Abstract
The pathogenesis of Alzheimer’s disease (AD) remains largely unclear. Exploring the genetic/epigenetic loci showing pleiotropic association with the neuropathologies of AD may greatly enhance understanding of the mechanisms underlying the development of AD. In this study, using data from the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP), we undertook a Mendelian randomization (MR) approach integrating GWAS and DNA methylation quantitative trait loci (mQTL) data to explore pleiotropic epigenetic loci for AD neuropathologies, including amyloid-β- (Aβ) load and tau-containing neurofibrillary tangle density. We performed GWAS of DNA methylation in brain tissues from 592 participants and mapped 60,595 cis-SNP-CpG pairs after correction for multiple-testing. By linking cis- mQTL with GWAS results for Aβ load and Tau tangles, we identified 47 CpGs showing pleiotropic association with Aβ load by MR analysis. We then used gene expression data from 537 individuals and performed quantitative trait methylation (QTM) analysis. We found that 18 of the 47 CpGs were in cis associated with 25 mRNAs/genes, comprising 41 unique CpG-mRNA/gene pairs. Our findings shed light on the role of DNA methylation in the pathogenesis of Aβ.
Keywords: Alzheimer’s disease, neuropathology, Mendelian randomization, DNA methylation, quantitative trait loci, quantitative trait methylation
Introduction
Alzheimer’s disease (AD), the most common neurodegenerative disease and a major cause of disability, affects patients’ quality of life and brings a tremendous economic burden to the society (Alzheimer’s Association, 2016; GBD 2017 DALYs and HALE Collaborators, 2018). Neuropathologically, AD brains contain an extracellular buildup composed of amyloid-β peptide (Aβ) and intraneuronal accumulation of neurofibrillary tangles (NFTs) (Congdon and Sigurdsson, 2018; Ising and Heneka, 2018; Jansen et al., 2019; Kumar et al., 2020). However, the exact neuropathogenesis of AD remains to be unclear. Therefore, it is important to further explore the pathological mechanisms underlying AD and identify genetic/epigenetic loci showing pleiotropic association of with AD neuropathologies.
Although genome-wide association studies (GWAS) have revealed the complex genetic architecture of AD (Jansen et al., 2019; Lambert et al., 2013), the identified genetic variants only accounted for a portion of the heritability (Escott-Price et al., 2017; Sims et al., 2020). DNA methylation is an epigenetic marker that has been reported to play a key role in many biological processes and diseases (Ahuja et al., 2016; Horvath, 2012; Klose and Bird, 2006). Previous methylome-wide association studies (MWASs) have been successful in identifying DNA methylation loci/regions associated with AD neuropathologies. For example, multiple differentially methylated probes (DMPs) in ABCA7, B1N1 and SORL1, differentially methylated regions (DMRs), and variably methylated regions (VMRs) were found to be associated with the burden of AD neuropathologies or pathological diagnosis of AD (De Jager et al., 2014; Huo et al., 2019; Lunnon et al., 2014; Smith et al., 2018; Watson et al., 2016; Yu et al., 2015). However, these findings could be subject to confounding factors and reverse causation that might influence the results. As a result, more studies are needed to explore pleiotropic DNA methylation loci for AD neuropathologies.
It is a major public health goal to identify modifiable causes of a disease/disorder in order to develop effective interventions or therapeutic strategies. However, risk factors identified by conventional observational epidemiology studies were often found to be misleading because the findings were subject to confounding, reverse causation and selection bias (Lawlor et al., 2004; Lawlor and Smith, 2006). Randomized controlled trials (RCTs) are often considered to be the gold standard to make pleiotropic inferences as all the characteristics, except the exposure of interest, are comparable among the groups (Bhide et al., 2018). However, conduction of RCTs is time-consuming, expensive, and in some cases, the allocation of exposure is unethical or impractical.
Mendelian randomization (MR) refers to methods that use proxy of modifiable risk factors to examine the pleiotropic effect of these risk factors on diseases/disorders by utilizing data from observational epidemiology studies without the need of conducting a conventional RCT (Emdin et al., 2017). MR uses instrumental variables (IVs) as the proxy to mimic randomization of individual to an exposure to ensure comparability of individuals with respect to known/unknown confounding factors, thereby enabling the estimation of a pleiotropic association with an outcome (Lawlor et al., 2008). Genetic variants are often used as the IVs because random allocation of alleles of genetic variants occur during gamete formation, well before the exposure or outcome. Estimation of the pleiotropic association can be made because the inherited genetic variants are independent of potentially confounding factors.
Recently, a novel analytical framework was applied to evaluate the pleiotropic association between DNA methylation levels and diseases that could minimize confounding and reverse causation through an MR approach integrating cis- DNA methylation quantitative trait loci (cis-mQTL) and GWAS data (Hannon et al., 2018; Hannon et al., 2017; Huan et al., 2019; Richardson et al., 2018; Richardson et al., 2017). Here, we adopted this novel MR approach to search for DNA methylation loci showing pleiotropic association with AD neuropathologies and to explore functional mechanisms underlying the association of genetic variants with AD neuropathologies.
Materials and methods
Study participants
Data for the present study were drawn from the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP), both of which are ongoing, prospective studies of brain aging and dementia in older individuals (Bennett et al., 2018; Bennett et al., 2012b). All participants were free of dementia at enrollment, and agreed to annual clinical evaluations and brain donation upon death. The clinical evaluation includes detailed neurologic examination and clinical classification of dementia and AD (Bennett et al., 2018). Postmortem human dorsolateral prefrontal cortex (DLPFC) tissues were obtained from deceased participants to measure DNA methylation, gene expression and neuropathological protein. More details regarding the design of ROSMAP study can be found in previous publications (Bennett et al., 2018; Bennett et al., 2012a; Bennett et al., 2012b).
Written informed consent was obtained from participants at the beginning of each study as was an Anatomical Gift Act. Participants also signed a repository consent to allow their data to be re-purposed. Both studies were approved by the Institutional Review Board of the Rush University Medical Center. The ethics approval was given in compliance with the Declaration of Helsinki. The clinical data can be requested at https://www.radc.rush.edu/ and the omics data can be requested at https://www.synapse.org/.
Assessment of neuropathological phenotypes
All the data used in our analyses were collected by ROSMAP study. The collection of whole blood and brain tissues was described previously (Bennett et al., 2018). Detailed procedures for postmortem brain examination and neuropathological phenotyping have been described previously (Yu et al., 2015). In brief, neuropathological hallmarks including amyloid-β (Aβ) and tau-containing neurofibrillary tangles (NFTs) were quantified across the brain in both studies. Aβ protein was identified by molecularly-specific immunohistochemistry and quantified by imaging analysis, with values being summarized as percent area occupied by Aβ. Aβ scores in eight regions (i.e., hippocampus, entorhinal cortex, midfrontal cortex, inferior temporal, angular gyrus, calcarine cortex, anterior cingulate cortex and superior frontal cortex) were averaged. Tau protein was identified by molecularly specific immunohistochemistry (antibodies to abnormally phosphorylated Tau protein, AT8). Cortical density (per mm2) was determined using systematic sampling. Tau scores in the same eight regions were averaged. Both indices were square root transformed to approach a normal distribution. More information is available at https://www.radc.rush.edu.
