Abstract
The Egyptian cotton leaf worm, Spodoptera littoralis (Boisd.), is a major agricultural lepidopterous pest causing extensive damage in a variety of crops including vegetable, cotton, fodder, and fiber crops. Heat shock protein (HSP) family members play important roles in protecting insects against environmental stressors. In this study, we characterized three putative heat shock proteins (SpliHsp70, SpliHsp90, and SpliHSF) from S. littoralis and analyzed their expression levels in response to heat, cold, ultraviolet irradiation, Bacillus thuringiensis, and Spodoptera littoralis nucleopolyhedrovirus treatments. Significant upregulation of SpliHsp70 was observed in female pupae, while the highest expression levels of SpliHsp90 and SpliHSF were found in female adults. Heat shock triggered increases in SpliHsp levels compared to cold treatment. SpliHsp90 exhibited the highest expression levels during the first 30 min of UV treatment. Both bacterial and viral pathogenic agents effected the regulation of Hsps in S. littoralis. These findings suggest that SpliHsp genes might play significant roles in the response to biotic and abiotic stress, as well as in the regulation of developmental stages.
Keywords: Heat shock protein, Spodoptera littoralis, Stress tolerance, Immune response
Introduction
Insects use a variety of tactics against stress. Regulation of protein expression is one of the defense mechanisms of insects against different stress signals. Heat shock proteins (HSPs) are chaperone proteins present in the cells of all organisms, which assist in the folding of nascent polypeptides. The formation of denatured polypeptides and proteins as a result of cellular stress leads to the upregulation of Hsp gene expression (Richards et al. 2017). Transcription of HSPs is regulated by the heat shock transcription factor (HSF) (Morimoto 1998).
The induction of HSP synthesis is a universal response upon exposure to various environmental conditions, including abiotic stresses such as heat (Joplin and Denlinger 1990; Mahroof et al. 2005; Zhang and Denlinger 2010; Tungjitwitayakul et al. 2015), cold (Rinehart et al. 2007; Tungjitwitayakul et al. 2015), radiation (Nguyen et al. 2009; Sang et al. 2012), insecticides (Li et al. 2017), oxidative stress (Lopez-Martinez et al. 2008), heavy metal (Braeckman et al. 1999; Sonoda et al. 2007; Shu et al. 2011), nutrient deficiencies (Benoit et al. 2011), dehydration (Benoit et al. 2010), and biotic stresses including virus (Lyupina et al. 2010), bacteria (Iryani et al. 2017), and fungus (Richards et al. 2017).
Insect HSPs are known to be responsive to elevated temperatures (Ritossa 1996), due to their increasing gene expression levels after exposure to heat (Zhao and Jones 2012). They are highly conserved proteins that also contribute to cold tolerance in insects (Rinehart et al. 2007). Most insects have evolved impressive strategies to enable their survival at sub-zero body temperatures (Renault et al. 2002; Lee and Denlinger 1991). In addition to temperature stress, insects fight with intrusive microorganisms by using their innate immune system, which comprises cellular and humoral responses (Wu and Yunhong 2018). Previous studies reported stimulation of immune responses by HSPs in insects against pathogens (Wojda and Jakubowicz 2007; Wojda et al. 2009; Dubovskiy et al. 2013; Richards et al. 2017). Ultraviolet (UV) irradiation is an additional abiotic stress, regularly experienced by insects, which can induce oxidative stress (Adams and Shick 2001) and potentially cause death by damaging nucleic acids (DNA and RNA) (Lah et al. 2012). Several studies have indicated that following UV exposure, expression of insect HSPs is strongly induced to help repair the DNA damage within the cell (Wang et al. 2014; Pan et al. 2018).
The Egyptian cotton leafworm, Spodoptera littoralis (Boisd.), (Lepidoptera: Noctuidae) is one of the most devastating phytophagous lepidopteran pests, which gives rise to extensive damage in many vegetable, fiber crops, and fodder, in countries in Southeast Asia and around the Mediterranean Basin (Balachowsky 1972; Sneh et al. 1981). Chemical-based control methods are still used as a management strategy for this polyphagous pest, though resistance has been developed to miscellaneous insecticides (Issa et al. 1984a, 1984b; El-Guindy et al. 1989; Rashwan et al. 1992). Therefore, it is crucial to understand Spodoptera littoralis’ physiology in order to develop alternative pest control strategies, which target the behavior, and physiology of the pest.
In this study, we characterized full-length cDNAs of the Heat shock protein 70 (SpliHsp70) and Heat shock protein 90 (SpliHsp90), and a partial cDNA of Heat shock transcription factor (SpliHSF) in S. littoralis. We construct phylogenetic trees based on amino acid sequences of SpliHsp70 and SpliHsp90 from various species, and examined expression profiles of SpliHsps in different tissues and developmental stages. We also analyzed the regulation of SpliHsp expression in response to thermal stress, radiation, and following infection with a gram-positive bacterium (Bacillus thuringiensis) and baculovirus (SpliNPV) over time.
Materials and methods
Biological material
S. littoralis laboratory cultures and dissected tissues were used from our previous study (Guz et al. 2013).
RACE PCR and sequencing analysis
Isolation of total RNA was carried out from sixth instar larvae using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s protocol. To prevent DNAse contamination, DNase digestion was applied to RNA using RNase-Free DNase (Ambion). Complementary DNAs (cDNAs) were synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s recommended protocol.
Contigs encoding SpliHSPs and SpliHSF were identified from transcriptome analysis of S. littoralis (Unpublished data). Gene-specific primers for RACE-PCR were designed using BLAST at the NCBI website to obtain full-length cDNA sequences of SpliHsp70 and SpliHsp90 (Table 1). Rapid amplification of cDNA ends (RACE) PCR technique was performed to amplify 5′ and 3′ ends of the SpliHsp70 and SpliHsp90 cDNAs using the 5′/3′ RACE Kit, 2nd Generation (Roche), according to the manufacturer’s recommended protocol. Sequencing reactions of SpliHSP70 and SpliHSP90 were carried out using DTCS Quick Start Kit (Beckman Coulter) and analyzed with CEQ 8800 Genetic Analysis System (Beckman Coulter, Fullerton, CA). Expasy MW/pI tool was utilized to calculate putative molecular weights and isoelectric points of proteins.
Table 1.
Primers used in this study
| Gene | Primer Sequence (5′-3′) | Application |
|---|---|---|
| SpliHsp70 |
F: CTTGTGAAAGGGCGAAGAGA R: GCTCTGGTGATGGATGTGTAG |
RT-qPCR |
| SpliHsp90 |
F: CGAGCCCATTGATGAGTATGT R: GTCTTCCTCACGCTTCTTCTT |
RT-qPCR |
| SpliHSF |
F: TCAGTGCTATGAAGCAGGAAA R: CAAGTGACATCAGGAATTGGATAA |
RT-qPCR |
| β-Actin |
F: ATC ATG TTC GAG ACC TTC AAC R: GCA CGA TTT CTC TCT CGG |
RT-qPCR |
| SpliHsp70 |
SP1: GCCACCTGGTTCTTGGCGGCA SP2: TGCCACCATCACTGACAACCTCGA SP3: GAGTCGTTGAAGTACGCTGGAACCG SP5: CTCCAACCAGCTGGCTGACAAGGA |
RACE-PCR |
| SpliHsp90 |
SP1: CTGTCCAGCTTTGATGGGTCGG SP2: GACGAGGTCAGCCTTGGTCATACC SP3: CTCCCACATGTACTGCTCGTCGTC SP5: CTGCCCTGCTGTCGTCTGGATTCA |
RACE-PCR |
Comparison of the deduced amino acid sequences of SpliHsp70, SpliHsp90, and SpliHSF were carried out using BLAST at the NCBI website and EXPASY. CLUSTAL OMEGA was used to align putative amino acid sequences. BOXSHADE was used to identify conserved regions.
