Summary/Abstract
Hematopoietic stem cells (HSCs) are used in the clinic to provide life-saving therapies to patients with a variety of hematological malignancies and disorders. Yet, serious deficiencies in our understanding of how HSCs develop and self-renew continue to limit our ability to make this therapy safer and more broadly available to those who have no available donor. Finding ways to expand HSCs and develop alternate sources of HSCs is an urgent priority. In the embryo, a critical transition in development of the blood system requires that newly emergent HSCs from the aorta-gonad-mesonephros (AGM) region migrate to the fetal liver where they aggressively self-renew and expand to numbers sufficient to sustain the adult long-term. This process of homing to the fetal liver is orchestrated by intrinsic regulators such as epigenetic modifications to the genome, expression of transcription factors, and adhesion molecule presentation, as well as sensing of extrinsic factors like chemokines, cytokines, and other molecules. Due to technical limitations in manipulating the fetal tissue microenvironment, mechanisms mediating the homing and expansion process remain incompletely understood. Importantly, HSC development is strictly dependent upon forces created by the flow of blood, and current experimental methods make the study of biophysical cues especially challenging. In the protocol presented herein, we address these limitations by designing a biomimetic ex vivo microfluidic model of the fetal liver that enables monitoring of HSC homing to and interaction with fetal liver niches under flow and matrix elasticity conditions typical during embryonic development. This model can be easily customized for the study of key microenvironmental factors and biophysical cues that support HSC homing and expansion.
Keywords: Biomechanical forces, Fetal liver, Hematopoietic stem cells, HSC expansion, Homing, Matrix elasticity, Shear stress
1. Introduction
Hematopoietic stem cells (HSCs) sustain nearly all lineages of the adult blood system in a process termed hematopoiesis. HSC transplantation is a curative therapy for a wide variety of blood disorders and cancers, most notably, leukemia. In fact, since the 1960s when allogeneic HSC transplantation was first attempted, the five-year survival rate for leukemia has increased from 14% to 65% in 2014 in the United States (1). Despite advances in clinical application of this therapy, significant gaps remain in our understanding of hematopoiesis and its regulation that limit success of clinical outcomes, placing patients at risk of infection, graft-versus-host disease, and other serious morbidities. Thus, there is a critical need to understand how the stem cell pool that sustains a life-long blood supply is established during development. Indeed, clues to the factors that drive HSC self-renewal and expansion can be found by exploring the various niches that nurture HSC specification and expansion during hematopoietic ontogeny.
During embryonic development, HSCs originate in the dorsal aorta of the aorta-gonad-mesonephros (AGM) region and migrate to the fetal liver, a region equivalent to the caudal hematopoietic tissue of fish (2). Homing to the fetal liver is a significant developmental transition in the life of an HSC that occurs at embryonic day 11 (E11) in mice and E28 in humans (3). Arrival in the fetal liver marks a period of aggressive expansion in the HSC pool (4), thus failure to properly home to the fetal liver niche results in hematopoietic collapse and embryonic death (5). HSCs leave the fetal liver around birth to populate the bone marrow, thereby establishing permanent residence in a distinctly different adult bone marrow niche that favors HSC maintenance rather than expansion (2). Self-renewal and expansion of HSCs is determined not only by intrinsic regulators, such as CD144 or β1 integrin adhesion molecules (6, 7), differential expression of transcription factors (8), epigenetic modifiers (9), and non-coding RNAs (10), but also by extrinsic signals presented within the specialized extracellular microenvironments of the niche (11). Indeed, arterial endothelial cells are implicated in forming initial associations with trafficking HSCs that enable lodgment and retention; whereas, peri-arterial Nestin+ NG2+ stromal cells in the portal vasculature provide proliferative signals to HSCs but do not impact homing behaviors (2, 12). Advances in imaging and genetic tools continue to shed light on the importance of interactions between HSCs and their niche (13).
A growing number of studies have revealed crucial roles for mechanical properties of the hematopoietic niche (14). HSC emergence in the AGM requires biomechanical forces created by blood flow, and embryos that lack a heartbeat fail to generate HSCs (15–17). The effects of flow, matrix stiffness, and other mechanical properties on HSC homing and expansion remain entirely unexplored in the fetal liver, in part due to the inaccessibility of the organ and difficulty in manipulating mechanical properties in vivo (18). In the protocol presented here, we develop a microfluidics-based ex vivo model to observe homing behavior of HSCs upon arrival at the fetal liver and implement design features that enable manipulation of fluid forces and matrix elasticity in the fetal liver niche. This biomimetic platform, akin to a fetal liver-on-a-chip, enables study of the dynamic interactions between HSCs and the fetal liver niche using two-photon microscopy. The system provides a tractable environment for understanding how biophysical features, ligand-receptor interactions, and intracellular signaling alter HSC lodgment and niche remodeling. Fluid flow conditions that are normally present in the embryonic vasculature and fetal liver such as pulsatile flow and frictional force, or shear stress, is readily adapted by adjusting flow rate and viscosity of the perfused media, as well as the height of the microfluidic channel. Similarly, matrix stiffness can be varied by adjusting the composition of the hydrogels that support the fetal liver tissue. Lastly, application of inhibitors, blocking antibodies, or genetic alterations can be applied to understand HSC trafficking and division. In summary, this ex vivo microfluidics model is highly customizable, and it enables study of the molecular and mechanical cues that impact HSC trafficking and the self-renewal behaviors that occur within the fetal liver niche.