Genotyping and genotype imputation
Genotyping was performed using the Affymetrix Genome-Wide Human SNP Array 6.0. Detailed information regarding the pipeline of quality control of the genotyping data was reported in a previous publication (De Jager et al., 2012; Ng et al., 2017). Dosages for all SNPs (>35 million) was imputed on the 1000 Genomes reference using BEAGLE 3.3.2. Imputed SNPs were filtered based on minor allele frequency (MAF) > 0.01 and imputation INFO score> 0.3, yielding 7,321,515 imputed SNPs that were used for further cis-mQTL mapping.
DNA methylation profiling
DNA methylation data were generated using the 450K Illumina array from DLPFC, and quality control was conducted as described previously (Ng et al., 2017). A total of 420,131 methylation sites (i.e., CpGs) remained after quality control, and all data had been adjusted for age at death, sex, and experimental batch (De Jager et al., 2014).
Gene expression data
Gene expression data were generated using RNA-seq from DLPFC at an average sequence depth of 90 million reads. Detailed description of the data generation and processing was described previously (Ng et al., 2017). Only highly expressed genes were kept (mean expression > 2log2(FPKM)), resulting in 55,889 mRNA and 50,999 expressed genes.
Statistical and bioinformatics analysis
Using data collected from ROSMAP study, we undertook an MR approach which included a series of analyses, as outlined below, to examine the pleiotropic association of DNA methylation with AD neuropathologies. We first performed a genome-wide mQTL analysis to identify SNPs associated with DNA methylation in participants with both genetic and DNA methylation data. The cis-mQTL analysis followed a similar approach to the one described in a previous study (Hannon et al., 2018). In brief, linear regression was performed to test the association between each SNP and CpG, with each individual CpG as the dependent variable and each individual SNP as the independent variable, adjusted for the first three principal components. In total, 7,321,515 imputed genetic variants against each of the 420,131 eligible CpGs were tested by using the R package “MatrixEQTL”. SNPs within 1 Mb of a CpG site showing significant association with the CpG (known as cis-mQTLs) were identified. We applied a conservative multiple testing correction to define cis-mQTLs (i.e., 0.05/number of SNP-CpG pairs=1.35×10−10) to reduce weak instrument bias in the MR analysis. In the case of multiple variants showing significant association with the same individual CpG, we chose the cis-mQTL variant with the smallest P-value as the instrumental variable (IV) for the CpG site. We then performed GWAS for AD neuropathologies, including amyloid-β- (Aβ) load and tau-containing neurofibrillary tangle density, adjusted for age at death, sex, education and the first three principal components. MR was undertaken with DNA methylation as the exposure, AD neuropathology as the outcome, and cis-mQTL as the IV. The analysis was done using the inverse-variance weighted (IVW) method as implemented in the mr_ivw function of the R package “MendelianRandomization” (Yavorska and Burgess, 2017). In the context of a single genetic variant as the IV, this is equivalent to a Wald ratio approach which allows distinct or overlapping samples to be used for the genetic association analysis in MR (Teumer, 2018). The weight was set to be delta to include the second-order term from the delta expansion in the calculation of the standard error of the estimate. We used the observed correlation between DNA methylation and the corresponding AD neuropathology to account for the correlation between genetic association with DNA methylation and genetic association AD neuropathologies due to sample overlap (Yavorska and Burgess, 2017) (see the supplementary file for the R and shell codes used for the analyses). We used false discovery rate (FDR) to adjust for multiple testing.
We performed the heterogeneity in dependent instruments (HEIDI) test, as provided in Summary-data-based Mendelian Randomization (SMR) (Zhu et al., 2016), to test the existence of linkage in the observed association. Rejection of the null hypothesis (i.e., PHEIDI<0.05) indicates that the observed association in MR might be due to two distinct genetic variants in high linkage disequilibrium with each other.
In addition, a cis- mRNA- quantitative trait methylation (QTM) analysis was performed to explore the association between the identified CpGs and gene expression in participants with both DNA methylation and gene expression data. A cis-CpG-mRNA pair was defined as the target CpG residing in ± 10 kb of the corresponding gene encoding the mRNA (FDR P<0.05). In brief, linear regression was performed to test the association between each CpG and mRNA, with each individual mRNA as the dependent variable and each individual CpG as the independent variable, adjusted for age at death, sex, education and postmortem interval.
The annotations of transcripts were based on the Affymetrix exon array S1.0 platforms. To functionally annotate putative transcripts, we conducted functional enrichment analysis using the functional annotation tool “Metascape” (Zhou et al., 2019) and the gene concept network analysis using the R package “clusterProfiler” (Yu et al., 2012) for the genes harboring the identified CpGs and the cis-associated genes, separately. Gene symbols corresponding to putative genes (P<0.05) were used as the input of the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
Data cleaning and statistical/bioinformatical analysis was performed using SAS version 9.4 (SAS Institute, Chicago, IL, USA), R version 4.0.0 (https://www.r-project.org/), PLINK 1.9 (https://www.cog-genomics.org/plink/1.9/) and SMR (https://cnsgenomics.com/software/smr/).
Results
The characteristics of the included participants
Fig. 1 is the flow chart showing the different stages of our analysis. Among the 1,708 participants who had genetic data, 1312 died during follow up (mean age at death: 89.61±6.42; male: 33.1%), and 1168 participants had brain neuropathology data, of whom 764 (65.4%) were diagnosed as having pathological AD. A total of 740 participants had methylation data and 542 participants had gene expression data. Pairwise association analyses between genetic variants and DNA methylation were performed for 592 participants who had both genotype and methylation data (mean age at death: 88.26±6.47; male: 36.4%) for further cis-mQTLs mapping and MR analysis. The cis paired association tests of the identified pleiotropic CpGs with mRNAs were performed in 537 participants (mean age at death: 88.44±6.70; male: 36.9%).
Figure 1. The flow chart of bioinformatical/statistical analysis.
ROS, Religious Orders Study; MAP, Rush Memory and Aging Project; SNP, single-nucleotide polymorphism; mQTL, DNA methylation quantitative trait loci; FDR, false discovery rate; MR, Mendelian randomization.
Putative pleiotropic CpGs for AD neuropathology
We identified more than 370 million cis- mQTL SNPs at P < 0.05 and found that 60,595 CpGs had at least one cis-mQTL SNP (P<1.35×10−10; Fig. 2). We therefore selected the cis-mQTL variant with the smallest P-value as the IV for each CpG in the MR testing.
Figure 2. Chicago plot for the association between cis-mQTLs and DNA methylation.
We identified a total of 370,257,499 cis- mQTL SNPs at P < 0.05 using the R package “MatrixEQTL”. In the plot, we only kept the cis- mQTL SNP having the smallest P-value for a CpG in the cases of multiple SNPs associated with the same CpG. Each point represents the association of a cis- mQTL SNP. The horizontal coordinate represents the location of a CpG within the chromosome, and the vertical coordinate is –log10 (P-value) if the association is positive and log10 (P-value) if the association is negative. The dashed line corresponds to the significance level using Bonferroni correction (1.35×10−10).