To construct phylogenetic trees, amino acid sequences of SpliHsp70 and SpliHsp90 from various species were aligned with MEGA6 (Tamura et al. 2013) suit using ClustalW (Higgins and Sharp 1988).
Expression analyses
RT-qPCR was used to investigate expression profiles of SpliHsp and SpliHSF genes, in different tissues and developmental stages of S. littoralis. Tissue-specific expression was carried out using isolated RNA from ovaries, fat body, Malpighian tubules, hemolymph, midgut, and nervous system. All the tissues from sixth instar larvae were dissected, whereas only the reproductive tract was dissected from adults. To analyze developmental stage-specific expression, total RNA was extracted from egg, neonate, second instar larvae, third instar larvae, fourth instar larvae, fifth instar larvae, sixth instar larvae, pupae, and 5-day-old female and male adults.
To examine gene expression patterns at different temperatures, third instar Spodoptera larvae were exposed to 25 °C (control), 0 °C and 42 °C for 1 h. In order to test the effects of UV irradiation on the expression of SpliHsps and SpliHSF, a UV light (X-15 N, Spectronics, USA) emitting UV in the 254 nm wavelength (UV-C) was used to irradiate third instar larvae at approximately 300 μW/cm2 and different time points (0.5, 1, 1.5, 2, and 3 h). Controls were subjected to the same handling procedures without UV exposure. Larvae were immediately frozen in liquid nitrogen after heat and irradiation treatments and stored at – 80 °C.
In order to check immune responsive gene expression profiles of SpliHsps and SpliHSF, recently emerged third instar larvae were challenged by 106 B. thuringiensis and SpliNPV cells, as previously described (Guz et al. 2013). Infected larvae were collected at 12, 24, 48, and 72 h post feeding (hpf). Three biological replicates were collected from the experimental and control groups in each time point, and held at – 80 °C. Total RNA was isolated from larvae using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA).
RT-qPCR analysis of SpliHsps and SpliHSF expressions
Six infected and age-matched control larvae were tested at the four time points post challenge (12, 24, 48, and 72 h) using RT-qPCR. Primer sequences are given in Table 1. At each time point, three biological replicates were collected from the experimental and control groups, and stored at – 80 °C (Guz et al. 2013). Three technical replicates were included for each biological replicate. cDNA was synthesized from total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s recommended protocol. RT-qPCR reactions were performed using LightCycler 480 SYBR Green I Master kit (Roche). Amplification conditions were as follows: an initial preincubation at 95 °C for 10 min, followed by amplification for 35 cycles at 95 °C for 10 s, 53 °C for 30 s, 72 °C for 1 s, and 1 cycle of cooling at 40 °C for 10 s with the LightCycler 480 (Roche). A standard curve was determined for each set of primers in order to measure the efficiency of each reaction. The SpliHSPs mRNA levels were normalized to β-actin levels (Guz et al. 2013). Developmental, tissue, and sex-specific expression were calculated using ΔCT method, which refers the difference in threshold cycle between SpliHSPs and β-actin genes. Relative expression levels of the SpliHSPs during the heat-cold shocks, UV-C radiation, B. thuringiensis, and SpliNPV infection were calculated using the relative quantitative method (2−ΔΔCt) (Livak and Schmittgen 2001). SPSS 16 software was used to perform statistical analyses and statistical significance was established using One-way ANOVA and Duncan’s multiple range tests. P value less than 0.05 was accepted statistically significant.
Results
Characterization of SpliHsp70, SpliHsp90, and SpliHSF cDNAs
The full-length SpliHsp70 cDNA (GenBank accession nr. KC787696.1) contains an ORF of 1965 bp encoding 654 amino acids with a molecular weight of 71.5 kDa and theoretical pI of 5.32. BLAST analysis revealed that the sequence of SpliHSP70 displayed 100% identity to HSP70 from Spodoptera litura and Helicoverpa armigera, 99.84% identity to HSP70 from Sesamia inferens, Mythimna separata, Agrotis ipsilon, and Xestia c-nigrum. The putative SpliHsp70 contains three Hsp70 family signatures, including IDLGTTYS (aa 10-17), IFDLGGGTFDVSIL (aa 198-211), and IVLVGGSTRIPKVQK (aa 335-349). Furthermore, two characteristic motifs of Hsp70 were found; the deduced ATP/GTP binding site motif AEAYLGKT (aa 132-139) (Saraste et al. 1990), and conserved EEVD motif at the end of SpliHsp70 (aa 650-653) C-terminus (Fig. 1).
Fig. 1.
Deduced amino acid sequence of Hsp70 in S. littoralis is shown with numbering beginning at the initiation methionine. Characteristic signature of amino acid sequence of Hsp70 are labeled
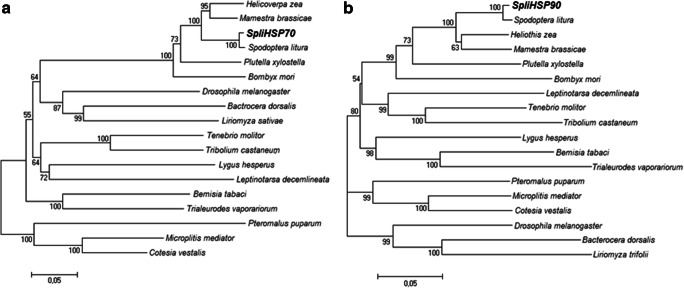
The phylogenetic tree constructed based on the alignment of the deduced amino acid sequences shows that insect Hsp70s are grouped in two main branches. One main branch includes Lepidoptera, Diptera, Coleoptera, and Hemiptera, and the other includes parasitic Hymenopteran Hsps. SpliHsp70 is clustered in Lepidopteran clade and phylogenetically close to Hsp70 from S. litura, which is also consistent with BLAST analysis. Hsps are clustered into their order clades (Fig. 4a).
Fig. 4.
