Abstract
Context
Though genome-wide association studies (GWASs) have identified hundreds of genetic variants associated with osteoporosis related traits, such as bone mineral density (BMD) and fracture, it remains a challenge to interpret their biological functions and underlying biological mechanisms.
Objective
Integrate diverse expression quantitative trait loci and splicing quantitative trait loci data with several powerful GWAS datasets to identify novel candidate genes associated with osteoporosis.
Design, Setting, and Participants
Here, we conducted a transcriptome-wide association study (TWAS) for total body BMD (TB-BMD) (n = 66 628 for discovery and 7697 for validation) and fracture (53 184 fracture cases and 373 611 controls for discovery and 37 857 cases and 227 116 controls for validation), respectively. We also conducted multi-SNP-based summarized mendelian randomization analysis to further validate our findings.
Results
In total, we detected 88 genes significantly associated with TB-BMD or fracture through expression or ribonucleic acid splicing. Summarized mendelian randomization analysis revealed that 78 of the significant genes may have potential causal effects on TB-BMD or fracture in at least 1 specific tissue. Among them, 64 genes have been reported in previous GWASs or TWASs for osteoporosis, such as ING3, CPED1, and WNT16, as well as 14 novel genes, such as DBF4B, GRN, TMUB2, and UNC93B1.
Conclusions
Overall, our findings provide novel insights into the pathogenesis mechanisms of osteoporosis and highlight the power of a TWAS to identify and prioritize potential causal genes.
Keywords: osteoporosis, fracture, TWAS, gene expressing, RNA splicing
Osteoporosis is one of the most common metabolic bone diseases, clinically characterized by low bone mineral density (BMD) and increased susceptibility to low trauma fractures (1). In the United States, it is estimated that more than 14 million individuals are afflicted by osteoporosis and more than 47 million individuals suffer from low bone mass (2). The annual direct costs from osteoporosis are expected to reach ~$25.3 billion by 2025 (3).
Genome-wide association studies (GWASs) have successfully identified hundreds of genetic variants associated with osteoporosis and related traits, such as BMD and fracture (4–8). However, it remains a challenge to interpret the biological functions and underlying biological mechanisms of these genetic loci. Many genetic variants play important roles in the regulation of gene expression, thus altering the abundance levels of 1 or multiple proteins (9–12). Alternative ribonucleic acid (RNA) splicing, which removes introns and joins exons in premessenger RNAs (mRNAs) to produce mature mRNAs, can also play important roles in the development of disease (13–15). For instance, some mutations may cause aberrant splicing of specific genes, leading to various skeletal diseases (16, 17). The relationships between RNA expression/splicing and disease phenotypes could be identified through measuring RNA expression/splicing directly, such as by RNA-seq, in specific tissue/cell samples. However, the sample sizes of such studies are typically limited due to specimen availability and cost, and therefore they may lack adequate statistical power to detect many important associations.
Recently, transcriptome-wide association studies (TWASs) have successfully enhanced the discovery of genetic risk loci for complex traits by integrating gene expression data from large transcriptome reference datasets, such as the Genotype-Tissue Expression project (GTEx) (18), with GWAS summary association statistics to identify associations between gene expression and phenotypic traits. S-PrediXcan (19) and S-MultiXcan (20) are commonly used TWAS approaches; S-PrediXcan trains linear models for the prediction of gene expression and assesses the association between predicted expression and traits in a single tissue, while S-MultiXcan uses multivariate regression to integrate the S-PrediXcan results from multitissues to test the joint effects of gene expression variation from multitissues. Compared with S-PrediXcan, S-MultiXcan leverages the substantial sharing of expression quantitative trait loci (eQTLs) across tissues and contexts to improve detection power to identify potential target genes (20). Osteoporosis is a complex disease related to multiple biological processes. For example, besides bone tissues itself, osteoporosis is also related to the immune system (21) and hormone levels (22). So, gene expressions in multitissues or cells have effects on osteoporosis etiology. Considering that, we used S-MultiXcan to assess the joint effect of gene expression across all the 48 tissues in GTEx.
The GEFOS Lifecourse GWAS, which investigates the genetic determinants of total body bone mineral density (TB-BMD) across various age strata from childhood to late adulthood, is the largest collection of existing GWAS samples for dual energy X-ray absorptiometry (DXA)-measured BMD (7). The TB-BMD and BMD of the lumbar spine (SPN) are commonly used to assess bone health in children and adolescents for the minimization of measurement errors resulting from changing areas in growing bones, while BMD of the SPN and hip are regularly used in the diagnosis of osteoporosis among adults (4). It has been shown that degenerative changes of the spine may lead to an elevation of SPN BMD and cause underdiagnosis of osteoporosis in elderly individuals (23). Therefore, TB-BMD is the most appropriate measure for studying BMD variation when combining individuals from childhood to old age (7). Furthermore, TB-BMD is closely related with BMD of the SPN and hip (24). As the clinical end point of osteoporosis, GWASs for fractures contribute to revealing genetic variations ultimately relevant to osteoporosis.