2. Materials
Appropriate alternatives may exist for all the products and vendors listed in this section.
2.1. Microfluidic Channel
Silicone sheet, 1/16 inch thick (Scientific Instrument Services, Inc, catalog # S10)
Single edge razor blade (VWR North American, catalog # 55411-055)
Medical Adhesive Spray, 3.8 oz (Hollister, catalog # 7730)
Sticky-Slide I0.8 Luer (IBIDI, catalog # 80198)
Plain glass microscope slides 25 × 75 × 1.0 mm (Fisher Scientific, catalog # 12-550-A3)
Hammer Driven Slot Hole punch 12 × 5 mm (Amazon, catalog # F0168-12×5mm-1P)
Kimwipes (Fisher Scientific, catalog # 06-666A)
Bind-silane (GE Healthcare, catalog # GE17-1330-01)
Acetic acid 2-methoxy-anthracen-9-yl ester (Millipore Sigma, catalog # R240184)
Sharpie ultra-fine point marker
Standard ruler
100% Ethanol
70% Ethanol
Paper towel
2.2. Polyacrylamide Hydrogels
Plastic petri dish 100 mm (Fisher Scientific, catalog # 09-720-500)
Cover glass 22 × 60 mm (Richard-Allan Scientific, catalog # 102260)
Sigmacote siliconizing reagent (Millipore Sigma, catalog # SL2)
40% Acrylamide (Bio-Rad, catalog # 1610140)
2% Bis-Acrylamide (Bio-Rad, catalog # 1610142)
Ammonium persulfate (APS; Millipore Sigma, catalog # 09913)
Tetramethylethylenediamine (TEMED; Millipore Sigma, catalog # T7024)
Bind-silane (GE Healthcare, catalog # GE17-1330-01)
Acetic acid 2-methoxy-anthracen-9-yl ester (Millipore Sigma, catalog # R240184)
Ultrapure or deionized water (DI water)
Kimwipes (Fisher Scientific, catalog # 06-666A)
Desiccator
2.3. Collagen Gel Coating
PBS (Thermo Fisher Scientific, catalog # 10-010-072)
Sulfo-SANPAH (Thermo Fisher Scientific, catalog # 22589)
Iscove’s Modified Dulbecco’s Medium (IMDM, without serum) (Thermo Fisher Scientific, catalog # 12440-053)
0.1 N Sodium hydroxide (NaOH; Millipore Sigma, catalog # 43617-1L)
Collagen type I, rat (3-4 mg/ml) (Corning, catalog # 354236)
Cell-Tak cell and tissue adhesive (Corning, catalog # CB40240)
0.1 M sodium bicarbonate (adjust to pH 8.0) (Millipore Sigma, catalog # 36486-1L)
1 N Sodium hydroxide (NaOH; Millipore Sigma, catalog # SX0607H-3)
High intensity UV lamp (λ=302-365 nm, intensity=37 mW)
Sterile ultrapure or deionized water
2.4. Fetal Liver Tissue
Mouse embryo from E12.5-E14.5 timed pregnant dam (C57BL/6) for fetal liver
PBS (Thermo Fisher Scientific, catalog # 10-010-072)
Pre-poured into 100 mm dish 4% low-melting point Agarose gel, 1 cm thick (Fisher Scientific, catalog # BP165-25)
150 mm bacteriological petri dish (Fisher Scientific, catalog # 08-757-148)
Krazy Glue pen (Krazy Glue, catalog # KG824)
Leica VT1000 S automatic vibrating blade microtome (VWR, catalog # 76001-016)
Camel hair round tip brush (Fisher Scientific, catalog # 03-670)
Kimwipes (Fisher Scientific, catalog # 06-666A)
70% Ethanol
8 inch Spatula, stainless steel with bent end (Lyon Scientific, catalog # SSFB08)
Culture media comprised of IMDM (ThermoFisher Scientific, catalog # 12440-053), 20% fetal bovine serum (R&D Systems, catalog # S10250) supplemented with L-glutamine (4 mM), 100 ng/ml SCF (Peprotech, catalog # 250-03), 100 ng/ml IL-3 (Peprotech, catalog # 213-13), and 100 ng/ml FLT3L (Peprotech, catalog # 250-31L)
2.5. AGM Dissociation for Isolation of Hematopoietic Stem and Progenitor Cells
Mouse embryos from E11.5 timed pregnant dams (Parent strains should harbor fluorescent hematopoietic stem cell reporter such as those available through the Jackson Laboratory Ly6a-GFP (B6.Cg-Tg(Ly6a-EGFP)G5Dzk/J), Alpha-catulin-GFP (Ctnnal1tm1.1Sjm/J), or Fgd5-mCherry (C57BL/6N-Fgd5tm1Djr/J)) for AGM regions
