Abstract
The measurement of UV-induced DNA damage as a dosimeter of exposure and predictor of skin cancer risk has been proposed by multiple groups. While UV-induced mutations and adducts are present in normal-appearing UV-exposed epidermis, sampling normal non-lesional skin requires non-invasive methods to extract epidermal DNA for analysis. Here we demonstrate the feasibility of such an approach, termed Surfactant-based Tissue Acquisition for Molecular Profiling (STAMP). Sampling in patients was performed using a felt-tip pen soaked in a mixture of surfactants (Brij-30/DPS). In mice, we show that the epidermis can be selectively removed without scarring, with complete healing within 2 weeks. We exposed Hairless mice to low-dose UV radiation over a period of 3 months, and serially sampled them through up to 2 months following the cessation of UV exposure, observing a progressive increase in a UV-signature mutational burden. To test whether STAMP could be applied to human patients, samples were collected from sun-exposed and sun-protected areas, which were then subjected to high-depth targeted exome sequencing. Extensive UV-driven mosaicism and substantially-increased mutational loads in sun-exposed vs. sun-protected areas were observed, suggesting that genomic measures, as an integrated readout of DNA damage, repair, and clonal expansion, may be informative markers of UV exposure.
INTRODUCTION
Skin cancer is the most common form of malignancy in humans, with significant associated morbidity and cost, and steadily increasing incidence (Rogers et al., 2015). Exposure to ultraviolet radiation (UVR) is thought to cause approximately 90% of keratinocyte carcinomas, causing malignant transformation and permissive alterations of the immune microenvironment (Griffin et al., 2016). UVR initiates a complex multistage process associated with accumulation of DNA photoproducts resulting in mutations, thus facilitating carcinogenesis (Brash, 2015). Among these events are C→T transitions or, more specifically characteristic of UVB-mediated DNA damage, CC→TT transitions (Brash, 2015). Such mutations frequently abrogate function of the TP53 tumor suppressor gene among others, rendering cells resistant to UV-induced apoptosis, whereupon they can acquire a growth advantage and expand as pre-malignant clones among normal keratinocytes while undergoing further genomic changes leading to full malignancy (Brash et al., 1996, Jonason et al., 1996, Ziegler et al., 1994). Furthermore, the expansion of such clones, whether under selection or not (Martincorena et al., 2015, Simons, 2016), provides an opportunity to detect mutations in otherwise normal appearing skin. Since it is now well established that UV-damaged epidermal keratinocytes in intact skin can harbor on the order of 5 mutations/Mb (Chitsazzadeh et al., 2016, Martincorena et al., 2015), this raises the possibility of using this mutational burden as a dosimeter of UV-induced damage and potentially skin cancer risk.
The utilization of biomarkers of skin carcinogenesis has not been well established, whereas they are used as standard of care in many other malignancies. We propose that sampling normal skin for biomarkers of UV exposure can lead to better understanding of the genomic alterations associated with initiation of skin cancer, which ultimately will aid in stratifying risk of skin cancer development, determination of effectiveness of current prevention methods, and better targeted therapies.
The interrogation of normal non-lesional skin requires non-invasive methods of sampling to extract epidermal DNA for analysis. Conventional sampling methods involve skin biopsies, which requires anesthesia and causes scarring and are there not suitable for assessing multiple sites of normal, non-lesional skin, especially in cosmetically-sensitive (and often sun-exposed) areas. Although there have been multiple other non-invasive methods, include tape stripping (Lacerenza et al., 2016, Wong et al., 2004), microbiopsy (Lin et al., 2013), and superficial scraping (Wang et al., 2018), reported yields appear to have been substantially lower. In this study, we validate a new non-invasive sampling technique for these purposes.
RESULTS
Non-invasive sampling using STAMP does not cause scarring
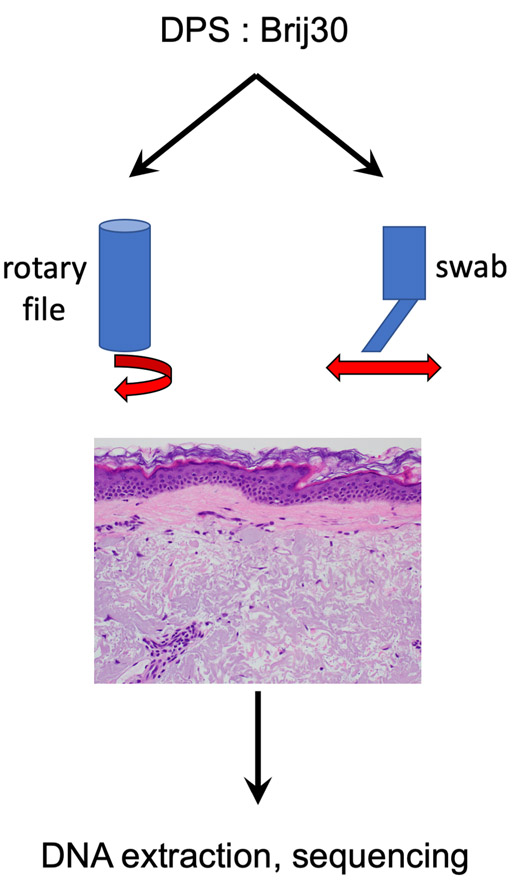
We have previously described a combination of surfactants, Brij-30 and DPS, which when combined in equimolar ratio (0.5% w/v), results in a mixture that enables the solubilization of tissue (Hwang et al., 2012, Hwang et al., 2013, Paliwal et al., 2012). Originally optimized and applied for epidermal protein recovery, we adapted it here for recovery of epidermal DNA. In brief, if coupled to ultrasound or mechanical energy even in the form of dermabrasion, the epidermis can be solubilized and DNA recovered and sequenced (Figure 1).