In this study, we applied S-MultiXcan using GWAS summary statistics to identify candidate genes for TB-BMD and fracture using pretrained prediction models for gene expression from 48 tissues in GTEx. We also applied S-PrediXcan to the same GWAS data to identify genes associated with these traits through RNA splicing using pretrained prediction models for RNA splicing derived in lymphoblastoid cell lines from the Genetic European Variation in Disease (GEUVADIS) Consortium. Lymphocytes are cells with immune recognition function, which can be divided into T lymphocytes (also known as T cells), B lymphocytes (also known as B cells) and natural killer (NK) cells (25). The interaction between the immune and skeletal systems has long been acknowledged. The immune and skeletal systems share various molecules, including cytokines, transcription factors, signaling molecules, and membrane receptors (21). For instance, lymphoblastoid interferon-a is a pleiotropic cytokine that modulates the cellular functions of both osteoblastic and osteoclastic lineages (26). Multi-SNP-based summarized mendelian randomization (SMR) analysis was used to explore potential causal effects of the identified genes in specific tissues. Finally, we constructed a protein–protein interaction (PPI) network and performed functional enrichment analyses to further examine potential biological functions and interactions of the osteoporosis candidate genes detected in our study. Then, TWAS results of significant genes were further compared with the mRNA expression profile analysis results of osteoporosis from published studies (including bone tissues, mesenchymal stem cells [MSCs] and B cells) (27–30). An overview of our study is provided in Fig. 1.
Figure 1.
Overview of our main analyses. Abbreviation: SMR, summarized mendelian randomization.
Materials and Methods
GWAS data for discovery
The GWAS summary statistics for TB-BMD were obtained from the GEFOS Lifecourse GWAS (http://www.gefos.org/?q = content/gefos-lifecourse-tb-bmd-gwas-results), which is a meta-analysis of 30 GWASs of TB-BMD, including 66 628 individuals from populations across the United States, Europe, and Australia (7). Most participants were acquired from population-based cohorts of European ancestry (86%), 2 cohorts comprised African American individuals (2%), and 4 other studies with admixed ancestry (14%), including both males and females. The study population includes individuals from 5 different age strata: 0–15 (N = 11 807), 15–30 (N = 4,180), 30–45 (N = 10 062), 45–60 (N = 18 805), and 60 or more (N = 22 504). The TB-BMD was corrected for age, weight, height, gender, and genetic principal components (derived from GWASs data), as well as any additional study-specific covariates (eg, recruiting center). More details for assessment, quality control, and association analysis can be found in the published study (7).
The GWAS summary statistics for fracture were obtained from the UK Biobank (http://www.gefos.org/?q = content/data-release-2018) (8), and the study population includes 53 184 fracture cases (60% female) and 373 611 controls (54% female) of British ancestry (aged 40–69). The fracture cases were identified through a hospital-based fracture diagnosis and questionnaire based on self-reported fractures within the past 5 years. The fracture sites included forearm, upper arm, hip, vertebrae, pelvis, femur, lower leg, patella, ankle, rib, scapula, sternum, clavicle, and other unspecified sites. Fractures of the skull, face, hands and feet, pathological fractures due to malignancy, atypical femoral fractures, and periprosthetic fractures were excluded (31).
GWAS data for validation
Since there are no other available independent GWASs for TB-BMD, we used GWAS data for DXA-measured BMD at other skeletal sites as partial validation, including femoral neck (FNK) and SPN BMD. The TB-BMD has significant correlations with SPN BMD and FNK BMD (24, 32). The replication data were obtained from a previous in-house meta-analysis of FNK and SPN-BMD in 7 samples of diverse ancestries (5). Of these 7 samples, the Framingham Heart Study overlaps with the GEFOS study. To keep the replication data independent from the discovery data, the Framingham Heart Study sample was excluded from the replication cohort. The remaining 6 GWAS samples were collected for meta-analysis of SPN BMD and FNK BMD. Three samples were from the in-house studies and the other 3were accessed through the NIH database of genotype and phenotype (dbGAP). All the samples were obtained under the approvals by the respective institutional ethics review boards, and all the participants provided written informed consent. Briefly, the first sample comprised 1000 unrelated subjects of European ancestry from the Omaha Osteoporosis Study. The second sample comprised 2286 unrelated subjects of European ancestry from the Kansas City Osteoporosis Study. The third sample comprised 1627 unrelated subjects of Chinese Han ancestry from the China Osteoporosis Study. The fourth sample was from the Indiana Fragility Study that was accessed through the dbGAP. The Indiana Fragility Study is a cross-sectional cohort comprising 1493 premenopausal sister pairs of European ancestry. Both the fifth and sixth samples were from the Women’s Health Initiative (WHI) observational study that was accessed through the dbGAP too. The WHI is a partial factorial randomized and longitudinal cohort with 12 000 genotyped women aged 50–79 years, of African American or Hispanic ancestry (33). The fifth sample comprised 845 subjects of African American ancestry (WHIAA), and the sixth sample comprised 446 subjects of Hispanic ancestry (WHI-HIS). In summary, we included 7697 individuals (76% female) of diverse ancestry from 6 independent GWAS samples for validation of TB-BMD associations. Population stratification was corrected by principal components derived from genome-wide genotype analysis (5, 34).
