Abstract
Intrinsic protein disorder is an interesting structural feature where fully functional proteins lack a three-dimensional structure in solution. In this work, we estimated the relative content of intrinsic protein disorder in 96 plant proteomes including monocots and eudicots. In this analysis, we found variation in the relative abundance of intrinsic protein disorder among these major clades; the relative level of disorder is higher in monocots than eudicots. In turn, there is an inverse relationship between the degree of intrinsic protein disorder and protein length, with smaller proteins being more disordered. The relative abundance of amino acids depends on intrinsic disorder and also varies among clades. Within the nucleus, intrinsically disordered proteins are more abundant than ordered proteins. Intrinsically disordered proteins are specialized in regulatory functions, nucleic acid binding, RNA processing, and in response to environmental stimuli. The implications of this on plants’ responses to their environment are discussed.
Keywords: Intrinsic disorder, Monocots, Eudicots, Stress, Nucleus
Introduction
The traditional structure-function paradigm states protein function depends on a well-defined three-dimensional structure. However, there are regions of proteins and even complete proteins that are fully functional even though they do not fold into secondary or tertiary structures in solution (Uversky 2011; Pancsa and Tompa 2012). These proteins are known as intrinsically disordered regions/proteins (IDRs/IDPs) and are present in all domains of life (Xue et al. 2010; Xue et al. 2012; Yruela et al. 2017). In eukaryotes, it is estimated that between 23 and 28% of proteins are highly disordered and more than 50% of eukaryotic proteins contain long IDRs (greater than 30 amino acids [aa]) (Xue et al. 2012). Structural disorder is considered to be significantly higher in eukaryotes than in prokaryotes, and it has been associated with organismic complexity (Ward et al. 2004; Liu et al. 2006; Xue et al. 2012; Peng et al. 2014; Yruela et al. 2017). Some gene families are particularly enriched in IDPs (Dai et al. 2016), and the total collection of IDPs/IDRs in a proteome is called the disordome (Zamora-Briseño et al. 2018).
IDPs/IDRs are characterized by their bias in aa composition, the low complexity in their sequences, and their low content of bulky hydrophobic aa (Romero et al. 2001; Wright and Dyson 2015). Several residues, known as order-promoting residues (W, C, F, I, Y, V, L, H, T, and N), are underrepresented, while they have an abundance of proline and polar and charged residues, known as disorder-promoting residues (K, E, P, S, Q, R, D, and M). Finally, the content of A and G is considered to be similar between IDPs and their ordered counterparts (Radivojac 2003). These characteristics in the primary structure give IDPs/IDRs a high net charge and a low average hydrophobicity (Uversky et al. 2000).
Intrinsic disorder promotes structural flexibility, and this flexibility allows fast transitions between different structural states, which promotes multispecific functions (Romero et al. 2001; Radivojac 2003; Uversky 2011; Sun et al. 2013; Covarrubias et al. 2017; Zamora-Briseño et al. 2018). IDPs/IDRs are associated with the regulation of transcription, signaling, and stress responses (Sun et al. 2013; Pietrosemoli et al. 2013).
The ubiquitous nature of IDPs in multiple cellular processes has encouraged the development of programs for intrinsic protein disorder prediction, which are based on the physicochemical attributes of these proteins. Some of these predictors have been shown to be highly reliable (Romero et al. 2001; Peng et al. 2006; Mészáros et al. 2009; Xue et al. 2010; Walsh et al. 2012; Dosztányi 2018). It is now possible to estimate the content of IDPs at the proteomic scale with high confidence (Walsh et al. 2012; Kurotani et al. 2014; Yruela and Contreras-Moreira 2012). This has made possible a significant number of studies aimed at answering questions about structural disorder at the genomic scale in a large number of models (Schad et al. 2011; Xue et al. 2012; Pietrosemoli et al. 2013; Peng et al. 2014). However, it is often difficult to compare results obtained from different studies and to produce generalizations from them, in part because each study uses different predictors (each with a different confidence level) and different criteria to estimate and classify structural disorder (Pancsa and Tompa 2012).
In plants, global-scale analyses of IDPs are limited to Arabidopsis thaliana and a few other plant models (Pancsa and Tompa 2012; Yruela and Contreras-Moreira 2012; Pietrosemoli et al. 2013; Kurotani et al. 2014; Vincent and Schnell 2016; Liu et al. 2017; Alvarez-Ponce et al. 2018). This limits the identification of biological roles of IDPs without homologous functions in other models. For example, plants have developed systems that allow them to adapt to the environment from which they cannot escape (Moore et al. 2008; Schad et al. 2011; Xue et al. 2012; Pietrosemoli et al. 2013; Peng et al. 2014). Since IDPs participate in signaling cascades and stress response processes, IDPs may be particularly important in plants’ development and adaptation to their environment (Kovacs et al. 2008; Pietrosemoli et al. 2013; Liu et al. 2017; Alvarez-Ponce et al. 2018; Zamora-Briseño et al. 2018). Furthermore, although conclusions derived from other models may be applicable to plants, this is not always the case. For example, in a study evaluating the correlation between the occurrence of post-translational modifications in IDPs/IDRs of plants, it was found that while phosphorylations, acetylations, and O-glycosylations show a preference for IDPs/IDRs as in animals, methylations occur preferentially in ordered regions (Kurotani et al. 2014).