Figure 1.
Surfactant-based Tissue Acquisition for Molecular Profiling (STAMP)
The STAMP procedure is accomplished by coupling mechanical energy in the form of rubbing the epidermis with a motorized rotary nail within a chamber full of the surfactant mixture (0.5% w/v each of DPS and Brij-30) or felt swab containing the solution.
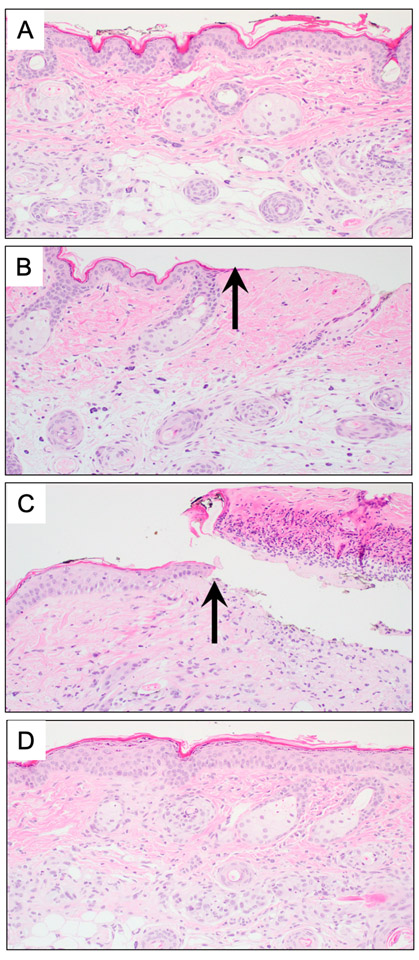
In order to demonstrate the feasibility of the non-scarring epidermal sampling, we applied STAMP using dermabrasion though the controlled application of a rotary nail file on Hairless mouse skin. The immediate post-sampling histology revealed skin stripped of the epidermis right up to the dermal-epidermal junction (Figure 2A-B). At 24 hours, a fibrinous exudate accumulates (Figure 2C), but within 2 weeks the wound is completely re-epithelialized, without evidence of fibrosis, expansion of the dermis, or reduction in the density of adnexal structures (Figure 2D).
Figure 2.
Skin sampled by STAMP does not scar
(A) Full thickness punch biopsy reveals unremarkable skin prior to sampling. (B) Immediately following samples, the epidermis is missing (arrow).
(C) At 24 hours post sampling, a fibrinous exudate accumulates.
(D) Two weeks following sampling, the epidermis has re-epithelialized, with no evidence of fibrosis or loss of adnexal structures.
STAMP enables high-sensitivity assessment of mutational burden in epidermis
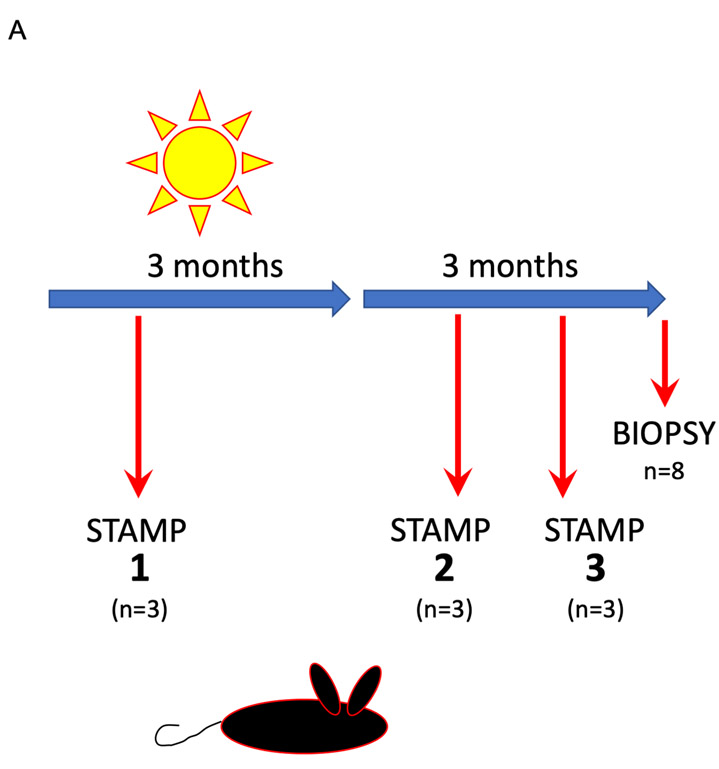
We next wished to understand whether increasing UV exposure in-vivo would result in detectable increases in mutations as sampled by either STAMP or full-thickness punch biopsy. We adapted our UV-driven model of cuSCC in Hairless mice (Chitsazzadeh et al., 2016, Vin et al., 2013) to sample skin progressively as the mice were exposed to increasing total doses of solar simulated radiation (Figure 3A) (Nghiem et al., 2002). In this model, we irradiate the mice over a three-month period thrice weekly for a total dose of 175 kJ/m2 of UVB at a standard erythema dose (SED) of 3.4 per session. With this exposure, all the mice develop papillomas within 4-8 weeks following cessation of irradiation, with a minority of lesions progressing to invasive carcinomas (Figure 3A) (Chitsazzadeh et al., 2016, Vin et al., 2014, Vin et al., 2013). We sampled three sets of three mice each with STAMP: set 1 at one month of UV exposure, set 2 at one month following completion of the three-month course of UV exposure and set 3 at two months following the completion of the three-month course of UV (Figure 3A). As a point of comparison, standard punch biopsies were taken from a 4th set of mice at three months post cessation of UV exposure. DNA was isolated and subjected to whole exome sequencing.