For fracture, we used GWAS data from the GEFOS ALLFX Fracture study (37 857 cases and 227 116 controls, aged 18–106 years, 69% female) (35) as partial validation. Fracture cases were defined as individuals (>18 years old) who had fractures at any skeletal site confirmed by medical, radiological, or questionnaire reports. Fractures of the fingers, toes, and skull as well as high trauma fractures were excluded whenever possible. The cohorts were predominantly of European descent and detailed information of the subjects, genotyping, imputation, and quality control can be found in the published report (35).
Transcriptomics panels for gene expression and RNA splicing
The transcriptomics panels for gene expression were downloaded from PredictDB (http://predictdb.org/). The prediction models were trained in 48 tissues, which have eQTL analysis data from the GTEx Consortium V7 (https://gtexportal.org/home/). We excluded the 5 gender-specific tissues (prostate, testis, uterus, ovary, and vagina) in our analyses, including prostate, testis, uterus, ovary, and vagina since our GWAS datasets consist of both males and females. For a given gene in a specific tissue, gene expression was trained with Elastic-Net linear models (19, 36). Only SNPs in the vicinity of each gene (within 1 Mb of the gene start or end, with minor allele frequency > 0.05 and in Hardy-Weinberg equilibrium P > 0.05) were used to estimate the expression levels of the corresponding gene.
The transcriptomics panel for RNA splicing was obtained from a study by Li et al (37). Genotype and expression data from the GEUVADIS study, which contains RNA sequencing data from lymphoblastoid cell lines of 465 individuals, were used to identify RNA splicing events and train the prediction models for RNA splicing. Similar to the prediction models for gene expression, the prediction models for RNA splicing were trained by fitting Elastic-Net linear models to each intron cluster using nearby SNPs.
Differently expressed genes identified through mRNA expression profile analysis
We included differently expressed genes identified through mRNA expression profile analysis for osteoporosis from 3 published studies (including bone tissues, MSCs, and B cells). The first study included 27 osteoporotic patients and 39 controls, totaling 66 subjects from postmenopausal women in the range of 50–85 years of age. Bone cells from osteoporotic and controls were isolated from transiliac bone biopsies (0.8 × 1.5 cm) containing cortical and trabecular bone. The differently expressed genes were identified at P-values < 0.01. A detailed description of sample characteristics, experimental design, statistical analysis, and quality control can be found in the previous study (27). The second study included 5 elderly patients (79–94 years old) suffering from osteoporosis and 5 age-matched controls. The MSCs of participants were isolated from the bone marrow of the femoral heads after total hip arthroplasty due to osteoarthritis and/or hip dysplasia. The differently expressed genes were identified at fold changes ≥ 2.0 and false discovery rates < 10% (28). The third study included 10 individuals with low BMD and 10 with high BMD, totaling 20 unrelated postmenopausal white women in the age range of 54–60 years old. The mRNA profiles were measured in B cells isolated from whole blood. The differently expressed genes were identified at Benjamini & Hochberg-adjusted false discovery rates q-values ≤ 0.05 (29).
Association tests between gene expression/splicing and BMD/fracture
We used S-PrediXcan (35) to evaluate the association between traits and imputed expression in each tissue. Specifically, the Z-score (, Wald statistic) of the association between predicted gene expression and the phenotype was assessed as
wheredenotes the weight of SNP in the prediction of the expression of gene; denotes the GWAS regression coefficients for SNP ; denotes the standard error of ; denotes the estimated variance of SNP ; anddenotes the estimated variance of the predicted expression of gene. The association P-value was computed as
where is the normal cumulative distribution function.
Next, we used S-MultiXcan to join the single-tissue association statistics obtained from S-PrediXcan. The details for the methods can be found in the primary literature (19, 20).
Multi-SNP-based SMR analyses
Summarized mendelian randomization utilizes genetic variants strongly associated with an exposure variable as an “instrument” to test for causal effects of the exposure (gene expression) on an outcome variable (TB-BMD/fracture). We included eQTL data from the 43 nongender-specific tissues from GTEx (V7) (excluding the 5 gender-specific tissues: prostate, testis, uterus, ovary, and vagina) and whole blood eQTL data from the eQTLGen Consortium (https://eqtlgen.org/index.html). Only cis-eQTLs (< 500 kb from the associated gene) with were selected as instrumental variables. To ensure that the instruments for the exposure are independent, the SNPs were pruned based on pairwise linkage disequilibrium (LD) using a weighted vertex coverage algorithm with an LD r2 threshold of 0.1. Since most individuals in the GWASs are from European ethnicity, a reference panel of European individuals from the 1000 genomes project (phase 3) was used for LD estimation (38). A significant association in a cis region detected by the SMR test may be caused by pleiotropy (ie, the exposure and the outcome are associated owing to a single shared genetic variant) or linkage (ie, there are 2 or more genetic variants in LD affecting the exposure and outcome independently). Therefore, we performed the heterogeneity in dependent instruments (HEIDI) test (39) to assess whether the association patterns across a cis region were caused by pleiotropy or linkage. The HEIDI test assumes only 1 causal variant (affecting both gene expression and a trait) in the cis-eQTL region, and the smaller the HEIDI P-value, the larger the probability of the observations being consistent with a model of linkage.