We undertook 60,595 tests to evaluate the pleiotropic associations between genetically determined DNA methylation and AD neuropathologies to explore putative pleiotropic effect of DNA methylation on AD neuropathology. MR analysis identified 47 pleiotropic associations between genetically determined CpGs and Aβ load, including 44 positive associations and 3 negative associations with Aβ load, but no significant pleiotropic association between genetically determined CpGs and Tau tangles (Table 1, Supplementary Table 1 and Fig. 3).
Table 1.
The characteristics of cis-mQTLs.
| CHR | SNP | CpG | Gene | Beta | SE | P-value |
|---|---|---|---|---|---|---|
| 2 | rs2373298 | cg04480325 | Intergenic | −0.039 | 0.005 | 1.92×10−16 |
| 2 | rs2373298 | cg04521224 | Intergenic | −0.026 | 0.003 | 5.18×10−15 |
| 2 | chr2:37987728 | cg06480171 | Intergenic | −0.017 | 0.002 | 9.71×10−13 |
| 2 | rs2373298 | cg24523650 | EIF2AK2 | −0.079 | 0.006 | 2.64×10−34 |
| 3 | rs3009045 | cg04863667 | Intergenic | −0.036 | 0.004 | 6.81×10−15 |
| 10 | rs1751949 | cg00730780 | KIAA1462 | −0.019 | 0.003 | 7.29×10−11 |
| 10 | rs2768668 | cg01062834 | KIAA1462 | −0.009 | 0.001 | 4.58×10−12 |
| 10 | rs2768668 | cg03325931 | BAMBI | −0.018 | 0.003 | 5.57×10−12 |
| 10 | rs2768668 | cg08461107 | KIAA1462 | 0.021 | 0.003 | 1.25×10−14 |
| 10 | rs2768668 | cg11614536 | KIAA1462 | −0.014 | 0.001 | 5.52×10−21 |
| 10 | rs71492582 | cg11902278 | SVIL | −0.034 | 0.003 | 1.19×10−28 |
| 10 | rs2768668 | cg16513467 | WAC | −0.007 | 0.001 | 6.98×10−15 |
| 10 | rs2768668 | cg17428896 | SVIL | −0.031 | 0.004 | 2.86×10−15 |
| 10 | rs71492582 | cg18975376 | BAMBI | −0.017 | 0.002 | 6.03×10−19 |
| 10 | rs1751947 | cg19359767 | Intergenic | −0.024 | 0.003 | 6.68×10−13 |
| 10 | rs71492582 | cg19826211 | KIAA1462 | −0.01 | 0.001 | 1.17×10−18 |
| 10 | rs71492581 | cg20628376 | Intergenic | −0.022 | 0.003 | 8.13×10−13 |
| 10 | rs71492581 | cg27036936 | KIAA1462 | −0.023 | 0.003 | 5.01×10−15 |
| 13 | rs10161769 | cg00473245 | RAB20 | −0.007 | 0.001 | 2.15×10−14 |
| 13 | rs10161769 | cg02742906 | Intergenic | 0.019 | 0.002 | 2.53×10−18 |
| 13 | rs10161769 | cg08152546 | Intergenic | 0.013 | 0.002 | 2.72×10−15 |
| 13 | rs10161769 | cg09469566 | Intergenic | 0.018 | 0.002 | 1.29×10−14 |
| 13 | rs10161769 | cg10372485 | CARS2 | 0.009 | 0.001 | 1.61×10−12 |
| 13 | rs10161769 | cg15182896 | Intergenic | 0.025 | 0.002 | 1.53×10−24 |
| 13 | rs10161769 | cg15270851 | CARS2 | 0.008 | 0.001 | 1.89×10−16 |
| 13 | rs10161769 | cg17488929 | Intergenic | 0.016 | 0.002 | 2.07×10−17 |
| 13 | rs10161769 | cg17633592 | Intergenic | 0.011 | 0.001 | 2.39×10−13 |
| 13 | rs10161769 | cg18541254 | Intergenic | 0.011 | 0.002 | 7.95×10−11 |
| 13 | rs10161769 | cg18973457 | Intergenic | 0.004 | 0.001 | 2.81×10−11 |
| 13 | rs10161769 | cg19246801 | Intergenic | 0.014 | 0.002 | 5.40×10−11 |
| 13 | rs10161769 | cg20829347 | SOX1 | 0.006 | 0.001 | 1.72×10−12 |
| 13 | rs10161769 | cg21054521 | Intergenic | 0.02 | 0.003 | 5.71×10−12 |
| 13 | rs10161769 | cg23119039 | COL4A2 | 0.029 | 0.003 | 7.25×10−17 |
| 13 | rs10161769 | cg23934404 | Intergenic | 0.012 | 0.002 | 2.89×10−13 |
| 13 | rs10161769 | cg24074965 | Intergenic | 0.013 | 0.002 | 1.49×10−14 |
| 13 | rs10161769 | cg25554685 | Intergenic | 0.012 | 0.001 | 5.13×10−16 |
| 17 | rs2159085 | cg03363653 | C17orf76 | −0.017 | 0.002 | 4.31×10−18 |
| 17 | rs2159085 | cg05090851 | UBB | −0.006 | 0.001 | 6.08×10−11 |
| 17 | rs2159085 | cg09105334 | Intergenic | −0.082 | 0.006 | 8.74×10−37 |
| 17 | rs2159085 | cg13180678 | CENPV | −0.016 | 0.002 | 3.42×10−12 |
| 17 | rs2159085 | cg18002814 | FAM18B2 | −0.014 | 0.002 | 2.31×10−11 |
| 17 | rs2159085 | cg20577663 | CDRT4 | −0.027 | 0.003 | 2.58×10−21 |
| 17 | rs2159085 | cg22229594 | PMP22 | −0.006 | 0.001 | 6.92×10−11 |
| 17 | rs2159085 | cg24443925 | NCOR1 | 0.012 | 0.001 | 3.08×10−20 |
| 17 | rs2159085 | cg26744387 | C17orf76 | −0.013 | 0.002 | 3.26×10−14 |
| 17 | rs2159085 | cg27131197 | Intergenic | −0.043 | 0.005 | 2.05×10−17 |
| 18 | rs56945077 | cg03671802 | RAX | 0.043 | 0.004 | 1.58×10−22 |
Analyses were performed using the R package “MatrixEQTL”, and multiple testings were used (i.e., 0.05/number of SNP-CpG pairs=1.35×10−10).
Beta is the estimated effect size.
CHR, chromosome; SNP, single-nucleotide polymorphism; mQTL: quantitative trait loci; SE, standard error of the effect size.
Figure 3. Forest plot for the estimated MR effects and 95% confidence intervals for the casual associations of CpGs with Aβ load.
Analyses were performed using the R package “MendelianRandomization”, and multiple testings were adjusted using false discovery rate.