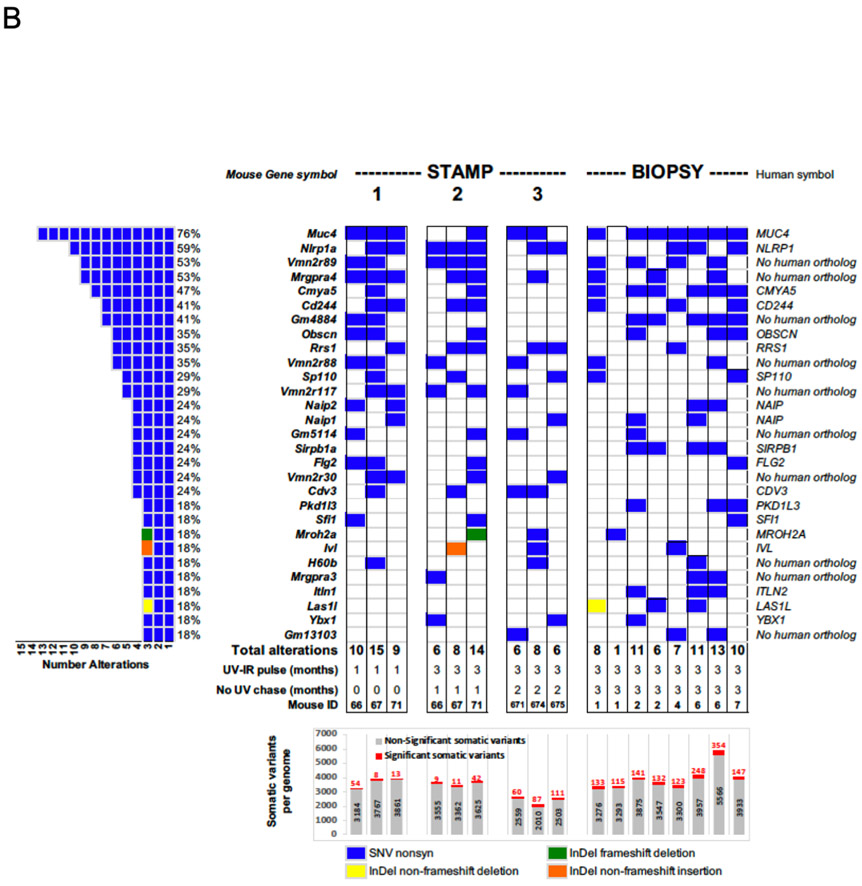
Figure 3.
Dose-dependent accumulation of mutations can be detected using whole-exome sequencing (WES) of skin samples obtained by STAMP in-vivo.
(A) The experimental scheme shows the irradiation schema used to produce papillomas and invasive SCC in Hairless mice. In brief, mice were irradiated thrice weekly over 3 months for a total dose of 175 kJ/m2 UVB (broadband). Mice were sampled at 1 month, 2 months, and 5 months using both STAMP and 6 months with standard full-thickness punch biopsy, DNA isolated and processed for whole exome sequencing at 72-140X coverage.
(B) Oncoprint showing the burden of mutations in the top 29 most frequently altered genes. Samples shown are from mice sampled by either STAMP (“STAMP”) or punch biopsy (“BIOPSY”). Each column in the center represents one mouse. There are three sets of STAMP samples (each n=3 mice). On the left is the total frequency of mutations observed in these 29 genes in descending order. Several of these genes have no apparent human orthologs or are unlikely to be expressed in skin (e.g. vomeronasal receptors). On the right, the gene names are listed with specific types of variants observed (single nucleotide vs. in-del). The STAMP columns are separated into sets of n=3 mice sampled at 1 month following the start of UV exposure (“1”), 1 month following the cessation of the 3-month course of UV (“2”), and 2 months following the cessation of UV (“3”). The BIOPSY columns represent n=9 full-thickness skin biopsies obtained at 3 months following the cessation of UV. Below, is listed the number of significant variants (red) over total variants (gray) across for each set of samples. The number of significant variants clearly rises over time, achieving an apparent plateau at the 5-6 month time point.
In order to easily visualize the data, we compiled a list of the 36 most frequently significantly mutated genes found to be altered at a frequency of at least 24%, plotting them in descending order of frequency (Figure 3B). Their mutation frequencies across samples and cohorts are listed (Figure 3B, left) and the mutational loads listed at the bottom, segregated by significance (numbers of total variants in gray, significant variants in red). As expected, there is a progressive increase in the number of statistically significant mutations with increasing total UVR exposure, which progresses beyond the time the UVR exposure was stopped (Figure 3B). By 5-6 months, sampled by STAMP and BIOPSY methods, the mutational loads appear to stabilize at approximately 3.3 mutations / Mb (range of 60 – 354 significant variants).