PBS (Thermo Fisher Scientific, catalog # 10-010-072)
Accutase cell detachment solution (Stem Cell Research, catalog # 07920)
Culture media comprised of IMDM (ThermoFisher Scientific, catalog # 12440-053), 20% fetal bovine serum (R&D Systems, catalog # S10250) supplemented with 4 mM L-glutamine (ThermoFisher Scientific, catalog # 25-030-081), 100 ng/ml SCF (Peprotech, catalog # 250-03), 100 ng/ml IL-3 (Peprotech, catalog # 213-13), and 100 ng/ml FLT3L (Peprotech, catalog # 250-31L)
DAPI (ThermoFisher Scientific, catalog # 62248), propidium iodide (ThermoFisher Scientific, catalog # P3566), or 7-AAD (ThermoFisher Scientific, catalog # A1310)
BD Falcon cell strainer, 70 μm mesh (Fisher Scientific, catalog # 08-771-2)
2.6. Fluidics Assembly and Imaging
Culture media comprised of IMDM (ThermoFisher Scientific, catalog # 12440-053), 20% fetal bovine serum (R&D Systems, catalog # S10250) supplemented with L-glutamine (4 mM), 100 ng/ml SCF (Peprotech, catalog # 250-03), 100 ng/ml IL-3 (Peprotech, catalog # 213-13), and 100 ng/ml FLT3L (Peprotech, catalog # 250-31L)
Three way stop cock, 2 female luer lock and one male luer lock (Qosina, catalog # 13813)
Silicone tubing, 1/16 ID (Fisher Scientific, catalog # 1118915G)
Female Luer x 1/16 hose barb, PVDF (Cole Parmer, catalog # T45512-00)
Elbow Luer connector (IBIDI, catalog # 10802)
Syringe, 10 ml (BD, catalog # 309604)
Two Syringe, 3 ml with Luer-Lok Tip (BD, catalog 309657)
Lab stand with base
Three prong extension clamp (Fisher catalog # 05-769-8Q)
Pipetman size P1000 with tips (Gilson)
Syringe pump PHD ULTRA 4400 with Remote (Harvard Apparatus, catalog # 703310)
Two-photon microscope
3. Methods
All procedures are carried out at room temperature unless mentioned otherwise.
3.1. Microfluidic Device Assembly
3.1.1. Microfluidic Channel Base Slide
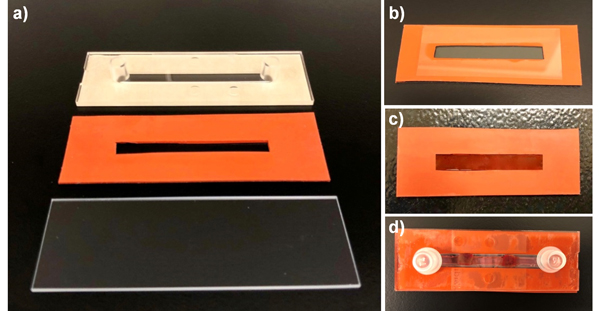
The microfluidic fetal liver-on-a-chip device is comprised of three primary structural components, including a glass slide, a slotted silicone sheet, and an IBIDI Sticky-Slide I0.8 Luer channel slide (Fig. 1a).
To prepare the silicone element of the device, use a single edge razor to cut out a rectangular shape from the silicone sheet to the same dimensions of the plain glass slide (25 × 75 mm2).
Take the cut-out piece of silicone and, using a ruler and Sharpie, measure 10 mm along the longest edge (75 mm). Draw a line across. From that same edge, measure 15 mm. Draw a line across. There will be two marker lines running in parallel separated by a width of 5 mm.
From the shortest edge (25 mm) measure 15 mm at both edges of the silicone sheet. Mark across only where the markings cross each other (see Note 1). This demarcates the boundaries of the channel where the polyacrylamide gel will be placed and represents an area of 5 mm x 45 mm referred to as the “channel area” below.
The “channel area” can be removed manually by cutting with a single edge razor producing a single long channel, as shown in Fig. 1a. Alternatively, a 12 × 5 mm slot hole punch can be used to remove smaller precise regions from the “channel area” on the silicone sheet by pressing down firmly on top of a hard-counter surface. Three slot hole punched areas can be produced along the “channel area” on the silicone sheet if using the hole punch.