MR, Mendelian randomization; CI, confidence interval
Of the 47 identified CpGs, 26 were located in the intragenic region with approximately 70% located in gene body (including 5’UTR, coding sequence [CDS], intron and 3’UTR) and 30% in the transcription start site (TSS), and the remaining 21 in intergenic regions (Supplementary Table 2). Examination by chromatin states revealed that most of the identified CpGs were in strong transcription and promoter sites. More information of the pleiotropic CpGs can be found in Supplementary Table 2.
Association of the pleiotropic CpGs with mRNAs
To further dissect the relationship between DNA methylation and Aβ load, we evaluated the influence of the methylation levels on gene expression. We identified 774 CpG-mRNA pairs using cis- mRNA-QTM analysis. Among the 47 CpGs, 18 CpGs were in cis associated with 25 mRNAs involved in 25 genes comprising 41 CpG-mRNA/gene pairs (Table 2). We found that 24 CpGs were positively associated with mRNAs, and 17 CpGs negatively associated with mRNAs.
Table 2.
The characteristics of cis-mRNA-QTMs.
| CHR | CpG | Gene_id | Gene | Beta | SE | P-value | Q-value |
|---|---|---|---|---|---|---|---|
| 17 | cg20577663 | ENSG00000170425.3 | ADORA2B | 17.54 | 3.14 | 3.85×10−8 | 2.22×10−5 |
| 17 | cg20577663 | ENSG00000109099.9 | PMP22 | 96.78 | 18.44 | 2.22×10−7 | 6.41×10−5 |
| 10 | cg18975376 | ENSG00000095739.7 | BAMBI | 11.44 | 2.29 | 8.59×10−7 | 1.65×10−4 |
| 10 | cg18975376 | ENSG00000150051.8 | MKX | −4.54 | 0.96 | 2.64×10−6 | 3.79×10−4 |
| 2 | cg04521224 | ENSG00000163171.6 | CDC42EP3 | −3.15 | 0.68 | 4.47×10−6 | 5.15×10−4 |
| 13 | cg25554685 | ENSG00000126218.7 | F10 | 3.73 | 0.82 | 7.25×10−6 | 6.96×10−4 |
| 2 | cg04521224 | ENSG00000115825.5 | PRKD3 | 3.17 | 0.71 | 1.14×10−5 | 9.36×10−4 |
| 17 | cg20577663 | ENSG00000175061.10 | C17orf76-AS1 | 170.58 | 41.27 | 4.16×10−5 | 3.00×10−3 |
| 2 | cg24523650 | ENSG00000115825.5 | PRKD3 | 1.52 | 0.38 | 7.33×10−5 | 4.69×10−3 |
| 17 | cg27131197 | ENSG00000141030.8 | COPS3 | −11.53 | 2.92 | 9.05×10−5 | 4.94×10−3 |
| 2 | cg04480325 | ENSG00000115825.5 | PRKD3 | 1.98 | 0.5 | 9.44×10−5 | 4.94×10−3 |
| 10 | cg18975376 | ENSG00000150054.12 | MPP7 | −3.93 | 1.01 | 1.12×10−4 | 5.37×10−3 |
| 2 | cg06480171 | ENSG00000003509.11 | C2orf56 | −5.52 | 1.43 | 1.35×10−4 | 5.97×10−3 |
| 2 | cg04480325 | ENSG00000115828.11 | QPCT | −5.88 | 1.59 | 2.42×10−4 | 9.95×10−3 |
| 13 | cg24074965 | ENSG00000139835.8 | GRTP1 | 3.7 | 1.05 | 4.9×10−4 | 0.019 |
| 13 | cg24074965 | ENSG00000185896.9 | LAMP1 | 45.14 | 12.98 | 5.45×10−4 | 0.019 |
| 17 | cg20577663 | ENSG00000170315.8 | UBB | −917.99 | 264.05 | 5.5×10−4 | 0.0198 |
| 2 | cg06480171 | ENSG00000163171.6 | CDC42EP3 | −3.89 | 1.12 | 5.82×10−4 | 0.019 |
| 10 | cg03325931 | ENSG00000150051.8 | MKX | −2.23 | 0.65 | 6.8×10−4 | 0.02 |
| 10 | cg01062834 | ENSG00000183621.10 | ZNF438 | 6.28 | 1.86 | 7.86×10−4 | 0.021 |
| 10 | cg19826211 | ENSG00000183621.10 | ZNF438 | 8.1 | 2.4 | 7.91×10−4 | 0.021 |
| 2 | cg04521224 | ENSG00000115828.11 | QPCT | −7.64 | 2.27 | 8.09×10−4 | 0.021 |
| 17 | cg27131197 | ENSG00000170425.3 | ADORA2B | 5.68 | 1.69 | 8.54×10−4 | 0.021 |
| 13 | cg23119039 | ENSG00000255874.1 | LINC00346 | 0.27 | 0.08 | 8.79×10−4 | 0.021 |
| 10 | cg20628376 | ENSG00000150051.8 | MKX | −1.78 | 0.54 | 1.06×10−3 | 0.022 |
| 13 | cg25554685 | ENSG00000057593.9 | F7 | −3.12 | 0.95 | 1.06×10−3 | 0.022 |
| 17 | cg13180678 | ENSG00000197566.5 | ZNF624 | 1.24 | 0.38 | 1.07×10−3 | 0.022 |
| 2 | cg04521224 | ENSG00000003509.11 | C2orf56 | −2.87 | 0.88 | 1.12×10−3 | 0.022 |
| 17 | cg24443925 | ENSG00000109099.9 | PMP22 | −130.38 | 39.93 | 1.16×10−3 | 0.022 |
| 17 | cg20577663 | ENSG00000175106.12 | FAM18B2 | 5.36 | 1.64 | 1.18×10−3 | 0.022 |
| 17 | cg18002814 | ENSG00000170425.3 | ADORA2B | 15.83 | 4.85 | 1.18×10−3 | 0.022 |
| 10 | cg03325931 | ENSG00000095739.7 | BAMBI | 5.01 | 1.57 | 1.50×10−3 | 0.027 |
| 17 | cg18002814 | ENSG00000108474.8 | PIGL | −10.7 | 3.37 | 1.57×10−3 | 0.027 |
| 10 | cg20628376 | ENSG00000095787.15 | WAC | 20.19 | 6.36 | 1.59×10−3 | 0.027 |
| 2 | cg06480171 | ENSG00000115825.5 | PRKD3 | 3.74 | 1.18 | 1.65×10−3 | 0.027 |
| 2 | cg06480171 | ENSG00000115828.11 | QPCT | −11.64 | 3.73 | 1.89×10−3 | 0.030 |
| 17 | cg18002814 | ENSG00000109099.9 | PMP22 | 86.49 | 28.41 | 2.45×10−3 | 0.038 |
| 13 | cg24074965 | ENSG00000057593.9 | F7 | −2.46 | 0.82 | 2.90×10−3 | 0.044 |
| 13 | cg10372485 | ENSG00000088448.9 | ANKRD10 | 27.55 | 9.26 | 3.06×10−3 | 0.045 |
| 2 | cg24523650 | ENSG00000115808.7 | STRN | 0.65 | 0.22 | 3.15×10−3 | 0.045 |
| 17 | cg13180678 | ENSG00000214941.3 | ZSWIM7 | 27.71 | 9.42 | 3.41×10−3 | 0.048 |
Results were obtained using linear regression, adjusting for age at death, sex, and education, and multiple testing were adjusted using false discovery rate (Q value< 0.05).
