Abstract
Primary mouse hepatocytes isolated from genetically defined and/or diverse lines and disease models are a valuable resource for studying the impact of genetic and environmental factors on drug response and disease. However, standard monolayer cultures result in a rapid decline in mouse hepatocyte viability and functionality. Therefore, we evaluated 3D spheroid methodology for long-term culture of primary mouse hepatocytes, initially to support investigations of drug-induced liver injury (DILI). Primary hepatocytes isolated from male and female C57BL/6J mice were used to generate spheroids by spontaneous self-aggregation in ultra-low attachment plates. Spheroids with well-defined perimeters were observed within 5 days after seeding and retained morphology, ATP, and albumin levels for an additional 2 weeks in culture. Global microarray profiling and quantitative targeted proteomics assessing 10 important drug metabolizing enzymes and transporters demonstrated maintenance of mRNA and protein levels in spheroids over time. Activities for 5 major P450 enzymes were also stable and comparable to activities previously reported for human hepatocyte spheroids. Time- and concentration-dependent decreases in ATP and albumin were observed in response to the DILI-causing drugs acetaminophen, fialuridine, AMG-009, and tolvaptan. Collectively, our results demonstrate successful long-term culture of mouse hepatocytes as spheroids and their utility to support investigations of DILI.
Keywords: drug-induced liver injury, primary mouse hepatocytes, 3D spheroids, gene expression, quantitative targeted absolute proteomics
INTRODUCTION
Primary human hepatocytes are the gold standard for studying hepatic drug response and disease in vitro. In particular, cultured human hepatocytes are an important tool for the preclinical evaluation of drug-induced liver injury (DILI), the most common cause of adverse drug reaction resulting in warnings and withdrawals of numerous medications (Watkins, 2011). As a result, significant effort has been invested to improve the long-term viability and phenotypic relevance of this model system. One promising approach is the cultivation of human hepatocytes in a 3D configuration, which has been shown to better preserve the in vivo phenotype due to the extensive formation of cell-cell contacts, reestablishment of cell polarity, and endogenous production of extracellular matrices (Messner et al., 2013; Tostoes et al., 2012). Recent studies have also demonstrated that hepatocytes in 3D spheroid cultures closely resemble the in vivo liver proteome, have bile canaliculi, and have stable liver-specific functionalities such as enzyme activity for at least 5 weeks of culture (Bell et al., 2016). Furthermore, spheroids allow for repeated drug exposures in long-term toxicity studies and are suitable to study a variety of mechanisms associated with DILI (Bell et al., 2016; Hendriks et al., 2016).
Genetically engineered mice, mouse models of liver disease, and mouse genetic reference populations are unique tools to study biology and pharmacology that would not otherwise be possible in human models. Therefore, primary mouse hepatocytes are an important resource to supplement human in vitro models and study the impact of genetic and environmental factors on hepatic drug disposition, toxicity, and disease. However, primary mouse hepatocytes are difficult to maintain in culture due to the higher metabolic rate and oxygen demand compared to human cells (Martinez et al., 2010; Swales et al., 1996). As a result, studies are limited to only a few days of exposure, which may not be sufficient to elicit the appropriate response (Atienzar et al., 2016). While several groups have reported the ability to culture primary mouse hepatocytes as spheroids (Chang and Hughes-Fulford, 2014; Vorrink et al., 2018), studies characterizing the impact of 3D culture on primary mouse hepatocytes are lacking. Only recently has the extensive characterization of a 3D mouse model been reported (Nudischer et al., 2020). The authors of this study describe the long-term viability and functionality of 3D liver spheroids generated by self-aggregation in ultra-low attachment (ULA) plates using freshly isolated cells from male C57Bl/6 mice. Cytotoxicity in response to both 2 and 8 days of exposure to DILI compounds is also shown (Nudischer et al., 2020).
In the present study, we also evaluate the impact of 3D spheroid methodology on the long-term morphology, viability, and functionality as well as the suitability of primary mouse hepatocytes for prolonged toxicity studies, but expand on the Nudischer et al. (2020) findings in several ways. Primary mouse hepatocytes were isolated from both male and female C57BL/6J mice, cryopreserved, and then used to generate spheroids using similar ULA methodology. Biochemical approaches, global gene expression profiling, and quantitative targeted absolute proteomics (QTAP) were performed on the spheroids cultured for 1, 7, and 14 days post spheroid formation. Cytotoxicity was measured in response to increasing concentrations of DILI drugs after 1, 7 and 14 days of exposure. Findings from this study similarly demonstrate improved phenotypic relevance of the 3D spheroid model for culturing mouse hepatocytes but also show the ability to use cryopreserved cells for spheroid formation, maintenance of an expanded set of important drug metabolizing enzymes and transporter (DMET) mRNAs, proteins, and activity levels, and comparison of spheroids generated from male and female hepatocytes. Furthermore, we show enhanced sensitivity to different DILI drugs in the mouse hepatocyte spheroids over time, which further supports the use of the 3D culture model for investigations of DILI.
MATERIALS AND METHODS
Hepatocyte isolation and cryopreservation
Male and female C57BL/6J mice, 12 weeks of age, were purchased from Jackson Laboratory and allowed to acclimate in-house for 7–10 days prior to study initiation. Water and food were provided ad libitum. Primary mouse hepatocytes were isolated using a two-step collagenase perfusion method as described in (Martinez et al., 2010). Hepatocytes were pooled from 3 mice for a single cryopreserved preparation (N=1) and a total of N=6 cryopreserved preparations (3 per sex) were made using 18 mice (9 males and 9 females) for the study. Throughout this manuscript, biological and technical replicates are designated as “N” and “n”, respectively. Hepatocyte preparations had a viability of 90% or more and were cryopreserved at a concentration of 5 million cells per vial in CryoPreserve solution (VitroPrep, Research Triangle Park, NC, USA). All studies were conducted in accordance with the guidelines for Animal Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the University of North Carolina, Chapel Hill, USA.
Hepatocyte spheroid culture
Cryopreserved hepatocytes were thawed in InVitroGRO CP Rodent Media (BioIVT, Westbury, NY, USA) supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA) and seeded at the optimized density of 1000 cells per well into round bottom, ULA 96-well plates (Corning, Corning, NY, USA) in 100 μL volume. Spheroids were switched to maintenance media (InVitroGRO CP Rodent Media without fetal bovine serum) on Day 4 and media was changed every 2–3 days (by removing and adding back 80 μL from 100 μL media per well). Well-defined spheroids were formed by spontaneous self-aggregation on culture day 5 and maintained until culture Day 19 as per the culture timelines in Supplementary Figure 1.
Spheroid morphology and size
Spheroid morphology was evaluated in n=1 well per time point from all N=6 preparations (3 per sex) using phase contrast microscopy photomicrographs captured at 10X magnification (Nikon Eclipse TS-100 Phase Contrast Microscope). Spheroid diameter was calculated using a total of 334 spheroids from a single male and a single female hepatocyte preparation over time: n=100 spheroids Day 1 (corresponds to culture Day 6, 43 male, 57 female), n=119 spheroids Day 7 (corresponds to culture Day 12, 66 male, 53 female) and n=115 spheroids Day 14 (corresponds to culture Day 19, 62 male, 53 female). Live spheroids were scanned at 10X using the CellInsight CX7 LZR High Content Screening Platform (Thermo Scientific, Waltham, MA, USA) in brightfield mode. Area was estimated using the HCS Studio Cell Analysis Software v2 (Thermo Scientific, Waltham, MA, USA) and used to calculate spheroid diameter as previously described (Ramaiahgari et al., 2017).
Histology
Approximately 150 spheroids from a single male hepatocyte preparation were collected from ULA plates into Eppendorf tubes on Days 1, 7 and 14 post spheroid formation, fixed in 10% neutral buffered formalin for 10 minutes at room temperature, washed with phosphate buffered saline and stored at 4 °C until sample preparation for processing, paraffin embedding, sectioning (5 μm) as previously described (Clayton et al., 2018). Spheroid sections were stained with hematoxylin and eosin (H&E) for morphological evaluation. Slides were scanned using an Aperio AT2 Digital Whole Slide Scanner (Leica Biosystems, Inc.). After scanning, images were captured using Aperio ImageScope v12.4.3.