Next, make the glass slide and the slotted silicone hydrophilic by immersing them in 10 ml of bind silane solution prepared by mixing 9.5 ml ethanol, 0.5 ml acetic acid, and 30 μl bind silane. Let it sit in a chemical fume hood for 15–20 min. Then spray both the slide and silicone with 70% ethanol and wipe dry with kimwipes.
To assemble the base slide, place the silicone sheet on top of a paper towel. Spray one side of the silicone sheet with two quick passes of the medical adhesive (see Note 2). The sprayed side of the silicone sheet should have a full layer of adhesive on the surface. Wait 2–3 min for the adhesive to set prior to attaching the silicone to the slide (see Note 3).
With gloved hands, pick up the silicone sheet and place one corner on top of the microscope slide to get correct alignment. Once positioned, place the remaining surface of the silicone sheet fully onto the slide and press firmly with your fingertips to adhere both together. Wait an additional 5 min for the silicone to adhere to the slide. If needed, place a waited flat object on top to compress the silicone and glass slide together. Slides are ready for the acrylamide gel.
The IBIDI sticky slide will be attached on the top of the base slide after liver tissue installation on the hydrogel (section 3.2).
Figure 1. Elements of the fetal liver on a chip device.
a) Three structural components of the device frame include one glass slide, one slotted piece of silicon sheet cut to the same size as the slide, and an IBIDI sticky slide. b) Base slide is assembled by attaching silicone piece to the slide, followed by pouring hydrogel into the channel within the silicon sheet. A hydrophobic glass slide is placed on top of the gel to allow uniform polymerization. c) Hydrogel is covered with sulfo-SANPAH. d) Assembly of the microfluidic slide is complete after sticking the IBIDI slide to the top of the silicone sheet.
3.1.2. Polyacrylamide Hydrogels
Before making the gels, prepare the hydrophobic cover glass by immersing it in Sigmacote. Put a rectangular cover glass (22 × 60 mm2) into a 100 mm plastic petri dish and add 1 ml Sigmacote to cover the surface of the glass and let it sit in a chemical hood at least for 15 min.
Pipette out the excess Sigmacote from petri dish and rinse the cover glass with ultrapure or deionized water (see Note 4).
Immediately, wipe the cover glass with kimwipes and put in a clean petri dish until used.
To prepare the gel, depending on the desired stiffness of polyacrylamide gels, combine the required amount of 40% Acrylamide, 2% Bis-Acrylamide, and deionized water in a 15 ml tube (Table 1).
Put the tube in a desiccator for at least one hour.
Immediately before pouring the gels into the base slide (section 3.1.1), add APS at 0.5% v/v of total solution (5 μl for 1 ml of solution) and TEMED at 0.05% v/v (0.5 μl for 1 ml of solution), and mix gently using a pipette (see Note 5).
Immediately pipette 300–350 μl of the gel into the channel of the silicon portion of the base slide (section 3.1.1) and put the hydrophobic cover glass on the top (Fig. 1b). Depending on stiffness of the gel, polymerization may take 10–60 min.
After polymerization (see Note 6), immerse the chamber in PBS and slowly remove the hydrophobic cover glass. Keep the base slide in PBS and proceed to the next step for collagen coating (see Note 7).
Table 1.
Composition and corresponding elastic properties of polyacrylamide gels
| Hydrogel elastic modulus (kPa) | Acrylamide | Bis-Acrylamide | To prepare 10 ml of polyacrylamide gel: | ||
|---|---|---|---|---|---|
| 40% Acrylamide (ml) | 2% Bis-Acrylamide (ml) | Ultrapure water (ml) | |||
| 0.02 ± 0.008 | 3% | 0.02% | 0.75 | 0.1 | 9.15 |
| 0.08 ± 0.047 | 3% | 0.05% | 0.75 | 0.25 | 9 |
| 0.09 ± 0.008 | 3% | 0.1% | 0.75 | 0.5 | 8.75 |
| 0.25 ± 0.05 | 4% | 0.075% | 1 | 0.375 | 8.625 |
| 0.43 ± 0.03 | 4% | 0.2% | 1 | 1 | 8 |
| 0.79 ± 0.15 | 5% | 0.1% | 1.25 | 0.5 | 8.25 |
| 0.9 ± 0.07 | 5% | 0.2% | 1.25 | 1 | 7.75 |
| 5.5 ± 0.29 | 10% | 0.15% | 2.5 | 0.75 | 6.75 |
| 9.5 ± 0.77 | 10% | 0.3% | 2.5 | 1.5 | 6 |
| 50.3 ± 4.2 | 12% | 0.6% | 3 | 3 | 4 |
3.1.3. Collagen Coating
Make 10 mM sulfo-SANPAH solution by adding 6 μl of 50 mg/ml sulfo-SANPAH stock into 94 μl sterile deionized water (see Note 8).