In this study, we predicted intrinsic disorder in 96 proteomes of plants. We found bias in the relative disordome content among the different clades analyzed, with significant differences between monocots and eudicots. Unlike other reports, we classified disorder predictions into four categories (0–25, 25–50, 50–75, and 75–100% of intrinsic protein disorder). Based on this criterion, we observed that protein roles depend on their disorder level. The disorder level affects the abundance of aa and influences protein size, its distribution in the cell, and protein functions. For these reasons, we considered that disordome may have major adaptive implications.
Materials and methods
In order to predict intrinsic protein disorder in plant proteomes, we downloaded proteomes available in the Ensembl Genomes (Howe et al. 2020) and Phytozome (Goodstein et al. 2012) genomic browsers and from NCBI. All sequences below 30 aa in length were removed, as well as all non-specific positions. For each proteome, disorder prediction was estimated in the Espritz program using “X-ray” and “Best sw” parameters (Walsh et al. 2012). Predictions were grouped into four categories of intrinsic protein disorder: 0–25, 25–50, 50–75, and 75–100%. We estimated the relative abundance of each disorder category for each species. A phylogenetic tree was constructed using PhyloT (https://phylot.biobyte.de/index.cgi) by using the scientific name of each species and results were visualized with iTOL v3.4 (Letunic and Bork 2019).
We estimated the abundance of each aa per intrinsic protein disorder category for monocots and eudicots. To find enriched ontological functions among each category, protein sequences were annotated with InterproScan5 (Jones et al. 2014). This allowed us to handle annotated proteins with a homogeneous criterion. Then, a random sub-sample of 25,000 proteins was taken from each category to be analyzed using the WEGO online program (Ye et al. 2006) and compare parental ontological terms that were significantly enriched by category (p < 0.001). The protein length and intrinsic disorder content of each category were compared between monocots and eudicots, using t test and Kruskal-Wallis, respectively. Statistical differences among them were determined in R (R Development Core Team 2016), and data were plotted using ggplot2 (Ginestet 2011). In addition, the binned data in the four categories of disorder were analyzed with a principal component analysis (PCA) biplot, calculated with the FactoMineR library (Lê et al. 2008) in the R environment. A linear discriminant analysis effect size (LEfSe) (Segata et al. 2011) was performed to detect the discriminant protein categories between the eudicots and monocots; the significance was stated at a p value < 0.05.
To examine in detail the association between intrinsic protein disorder in both biological processes and cellular location, the A. thaliana proteome was submitted to a GO enrichment analysis using ShinyGO v0.61 (Ge et al. 2020). Before the analysis, the Arabidopsis proteins were allotted according to their disorder category (a p < 0.001 was used to define significantly enriched terms).
Results
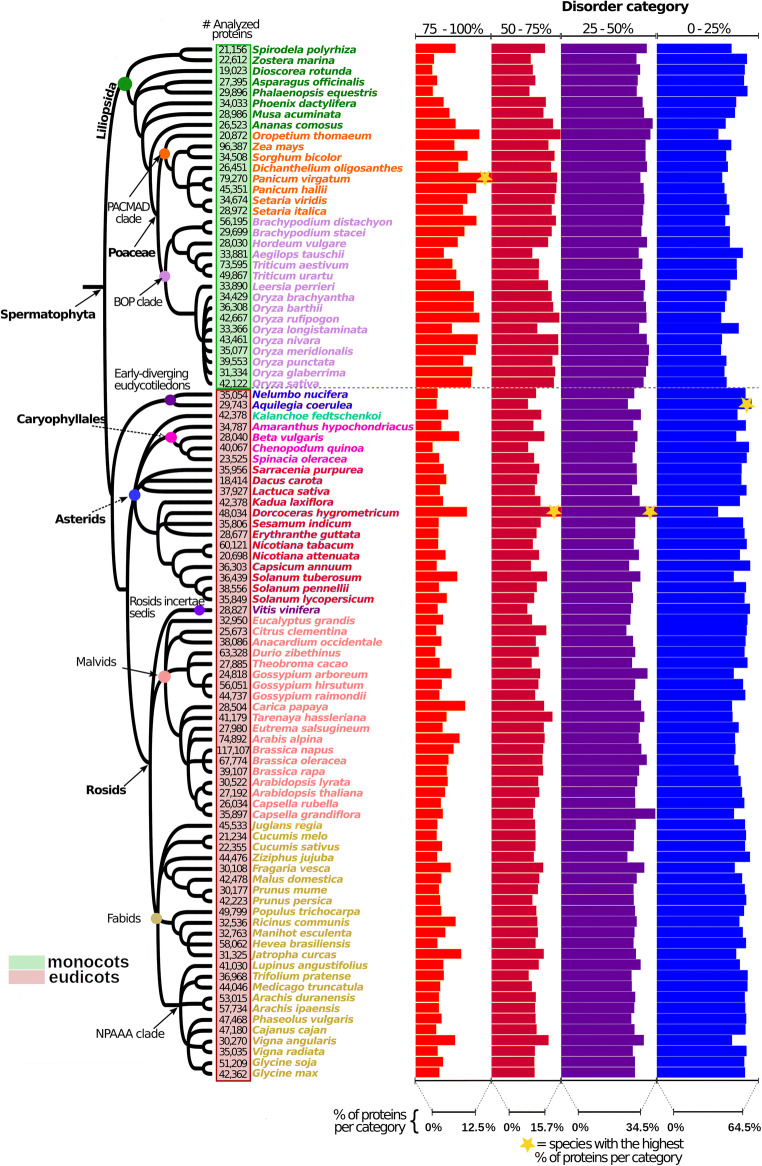
Intrinsic protein disorder predictions showed that the proportion of intrinsic disorder is greater in some clades (Fig. 1). In monocots, the Poaceae family (both BOP and PACMAD clades) showed the highest proportion of proteins with highly disordered proteins (> 75%), compared with the group of Embryophyta. In eudicots, there was a higher proportion of IDPs < 50%. Some exceptions in this group, with a higher proportion of IDPs > 75% (compared with other eudicots), were D. hygrometricum (resurrection plant from the Asterids), Carica papaya (a drought-tolerant plant from the Malvids), and Jatropha curcas (a drought-tolerant plant from the Fabids).
