Abstract
Saccharomyces cerevisiae, Baker’s yeast, is the industrial workhorse for producing ethanol and the subject of substantial metabolic engineering research in both industry and academia. S. cerevisiae has been used to demonstrate production of a wide range of chemical products from glucose. However, in many cases, the demonstrations report titers and yields that fall below thresholds for industrial feasibility. Ethanol synthesis is a central part of S. cerevisiae metabolism and redirecting flux to other products remains a barrier to industrialize strains for producing other molecules. Removing ethanol producing pathways leads to poor fitness, such as impaired growth on glucose. Here, we review metabolic engineering efforts aimed at restoring growth in non-ethanol producing strains with emphasis on relieving glucose repression associated with the Crabtree effect and rewiring metabolism to provide access to critical cellular building blocks. Substantial progress has been made in the past decade, but many opportunities for improvement remain.
Keywords: Yeast, Saccharomyces cerevisiae, Crabtree-Warburg effect, metabolic engineering, pyruvate decarboxylase deficient, glucose, ethanol, acetyl-CoA, adaptive laboratory evolution
The yeast Saccharomyces cerevisiae has long been an important species to humans, from its use in brewing and baking to its role as a model organism for studying eukaryotic biology. Over the past few decades, S. cerevisiae has also gained prominence as a platform for synthesizing chemical products due to several attractive attributes: a well-developed genetic toolkit [1,2]; fast growth (doubling time ~90 min [3]); and tolerance to a variety of industrial stressors [4]. To date, S. cerevisiae has been engineered to produce molecules used in a wide variety of applications including: biofuels, pharmaceuticals, food additives, beauty agents, bulk-chemicals and specialty-chemicals [5,6]. In order for these products to be produced economically, high titers, rates, and yields must be achieved in cultures grown on low-cost feedstocks. This means that metabolic engineering strategies must overpower the dominant native flux to ethanol. In other organisms, such as Escherichia coli, this can be simply accomplished by deleting the genes involved in ethanol biosynthesis [7]. Unfortunately, ethanol production is integrated with many central aspects of S. cerevisiae biology including regulation, substrate uptake, and energy generation. As such, substantial efforts have been undertaken in the metabolic engineering community to break S. cerevisiae’s reliance on and preference for ethanol production. These efforts are the subject of this article.
The significance of ethanol production is clearly seen in the Crabtree-Warburg effect, a central feature of S. cerevisiae metabolism. Under glucose rich conditions, Crabtree-positive yeast ferment sugars into ethanol even under aerobic conditions. The Crabtree effect, thoroughly reviewed elsewhere [8–10], is triggered by overflow metabolism at the pyruvate node (short-term effect) and by glucose-driven repression of respiratory enzymes (long-term effect). The evolutionary driving force behind the Crabtree effect is still debated [10,11], but the leading theory opines that it is an economical approach to maximize growth (biomass generation), while minimizing resource allocation to protein synthesis [12]. The theory is supported by the fact that the catalytic capacity of fermentation is higher than respiration (more ATP is produced per protein mass) [12]. Simulations have also captured the Crabtree effect and overflow metabolism by using an enzyme-constraint based approach to optimize biomass generation in a genome-scale metabolic model [12,13]. While advantageous in the natural world, these effects are not helpful to industrial chemical producing yeasts. Ethanol produced either via fermentation or the Crabtree effect reduces flux to desired chemical products and therefore needs to be by-passed to engineer industrially viable biocatalysts.
Efforts to eliminate ethanol production have proven challenging since ethanol synthesis plays an essential role in redox balance (i.e. reoxidizing NADH produced in glycolysis) and other aspects of S. cerevisiae biology. Non-ethanol producing strains have been created by deleting either the complete set of three pyruvate decarboxylases (PDC1, PDC5, and PDC6) [14] or the set of six alcohol dehydrogenases (ADH1, ADH2, ADH3, ADH4, ADH5, and SFA1) [15], which catalyze the final two catalytic steps in ethanol production respectively (Figure 1). Strains lacking these enzymes produce no ethanol but also experience severe physiological defects [16]. Complete deletion of Pdc activity prevents the production of acetaldehyde which in addition to being the substrate for ethanol production is also the primary precursor of cytosolic acetyl-CoA. Without supplementation of ethanol, acetate, or heterologous pathways discussed below, cells cannot produce sufficient acetyl-CoA flux to support fatty acid biosynthesis and other biosynthetic requirements. When supplemented, strains still grow slowly in part because cells are unable to rapidly replenish the NAD+ supply required for glucose catabolism. In the presence of high glucose concentrations, this redox imbalance is exacerbated by native regulation of key catabolic components. In summary, rerouting carbon flux away from ethanol requires an alternate, balanced, high-flux NADH-oxidizing pathway, an alternative cytosolic acetyl-CoA generation pathway, and fine-tuning of internal metabolic regulation.