Biochemical assays
Cellular ATP was measured in n=8 individual spheroids per time point from N=6 preparations (3 per sex) using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) according to manufacturer’s instructions. Media (75 μL out of 100 μL per well) was collected from same wells used for cellular ATP measurements and stored at −80 °C for albumin measurements. Albumin content in the media was quantified using a mouse albumin ELISA quantification set (Bethyl Laboratories, Montgomery, TX, USA) per manufacturer’s instructions as previously described (Norona et al., 2016). Raw data was collected using a SpectraMax M3 microplate reader (Molecular Devices, San Jose, CA, USA). A standard curve was used to calculate ATP (nM) and albumin (ng/mL) for all samples.
Cytochrome P450 enzyme activity
A total of 8 spheroids per time point from N=4 preparations (2 per sex) were incubated with a mixture of probe substrates containing midazolam (10 μM), dextromethorphan (15 μM), phenacetin (100 μM), amodiaquine (10 μM) and tolbutamide (100 μM) (Sigma, St. Louis, MO, USA) for 4 hours. Supernatants were pooled from n=8 spheroids for each time point and frozen. Formed metabolites (1-OH- midazolam, dextrorphan, acetaminophen, desethyl-amodiaquine and OH-tolbutamide) were quantified by LC-MS/MS (Biotranex, NJ, USA).
Gene expression profiling
Total cellular RNA was isolated from approximately 200 spheroids pooled per time point from N=4 preparations (2 per sex) using an RNeasy Mini Kit (Qiagen, Germantown, MD, USA). The quantity and integrity of the RNA was evaluated spectrophotometrically with an Agilent 2200 TapeStation. Double-stranded cDNA was synthesized using the GeneChip WT PLUS Reagent Kit (Affymetrix, Santa Clara, CA, USA). Labeled cRNA was fragmented and prepared for hybridization on Affymetrix Clariom S HT, mouse arrays. The Affymetrix Gene Titan system was used to perform the hybridization, washing and scanning of the arrays. Affymetrix CEL files were normalized using Robust Multi-array Average method with a log base 2 transformation (Irizarry et al., 2003). Gene expression analysis was performed as previously described (Mosedale et al., 2018a; Mosedale et al., 2018b). Briefly, data summarization and QC was performed using the Transcriptome Analysis Console software version 4.0 (ThermoFisher, Waltham, MA, USA). A filtering step was performed to remove low expression probe sets resulting in 13,081 total mRNAs detected. Pathways enriched among statistically significant, differentially expressed genes in the data were identified using the Tox Analysis module in Ingenuity Pathway Analysis (Ingenuity Systems; Build version: exported; Content version: 51963813). Gene expression data generated for this manuscript can be downloaded in its entirety from the Gene Expression Omnibus repository under the accession number GSE152173.
Quantitative targeted absolute proteomics
QTAP was performed using approximately 100 spheroids per time point from N=6 preparations (3 per sex) by nanoLC-MS/MS as previously described with slight modifications (Fallon et al., 2016; Khatri et al., 2019; Ramsby and Makowski, 1999). Briefly, samples (20 μg protein) were evaporated to dryness, denatured, reduced, blocked with iodoacetamide and spiked with 1 pmol of stable isotope labeled peptide standards before trypsin digestion. Clean up was with solid phase extraction (SPE) Strata™-X 33 μm Polymeric Reversed Phase cartridges (Phenomenex, Torrance, CA, USA). Analysis was performed on a nanoACQUITY (Waters, Milford, MA, USA) interfaced with a SCIEX QTRAP 5500 hybrid mass spectrometer operated in the MRM mode and equipped with a NanoSpray III source. System control was via Analyst 1.5 software (SCIEX, Framingham, MA, USA) and nanoACQUITY UPLC Console. Peaks were integrated using MultiQuant 2.0.2 (SCIEX, Framingham, MA, USA) with a limit of quantification set at 0.1 pmol/mg protein for all peptides. Peptide sequences targeted for analysis are included in Supplementary Table 1.
Drug treatments
Drug treatments were performed in n=6 spheroids per time point from a single male preparation (N=1, pooled hepatocytes from 3 mice). ATP and albumin data were generated from at least n=3–6 spheroids per time point, drug and concentration, except for a single concentration on day 14 for both acetaminophen (APAP) and tolvaptan (for which the data was obtained from n=2 spheroids due to spheroid loss during repeated media changes over time). APAP, fialuridine, and tolvaptan were purchased from Sigma (St. Louis, MO, USA) and AMG-009 was purchased from MedKoo Biosciences (Morrisville, NC, USA). Stock solutions of chemicals were prepared in DMSO at 500X final concentration and diluted in culture media to create dosing solutions with DMSO at 0.2% v/v. Dosing solutions of APAP were prepared directly in culture media. The highest concentration for each drug was based on the maximum solubility limit determined either experimentally or from manufacturer’s product sheet. As in baseline cultures, media was changed and fresh compound added every 2–3 days for the length of drug exposure.
Statistical analyses
Statistical analyses for biochemical, morphological, and toxicity endpoints were performed using GraphPad Prism statistical software version 8.0 (GraphPad Software Inc.). Mean values of biochemical and enzyme activity assays for all time points were compared using a one-way analysis of variance (ANOVA). Comparisons between Days 7 and 14 to Day 1 were performed using a Dunnett’s multiple comparison test with p<0.05 considered statistically significant. For drug cytotoxicity assessments, data was normalized (to respective vehicle control on each day) and dose response curves were generated using log (drug concentration) vs normalized response with a 3PL fit. This model forces the curve to run from 100% down to 0% and determines the EC50 as the concentration that provokes a response equal to 50%. Gene expression and proteomic analyses were performed using Partek Genomics Suite version 7.0 (Partek Inc.). For the gene expression data, an initial analysis was performed with all 13,081 mRNAs detected and then a targeted analysis was performed with just the mouse orthologues of 10 key DMET mRNAs known to be important in drug metabolism and toxicity in humans (Supplementary Table 1). For all analyses, differences were identified using an ANOVA model with linear contrasts. Preparation was included as random factor in the analysis. Probability values were adjusted for multiple comparisons using a false discovery rate of 5% (FDR = 0.05) (Benjamini and Hochberg, 1995). An absolute value fold change (FC) cutoff of >2 was also used as noted below.
RESULTS
Primary mouse hepatocyte spheroids maintain morphology, viability, and function over time in culture
Primary mouse hepatocyte spheroids with well-defined perimeters were formed within 5 days after seeding and appropriate morphology was maintained for an additional 2 weeks in culture (Figures 1A and B). No significant difference in ATP levels, albumin production or spheroid diameter was observed in spheroids over time (Figures 1C, D and E). No sex effects were observed in morphology, ATP, albumin, or spheroid diameter at any time in culture (Supplementary Figure 2).
Figure 1.