Beta is the estimated effect size.
CHR, chromosome; QTM: quantitative trait methylation; SE, standard error.
Pathway analysis/Functional informatics
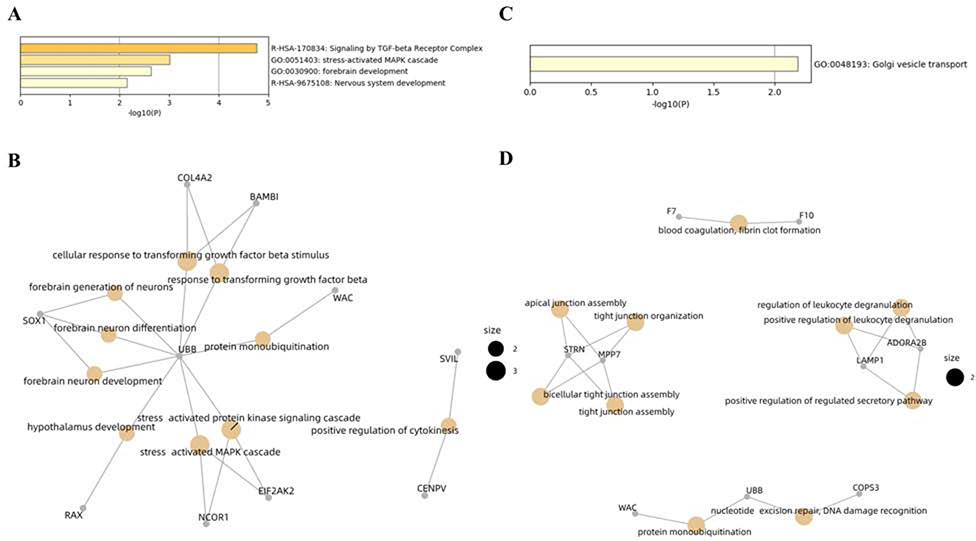
GO enrichment analysis of biological process, molecular function, and cellular component pathways showed that the genes harboring the identified CpGs were involved in four GO terms, including nervous system development (R-HSA-9675108), forebrain development (GO:0030900), signaling by TGF-beta Receptor complex (R-HSA-170834), and stress-activated MAPK cascade (GO:0051403; Figure 4A). Concept network analysis of genes harboring the identified CpGs revealed multiple domains related with neuron generation, development and differentiation (Fig. 4B). GO enrichment analysis of cis-associated genes identified only one significant GO term of Golgi vesicle transport (GO:0048193; Fig. 4C). Concept network analysis of cis-associated genes revealed domains associated with leukocyte deregulation and DNA damage recognition (Fig. 4D). More information can be found in Figure 4 and Supplementary Table 3–4.
Figure 4. Functional enrichment and gene concept network analysis.
A) Enriched GO terms based on genes harboring the identified CpGs; B) Concept network analysis of the genes harboring the identified CpGs; C) Enriched GO terms based on cis-associated genes; and D) Concept network analysis of the cis associated genes.
GO, gene ontology
Discussion
In this study, we integrated GWAS and mQTL data in MR analysis to explore putative pleiotropic DNA methylation loci for AD neuropathology. We identified 47 pleiotropic DNA methylation loci for Aβ load. Among the 47 CpGs, 18 CpGs were associated in cis with 25 mRNAs involved in 25 genes comprising 41 CpG-mRNA/gene pairs. To the best of our knowledge, this is the first study to examine putative pleiotropic epigenetic loci for AD neuropathology through an MR approach.
Evidence is accumulating that DNA methylation is related to AD (De Jager et al., 2014; Huo et al., 2019; Lunnon et al., 2014; Smith et al., 2018; Yu et al., 2015). However, previous studies focusing on exploring the association between differential DNA methylation and AD are prone to confounding and reverse causation (Dekkers et al., 2016; Huo et al., 2019; Wahl et al., 2017). As a result, some disease-associated CpGs from MWAS may reflect the influence of the diseases on DNA methylation rather than the pleiotropic effects of DNA methylation on the diseases. By contrast, MR incorporates cis-mQTL information into GWAS analyses and has the potential to increase the power of GWAS in identifying pleiotropic loci associated with complex diseases (Hannon et al., 2018; Hannon et al., 2017; Richardson et al., 2017). This novel analytical framework that integrates GWAS and mQTL data in MR analysis enabled us to identify multiple epigenetic loci that showed pleiotropic relationship with Aβ load, indicating that DNA methylation resides along the pleiotropic pathway linking genetic variants with AD neuropathology.
Some of the casual CpGs identified in our study have been reported to be associated with Aβ load or AD related risk factors. For example, the epigenetic locus cg24523650 in EIF2AK2 was associated with Aβ load by a previous EWAS study of AD neuropathology (Huo et al., 2019). Two other methylation loci, cg08152546 and cg13180678 which reside in or are close to CENPV, were associated with aging (Florath et al., 2014; Weidner et al., 2014), a major risk factor for AD. In addition, we identified several pleiotropic epigenetic loci, such as cg00473245 in RAB20 and cg27036936 in KIAA1462, that were associated with metabolism-related disorders such as metabolic syndrome, obesity and diabetes (Akinyemiju et al., 2018; Nilsson et al., 2014). Increasing evidence suggests epidemiological and pathological links between AD and metabolic traits (Jansen et al., 2019; Sims et al., 2020). Previous research identified common genetic architectures shared between AD and metabolic traits, and shed light on molecular mechanisms underlying the association between AD and metabolic dysregulation (Zhu et al., 2019). Whether and how these identified epigenetic loci affect the risk of AD via metabolic pathways warrants further investigation.
Some of the genes harboring the identified CpGs were reported to be involved in the development of the nervous system or AD neuropathologies. For example, we identified cg20829347 in SOX1 as a pleiotropic epigenetic site for Aβ load. SOX1 encodes a member of the SOX (SRY-related HMG-box) family of transcription factors, and was reported to be involved in the regulation of embryonic development and in the determination of cell fate (Berger et al., 2016). SOX1 plays an important role in regulating neurogenesis in the nervous system (Kan et al., 2004). In addition, we found cg05090851 in UBB as a pleiotropic epigenetic site for Aβ load. UBB encodes ubiquitin, which has a major role in targeting cellular proteins for degradation and is also involved in the maintenance of chromatin structure, regulation of gene expression, and stress response (Grumati and Dikic, 2018). An aberrant form of ubiquitin was found to be accumulated in brain tissues of AD patients (Munari et al., 2018).