Phylogenetic analysis of heat shock proteins (Hsps) from various insects. a The species and GenBank accession numbers of the Hsp70 sequences used to construct the phylogenetic tree are as follows: H. zea ACV32640.1; M. brassicae BAF03555.1; S. litura, ADV03160.1; P. xylostella ADK39311.1; B. mori BAF69068.1; D. melanogaster NP_731651.1; B. dorsalis XP_011208284.1; L. sativae AAW32099.2; T. molitor AFE88580.1; T. castaneum XP_015834944.1; L. hesperus JAQ04155.1; L. decemlineata XP_023021706.1; B. tabaci ADO14473.1; T. vaporariorum ACH85201.1; P. puparum ACO57618.1; M. mediator ABV55505.1; C. vestalis AGF34718.1. b The species and GenBank accession numbers of the Hsp90 sequences used to construct the phylogenetic tree are as follows: S. litura, ADM26738.1; H. zea ACV32639.1; M. brassicae BAF03554.1; P. xylostella AHA36864.1; B. mori BAB41209.1; L. decemlineata AHB18587.1; T. molitor AFN02498.1; T. molitor AHF20221.1; T. castaneum AHF20221.1; L. hesperus AFX84559.1; B. tabaci ADO14474.1; T. vaporariorum ACH85202.1; P. puparum ACO57617.1; M. mediator ABV55506.1; C. vestalis AGF34719.1; D. melanogaster NP_001261362.1; B. dorsalis XP_011212529.1; L. trifolii ARQ84029.1
The full length of SpliHsp90 cDNA (GenBank accession nr. KC787695.1) contains an ORF of 2154 bp encoding a 717 amino acid with a molecular weight of approximately 83 kDa and theoretical pI of 5.00. BLAST analysis revealed that the deduced sequence of SpliHSP90 displayed 99.86% identity to HSP70 from S. litura and Spodoptera frugiperda, 99.58% identity to HSP90 from H. armigera, 99.44% identity to HSP90 from S. exigua and Helicoverpa zea, and 99.30% identity to HSP90 from Helicoverpa assulta. Five Hsp90 protein family signatures (NKEIFLRELISN(S/A)SDALDKIR (aa 48-68), LGTIA(K/R)SGT (aa 115-123), IGQFGVGFYS(A/C)(Y/F)LVA(E/D) (aa 139-154), IKLYVRRVFI (aa 360-369), and GVVDS(E/D)DLPLN(I/V)SRE (aa 386-400)) were found in SpliHsp90 (Gupta 1995). Additionally, MEEVD conserved pentapeptide was detected at the C-terminal region (Fig. 2).
Fig. 2.
Deduced amino acid sequence of HSP90 in S. littoralis is shown with numbering beginning at the initiation methionine. Five characteristic signatures of amino acid sequence of HSP90 are labeled
Based on the constructed phylogenetic tree, the deduced amino acid sequence of SpliHsp90 is phylogenetically close to Hsp90 from S. litura in the first main branch, which is consistent with BLAST analysis. The first main branch consists of Lepidopteran, Coleopteran, and Hemipteran Hsps; the second main branch includes parasitic Hymenoptera; and the third branch includes Dipteran Hsps (Fig. 4b).
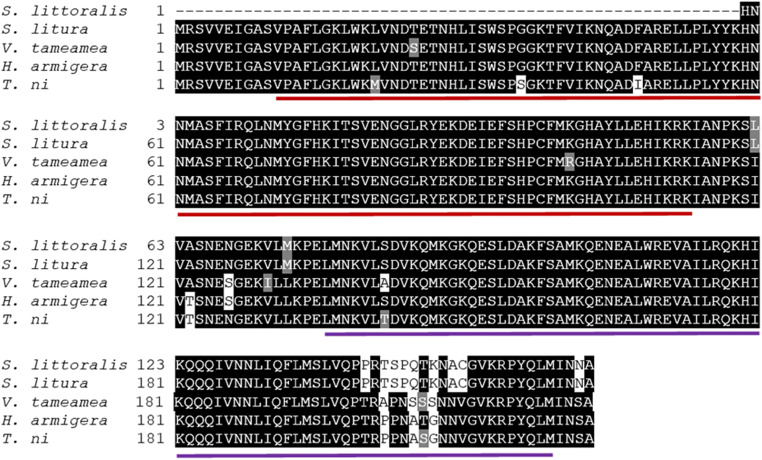
The length of partial SpliHSF was 497 bp (GenBank accession nr. KC787697.1). A heat shock transcription factor conserved domain family signature was found in SpliHSF using CD-search tool (Marchler-Bauer et al. 2017). BLAST analysis revealed that the partial cDNA sequence of SpliHSF displayed 100% identity to HSF from S. litura and S. frugiperda, 92.12% identity to HSF from H. armigera, and 91.52% identity to HSF from Trichoplusia ni. One DNA binding domain (DBD), two hydrophobic heptad repeat domains (HR-A/B and HR-C), and one C-terminal trans-activation domain (CTAD) were found in the SpliHSF sequence (Fig. 3).
Fig. 3.
Multiple alignment of the S. littoralis HSF partial amino acid sequence with other insect species. Conserved amino acids are shown in black. The GenBank accession numbers of the HSF sequences are as follows: S. litura XP_022822206, H. armigera XP_021188844, Vanessa tameamea XP_026493788, and T. ni XP_026732073. DBD and HR-A/B domains are underlined with red and purple, respectively
Developmental, tissue, and sex-specific expression of SpliHsps
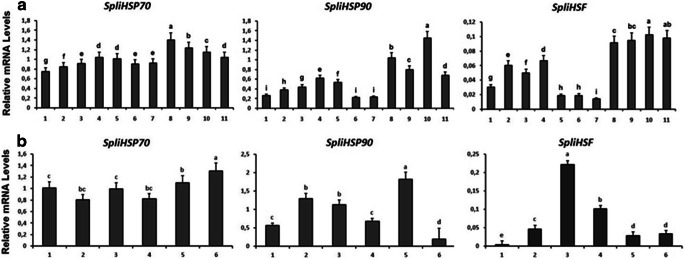
RT-qPCR analysis showed that SpliHsp70, SpliHsp90, and SpliHSF were expressed throughout all the stages of leaf worm’s life cycle (Fig. 5a). Significant upregulation of SpliHsp70 was observed in female pupae, with high expression levels also observed in male pupae and female adults. The highest expression levels for SpliHsp90 and SpliHSF were found in female adults. The expression level of SpliHsp90 increased progressively throughout development and showed a significantly higher level of expression in females than in males at the adult stage. SpliHSF transcript levels were statistically significant higher in pupal and adult stages of both males and females compared to the other developmental stages (Fig. 5).
Fig. 5.
RT-qPCR analysis of developmental (a) and tissue specific (b) expression profiles of SpliHsps and SpliHSF. a 1. Egg, 2. Neonate, 3. 2nd instar larvae, 4. 3rd instar larvae, 5. 4th instar larvae, 6. 5th instar larvae, 7. 6th instar larvae, 8. female pupae, 9. male pupae, 10. female adult, 11. male adult. b 1. Fat body, 2. hemolymph, 3. nervous system, 4. Malpighian tubules, 5. reproductive tract, 6. midgut. Developmental, tissue and sex specific expression levels were normalized to β-actin levels using ΔCT method
Tissue-specific expression is shown in Fig. 5b. SpliHsp70 was highly expressed in the midgut and reproductive tract. Interestingly, SpliHsp90 had higher expression levels in the reproductive tract than SpliHsp70. SpliHSF was predominantly expressed in nervous system of S. littoralis.
