Abstract
The mammalian skin is equipped with a highly dynamic stratified epithelium. The maintenance and regeneration of this epithelium is supported by basally located keratinocytes which display stem cell properties, including lifelong proliferative potential and the ability to undergo diverse differentiation trajectories. Keratinocytes support not just the surface of the skin, called the epidermis, but also a range of ectodermal structures including hair follicles, sebaceous glands, and sweat glands. Recent studies have shed light on the hitherto underappreciated heterogeneity of keratinocytes by employing state-of-the-art imaging technologies and single-cell genomic approaches. In this mini review, we highlight major recent discoveries that illuminate the dynamics and cellular mechanisms that govern keratinocyte differentiation in the live mammalian skin, and discuss the broader implications of these findings for our understanding of epithelial and stem cell biology in general.
Introduction
The skin is the primary interface between the body and the external environment. As such, the tissue is primarily responsible for maintaining a watertight barrier and mounting an effective regenerative response after injury. The skin consists of an outermost layer, the epidermis, which is characterized by its layered organization, and is interspersed with appendages, such as hair follicles, that serve specific roles in barrier formation, thermoregulation, and innate immunity. The structural and functional diversity of the skin and its associated appendages is reflected in the diverse self-renewal and differentiation patterns of their stem cells, as well as differences in gene expression. Despite the compartmentalized activities of these keratinocytes during homeostasis, it is well documented that these cells display remarkable plasticity during wound repair and are capable of regenerating other epithelial structures after injury. To elucidate the mechanisms of keratinocyte differentiation, cutting-edge experimental approaches, including intravital imaging and single-cell sequencing, have been recently employed [1–3] (Figure 1). Here we discuss how these powerful techniques have begun to elucidate the heterogeneity and dynamic cellular behaviors that keratinocytes display in vivo, fueling exciting new hypotheses for the intrinsic and extrinsic nature of their regulatory framework.
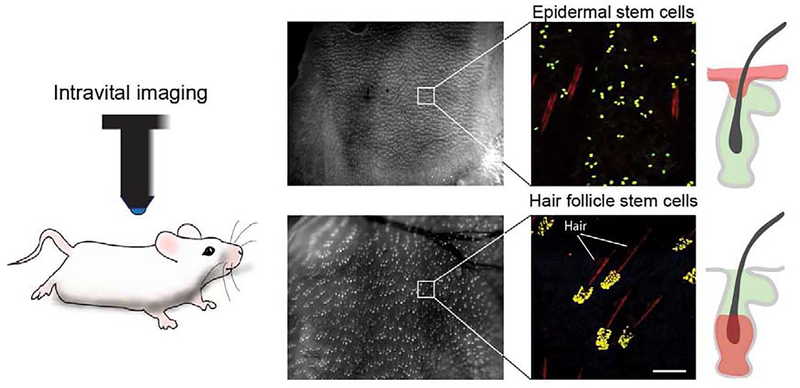
Figure 1. Genetic labeling and visualization of single keratinocytes in live mouse skin.
Representative low and high magnification images demonstrating high-throughput visualization of single-labeled stem cells in the mouse skin, by intravital imaging. Cells in the epidermis and hair follicles can be differentially labeled using cell-type specific inducible drivers. Scale bar 100μm.