Figure 1.
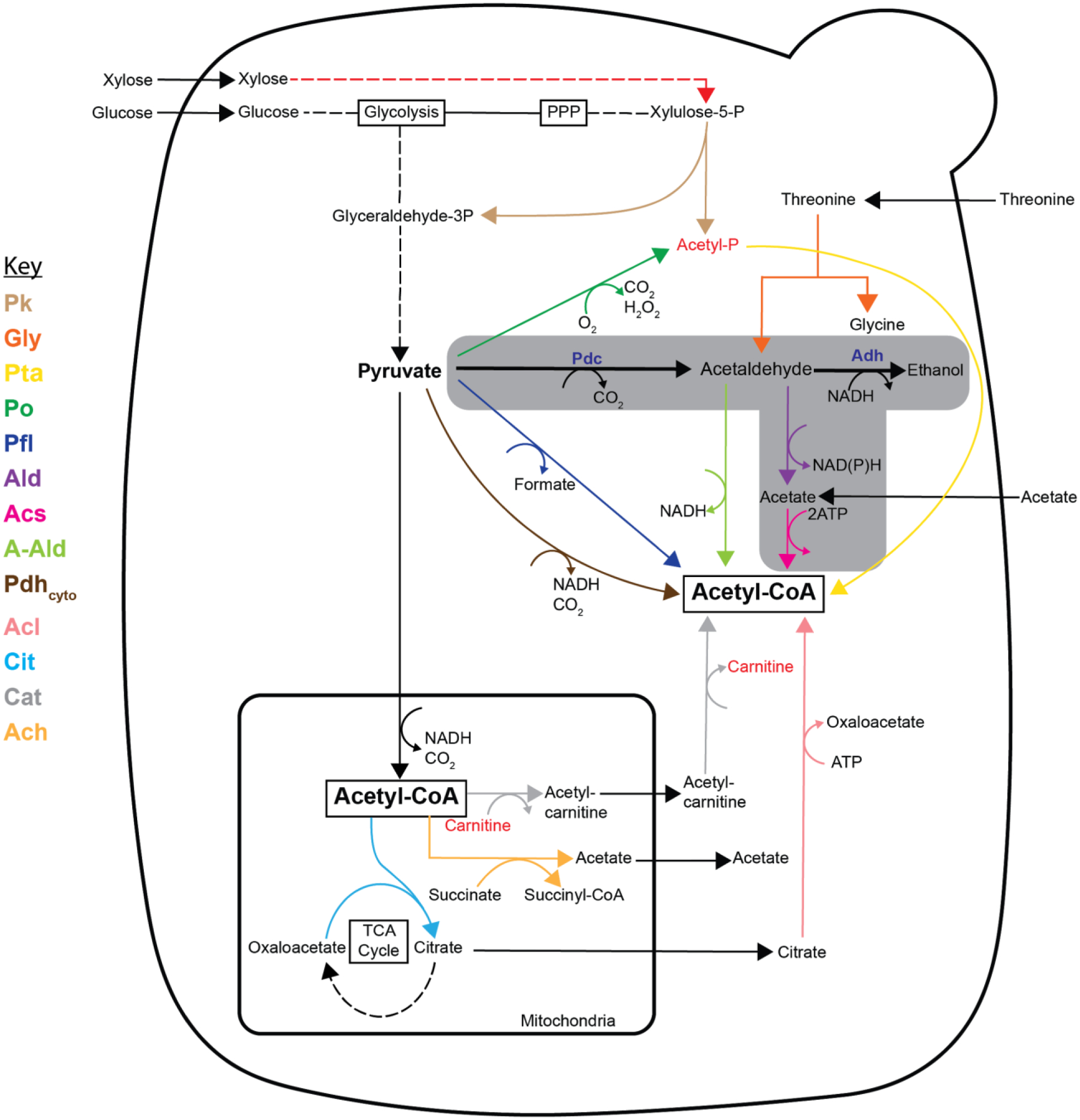
Metabolic map of native and heterologous acetyl-CoA biosynthetic pathways in S. cerevisiae. Naturally, cytosolic acetyl-CoA is derived from the pyruvate dehydrogenase (Pdh) bypass consisting of pyruvate decarboxylase (Pdc), acetaldehyde dehydrogenase (Ald, purple), and acetyl-CoA synthetase (Acs, pink); ethanol is also derived from an intermediate in this bypass (grayed box). Other native metabolic pathways include threonine aldolase (Gly, dark orange) and the mitochondrial shuttle system consisting of a CoA-transferase (Ach1, light orange). Heterologous tested pathways for the synthesis of cytosolic acetyl-CoA include the following: phosphoketolase (Pk, tan), phosphotransacetylase (Pta, yellow), pyruvate oxidase (Po, dark green), pyruvate-formate lyase (Pfl, dark blue), acetylating aldehyde dehydrogenase (A-Ald, light green), pyruvate dehydrogenase (Pdhcyto, brown), citrate lyase (Acl, light pink), citrate synthase (Cit, light blue), and carnitine acetyl-CoA transferase (Cat, gray). Non-native metabolites (acetyl-P and carnitine) and pathway (xylose catabolism) are indicated in red.
This review highlights the recent advances towards abolishing the Crabtree effect in S. cerevisiae and redirecting flux to non-ethanol products. We summarize strategies to restore redox balance through respiration and alternative pyruvate-derived products as well as strategies to regenerate cytosolic acetyl-CoA in Pdc− strains. In addition, we tabulated recent examples of how these strategies have been successfully implemented in chemical producing strains. This review serves as a guide for understanding the facets of S. cerevisiae’s ethanol-dependencies, the strategies that have been publicly disclosed for circumventing these obstacles, and the remaining challenges to using S. cerevisiae for producing chemical products beyond ethanol.
1. Cytosolic acetyl-CoA generation in Pdc− S. cerevisiae
Pdc− strains are auxotrophic for C2-compounds due to their inability to produce cytosolic acetyl-CoA with the native pyruvate dehydrogenase (Pdh) bypass consisting of pyruvate decarboxylase (Pdc), acetaldehyde dehydrogenase (Ald), and acetyl-CoA synthetase (Acs) (Figure 1). Acetyl-CoA supply is further complicated by the fact that in eukaryotes it cannot transverse the membranes of subcellular compartments such that mitochondria-made acetyl-CoA cannot be directly exported to the cytosol. Some reports propose that evolved Pdc− strains circumvent this barrier through use of a CoA-transferase, Ach1p, that converts acetyl-CoA into acetate which can cross into the cytosol and subsequently be reactivated as acetyl-CoA at the cost of ATP [17]. Another report suggests that the native shuttle system is not sufficient [18]. Alternatively, the C2-auxotrophy can be circumvented by providing a heterologous synthesis pathway or by supplementation with a C2-compound (ethanol or acetate). Alternative acetyl-CoA producing pathways include: pyruvate-formate lyase (Pfl), acetylating acetaldehyde dehydrogenase (A-Ald), cytosolic-pyruvate dehydrogenase (Pdhcyto), pyruvate oxidase (Po)/phosphotransacetylase (Pta), phosphoketolase (Pk)/phosphotransacetylase (Pta), threonine aldolase (Gly), carnitine shuttle (Cat), and citrate-oxaloacetate shuttle (Cit/Acl) (Figure 1) [19,20]. The genetics and biochemistry of these pathways is thoroughly reviewed in reference [20]. While many alternative pathways exist, heterologous expression by itself is not sufficient to restore growth in a Pdc− strain to wild-type levels. As discussed below, additional modifications are necessary, but overcoming acetyl-CoA auxotrophy is a critical first step.
2. Balancing NAD+ regeneration with glycolysis in the absence of ethanol synthesis
It is widely known that ethanol is produced during yeast fermentation in order to regenerate the NAD+ needed to enable glycolysis. Analogously, when oxygen is available, non-ethanol producing yeast grown in glucose-limited conditions regenerate NAD+ through oxidative phosphorylation, which can support a low glycolytic flux. However, at high glucose concentrations, genes involved in oxidative phosphorylation are downregulated while genes involved in glucose transport and glycolysis are upregulated [21]. This phenomenon is controlled by the three glucose sensing systems (Rgt2p/Snf3p, Snf1p/Mig1p, and cAMP/PKA; see Figure 3 of reference [22] for a detailed depiction) and is associated with the Crabtree effect [9,22]. Elevated rates of glucose import ultimately lead to an increased glycolytic flux above the catalytic capacity of the respiratory chain, resulting in insufficient NAD+ regeneration and a bottleneck in the catabolism of glucose [8]. The bottleneck is caused in part by the indirect repression of the mitochondrial pyruvate dehydrogenase (Pdh) by Mig1p [23]. Thus, under excess glucose conditions, non-ethanol producing strains do not grow due to the NADH/NAD+ redox imbalance caused by a metabolic bottleneck at the pyruvate node. Strategies to overcome the imbalance include relieving glucose repression caused by the Crabtree-effect, restricting glucose uptake, downregulating glycolytic flux, providing alternative redox sinks, and throttling the delivery of glucose to cells with fed-batch bioreactors [24].