Functional annotation analysis
The PPI network was constructed using the web-based tool STRING (https://string-db.org/) (40) under default settings and visualized by Cytoscape software (41). GO and KEGG pathway analyses were carried out by using the DAVID (https://david.ncifcrf.gov/) tool (42) under default settings.
Results
Genes associated with TB-BMD/fracture through expression/RNA splicing
In our study, we performed S-MultiXcan to test gene expression across 48 GTEx tissues in the expression level, including 25 841 genes in total. Considering that the prediction models for RNA splicing were trained in a single tissue—the lymphoblastoid cell line—we performed S-PrediXcan to assess the association between genes and the traits at the RNA splicing level for 45 823 RNA splicing events. Only genes with P-values smaller than the Bonferroni-corrected threshold ( for gene expression and for RNA splicing) in the discovery dataset and with P-values < 0.05 in the validation dataset were retained for further analysis. To evaluate the significance of associations between genes and traits across both the discovery and validation, so as to obtain the overall significance of these genes in the study samples, we performed a meta-analysis with Fisher’s method in R package metap (43).
In the discovery dataset, we detected a total of 148 genes significantly associated with TB-BMD and 38 genes significantly associated with fracture through gene expression, such as ING3 (for TB-BMD, for fractures), CPED1 (for TB-BMD, for fractures), and WNT16 (for TB-BMD, for fractures).
Among the 148 genes significantly associated with TB-BMD through gene expression, 46 genes showed association (), with SPN BMD or FNK BMD (34 genes for SPN BMD and 30 genes for FNK BMD) in the validation dataset (Table 1). Among the 38 significant genes for fracture, 30 were replicated in the validation dataset (Table 1). Through RNA splicing, we identified 32 genes significantly associated with TB-BMD in the discovery dataset, and 11 of them were successfully replicated, such as USP48, KLHL8, and TMEM161B (Table 2). For fracture, we identified 10 genes significantly associated with the phenotype through RNA splicing in the discovery dataset, and replicated 8 genes, such as CTBP1-AS2, MAEA, and MARK3 (Table 2).
Table 1.
Genes Associated With BMD/Fracture Through Gene Expression
| Ensemble ID | Gene Name | Gene Type | CHR | Meta-P | Significant Trait | ||
|---|---|---|---|---|---|---|---|
| TB-BMD & SPN BMD | TB-BMD & FNK BMD | Fracture | |||||
| ENSG00000271420 | RP5-1057J7.7 | lincRNA | 1 | 2.84E-12 | 5.48E-13 | 2.79E-01 | BMD |
| ENSG00000116729 | WLS | protein coding | 1 | 3.18E-16 | 8.44E-17 | 8.61E-06 | BMD |
| ENSG00000024526 | DEPDC1 | protein coding | 1 | 4.58E-10 | 2.98E-09 | 9.39E-02 | BMD |
| ENSG00000180875 | GREM2 | protein coding | 1 | 7.77E-12 | 7.83E-11 | 2.82E-03 | BMD |
| ENSG00000183023 | SLC8A1 | protein coding | 2 | 1.74E-02 | 8.77E-02 | 7.81E-09 | Fracture |
| ENSG00000162944 | RFTN2 | protein coding | 2 | 3.36E-08 | 3.87E-07 | 1.48E-01 | BMD |
| ENSG00000234270 | RPL36P20 | pseudogene | 3 | 6.66E-09 | 2.23E-07 | 4.56E-02 | BMD |
| ENSG00000114268 | PFKFB4 | protein coding | 3 | 2.15E-09 | 3.52E-08 | 1.28E-01 | BMD |
| ENSG00000163659 | TIPARP | protein coding | 3 | 1.54E-07 | 2.58E-08 | 3.83E-01 | BMD |