Immediately, add 100 μl of prepared sulfo-SANPAH solution on the polyacrylamide gel in the base slide (section 3.1.2) and swirl from side to side to coat the gel surface evenly (Fig. 1c).
Place the base slide under a UV lamp (λ=302–365 nm, intensity=37 mW) for 5 min and proceed to prepare the collagen solution.
For collagen coating, prepare 1 ml of collagen solution on ice with a concentration of 1 mg/ml by mixing in the following order: 692 μl IMDM (no serum), 58 μl 0.1N sodium hydroxide, and 250 μl 3–4 mg/ml rat tail collagen. Keep on ice until ready to transfer to the polyacrylamide gel.
When the 5 min treatment with sulfo-SANPAH is complete, rinse the base slide 2x with PBS to remove excess sulfo-SANPAH from top of the gel.
Immediately add 100 μl of collagen solution on the gel and make sure to completely and evenly cover the surface.
Keep the base slide at 4°C overnight or in an incubator at 37°C for 2 hours (if needed the same day) until the collagen polymerizes (see Note 9).
After collagen incubation, wash the base slide with sterile PBS to remove excess collagen solution.
Prepare 150 μl of Cell-Tak adhesive solution by mixing 99 μl 0.1 M sodium bicarbonate (be sure to pH to 8.0 before use), 34 μl Cell-Tak, and 17 μl 1 N sodium hydroxide.
Apply 100 μl Cell-Tak solution to the top of the collagen gel for at least 20 min at either room temperature or 37°C. Freshly prepared Cell-Tak solution should be applied within 10 min to ensure efficient adsorption to the collagen gel, equivalent to a concentration of approximately 3.5 μg/cm2.
3.2. Fetal Liver-On-A-Chip Assembly
Sacrifice the E12.5-E14.5 pregnant dam by CO2 inhalation and cervical dislocation. Spray the abdomen with 70% ethanol for disinfection. Open the abdomen with dissecting scissors to remove the uterus. Wash the uterus in 1x PBS and transfer to a culture plate filled with PBS. Dissect the embryo(s) from the uterus under sterile conditions using a dissecting microscope and 70% ethanol-sterilized tools.
Dissect the region of the embryo containing fetal liver (red in color, caudal to the heart) by holding the body of the embryo with one forceps and, with another forceps, gently decapitating the head and removing the lower torso and tail, exposing the rostral portion of the fetal liver.
Place the portion of the embryo torso containing the fetal liver into fresh PBS in a clean dish.
Assemble a small block of agarose gel (approximate 1.2 × 1.2 cm2) on the chuck of the vibratome using super glue. Secure the embryonic tissue alongside the agarose block by gluing it directly to the chuck. The dorsal torso portion of the embryo should face the blade during sectioning (see Note 10).
Place chuck on vibratome, submerged in PBS and section thin slices (100–200 μm) of the fetal liver. Collect live fetal liver slices into a specimen dish containing clean PBS on ice.
Carefully scoop up the fetal liver with a bent end spatula and brush, then place the liver, along with PBS, on top of the gel. Remove excess PBS by pipette and or a rolled Kimwipe tissue to soak up excess liquid to allow the liver to flatten and adhere to the gel. Work quickly to keep tissue moist.
Once the liver sections have flattened on the gel, allow the tissue to adhere further by overlaying PBS on the channel. Place the slide into a fresh 150 mm bacteriological dish with lid and transfer to a 37°C incubator for 5 minutes. To allow the tissue additional time to attach, the slide can then be submerged in a smaller dish containing media for longer periods of time until ready to assemble the microfluidic sticky slide.
Blot dry the side areas of the silicone base surface which will make contact with the top of the IBIDI sticky slide. The channel portion of the sticky slide will align on top of the fetal liver/gel coat area.
Remove the paper backing from the sticky slide and carefully place on top of the silicone base slide. Firmly press the sticky areas until visible adherence is noted onto the silicone sheet. The channel space must align with the tissue and should not compress the fetal liver sections.
Once the sticky slide is secure, gently pipette 250 μl of warm culture media into the reservoir ports to fill the channel. Check for any media leaks on the sides of the slide.
Cap the two reservoir ports on top of the slide (Fig. 1d). Transfer slide into a 150 mm bacteriological dish with lid and place inside the tissue culture 37°C incubator until the HSCs are prepared.
3.3. HSC Isolation
Sacrifice the E11.5 pregnant dam(s), isolate uteri, and recover embryos as detailed for fetal liver isolation above.
Dissect the AGM region from embryos in cold PBS and transfer each by pipette into a 15 ml-capacity conical tube. The end of a p200 or p1000 pipette tip can be cut off to create a larger opening, thus minimizing disruption of the AGM tissue.
Remove excess PBS from the tube and add 3–4 ml Accutase solution to the AGM tissue from approximately 10 embryos.
Incubate tissues on a plate shaker for 20 min at room temperature.
Gently pipette solution 5 times and shake for 3–5 more min.