Fig. 1.
Distribution of disordered proteins among plant proteomes. Species on the tree are grouped into two major clades by colored squares next to the tips, as follows: monocots (green square) and eudicots (red square). The relative abundance of disorder was not constant among the analyzed clades
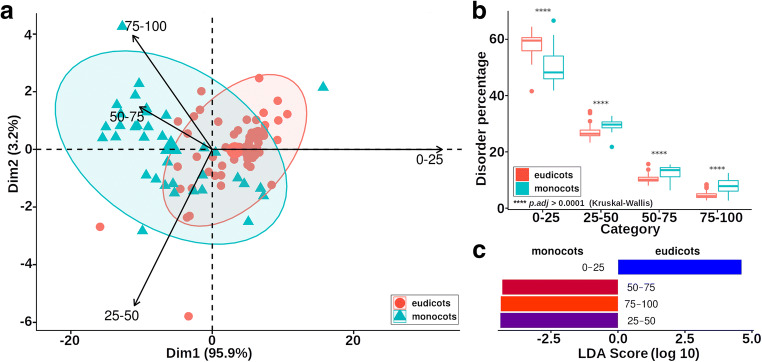
Using PCA analysis, we observed that proteomes of monocots and dicots can be separated according to the relative abundance of the disorder content (Fig. 2a). In each case, the relative content of proteins with a disorder of at least 25% was statistically higher in monocots than eudicots. In contrast, eudicots had a higher relative content of ordered proteins (0–25% disorder) and monocots (Fig. 2b). In general, monocot proteomes had higher intrinsic disorder than eudicots (Fig. 2c). Interestingly, proteins with higher disorder level tended to be smaller, regardless of their clade (Fig. 3).
Fig. 2.
Comparison between the proteomes of monocots and dicots considering the disorder abundance. a PCA biplot of the four categories of disordered proteins between eudicots and monocots. Vectors are plotted towards the direction of its major abundance in the samples. b Paired comparisons reveal differences in the relative protein content of each disorder category. c LEfSe analysis results. The category 0–25 is better represented in the eudicots, while the categories 25–50, 50–75, and 75–100 are better represented in the monocots
Fig. 3.
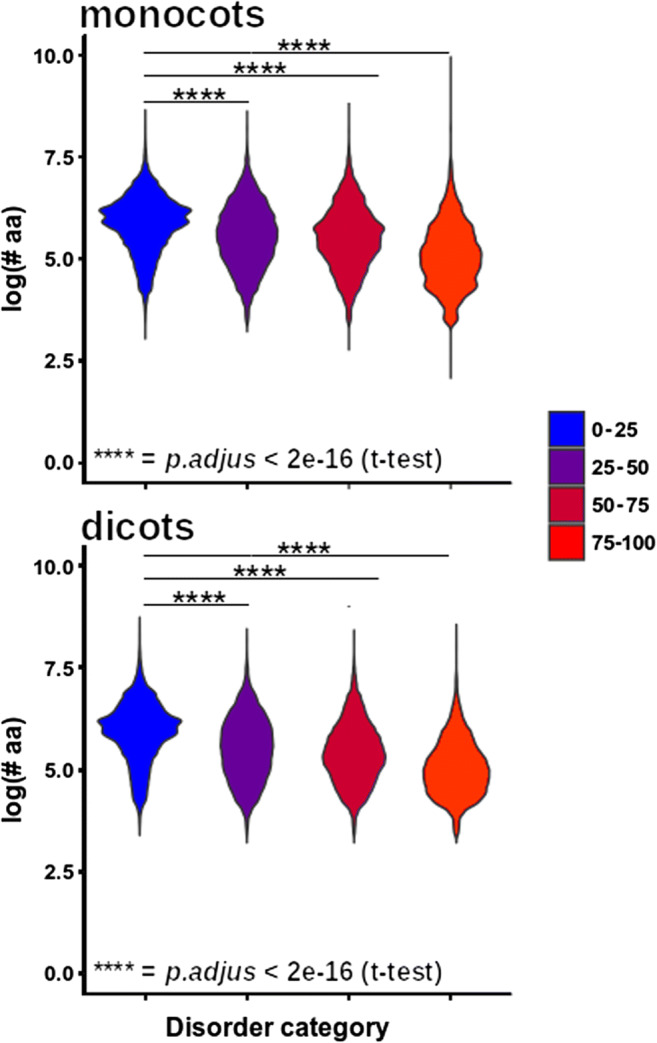
Relationship between level of intrinsic protein disorder and protein length. At a higher level of disorder, the length of the proteins tended to decrease. This pattern was observed for all clades. Post hoc comparisons were of each category against the 0–25 category using Student’s t test. ****p < 0.0001