The Mammalian Epidermis: Mechanisms of Keratinocyte Fate and Differentiation
The epidermis is a stratified epithelium composed of morphologically distinct cellular layers that reflect the terminal differentiation process of keratinocytes. After exiting the cell cycle and committing to differentiation, keratinocytes leave the innermost basal layer and undergo an upwards-directed transit into the more superficial spinous, granular, and cornified layers (Figure 2). This unique terminal differentiation process culminates in a stereotypical form of cell death, termed cornification, after which the cells are eventually shed from the surface of the epidermis. Thus, cell divisions in the basal layer, the only layer with proliferative potential, must be regulated to offset cell loss through shedding. The identity of basal layer keratinocytes and their organization into distinct long-lived stem cell and transient progenitor populations has been the subject of intense research [4–7]. Live imaging combined with the use of inducible and light-modulated fluorescent reporters have enabled unbiased tracking of individual basal keratinocytes in vivo over multiple generations. These experiments demonstrated that the fates of epidermal daughter cells are not pre-determined, and appear to have lifetimes that are coupled [8,9]. How epidermal maintenance is achieved has remained an outstanding question in the field with much of the debate contesting a stochastic common progenitor model of maintenance versus a hierarchical stem cell paradigm. Recent studies that combined lineage tracing with cell-cycle distribution analyses provided further support for a model of epidermal homeostasis achieved by an equipotent keratinocyte population [10,11*]. Moreover, it seems the common progenitor model may also hold true during postnatal growth of the epithelium. In applying quantitative clonal analysis and single-cell transcriptomics, investigators have defined the cellular dynamics that support the organized growth of the expanding epithelium. This work revealed that a single population of keratinocyte progenitors, whose fates are uniformly imbalanced towards self-renewal, mediate postnatal development of the mouse tail and paw epidermis [12**].
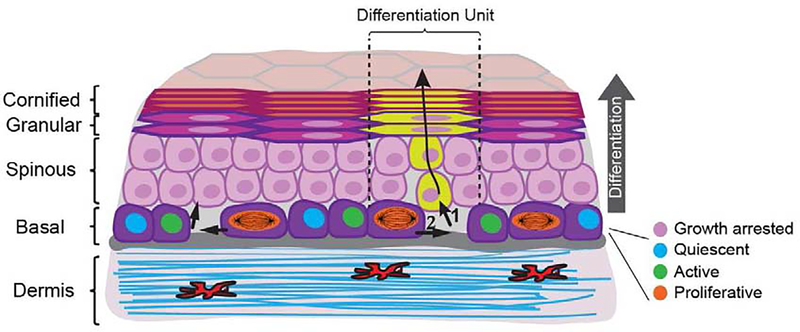
Figure 2. Keratinocyte dynamics during epidermal homeostasis.
Mapping the time and location of self-renewal and differentiation events in large epidermal regions by longitudinal live imaging, revealed that keratinocyte fate choices are locally coordinated. As epidermal stem cells differentiate and exit the basal layer (1), neighboring epidermal stem cells are prompted to self-renew in response to the local decreases in cell number and density (2). Differentiation then proceeds along individual epidermal differentiation units (EDUs) defined by the squamous cells in the granular and cornified layers. Upon terminal differentiation and desquamation, a cell is immediately replaced by a superseding keratinocyte within the same unit.
Keratinocytes contribute to the skin barrier, even before their cornification, by forming tight junctions in the granular layer of the epidermis [13]. But how the epidermis maintains barrier integrity and stable cellular connections while it undergoes significant cellular turnover has been an open question. Tracking individual epidermal cells, in vivo, during their differentiation revealed that neighboring keratinocytes transit towards the top layers independently from each other rather than stratifying in unison as whole layers [8]. More refined analyses have revealed that keratinocytes in the granular layer form transiting columns. As cells move upwards through the granular layer, they are replaced by newly formed granular cells that fill the space of the preceding cell while assuming the shape of a tetrakaidecahedron [14]. This shape enables the immediate formation of three-way tight junction contacts with neighboring keratinocytes in the granular layer, thus ensuring barrier stability. E-cadherin appears to play a critical dual role in this process as well by inhibiting tight junctions in the early stages of keratinocyte differentiation and promoting their formation when cells reach the second granular layer [15].
During cornification, terminally differentiating keratinocytes in the top layers of the epidermis undergo structural changes that are critical for epidermal barrier function [16]. 2-photon live imaging experiments in human organotypic skin cultures have resolved some of the structural changes that occur during cornification. The thickening and flattening of keratinocytes during their transition from the granular to the top cornified layer of the epidermis is preceded by the cessation of intracellular vesicle motility and mitochondrial depolarization [17]. Furthermore, intravital imaging in mouse skin showed that intracellular calcium is transiently elevated in keratinocytes prior to their transition from the granular to the cornified layer, suggesting a role for calcium signaling in the cornification process [18].