Importantly, there is remarkable consistency in the ability to detect recurrently-mutated genes Of the 29 top mutated genes, many do not have human orthologs and many are not expressed in keratinocytes (Figure 3B), including some vomeronasal (olfactory) receptor genes suggesting that at this level, clones can be easily detected but may not necessarily reflect strong selection for expansion of cells harboring mutations in cancer-related (tumor suppressor) genes or proto-oncogenes but may instead reflect lack of repair at these loci. Thus, at this depth of sequencing of the UV-exposed samples (approximately 72-140X), it is apparent that consistently recurrently-mutated genes in UV-exposed (non-lesional) skin can be readily identified and potentially used as a gauge of prior UV exposure and resultant mutations.
As it is well-established that mutations in cancer-related genes can be observed in chronically UV-exposed skin (Chitsazzadeh et al., 2016, Martincorena et al., 2015), we then focused on whether genes known to be mutated in cuSCC were also mutated in our dataset. We found mutations in Trp53, Notch family, Fat family, and Kmt2 family members at a low frequency (6-16 variants) in the STAMP samples (n=3 per set), with variant allele frequencies ranging from 10-84% (Supplementary Table 1). The BIOPSY samples (n=6) harbored a total of 41 distinct variants in these cuSCC-related genes and gene families with similar variant allele frequencies of 10-93% (Supplementary Table 1). This coincides with a decrease in the frequency of mutations observed in the top 29-gene panel (Figure 3B), suggestive of the emergence of true selective advantages conferred by pathogenic mutations in known drivers of cuSCC development.
Taken together, these findings suggest that in this model, mutational loads approach previously reported loads observed in chronically UV-exposed human skin within 3 months after completion of the 3-month course of UV irradiation. In our experience, all exposed mice by this point have developed papillomas and most have developed invasive cuSCC (Chitsazzadeh et al., 2016). Although it is also likely that the sensitivity of detecting these mutations in epidermal keratinocytes is heightened by sampling epidermis only with STAMP, as opposed to what is sampled in full-thickness biopsies, the consistency of observing mutations in overlapping genes shows that non-invasive STAMP likely at least matches the sensitivity of full-thickness punch biopsies.
STAMP enables sampling of human epidermis in-vivo and highlights significant mutational differences between sun-exposed and non-exposed skin
We then elected to test this in human subjects by sampling skin from multiple sites on organ transplant recipients because this group of patients is highly predisposed to developing cuSCC, with a risk of over 100-fold greater than that of the general population (Rangwala and Tsai, 2011). We reasoned that mutational burdens would be higher in sun-exposed areas relative to non-sun-exposed areas and sampled a total of 15 areas (approximately 0.8-1.8 cm2) from 7 immunosuppressed male organ transplant recipients aged 56-73 (Supplementary Table 2). Sampling was performed by wetting an angled felt-tip pen (Supplementary Figure S1A) wet with surfactant and rubbing the skin for 3 minutes. A small abrasion is seen almost immediately following this, with resultant surrounding erythema within 6 hours and crust at 24 hours (Supplementary Figure S1B). Histology of an area of peritumoral normal skin removed at Mohs surgery was subjected to STAMP ex-vivo and confirmed the ability of this procedure to remove epidermis in human skin (Supplementary Figure S1C). No adverse effects were noted in 5 of the patients seen in the subsequent 3-4 months. Sun-exposed areas consisted only of areas with clinically-evident dermatoheliosis. Total DNA yields following standard column-based purification (Purelink) ranged from 7.2 to 324.0 ng (Supplementary Table 2), with DNA integrity (DIN) in the range of 6.1 to 8.1 (Supplementary Figure S2).
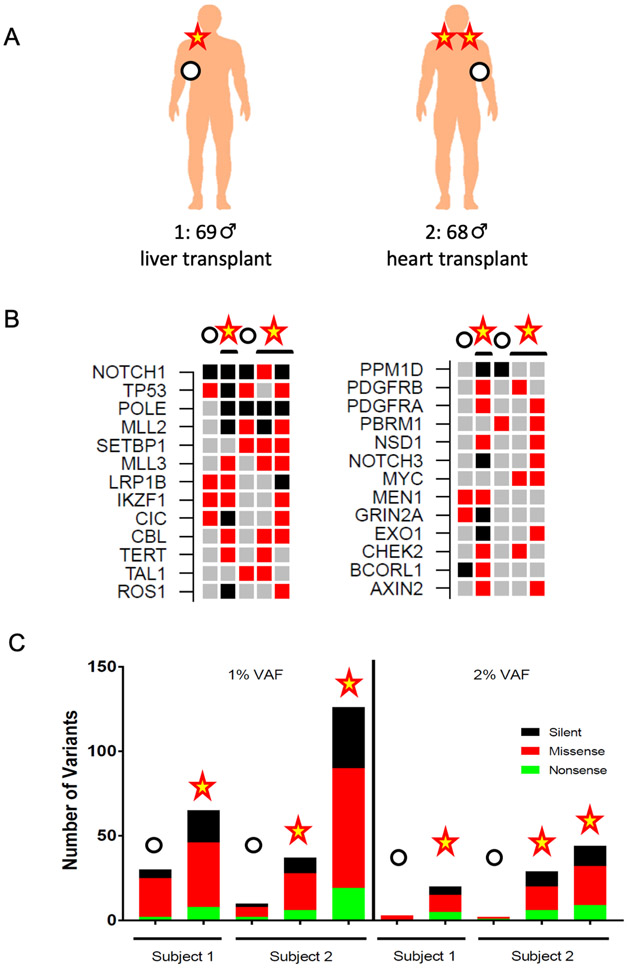
Based on multiple prior reports, we assumed that the sizes of clones in human skin would be relatively small, on the order of several hundred cells (Brash et al., 1991, Jonason et al., 1996). Therefore, we elected to use a more cost-effective targeted exome sequencing approach at high depth (>3000X) to ensure we could identify evidence for clones harboring mutations. Having established a range of possible yields using STAMP, we then performed targeted exome sequencing (QIAseq Cancer Panel) on two of the seven organ transplant recipients: Subject 1 was a 69-year old man who received an orthotopic liver transplant in 2017 with a history of melanoma and basal cell carcinoma. He was sampled in two areas (sun-protected right lateral upper chest / axilla; sun-exposed right lower neck). Subject 2 was a 68-year old man who received a heart transplant in 2016 and had a history of squamous and basal cell carcinomas. He was sampled in three areas (sun-protected left lateral chest; sun-exposed left and right upper shoulders) (Figure 4A). DNA from these 5 sites were isolated, purified and sequenced on the QIAseq platform matched to germline DNA isolated from saliva.