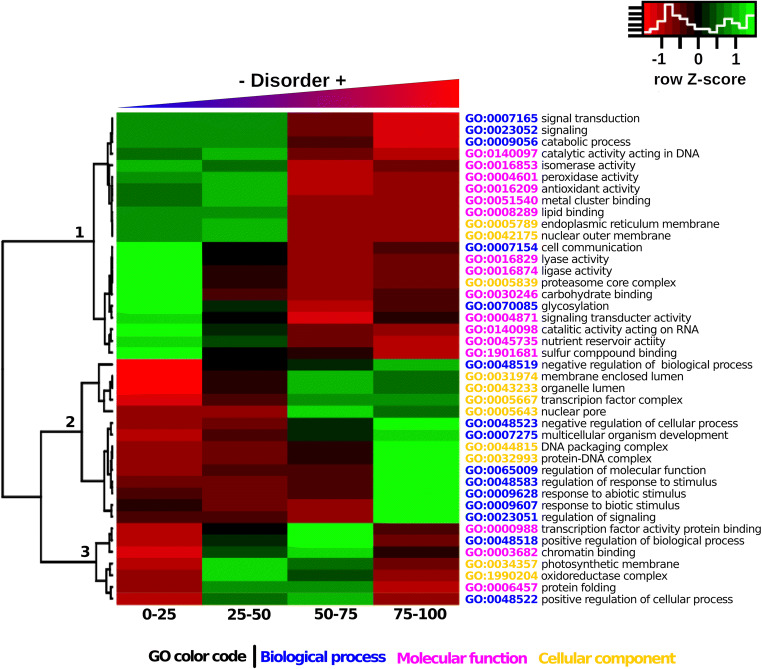
As expected, the proportion of hydrophobic amino acids was higher for more structured proteins and maintained fairly uniform compared with that of small, hydrophilic, and charged amino acids (Fig. 4). In addition, we found that each disorder category was enriched in different GO terms (Fig. 5). For example, the least disordered proteins (< 25% intrinsic disorder) are enriched in catalytic functions (clade 1, Fig. 5), while the most disordered proteins (> 75% disorder) were enriched with GO terms associated with responses to biotic and abiotic environmental stimuli, as well regulatory processes (clade 2 in Fig. 5). This coincides with results obtained in the ontology analysis of the A. thaliana proteome. There was a clear separation between biological processes in which each category of intrinsic disorder was enriched (Fig. 6). This was more evident the higher the degree of disorder.
Fig. 4.
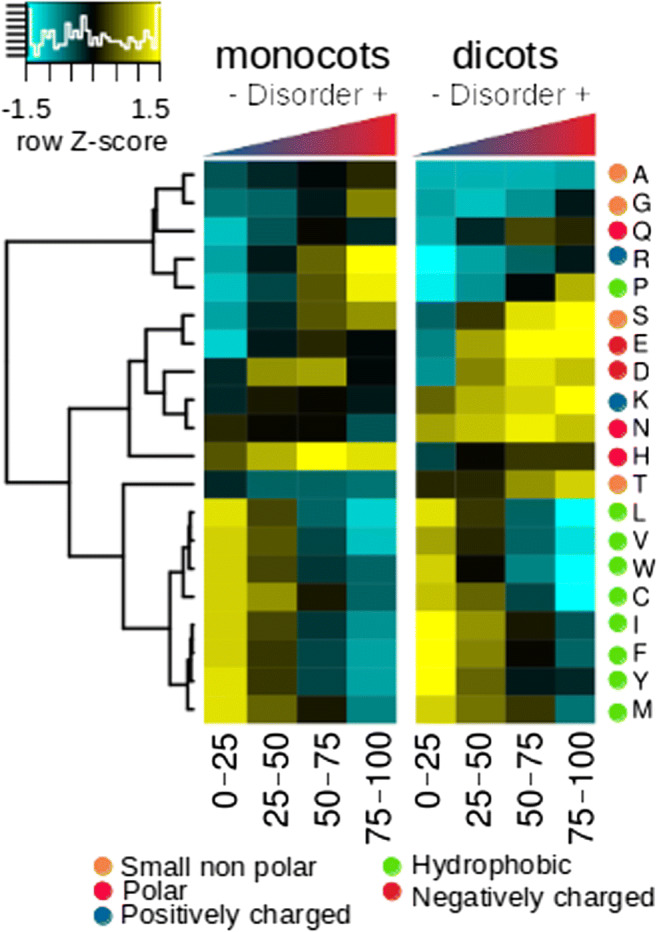
A heat map of the percentage composition of the amino acids that make up the proteins according to the level of intrinsic disorder. The hydrophobic amino acids separated perfectly from the rest, with the exception of P. These amino acids were enriched in the most structured proteins and had a more uniform abundance pattern compared with the rest of the amino acids. The disorder-promoting amino acids were enriched (K, E, P, S, Q, R, D) in the proteins with the highest levels of disorder, with the exception of M, which did not seem to follow this typical behavior. The enrichment in the relative abundance of these amino acids was not constant between clades, which seems to be influenced by taxonomic factors
Fig. 5.