2.1. Adaptive Laboratory Evolution
Adaptive laboratory evolution (ALE) has been widely used to enhance growth of Pdc− strains on glucose since the early 2000s. Recent efforts have tried to understand the role and optimize the function of ALE-created mutations in regulating glucose catabolism, ethanol production, and the Crabtree effect. Through multiple studies, researchers identified mutations or deletions in MTH1, a transcriptional repressor in the Rgt2p/Snf3p glucose-sensing cascade involved in hexose transporter (Hxt) expression [16,18,25,26]. These mutations reduce expression of the Hxts, thereby limiting glucose uptake and preventing the respiratory chain from being overloaded. Cells carrying a mth1 mutation can grow on glucose because glucose-repression is alleviated, however, growth comes at a reduced rate (less than 30% of Pdc+ strains), which is insufficient for industrial purposes. In an attempt to overcome the growth rate limitation, researchers performed extra rounds of ALE starting with strains carrying the mth1 mutation and isolated a strain with a higher growth rate when grown on 2% glucose. The resulting strain had a mutation in YAK1 and a growth rate that increased 2.5 fold over the base strain, albeit at rates still below the Pdc+ parent [27]. YAK1 encodes a serine/threonine protein kinase involved in regulating cell growth in the presence of glucose. In a separate study, downregulating PYK1, a.k.a. CDC19, which encodes the major pyruvate kinase, was also shown to relieve glucose repression in the context of another Pdc− strain [28]. Pyk1p is considered a control node in glycolysis and its activity is tightly regulated by fructose-1,6-biphosphate (FBP) concentrations. Engineering the allosteric sites on Pyk1p could be an alternative strategy to the regulatory approaches discovered in the evolutionary studies. While ALE has provided the initial insights into the importance of the regulatory network in allowing Pdc− strains to grow on glucose, the problem is not yet solved. Evolved mutants often have undesirable characteristics, such as lower than wild-type growth, that must be overcome before these strains can become useful platforms for bioproduction.
2.2. Rational re-wiring of native sugar metabolism regulation
Increasing the catalytic capacity of the respiratory chain by rational engineering is also a valid yet underdeveloped strategy for enabling Pdc− strains to grow on glucose. The signaling and regulation involved in the Crabtree effect have been heavily studied, and recent work has shown that the Snf1p/Mig1p glucose repression pathway [29] as well as the ratio of glucose-6-phosphate (G6P) to FBP [30] is directly linked to the Crabtree phenotype. The Snf1p signaling pathway negatively regulates genes associated with the uptake and catabolism of non-glucose sugars and expression of gluconeogenesis, respiratory, and Hxt genes by direct interaction with many transcription factors such as Mig1p, Rgt1, Msn2, and Cat8 [9,23]. Deletion of SNF1, HXK2, or MIG1 increases respiratory capacity and reduces overflow metabolism, suggesting that these regulators are promising targets for shifting production from ethanol to other compounds [29,31]. This strategy is supported by increased transcript levels of Mig1p-mediated repressed genes in Pdc− strains that have undergone ALE [26]. Tps1p is also an important glycolytic regulator that has been recently linked to Snf1p signaling and may provide another engineering target [32]. The intentional rewiring of regulation is a promising strategy for increasing the growth rates of Pdc− strains by increasing the respiratory capacity rather than limiting sugar metabolism. Furthermore, it could allow for intentional tuning of metabolism while minimizing undesirable effects. Alternatively, non-Crabtree yeasts such as Kluyveromyces. lactis, which natively contain the desired regulatory structure, could be engineered to produce desired products.
2.3. NADH-oxidation coupled to pyruvate-derived metabolites
While strategies that reduce glucose import lead to improved growth, the glucose utilization rate is an important parameter in industrial bioprocessing that should be as large as possible. Alternatively, the NADH/NAD+ redox balance in Pdc− strains can be maintained by producing products that require the same amount of reducing equivalents as ethanol. Pyruvate is a common metabolic node from which these products are derived (Figure 2). Products such as 2,3-butanediol [25,27,33–36] and lactate [37–41] can directly replace ethanol fermentation and each product has been produced in substantial quantities in Pdc− strains. Malate production is also redox-balanced with glycolysis, but requires ATP [42]. Other desirable products such as isobutanol and free fatty acids require NADPH reducing equivalents, necessitating a means for converting glycolytic NADH into NADPH [28,43]. This can be overcome by introducing a transhydrogenase reaction (NADPH + NAD+ ⇌ NADP+ + NADH) into S. cerevisiae [28,44,45]. Heterologous expression of an E. coli transhydrogenase in Pdc− S. cerevisiae has been shown to increase growth and pyruvate production [46]. Similarly, relocalization of S. cerevisiae malic enzyme to the cytosol creates a transhydrogenase cycle that has been shown to improve isobutanol production [47]. Alternatively, diverting carbon from glycolysis into the pentose phosphate pathway makes more NADPH available and can increase both free fatty acid [28] and isobutanol [48] production. Engineering NADPH-dependent enzymes to instead utilize NADH is another approach and has been demonstrated in isobutanol production, though with limited effect on production in S. cerevisiae [43,49].
Figure 2.