Expression profiles of SpliHsps during the heat and cold shocks
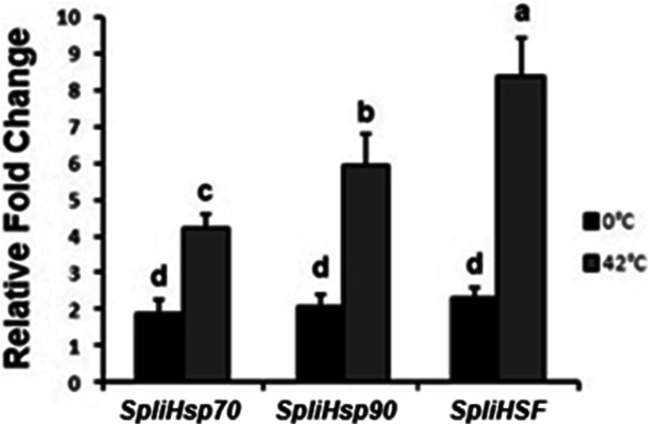
Expression levels of SpliHsps and SpliHSF genes under temperature stress are shown in Fig. 6. Under high-temperature stress (42 °C), all genes were significantly upregulated and the highest expression was observed in SpliHSF when compared to control (25 °C). No significant difference has been observed in the genes we analyzed during cold shock. Under cold stress (0 °C) expression levels were downregulated when compared to the heat shock treatment.
Fig. 6.
SpliHsp and SpliHSF mRNA expression after 1 h heat (42 °C) and cold (0 °C) shock treatments. The bars represent the 2−ΔΔCt method normalized to the β-actin gene expression in three heat/cold treated versus non-treated ones (25 °C)
Expression profiles of SpliHsps in response to UV-C radiation
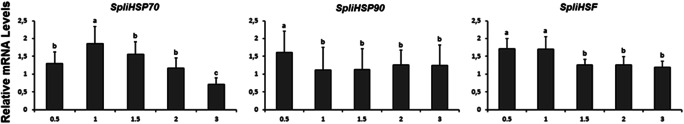
SpliHsp gene expression profiles were evaluated in third instar larvae exposed to UV-C radiation at different time points (0.5, 1, 1.5, 2, and 3 h). SpliHsp90 exhibited the highest expression at first 30 min of exposure, after which expression of this gene showed tendency to decrease over time (Fig. 7). SpliHsp70 and SpliHSF were expressed at all time points, with the highest levels occurring after 60 min of exposure for SpliHsp70, and 30–60 min of exposure for SpliHSF.
Fig. 7.
Effects of UV-C irradiation on mRNA levels of Hsps and HSF of S. littoralis larvae following different exposure durations. Relative expression of SpliHSPs was analyzed using β-actin reference gene for qPCR normalization in three irradiated larvae versus non-treated ones
SpliHsp expressions in response to B. thuringiensis and SpliNPV infection
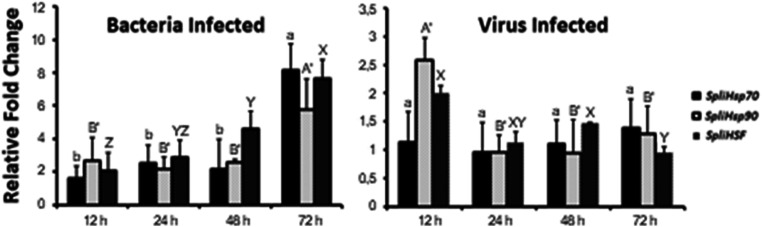
The expression of SpliHsp in response to B. thuringiensis and SpliNPV infection was analyzed using RT-qPCR in infected third instar larvae (Fig. 8). Both bacterial and viral exposure affected the regulation of Hsps in S. littoralis. The results indicated that all transcripts were upregulated following bacterial infection, with the highest expression levels for all transcripts observed at 72 h post infection. In response to SpliNPV infection, SpliHsps were also upregulated and expression values presented similar profiles in the three transcripts. SpliHsp90 had the most significant expression levels upon viral infection, with the highest expression level found at 12 h post infection. SpliHSF showed higher expression levels at 12 h post infection; however, there was no statistically significant difference detected among the 12, 24, and 48 h groups compared to untreated samples. Over the 72-h exposure period, HSP and HSF expression levels were higher upon exposure to bacterial infection, compared to viral infection. In addition, expression of all transcripts was upregulated faster following exposure to B. thuringiensis compared to SpliNPV, potentially an earlier immune response to bacterial infection.
Fig. 8.
Expression profiles of SpliHsp and SpliHSF genes of bacteria (B. thuringiensis) and baculovirus (SpliNPV) infected larvae. Relative expression of SpliHSPs was analyzed using β-actin reference gene for qPCR normalization in three infected larvae with B. thuringiensis and SpliNPV versus non-infected ones
Discussion
Insects use their complex defense system to adapt to environmental stress and induce their stress proteins as a universal response against several factors, such as insecticides, microbial agents, extreme temperatures, etc. Expression profiles of the heat-shock proteins in Spodoptera species in response to UV, heat shock, cold shock, heavy metals, and virus and bacterial infection have been previously reported for S. frugiperda (Landais et al. 2001; Lyupina et al. 2010), Spodoptera exigua (Xu et al. 2011; Jiang et al. 2012), and S. litura (Shen et al. 2011; Shu et al. 2011). To our knowledge, there is no information about transcriptional regulation of heat shock proteins in S. littoralis.
In the present study, we identified two full-length cDNAs encoding SpliHsp70 and SpliHsp90, and a partial cDNA sequence of SpliHSF from S. littoralis. SpliHsp70, SpliHsp90, and SpliHSF showed high similarity to other known insect Hsps (100% identity) indicating that these large Hsps are highly conserved when compared with small Hsp gene families (Denlinger et al. 2001). The main signatures of SpliHsp70, SpliHsp90, and SpliHSF were predicted in the deduced amino acid sequences of Hsp70, Hsp90, and HSF proteins. IDLGTTYS, IFDLGGGTFDVSLLT, and VVLVGGSTRTRKVQS are the typical signature sequences of SpliHsp70, and AEAYLGKT is the putative ATP/GTP binding site motif (Gupta and Singh 1994; Zhang and Denlinger 2010; Zhang et al. 2015), of the deduced amino acid sequence. The conserved EVVD motif found in SpliHsp70 helps to recognize the tetratricopeptide (TPR) domain of the Hsp90/70 organizing protein (HOP) and suggests that SpliHsp70 is a cytosolic homolog (Chen et al. 2015). The EEVD motif is considered to mediate interactions with chaperone cofactors and is included in binding with other co-chaperones (Boorstein et al. 1994; Freeman et al. 1995; Johnson et al. 1998). Early studies demonstrated that deletion of the EEVD motif affected ATPase activity (Junprung et al. 2019). The SpliHsp90 protein has five conserved signature sequences including NKEIFLRELISNSSDALDKIR, LGTIAKSGT, IGQFGVGFYSCYLVAD, IKLYVRRVFI, and GVVDSEDLPLNISRE (Gupta 1995). The conserved “MEEVD” pentapeptide located at the C-terminus of SpliHsp90 demonstrates that SpliHsp90 is a cytosolic homolog, similar to SpliHsp70 (Chen et al. 2015; Gupta 1995). The characteristic domains and conserved motifs of SpliHsp90 emphasize that SpliHsp90 is functional and belongs to the Hsp90 family. Evolutionary conserved domains of insect HSFs, as described by Chen et al. (2018), one DNA binding domain (DBD), two hydrophobic heptad repeat domain (HR-A/B and HR-C), and one C-terminal trans-activation domain (CTAD), were found in the SpliHSF sequence. It is considered that the amino-terminal helix-turn-helix DBD is the most conserved functional domain of HSFs throughout evolution (Pirkkala et al. 2001; Chen et al. 2018). While active HSF trimerization formation is facilitated by HR-A/B, inhibition of the formation of trimerization is carried out by HR-C. The last domain, CTAD, includes hydrophobic and acidic residues (Chen et al. 2018). In the present study, the partial amino acid sequence of SpliHSF contains DBD and HR-A/B domains, suggesting that SpliHSF is functional, evolutionarily conserved, and part of the HSF family.