Single-cell transcriptomic analyses of mouse and human basal epidermal keratinocytes paint a rich picture of a heterogeneous cellular landscape [19,20]. These data have provided a new perspective into the transcriptional states of basal keratinocytes during epidermal lineage specification. Single-cell RNA sequencing (scRNA-seq) in mouse epidermis have defined four unique transcriptional profiles among basal layer keratinocytes, which consist of one proliferative and three non-proliferative states [19]. A combination of lineage fate prediction analyses revealed these transcriptional states to be transitional, likely existing in a differentiation hierarchy [19]. Similar analyses of human epidermal keratinocytes also identified four transcriptionally diverse basal keratinocyte states. Furthermore, these transcriptional states appeared to correlate to distinct locations within the basal layer at the top and bottom of rete ridges; epithelial extensions unique to the human epidermis [20]. These data suggest a linear hierarchy among basal epidermal keratinocytes. Future mechanistic studies will reveal the specific genes that govern the fate transitions of epidermal keratinocytes and illuminate the factors that regulate their expression.
It has become increasingly evident that keratinocytes in the basal layer of the epidermis are transcriptionally diverse and exist in different stages of the cell cycle. However, it is not clear how these cells coordinate their fate decisions to maintain tissue-wide homeostatic balance. Positioning the mitotic spindle perpendicular to the basement membrane in dividing keratinocytes is a plausible mechanism by which to achieve asymmetric fates. Although this mechanism occurs during development [21–23], most cell divisions in the adult epidermis appear to be planar [8,24]. Furthermore, there is evidence that epidermal cell fate may be regulated through keratinocyte mechanics rather than spindle orientation [25]. In one of these processes, the polarity protein Par3 was found to regulate epidermal cell fate by coupling Rho/actomyosin contractility to genome integrity and mitotic outcomes.
The importance of tissue mechanics has also been underscored by a recently developed in vivo model that has elucidated a mechanism by which stretching of the skin induces a response at the single cell level [26**]. Under mechanical tension, epidermal stem cells transiently shift their fates towards self-renewal. Consistent with this, intravital imaging studies in the mouse that spatiotemporally mapped local differentiation dynamics of epidermal keratinocytes uncovered a coordinated fate response. Local decreases in cell number/density, caused by keratinocyte differentiation and basal-layer exit, prompted a compensatory self-renewal response by neighboring basal keratinocytes [27]. Using a similar live imaging approach, more recent investigations have revealed epidermal keratinocytes also couple their growth with cell-cycle progression. These studies proposed a “sizer” mechanism acts in the G1 phase to induce cell division once a certain cell volume is reached [28**]. Another study captured spatially segregated pulses of ERK activity in both human and mouse epidermal keratinocytes by live imaging, implicating intracellular signaling cascades in the regulation of cell fate [29]. These exciting new findings further highlight the rich network of cell intrinsic and extrinsic mechanisms that regulate keratinocyte dynamics in the mammalian epidermis. Future work will uncover how these are integrated at the cellular and tissue level to ensure robustness in maintaining homeostatic balance.
When the skin barrier becomes disrupted after injury, a wound-healing response is initiated. Recent investigations in the intestinal epithelium revealed induction of a fetal-like transcriptional program after injury [30,31]. However, it is not clear whether reactivation of an embryonic gene signature occurs, or is necessary, during epidermal wound repair in mice. In vivo studies of small wounds (< 1cm in diameter) revealed that epidermal cells at the wound edge begin to display an embryonic gene signature. Among these genes, Sox11 was found to be significantly enriched at the wound edge. In the absence of Sox11 and its closest relative, Sox4, wound edge epidermal cells failed to upregulate an embryonic gene signature. This resulted in premature cell differentiation and failure of cells to migrate and re-epithelialize [32*]. Moreover, disruption of the skin barrier due to epidermal loss of ceramide synthase 4 (CerS4) induces a repair response reminiscent of the molecular programs that establish the barrier during skin morphogenesis [33]. These data provide strong evidence that reversion to an embryonic-like state is critical for wound repair.