Figure 4.
In-vivo STAMP of sun-exposed, at-risk human skin reveals high burden of mutations
(A) Two male immunocompromised organ transplant recipients with a history of skin cancer and clinically-evident dermatoheliosis were sampled in both sun-exposed and sun-protected areas and DNA isolated and subjected to targeted exome sequencing to a median depth of 3,000X.
(B) Oncoprint of cancer-related genes in sun-exposed vs. sun-protected samples from high-risk organ transplant recipients. The 26 most frequently altered genes in the QIAseq panel are listed in order of decreasing frequency (top to bottom / left to right). Among the most frequently mutated genes are TP53, NOTCH1, MLL2/3, as previously reported. Red boxes denote non-frameshift, non-synonymous variants and black boxes denote frameshift, stopgain, stoploss, or splicing variants.
(C) Somatic single-nucleotide variant burdens using both 1% and 2% variant allele cutoffs in sun-exposed and sun-protected samples from high-risk organ transplant recipients.
Consistent with prior published results, there was significant enrichment for mutations in genes known to be altered in cutaneous SCC (Chitsazzadeh et al., 2016, Martincorena et al., 2015) (Figure 4B). These include most prominently, NOTCH1, TP53, KMT2 family members among others. At a 2% VAF, there is a very clear delineation between sun-exposed vs. sun-protected areas (Figure 4C). The range of distinction between sun-exposed and sun-protected areas suggests a dynamic range of 6.7 to 22-fold, which may eventually enable discernment of finer gradations of chronic UV exposure in skin.
DISCUSSION
Here we report the ability of STAMP to successfully harvest DNA non-invasively from epidermis and validate its use in-vivo in human skin. We used a simple and non-invasive method for obtaining DNA from normal non-lesional skin suitable for standard whole exome and targeted exome sequencing. The method is based on use of two commercially available surfactants that solubilize tissue and preserve DNA, RNA, and protein biological functions (Hwang et al., 2012, Hwang et al., 2013, Paliwal et al., 2012). Importantly, our data demonstrate the feasibility of using this method to obtain and sequence epidermal DNA in-vivo. Other published methods of sampling have been described and used, including tape stripping (Lacerenza et al., 2016, Wong et al., 2004), microbiopsy (Lin et al., 2013), and superficial scraping (Wang et al., 2018), but have yields that are substantially lower.
We have demonstrated that STAMP successfully samples epidermis non-invasively, without causing scarring (Fig. 2). In our mouse model of UV-driven cuSCC, a dose-dependent rise in mutational load is seen using serial sampling of mice across time. Importantly, the number of statistically significant variants (SNV and in-del) rose, stabilizing to almost 4 mutations / Mb, in line with previously-reported results on chronically exposed human skin (Chitsazzadeh et al., 2016, Martincorena et al., 2015).
The top 29 statistically significantly mutated genes are enriched for either ones not expected to be expressed in epidermis, that have no human analogues, or that have not been implicated in cuSCC, but rather are more likely to represent genes not subject to transcription-coupled repair (Zheng et al., 2014). This demonstrates and further confirms the non-uniform distribution of mutations across the genome, a characteristic of UV-induced DNA damage which has been demonstrated in melanocytes recently (Premi et al., 2019). Furthermore, it suggests that targeted sequencing could potentially be used to measure prior UV exposure if genomic sites are selected judiciously.
In the cases where we sampled from high-risk patients followed for skin cancer surveillance following solid organ transplantation, we targeted relatively sun-exposed and sun-protected sites, acknowledging that we did not have exposure data for patients, and only the appearance of dermatoheliosis to guide site selection. While the dynamic range of mutations in this limited cohort appears wide, it remains to be seen how well this ultimately correlates with prior exposure and subsequent risk of cancer and how site-specific these measures can be. The enumeration of mutations reflects a record of UV-induced damage filtered through nucleotide excision and transcription-coupled repair, clonal dynamics, immune surveillance and other factors, so any variation in these elements also contributes to the mutational record in different ways across patients and sites.
Finally, we show as a means of contrasting UV-induced mutation loads across multiple anatomic sites, the potential for establishing comprehensive spatial maps of genomic evidence of UV-induced DNA damage which can be coupled to objective risk assessment. This could serve as a means of personalized risk prediction and cancer prevention.