The expression of insect Hsps varies among different developmental stages, depending on species (Xu et al. 2011; Jiang et al. 2012; Sonoda et al. 2006). Shu et al. (2011) analyzed expression levels of Hsp70 (Slhsp70) and Hsp90 (Slhsp90) in S. litura from four different life stages including fifth instar larvae, sixth instar larvae, pupae, and adult. The highest levels of Slhsp70/90 expression were detected in adults, Hsp70 expression in pupae was remarkably lower than in adults, and the lowest expression levels were found in fifth instar larvae. Moreover, the highest level of Slhsp90 expression was also detected in adults, which was remarkably higher than sixth instar larvae, pupae and the fifth instar larvae. It was also shown that S. litura Hsp70 was involved in developmental processes, being upregulated around pupation, suggesting that Hsp70 and sHsps might be the main players in insect midgut metamorphosis (Gu et al. 2012). In our study, the highest expression levels of SpliHsp70 and SpliHsp90 were observed in female pupae and female adults respectively. Expression levels of SpliHsp70 in male pupae and female adults were also high. Among developmental stages, the lowest expression levels of SpliHsp70/90 were detected in egg and in fifth/sixth instar larvae. SpliHsp70, SpliHsp90, and SpliHSF were expressed throughout S. littoralis development, suggesting that these genes might be expressed to deal with environmental stress and pupal adult transition (Feder and Hofmann 1999). Inhibition of HSF function gives rise to rapid aging and short life span, overexpression of it leads to extended life span in Caenorhabditis elegans (Garigan et al. 2002; Hsu et al. 2003; Morley and Morimoto 2004). The higher SpliHsp expression levels observed in this study at the beginning of the larval and throughout the pupal stages could imply that these genes are crucial in terms of survival against environmental conditions and regulation of growth and aging in the Spodoptera genus. In addition, the highest expression levels of SpliHsp90 and SpliHSF in female adults could indicate important roles in female reproduction, since temperature is one of the main factors that determine the various life activities including development and reproduction.
Although Hsp genes are expressed in all insect tissues, they have different expression levels, which are specific to the type of tissue and species. The expression of SpliHsp70 was detected in all tissues including fat body, hemolymph, nervous system, Malpighian tubules, reproductive tract, and midgut. Similarly, Hsp70 was specifically expressed in fat body, midgut, epidermis, spermary, and trachea in S. exigua (Xu et al. 2011). Although some Hsps are considered as tissue-specific genes (Sharma et al. 2006), SpliHsp70 is not regulated in a tissue-specific manner for S. littoralis and S. exigua. It has been stated that Hsp70 is involved in insect midgut metamorphosis in S. litura (Gu et al. 2012). In this study, the highest SpliHsp70 expression levels were found in midgut, which should be further analyzed using midgut tissues from different developmental time points. On the other hand, the highest SpliHsp90 expression levels were found in the reproductive tract of S. littoralis, a pattern which is consistent with that described in Bombyx mori (Saravanakumar et al. 2008). SpliHsp90 might protect the reproductive tract cells from environmental stress and sustain their regeneration, and it might be involved in reproduction, fecundity, and survival of S. littoralis. Protection of the insect immune system against foreign invaders is sustained by humoral and cellular responses and hemocytes, which phagocytize microorganisms (Williams 2007). HSP levels are known to be elevated when non-native proteins exist in the cells (Krebs and Feder 1997). After immune challenge, HSP70 and HSP82 were upregulated in the hemolymph of D. melanogaster and B. mori (de Morais et al. 2005; Song et al. 2006). In our study, mRNA transcripts were detected in the hemolymph of S. littoralis, which might be involved in an immune defence mechanism. In various insects, heat shock proteins have been detected in Malpighian tubules (Shen et al. 2011). Furthermore, after heat shock treatment, expression of different HSPs, including small HSPs, was observed in Malpighian tubules of D. melanogaster and Lucilia cuprina (Lakhotia and Singh 1989; Tiwari et al. 1995; Lakhotia and Singh 1996; Xu et al. 2011). Malpighian tubules are excretory organs and have a role in the re-absorption of water or catabolism of the toxic substances, thus the prevalence of SpliHSPs in Malpighian tubules suggests a potential role in heat tolerance and water loss in insects. Heat shock transcription factors play vital roles in the regulation of tolerance to various temperatures. Besides their role in the regulation of longevity in various tissues of C. elegans (Morley and Morimoto 2004), it has been reported that HSFs have major roles in brain development and regulation of neuronal functions in mammals (Gomez-Pastor et al. 2018). The high expression levels observed in the nervous system suggest that SpliHSF might have different roles in S. littoralis physiology, although analysis of stage-specific expression in this tissue is required before further conclusions can be drawn.
The Spodoptera genus is considered freeze-susceptible, but different species within the genus may present differences in cold tolerance mechanisms (Kim and Kim 1997). Using RNA interference, Hsp70 was shown to be the predominant contributor to thermotolerance in S. exigua (Choi et al. 2014). In fact, a thousand-fold Hsp70 upregulation has been detected in different insects in response to heat or cold shock (Velazquez et al. 1983; Huang and Kang 2007). In this study, SpliHsp70, SpliHsp90, and SpliHSF demonstrated similar expression profiles against cold shock and heat stress. The results suggest that S. littoralis is more tolerant to heat than cold stress and that upregulation of SpliHsps may play a crucial role in thermotolerance of S. littoralis.