Keratinocyte Dynamics in Epidermal Appendages
The mammalian skin is densely populated by epidermal appendages including hair follicles which are visible on the surface of the skin. These self-contained mini organs are specified during embryonic development and periodically generate hair fibers that assist in protection and thermoregulation. The cyclic growth of hair fibers is powered by a heterogeneous pool of resident keratinocytes, commonly referred to as hair follicle stem cells (HFSC). Live imaging experiments combined with lineage tracing and single-cell genomic analyses have revealed the hierarchical organization of stem cells and their progeny within this organ [34–38*] (Figure 3). Such work has also extended to the sebaceous glands; bi-lobed structures associated with the upper portion of the hair follicle. Sebaceous glands produce and secrete waxes and lipids critical to hair and skin physiology. Recent in vivo studies have begun to dissect the cellular mechanisms by which lipid-secreting sebocytes that are lost during holocrine secretion are replenished [39,40]. However, the specific genes and environmental factors that define the sebaceous gland progenitors remain to be fully resolved.
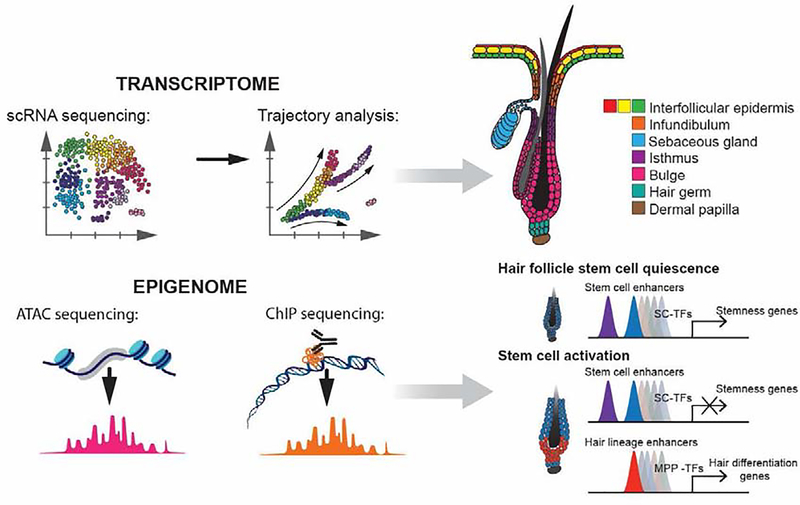
Figure 3. Resolving the cellular heterogeneity and regulation of hair growth by high-throughput next-generation sequencing.
Advancements in modern sequencing techniques provide a unique opportunity to explore tissue composition and cellular states with unprecedented detail. The application of such innovative approaches has begun to scratch the surface towards a holistic understanding of epidermal and hair follicle composition and development [37, 50]. Abbreviations: scRNA – single-cell RNA, ATAC- Assay for Transposase-Accessible Chromatin, ChIP - chromatin immunoprecipitation, SC- stem cell, TF - transcription factor, MPP - multipotent progenitor.
The first step in hair regeneration is the activation of HFSCs. This is required for progenitor expansion and to induce necessary morphogenetic changes to accommodate the subsequent construction and growth of the hair fiber. The mesenchymal dermal papilla is the primary niche that regulates HFSC activity. To initiate hair regeneration, the dermal papilla downregulates inhibitory cues that maintain HFSCs in quiescence [41–44]. In addition to the dermal papilla, a complex network of local and systemic regulatory factors plays a significant role in the timing and amplitude of hair regeneration. Recent work has uncovered a previously unappreciated involvement of the sympathetic nervous system in the activation of HFSCs by neurotransmitters [45,46]. Moreover, endothelial cells from the local vasculature appear to also modulate the timing of activation and hair regeneration through BMP signaling [47].