MATERIALS & METHODS
Surfactant
We have previously described a combination of two surfactants, the non-ionic Brij-30 and the zwitterionic N-decyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (DPS), which when combined in equimolar ratio (0.5% w/v each), results in a mixture that enables the solubilization of tissue (Hwang et al., 2012, Hwang et al., 2013, Paliwal et al., 2012). Originally optimized and applied for epidermal protein recovery, we adapted it here for recovery of epidermal DNA. In brief, if coupled to ultrasound or mechanical energy even in the form of dermabrasion, the epidermis can be solubilized, and, if the dermis is not breached, no scarring results. This originally termed Surfactant-based Tissue Acquisition for Molecular Profiling (STAMP).
Mouse model of UV-induced cSCC
Hairless mice (SKH-1E, Charles River) were exposed to solar simulated light (Newport 94082A; approx. 285 to 4000 nm) thrice weekly over a period of 3 months for a cumulative total dose of 175 kJ/m2 of broadband UVB as dosed using the broadband UVB Radiometer IL73-BB (International Light Technologies). Each dose averaged 3.4 standard erythema dose (SED) (Diffey et al., 1997). This is a spontaneous cuSCC mouse model with no genetically-engineered cuSCC-predisposing mutations (Chitsazzadeh et al., 2016). Typically, mice begin to develop tumors within 1-2 months following the cessation of irradiation though the samples examined here were strictly normal, UV-exposed, non-lesional skin. These tumors bear UV signature mutations and show mutations in Trp53, Notch1-2 and Fat1 with high frequency (Tsai Lab, unpublished), akin to human cuSCC (Knatko et al., 2017).
Skin epithelial sampling (rotary file, mouse)
Studies were performed under IACUC approval at both MD Anderson Cancer Center (06-09-06332) and Moffitt Cancer Center (IS 2403). Mice were briefly anesthetized with isoflurane, a small double-walled glass chamber (inner diameter 1.3 cm; outer diameter 2.4 cm) placed on the back and held in place using gentle suction. DXB (approximately 250-400 μL) was deposited centrally, the flat abrasive tip (0.5 cm) of an electric rotary nail file (Mani-Pro) submerged and placed in contact with epidermis and turned on to solubilize superficial epidermis over an area of 1.33 cm2 for 30-60 sec. Biopsy (4 mm punch) mouse tissues were obtained at necropsy after the indicated pulse and chase time.
Patient selection
Patients were consented during standard of care dermatology appointments for routine skin examination (N. Patel) under IRB-approved Protocol MCC #19076. All patients were organ transplant recipients on immunosuppressive therapy and under surveillance for a history of skin cancer. The patient from which histology collected from discarded tissue following Mohs surgery was consented under IRB-approved Protocol MCC # 18934.
Skin epithelial sampling (STAMP, human)
Skin was swabbed with cosmetic pens with tips soaked in DXB solution. Swabbing was done in circular motion for three minutes, applying light pressure, over a 1.0 to 1.5 cm diameter (thus covering an area of 0.79 to 1.77 cm2). While erythema was noted, there was never any bleeding or hyperpigmentation at follow-up over 2 months. DXB solution (40 μL) was pipetted and used to wet the pen tip with the sample. The tip was pressed against the interior of a microcentrifuge tube to squeeze as much liquid as possible and the tip was then rubbed against the opening of the tube to remove any attached cells. The released epithelial cells were immediately labeled and stored on ice. In a separate microcentrifuge tube, the tip was cut off with a blade and 40 microliters of DXB solution was added to preserve any possible left epithelial cells. The samples were later frozen in −20 deg C. Samples collected represented two distinct sun-exposure sites: those areas usually exposed to the sun (neck, shoulder) and those rarely exposed to the sun (lateral chest under the arm). Tubes were labeled as S-sun exposed and N- sun unexposed accordingly.
DNA extraction
The sample was mixed with 2 μL of Proteinase K and 180 μL of digestion buffer and resuspended, kept at 55 deg C, and rocked overnight. The Invitrogen Purelink DNA kit was used to extract the DNA from the samples per manufacturer’s instructions. Columns were eluted in a final volume of 60 μL.
Mouse exome sequencing
Purified DNA was subjected to sequencing as previously reported on the Illumina HiSeq platform (Chitsazzadeh et al., 2016). The NCBI SRA submission Bioprojects for the STAMP and BIOPSY studies are as follows: STAMP Bioprojects: PRJNA564608 and PRJNA564610, BIOPSY Bioproject: PRJNA564607. A mouse exome pipeline was used to map reads to the mm9 reference genome (Mann et al., 2015). Alignment of the raw reads was performed with bwa followed by samtools to fix mate pairs and sort files. Individual files were marked for PCR duplications, converted to bam file format and used to generate mpileup for each specimen. Tumor and normal specimen pairs were defined, run through varscan2, and then annovar to define somatic variants and their positions within the mouse genome. Varscan2 was run on the mpileup files with preset parameters, however our defined parameters more stringent than the preset values were also included: --min-coverage 10 --min-coverage-normal 10 --min-coverage-tumor 10 --min-var-freq 0.1, including a minimum coverage required at least 10 supporting reads (minimum read depth at a position to make a call; minimum coverage in normal and tumor to call somatic variants) and a minimum variant frequency of 0.10 was used. The additional ‘manual’ filtering steps, included those we previously discussed and set to remake the oncoprint, occurred after variants calls were annotated.