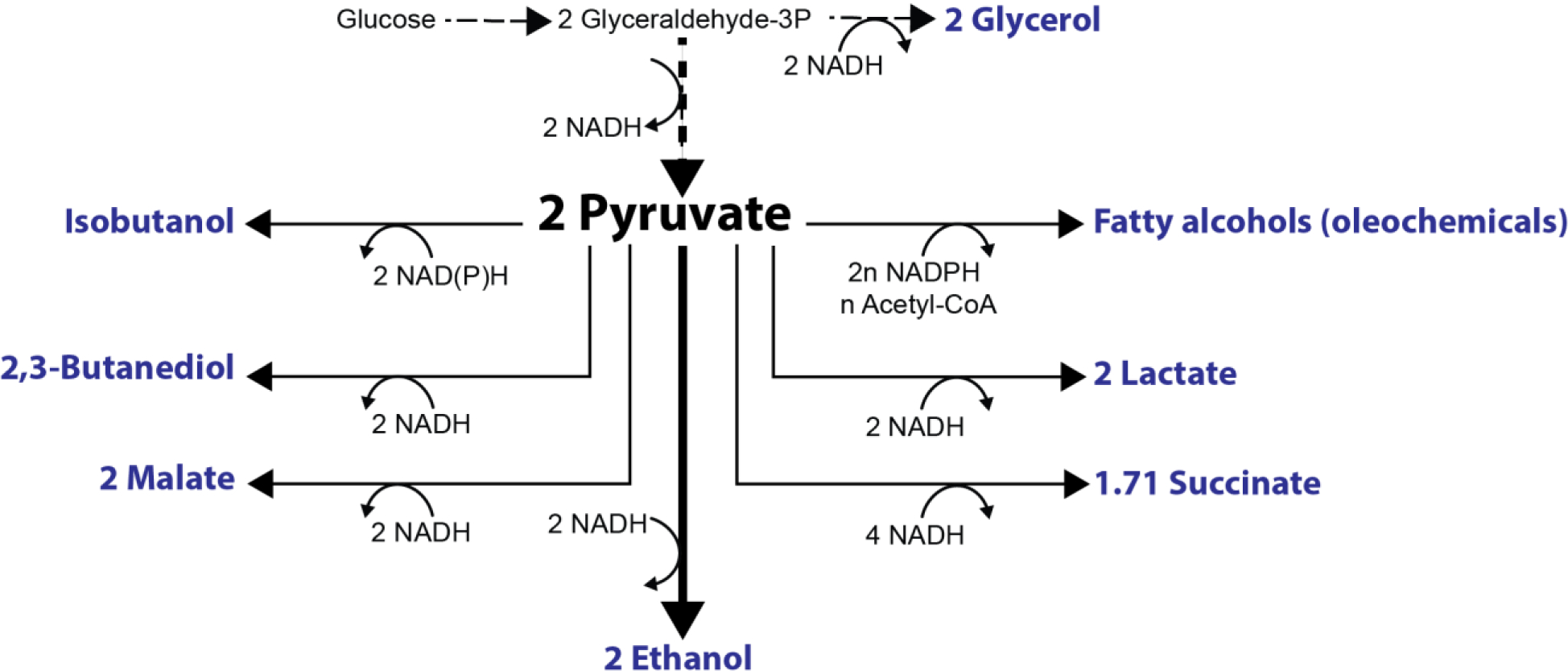
Schematic depicting pyruvate-derived products. Glycerol, malate, lactate, and succinate [74] production require NADH reducing equivalents and can theoretically replace ethanol fermentation while isobutanol and fatty alcohols/other oleochemicals require NAD(P)H reducing equivalents.
2.4. Regenerating NAD+ for anabolism
Though replacing ethanol with other products can balance glycolytic NADH generation, S. cerevisiae anabolism also produces NADH, which is normally balanced by glycerol production [50]. Glycerol production in S. cerevisiae can divert up to 8.5% of carbon, making its removal via GPD1 and GPD2 deletion a common metabolic engineering strategy, even when ethanol is the desired product [51]. However, glycerol is important in S. cerevisiae osmotic tolerance so completely blocking its synthesis is often undesirable [52]. Glycerol formation can be reduced by providing other routes to oxidize NADH, such as through succinate production, which requires two reducing equivalents per pyruvate. Succinate has been produced in Pdc− S. cerevisiae in significant amounts, but only under aerobic conditions [53,54]. Other NADH oxidizing routes include heterologous expression of a water-forming NADH oxidase [33,36] or an alternative oxidase [8]. These enzymes require aerobic conditions to function, and thus, it is common to see them used under semi-aerobic conditions with a fermentation product. Carbon fixation is also a common sink of excess reducing power in photoautotrophs. When Calvin-cycle enzymes RuBisCo and phosphoribulokinase were heterologously expressed in S. cerevisiae, it coupled NADH oxidation with CO2 reduction and significantly decreased glycerol production [55]. Acetate, which is present in high amounts in lignocellulosic hydrolysates, can also be fermented into ethanol to oxidize residual NADH via Ald and Adh and reduce glycerol formation (Figure 2) [56]. In many cases, it may be possible to couple NADH recycling with production of a desired product. In others, feedstocks may need to be optimized such that redox balance, ATP generation, and product synthesis are optimized.
3. Metabolic rewiring to reduce ethanol production without its complete elimination
As discussed above, eliminating ethanol production in yeast is difficult, especially if the adverse impacts on physiology limit desired applications. For this reason, researchers have also explored reducing, but not eliminating, ethanol production. Successful strategies include controlling glycolytic flux with a metabolic circuit, utilizing a reduced activity Pdc from a Crabtree-negative yeast [35,36], or co-catabolizing non-preferred carbon sources, such as xylose or glycerol.
3.1. Redirecting flux using metabolic circuits
Controlling when and where carbon flux is directed in a fermentation process can be achieved with dynamic control strategies, including inducible-metabolic valves [57–59]. In practice, the fermentation occurs in alternating decoupled phases (growth and production). During the growth phase, biomass generation is promoted via NAD+ regeneration through ethanol fermentation as flux is directed through glycolysis. During the production phase, ethanol flux is restricted and resources are directed towards a product of interest. This switch is achieved by turning on transcription of pathway genes using either a chemical inducer or light via an optogenetics switch. The balance between growth and product synthesis has been successfully demonstrated with isobutanol production [60,61]. Expansion of this idea will depend on finding ways to minimize the amount of time cells spend in the biomass generation phase such that maximum titers, rates, and yields of products can be achieved.
3.2. Non-preferred carbon sources
Many of the challenges to removing ethanol production depend on glucose being present. Using alternative carbon sources, such as xylose or glycerol that do not induce the Crabtree effect, is another way to decrease ethanol production [62–64]. S. cerevisiae cannot naturally assimilate xylose, but xylose catabolism has been extensively studied and introduced into yeast by many groups [65,66]. In one case, isobutanol producing strains generated 2-fold less ethanol and 6-fold more isobutanol when grown on xylose instead of glucose [63]. Improved performance is not restricted to xylose. In another case, a strain produced ~1.5-fold more 1,2-propanediol than ethanol when grown on glycerol [67]. Recently, there has also been interest in the generation of xylose-utilizing Pdc− strains for generating pyruvate-derived products, but the strains suffer from low productivities due to slow-growth [68–70]. While catabolism of xylose and glycerol has proved beneficial in decreasing ethanol production in defined media on a lab scale, the ultimate application will be growth of Pdc- strains on lignocellulosic hydrolysates containing mixtures of sugars, organic acids, and inhibitors.
4. Recent engineering strategies to increase production of non-ethanol products
The metabolic engineering community has leveraged many of the strategies described above to improve the production of non-ethanol products. In Table 1, we summarize recent studies by tabulating the approaches used and commonly reported performance metrics for several key products. Additional insight can be obtained in the highlighted references.
Table 1.