Following HFSC activation and the initial expansion of the progenitor pool, several differentiated hair lineages emerge from micro-niches along the interface between the epithelium and dermal papilla [34]. These lineages collectively build and facilitate growth of the newly formed hair fiber. Defining the point at which stem cells and/or their progeny become irreversibly fated to terminally differentiate has been a central question in the field. Recently, single-labeled HFSCs were followed in real-time by 2-photon live imaging. This revealed that differentiated hair lineage fates resulted from the spatial priming of HFSCs. In other words, the lineage fates of HFSCs are determined based on their initial location within the niche [48**]. Furthermore, the progenitors of HFSCs were also revealed to be capable of flexibly adopting new fates in the absence of an injury stimulus. This finding is especially interesting as it suggests hair progenitors retain stem cell characteristics during homeostasis with fate commitment occurring later than previously thought, to ensure robust differentiation.
Chromatin organization is intimately linked to cell fate regulation. ATAC-seq was recently used to investigate how the chromatin landscape is altered to achieve lineage specification in keratinocytes [49–52]. Overlaying lineage-specific transcriptomes with these chromatin landscapes revealed putative master transcription factors and chromatin-associated regulatory elements. Together, the spatial and temporal emergence of these signaling features drives hair follicle lineage diversity and specification during hair growth. Furthermore, coupling these analyses with new and previously published chromatin immunoprecipitation sequencing datasets further informed on the cooperative roles between WNT and BMP signaling effectors with key transcription factors in regulating the hair lineage cascade [49,50*].
Hair follicle-derived keratinocytes contribute to cutaneous wound repair and long-term maintenance of the healed epidermis [53–56]. Yet how HFSCs and their progeny adapt at the molecular level to contribute to wound repair was not clear. By using scRNA-seq on individual lineage-traced cells, investigators have underpinned the molecular adaptations that occur in different epidermal stem cell populations during wound repair. Prior to exiting their niche to contribute to wound repair, HFSCs upregulate an epidermal-like signature with rapid remodeling of receptor gene expression to enable interactions with wound-derived ligands from the stroma [57]. As these cells actively contribute to wound repair, they gradually lose their stem cell identity and become a part of the epidermal lineage. The transient manifestation of both hair follicle and epidermal signature genes in hair keratinocytes may be critical to achieve plasticity necessary for wound repair and also support the recently proposed concept of “lineage infidelity” [58].
Conclusion
In the past few years, in vivo imaging approaches, together with modern sequencing technologies, have yielded new insights and provided greater clarity into the spatiotemporal regulations which control epithelial differentiation in the skin epidermis and hair appendage during homeostasis and wound repair. With this knowledge, future studies can elucidate the multiplicity of external inputs that drive chromatin remodeling to achieve lineage-restricted fates and explore the intimate crosstalk between stem cells and their niches.
Acknowledgments:
We acknowledge the support of the Department of Dermatology, the Department of Cell and Developmental Biology, the Institute for Regenerative Medicine, and the entire stem cell community at Penn. G.R. was supported by training grant T32GM007229 from NIH/NIGMS. P.R. was supported by grants from NIH/NEI (R01EY030599) and from the American Cancer Society (RSG1803101DCC).