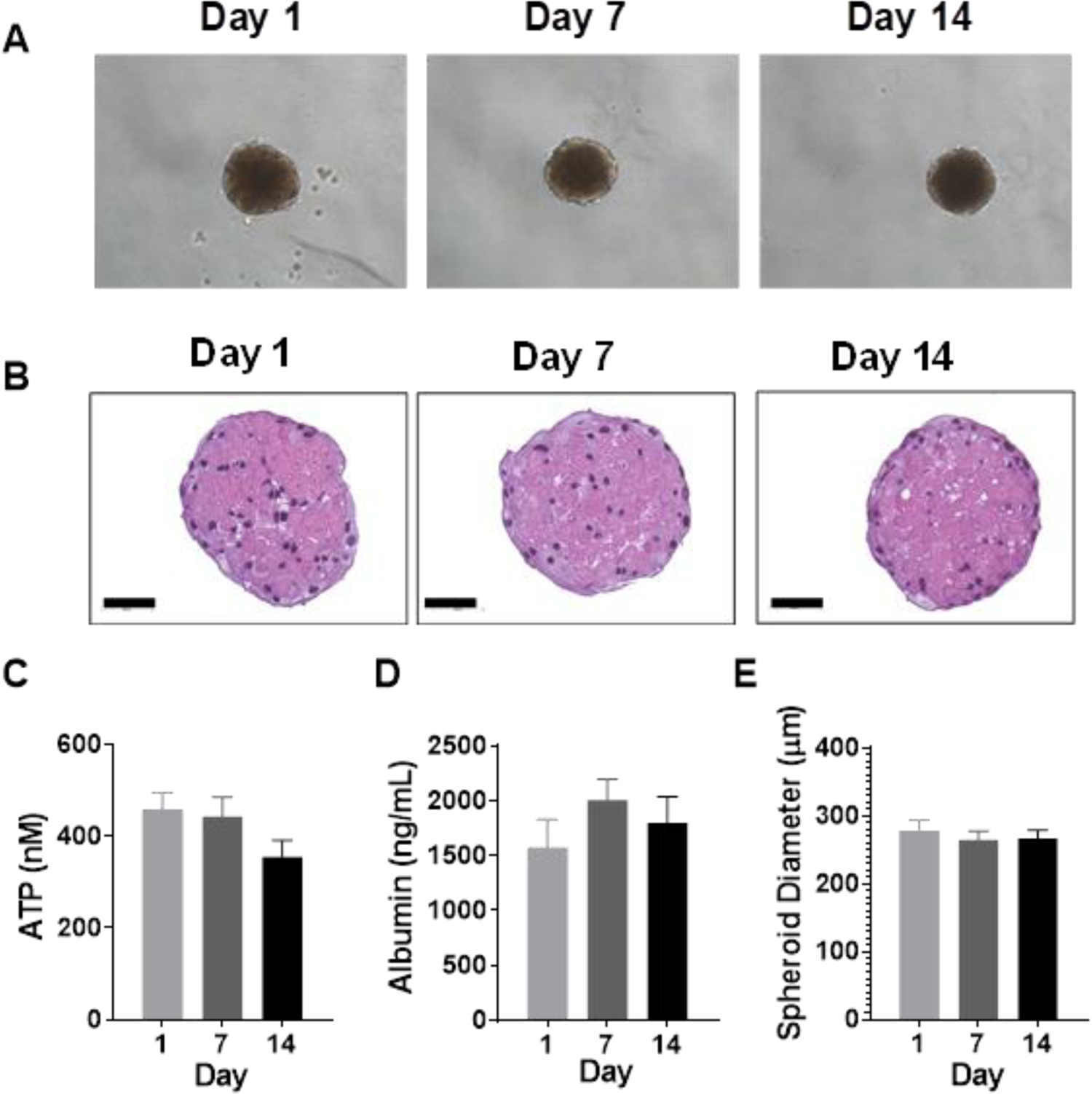
(A) Representative phase-contrast photomicrographs of spheroids at 10X magnification on Days 1, 7, and 14 post spheroid formation. (B) Representative H&E staining of spheroids over time. Bars indicate 50 μm. (C) ATP, (D) albumin, and (E) spheroid diameter measured over time. Data represent the mean+SEM from N=6 preparations (3 per sex) for ATP and albumin, and N=2 preparations (1 per sex) for spheroid size. No significant differences were observed in baseline ATP, albumin, or spheroid diameter over time as determined by repeated measures ANOVA with Dunnett’s multiple comparison test comparing Days 7 and 14 to Day 1.
Levels of key drug metabolizing enzymes and transporter mRNAs are constant over time in spheroid culture
Only 287 of 13,081 total mRNAs detected (2.2%) were differentially expressed in spheroids over time. The largest number of changes were observed in spheroids on Day 14, with 159 mRNAs increased and 123 mRNAs decreased compared to spheroids on Day 1 (Figure 2A). Pathways enriched among these upregulated and downregulated genes are shown in Figure 2B. We also performed a targeted analysis of the mouse orthologues of 10 key DMET mRNAs known to be important in drug metabolism and toxicity in humans (Supplementary Table 1). No significant differences were observed in the levels of any of the DMET transcripts over time in spheroid culture (Figure 3). However, 2 of these 10 mRNAs were higher in spheroids from females than males on specific days of analysis including Cyp2c29 and Cyp3a11 (Supplementary Figure 3).
Figure 2.
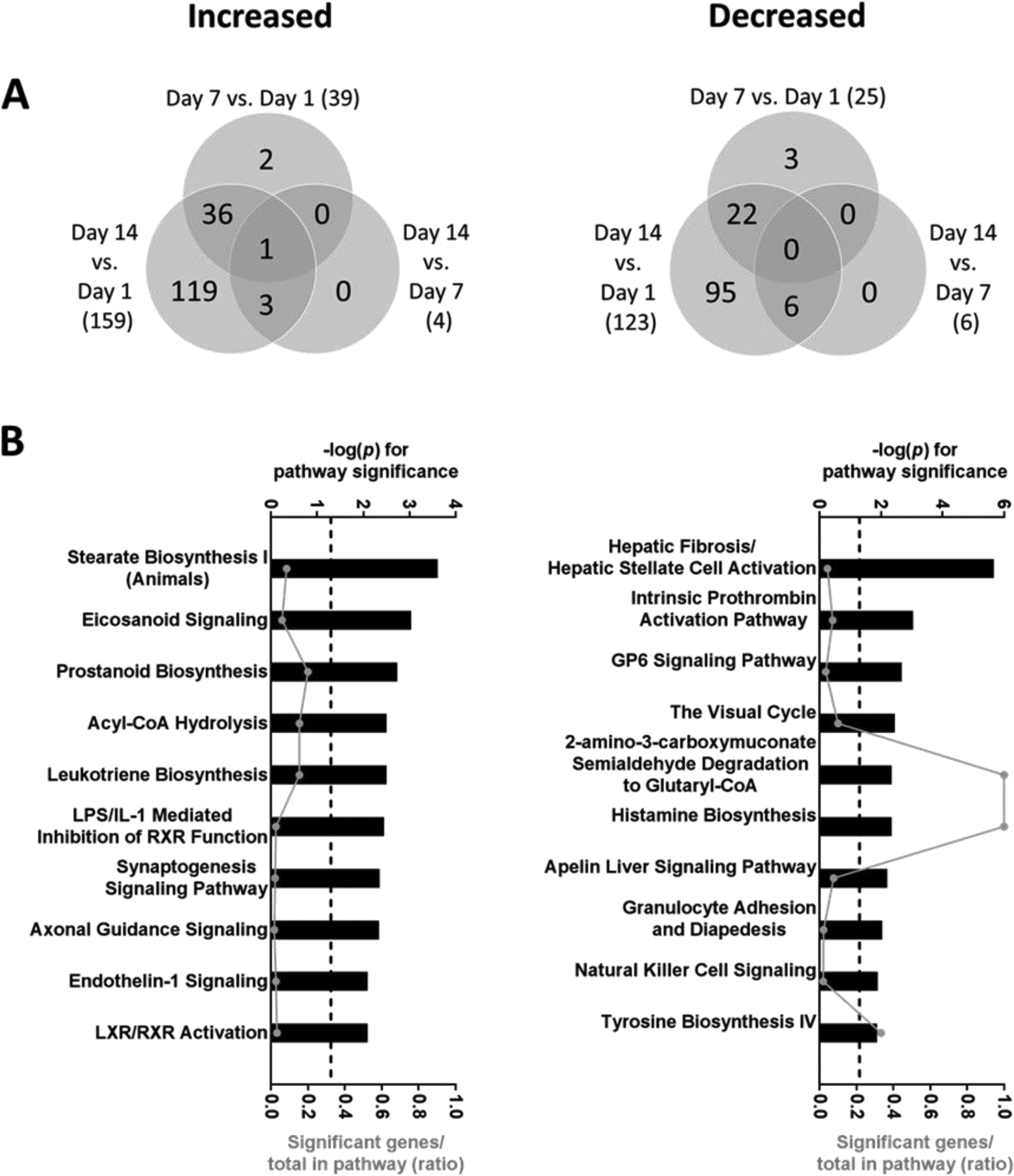
(A) Venn diagrams showing overlap of genes significantly increased or decreased on Days 7 and 14 compared to Days 1 and 7 post spheroid formation. (B) Top 10 most significantly enriched pathways among transcripts that were increased or decreased on Day 14 vs. Day 1. Pathway significance is plotted on the upper x-axis and represented by the bars on the graphs. The ratio of significant genes to total genes in the pathway is plotted on the lower x-axis and represented by the dots (connected by a line) on the graphs. The dashed line represents a threshold for significance set at −log10(p) >1.3. Differentially expressed genes were determined by ANOVA with linear contrasts (FDR p<0.05 and lFCl>2). Data was obtained from N=4 preparations (2 per sex).