| ENSG00000182903 | ZNF721 | protein coding | 4 | 2.05E-03 | 7.41E-05 | 8.63E-09 | Fracture |
| ENSG00000178950 | GAK | protein coding | 4 | 1.12E-08 | 9.17E-09 | 5.03E-08 | Fracture |
| ENSG00000127415 | IDUA | protein coding | 4 | 4.96E-09 | 2.01E-10 | 8.67E-10 | Both |
| ENSG00000127418 | FGFRL1 | protein coding | 4 | 1.58E-08 | 1.02E-09 | 1.59E-15 | Both |
| ENSG00000178222 | RNF212 | protein coding | 4 | 5.11E-07 | 6.52E-08 | 2.68E-11 | Fracture |
| ENSG00000159674 | SPON2 | protein coding | 4 | 1.48E-05 | 9.90E-07 | 2.63E-09 | Both |
| ENSG00000163945 | UVSSA | protein coding | 4 | 3.57E-04 | 4.69E-05 | 4.89E-12 | Fracture |
| ENSG00000179979 | CRIPAK | protein coding | 4 | 2.01E-03 | 1.89E-03 | 1.33E-13 | Fracture |
| ENSG00000029559 | IBSP | protein coding | 4 | 5.10E-12 | 4.37E-13 | 7.87E-03 | BMD |
| ENSG00000152595 | MEPE | protein coding | 4 | 9.23E-21 | 3.21E-18 | 4.95E-01 | BMD |
| ENSG00000118785 | SPP1 | protein coding | 4 | 2.83E-17 | 1.46E-16 | 2.06E-02 | BMD |
| ENSG00000134982 | APC | protein coding | 5 | 1.44E-07 | 1.24E-07 | 1.24E-02 | BMD |
| ENSG00000219747 | RP1-292B18.1 | pseudogene | 6 | 5.89E-29 | 1.28E-25 | 4.90E-16 | Both |
| ENSG00000120262 | CCDC170 | protein coding | 6 | 9.06E-36 | 5.43E-33 | 2.53E-14 | Both |
| ENSG00000006377 | DLX6 | protein coding | 7 | 1.66E-16 | 4.06E-17 | 1.46E-07 | BMD |
| ENSG00000105880 | DLX5 | protein coding | 7 | 4.14E-18 | 8.42E-19 | 2.12E-04 | BMD |
| ENSG00000071243 | ING3 | protein coding | 7 | 2.24E-63 | 7.04E-63 | 6.03E-20 | Both |
| ENSG00000106034 | CPED1 | protein coding | 7 | 1.25E-59 | 6.59E-60 | 1.10E-21 | Fracture |
| ENSG00000002745 | WNT16 | protein coding | 7 | 3.24E-51 | 1.90E-50 | 2.26E-52 | Both |
| ENSG00000196937 | FAM3C | protein coding | 7 | 7.36E-24 | 1.28E-22 | 2.91E-11 | Both |
| ENSG00000184374 | COLEC10 | protein coding | 8 | 5.31E-26 | 1.75E-24 | 2.91E-01 | BMD |
| ENSG00000186466 | AQP7P1 | pseudogene | 9 | 2.56E-08 | 3.52E-15 | 7.75E-01 | BMD |
| ENSG00000110693 | SOX6 | protein coding | 11 | 1.67E-13 | 4.37E-14 | 2.49E-04 | BMD |
| ENSG00000148943 | LIN7C | protein coding | 11 | 8.91E-15 | 2.41E-13 | 2.49E-01 | BMD |
| ENSG00000176697 | BDNF | protein coding | 11 | 1.92E-08 | 3.15E-07 | 7.88E-01 | BMD |
| ENSG00000175213 | ZNF408 | protein coding | 11 | 1.63E-14 | 3.45E-16 | 2.94E-05 | BMD |
| ENSG00000180210 | F2 | protein coding | 11 | 6.48E-09 | 1.63E-10 | 6.44E-07 | BMD |
| ENSG00000025434 | NR1H3 | protein coding | 11 | 5.64E-06 | 8.52E-08 | 6.44E-04 | BMD |
| ENSG00000165917 | RAPSN | protein coding | 11 | 2.20E-12 | 3.23E-14 | 1.00E-07 | Both |
| ENSG00000110697 | PITPNM1 | protein coding | 11 | 7.35E-34 | 1.63E-31 | 7.39E-10 | Both |
| ENSG00000255147 | RP11-655M14.4 | pseudogene | 11 | 2.06E-12 | 6.00E-11 | 4.67E-08 | BMD |
| ENSG00000110057 | UNC93B1 | protein coding | 11 | 1.72E-27 | 3.80E-26 | 6.66E-08 | BMD |
| ENSG00000110719 | TCIRG1 | protein coding | 11 | 3.63E-08 | 2.75E-07 | 2.52E-08 | BMD |
| ENSG00000132749 | MTL5 | protein coding | 11 | 2.52E-13 | 1.49E-11 | 2.89E-04 | BMD |
| ENSG00000139055 | ERP27 | protein coding | 12 | 5.42E-02 | 8.94E-02 | 1.28E-09 | Fracture |
| ENSG00000135476 | ESPL1 | protein coding | 12 | 7.33E-16 | 3.08E-15 | 2.78E-04 | BMD |
| ENSG00000123349 | PFDN5 | protein coding | 12 | 6.83E-13 | 3.47E-12 | 6.16E-02 | BMD |
| ENSG00000170374 | SP7 | protein coding | 12 | 4.82E-16 | 2.57E-15 | 4.75E-04 | BMD |