Footnotes
Competing interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huang S, Rompolas P: Two-photon microscopy for intracutaneous imaging of stem cell activity in mice. Exp Dermatol 2017, 26:379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obeidy P, Tong PL, Weninger W: Research Techniques Made Simple: Two-Photon Intravital Imaging of the Skin. J Invest Dermatol 2018, 138:720–725. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Yang B, Udo-Inyang I, Ji S, Ozog D, Zhou L, Mi QS: Research Techniques Made Simple: Single-Cell RNA Sequencing and its Applications in Dermatology. J Invest Dermatol 2018, 138:1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH: A single type of progenitor cell maintains normal epidermis. Nature 2007, 446:185–189. [DOI] [PubMed] [Google Scholar]
- 5.Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohée S, Sotiropoulou PA, Simons BD, Blanpain C: Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 2012, 489:257–262. [DOI] [PubMed] [Google Scholar]
- 6.Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T: Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol 2016, 18:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinley KL, Castillo-Azofeifa D, Klein OD: Tools and Concepts for Interrogating and Defining Cellular Identity. Cell Stem Cell 2020, 26:632–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V: Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 2016, 352:1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrelly O, Kuri P, Rompolas P: In vivo genetic alteration and lineage tracing of single stem cells by live imaging. Methods Mol Biol 2019, 1879:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marques-Pereira JP, Leblond CP: Mitosis and differentiation in the stratified squamous epithelium of the rat esophagus. Am J Anat 1965, 117:73–87. [DOI] [PubMed] [Google Scholar]
- 11.Piedrafita G, Kostiou V, Wabik A, Colom B, Fernandez-Antoran D, Herms A, Murai K, Hall BA, Jones PH: A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nat Commun 2020, 11:1–15.* Combining lineage tracing with cell cycle distribution analysis provided data which support a single progenitor model of epidermal homeostasis. The authors proposed that cell outcomes to differentiate or self-renew are unpredictable, but occur on average with an equal likelihood.
- 12.Dekoninck S, Hannezo E, Sifrim A, Miroshnikova YA, Aragona M, Malfait M, Gargouri S, de Neunheuser C, Dubois C, Voet T, et al. : Defining the Design Principles of Skin Epidermis Postnatal Growth. Cell 2020, 181:604–620.** This study used a multidisciplinary approach to resolve the cellular dynamics that mediate postnatal expansion of epidermal paw and tail skin. Epidermal expansion is mediated by a single population of developmental progenitors that exhibit a fixed fate imbalance towards self-renewal to facilitate organ growth.
- 13.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S: Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J Cell Biol 2002, 156:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokouchi M, Atsugi T, Van Logtestijn M, Tanaka RJ, Kajimura M, Suematsu M, Furuse M, Amagai M, Kubo A: Epidermal cell turnover across tight junctions based on Kelvin’s tetrakaidecahedron cell shape. Elife 2016, 5:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rübsam M, Mertz AF, Kubo A, Marg S, Jüngst C, Goranci-Buzhala G, Schauss AC, Horsley V, Dufresne ER, Moser M, et al. : E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat Commun 2017, 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckhart L, Lippens S, Tschachler E, Declercq W: Cell death by cornification. Biochim Biophys Acta - Mol Cell Res 2013, 1833:3471–3480. [DOI] [PubMed] [Google Scholar]
- 17.Ipponjima S, Umino Y, Nagayama M, Denda M: Live imaging of alterations in cellular morphology and organelles during cornification using an epidermal equivalent model. Sci Rep 2020, 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata T, Honda T, Egawa G, Yamamoto Y, Ichijo R, Toyoshima F, Dainichi T, Kabashima K: Transient elevation of cytoplasmic calcium ion concentration at a single cell level precedes morphological changes of epidermal keratinocytes during cornification. Sci Rep 2018, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, et al. : Defining Epidermal Basal Cell States during Skin Homeostasis and Wound Healing Using Single-Cell Transcriptomics. Cell Rep 2020, 30:3932–3947.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Drummond M, Guerrero-Juarez C, Tarapore E, MacLean A, Stabell A, Wu S, Gutierrez G, That B, Benavente C, et al. : Single cell transcriptomics of human epidermis reveals basal stem cell transition states. Nat Commun 2019, doi: 10.1101/784579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattarai SR, Begum S, Popow R, Ezratty EJ: The ciliary GTPase Arl3 maintains tissue architecture by directing planar spindle orientation during epidermal morphogenesis. Dev 2019, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lough KJ, Byrd KM, Descovich CP, Spitzer DC, Bergman AJ, Beaudoin GM, Reichardt LF, Williams SE: Telophase correction refines division orientation in stratified epithelia. Elife 2019, 8:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Box K, Joyce BW, Devenport D: Epithelial geometry regulates spindle orientation and progenitor fate during formation of the mammalian epidermis. Elife 2019, 8:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ipponjima S, Hibi T, Nemoto T: Three-dimensional analysis of cell division orientation in epidermal basal layer using intravital two-photon microscopy. PLoS One 2016, 11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dias Gomes M, Letzian S, Saynisch M, Iden S: Polarity signaling ensures epidermal homeostasis by coupling cellular mechanics and genomic integrity. Nat Commun 2019, 10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aragona M, Sifrim A, Malfait M, Song Y, Van Herck J, Dekoninck S, Gargouri S, Lapouge G, Swedlund B, Dubois C, et al. : Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 2020, 584(7820):268–273.** This study represents an elegant use of a tractable mouse model to dissect the mechanism by which mechanical stretching induces expansive skin growth. The authors uncovered an adaptive mechanism of cytoskeletal re-organization that alters the fate of individual keratinocytes through the transcription factors YAP, TAZ, and MAL.