Targeted exome sequencing
Following the QC of the DNA samples, we used 40 ng of high-quality DNA as template for the QIAseq targeted DNA library prep. Targeted DNA sequencing was performed using QIAseq Comprehensive Cancer Panel. Variant calling was conducted in individual samples (sun exposed, non-sun exposed, and saliva) using QIAGEN’s smCounter2 pipeline (Xu et al., 2019). Somatic variants in sun and non-sun exposed samples were determined by removing variants that were present within paired saliva sample. To infer somatic variants specific to sun exposed sample, somatic variants present in both sun and non-sun exposed samples were removed from sun exposed sample. The adjusted somatic variants in sun-exposed sample were further filtered for common variants by excluding those with an allele frequency >1% in 1000 Genomes or ExAC data. The variant allele cutoffs were determined empirically by assessing the numbers of variants detected at a 1% vs. 2% cutoff and the ability to display discriminatory power between samples from exposed vs. unexposed skin.
Saliva sampling
For saliva sampling and DNA extraction, the DNA Genotek protocol for manual purification of DNA from 0.5 ml sample was followed.
Patient Selection
Because of the well-established elevated risk of immunosuppressed organ transplant recipients for developing skin cancer, we chose to focus on this population for our initial trial. Patents were consented and enrolled to the study under an IRB-approved protocol for swabbing (MCC #19076) and ex-vivo assessment of STAMP for histology (MCC #18934).
Data Availability
The NCBI SRA submission Bioprojects for the STAMP and BIOPSY studies are as follows: STAMP Bioprojects: PRJNA564608 and PRJNA564610, BIOPSY Bioproject: PRJNA564607. The processed output from the QIAseq panel (human targeted exome sequencing) is included in Supplementary Table 2.
The code used for analysis has been deposited in GitHub and is now freely accessible: https://gist.github.com/bkben1/b5584fd851fc14842a5597a7e15a991a
Supplementary Material
Supplementary Figure 1. Clinical and histologic features of STAMP on human skin.
(A)The felt tip pen used for swabbing consists of an angled absorbent tip.
(B)The clinical appearance of human skin before and immediately following application of DXB for 3 minutes with gentle pressure from a wetted felt tip pen through 24 hours. The dotted oval around the “immediate” panel denotes the approximate extent of the rubbing. The scale bar is 1 cm. Note the linear scar present at the inferior aspect of the site for reference.
(C) The histologic appearance of human rubbed with DXB ex-vivo shows removal of epidermis with an intact dermal papilla on the left.
Supplementary Figure 2. DNA integrity of STAMP-sampled epidermis.
The DNA integrity as measured by Tapestation for several of the STAMP samples from patients is shown. All samples that were ultimately sequenced had major peaks at 7 kb and above with DIN ranging from 6.1 to 8.1.
Supplementary Table 1. The list of variants observed in cuSCC-related genes Trp53, Notch family, Fat family, and Kmt2 family members. Each tab represents the cohort of mice corresponding to each time point.
Supplementary Table 2. The immunocompromised patient cohort and relevant clinical features described in Tab 1. Sample ID / Key is in the Tab 2. Tabs 3-11 detail the processed data including filtering through the 1000 Genomes project data. The list of numbers of variants detected at 1% and 2% variant allele frequency (VAF) levels in the QIAseq targeted exome sequencing panel for both sun-exposed and sun-protected skin from high-risk organ transplant recipients (Figure 4; Tab 12).
ACKNOWLEDGEMENTS
This study was supported by the NIH/NCI R01CA194617 (KYT) and R21CA191133 (KYT, SM), NIH/NIEHS R01 CA240602, ES030562 (KYT). This work has been supported in part by the Next Generation Sequencing Core – Smithville (CPRIT RP170002) and South Campus Vivarium at the University of Texas MD Anderson Cancer Center, an NCI designated Comprehensive Cancer Center (P30CA16672), as well as the Molecular Genomics Core and Vivarium at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30CA076292).
Footnotes
CONFLICTS OF INTEREST
SM and KYT report equity and prior research support from DXB Biosciences / Clearista as well co-inventorship on “Compositions for solubilizing cells and/or tissue”, U.S. Patent number: 9814422. These relationships did not impact funding, study design, or interpretation of results reported.