GO enrichment analysis of each intrinsic protein disorder category. Highly disordered proteins (clade 2) were enriched in ontologies related to the regulation of nucleus activities, regulation of metabolic processes, response to biotic and abiotic stimuli, and signaling, while ordered proteins were enriched among catalytic activities (clade 1)
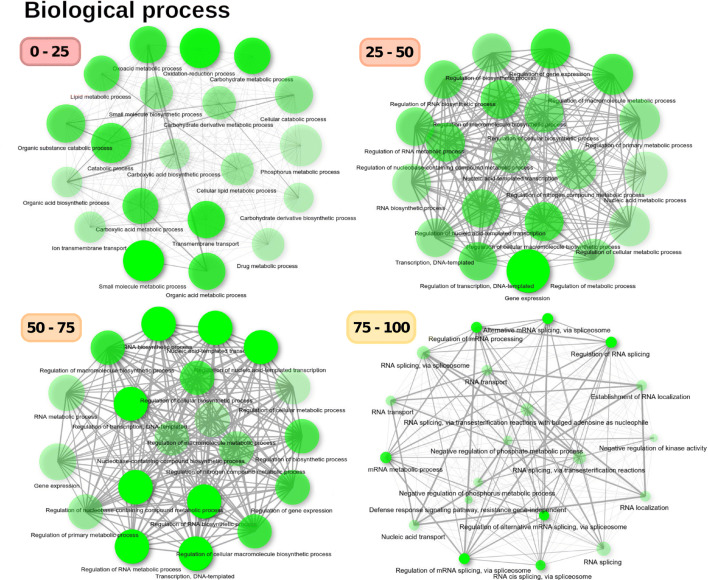
Fig. 6.
GO enrichment analysis of biological components for the A. thaliana proteome divided into disorder categories. Biological processes varied among categories
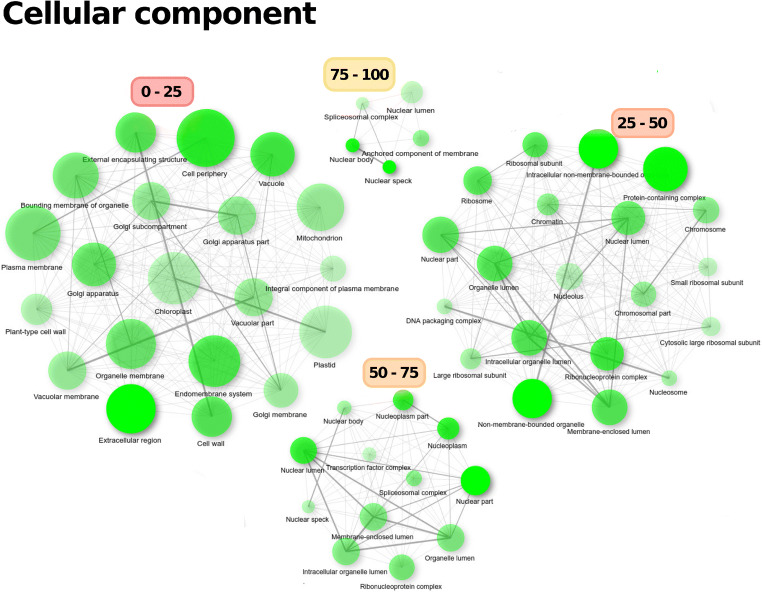
Proteins with 0–25% intrinsic disorder were enriched in biosynthetic processes, such as lipid metabolism processes, catabolic processes or processes associated with phosphorous metabolism, and membrane transport and ion transport through membranes. As relative disorder increased, there was enrichment of biological regulatory processes, such as regulation of gene expression, regulation of transcription, or regulation of metabolic processes (categories 25–50 and 25–75%). Highly disordered proteins (> 75% intrinsic disorder) were specialized in biological processes associated with RNA regulation and RNA processing, such as alternative splicing and RNA transport, as well as negative regulation of metabolic processes and kinase activity (Fig. 6). These observed ontologies correlated with the enrichment of cellular components depicted in Fig. 7. There was a clear separation of GO-associated terms based on their disorder level. Thus, highly ordered proteins (< 25%) were widely enriched in ontological terms associated with various sub-cellular spaces, such as the mitochondria, Golgi apparatus, chloroplasts, or cell wall, but the nucleus was not enriched. In proteins with 25–50% disorder, enriched GO terms included the ribosomes, nucleosome, chromatin, nuclear lumen, nucleolus, or non-membrane-bound organelles. However, as intrinsic protein disorder increased, the diversity of enriched sub-cellular spaces decreased, while the nucleus was enriched. Thus, proteins with > 50% disorder were exclusively enriched in GO terms associated with the nucleus, particularly the nucleolus, spliceosome complex, nuclear pore, and transcription complex. In the 75–100% category, nuclear lumen, splicing complex, or nuclear body were enriched. Thus, the distribution of proteins within cells was associated with their level of disorder, with the nucleus enriched in IDPs.
Fig. 7.
GO enrichment analysis of cellular components of the A. thaliana proteome divided into disorder categories. The distribution of the proteins according to their level of disorder in the cell was not random; while the nucleus was enriched in disordered proteins, proteins with less than 25% disorder were more broadly distributed in the cell and were not enriched in the nucleus
Discussion
Studies aimed at understanding the roles of intrinsic protein disorder in plants are still scarce considering the overall number of reports on this topic (Zhang et al. 2018; Zamora-Briseño et al. 2019). In turn, most of these evaluations are not specifically on plants (Xue et al. 2012; Peng et al. 2014) or have been focused on studying very few models (Pazos et al. 2013; Kurotani et al. 2014; Choura et al. 2019). General conclusions obtained in these studies are highly valuable and deserve to be corroborated. For this reason, in this work, we carried out an extensive analysis of the distribution of intrinsic protein disorder by analyzing proteins from the proteomes of 96 plants.