Figure 3.
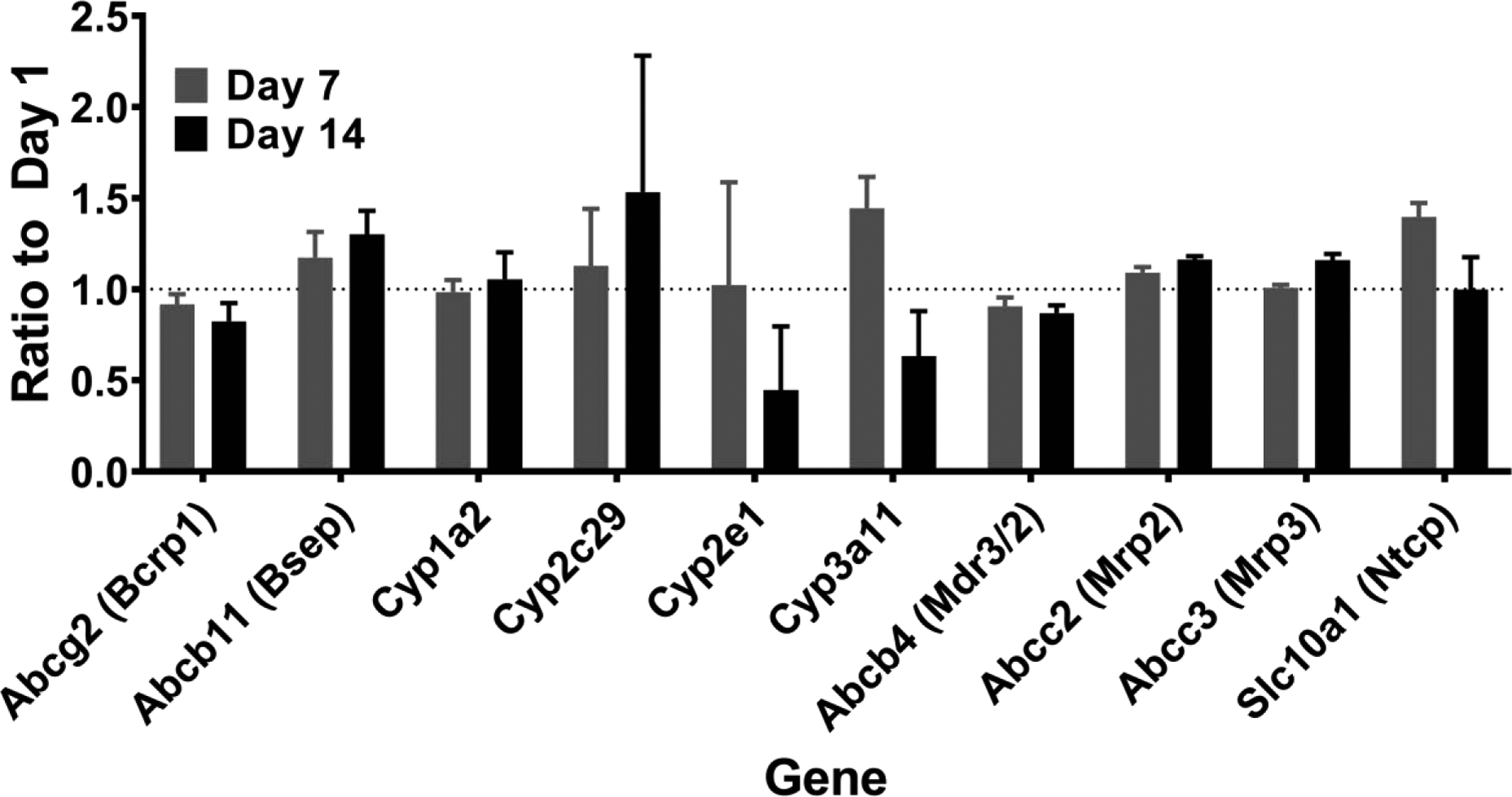
Ratio of mRNA levels for 10 key drug metabolizing enzymes and transporters on Days 1, 7 and 14 compared to Day 1 post spheroid formation. Data represent the geometric mean+SD of the ratio from repeated measurements of N=4 preparations (2 per sex). No differences over time were determined by ANOVA with linear contrasts (FDR p<0.05 and lFCl>2). Corresponding proteins, if different than the gene name, are listed within brackets.
Levels of key drug metabolizing enzymes and transporter proteins are constant over time in spheroid culture
The same 10 key DMET genes were also measured at the protein level by QTAP. Only Cyp2e1 protein showed a significant decline over time in culture (Figure 4). However, 3 of these 10 proteins were higher in spheroids from females than males on specific days of analysis including Cyp2c29, Cyp3a11, and Ntcp (Supplementary Figure 4).
Figure 4.
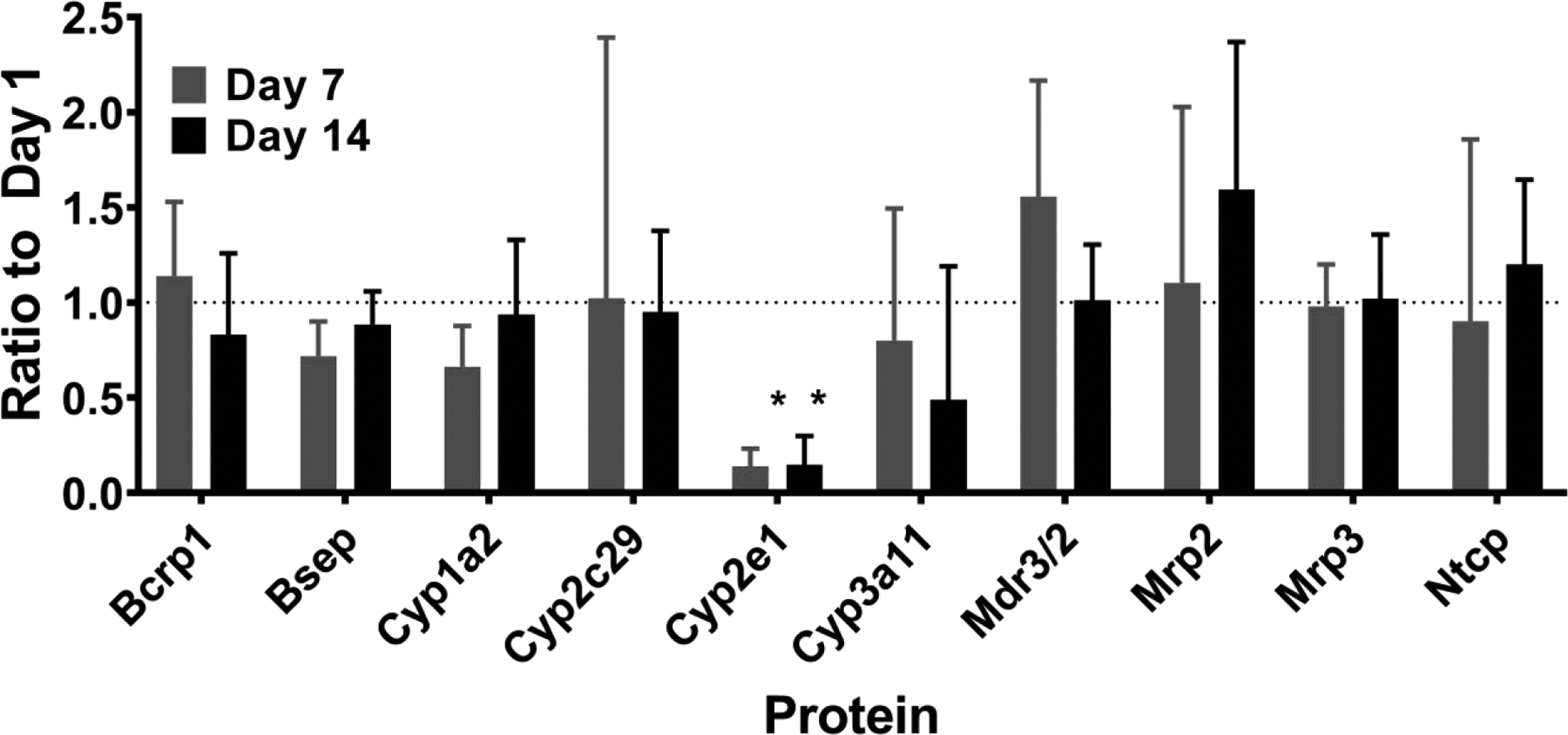
Ratio of protein levels for 10 key drug metabolizing enzymes and transporters on Days 1, 7 and 14 compared to Day 1 post spheroid formation. Data represent the geometric mean+SD of the ratio from repeated measurements of N=6 preparations (3 per sex). * indicates FDR p<0.05 and lFCl>2 as determined by ANOVA with linear contrasts comparing Days 7 and 14 to Day 1.