Although it is widely hypothesized that DNA methylation could influence gene expression, its relationship with transcriptional activity in influencing AD neuropathology is not fully understood. We found that two of the identified CpGs (cg18975376 and cg03325931) were associated with the expression of BAMBI which encodes a transmembrane glycoprotein related to transforming growth factor beta type I (TGF-β1) family (Bai et al., 2017). It was found that impairment of TGF-β1 signaling was associated with exacerbated Aβ deposition and neurofibrillary tangle formation (Estrada et al., 2018). In addition, many of the identified pleiotropic CpGs for Aβ load showed in cis association with genes that might be involved in AD neuropathologies. For example, cg04480325, cg04521224 and cg06480171, located in the intergenic region, were associated in cis with the expression of QPCT which encodes human pituitary glutaminyl cyclase and is responsible for the presence of pyroglutamyl residues in many neuroendocrine peptides. It was found that lowering QPCT expression could reduce the amount of pyroglutamate-amyloid-β, a major constituent of Aβ deposits in human AD (Alexandru et al., 2011).
We also found that some of identified pleiotropic CpGs were in cis associated with genes which have not been reported to be involved in the pathogenesis of AD. For example, cg20577663 in CDRT4 was in cis associated with the expression of ADORA2B which encodes an adenosine receptor. Adenosine receptor signaling plays important roles in normal physiology, and was reported to be involved in cardiovascular diseases (Geldenhuys et al., 2017; Gile and Eckle, 2016; Novotný, 2015). In addition, cg24523650 in EIF2AK2 was in cis associated with the expression of STRN which encodes striatin, a cell–cell junctional protein involved in maintaining correct cell adhesion (Lahav-Ariel et al., 2019). More studies are needed to reveal the exact function of these genes in the development of AD neuropathology.
Our study has some strengths. The Religious Orders Study and the Rush Memory and Aging projects have similar procedures in the collection of ‘omics’ data and pathologic data, which allows various types of data to be merged for analyses that may have small to moderate effects. The MR approach greatly reduced confounding bias and reverse causation. Therefore, the identified DNA methylation loci likely represent some ‘true’ pleiotropic epigenetic markers for AD neuropathology. The ROSMAP studies have a very high rate of participation in the follow up and autopsy, which reduced bias from selective attrition. We only included study participants of European ancestry in our analyses and controlled for the first three principal components the cis-mQTL analysis; therefore, population stratification is less likely to be a problem.
Our study also has some limitations. Our MR analysis was based on the following four assumptions: 1) the genotype is associated with DNA methylation; 2) the genotype is not associated with confounding factors that bias the associations between DNA methylation and amyloid-β load; 3) the genotype is related to amyloid-β load only via its association with DNA methylation; and 4) all the associations are linear and unaffected by statistical interaction. In the cis-mQTL analysis, we filtered genetic variants with conservative multiple testing, and chose the genetic variant with the smallest P-value for each CpG, minimizing the concern of weak IV (assumption 1). Non-association of genotype with confounders is often based on the biological belief that the genotype will not be associated with socioeconomic and behavioral characteristics that commonly confound the effects of exposure (i.e., DNA methylation) on the outcome (i.e., amyloid-β load) (Lawlor et al., 2008). Moreover, we used data from the same studies in obtaining both genotype-DNA methylation and genotype- amyloid-β load estimates, further alleviating the concern about violation of assumption 2 (Lawlor et al., 2008). Regarding assumption 3, we acknowledge that with the use of single-variant-based MR method, we could not rule out the possibility of horizontal pleiotropy which could distort the MR results. A recent study found that horizontal pleiotropy was detectable in approximately 50% of significant causal relationships in MR, and could induce severe distortions in MR estimates as well as around 10% of false positive causal relationships (Verbanck et al., 2018). We performed HEIDI test and found that of the 47 identified CpGs, the test was available for 16 CpGs. The HEIDI test was not significant for most CpGs except one (cg03671802 in RAX; Supplementary Table 1), indicating that these observed associations were less likely subject to horizontal pleiotropy. However, caution should still be executed in interpreting the MR results because the HEIDI test was not feasible for the remaining identified CpGs. It is often difficult to validate the linearity assumption (Lawlor et al., 2008). Violation of the assumption is not essential when the purpose is to test the null hypothesis of no effect of the exposure (e.g., DNA methylation) on the outcome (e.g., amyloid-β load), but can cause problems when the purpose is to estimate the size of the effect. We adopted correction for multiple testing to reduce false positive rate; however, we may have missed important SNPs or methylation loci. Moreover, our pathway analysis only included pleiotropic methylation loci in association with gene expression, and we did not consider other methylation loci that could also affect the development of Aβ. More studies are needed to systematically explore the role of DNA methylation in influencing transcriptional activity in the neuropathogenesis of AD.
In conclusion, by performing multi-stage analyses incorporating GWAS and mQTL data through an MR approach, we identified multiple pleiotropic DNA methylation loci for Aβ load. Our findings shed light on the role of DNA methylation in the pathogenesis of Aβ load. Future studies are needed to validate our findings and elucidate the exact functions of the identified epigenetic loci and the associated genes in the development of AD neuropathology.
Supplementary Material
Supplementary Table 1. Pleiotropic estimates of the association between DNA methylation and Aβ load.
Analyses were performed using the R package “MendelianRandomization”, and multiple testings were adjusted using false discovery rate (Q value< 0.05). PMR means P value for CpG-trait association from the MR test, PHEIDI means P value from HEIDI test to indicate whether the CpG-trait association is due to a single shared genetic variant (the HEIDI test needs more than three genetic variant based on cis-mQTLs P-value <1.35×10−10, otherwise HEIDI test has little power to detect heterogeneity and possibly generates misleading result).
MR, Mendelian randomization; NA, not available, SNP, single-nucleotide polymorphism; SE, standard error.
Supplementary Table 2. Basic information of the identified pleiotropic CpGs.
Supplementary Table 3. Functional enrichment analysis of the genes harboring the identified CpGs.
Supplementary Table 4. Functional enrichment analysis of the cis associated genes.
Highlights.
We aimed to search for pleiotropic DNA methylation loci for neuropathologies of AD.
We adopted a Mendelian randomization approach integrating GWAS and DNA methylation quantitative trait loci data.
We identified 47 pleiotropic CpGs for Aβ load but none for neurofibrillary tangles.
Some CpGs were found related with Aβ load and others represent novel epigenetic loci.
Many identified CpGs were associated in cis with mRNAs involved in nervous system or AD neuropathologies.
Acknowledgments
Funding
The study was supported by the National Natural Science Foundation of China (Grant No. 81771493), NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG36042, U01AG61356 and 1RF1AG064312-01. Di Liu was supported by China Scholarship Council (CSC 201908110339).
The authors confirmed that all authors have reviewed the contents of the article being submitted, approved its contents, and validated the accuracy of the data.
Footnotes
Disclosure
The authors have no conflict of interest to report.
No author is affiliated with an institution that has contracts relating to this research through which it or any other organization may stand to gain financially now or in the future.
There are no any other agreements of authors or their institutions that could be seen as involving a financial interest in this work.