Some previous estimations of the variation in relative intrinsic protein disorder content among plant clades have yielded contradictory results. First, it was found that the relative content of intrinsic protein disorder does not vary between monocots and dicots (Yruela and Contreras-Moreira 2012). However, it was later found that the relative content of protein intrinsic disorder is different between them (Kurotani et al. 2014; Choura et al. 2019). These differences are likely due to the small sample size used, as well as differences in the criteria used to estimate intrinsic protein disorder.
We compared the relative content of intrinsic protein disorder among different plant clades. For categories of disorder greater than 25%, we found that intrinsic protein disorder content is higher in monocots (specifically the Poaceae family) and eudicots, with the opposite trend in the 0–25% disorder category. For comparative purposes, we considered that this category is mainly composed of structured proteins, while the other three categories are composed of intrinsically disordered proteins with different levels of disorder.
Although the definition of the four disorder categories was not based on any a priori biological criterion, it allowed us to observe a clear relationship between proteins’ level of intrinsic disorder and their functions. We consider that cataloging proteins into ordered versus disordered is overly simplistic. In other words, it is important to determine not only whether or not a protein is intrinsically disordered but also their degree of disorder, since this is associated with function. In some ways, it attempts to capture part of the different intrinsic disorder flavors described for IDPs (Dunker et al. 2008; Walsh et al. 2012; Forcelloni and Giansanti 2020).
It is interesting that as intrinsic disorder increases, there is a decrease in protein length. This negative correlation between intrinsic disorder and protein length has been previously reported and is generally accepted (Howell et al. 2012; Peng et al. 2014; Afanasyeva et al. 2018; Zamora-Briseño et al. 2019). This is expected given the biased aa composition of IDPs because in some way the occurrence of amino acids is associated with protein length (Carugo 2008). Since longer proteins tend to be more conserved than small proteins (Lipman et al. 2002), more disordered proteins must be less conserved. It is known that amino acid changes are faster for proteins with higher proportions of aa exposed to the solvent, as occurs with IDPs (Lin et al. 2007). Moreover, IDPs have a higher mutational rate than globular proteins and have a high tolerance to mutations (Brown et al. 2002; Forcelloni and Giansanti 2020). This suggests that disorder-promoting aa are subjected to reduced evolutionary constraints (relaxed evolutionary forces at these sites) and therefore have a higher mutation rate than order-promoting aa (with stronger evolutionary constraints to keep their function). This explains why the former are clearly separated on the heat map, with a more conserved distribution pattern among clades compared with disorder-promoting aa.
The relative abundance of aa apparently differs among the clades. In general, it is considered that compared with structured proteins, IDPs show a reduction in their contents of C, W, Y, F, I, V, and L, at the same time as being significantly enriched in M, K, R, S, Q, P, and E (Dunker et al. 2008). This general rule does not seem to follow the same pattern in plants because M is not enriched in any of the clades. Furthermore, A and G are enriched in IDPs of algae and monocot aa, but these aa are not usually considered enriched in IDPs (Radivojac 2003). Furthermore, the enrichment of disorder-promoting aa seems to differ among clades.
Since there is a positive correlation between genetic recombination rate and protein disorder frequency in plants, it has been proposed that genetic recombination could be considered an evolutionary force that contributes to structural disorder in proteins (Yruela and Contreras-Moreira 2013). The fact that IDPs have a higher recombination rate, higher mutability is of particular interest in plant adaptation to challenging environmental conditions. According to our GO enrichment analysis, highly intrinsically disordered proteins are enriched in the response to biotic and abiotic stimuli. Moreover, intrinsic disorder is higher in young genes and in genes created de novo in alternative reading frames, as well as in orphan genes of several non-plant species (Rancurel et al. 2009; Mukherjee et al. 2015; Wilson et al. 2017). So, it is possible that proteins encoded by young and orphan genes in plants possess a higher degree of disorder, which must be answered in the future.
Highly disordered proteins (> 75% intrinsic disorder) are particularly enriched in the regulation and transport of RNA, as well as RNA splicing. This is consistent with previous data indicating that a large number of proteins that bind to RNA exhibit broad IDRs. For example, it is estimated that more than 50% of amino acid residues of RNA chaperones occur in IDRs (Tompa and Csermely 2004). This has wide-reaching consequences. For example, alternative splicing is a very important process for stress-induced responses in plants, which can modulate the phenotypic traits of plants and can contribute to their adaptations to different environmental stressors (Mastrangelo et al. 2012; Ling et al. 2019).
In addition, intrinsic disorder influences the sub-cellular localization of proteins; IDPs are enriched in the nucleus, as has been suggested for other non-plant models (Frege and Uversky 2015; Skupien-Rabian et al. 2015). This is very reasonable considering that IDPs have functional specialization (Vincent and Schnell 2016; Deiana et al. 2019) and such functional specialization also depends on protein length (Howell et al. 2012). Considering that monocots possess a higher proportion of IDPs than eudicots, it is feasible that the proportion of nuclear proteins is also higher. In comparative terms, we expected that monocots would possess a higher proportion of nuclear proteins than eudicots.