| ENSG00000260492 | RP11-686F15.3 | lincRNA | 12 | 5.59E-11 | 3.88E-10 | 3.59E-03 | BMD |
| ENSG00000240631 | RPL30P12 | pseudogene | 12 | 4.98E-14 | 7.91E-15 | 3.77E-04 | BMD |
| ENSG00000151135 | C12orf23 | protein coding | 12 | 1.00E-10 | 8.83E-11 | 1.73E-04 | BMD |
| ENSG00000166949 | SMAD3 | protein coding | 15 | 9.61E-11 | 1.27E-09 | 1.80E-07 | BMD |
| ENSG00000103126 | AXIN1 | protein coding | 16 | 2.99E-09 | 1.40E-09 | 8.78E-02 | BMD |
| ENSG00000108784 | NAGLU | protein coding | 17 | 3.90E-09 | 7.10E-09 | 5.90E-04 | BMD |
| ENSG00000260105 | AOC4P | pseudogene | 17 | 1.05E-01 | 2.31E-01 | 3.07E-18 | Fracture |
| ENSG00000266967 | AARSD1 | protein coding | 17 | 3.15E-02 | 2.43E-02 | 5.68E-11 | Fracture |
| ENSG00000175906 | ARL4D | protein coding | 17 | 1.22E-06 | 1.34E-05 | 1.07E-17 | Both |
| ENSG00000067596 | DHX8 | protein coding | 17 | 7.26E-07 | 2.62E-06 | 9.26E-13 | Fracture |
| ENSG00000167941 | SOST | protein coding | 17 | 5.17E-06 | 1.57E-05 | 3.03E-13 | Fracture |
| ENSG00000108840 | HDAC5 | protein coding | 17 | 3.60E-05 | 9.44E-06 | 4.77E-13 | Fracture |
| ENSG00000125319 | C17orf53 | protein coding | 17 | 9.14E-04 | 2.79E-03 | 3.44E-12 | Fracture |
| ENSG00000161664 | ASB16 | protein coding | 17 | 1.18E-05 | 5.53E-06 | 3.17E-14 | Fracture |
| ENSG00000168591 | TMUB2 | protein coding | 17 | 4.06E-04 | 1.37E-03 | 1.17E-08 | Fracture |
| ENSG00000030582 | GRN | protein coding | 17 | 1.27E-05 | 3.51E-05 | 4.28E-08 | Fracture |
| ENSG00000161682 | FAM171A2 | protein coding | 17 | 9.44E-05 | 5.81E-05 | 1.49E-07 | Fracture |
| ENSG00000225190 | PLEKHM1 | protein coding | 17 | 3.66E-01 | 4.71E-01 | 1.40E-08 | Fracture |
Abbreviations: BMD, bone mineral density; FNK BMD, femoral neck BMD; SPN BMD, lumbar spine BMD; TB-BMD, total body BMD.
Table 2.
Genes Associated with BMD/Fracture Through RNA Splicing
| Ensemble ID | Gene Name | Gene Type | Alternative Excised Intron | Meta-P | Significant Trait | ||
|---|---|---|---|---|---|---|---|
| TB-BMD & SPN BMD | TB-BMD & FNK BMD | Fracture | |||||
| ENSG00000090686 | USP48 | protein coding | chr1: 22 073 642:22 079 020 | 1.39E-07 | 1.26E-08 | 2.32E-07 | BMD |
| ENSG00000196810 | CTBP1-AS2 | antisense | chr4: 1 244 138:1 244 684 | 9.10E-04 | 9.02E-04 | 2.11E-09 | Fracture |
| ENSG00000090316 | MAEA | protein coding | chr4: 1 283 770:1 291 744 | 1.09E-04 | 1.07E-04 | 6.08E-10 | Fracture |
| ENSG00000145332 | KLHL8 | protein coding | chr4: 88 116 842:88 141 570 | 1.43E-07 | 4.07E-08 | 6.48E-02 | BMD |
| ENSG00000164180 | TMEM161B | protein coding | chr5: 87 524 345:87 527 536 | 3.51E-14 | 3.38E-18 | 7.64E-02 | BMD |
| ENSG00000155906 | RMND1 | protein coding | chr6: 151 757 689:151 773 151 | 4.01E-09 | 4.64E-08 | 4.35E-05 | BMD |
| ENSG00000033050 | ABCF2 | protein coding | chr7: 150 915 959:150 916 150 | 1.26E-11 | 4.48E-11 | 6.69E-03 | BMD |
| ENSG00000033050 | ABCF2 | protein coding | chr7: 150 916 245:150 918 664 | 1.82E-12 | 1.32E-11 | 3.30E-03 | BMD |
| ENSG00000107164 | FUBP3 | protein coding | chr9: 133 470 975:133 471 924 | 2.65E-10 | 5.10E-11 | 2.08E-06 | BMD |
| ENSG00000149091 | DGKZ | protein coding | chr11: 46 391 100:46 391 491 | 9.94E-09 | 9.82E-10 | 1.68E-03 | BMD |
| ENSG00000149179 | C11orf49 | protein coding | chr11: 47 008 861:47 044 890 | 6.07E-08 | 1.81E-08 | 9.96E-02 | BMD |