Summary of strategies for non-ethanol chemical production in S. cerevisiae
| Reference | Pdc− phenotype | Relieve Crabtree effect through ALE | Alternate acetyl-CoA routes | NAD+ regeneration | Metabolic control of ethanol production | Alternate carbon source | Product(s) | Yield (g/g carbon source) | Titer (g/L) | Conditions (carbon source, medium, batch/fed-batch, vessel, conditions) |
|---|---|---|---|---|---|---|---|---|---|---|
| [71] | ✓ | ✓ | ✓ | Acetate | 0.14a | < 0.5 | Glucose, minimal medium, batch, flask, aerobic | |||
| Pyruvate | 0.007a | < 0.5 | ||||||||
| [25] | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.28 | 96.2 | Glucose, YP medium, fed-batch, bioreactor, aerobic | |||
| Glycerol | 0.09a | >30c | ||||||||
| Acetoin | 0.01a | ~5c | ||||||||
| [33] | ✓ | ✓ | 2,3-Butanediol | 0.407 | 72.91 | Glucose, YP medium, fed-batch, flask, aerobic | ||||
| Acetoin | 0.01a | 1.38 | ||||||||
| [34] | ✓ | ✓ | 2,3-Butanediol | 0.359 | 32.31c | Glucose + ethanol, minimal media, batch, bioreactor, oxygen-limited | ||||
| Glycerol | 0.069 | 6.21c | ||||||||
| Acetoin | 0.052 | 4.68c | ||||||||
| [35] | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.404 | 154.3 | Glucose, YP medium, fed-batch, bioreactor, full aeration followed by oxygen limitation | |||
| Glycerol | 0.088 | 33.5c | ||||||||
| Acetate | 0.006a | 2.3 | ||||||||
| [36] | ✓ | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.462 | 108.6 | Glucose, YP medium, fed-batch, bioreactor, full aeration followed by oxygen limitation | ||
| Acetoin | 0.02a | 4.6 | ||||||||
| Acetate | 0.0004a | 0.1 | ||||||||
| [27] | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.27 | 81.0 | Glucose, synthetic defined medium, fed-batch, flask, mild aerobic | |||
| Glycerol | 0.239a | 71.8 | ||||||||
| Succinate | 0.013a | 4.0 | ||||||||
| [72] | ✓ | ✓ | n-Butanol | 0.012 | 0.130 | Glucose + pantothenate, SMD medium, flask, semi-anaerobic | ||||
| Ethanol | 0.10 | ~1c | ||||||||
| Glycerol | 0.17 | ~2c | ||||||||
| [43] | ✓ | ✓ | ✓ | Isobutanol | 0.0074a | b | Glucose, SMG medium + Tween-80 + ergosterol, batch, serum bottle, microaerobic | |||
| Ethanol | 0.033a | b | ||||||||
| Glycerol | 0.26a | b | ||||||||
| Pyruvate | 0.02a | b | ||||||||
| 2,3-butanediol | 0.325a | b | ||||||||
| Dihydroxyisovalerate | 0.05a | b | ||||||||
| Isobutyrate | 0.03a | b | ||||||||
| [61] | ✓ | ✓ | Isobutanol | 0.0535 | 8.49 | Glucose, SC-Ura, fed-batch, bioreactor, microaerobic | ||||
| 2-Methyl-1-butanol | 0.01417 | 2.38 | ||||||||
| Ethanol | 0.187 | 39.8 | ||||||||
| Glycerol | b | b | ||||||||
| [28] | ✓ | ✓ | ✓ | ✓ | Free fatty acids | 0.1 | 25 | Glucose, minimal N₂-limited medium, fed-batch, bioreactor, aerobic | ||
| Glycerol | b | b | ||||||||
| [42] | ✓ | ✓ | ✓ | Malic acid | 0.31a | 59 | Glucose, synthetic medium, batch, flask, aerobic | |||
| Succinate | 0.04a | 8 | ||||||||
| Glycerol | 0.13a | 25 | ||||||||
| Pyruvate | 0.02a | 3 | ||||||||
| Fumarate | 0.01a | 2 | ||||||||
| [54] | ✓ | ✓ | ✓ | Succinic acid | 0.044a | 2.2 | Glucose + formate, mineral salt medium, deep-well plate, microaerobic | |||
| Pyruvate | 0.364a | 18.2 | ||||||||
| Malate | 0.022a | 1.1 | ||||||||
| Glycerol | 0.076a | 3.8 | ||||||||
| [60] | ✓ | ✓ | Gluconate | b | 2.31 | Glucose, synthetic defined, batch, culture tube, semi-anaerobic | ||||
| Ethanol | b | 1.01 | ||||||||
| Glycerol | b | b | ||||||||
| [41] | ✓ | ✓ | Lactic acid | 0.6 | 80 | Glucose, YP medium, fed-batch, bioreactor, aerobic | ||||
| Ethanol | 0.01 | 1.6 | ||||||||
| Glycerol | 0.02 | 2.6 | ||||||||
| [40] | ✓ | Lactic acid | 0.8 | 112.0 | Glucose, YP medium, fed-batch, flask, aerobic | |||||
| Ethanol | 0.02a | 2.6 | ||||||||
| [69] | ✓ | ✓ | ✓ | Lactic acid | 0.43a | 7.8 | 2-stage sugar consumption (ethanol then xylose), YP medium, batch, bioreactor, aerobic followed by microaerophilic | |||
| Acetate | 0.16a | 3c | ||||||||
| Xylitol | 0.01a | 0.17c | ||||||||
| [70] | ✓ | ✓ | Lactic acid | 0.67 | 60 | Xylose, YP medium, batch, flask, aerobic | ||||
| Ethanol | 0.01a | <1 | ||||||||
| Xylitol | 0.01a | <1 | ||||||||
| Glycerol | 0.01a | <1 | ||||||||
| Acetate | 0.01a | <1 | ||||||||
| [68] | ✓ | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.46a | 96.8 | Xylose + glucose, YP medium, fed-batch, bioreactor, microaerobic | ||
| Glycerol | 0.11a | 23.3 | ||||||||
| Ethanol | 0.08a | 16.3 | ||||||||
| Xylitol | 0.03a | 5.9 | ||||||||
| [73] | ✓ | ✓ | ✓ | 2,3-Butanediol | 0.26a | 43.6 | Xylose + glucose, YP medium, fed-batch, bioreactor, microaerobic | |||
| Glycerol | 0.26a | 45 | ||||||||
| Xylitol | 0.03a | 4.7 | ||||||||
| [63] | ✓ | ✓ | Isobutanol | 0.026a | 2.6 | Xylose, nutrient-rich Verduyn medium, fed-batch, bioreactor, aerobic | ||||
| Ethanol | 0.275a | ~27.5c | ||||||||
| Glycerol | 0.10a | ~10 g/Lc | ||||||||
| [62] | ✓ | ✓ | Isobutanol | 0.0196 | 3.10 | Xylose, synthetic medium, fed-batch, conical tube, semi-anaerobic | ||||
| 2-Methyl-1-butanol | 0.0053 | 0.79 | ||||||||
| Ethanol | 0.32a | <50c | ||||||||
| Glycerol | b | b | ||||||||
| [67] | ✓ | ✓ | 1,2-Propanediol | 0.129 | 4.3 | Glycerol, YP medium, batch, bioreactor, aerobic | ||||
| Ethanol | ~0a | ~0c |
Bold = product of interest
Italics = by-product
Absence of ethanol or glycerol in a row indicates it was not detected or that the yield or titer was <0.01
Yield not directly reported. Yield is either read from a graph, converted from a molar yield, calculated as g/g consumed carbon source, or calculated by dividing production rate by carbon source uptake rate.