Finally, given that a large part of the proteome with unknown functions (dark proteome) is enriched in IDPs (Bhowmick et al. 2016), it can be inferred that a large part of the disordome represents a reservoir of potential functions involved in the stress response that are waiting to be discovered. This may be exploited for biotechnological purposes, particularly those aimed at increasing resistance to environmental stressors. Thus, we consider that the characterization of genes that encode IDPs with unknown functions and that respond to stress could lead to the discovery of new mechanisms of stress response in plants.
Conclusion
This study represents the most extensive analysis of intrinsic protein disorder in plants to date. It is evident that the level of intrinsic disorder actively influences several functional characteristics of proteins beyond their lack of folding. In plants, the involvement of intrinsic disorder in environmental adaptation processes is of particular importance and represents a promising opportunity for discovery.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Afanasyeva A, Bockwoldt M, Cooney CR, Heiland I, Gossmann TI. Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 2018;28(7):975–998. doi: 10.1101/gr.232645.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Ruiz-González MX, Vera-Sirera F, Feyertag F, Perez-Amador M, Fares M. Arabidopsis heat stress-induced proteins are enriched in electrostatically charged amino acids and intrinsically disordered regions. Int J Mol Sci. 2018;19(8):2276. doi: 10.3390/ijms19082276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick A, Brookes DH, Yost SR, Dyson HJ, Forman-Kay JD, Gunter D, Head-Gordon M, Hura GL, Pande VS, Wemmer DE, Wright PE, Head-Gordon T. Finding our way in the dark proteome. J Am Chem Soc. 2016;138:9730–9742. doi: 10.1021/jacs.6b06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Keith Dunker A. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55(1):104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- Carugo O. Amino acid composition and protein dimension. Protein Sci. 2008;17:2187–2191. doi: 10.1110/ps.037762.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choura M, Ebel C, Hanin M (2019) Genomic analysis of intrinsically disordered proteins in cereals: from mining to meaning. Gene:143984. 10.1016/j.gene.2019.143984 [DOI] [PubMed]
- Covarrubias AA, Cuevas-Velazquez CL, Romero-Pérez PS, Rendón-Luna DF, Chater CCC. Structural disorder in plant proteins: where plasticity meets sessility. Cell Mol Life Sci. 2017;74:3119–3147. doi: 10.1007/s00018-017-2557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Liu H, Zhou J, Huang K. Selenoprotein R protects human lens epithelial cells against D-galactose-induced apoptosis by regulating oxidative stress and endoplasmic reticulum stress. Int J Mol Sci. 2016;17(2):231–250. doi: 10.3390/ijms17020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiana A, Forcelloni S, Porrello A, Giansanti A. Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS One. 2019;14(8):e0217889. doi: 10.1371/journal.pone.0217889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018;27:331–340. doi: 10.1002/pro.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Forcelloni S, Giansanti A. Evolutionary forces and codon bias in different flavors of intrinsic disorder in the human proteome. J Mol Evol. 2020;88(2):164–178. doi: 10.1007/s00239-019-09921-4. [DOI] [PubMed] [Google Scholar]
- Frege T, Uversky VN. Intrinsically disordered proteins in the nucleus of human cells. Biochem Biophys Reports. 2015;1:33–51. doi: 10.1016/j.bbrep.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Jung D, Yao R. ShinyGO: a graphical enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet C. ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A (Statistics Soc) 2011;174(1):245–246. doi: 10.1111/j.1467-985x.2010.00676_9.x. [DOI] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(1):D1178–DD118. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J, Alvarez-Jarreta J, Barba M, Bolser DM, Cambell L, Carbajo M, Chakiachvili M, Christensen M, Cummins C, Cuzick A, Davis P, Fexova S, Gall A, George N, Gil L, Gupta P, Hammond-Kosack KE, Haskell E, Hunt SE, Jaiswal P, Janacek SH, Kersey PJ, Langridge N, Maheswari U, Maurel T, McDowall MD, Moore B, Muffato M, Naamati G, Naithani S, Olson A, Papatheodorou I, Patricio M, Paulini M, Pedro H, Perry E, Preece J, Rosello M, Russell M, Sitnik V, Staines DM, Stein J, Tello-Ruiz MK, Trevanion SJ, Urban M, Wei S, Ware D, Williams G, Yates AD, Flicek P. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020;48(D1):D689–D695. doi: 10.1093/nar/gkz890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MH, Green R, Killeen A, Wedderburn L, Picascio V, Rabionet A, Peng Z, Larina MV, Xue B, Kurgan LA, Uversky VN. Not that rigid midgets and not so flexible giants: on the abundance and roles of intrinsic disorder in short and long proteins. J Biol Syst. 2012;20(4):471–511. doi: 10.1142/S0218339012400086. [DOI] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs D, Agoston B, Tompa P. Disordered plant LEA proteins as molecular chaperones. Plant Signal Behav. 2008;3:710–713. doi: 10.4161/psb.3.9.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani A, Tokmakov AA, Kuroda Y, Fukami Y, Shinozaki K, Sakurai T. Correlations between predicted protein disorder and post-translational modifications. Bioinformatics. 2014;30:1095–1103. doi: 10.1093/bioinformatics/btt762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. Journal of statistical software. 2008;25(1):1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1):W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed]