REFERENCES
- Brash DE. UV signature mutations. Photochem Photobiol 2015;91(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, et al. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A 1991;88(22):10124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Ziegler A, Jonason AS, Simon JA, Kunala S, Leffell DJ. Sunlight and sunburn in human skin cancer: p53, apoptosis, and tumor promotion. J Investig Dermatol Symp Proc 1996;1(2):136–42. [PubMed] [Google Scholar]
- Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 2016;7:12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffey BL, Jansen CT, Urbach F, Wulf HC. The standard erythema dose: a new photobiological concept. Photodermatol Photoimmunol Photomed 1997;13(1–2):64–6. [DOI] [PubMed] [Google Scholar]
- Griffin LL, Ali FR, Lear JT. Non-melanoma skin cancer Clinical medicine (London, England: ) 2016;16(1):62–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang BH, Doshi N, Tsai KY, Mitragotri S. A reagent to facilitate protein recovery from cells and tissues. Drug Deliv Transl Res 2012;2(5):297–304. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Tsai KY, Mitragotri S. Optimized lysis buffer reagents for solubilization and preservation of proteins from cells and tissues. Drug Delivery and Translational Research 2013. [DOI] [PubMed] [Google Scholar]
- Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A 1996;93(24):14025–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knatko EV, Praslicka B, Higgins M, Evans A, Purdie KJ, Harwood CA, et al. Whole-Exome Sequencing Validates a Preclinical Mouse Model for the Prevention and Treatment of Cutaneous Squamous Cell Carcinoma Cancer Prev Res (Phila: ) 2017;10(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerenza D, Aneli S, Omedei M, Gino S, Pasino S, Berchialla P, et al. A molecular exploration of human DNA/RNA co-extracted from the palmar surface of the hands and fingers. Forensic Sci Int Genet 2016;22:44–53. [DOI] [PubMed] [Google Scholar]
- Lin LL, Prow TW, Raphael AP, Harrold Iii RL, Primiero CA, Ansaldo AB, et al. Microbiopsy engineered for minimally invasive and suture-free sub-millimetre skin sampling. F1000Res 2013;2:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MB, Black MA, Jones DJ, Ward JM, Yew CC, Newberg JY, et al. Transposon mutagenesis identifies genetic drivers of Braf(V600E) melanoma. Nat Genet 2015;47(5):486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348(6237):880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem DX, Walterscheid JP, Kazimi N, Ullrich SE. Ultraviolet radiation-induced immunosuppression of delayed-type hypersensitivity in mice. Methods 2002;28(1):25–33. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Hwang BH, Tsai KY, Mitragotri S. Diagnostic Opportunities Based on Skin Biomarkers. Eur J Pharm Sci 2012. [DOI] [PubMed] [Google Scholar]
- Premi S, Han L, Mehta S, Knight J, Zhao D, Palmatier MA, et al. Genomic sites hypersensitive to ultraviolet radiation. Proc Natl Acad Sci U S A 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala S, Tsai KY. Roles of the immune system in skin cancer. Br J Dermatol 2011;165(5):953–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA dermatology 2015;151(10):1081–6. [DOI] [PubMed] [Google Scholar]
- Simons BD. Deep sequencing as a probe of normal stem cell fate and preneoplasia in human epidermis. Proc Natl Acad Sci U S A 2016;113(1):128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vin H, Ching G, Ojeda SS, Adelmann CH, Chitsazzadeh V, Dwyer DW, et al. Sorafenib Suppresses JNK-Dependent Apoptosis through Inhibition of ZAK. Mol Cancer Ther 2014;13(1):221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vin H, Ojeda SS, Ching G, Leung ML, Chitsazzadeh V, Dwyer DW, et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. Elife 2013;2:e00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Masaki T, Khan SG, Tamura D, Kuschal C, Rogers M, et al. Four-dimensional, dynamic mosaicism is a hallmark of normal human skin that permits mapping of the organization and patterning of human epidermis during terminal differentiation. PLoS One 2018;13(6):e0198011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R, Tran V, Morhenn V, Hung SP, Andersen B, Ito E, et al. Use of RT-PCR and DNA microarrays to characterize RNA recovered by non-invasive tape harvesting of normal and inflamed skin. J Invest Dermatol 2004;123(1):159–67. [DOI] [PubMed] [Google Scholar]
- Xu C, Gu X, Padmanabhan R, Wu Z, Peng Q, DiCarlo J, et al. smCounter2: an accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics 2019;35(8):1299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CL, Wang NJ, Chung J, Moslehi H, Sanborn JZ, Hur JS, et al. Transcription restores DNA repair to heterochromatin, determining regional mutation rates in cancer genomes. Cell Rep 2014;9(4):1228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, et al. Sunburn and p53 in the onset of skin cancer. Nature 1994;372(6508):773–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Clinical and histologic features of STAMP on human skin.
(A)The felt tip pen used for swabbing consists of an angled absorbent tip.
(B)The clinical appearance of human skin before and immediately following application of DXB for 3 minutes with gentle pressure from a wetted felt tip pen through 24 hours. The dotted oval around the “immediate” panel denotes the approximate extent of the rubbing. The scale bar is 1 cm. Note the linear scar present at the inferior aspect of the site for reference.
(C) The histologic appearance of human rubbed with DXB ex-vivo shows removal of epidermis with an intact dermal papilla on the left.
Supplementary Figure 2. DNA integrity of STAMP-sampled epidermis.
The DNA integrity as measured by Tapestation for several of the STAMP samples from patients is shown. All samples that were ultimately sequenced had major peaks at 7 kb and above with DIN ranging from 6.1 to 8.1.
Supplementary Table 1. The list of variants observed in cuSCC-related genes Trp53, Notch family, Fat family, and Kmt2 family members. Each tab represents the cohort of mice corresponding to each time point.
Supplementary Table 2. The immunocompromised patient cohort and relevant clinical features described in Tab 1. Sample ID / Key is in the Tab 2. Tabs 3-11 detail the processed data including filtering through the 1000 Genomes project data. The list of numbers of variants detected at 1% and 2% variant allele frequency (VAF) levels in the QIAseq targeted exome sequencing panel for both sun-exposed and sun-protected skin from high-risk organ transplant recipients (Figure 4; Tab 12).
Data Availability Statement
The NCBI SRA submission Bioprojects for the STAMP and BIOPSY studies are as follows: STAMP Bioprojects: PRJNA564608 and PRJNA564610, BIOPSY Bioproject: PRJNA564607. The processed output from the QIAseq panel (human targeted exome sequencing) is included in Supplementary Table 2.
The code used for analysis has been deposited in GitHub and is now freely accessible: https://gist.github.com/bkben1/b5584fd851fc14842a5597a7e15a991a