After incubation, pipette 10 more times gently to generate a single cell suspension. Add 4 ml culture medium and mix.
Centrifuge cells at 600–700 rcf for 5 min and resuspend in medium at a final concentration suitable for sorting on a flow cytometer. Approximately 5 × 106 cells/ml is a reasonable starting point for most cytometers, but the volume of the solution can be adjusted if clogging occurs or cell purity after the sort is unacceptably low.
Add at a viability dye to a concentration appropriate for live-dead discrimination, such as DAPI (500–1000 ng/ml), propidium iodide (1 μg/ml), or 7-AAD (2.5 μm).
Filter the cell suspension through a 70-μm nylon cell strainer just prior to sorting.
Using a flow cytometer capable of sorting, enrich for live cells harboring the HSC reporter based upon positivity for GFP or mCherry and negativity for the viability dye (DAPI, propidium iodide, or 7-AAD). Sort the selected HSC population into culture media. After sorting is complete, test the purity of the sort by acquiring events from the sorted sample to ensure that HSC enrichment was successful.
Centrifuge sorted cells at 600–700 rcf for 5–10 min and resuspend in warm culture medium at a final concentration of between 5 × 104 to 1 × 105 cells/ml. Cell suspension can then be transferred to a 15 ml conical centrifuge tube for storage while waiting for assembly of the syringe pump, followed by perfusion of the cells into the microfluidic channel and live imaging by two-photon microscopy.
3.4. Fluidics Assembly and Imaging
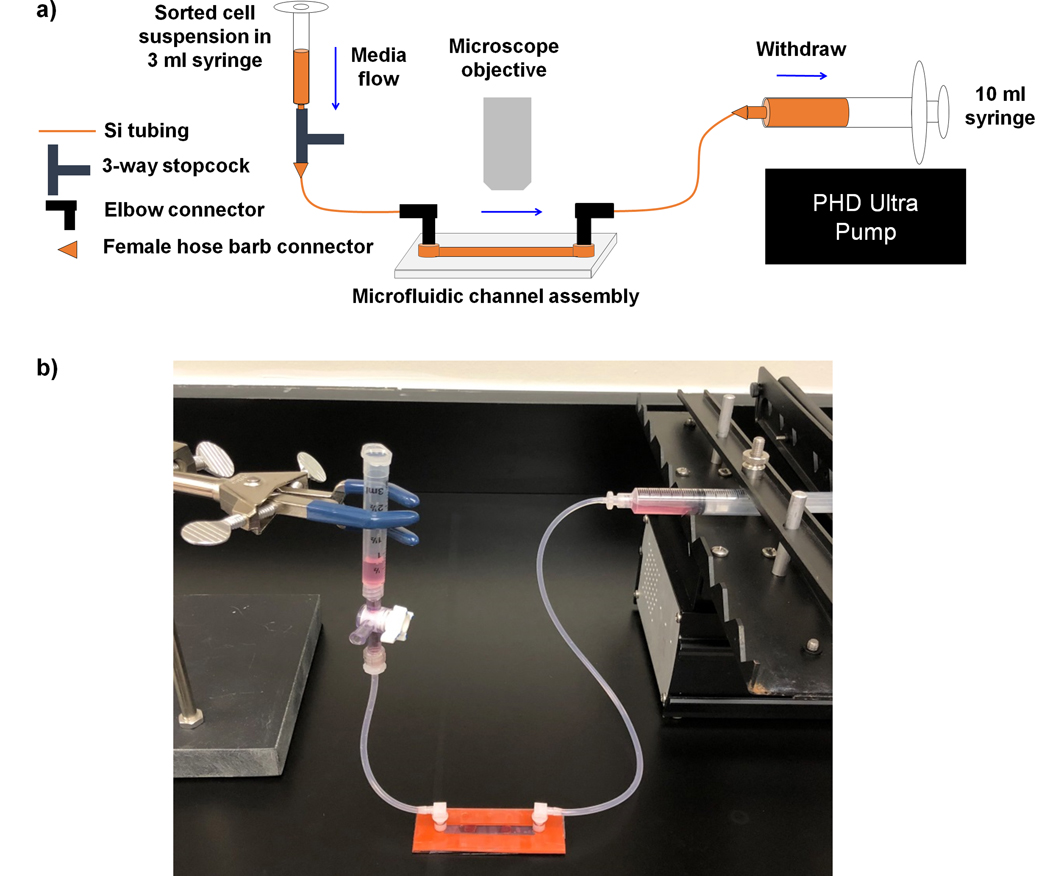
Gather together components needed for connection of the fetal liver-on-a-chip to the fluid pump and syringe reservoir for the HSCs. Final assembly is depicted in Fig. 2, but should be in close proximity to the two-photon microscope for analysis of HSC movement and interactions with the fetal liver.