Activity levels of most key cytochrome P450 enzymes are stable over time in spheroid culture
Enzyme activity was evaluated by measuring the formation of metabolites from probe substrates corresponding to five major human cytochrome P450 enzymes: CYP1A2 (phenacetin), CYP2C8 (amodiaquine), CYP2C9 (tolbutamide), CYP2D6 (dextromethorphan) and CYP3A4 (midazolam) as previously described for human hepatocyte spheroids (Bell et al., 2016). No significant differences were observed in the levels of any metabolites formed over time in spheroid culture (Figure 5). However, levels of metabolite formation from phenacetin and midazolam were higher in females than males on Day 1 and 7, respectively (Supplementary Figure 5).
Figure 5.
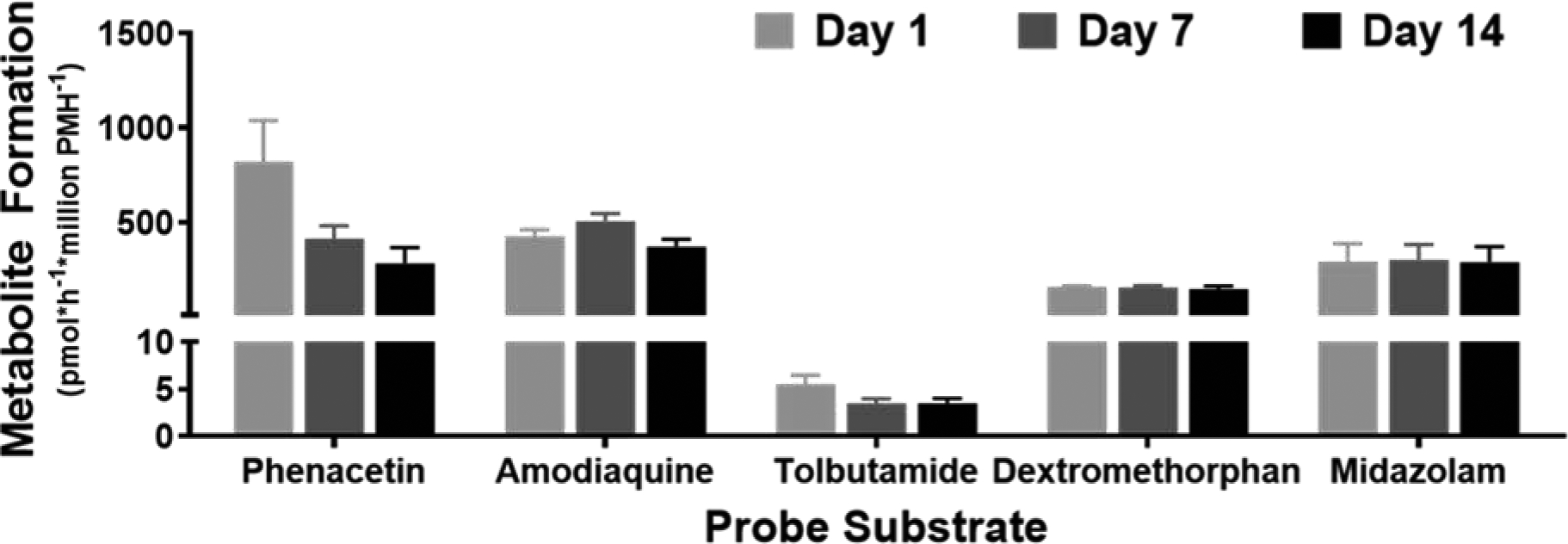
Metabolite levels measured after a 4-hour incubation with cytochrome P450 substrate cocktail. Data are presented as mean+SEM from N=4 preparations (2 per sex). No significant differences were observed by repeated measures ANOVA with Dunnett’s multiple comparison test comparing Days 7 and 14 to Day 1 for each metabolite.
Hepatotoxicity to DILI drugs was increased with prolonged drug exposures using the mouse spheroid model
Decreased viability (ATP) and functionality (albumin) were observed in spheroids treated with increasing concentrations of four major DILI drugs that operate via different mechanisms of toxicity: APAP, fialuridine, tolvaptan, and AMG-009 (Figure 6). EC50 values for each endpoint were generally decreased (below 100-fold plasma Cmax concentrations) in response to increased length of exposure with each drug (Bowsher et al., 1994; Ryan et al., 2018; Sevilla-Tirado et al., 2003; Shoaf et al., 2007). EC50 values for albumin responses were lower than ATP, although the variability with albumin measurements was higher.
Figure 6.