- Lin YS, Hsu WL, Hwang JK, Li WH. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 2007;24(4):1005–1011. doi: 10.1093/molbev/msm019. [DOI] [PubMed] [Google Scholar]
- Ling Z, Brockmöller T, Baldwin IT, Xu S (2019) Evolution of alternative splicing in eudicots. Front Plant Sci 10. 10.3389/fpls.2019.00707 [DOI] [PMC free article] [PubMed]
- Lipman DJ, Souvorov A, Koonin EV, Panchenko AR, Tatusova TA. The relationship of protein conservation and sequence length. BMC Evol Biol. 2002;2:20. doi: 10.1186/1471-2148-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu J, Sun N, Tu C, Shi X, Cheng H, Liu S, Li S, Wang Y, Zheng Y, Uversky VN. Intrinsically disordered proteins as important players during desiccation stress of soybean radicles. J Proteome Res. 2017;16:2393–2409. doi: 10.1021/acs.jproteome.6b01045. [DOI] [PubMed] [Google Scholar]
- Mastrangelo AM, Marone D, Laidò G, De Leonardis AM, De Vita P. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185-186:40–49. doi: 10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5(5):e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Vicré-Gibouin M, Farrant JM, Driouich A. Adaptations of higher plant cell walls to water loss: drought vs desiccation. Physiol Plant. 2008;124:336–342. doi: 10.1111/j.1399-3054.2008.01134.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Panda A, Ghosh TC. Elucidating evolutionary features and functional implications of orphan genes in Leishmania major. Infect Genet Evol. 2015;32:330–337. doi: 10.1016/j.meegid.2015.03.031. [DOI] [PubMed] [Google Scholar]
- Pancsa R, Tompa P. Structural disorder in eukaryotes. PLoS One. 2012;7(4):e3468–e34687. doi: 10.1371/journal.pone.0034687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Pietrosemoli N, García-Martín JA, Solano R (2013) Protein intrinsic disorder in plants. Front Plant Sci 4. 10.3389/fpls.2013.00363 [DOI] [PMC free article] [PubMed]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein in intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2014;72:137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrosemoli N, García-Martín JA, Solano R, Pazos F. Genome-wide analysis of protein disorder in Arabidopsis thaliana: implications for plant environmental adaptation. PLoS One. 2013;8(2):e55524. doi: 10.1371/journal.pone.0055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2016) R: a language and environment for statistical computing. R Found Stat Comput. 10.1007/978-3-540-74686-7
- Radivojac P. Protein flexibility and intrinsic disorder. Protein Sci. 2003;13(1):71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins Struct Funct Genet. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Schad E, Tompa P, Hegyi H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011;12(12):R120. doi: 10.1186/gb-2011-12-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):1–18. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skupien-Rabian B, Jankowska U, Swiderska B, Lukasiewicz S, Ryszawy D, Dziedzicka-Wasylewska M, Kedracka-Krok S. Proteomic and bioinformatic analysis of a nuclear intrinsically disordered proteome. J Proteome. 2015;130:76–84. doi: 10.1016/j.jprot.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Sun X, Rikkerink EHA, Jones WT, Uversky VN. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell. 2013;25(1):38–55. doi: 10.1105/tpc.112.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. ASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43(8):1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Genet. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Vincent M, Schnell S. A collection of intrinsic disorder characterizations from eukaryotic proteomes. Sci Data. 2016;3:160045. doi: 10.1038/sdata.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I, Martin AJM, Di Domenico T, Tosatto SCE. Espritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28(4):503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wilson B, Foy S, Neme R, Masel J. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat Ecol Evol. 2017;1:0146. doi: 10.1038/s41559-017-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Williams RW, Oldfield CJ, Dunker K, Uversky VN. Archaic chaos : intrinsically disordered proteins in Archaea. BMC Syst Biol. 2010;4:S1. doi: 10.1186/1752-0509-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang Jing Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;3:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I, Contreras-Moreira B. Protein disorder in plants: a view from the chloroplast. BMC Plant Biol. 2012;12(1):165. doi: 10.1186/1471-2229-12-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I, Contreras-Moreira B (2013) Genetic recombination is associated with intrinsic disorder in plant proteomes. BMC Genom:14. 10.1186/1471-2164-14-772 [DOI] [PMC free article] [PubMed]
- Yruela I, Oldfield CJ, Niklas KJ, Dunker AK. Evidence for a strong correlation between transcription factor protein disorder and organismic complexity. Genome Biol Evol. 2017;9:1248–1265. doi: 10.1093/gbe/evx073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Briseño JA, Reyes-Hernández SJ, Zapata LCR. Does water stress promote the proteome-wide adjustment of intrinsically disordered proteins in plants? Cell Stress Chaperones. 2018;23(5):807–812. doi: 10.1007/s12192-018-0918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Briseño JA, Pereira-Santana A, Reyes-Hernández SJ, Castaño E, Rodríguez-Zapata LC. Global dynamics in protein disorder during maize seed development. Genes (Basel) 2019;10(7):pii: E502. doi: 10.3390/genes10070502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Launay H, Schramm A, Lebrun R, Gontero B. Exploring intrisincally disordered proteins in Chlamydomonas reinhardtii. Sci Rep. 2018;8(1):6805. doi: 10.1038/s41598-018-24772-7. [DOI] [PMC free article] [PubMed] [Google Scholar]