The data contained in the manuscript being submitted have not been previously published, have not been submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
Data for the present study were drawn from the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP). Both studies were approved by an Institutional Review Board of the Rush University Medical Center. The ethics approval was given in compliance with the Declaration of Helsinki. Data can be requested at www.radc.rush.edu and https://www.synapse.org/.
All authors have reviewed the contents of the manuscript being submitted, approved its contents and validated the accuracy of the data
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahuja N, Sharma AR, Baylin SB, 2016. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annual Review of Medicine 67(1), 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinyemiju T, Do AN, Patki A, Aslibekyan S, Zhi D, Hidalgo B, Tiwari HK, Absher D, Geng X, Arnett DK, Irvin MR, 2018. Epigenome-wide association study of metabolic syndrome in African-American adults. Clin Epigenetics 10, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru A, Jagla W, Graubner S, Becker A, Bäuscher C, Kohlmann S, Sedlmeier R, Raber KA, Cynis H, Rönicke R, Reymann KG, Petrasch-Parwez E, Hartlage-Rübsamen M, Waniek A, Rossner S, Schilling S, Osmand AP, Demuth HU, von Hörsten S, 2011. Selective hippocampal neurodegeneration in transgenic mice expressing small amounts of truncated Aβ is induced by pyroglutamate-Aβ formation. The Journal of neuroscience : the official journal of the Society for Neuroscience 31(36), 12790–12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Association, 2016. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement 12(4), 459–509. [DOI] [PubMed] [Google Scholar]
- Bai L, Chu G, Wang W, Xiang A, Yang G, 2017. BAMBI promotes porcine granulosa cell steroidogenesis involving TGF-β signaling. Theriogenology 100, 24–31. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA, 2018. Religious Orders Study and Rush Memory and Aging Project. J Alzheimers Dis 64(s1), S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS, 2012a. Overview and findings from the religious orders study. Curr Alzheimer Res 9(6), 628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS, 2012b. Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res 9(6), 646–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Dersch R, Ruthardt E, Rasiah C, Rauer S, Stich O, 2016. Prevalence of anti-SOX1 reactivity in various neurological disorders. J Neurol Sci 369, 342–346. [DOI] [PubMed] [Google Scholar]
- Bhide A, Shah PS, Acharya G, 2018. A simplified guide to randomized controlled trials. Acta obstetricia et gynecologica Scandinavica 97(4), 380–387. [DOI] [PubMed] [Google Scholar]
- Congdon EE, Sigurdsson EM, 2018. Tau-targeting therapies for Alzheimer disease. Nat Rev Neurol 14(7), 399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, Yu L, Leurgans SE, Tran D, Aubin C, Anderson CD, Biffi A, Corneveaux JJ, Huentelman MJ, Rosand J, Daly MJ, Myers AJ, Reiman EM, Bennett DA, Evans DA, 2012. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging 33(5), 1017.e1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe C, Tang A, Raj T, Replogle J, Brodeur W, Gabriel S, Chai HS, Younkin C, Younkin SG, Zou F, Szyf M, Epstein CB, Schneider JA, Bernstein BE, Meissner A, Ertekin-Taner N, Chibnik LB, Kellis M, Mill J, Bennett DA, 2014. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17(9), 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, Mei H, Zhernakova DV, van den Berg LH, Deelen J, van Dongen J, van Heemst D, Hofman A, Hottenga JJ, van der Kallen CJ, Schalkwijk CG, Stehouwer CD, Tigchelaar EF, Uitterlinden AG, Willemsen G, Zhernakova A, Franke L, T HPA, Jansen R, van Meurs J, Boomsma DI, van Duijn CM, van Greevenbroek MM, Veldink JH, Wijmenga C, van Zwet EW, Slagboom PE, Jukema JW, Heijmans BT, 2016. Blood lipids influence DNA methylation in circulating cells. Genome Biol 17(1), 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emdin CA, Khera AV, Kathiresan S, 2017. Mendelian Randomization. Jama 318(19), 1925–1926. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Shoai M, Pither R, Williams J, Hardy J, 2017. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging 49, 14.e217–214.e211. [DOI] [PubMed] [Google Scholar]
- Estrada LD, Oliveira-Cruz L, Cabrera D, 2018. Transforming Growth Factor Beta Type I Role in Neurodegeneration: Implications for Alzheimeŕs Disease. Current protein & peptide science 19(12), 1180–1188. [DOI] [PubMed] [Google Scholar]
- Florath I, Butterbach K, Muller H, Bewerunge-Hudler M, Brenner H, 2014. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet 23(5), 1186–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2017 DALYs and HALE Collaborators, 2018. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392(10159), 1859–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys WJ, Hanif A, Yun J, Nayeem MA, 2017. Exploring Adenosine Receptor Ligands: Potential Role in the Treatment of Cardiovascular Diseases. Molecules (Basel, Switzerland) 22(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gile J, Eckle T, 2016. ADORA2b Signaling in Cardioprotection. Journal of nature and science 2(10). [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Dikic I, 2018. Ubiquitin signaling and autophagy. The Journal of biological chemistry 293(15), 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Gorrie-Stone TJ, Smart MC, Burrage J, Hughes A, Bao Y, Kumari M, Schalkwyk LC, Mill J, 2018. Leveraging DNA-Methylation Quantitative-Trait Loci to Characterize the Relationship between Methylomic Variation, Gene Expression, and Complex Traits. Am J Hum Genet 103(5), 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Weedon M, Bray N, O’Donovan M, Mill J, 2017. Pleiotropic Effects of Trait-Associated Genetic Variation on DNA Methylation: Utility for Refining GWAS Loci. Am J Hum Genet 100(6), 954–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, 2012. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biology 13(10), R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, Yao C, Liu C, Ma J, Richard M, Agha G, Guan W, Almli LM, Conneely KN, Keefe J, Hwang SJ, Johnson AD, Fornage M, Liang L, Levy D, 2019. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 10(1), 4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Z, Zhu Y, Yu L, Yang J, De Jager P, Bennett DA, Zhao J, 2019. DNA methylation variability in Alzheimer’s disease. Neurobiol Aging 76, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising C, Heneka MT, 2018. Functional and structural damage of neurons by innate immune mechanisms during neurodegeneration. Cell Death Dis 9(2), 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hagg S, Athanasiu L, Voyle N, Proitsi P, Witoelar A, Stringer S, Aarsland D, Almdahl IS, Andersen F, Bergh S, Bettella F, Bjornsson S, Braekhus A, Brathen G, de Leeuw C, Desikan RS, Djurovic S, Dumitrescu L, Fladby T, Hohman TJ, Jonsson PV, Kiddle SJ, Rongve A, Saltvedt I, Sando SB, Selbaek G, Shoai M, Skene NG, Snaedal J, Stordal E, Ulstein ID, Wang Y, White LR, Hardy J, Hjerling-Leffler J, Sullivan PF, van der Flier WM, Dobson R, Davis LK, Stefansson H, Stefansson K, Pedersen NL, Ripke S, Andreassen OA, Posthuma D, 2019. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51(3), 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA, 2004. Sox1 acts through multiple independent pathways to promote neurogenesis. Developmental biology 269(2), 580–594. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP, 2006. Genomic DNA methylation: the mark and its mediators. Trends in Biochemical Sciences 31(2), 89–97. [DOI] [PubMed] [Google Scholar]