Ultraviolet C (UV-C) radiation is the most active and shortest wavelength of UV (100–280 nm), which has great impact on organisms and can cause cellular damage (Pattison and Davies 2006). Recent studies implied that Hsps could be prompted by exposure to radiation including UV light (Sang et al. 2012), which might be a significant abiotic factor for insects feeding on plants (Zhao and Jones 2012). In this study, UV-C exposure had no influence on the survival of S. littoralis, whereas expression levels of SpliHsp70, SpliHsp90, and SpliHSF were induced by UV-C radiation. SpliHsp70 displayed the highest increase followed by SpliHSF, which is consistent with the reports that HSP70’s transcriptional and translational levels were immediately elevated after exposure to UV (Datkhile et al. 2011; Sang et al. 2012; Wang et al. 2014; Pan et al. 2018). However, in Chouioia cuneae, the response of Hsp90 to UV irradiation represented the highest expression level rather than Hsp70 (Pan et al. 2018). The significant expression levels of SpliHsp90 and SpliHSF occurred after 0.5 h of exposure appear to play a role in protecting cells from damage generated by UV damage. After 0.5 h of UV exposure, expression of SpliHsp90 decreased significantly, whereas it remained stable for SpliHSF. On the other hand, maximum expression of SpliHsp70 peaked after 1 h, before decreased gradually up to 3 h, a pattern that might be related to the negative feedback on the regulation mechanism of Hsp expression, which helps avoid the accumulation of hazardous substances such as chemicals including heavy metals, ethanol, and other contaminants (Sang et al. 2012). The results implied that these genes could take a part in defense mechanism in S. littoralis, which provides cell protection against UV-C light (Simon et al. 1995).
The humoral immune responses of insects generally involve the release of AMPs through three main pathways (Tsakas and Marmaras 2010); the Toll, the Imd, and the Jak-Stat pathways (Rosales 2017). In Drosophila, Gram-positive bacteria and fungi mainly induce the Toll signaling pathway, while Gram-negative bacteria activate the Imd pathway and antiviral immunity is mediated by the Jak-Stat pathway (Michel et al. 2001; Dostert et al. 2005; Kemp et al. 2013; Rosales 2017). Insect heat shock proteins greatly influence the functioning of the immune system (Wrońska and Boguś 2020). In this study, we showed that SpliHsps were upregulated in response to challenge with B. thuringiensis and SpliNPV, with 106 B. thuringiensis and 3000 OB of SpliNPV inoculation being enough to induce SpliHsps expression. Furthermore, expression of the different SpliHsp genes upon exposure to these microbial pathogens was dependent on time and infection-type. Major protection by three SpliHsp genes against B. thuringiensis and SpliNPV occurred 72 h and 12 h after treatment, respectively. SpliHsp genes could therefore act as an immune response mediator in S. littoralis.
In conclusion, we characterized Hsp and HSF genes from an important lepidopteran pest, S. littoralis. The genes analyzed in this study showed different expression profiles in response to elevated temperatures, UV light, and pathogen stress, as well as during different developmental stages and in different tissues. Further work should investigate the effect of the daily and seasonal temperature changes on SpliHsps and SpliHSF expression, and the relationship between gene expression and thermal tolerance in wild populations of S. littoralis. Understanding these processes might be useful to estimate the distribution and outbreaks of the Egyptian cotton leaf worm. Furthermore, the present results could offer opportunities to analyze detailed function of Hsp genes via RNA interference.
Author contributions
Conceptualization: [Nurper Guz, Asli Dageri]; Methodology: [Nurper Guz, Asli Dageri, Boran Altincicek]; Formal analysis and investigation: [Nurper Guz, Asli Dageri, Boran Altincicek]; Writing—original draft preparation: [Nurper Guz, Asli Dageri, Boran Altincicek, Serap Aksoy]; Writing—review and editing: [Nurper Guz, Asli Dageri, Boran Altincicek, Serap Aksoy]; Funding acquisition: [Nurper Guz]; Resources: [Nurper Guz]; Supervision: [Boran Altincicek, Serap Aksoy]; Data curation: [Nurper Guz, Asli Dageri, Boran Altincicek, Serap Aksoy]; Project Administration: [Nurper Guz]; Software: [Nurper Guz, Asli Dageri]; Validation: [Nurper Guz, Asli Dageri, Boran Altincicek, Serap Aksoy]; Visualization: [Nurper Guz, Asli Dageri].
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams NL, Shick JM. Mycosporine-like amino acids prevent UVB-induced abnormalities during early development of the green sea urchin Strongylocentrotus droebachiensis. Mar Biol. 2001;138:267–280. [Google Scholar]
- Balachowsky AS. Entomologie appliquée à l'agriculture. 2. Paris: Lépidoptères; 1972. pp. 1131–1133. [Google Scholar]
- Benoit JB, Lopez-Martinez G, Phillips ZP, Patrick KR, Denlinger DL. Heat shock proteins contribute to mosquito dehydration tolerance. J Insect Physiol. 2010;56:151–156. doi: 10.1016/j.jinsphys.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit JB, Lopez-Martinez G, Patrick KR, Phillips ZP, Krause TB, Denlinger DL. Drinking a hot blood meal elicits a protective heat shock response in mosquitoes. Proc Natl Acad Sci U S A. 2011;108:8026–8029. doi: 10.1073/pnas.1105195108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Braeckman B, Smagghe G, Brutsaert N, Cornelis R, Raes H. Cadmium uptake and defense mechanism in insect cells. Environ Res. 1999;80:231–243. doi: 10.1006/enrs.1998.3897. [DOI] [PubMed] [Google Scholar]
- Chen W, Li D, Zhang M, Zhao Y, Wu W, Zhang G. Cloning and differential expression of five heat shock protein genes associated with thermal stress and development in the polyphagous predatory mite Neoseiulus cucumeris (Acari: Phytoseiidae) Exp Appl Acarol. 2015;67:65–85. doi: 10.1007/s10493-015-9933-0. [DOI] [PubMed] [Google Scholar]
- Chen W, Geng SL, Song Z, Li YJ, Wang H, Cao JY. Alternative splicing and expression analysis of HSF1 in diapause pupal brains in the cotton bollworm, Helicoverpa armigera. Pest Manag Sci. 2018;75:1258–1269. doi: 10.1002/ps.5238. [DOI] [PubMed] [Google Scholar]
- Choi BG, Hepat R, Kim Y. RNA interference of a heat shock protein, Hsp70, loses its protection role in indirect chilling injury to the beet armyworm, Spodoptera exigua. Comp Biochem Physiol A Mol Integr Physiol. 2014;168:90–95. doi: 10.1016/j.cbpa.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Datkhile KD, Mukhopadhyaya R, Dongre TK, Nath BB. Hsp70 expression in Chironomus ramosus exposed to gamma radiation. Int J Radiat Biol. 2011;87:213–221. doi: 10.3109/09553002.2010.518215. [DOI] [PubMed] [Google Scholar]
- de Morais GS, Vitorino R, Domingues R, Tomer K, Correia AF, Amado F, Domingues P. Proteomics of immune-challenged Drosophila melanogaster larvae hemolymph. Biochem Biophys Res Commun. 2005;328(1):106–115. doi: 10.1016/j.bbrc.2004.12.135. [DOI] [PubMed] [Google Scholar]
- Denlinger DL, Rinehart JP, Yocum GD, Denlinger DL, Giebultowicz JM, Saunders DS (2001) Stress proteins: a role in insect diapause? In Insect timing: circadian rhythmicity to seasonality, Elsevier Science BV., Amsterdam, pp. 155-171