| ENSG00000151135 | C12orf23 | protein coding | chr12: 107 349 843:107 350 860 | 6.31E-14 | 1.77E-14 | 1.05E-03 | BMD |
| ENSG00000151135 | C12orf23 | protein coding | chr12: 107 349 843:107 360 889 | 5.16E-13 | 5.32E-13 | 4.57E-02 | BMD |
| ENSG00000151135 | C12orf23 | protein coding | chr12: 107 350 927:107 360 889 | 5.00E-10 | 2.52E-10 | 3.40E-01 | BMD |
| ENSG00000135476 | ESPL1 | protein coding | chr12: 53 662 631:53 662 808 | 6.07E-17 | 1.33E-15 | 4.51E-05 | BMD |
| ENSG00000135476 | ESPL1 | protein coding | chr12: 53 662 712:53 662 808 | 1.11E-17 | 2.33E-16 | 2.20E-05 | BMD |
| ENSG00000135390 | ATP5G2 | protein coding | chr12: 54 036 648:54 062 932 | 8.65E-10 | 4.25E-08 | 1.77E-02 | BMD |
| ENSG00000075413 | MARK3 | protein coding | chr14: 103 918 320:103 921 656 | 3.89E-06 | 5.49E-06 | 3.25E-08 | Fracture |
| ENSG00000108797 | CNTNAP1 | protein coding | chr17: 40 834 592:40 835 839 | 4.96E-01 | 8.84E-01 | 1.27E-07 | Fracture |
| ENSG00000012048 | BRCA1 | protein coding | chr17: 41 276 132:41 277 288 | 8.34E-07 | 1.19E-06 | 1.58E-11 | Fracture |
| ENSG00000188554 | NBR1 | protein coding | chr17: 41 323 319:41 327 808 | 7.90E-07 | 5.10E-07 | 1.28E-13 | Fracture |
| ENSG00000188554 | NBR1 | protein coding | chr17: 41 341 819:41 343 389 | 1.51E-01 | 2.39E-01 | 9.46E-08 | Fracture |
| ENSG00000161692 | DBF4B | protein coding | chr17: 42 808 383:42 811 458 | 3.68E-07 | 2.05E-06 | 6.93E-12 | Fracture |
| ENSG00000214425 | LRRC37A4P | pseudogene | chr17: 43 592 891:43 598 673 | 1.90E-02 | 2.44E-02 | 1.01E-07 | Fracture |
Abbreviations: BMD, bone mineral density; FNK BMD, femoral neck BMD; SPN BMD, lumbar spine BMD; TB-BMD, total body BMD.
Taken together, we identified and replicated 55 genes significantly associated with TB-BMD and 38 genes significantly associated with fractures through gene expression or RNA splicing, with a total of 82 unique genes. Among them, 11 genes were associated with both TB-BMD and fractures: ARL4D, CCDC170, FAM3C, FGFRL1, IDUA, ING3, PITPNM1, RAPSN, RP1-292B18.1, SPON2, and WNT16. Interestingly, we noted that only 2 genes, namely C12orf23 and ESPL1, exhibited significant association with TB-BMD through both gene expression and RNA splicing.
Since our GWAS datasets included both males and females, the gender-specific tissues in the reference panels may cause bias. So, we repeated the above S-MultiXcan analysis with GTEx reference panels excluding the 5 gender-specific tissues (ie, prostate, testis, uterus, ovary, and vagina). We totally detected 136 genes significantly associated with TB-BMD, and 44 of them were validated in GWAS for SPN BMD or FNK BMD. For fracture, we detected a total of 41 genes in discovery, and 31 of them were validated. Among the 46 significant genes for BMD and the 30 significant genes for fracture (65 unique genes in total) in the above S-MultiXcan results based on all of 48 GTEx tissues, 51 genes were also detected in S-MultiXcan results based on the 43 nongender-specific GTEx tissues, and 3 genes (GRN, WLS, and PLEKHM1) reached significant levels in discovery but were not validated in validation, while the remaining 11 genes did not reach significant levels. We also detected 6 extra genes (G6PC3, POU6F2, LINC00910, NAGS, PKDCC, and POU6F2) in S-MultiXcan analysis based on the 43 nongender-specific GTEx tissues. We also included these 6 genes in our further analyses (totally 88 unique genes).