- Kumar D, Sharma A, Sharma L, 2020. A comprehensive review of Alzheimer’s association with related proteins: Pathological role and therapeutic significance. Curr Neuropharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav-Ariel L, Caspi M, Nadar-Ponniah PT, Zelikson N, Hofmann I, Hanson KK, Franke WW, Sklan EH, Avraham KB, Rosin-Arbesfeld R, 2019. Striatin is a novel modulator of cell adhesion. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 33(4), 4729–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW., Yu L., Tsolaki M., Bossù P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fo NC., Hard J., Deniz Naranjo MC., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthew F., Moebus S., Mecocci P., Del Zompo M., Maier W., Hampel H., Pilotto A., Bullido M., Panza F., Caffarra P., Nacmias B., Gilbert JR., Mayhau M., Lannefelt L., Hakonarson H., Pichler S., Carrasquillo MM., Ingelsson M., Beekly D., Alvarez V., Zou F., Valladares O., Younkin SG., Cot E., Hamilton-Nelson KL., G W., Razquin C., Pastor P., Mateo I., Owen MJ., Fabe KM., Jonsso PV., Combarro O., O’Donova MC., Cantwel LB., Soinine H., Blacke D., Mea S., Mosley TH Jr., Bennett DA., Harris TB., Fratiglioni L., Holme C., de Bruij RF., Passmor P., Montine TJ., Bettens K., Rotter JI., Brice A., Morgan K., Foroud TM., Kukul WA., Hannequi D., Powel JF., Nalls MA., Ritchie K., Lunetta KL., Kauwe JS., Boerwinkle E., Riemenschneider M., Boada M., Hiltuenen M., Martin ER., Schmidt R., Rujescu D., Wang LS., Dartigues JF., Mayeux R., Tzourio C., Hofman A., Nöthe MM., Graff C., Psaty BM., Jones L., Haines JL., Holman PA., Lathrop M., Pericak-Vance MA., Launer LJ., Farre LA., van Duijn CM., Van Broeckhoven C., Moskvina V., Seshadri S., Williams J., Schellenberg GD., Amouyel P., 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Davey Smith G, Kundu D, Bruckdorfer KR, Ebrahim S, 2004. Those confounded vitamins: what can we learn from the differences between observational versus randomised trial evidence? Lancet 363(9422), 1724–1727. [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JAC, Nic T, George DS, 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in Medicine 27(8), 1133. [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Smith GD, 2006. Cardiovascular risk and hormone replacement therapy. Current opinion in obstetrics & gynecology 18(6), 658–665. [DOI] [PubMed] [Google Scholar]
- Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, Macdonald R, Condliffe D, Harries LW, Katsel P, Haroutunian V, Kaminsky Z, Joachim C, Powell J, Lovestone S, Bennett DA, Schalkwyk LC, Mill J, 2014. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci 17(9), 1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munari F, Bortot A, Assfalg M, D’Onofrio M, 2018. Alzheimer’s disease-associated ubiquitin mutant Ubb(+1): Properties of the carboxy-terminal domain and its influence on biomolecular interactions. Int J Biol Macromol 108, 24–31. [DOI] [PubMed] [Google Scholar]
- Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, Xu J, Yu L, Gaiteri C, Bennett DA, Mostafavi S, De Jager PL, 2017. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci 20(10), 1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, Poulsen P, Ribel-Madsen R, Pedersen NL, Almgren P, Fadista J, Ronn T, Klarlund PB, Scheele C, Vaag A, Ling C, 2014. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63(9), 2962–2976. [DOI] [PubMed] [Google Scholar]
- Novotný J, 2015. [Adenosine and its role in physiology]. Ceskoslovenska fysiologie 64(1), 35–44. [PubMed] [Google Scholar]
- Richardson TG, Haycock PC, Zheng J, Timpson NJ, Gaunt TR, Davey SG, Relton CL, Hemani G, 2018. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum Mol Genet 27(18), 3293–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TG, Zheng J, Davey SG, Timpson NJ, Gaunt TR, Relton CL, Hemani G, 2017. Mendelian Randomization Analysis Identifies CpG Sites as Putative Mediators for Genetic Influences on Cardiovascular Disease Risk. Am J Hum Genet 101(4), 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R, Hill M, Williams J, 2020. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci 23(3), 311–322. [DOI] [PubMed] [Google Scholar]
- Smith RG, Hannon E, De Jager PL, Chibnik L, Lott SJ, Condliffe D, Smith AR, Haroutunian V, Troakes C, Al-Sarraj S, Bennett DA, Powell J, Lovestone S, Schalkwyk L, Mill J, Lunnon K, 2018. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement 14(12), 1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teumer A, 2018. Common Methods for Performing Mendelian Randomization. Frontiers in cardiovascular medicine 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbanck M, Chen CY, Neale B, Do R, 2018. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50(5), 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, Tan S, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, de Craen AJ, de Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, van Duijn C, van Meurs JB, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, van der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Jarvelin MR, Bollati V, Soong R, Spector TD, Scott J, McCarthy MI, Elliott P, Bell JT, Matullo G, Gieger C, Kooner JS, Grallert H, Chambers JC, 2017. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541(7635), 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL, Haroutunian V, Sharp AJ, 2016. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med 8(1), 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jockel KH, Erbel R, Muhleisen TW, Zenke M, Brummendorf TH, Wagner W, 2014. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol 15(2), R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavorska OO, Burgess S, 2017. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 46(6), 1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY, 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5), 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, Kozubek J, Obholzer N, Leurgans SE, Schneider JA, Meissner A, De Jager PL, Bennett DA, 2015. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 72(1), 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1), 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Lin Y, Li X, Driver JA, Liang L, 2019. Shared genetic architecture between metabolic traits and Alzheimer’s disease: a large-scale genome-wide cross-trait analysis. Hum Genet 138(3), 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, Yang J, 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5), 481–487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Pleiotropic estimates of the association between DNA methylation and Aβ load.
Analyses were performed using the R package “MendelianRandomization”, and multiple testings were adjusted using false discovery rate (Q value< 0.05). PMR means P value for CpG-trait association from the MR test, PHEIDI means P value from HEIDI test to indicate whether the CpG-trait association is due to a single shared genetic variant (the HEIDI test needs more than three genetic variant based on cis-mQTLs P-value <1.35×10−10, otherwise HEIDI test has little power to detect heterogeneity and possibly generates misleading result).
MR, Mendelian randomization; NA, not available, SNP, single-nucleotide polymorphism; SE, standard error.
Supplementary Table 2. Basic information of the identified pleiotropic CpGs.
Supplementary Table 3. Functional enrichment analysis of the genes harboring the identified CpGs.
Supplementary Table 4. Functional enrichment analysis of the cis associated genes.