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- Dubovskiy IM, Whitten MM, Yaroslavtseva ON, Greig C, Kryukov VY, Grizanova EV, et al. Can insects develop resistance to insect pathogenic fungi? PLoS One. 2013;8:e60248. doi: 10.1371/journal.pone.0060248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Guindy MA, Keddis NE, Abd El-Satter MM, Ghonieim YA. Status of resistance to pesticides in the cotton leaf worm S. littoralis (Boisd.), under the present Egyptian cotton pest control programme. In Proc 1st Int Conf Ent. 1989;11:453–462. [Google Scholar]
- Feder EM, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Huang LX, Shen Y, Huang LH, Feng QL. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cut worm, Spodoptera litura. Insect Mol Biol. 2012;21:535–543. doi: 10.1111/j.1365-2583.2012.01158.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol. 1994;4:1104–1114. doi: 10.1016/s0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- Guz N, Dağeri A, Erdoğan T, Mousavi M, Bayram Ş, Gürkan MO. Transcriptional profiling of transferrin gene from Egyptian cotton leafworm, Spodoptera littoralis. Turk J Biol. 2013;37:582–590. [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Huang LH, Kang L. Cloning and interspecific altered expression of heat shock protein genes in two leaf miner species in response to thermal stress. Insect Mol Biol. 2007;16:491–500. doi: 10.1111/j.1365-2583.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Iryani MTM, MacRae TH, Panchakshari S, Tan J, Bossier P, Wahid MEA, Sung YY. Knockdown of heat shock protein 70 (Hsp70) by RNAi reduces the tolerance of Artemia franciscana nauplii to heat and bacterial infection. J Exp Mar Biol Ecol. 2017;487:106–112. [Google Scholar]
- Issa YH, Keddis ME, Ayad FA, Abdel-Sattar MM, El-Guindy MA. Survey of resistance to organophosphorous insecticides in field strains of the cotton leafworm Spodoptera littoralis (Boisd.) during 1980-1984 cotton growing seasons. Bulletin of the Entomological Society of Egypt. Economic Series. 1984;14:399–404. [Google Scholar]
- Issa YH, Keddis ME, Abdel-Sattar MA, Ayad FA, El-Guindy MA. Survey of resistance to pyrethroids in field strains of the cotton leafworm Spodoptera littoralis (Boisd.) during 1980-1984 cotton growing seasons. Bulletin of the Entomological Society of Egypt. Economic Series. 1984;14:405–411. [Google Scholar]
- Jiang X, Zhai H, Wang L, Luo L, Sappington TW, Zhang L. Cloning of the heat shock protein 90 and 70 genes from the beet armyworm, Spodoptera exigua, and expression characteristics in relation to thermal stress and development. Cell Stress Chaperones. 2012;17:67–80. doi: 10.1007/s12192-011-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- Joplin KH, Denlinger DL. Developmental and tissue specific control of the heat shock induced 70 kDa related proteins in the flesh fly, Sarcophaga crassipalpis. J Insect Physiol. 1990;36:239–249. [Google Scholar]
- Junprung W, Norouzitallab P, De Vos S, Tassanakajon A, Viet DN, Van Stappen G, Bossier P. Sequence and expression analysis of HSP70 family genes in Artemia franciscana. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-44884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, Pfeffer S, Hoffmann JA, Imler JL. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim N. Cold hardiness in Spodoptera exigua (Lepidoptera: Noctuidae) Environ Entomol. 1997;26:1117–1123. [Google Scholar]
- Krebs RA, Feder ME. Tissue-specific variation in Hsp70 expression and thermal damage in Drosophila melanogaster larvae. J Exp Biol. 1997;200(14):2007–2015. doi: 10.1242/jeb.200.14.2007. [DOI] [PubMed] [Google Scholar]
- Lah EFC, Musa RNAR, Ming HT. Effect of germicidal UV-C light (254 nm) on eggs and adult of house dustmites, Dermatophagoides pteronyssinus and Dermatophagoides farinae (Astigmata: Pyroglyhidae) Asian Pac J Trop Biomed. 2012;2:679–683. doi: 10.1016/S2221-1691(12)60209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhotia SC, Singh AK. A novel set of heat shock polypeptides in Malpighian tubules of Drosophila melanogaster. J Genet. 1989;68:129–137. [Google Scholar]
- Lakhotia SC, Singh BN. Synthesis of a ubiquitously present new HSP60 family protein is enhanced by heat shock only in the Malpighian tubules of Drosophila. Experientia. 1996;52:751–756. doi: 10.1007/BF01923984. [DOI] [PubMed] [Google Scholar]
- Landais I, Pommet JM, Mita K, Nohata J, Gimenez S, Fournier P, Devauchelle G, Duonor-Cerutti M, Ogliastro M. Characterization of the cDNA encoding the 90 kDa heat-shock protein in the Lepidoptera Bombyx mori and Spodoptera frugiperda. Gene. 2001;271:223–231. doi: 10.1016/s0378-1119(01)00523-6. [DOI] [PubMed] [Google Scholar]
- Lee RE, Denlinger DL. Insects at Low Temperature. New York: Chapman and Hall; 1991. [Google Scholar]
- Li Y, Zhao Q, Duan X, Song C, Chen M. Transcription of four Rhopalosiphum padi (L.) heat shock protein genes and their responses to heat stress and insecticide exposure. Comp Biochem Physiol A Mol Integr Physiol. 2017;205:48–57. doi: 10.1016/j.cbpa.2016.12.021. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Martinez G, Elnitsky MA, Benoit JB, Lee RE, Jr, Denlinger DL. High resistance to oxidative damage in the Antarctic midge Belgica antarctica, and developmentally linked expression of genes encoding superoxide dismutase, catalase and heat shock proteins. Insect Biochem Mol Biol. 2008;38:796–804. doi: 10.1016/j.ibmb.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Lyupina YV, Dmitrieva SB, Timokhova AV, Beljelarskaya SN, Zatsepina OG, Evgen'ev MB, Mikhailov VS. An important role of the heat shock response in infected cells for replication of baculoviruses. Virology. 2010;406:336–341. doi: 10.1016/j.virol.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Mahroof R, Zhu KY, Neven L, Subramanyam B, Bai J. Expression patterns of three heat shock protein 70 genes among developmental stages of the red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae) Comp Biochem Physiol A Mol Integr Physiol. 2005;141:247–256. doi: 10.1016/j.cbpb.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:200–203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTA, Michaud D, Cloutier C. A proteomic analysis of the aphid Macrosiphum euphorbiae under heat and radiation stress. Insect Biochem Mol Biol. 2009;39:20–30. doi: 10.1016/j.ibmb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Pan LN, Wang FZ, Zhang XY, Zhao YN, Zhu GP, Li M. Identification and characterization of heat shock proteins in a parasitic wasp Chouioia cuneae (Hymenoptera: Eulophidae) Entomol Res. 2018;48:145–155. [Google Scholar]
- Pattison DI, Davies MJ (2006) Actions of ultraviolet light on cellular structures. In Cancer: cell structures, carcinogens and genomic instability, Birkhäuser Basel pp. 131-157 [DOI] [PubMed]
- Pirkkala L, Nykanen P, Sistonen LEA. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rashwan MH, Elbaramawy ZA, El-Sheikh AE, Radwan HSA. The onset of organophosphate and carabamate resistance among lower Egypt population of the cotton leafworm S. littoralis (Boisd) Bull Ent Soc Egypt Econ Ser. 1992;19:211–220. [Google Scholar]