Pre-cut one 12 inch and one 4 inch piece of silicone tubing (see Note 11). To each tubing, insert one elbow luer connector (see Note 12). To the 12 inch tubing, insert a female luer hose barbed connector. This will connect to the syringe/pump assembly. To the 4 inch tubing, attach another female luer hose barbed connector, which will attach to the three way stop cock. Once the tubing is assembled, autoclave for sterility prior to shearing setup.
The pump and slide assembly will be setup next to the two-photon microscope. Place the PHD ULTRA 4400 remote to the left side of the microscope. Assemble the 10 ml syringe onto the pump remote. Connect the 12 inch silicone tubing with female luer connector to the end of the 10 ml syringe. Push the syringe plunger toward the end at the male/female luer connection.
Take the microfluidic channel (section 3.2) out of the incubator and place it on the stage of the microscope. Secure the slide if needed with either lab tape applied to the edges of the slide and stage or by use of the stage clips (see Note 13). Remove one port cap nearest the syringe pump and apply the elbow connector end to the port (see Notes 14–15).
Preassemble one 3 ml syringe. Remove the plunger completely. The 3 ml syringe will act as a reservoir. Attach the syringe to the 3 way stop cock at the female luer lock opposite from the male luer lock. Turn the swivel knob to the off position toward the syringe (see Note 16).
At the male luer lock of the 3 way stop cock, attach the female luer connector from the 4-inch silicone tubing. Tighten up the connection.
The syringe and 3 way stop cock will be in a vertical position. Use a standard lab stand with three prong clamp extension to hold the syringe upright.
Prefill a second 3 ml syringe with warm culture media (~1.5 ml). On the 3 way stop cock, there is a second female luer lock perpendicular. Insert the syringe with media and slowly inject media to pre-fill the 4-inch silicone tubing, up to the elbow connector end, allowing for a small droplet to form at the orifice.
Proceed with attaching the elbow connector to the slide port (closest to the 3 ml syringe reservoir) (see Note 17). The slide assembly and tubing connection is complete. Leave the syringe in the side luer lock.
Set up the two-photon microscope with parameters appropriate for live image acquisition.
Turn on the syringe pump and set the run parameters to “withdraw” at a shear rate of 1 dyne/cm2 (0.76 ml/min) (see Note 18).
Recheck the whole assembly prior to starting the shear.
At the 3 ml syringe assembly, re-suspend and pipette in the sorted cell suspension (~1 ml of 10,000–50,000 cells) into the 3 ml syringe reservoir.
Quickly turn the swivel knob OFF position toward the perpendicular female luer lock of the 3 way stop cock. The cell suspension is ready for fluidic flow.
Initiate the syringe pump to begin fluid withdrawal, while simultaneously turning on the microscope acquisition.
The cell suspension will begin flowing down the syringe through the 4-inch tubing making its way to the microfluidic channel.
Observe the fluid level at the 3 ml syringe assembly. As it nears to a quarter full (of the original 1 ml volume) quickly pipette in 1 ml of warm culture media. Repeat this two more times (for a total of 3 ml of media added).
At this point, begin adding a final 3 ml of media into the 3 ml syringe reservoir (see Note 19).
Allow the media to continue flowing through the 4-inch tubing, until nearing the elbow connector.
Immediately stop the syringe pump and terminate all imaging acquisition.
Figure 2. Overview of the microfluidic assembly that enables modeling of HSC homing to fetal liver.
a) Schematic diagram of the biomimetic platform that depicts key fluid paths, and b) photograph of the assembled fluid flow system and microfluidic device.
Acknowledgments
This work was supported by the American Society of Hematology Scholar Award and a grant from the National Institutes of Health (R01DK111599) to P.L.W.
4. Notes
What is being designed is the channel area of an IBIDI single channel slide.
Maintain the spray can of adhesive 10–12 inches from the silicone sheet while spraying.
The silicone sheet will contort some due to the adhesive.
Excess Sigmacoat can be reused if it does not turn cloudy.
When mixing the gel, be very gentle to avoid making bubbles.
Depending on the stiffness of the polyacrylamide gel and thickness it may take around 10–20 min to fully polymerize. You can make sure the gel has polymerized by examining the amount remaining in the 15 ml tube.
If not used immediately, the chamber with polyacrylamide gel can be wrapped with parafilm and kept at 4°C for a few days.
To prepare and preserve the crosslinker efficiency of sulfo-SANPAH, dissolve sulfo-SANPAH in sterile DMSO at 50 mg/ml. Aliquot 20 μl of the stock solution into centrifuge tubes and flash freeze in liquid nitrogen or dry ice. Aliquots can be stored at −80°C for several months.
Collagen coated hydrogels can be wrapped in foil and kept in 4°C overnight, if not used on the same day.
It is important to adhere the rostral base at the fetal liver to the chuck in order to anchor this tissue while sectioning slices preventing any form of tissue movement while sectioning.