Not reported
Titer not directly reported. Titer is either read from a graph, converted from molar concentration, or calculated as g/L fed carbon source, which is quantified by multiplying the reported yield (g/g carbon source) by the carbon source concentration (g/L).
5. Conclusion/perspectives
Redirecting flux away from ethanol production in yeast remains a challenging endeavor. Recent work shows that enhancing carbon flux through non-ethanol pathways can be achieved by several metabolic engineering strategies: 1.) relieving the Crabtree effect through regulatory modifications, 2.) regenerating NAD+ by producing alternative reduced products, and 3.) providing routes for synthesizing cytosolic acetyl-CoA. In many cases, it has been far easier to throttle down ethanol production, rather than to completely eliminate it. Production of alternative chemical products in yeast has provided the scientific and economic motivation to deepen our understanding of the Crabtree effect and identify strategies for circumventing it. Deleting ethanol synthesis remains hard, but a promising strategy is emerging. This approach starts by providing alternative pathways for synthesis of acetyl-CoA, oxidation of NADH, and circumventing the Crabtree effect. Next, eliminate ethanol production by deleting all three PDC genes or control ethanol production with advanced genetic circuits. Lastly, the selective pressure to grow, particularly under fermentation conditions, provides a driving force for applying ALE to optimize strain performance. These steps have likely been applied in industry to optimize production of several commercially available products (e.g. lactate, isobutanol, …), but the secrets of the evolved strains remain to be elucidated by academic researchers. Herein, we have described several substantial advances in the academic literature. Each has room for further improvement. The pursuit of these improvements will hopefully deepen our understanding of regulation (particularly of central metabolism and substrate uptake), establish new tools for manipulating metabolism, and establish publicly available strains that can be rapidly engineered to produce any of the remaining chemical products desired by society.
Acknowledgements:
This material is based upon work supported in part by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018409 and in part by the Center for Bioenergy and Bioproducts Innovation, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018420). Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the U.S. Department of Energy
Bibliography
- 1.Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnol Adv 2018, 36:1870–1881. [DOI] [PubMed] [Google Scholar]
- 2.Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications. Metab Eng 2018, 50:85–108. [DOI] [PubMed] [Google Scholar]
- 3.Salari R, Salari R: Investigation of the best Saccharomyces cerevisiae growth condition. Electronic physician 2017, 9:3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIlwain SJ, Peris D, Sardi M, Moskvin OV, Zhan F, Myers KS, Riley NM, Buzzell A, Parreiras LS, Ong IM, et al. : Genome Sequence and Analysis of a Stress-Tolerant, Wild-Derived Strain of Saccharomyces cerevisiae Used in Biofuels Research. G3: Genes, Genomes, Genetics 2016, 6:1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jullesson D, David F, Pfleger B, Nielsen J: Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv 2015, 33:1395–1402. [DOI] [PubMed] [Google Scholar]
- 6.Hong K-K, Nielsen J: Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci 2012, 69:2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehrer CR, Incha MR, Politz MC, Pfleger BF: Anaerobic Production of Medium-Chain Fatty Alcohols via a β-Reduction Pathway. Metab Eng 2018, 48:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J: Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2007, 104:2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kayikci Ö, Nielsen J: Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res 2015, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Alteriis E, Cartenì F, Parascandola P, Serpa J, Mazzoleni S: Revisiting the Crabtree/Warburg effect in a dynamic perspective: a fitness advantage against sugar-induced cell death. Cell Cycle 2018, 17:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Review discussing and interpreting the proposed mechanisms and theories behind the Crabtree-Warburg effect. Detailing the short/long term effect and sugar-induced cell death.
- 11.Pfeiffer T, Morley A: An evolutionary perspective on the Crabtree effect. Front Mol Biosci 2014, 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson A, Nielsen J: Metabolic Trade-offs in Yeast are Caused by F1F0-ATP synthase. Sci Rep 2016, 6:22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez BJ, Zhang C, Nilsson A, Lahtvee P-J, Kerkhoven EJ, Nielsen J: Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol Syst Biol 2017, 13:935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flikweert MT, Van Der Zanden L, Janssen WM, Steensma HY, Van Dijken JP, Pronk JT: Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 1996, 12:247–257. [DOI] [PubMed] [Google Scholar]
- 15.Ida Y, Furusawa C, Hirasawa T, Shimizu H: Stable disruption of ethanol production by deletion of the genes encoding alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. J Biosci Bioeng 2012, 113:192–195. [DOI] [PubMed] [Google Scholar]
- 16.Oud B, Flores C-L, Gancedo C, Zhang X, Trueheart J, Daran J-M, Pronk JT, van Maris AJA: An internal deletion in MTH1 enables growth on glucose of pyruvate-decarboxylase negative, non-fermentative Saccharomyces cerevisiae. Microb Cell Fact 2012, 11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rossum HM, Kozak BU, Niemeijer MS, Duine HJ, Luttik MAH, Boer VM, Kötter P, Daran J-MG, van Maris AJA, Pronk JT: Alternative reactions at the interface of glycolysis and citric acid cycle in Saccharomyces cerevisiae. FEMS Yeast Res 2016, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Liu G, Engqvist MKM, Krivoruchko A, Hallström BM, Chen Y, Siewers V, Nielsen J: Adaptive mutations in sugar metabolism restore growth on glucose in a pyruvate decarboxylase negative yeast strain. Microb Cell Fact 2015, 14:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Fan J, Wang C, Li C, Zhou X: Enhanced β-Amyrin Synthesis in Saccharomyces cerevisiae by Coupling An Optimal Acetyl-CoA Supply Pathway. J Agric Food Chem 2019, 67:3723–3732. [DOI] [PubMed] [Google Scholar]; *Authors generated a strain with a high flux towards cytosolic acetyl-CoA for the production of β-amyrin at high titers (279.0 +/− 13.0 mg/L) in a fed-batch system. This was achieved by combining multiple heterologous acetyl-CoA supply routes and eliminating competing pathways in a β-amyrin producing strain.
