion of Enterobacteriaceae. The other authors have no additional financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. (2014). Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 147(5), 1055–1063 e1058. Published online 2014/07/22 DOI: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 338(6103), 120–123. Published online 2012/08/21 DOI: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T, and Brondsted L (1994). Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 176(17), 5414–5422. Published online 1994/09/01 DOI: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T, Knudsen K, Heerfordt L, and Brondsted L (1997). Effects of sigmaS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J Bacteriol. 179(7), 2141–2146. Published online 1997/04/01 DOI: 10.1128/jb.179.7.2141-2146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviello G, and Knaus UG (2018). NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol. 11(4), 1011–1023. Published online 2018/05/11 DOI: 10.1038/s41385-018-0021-8. [DOI] [PubMed] [Google Scholar]
- Banfi B, Clark RA, Steger K, and Krause KH (2003). Two novel proteins activate superoxide generation by the NADPH oxidase NOX1. J Biol Chem. 278(6), 3510–3513. Published online 2002/12/11 DOI: 10.1074/jbc.C200613200. [DOI] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, and Salzman N (2008). Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 76(3), 907–915. DOI: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, and de Mattos MJ (2009). Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J Bacteriol. 191(17), 5510–5517. Published online 2009/06/23 DOI: 10.1128/JB.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ Jr., Roth J, and Ames BN (1968). Procedure for identifying nonsense mutations. J Bacteriol. 96(1), 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Forte E, Davletshin A, Mastronicola D, Sarti P, and Giuffre A (2013). Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett. 587(14), 2214–2218. Published online 2013/06/04 DOI: 10.1016/j.febslet.2013.05.047. [DOI] [PubMed] [Google Scholar]
- Brondsted L, and Atlung T (1996). Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 178(6), 1556–1564. Published online 1996/03/01 DOI: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss MX, Olsan EE, Rivera-Chavez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, et al. (2017). Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 357(6351), 570–575. Published online 2017/08/12 DOI: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EL, Kao DJ, and Colgan SP (2016). Neutrophils and the inflammatory tissue microenvironment in the mucosa. Immunol Rev. 273(1), 112–120. Published online 2016/08/26 DOI: 10.1111/imr.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, Freund JN, Cazals-Hatem D, Bouhnik Y, Woerther PL, et al. (2010). NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol. 30(11), 2636–2650. Published online 2010/03/31 DOI: 10.1128/MCB.01194-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnutte JT, Berkow RL, Roberts RL, Shurin SB, and Scott PJ (1988). Chronic granulomatous disease due to a defect in the cytosolic factor required for nicotinamide adenine dinucleotide phosphate oxidase activation. J Clin Invest. 81(2), 606–610. Published online 1988/02/01 DOI: 10.1172/JCI113360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa J, Fsihi H, Marck C, Dion M, Kieffer-Bontemps M, and Boquet PL (1991). A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol Gen Genet. 229(3), 341–352. Published online 1991/10/01 DOI: 10.1007/bf00267454. [DOI] [PubMed] [Google Scholar]
- Dinauer MC, Orkin SH, Brown R, Jesaitis AJ, and Parkos CA (1987). The glycoprotein encoded by the X-linked chronic granulomatous disease locus is a component of the neutrophil cytochrome b complex. Nature. 327(6124), 717–720. Published online 1987/06/01 DOI: 10.1038/327717a0. [DOI] [PubMed] [Google Scholar]
- Eissa N, Hussein H, Wang H, Rabbi MF, Bernstein CN, and Ghia JE (2016). Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis. PLoS One. 11(5), e0156289 Published online 2016/06/01 DOI: 10.1371/journal.pone.0156289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, and Pace NR (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 104(34), 13780–13785. DOI: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, and Glimcher LH (2007). Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131(1), 33–45. Published online 2007/10/10 DOI: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, et al. (2018). Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe. 23(1), 54–64 e56. Published online 2017/12/26 DOI: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, and Baumler AJ (2008). T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun. 76(5), 2008–2017. Published online 2008/03/19 DOI: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, and Dobrindt U (2004). Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 186(16), 5432–5441. Published online 2004/08/05 DOI: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale LP, Gottfried MR, and Swidsinski A (2005). Piroxicam treatment of IL-10-deficient mice enhances colonic epithelial apoptosis and mucosal exposure to intestinal bacteria. Inflamm Bowel Dis. 11(12), 1060–1069. Published online 2005/11/25 DOI: 10.1097/01.mib.0000187582.90423.bc. [DOI] [PubMed] [Google Scholar]
- Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Buttner L, Smoot MP, Behrendt CL, et al. (2017). Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 21(2), 208–219. Published online 2017/02/10 DOI: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA (2019). Where in the world do bacteria experience oxidative stress? Environ Microbiol. 21(2), 521–530. Published online 2018/10/12 DOI: 10.1111/1462-2920.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Kuwano Y, Teshima-Kondo S, Takeya R, Sumimoto H, Kishi K, Tsunawaki S, Hirayama T, and Rokutan K (2004). Role of nicotinamide adenine dinucleotide phosphate oxidase 1 in oxidative burst response to Toll-like receptor 5 signaling in large intestinal epithelial cells. J Immunol. 172(5), 3051–3058. Published online 2004/02/24 DOI: 10.4049/jimmunol.172.5.3051. [DOI] [PubMed] [Google Scholar]
- Khademian M, and Imlay JA (2017). Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci U S A. 114(33), E6922–E6931. Published online 2017/07/12 DOI: 10.1073/pnas.1701587114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, Dougan G, Baumler AJ, Roberts M, and Williams PH (1999). Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Appl Environ Microbiol. 65(4), 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konior A, Schramm A, Czesnikiewicz-Guzik M, and Guzik TJ (2014). NADPH oxidases in vascular pathology. Antioxid Redox Signal. 20(17), 2794–2814. Published online 2013/11/05 DOI: 10.1089/ars.2013.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasaro M, Liu Z, Bishar R, Kelly K, Chattopadhyay S, Paul S, Sokurenko E, Zhu J, and Goulian M (2014). Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J Bacteriol. 196(9), 1723–1732. Published online 2014/02/25 DOI: 10.1128/JB.01296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, Hilgarth RS, Kundu K, Murthy N, Kusters D, et al. (2013). Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest. 123(1), 443–454. Published online 2012/12/18 DOI: 10.1172/JCI65831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Membrillo-Hernandez J, Poole RK, and Cook GM (2000). Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek. 78(1), 23–31. Published online 2000/10/04 DOI: 10.1023/a:1002779201379. [DOI] [PubMed] [Google Scholar]
- Litvak Y, Byndloss MX, and Baumler AJ (2018). Colonocyte metabolism shapes the gut microbiota. Science. 362(6418). Published online 2018/12/01 DOI: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, and Finlay BB (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2(3), 204 Published online 2007/11/23. [DOI] [PubMed] [Google Scholar]
- Maehara Y, Miyano K, Yuzawa S, Akimoto R, Takeya R, and Sumimoto H (2010). A conserved region between the TPR and activation domains of p67phox participates in activation of the phagocyte NADPH oxidase. J Biol Chem. 285(41), 31435–31445. Published online 2010/08/04 DOI: 10.1074/jbc.M110.161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran CJ (2017). Very early onset inflammatory bowel disease. Semin Pediatr Surg. 26(6), 356–359. Published online 017/11/12 DOI: 10.1053/j.sempedsurg.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Normann E, Fahlen A, Engstrand L, and Lilja HE (2013). Intestinal microbial profiles in extremely preterm infants with and without necrotizing enterocolitis. Acta Paediatr. 102(2), 129–136. Published online 2012/10/23 DOI: 10.1111/apa.12059. [DOI] [PubMed] [Google Scholar]
- Pal D, Venkova-Canova T, Srivastava P, and Chattoraj DK (2005). Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J Bacteriol. 187(21), 7167–7175. Published online 2005/10/21 DOI: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panday A, Sahoo MK, Osorio D, and Batra S (2015). NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 12(1), 5–23. Published online 2014/09/30 DOI: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DM, Akhtar MS, Ansari AZ, Landick R, and Kiley PJ (2013). The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet. 9(10), e1003839 Published online 2013/10/23 DOI: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Zeng MY, Caruso R, and Nunez G (2017). Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 279(1), 70–89. Published online 2017/09/01 DOI: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircalabioru G, Aviello G, Kubica M, Zhdanov A, Paclet MH, Brennan L, Hertzberger R, Papkovsky D, Bourke B, and Knaus UG (2016). Defensive Mutualism Rescues NADPH Oxidase Inactivation in Gut Infection. Cell Host Microbe. 