- 27.Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM, Greco V: Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell 2018, 23:677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie S, Skotheim JM: A G1 Sizer Coordinates Growth and Division in the Mouse Epidermis. Curr Biol 2020, 30:916–924.**First in vivo study to report a mechanism for cell size regulation in a mammalian organ. Through longitudinal quantification of epidermal keratinocyte growth, the authors show that stem cells in the epidermis couple their growth with cell-cycle progression through a G1 sizer.
- 29.Hiratsuka T, Bordeu I, Pruessner G, Watt FM: Regulation of ERK basal and pulsatile activity control proliferation and exit from the stem cell compartment in mammalian epidermis. Proc Natl Acad Sci U S A 2020, 117:17796–17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, De Sauvage FJ, Locksley RM, Klein OD: Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 2018, 559:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, et al. : YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 2018, 22:35–49.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao Q, Hill MC, Chen F, Mo Q, Ku AT, Ramos C, Sock E, Lefebvre V, Nguyen H: SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat Commun 2019, 10:1–20.* Bulk RNA-seq reveals conversion of epidermal wound cells to an embryonic-like state dictated by SOX11 and SOX4. These genes play a critical role in wound repair by reactivating an embryonic gene program while loss of these genes leads to impaired wound healing.
- 33.Peters F, Tellkamp F, Brodesser S, et al. Murine Epidermal Ceramide Synthase 4 Is a Key Regulator of Skin Barrier Homeostasis. J Invest Dermatol 2020, (20)30160–3. doi: 10.1016/j.jid.2020.02.006 [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E: Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 2017, 169:483–496.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi R, Grzenda A, Allison TF, Rawnsley J, Balin SJ, Sabri S, Plath K, Lowry WE: Defining Transcriptional Signatures of Human Hair Follicle Cell States. J Invest Dermatol 2020, 140:764–-773.e4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rompolas P, Mesa KR, Greco V: Spatial organization within a niche as a determinant of stem-cell fate. Nature 2013, 502:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, Kasper M: Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 2016, 3:221–237.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, Kasper M: The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 2020, 26:441–457.e7.* By combining single-cell RNA-seq with single-molecule RNA FISH, this study provides a comprehensive cell atlas of full thickness mouse skin that defines gene expression profiles and spatial locations of 56 epithelial and stromal cell populations and states during hair growth and rest. The unprecedented molecular detail in this investigation reveals cell types and states that coordinate hair growth, progenitor lineage commitment, fibroblast heterogeneity, and the capacity for epithelial-stromal interactions.