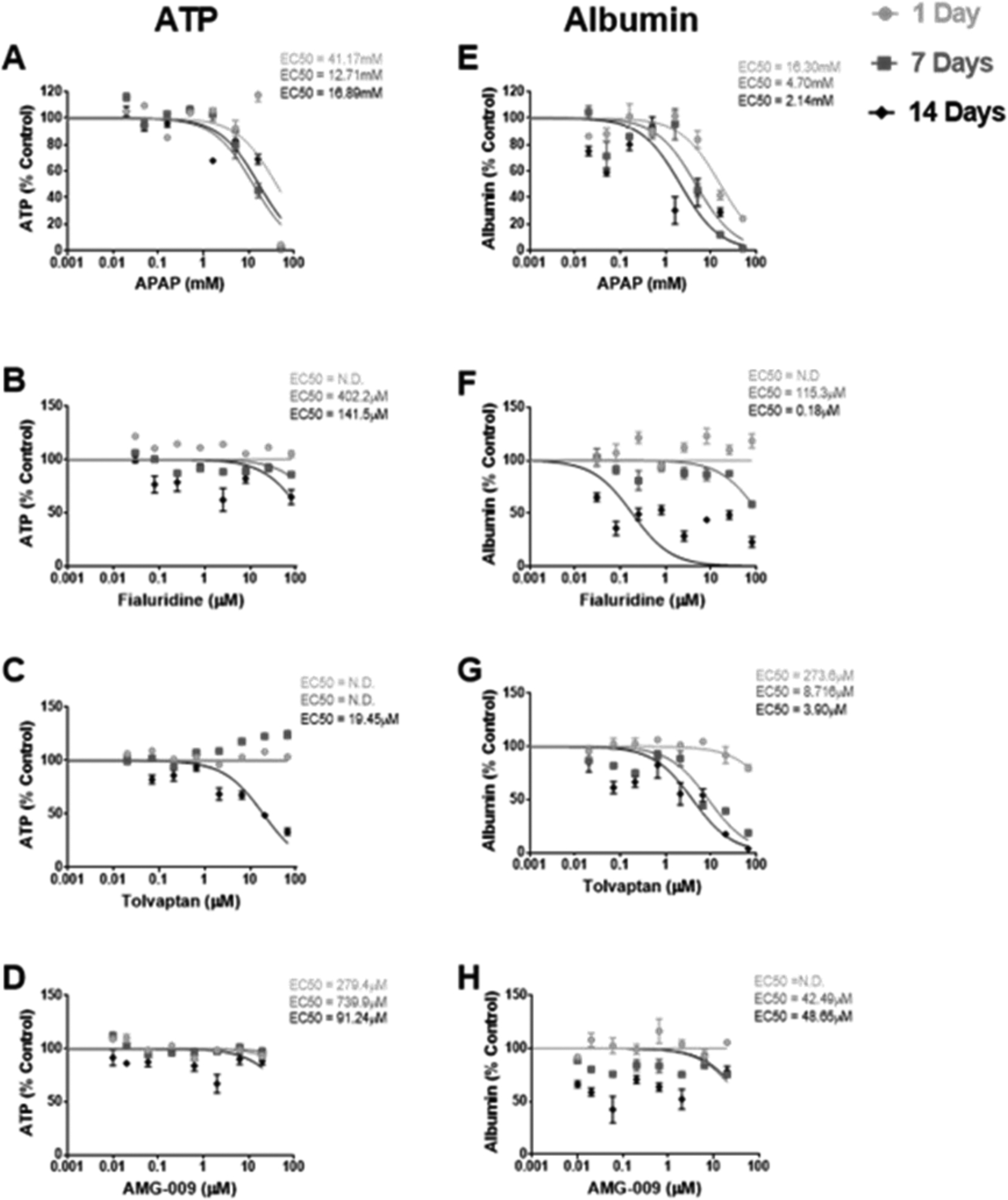
Dose-response curves for ATP and albumin as % control in response to 1, 7, and 14 days of treatment with increasing concentrations of (A,E) acetaminophen (APAP), (B,F) fialuridine, (C,G) tolvaptan, and (D,H) AMG-009. The concentration at which a 50% effect was observed (EC50) for each treatment duration is reported on the respective graph. Data is represented as mean±SEM for n=3–6 spheroids (from a single male preparation) per concentration, time point, and drug and corresponding least squares curve fit of log (drug concentration) vs. normalized response. n=2 spheroids were used for a single concentration and time point (day 14) for APAP and tolvaptan due to spheroid loss during repeated media changes over time.
DISCUSSION
Extensive characterization of primary human hepatocyte spheroids (Bell et al., 2018; Bell et al., 2016; Messner et al., 2018) has facilitated their adoption for preclinical toxicity testing (Proctor et al., 2017; Vorrink et al., 2018) and to study mechanisms of DILI (Bell et al., 2016; Hendriks et al., 2016; Jiang et al., 2019; Nautiyal et al., 2020). Although human hepatocyte models are important for translational work, genetically engineered mice, mouse models of liver disease, and mouse genetic reference populations provide unique tools to study the impact of genetic and environmental factors on hepatic drug disposition, toxicity, and disease that would not otherwise be possible in human models. Rodent models offer further advantages over human cell-based models in the in vitro setting as they provide a reliable, reproducible, and cost-effective supply of cells resulting in consistent outcomes and responses. Using the spheroid approach will also substantially increase the power (number of replicates), content (number of endpoints), and throughput (speed) of these studies while decreasing the need for live animal studies. To this end, the first report of an extensive characterization of a 3D mouse model was recently published (Nudischer et al., 2020). The authors describe cultivation of 3D liver spheroids containing freshly isolated cells derived from male C57Bl/6J mice (Nudischer et al., 2020). Our study adds to this work by including a comparison of phenotypic differences between cells from male and female mice as well as a more extensive characterization of the gene expression profile, DMET protein levels, and P450 enzyme activity. Furthermore, hepatocytes in our study were cryopreserved prior to use, which would facilitate convenience and the ability to culture hepatocytes from multiple mouse models simultaneously in future studies.
Similar to Nudischer et al. (2020), mouse spheroids in our study were generated by spontaneous self-aggregation in ULA plates. This is a simple, rapid, cost-effective and efficient method for generating and culturing 3D spheroids, and there is an abundance of data characterizing spheroids generated using this approach (Bell et al., 2016; Bell et al., 2017; Jensen and Teng, 2020; Ramaiahgari et al., 2017; Vorrink et al., 2017). Other methods, such as, scaffold-based approaches or use of magnetic field, hanging-drop or bioreactors have also been utilized for generating 3D spheroids (Desai et al., 2017; Godoy et al., 2013; Messner et al., 2018; Tostoes et al., 2012). However, these approaches lack reproducibility (due to lot-to-lot differences in various scaffolds and their interference with cells, drugs or the assay), scalability (not amenable to high-throughput applications in bioreactor-based approach) and require specialized or proprietary equipment(s) and/or culture techniques (e.g. magnetic field, hanging-drop). Using ULA methodology, we generated primary mouse hepatocyte spheroids of uniform size with well-defined morphology, and under 300 μm to ensure supply of nutrients and oxygen to all cells within the spheroid. Spheroids maintained a stable phenotype for 2 weeks post formation, which was the longest time point tested in this study. Nudischer et al. (2020) demonstrated viability for a total of 24 days in culture, but it is likely that mouse hepatocytes could be maintained in spheroid culture for longer periods of time as has been previously demonstrated with human hepatocyte spheroids (Bell et al., 2016).
We expanded on the findings from Nudischer et al. (2020) to show that the global gene expression profile and levels of 10 key DMET genes and proteins were also maintained in hepatocyte spheroids over time in culture. This suggests that the spheroid approach helps to preserve the phenotype of mouse hepatocytes better than more traditional 2D culture formats (Godoy et al., 2016). The pathways enriched among the small number of genes that were increased over time in spheroid culture included signaling molecules and genes involved in lipid metabolism, which may be due to reestablishment of cell-cell interactions but may also reflect a small degree of cellular stress that occurs over time in culture (Cassim et al., 2017). Interestingly, the pathways enriched among the small number of genes that were decreased overtime largely reflect a loss of type 1 collagen gene expression, which would likely result from the loss or inactivation of contaminating stellate cells and/or portal fibroblasts (Liu et al., 2013).
We also observed sex differences in the expression of some DMET genes and proteins in spheroids, which is consistent with studies in native mouse liver. At the gene expression level, we observed female-predominant expression of Cyp2c29 and Cyp3a11 which is in line with previous reports of these mRNAs in mouse liver (Lofgren et al., 2009; Lu et al., 2013). CYP3A4, the human orthologue of Cyp3a11, is also female predominant in human liver (Waxman and Holloway, 2009), but CYP2C19, the human orthologue of Cyp2c29 (Pan et al., 2016), is reportedly higher in liver tissue from males than females (Lofgren et al., 2008), indicating some species-specific differences. At the protein level, we observed female-predominant expression of Cyp2c29 and Cyp3a11, as well as Ntcp. Again, these findings are in line with previous reports of these proteins and their orthologues in mouse and human liver (Cheng et al., 2007; Waxman and Holloway, 2009), except for Cyp2c29 for which both the mouse protein and the human orthologue CYP2C19 are reportedly higher in livers from males than females (Chen et al., 2018; Lofgren et al., 2008; Shirasaka et al., 2016). Taken together, this indicates that some sex effects may be retained in vitro.
Importantly, we found that activities of most key cytochrome P450 enzymes were both measurable and stable over time in mouse spheroid culture, consistent with DMET mRNA and protein expression, and the overall trends were comparable to those reported in human spheroids (Bell et al., 2018). We also observed some sex differences in enzyme activity, with higher acetaminophen and OH-midazolam observed in spheroids from females than males at various time points in culture. Higher OH-midazolam formation in females than males is consistent with reports in both mouse (Cyp3a11) and human (CYP3A4) livers (Down et al., 2007; Waxman and Holloway, 2009), but acetaminophen formation from phenacetin (Cyp1a2) is reported to be higher in males than females in humans (Waxman and Holloway, 2009). Again, some sex effects may be retained in vitro and thus the use of spheroids from both male and female mice may be considered to better represent the sex differences in drug metabolism seen in humans.