- 20.van Rossum HM, Kozak BU, Pronk JT, van Maris AJA: Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab Eng 2016, 36:99–115. [DOI] [PubMed] [Google Scholar]
- 21.Gancedo JM: The early steps of glucose signalling in yeast. FEMS Microbiol Rev 2008, 32:673–704. [DOI] [PubMed] [Google Scholar]
- 22.Kim J-H, Roy A, Jouandot D 2nd, Cho KH: The glucose signaling network in yeast. Biochim Biophys Acta 2013, 1830:5204–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein CJ, Olsson L, Nielsen J: Glucose control in Saccharomyces cerevisiae: the role of Mig1 in metabolic functions. Microbiology 1998, 144 ( Pt 1):13–24. [DOI] [PubMed] [Google Scholar]
- 24.Habegger L, Rodrigues Crespo K, Dabros M: Preventing Overflow Metabolism in Crabtree-Positive Microorganisms through On-Line Monitoring and Control of Fed-Batch Fermentations. Fermentation 2018, 4:79. [Google Scholar]
- 25.Kim S-J, Seo S-O, Jin Y-S, Seo J-H: Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresour Technol 2013, 146:274–281. [DOI] [PubMed] [Google Scholar]
- 26.Van Maris AJA, Geertman J-MA, Vermeulen A, Groothuizen MK, Winkler AA, Piper MDW, Van Dijken JP, Pronk JT: Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microbiol 2004, 70:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii J, Morita K, Ida K, Kato H, Kinoshita S, Hataya S, Shimizu H, Kondo A, Matsuda F: A pyruvate carbon flux tugging strategy for increasing 2,3-butanediol production and reducing ethanol subgeneration in the yeast. Biotechnol Biofuels 2018, 11:180. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Demonstrated that NAD+ regeneration through 2,3-butanediol production can replace ethanol fermentation in an evolved Pdc- S. cerevisiae strain. The best strain in the study produced 81 g/L of 2,3-butanediol by pulling pyruvate flux to 2-acetolactate, the first metabolite in the 2,3-butanediol pathway, by heterologously expressing an acetolactate synthase variant from Lactobacillus plantarum and co-expressing an endogenous butanediol dehydrogenase.
- 28.Yu T, Zhou YJ, Huang M, Liu Q, Pereira R, David F, Nielsen J: Reprogramming Yeast Metabolism from Alcoholic Fermentation to Lipogenesis. Cell 2018, 174:1549–1558.e14. [DOI] [PubMed] [Google Scholar]; **Yu and co-workers rewired yeast metabolism to enhance carbon flux to free fatty acids instead of ethanol by rational metabolic engineering and ALE in a design-build-evolve-test cycle. Free fatty acids were produced at high titers of 25.0 g/L in fed-batch fermentation by enhancing carbon flux to cytosolic acetyl-CoA, increasing the NADPH supply, balancing ATP levels, eliminating ethanol fermentation, and relieving the Crabtree-effect.
- 29.Martinez-Ortiz C, Carrillo-Garmendia A, Correa-Romero BF, Canizal-García M, González-Hernández JC, Regalado-Gonzalez C, Olivares-Marin IK, Madrigal-Perez LA: SNF1 controls the glycolytic flux and mitochondrial respiration. Yeast 2019, 36:487–494. [DOI] [PubMed] [Google Scholar]
- 30.Rosas Lemus M, Roussarie E, Hammad N, Mougeolle A, Ransac S, Issa R, Mazat J-P, Uribe-Carvajal S, Rigoulet M, Devin A: The role of glycolysis-derived hexose phosphates in the induction of the Crabtree effect. J Biol Chem 2018, 293:12843–12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek S-H, Kwon EY, Kim S-Y, Hahn J-S: GSF2 deletion increases lactic acid production by alleviating glucose repression in Saccharomyces cerevisiae. Sci Rep 2016, 6:34812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vicente RL, Spina L, Gómez JPL, Dejean S, Parrou J-L, François JM: Trehalose-6-phosphate promotes fermentation and glucose repression in Saccharomyces cerevisiae. Microb Cell Fact 2018, 5:444–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S, Hahn J-S: Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 2015, 31:94–101. [DOI] [PubMed] [Google Scholar]
- 34.Kim J-W, Seo S-O, Zhang G-C, Jin Y-S, Seo J-H: Expression of Lactococcus lactis NADH oxidase increases 2,3-butanediol production in Pdc-deficient Saccharomyces cerevisiae. Bioresour Technol 2015, 191:512–519. [DOI] [PubMed] [Google Scholar]
- 35.Kim J-W, Kim J, Seo S-O, Kim KH, Jin Y-S, Seo J-H: Enhanced production of 2,3-butanediol by engineered Saccharomyces cerevisiae through fine-tuning of pyruvate decarboxylase and NADH oxidase activities. Biotechnol Biofuels 2016, 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J-W, Lee Y-G, Kim S-J, Jin Y-S, Seo J-H: Deletion of glycerol-3-phosphate dehydrogenase genes improved 2,3-butanediol production by reducing glycerol production in pyruvate decarboxylase-deficient Saccharomyces cerevisiae. J Biotechnol 2019, 304:31–37. [DOI] [PubMed] [Google Scholar]
- 37.Ida Y, Hirasawa T, Furusawa C, Shimizu H: Utilization of Saccharomyces cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for anaerobic bioproduction. Appl Microbiol Biotechnol 2013, 97:4811–4819. [DOI] [PubMed] [Google Scholar]
- 38.Hirasawa T, Ida Y, Furuasawa C, Shimizu H: Potential of a Saccharomyces cerevisiae recombinant strain lacking ethanol and glycerol biosynthesis pathways in efficient anaerobic bioproduction. Bioengineered 2014, 5:123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Maris AJA, Winkler AA, Porro D, van Dijken JP, Pronk JT: Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microbiol 2004, 70:2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek S-H, Kwon EY, Kim YH, Hahn J-S: Metabolic engineering and adaptive evolution for efficient production of D -lactic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 2015, 100:2737–2748. [DOI] [PubMed] [Google Scholar]
- 41.Zhong W, Yang M, Mu T, Wu F, Hao X, Chen R, Sharshar MM, Thygesen A, Wang Q, Xing J: Systematically redesigning and optimizing the expression of D-lactate dehydrogenase efficiently produces high-optical-purity D-lactic acid in Saccharomyces cerevisiae. Biochem Eng J 2019, 144:217–226. [Google Scholar]
- 42.Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman J-MA, van Dijken JP, Pronk JT, van Maris AJA: Malic Acid Production by Saccharomyces cerevisiae: Engineering of Pyruvate Carboxylation, Oxaloacetate Reduction, and Malate Export. Appl Environ Microbiol 2008, 74:2766–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milne N, Wahl SA, van Maris AJA, Pronk JT, Daran JM: Excessive by-product formation: A key contributor to low isobutanol yields of engineered strains. Metab Eng Commun 2016, 3:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi A, Zhu X, Lu J, Zhang X, Ma Y: Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng 2013, 16:1–10. [DOI] [PubMed] [Google Scholar]
- 45.Ida K, Ishii J, Matsuda F, Kondo T, Kondo A: Eliminating the isoleucine biosynthetic pathway to reduce competitive carbon outflow during isobutanol production by Saccharomyces cerevisiae. Microb Cell Fact 2015, 14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Gao C, Wang Q, Liang Q, Qi Q: Production of pyruvate in Saccharomyces cerevisiae through adaptive evolution and rational cofactor metabolic engineering. Biochem Eng J 2012, 67:126–131. [Google Scholar]