Tubing lengths might need to be longer based on accessibility of the microscope stage.
These elbow connectors will insert into the slide reservoir ports of the IBIDI slide.
Be sure the slide reservoir ports are near topped off with warm media.
Placement of this elbow connector from the 10 ml syringe will prevent any back flow due to the closed space.
Some small amount of media may spill out. Use a twisted Kimwipe to absorb the spill.
The sorted cell suspension will eventually be aliquoted into the 3 ml syringe reservoir, which is why the swivel knob MUST be at the OFF position toward the syringe.
Some small amounts of media might spill out during connection.
Be sure to have a 50 ml conical of warm IMDM media with a p1000 pipetman and tip.
Final amount of media added is 6 ml.
References
- 1.Bailey C, Richardson LC, Allemani C, et al. (2018) Adult leukemia survival trends in the United States by subtype: A population-based registry study of 370,994 patients diagnosed during 1995–2009. Cancer 124:3856–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamplin OJ, Durand EM, Carr LA, et al. (2015) Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S and Lodish HF (2010) Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc Natl Acad Sci U S A 107:7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ema H and Nakauchi H (2000) Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95:2284–2288 [PubMed] [Google Scholar]
- 5.Ghiaur G, Ferkowicz MJ, Milsom MD, et al. (2008) Rac1 is essential for intraembryonic hematopoiesis and for the initial seeding of fetal liver with definitive hematopoietic progenitor cells. Blood 111:3313–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim I, Yilmaz ÖH, and Morrison SJ (2005) CD144 (VE-cadherin) is transiently expressed by fetal liver hematopoietic stem cells. Blood 106:903–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch E, Iglesias A, Potocnik AJJ, et al. (1996) Impaired migration but not differentiation of haematopoietic stem cells in the absence of beta1 integrins. Nature 380:171–175 [DOI] [PubMed] [Google Scholar]
- 8.Manesia JK, Franch M, Tabas-Madrid D, et al. (2017) Distinct Molecular Signature of Murine Fetal Liver and Adult Hematopoietic Stem Cells Identify Novel Regulators of Hematopoietic Stem Cell Function. Stem Cells Dev 26:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teitell MA and Mikkola HKA (2006) Transcriptional activators, repressors, and epigenetic modifiers controlling hematopoietic stem cell development. Pediatr Res 59:33–39 [DOI] [PubMed] [Google Scholar]
- 10.Sommerkamp P, Renders S, Ladel L, et al. (2019) The long non-coding RNA Meg3 is dispensable for hematopoietic stem cells. Sci Rep 9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayani H (2016) The regulation of hematopoietic stem cell populations. F1000 Res 5:1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan JA, Mendelson A, Kunisaki Y, et al. (2016) Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351:176–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph C, Quach JM, Walkley CR, et al. (2013) Deciphering Hematopoietic Stem Cells in Their Niches: A Critical Appraisal of Genetic Models, Lineage Tracing, and Imaging Strategies. Cell Stem Cell 13:520–533 [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Zhang C, Li J, et al. (2019) The physical microenvironment of hematopoietic stem cells and its emerging roles in engineering applications. Stem Cell Res Ther 10:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamo L, Naveiras O, Wenzel PL, et al. (2009) Biomechanical forces promote embryonic haematopoiesis. Nature 459:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.North TE, Goessling W, Peeters M, et al. (2009) Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell 137:736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz MF, Li N, Lee HJ, et al. (2015) Biomechanical Forces Promote Blood Development Through Prostaglandin E2 and the cAMP-PKA Signaling Axis. J Exp Med 212:665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai HA, Shen CN, and Chang YC (2012) Use of surface properties to control the growth and differentiation of mouse fetal liver stem/progenitor cell colonies. Biomacromolecules 13:3483–3493 [DOI] [PubMed] [Google Scholar]
- 19.Ulrich TA, Juan Pardo EM De, and Kumar S (2009) The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res 69:4167–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha K, Kim J, Irwin E, et al. (2010) Surface creasing instability of soft polyacrylamide cell culture substrates. Biophys J 99:94–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frye M, Taddei A, Dierkes C, et al. (2018) Matrix stiffness controls lymphatic vessel formation through regulation of a GATA2-dependent transcriptional program. Nat Commun 9:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagasaka A, Shinoda T, Kawaue T, et al. (2016) Differences in the mechanical properties of the developing cerebral cortical proliferative zone between mice and ferrets at both the tissue and single-cell levels. Front Cell Dev Biol 4:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevalier NR, Gazquez E, Bidault L, et al. (2016) How Tissue Mechanical Properties Affect Enteric Neural Crest Cell Migration. Sci Rep 6:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwashita M, Kataoka N, Toida K, et al. (2014) Systematic profiling of spatiotemporal tissue and cellular stiffness in the developing brain. Dev 141:3793–3798 [DOI] [PubMed] [Google Scholar]