- 39.Andersen MS, Hannezo E, Ulyanchenko S, Estrach S, Antoku Y, Pisano S, Boonekamp KE, Sendrup S, Maimets M, Pedersen MT, et al. : Tracing the cellular dynamics of sebaceous gland development in normal and perturbed states. Nat Cell Biol 2019, 21:924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veniaminova NA, Grachtchouk M, Doane OJ, Peterson JK, Quigley DA, Lull MV., Pyrozhenko DV, Nair RR, Patrick MT, Balmain A, et al. : Niche-Specific Factors Dynamically Regulate Sebaceous Gland Stem Cells in the Skin. Dev Cell 2019, 51:326–340.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, Greco V: Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 2012, 487:496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avigad Laron E, Aamar E, Enshell-Seijffers D: The Mesenchymal Niche of the Hair Follicle Induces Regeneration by Releasing Primed Progenitors from Inhibitory Effects of Quiescent Stem Cells. Cell Rep 2018, 24:909–921.e3. [DOI] [PubMed] [Google Scholar]
- 43.Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, et al. : Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Rep 2016, 14:3001–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Z, Jiang K, Xu Z, Huang H, Qian N, Lu Z, Chen D, Di R, Yuan T, Du Z, et al. : Hoxc-Dependent Mesenchymal Niche Heterogeneity Drives Regional Hair Follicle Regeneration. Cell Stem Cell 2018, 23:487–500.e6. [DOI] [PubMed] [Google Scholar]
- 45.Fan SMY, Chang YT, Chen CL, Wang WH, Pan MK, Chen WP, Huang WY, Xu Z, Huang HE, Chen T, et al. : External light activates hair follicle stem cells through eyes via an ipRGC–SCN–sympathetic neural pathway. Proc Natl Acad Sci U S A 2018, 115:E6880–E6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shwartz Y, Gonzalez-Celeiro M, Chen CL, Pasolli HA, Sheu SH, Fan SMY, Shamsi F, Assaad S, Lin ETY, Zhang B, et al. : Cell Types Promoting Goosebumps Form a Niche to Regulate Hair Follicle Stem Cells. Cell 2020, 182:578–593.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li KN, Jain P, He CH, Eun FC, Kang S, Tumbar T: Skin vasculature and hair follicle cross- talking associated with stem cell activation and tissue homeostasis. Elife 2019, 8:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin T, Gonzalez D, Rompolas P, Greco V: Flexible fate determination ensures robust differentiation in the hair follicle. Nat Cell Biol 2018, 20:1361–1369.** Live imaging of single stem cells in hair follicles of live mice revealed fate determination during homeostasis to be unexpectedly flexible. Stem cells in the hair germ are spatially primed to generate lineage-specific progenitors but can flexibly establish all differentiation lineages while matrix progenitor fates are continuously channeled as a consequence of their unexpected dynamic relocation around the dermal papilla.
- 49.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. : Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 2015, 521:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adam RC, Yang H, Ge Y, Lien WH, Wang P, Zhao Y, Polak L, Levorse J, Baksh SC, Zheng D, et al. : Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell Stem Cell 2018, 22:398–413.* This study combined in vivo chromatin and genetic analyses to investigate how signaling from the microenvironment interfaces with chromatin to drive lineage diversification of hair follicle stem cells. Diverse fates are acquired as a result of temporally layered signaling effectors atop chromatin platforms established by simultaneous BMP-inhibitory and WNT signaling.
- 51.Yadav T, Quivy JP, Almouzni G: Chromatin plasticity: A versatile landscape that underlies cell fate and identity. Science 2018, 361:1332–1336. [DOI] [PubMed] [Google Scholar]
- 52.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ: Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G: Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 2005, 11:1351–1354. [DOI] [PubMed] [Google Scholar]
- 54.Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA: Epidermal stem cells arise from the hair follicle after wounding. FASEB J 2007, 21:1358–1366. [DOI] [PubMed] [Google Scholar]
- 55.Page ME, Lombard P, Ng F, Göttgens B, Jensen KB: The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 2013, 13:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vagnozzi AN, Reiter JF, Wong SY: Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle 2015, 14:3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joost S, Jacob T, Sun X, Annusver K, La Manno G, Sur I, Kasper M: Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep 2018, 25:585–597.e7. [DOI] [PubMed] [Google Scholar]
- 58.Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CPJ, Polak L, Yuan S, Elemento O, et al. : Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell 2017, 169:636–650.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]