Finally, we also observed that longer drug exposures caused a leftward shift in dose-response curves for all four DILI drugs tested, similar to what has been previously reported for human hepatocyte spheroids (Bell et al., 2018; Bell et al., 2016; Bell et al., 2017), and recently for mouse hepatocyte spheroids as well (Nudischer et al., 2020; Vorrink et al., 2018). We expand on these findings by including the measurement of albumin in addition to ATP and by assessing different DILI drugs in the mouse model. Interestingly, albumin was found to be a more sensitive endpoint than ATP as seen in other organotypic formats such as the bioprinted liver and micropatterned co-cultures (Nguyen et al., 2016; Ware et al., 2015) but can often be more variable. The human equilibrative nucleoside transporter required for mitochondrial transport and toxicity of fialuridine is lacking in mouse (Lee et al., 2006), which makes our observations of toxicity for this drug somewhat curious. However, fialuridine toxicity has been observed in mice at high concentrations (Manning FJ and Swartz M, 1995) and may suggest a different mechanism is involved in our study as well. The lack of a more robust effect of AMG-009 (bile acid-mediated toxicity) could be due to lack of bile acids in media. Only 5% of bile acids are synthesized de novo in the liver as most of the bile acids are recirculated back via the enterohepatic pathway which is missing in the in vitro system (Chiang and Ferrell, 2018). To address this issue, we are exploring the addition of a physiologically-relevant pool of bile acids to our media formulation as previously reported for human hepatocyte spheroids (Hendriks et al., 2016).
In conclusion, we describe the successful long-term cultivation of primary mouse hepatocytes as 3D spheroids that are suitable for use in toxicity studies with prolonged drug exposures. Our data adds to the recent report from Nudischer et al. (2020) by using cryopreserved cells, demonstrating phenotypic comparison between cells from male and female mice, and including a more extensive characterization of the gene expression profile, DMET protein levels, and cytochrome P450 enzyme activity over time. Primary mouse hepatocytes are a valuable resource for studying the impact of genetic and environmental factors on drug response and disease and the 3D spheroid approach will allow for the simultaneous evaluation of multiple mechanistic endpoints at multiple concentrations and time points using substantially fewer cells, and, as a result, fewer animals, than traditional 2D culture.
Supplementary Material
3D spheroids were generated from primary mouse hepatocytes in ULA plates
Morphology, ATP, and albumin levels are stable for 2 weeks post spheroid formation
Global mRNA profile, DMET protein level, and P450 enzyme activities are also stable
Time- and concentration-dependent toxicity responses to DILI drugs were observed
ACKNOWLEDGMENTS
This work was supported by the Burroughs Wellcome Fund Innovation in Regulatory Science Award and the National Institutes of Health Award Number R21OD028216 to MM. We would like to thank Magnus Ingelman-Sundberg and Sabine U. Vorrink (Karolinska Institutet) for providing training and guidance on 3D culture methodology, Sreenivasa Ramaiahgari (National Toxicology Program) for help with the 3D culture techniques, Michael Vernon (UNC Functional Genomics Core) for running the gene expression microarrays, and Paul Watkins (UNC Eshelman School of Pharmacy) for guidance on the DILI-causing drugs used in these experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY DATA DESCRIPTION
Full names, IDs, and probe information for the 10 key DMET mRNAs and proteins measured are described in Supplementary Table 1. Spheroid culture timelines for all endpoints are shown in Supplementary Figure 1. Comparisons of baseline ATP, albumin, size, and morphology, DMET mRNAs and proteins, and cytochrome P450 enzyme activity in spheroids generated from male and female mouse hepatocytes are shown in Supplementary Figures 2–5.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- Atienzar FA, Blomme EA, Chen M, Hewitt P, Kenna JG, Labbe G, Moulin F, Pognan F, Roth AB, Suter-Dick L, et al. (2016). Key Challenges and Opportunities Associated with the Use of In Vitro Models to Detect Human DILI: Integrated Risk Assessment and Mitigation Plans. Biomed Res Int 2016, 9737920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Dankers ACA, Lauschke VM, Sison-Young R, Jenkins R, Rowe C, Goldring CE, Park K, Regan SL, Walker T, et al. (2018). Comparison of Hepatic 2D Sandwich Cultures and 3D Spheroids for Long-term Toxicity Applications: A Multicenter Study. Toxicol Sci 162, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Hendriks DF, Moro SM, Ellis E, Walsh J, Renblom A, Fredriksson Puigvert L, Dankers AC, Jacobs F, Snoeys J, et al. (2016). Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 6, 25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Lauschke VM, Vorrink SU, Palmgren H, Duffin R, Andersson TB, and Ingelman-Sundberg M (2017). Transcriptional, Functional, and Mechanistic Comparisons of Stem Cell-Derived Hepatocytes, HepaRG Cells, and Three-Dimensional Human Hepatocyte Spheroids as Predictive In Vitro Systems for Drug-Induced Liver Injury. Drug Metab Dispos 45, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society 57, 289–300. [Google Scholar]
- Bowsher RR, Compton JA, Kirkwood JA, Place GD, Jones CD, Mabry TE, Hyslop DL, Hatcher BL, and DeSante KA (1994). Sensitive and specific radioimmunoassay for fialuridine: initial assessment of pharmacokinetics after single oral doses to healthy volunteers. Antimicrob Agents Chemother 38, 2134–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassim S, Raymond VA, Lapierre P, and Bilodeau M (2017). From in vivo to in vitro: Major metabolic alterations take place in hepatocytes during and following isolation. PLoS One 12, e0190366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TT, and Hughes-Fulford M (2014). Molecular mechanisms underlying the enhanced functions of three-dimensional hepatocyte aggregates. Biomaterials 35, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Zhang QS, Li XY, Gong X, Ruan YJ, Zeng SJ, Lu LL, Qi XX, Wang Y, Hu M, et al. (2018). Tissue Distribution and Gender-Specific Protein Expression of Cytochrome P450 in five Mouse Genotypes with a Background of FVB. Pharm Res 35, 114. [DOI] [PubMed] [Google Scholar]
- Cheng X, Buckley D, and Klaassen CD (2007). Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem Pharmacol 74, 1665–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JYL, and Ferrell JM (2018). Bile Acid Metabolism in Liver Pathobiology. Gene Expr 18, 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton NP, Burwell A, Jensen H, Williams BF, Brown QD, Ovwigho P, Ramaiahgari S, Hermon T, and Dixon D (2018). Preparation of Three-dimensional (3-D) Human Liver (HepaRG) Cultures for Histochemical and Immunohistochemical Staining and Light Microscopic Evaluation. Toxicol Pathol 46, 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PK, Tseng H, and Souza GR (2017). Assembly of Hepatocyte Spheroids Using Magnetic 3D Cell Culture for CYP450 Inhibition/Induction. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down MJ, Arkle S, and Mills JJ (2007). Regulation and induction of CYP3A11, CYP3A13 and CYP3A25 in C57BL/6J mouse liver. Arch Biochem Biophys 457, 105–110. [DOI] [PubMed] [Google Scholar]
- Fallon JK, Smith PC, Xia CQ, and Kim MS (2016). Quantification of Four Efflux Drug Transporters in Liver and Kidney Across Species Using Targeted Quantitative Proteomics by Isotope Dilution NanoLC-MS/MS. Pharm Res 33, 2280–2288. [DOI] [PubMed] [Google Scholar]
- Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, et al. (2013). Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87, 1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy P, Widera A, Schmidt-Heck W, Campos G, Meyer C, Cadenas C, Reif R, Stober R, Hammad S, Putter L, et al. (2016). Gene network activity in cultivated primary hepatocytes is highly similar to diseased mammalian liver tissue. Arch Toxicol 90, 2513–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks DF, Fredriksson Puigvert L, Messner S, Mortiz W, and Ingelman-Sundberg M (2016). Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci Rep 6, 35434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, and Speed TP (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jensen C, and Teng Y (2020). Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front Mol Biosci 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Messner S, Kelm JM, van Herwijnen M, Jennen DGJ, Kleinjans JC, and de Kok TM (2019). Human 3D multicellular microtissues: An upgraded model for the in vitro mechanistic investigation of inflammation-associated drug toxicity. Toxicol Lett 312, 34–44. [DOI] [PubMed] [Google Scholar]
- Khatri R, Fallon JK, Rementer RJB, Kulick NT, Lee CR, and Smith PC (2019). Targeted quantitative proteomic analysis of drug metabolizing enzymes and transporters by nano LC-MS/MS in the sandwich cultured human hepatocyte model. J Pharmacol Toxicol Methods 98, 106590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EW, Lai Y, Zhang H, and Unadkat JD (2006). Identification of the mitochondrial targeting signal of the human equilibrative nucleoside transporter 1 (hENT1): implications for interspecies differences in mitochondrial toxicity of fialuridine. J Biol Chem 281, 16700–16706. [DOI] [PubMed] [Google Scholar]
- Liu X, Xu J, Brenner DA, and Kisseleva T (2013). Reversibility of Liver Fibrosis and Inactivation of Fibrogenic Myofibroblasts. Curr Pathobiol Rep 1, 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren S, Baldwin RM, Carleros M, Terelius Y, Fransson-Steen R, Mwinyi J, Waxman DJ, and Ingelman-Sundberg M (2009). Regulation of human CYP2C18 and CYP2C19 in transgenic mice: influence of castration, testosterone, and growth hormone. Drug Metab Dispos 37, 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren S, Baldwin RM, Hiratsuka M, Lindqvist A, Carlberg A, Sim SC, Schulke M, Snait M, Edenro A, Fransson-Steen R, et al. (2008). Generation of mice transgenic for human CYP2C18 and CYP2C19: characterization of the sexually dimorphic gene and enzyme expression. Drug Metab Dispos 36, 955–962. [DOI] [PubMed] [Google Scholar]
- Lu YF, Jin T, Xu Y, Zhang D, Wu Q, Zhang YK, and Liu J (2013). Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol Int 30, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning FJ, and Swartz M (1995). FIAC and FIAU Preclinical Toxicity Studies. In Review of the Fialuridine (FIAU) Clinical Trials, Manning FJ, and e. Swartz M, eds. (Washington (DC: ): National Academies Press; https://www.ncbi.nlm.nih.gov/books/NBK232073/). [PubMed] [Google Scholar]
- Martinez SM, Bradford BU, Soldatow VY, Kosyk O, Sandot A, Witek R, Kaiser R, Stewart T, Amaral K, Freeman K, et al. (2010). Evaluation of an in vitro toxicogenetic mouse model for hepatotoxicity. Toxicol Appl Pharmacol 249, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Agarkova I, Moritz W, and Kelm JM (2013). Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol 87, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Fredriksson L, Lauschke VM, Roessger K, Escher C, Bober M, Kelm JM, Ingelman-Sundberg M, and Moritz W (2018). Transcriptomic, Proteomic, and Functional Long-Term Characterization of Multicellular Three-Dimensional Human Liver Microtissues. APPLIED IN VITRO TOXICOLOGY 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M, Button D, Jackson JP, Freeman KM, Brouwer KR, Caggiano AO, Eisen A, Iaci JF, Parry TJ, Stanulis R, et al. (2018a). Transient Changes in Hepatic Physiology That Alter Bilirubin and Bile Acid Transport May Explain Elevations in Liver Chemistries Observed in Clinical Trials of GGF2 (Cimaglermin Alfa). Toxicol Sci 161, 401–411. [DOI] [PubMed] [Google Scholar]
- Mosedale M, Eaddy JS, Trask OJ Jr., Holman NS, Wolf KK, LeCluyse E, Ware BR, Khetani SR, Lu J, Brock WJ, et al. (2018b). miR-122 Release in Exosomes Precedes Overt Tolvaptan-Induced Necrosis in a Primary Human Hepatocyte Micropatterned Coculture Model. Toxicol Sci 161, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal M, Bauch C, Walker P, Watkins PB, and Mosedale M (2020). Fit-For-Purpose Hepatocyte Models Enable the Identification of Early Events Contributing to Idelalisib-Induced Liver Injury. APPLIED IN VITRO TOXICOLOGY Submitted. [Google Scholar]
- Norona LM, Nguyen DG, Gerber DA, Presnell SC, and LeCluyse EL (2016). Editor’s Highlight: Modeling Compound-Induced Fibrogenesis In Vitro Using Three-Dimensional Bioprinted Human Liver Tissues. Toxicol Sci 154, 354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudischer R, Renggli K, Hierlemann A, Roth AB, and Bertinetti-Lapatki C (2020). Characterization of a long-term mouse primary liver 3D tissue model recapitulating innate-immune responses and drug-induced liver toxicity. PLoS One 15, e0235745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ST, Xue D, Li ZL, Zhou ZW, He ZX, Yang Y, Yang T, Qiu JX, and Zhou SF (2016). Computational Identification of the Paralogs and Orthologs of Human Cytochrome P450 Superfamily and the Implication in Drug Discovery. Int J Mol Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor WR, Foster AJ, Vogt J, Summers C, Middleton B, Pilling MA, Shienson D, Kijanska M, Strobel S, Kelm JM, et al. (2017). Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch Toxicol 91, 2849–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari SC, Waidyanatha S, Dixon D, DeVito MJ, Paules RS, and Ferguson SS (2017). Three-Dimensional (3D) HepaRG Spheroid Model With Physiologically Relevant Xenobiotic Metabolism Competence and Hepatocyte Functionality for Liver Toxicity Screening. Toxicol Sci 160, 189–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsby ML, and Makowski GS (1999). Differential detergent fractionation of eukaryotic cells. Analysis by two-dimensional gel electrophoresis. Methods Mol Biol 112, 53–66. [DOI] [PubMed] [Google Scholar]
- Ryan J, Morgan RE, Chen Y, Volak LP, Dunn RT 2nd, and Dunn KW (2018). Intravital Multiphoton Microscopy with Fluorescent Bile Salts in Rats as an In Vivo Biomarker for Hepatobiliary Transport Inhibition. Drug Metab Dispos 46, 704–718. [DOI] [PubMed] [Google Scholar]
- Sevilla-Tirado FJ, Gonzalez-Vallejo EB, Leary AC, Breedt HJ, Hyde VJ, and Fernandez-Hernando N (2003). Bioavailability of two new formulations of paracetamol, compared with three marketed formulations, in healthy volunteers. Methods Find Exp Clin Pharmacol 25, 531–535. [DOI] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016). Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaf SE, Wang Z, Bricmont P, and Mallikaarjun S (2007). Pharmacokinetics, pharmacodynamics, and safety of tolvaptan, a nonpeptide AVP antagonist, during ascending single-dose studies in healthy subjects. J Clin Pharmacol 47, 1498–1507. [DOI] [PubMed] [Google Scholar]
- Swales NJ, Johnson T, and Caldwell J (1996). Cryopreservation of rat and mouse hepatocytes. II. Assessment of metabolic capacity using testosterone metabolism. Drug Metab Dispos 24, 1224–1230. [PubMed] [Google Scholar]
- Tostoes RM, Leite SB, Serra M, Jensen J, Bjorquist P, Carrondo MJ, Brito C, and Alves PM (2012). Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology 55, 1227–1236. [DOI] [PubMed] [Google Scholar]
- Vorrink SU, Ullah S, Schmidt S, Nandania J, Velagapudi V, Beck O, Ingelman-Sundberg M, and Lauschke VM (2017). Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J 31, 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorrink SU, Zhou Y, Ingelman-Sundberg M, and Lauschke VM (2018). Prediction of Drug-Induced Hepatotoxicity Using Long-Term Stable Primary Hepatic 3D Spheroid Cultures in Chemically Defined Conditions. Toxicol Sci 163, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PB (2011). Drug safety sciences and the bottleneck in drug development. Clin Pharmacol Ther 89, 788–790. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, and Holloway MG (2009). Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