- 47.Matsuda F, Ishii J, Kondo T, Ida K, Tezuka H, Kondo A: Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb Cell Fact 2013, 12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng R, Li J, Zhang A: Improving isobutanol titers in Saccharomyces cerevisiae with over-expressing NADPH-specific glucose-6-phosphate dehydrogenase (Zwf1). Ann Microbiol 2017, 67:785–791. [Google Scholar]
- 49.Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MMY, Arnold FH: Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab Eng 2011, 13:345–352. [DOI] [PubMed] [Google Scholar]
- 50.Naghshbandi MP, Tabatabaei M, Aghbashlo M, Gupta VK, Sulaiman A, Karimi K, Moghimi H, Maleki M: Progress toward improving ethanol production through decreased glycerol generation in Saccharomyces cerevisiae by metabolic and genetic engineering approaches. Renewable Sustainable Energy Rev 2019, 115:109353. [Google Scholar]
- 51.Jouhten P, Rintala E, Huuskonen A, Tamminen A, Toivari M, Wiebe M, Ruohonen L, Penttilä M, Maaheimo H: Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae CEN.PK113-1A. BMC Syst Biol 2008, 2:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nevoigt E, Stahl U: Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 1997, 21:231–241. [DOI] [PubMed] [Google Scholar]
- 53.Yan D, Wang C, Zhou J, Liu Y, Yang M, Xing J: Construction of reductive pathway in Saccharomyces cerevisiae for effective succinic acid fermentation at low pH value. Bioresour Technol 2014, 156:232–239. [DOI] [PubMed] [Google Scholar]
- 54.Zahoor A, Küttner FTF, Blank LM, Ebert BE: Evaluation of pyruvate decarboxylase-negative strains for the production of succinic acid. Eng Life Sci 2019, 19:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papapetridis I, Goudriaan M, Vázquez Vitali M, de Keijzer NA, van den Broek M, van Maris AJA, Pronk JT: Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol Biofuels 2018, 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guadalupe Medina V, Almering MJH, van Maris AJA, Pronk JT: Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol 2010, 76:190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venayak N, von Kamp A, Klamt S, Mahadevan R: MoVE identifies metabolic valves to switch between phenotypic states. Nat Commun 2018, 9:5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flux controlling technology for central carbon metabolism for efficient microbial bio-production. Curr Opin Biotechnol 2020, 64:169–174. [DOI] [PubMed] [Google Scholar]
- 59.Lalwani MA, Zhao EM, Avalos JL: Current and future modalities of dynamic control in metabolic engineering. Curr Opin Biotechnol 2018, 52:56–65. [DOI] [PubMed] [Google Scholar]
- 60.Tan SZ, Manchester S, Prather KLJ: Controlling Central Carbon Metabolism for Improved Pathway Yields in Saccharomyces cerevisiae. ACS Synth Biol 2016, 5:116–124. [DOI] [PubMed] [Google Scholar]
- 61.Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, Avalos JL: Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 2018, 555:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study combines two circuits, OptoEXP (light-induced) and OptoINVRT (dark-induced), to enable a switch between a cellular growth phase with ethanol fermentation and an isobutanol production phase. Through optimizing the light schedule and co-expressing a mitochondrial compartmentalized isobutanol pathway, the study demonstrated 8.49 g/L isobutanol production in a fed-batch fermentation, the highest reported isobutanol titer produced by yeast to date.
- 62.Zhang Y, Lane S, Chen J-M, Hammer SK, Luttinger J, Yang L, Jin Y-S, Avalos JL: Xylose utilization stimulates mitochondrial production of isobutanol and 2-methyl-1-butanol in Saccharomyces cerevisiae. Biotechnol Biofuels 2019, 12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lane S, Zhang Y, Yun EJ, Ziolkowski L, Zhang G, Jin Y-S, Avalos JL: Xylose assimilation enhances the production of isobutanol in engineered Saccharomyces cerevisiae. Biotechnol Bioeng 2020, 117:372–381. [DOI] [PubMed] [Google Scholar]; *This study demonstrated the advantages of using xylose as a carbon source over other sugars like glucose and galactose. In a fermentation with a strain harboring the mitochondrial compartmentalized pathway, more isobutanol and less ethanol was produced compared to the other sugars.
- 64.Jin Y-S, Laplaza JM, Jeffries TW: Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl Environ Microbiol 2004, 70:6816–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwak S, Jo JH, Yun EJ, Jin Y-S, Seo J-H: Production of biofuels and chemicals from xylose using native and engineered yeast strains. Biotechnol Adv 2019, 37:271–283. [DOI] [PubMed] [Google Scholar]
- 66.Sato TK, Tremaine M, Parreiras LS, Hebert AS, Myers KS, Higbee AJ, Sardi M, McIlwain SJ, Ong IM, Breuer RJ, et al. : Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae. PLoS Genet 2016, 12:e1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Islam Z-U, Klein M, Aßkamp MR, Ødum ASR, Nevoigt E: A modular metabolic engineering approach for the production of 1,2-propanediol from glycerol by Saccharomyces cerevisiae. Metab Eng 2017, 44:223–235. [DOI] [PubMed] [Google Scholar]
- 68.Kim SJ, Sim HJ, Kim JW, Lee YG, Park YC, Seo JH: Enhanced production of 2,3-butanediol from xylose by combinatorial engineering of xylose metabolic pathway and cofactor regeneration in pyruvate decarboxylase-deficient Saccharomyces cerevisiae. Bioresour Technol 2017, 245. [DOI] [PubMed] [Google Scholar]
- 69.Novy V, Brunner B, Nidetzky B: l -Lactic acid production from glucose and xylose with engineered strains of Saccharomyces cerevisiae : aeration and carbon source influence yields and productivities. Microb Cell Fact 2018, 17:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner TL, Zhang G-C, Kim SR, Subramaniam V, Steffen D, Skory CD, Jang JY, Yu BJ, Jin Y-S: Lactic acid production from xylose by engineered Saccharomyces cerevisiae without PDC or ADH deletion. Appl Microbiol Biotechnol 2015, 99:8023–8033. [DOI] [PubMed] [Google Scholar]
- 71.Dai Z, Huang M, Chen Y, Siewers V, Nielsen J: Global rewiring of cellular metabolism renders Saccharomyces cerevisiae Crabtree negative. Nat Commun 2018, 9:3059. [DOI] [PMC free article] [PubMed] [Google Scholar]; **In this study, a Crabtree-negative S. cerevisiae was created by combining ALE with a rationally designed acetyl-CoA synthesis pathway (Po and Pta) to reduce the growth defect seen in Pdc- strains grown on glucose. More specifically, the evolved mutant carried mutations in MED2, which encodes a component of the RNA polymerase II mediator complex, and GPD1, which encodes an enzyme in glycerol biosynthesis. Using reverse engineering they generated a strain that had a growth rate of 0.218 h−1, which is the highest reported for a Pdc− strain grown on glucose medium.
- 72.Schadeweg V, Boles E: n-Butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA. Biotechnol Biofuels 2016, 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Production of 2,3-butanediol from xylose by engineered Saccharomyces cerevisiae. J Biotechnol 2014, 192:376–382. [DOI] [PubMed] [Google Scholar]
- 74.Abbott DA, Zelle RM, Pronk JT, van Maris AJA: Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: current status and challenges. FEMS Yeast Res 2009, 9:1123–1136. [DOI] [PubMed] [Google Scholar]