19(5), 651–663. Published online 2016/05/14 DOI: 10.1016/j.chom.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 118(2), 229–241. Published online 2004/07/21 DOI: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. (2006). Escherichia coli K-12: a cooperatively developed annotation snapshot−-2005. Nucleic Acids Res. 34(1), 1–9. Published online 2006/01/07 DOI: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, et al. (2016). Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 19(4), 443–454. Published online 2016/04/15 DOI: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KN, Amstad P, Cerutti P, and Baeuerle PA (1995). The roles of hydrogen peroxide and superoxide as messengers in the activation of transcription factor NF-kappa B. Chem Biol. 2(1), 13–22. Published online 1995/01/01 DOI: 10.1016/1074-5521(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Shin NR, Whon TW, and Bae JW (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33(9), 496–503. Published online 2015/07/27 DOI: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1, pp. 784–791. [Google Scholar]
- Sousa PMF, Videira MAM, Bohn A, Hood BL, Conrads TP, Goulao LF, and Melo AMP (2012). The aerobic respiratory chain of Escherichia coli: from genes to supercomplexes. Microbiology. 158(Pt 9), 2408–2418. Published online 2012/06/16 DOI: 10.1099/mic.0.056531-0. [DOI] [PubMed] [Google Scholar]
- Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, et al. (2017). An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe. 22(3), 291–301 e296. Published online 2017/08/29 DOI: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5(10), 2177–2189. DOI: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturr MG, Krulwich TA, and Hicks DB (1996). Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a delta cyo delta cyd strain complemented by genes from Bacillus firmus OF4. J Bacteriol. 178(6), 1742–1749. Published online 1996/03/01 DOI: 10.1128/jb.178.6.1742-1749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, and Lambeth JD (1999). Cell transformation by the superoxide-generating oxidase Mox1. Nature. 401(6748), 79–82. Published online 1999/09/15 DOI: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- Taylor CT, and Colgan SP (2017). Regulation of immunity and inflammation by hypoxia in immunological niches. Nat Rev Immunol. 17(12), 774–785. Published online 2017/10/04 DOI: 10.1038/nri.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajah JR, Chang J, Goettel JA, Verkman AS, and Lencer WI (2017). Aquaporin-3 mediates hydrogen peroxide-dependent responses to environmental stress in colonic epithelia. Proc Natl Acad Sci U S A. 114(3), 568–573. Published online 2017/01/05 DOI: 10.1073/pnas.1612921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOrsdel CE, Bhatt S, Allen RJ, Brenner EP, Hobson JJ, Jamil A, Haynes BM, Genson AM, and Hemm MR (2013). The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J Bacteriol. 195(16), 3640–3650. Published online 2013/06/12 DOI: 10.1128/JB.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez OC, Lederer HM, and Rombeau JL (1997). Butyrate and the colonocyte. Production, absorption, metabolism, and therapeutic implications. Adv Exp Med Biol. 427, 123–134. Published online 1997/01/01. [PubMed] [Google Scholar]
- Voltan S, Martines D, Elli M, Brun P, Longo S, Porzionato A, Macchi V, D’Inca R, Scarpa M, Palu G, et al. (2008). Lactobacillus crispatus M247-derived H2O2 acts as a signal transducing molecule activating peroxisome proliferator activated receptor-gamma in the intestinal mucosa. Gastroenterology. 135(4), 1216–1227. Published online 2008/08/30 DOI: 10.1053/j.gastro.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 5(193), 193ra191 Published online 2013/07/12 DOI: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Delaney JM, Fayet O, Lipinska B, Yamamoto T, and Georgopoulos C (1992). arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J Bacteriol. 174(20), 6554–6562. Published online 1992/10/01 DOI: 10.1128/jb.174.20.6554-6562.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF, and Kushner SR (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 100, 195–199. Published online 1991/04/01. [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 467(7314), 426–429. DOI: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 339(6120), 708–711. Published online 2013/02/09 DOI: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Buttner L, de Lima Romao E, Behrendt CL, Lopez CA, et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature. 553(7687), 208–211. Published online 2018/01/13 DOI: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets or code.
No large datasets or code were generated as part of this study.