- Renault D, Salin C, Vannier G, Vernon P. Survival at low temperatures in insects: what is the ecological significance of the supercooling point? CryoLetters. 2002;23:217–228. [PubMed] [Google Scholar]
- Richards EH, Dani MP, Lu Y, Butt T, Weaver RJ. Effect of stress on heat shock protein levels, immune response and survival to fungal infection of Mamestra brassicae larvae. J Insect Physiol. 2017;96:53–63. doi: 10.1016/j.jinsphys.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SAL, Denlinger DL. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. PNAS. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. Discovery of the heat shock response. Cell Stress Chaperones. 1996;1:97–98. doi: 10.1379/1466-1268(1996)001<0097:dothsr>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales C (2017) Cellular and molecular mechanisms of insect immunity. Insect Physiology and Ecology. InTeach Publication, CCBY: London, pp. 179-212
- Sang W, Ma WH, Qiu L, Zhu ZH, Lei CL. The involvement of heat shock protein and cytochrome P450 genes in response to UV-A exposure in the beetle Tribolium castaneum. J Insect Physiol. 2012;58:830–836. doi: 10.1016/j.jinsphys.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP-and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Saravanakumar R, Ponnuvel KM, Qadri SMH. Expression of metabolic enzyme genes and heat-shock protein genes during embryonic development in diapause and non-diapause egg of multivoltine silkworm Bombyx mori. Biologia. 2008;63:737–744. [Google Scholar]
- Sharma S, Reddy PVJ, Rohilla MS, Tiwari PK. Expression of HSP60 homologue in sheep blowfly Lucilia cuprina during development and heat stress. J Therm Biol. 2006;31:546–555. [Google Scholar]
- Shen Y, Gu J, Huang LH, Zheng SC, Liu L, Xu WH, Feng QL, Kang L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J Insect Physiol. 2011;57:908–914. doi: 10.1016/j.jinsphys.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Shu Y, Du Y, Wang J. Molecular characterization and expression patterns of Spodoptera litura heat shock protein 70/90, and their response to zinc stress. Comp Biochem Physiol A Mol Integr Physiol. 2011;158:102–110. doi: 10.1016/j.cbpa.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Simon MM, Reikerstorfer A, Schwarz A, Krone C, Luger TA, Jäättelä M, Schwarz T. Heat shock protein 70 overexpression affects the response to ultraviolet light in murine fibroblasts. Evidence for increased cell viability and suppression of cytokine release. J Clin Investig. 1995;95:926. doi: 10.1172/JCI117800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneh B, Schuster S, Broza M. Insecticidal activity of Bacillus thuringiensis strains against the egyptian cotton leaf worm Spodoptera littoralis [Lep.: Nocutidae] Entomophaga. 1981;26:179–190. [Google Scholar]
- Song KH, Jung SJ, Seo YR, Kang SW, Han SS. Identification of up-regulated proteins in the hemolymph of immunized Bombyx mori larvae. Comp Biochem Phys D. 2006;1:260–266. doi: 10.1016/j.cbd.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ashfaq M, Tsumuki H. Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19. 5) from the diamondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch Insect Biochem Physiol: Published in Collaboration with the Entomological Society of America. 2006;62:80–90. doi: 10.1002/arch.20124. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ashfaq M, Tsumuki H. A comparison of heat shock protein genes from cultured cells of the cabbage armyworm, Mamestra brassicae, in response to heavy metals. Arch Insect Biochem Physiol: Published in Collaboration with the Entomological Society of America. 2007;65:210–222. doi: 10.1002/arch.20178. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari PK, Mohan DRK, Joshi A. Developmental study of thermotolerance and the heat shock response in Lucilia cuprina (Weidemann) J Biosci. 1995;20:341–354. [Google Scholar]
- Tsakas S, Marmaras VJ. Insect immunity and its signalling: an overview. Invertebr Surviv J. 2010;2010(7):228–238. [Google Scholar]
- Tungjitwitayakul J, Tatun N, Vajarasathira B, Sakurai S. Expression of heat shock protein genes in different developmental stages and after temperature stress in the maize weevil (Coleoptera: Curculionidae) J Insect Physiol. 2015;108:1313–1323. doi: 10.1093/jee/tov051. [DOI] [PubMed] [Google Scholar]
- Velazquez JM, Sonoda S, Bugaisky G, Lindquist S. Is the major Drosophila heat shock protein present in cells that have not been heat shocked? J Cell Biol. 1983;96:586–290. doi: 10.1083/jcb.96.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Zhou LJ, Zhu ZH, Ma WH, Lei CL. Differential temporal expression profiles of heat shock protein genes in Drosophila melanogaster (Diptera: Drosophilidae) under ultraviolet A radiation stress. Environ Entomol. 2014;43:1427–1434. doi: 10.1603/EN13240. [DOI] [PubMed] [Google Scholar]
- Williams MJ. Drosophila hemopoiesis and cellular immunity. J Immunol. 2007;178(8):4711–4716. doi: 10.4049/jimmunol.178.8.4711. [DOI] [PubMed] [Google Scholar]
- Wojda I, Jakubowicz T. Humoral immune response upon mild heat-shock conditions in Galleria mellonella larvae. J Insect Physiol. 2007;53:1134–1144. doi: 10.1016/j.jinsphys.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Wojda I, Kowalski P, Jakubowicz T. Humoral immune response of Galleria mellonella larvae after infection by Beauveria bassiana under optimal and heat-shock conditions. J Insect Physiol. 2009;55:525–531. doi: 10.1016/j.jinsphys.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Wrońska AK, Boguś MI. Heat shock proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (Lepidoptera) hemolymph are affected by infection with Conidiobolus coronatus (Entomophthorales) PLoS One. 2020;15:e0228556. doi: 10.1371/journal.pone.0228556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yunhong Y. Transcriptome analysis of differentially expressed genes involved in innate immunity following Bacillus thuringiensis challenge in Bombyx mori larvae. Mol Immunol. 2018;103:220–228. doi: 10.1016/j.molimm.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Xu Q, Zou Q, Zheng H, Zhang F, Tang B, Wang S. Three heat shock proteins from Spodoptera exigua: gene cloning, characterization and comparative stress response during heat and cold shocks. Comp Biochem Physiol B: Biochem Mol Biol. 2011;159:92–102. doi: 10.1016/j.cbpb.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Denlinger DL. Molecular characterization of heat shock protein 90, 70 and 70 cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J Insect Physiol. 2010;56:138–150. doi: 10.1016/j.jinsphys.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Wang KF, Jing YP, Zhuang HM, Wu G. Identification of heat shock protein genes hsp70s and hsc70 and their associated mRNA expression under heat stress in insecticide-resistant and susceptible diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) Eur J Entomol. 2015;112:215–226. [Google Scholar]
- Zhao L, Jones WA. Expression of heat shock protein genes in insect stress responses. Invertebr Surviv J. 2012;9:93–101. [Google Scholar]