Causal association between significant genes and TB-BMD/fracture
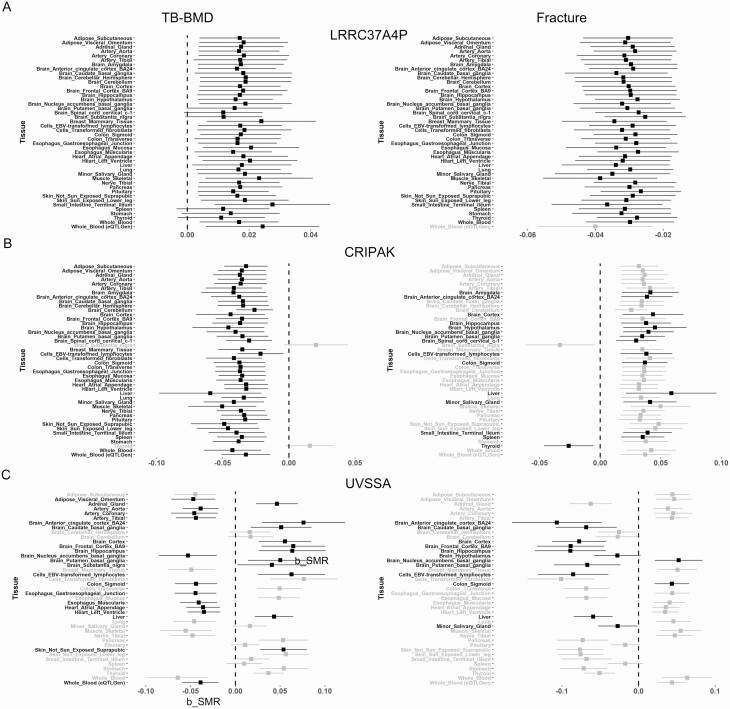
To examine whether there was evidence for a causal relationship between significant genes detected in our analyses and TB-BMD/fracture, we conducted multi-SNP-based SMR analyses (39, 44) for the 88 replicated genes identified above. The associations with in SMR analysis were defined as causal associations. We filtered out genes with in the HEIDI test. In total, we identified 1320 significant associations between genes and TB-BMD/fracture, and 954 of these associations (78 unique genes) remained after the HEIDI filtering in various tissues. Interestingly, most associations appeared to be detected in multitissues. For instance, LRRC37A4P was significantly associated with TB-BMD and fracture in all tissues, and most of these associations were retained after the HEIDI filtering (Fig. 2A). CRIPAK was also associated with both traits across all tissues, but only associations in 19 tissues remained after HEIDI filtering when testing the association with fracture (Fig. 2B). Additionally, the direction of effects of LRRC37A4P and CRIPAK on the traits showed rather good consistency in different tissues. In contrast, some genes, such as UVSSA, which was associated with both TB-BMD and fracture in 41 tissues, showed strong heterogeneity of effects on the traits in different tissues (Fig. 2C), suggesting the tissue-dependency of associations for these genes.
Figure 2.
Forest plots for tissue-dependent effects identified in SMR analysis of 3 TB-BMD/fracture-associated genes. Black dots represent that the gene was associated with the trait in SMR analysis and survived the HEIDI filtering in the specific tissue, while the grey dots represent that the gene was associated with the trait in SMR analysis but was filtered out in the HEIDI analysis. A: LRRC37A4P was associated with TB-BMD and fracture across all 43 GTEx tissues and whole blood of eQTLGen and the direction of effects showed good consistency in different tissues. B: CRIPAK was also associated with both traits across all 43 GTEx tissues and whole blood of eQTLGen, but only survived the HEIDI filtering in 19 tissues in the analysis for fracture. The direction of effects also showed good consistency in different tissues. C: UVSSA was associated with both TB-BMD and fracture in 41 tissues, but showed strong heterogeneity of effect on the trait in different tissues. HEIDI, the heterogeneity in dependent instruments; SMR, summarized Mendelian randomization; TB-BMD, total body BMD.
In our further analysis, we focused on the 78 genes with significant signals, which remained after HEIDI filtering in the SMR analysis. We compared our results with previous GWASs (GWAS catalog, https://www.ebi.ac.uk/gwas/, accessed on November 17, 2019) and TWASs (TWAS hub and UKBBeta) for bone-related traits. The TWAS hub (http://twas-hub.org/about/, accessed on November 17, 2019) is an interactive browser of TWAS results from hundreds of complex traits and dozens of expression studies, which was generated using the FUSION software (45), while UKBBeta (http://apps.hakyimlab.org/metabeta/, accessed on November 17, 2019) includes PrediXcan results based on Neale Lab’s UKBB results and 44 tissue models from the GTEx V6p release. Taken together, we identified 14 genes that were not reported by previous GWASs nor TWASs, thus they were considered as novel putative osteoporosis-related genes (Table 3). We compared our results with a recent TWAS (46), in which the authors performed single-tissue-based method FUSION (45) to test the association between gene expression and SPN BMD, FNK BMD, as well as forearm (FA) BMD in muscle skeleton and peripheral blood separately. Among the 88 significant genes, we only detected 1 common gene (FAM3C) shared with their study.
Table 3.
Fourteen Novel Putative Osteoporosis-related Genes Identified in Our Analysis
| Ensemble ID | Gene Name | Gene Type | CHR | Start | End | BMD | Fracture | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Expression | Splicing | Expression | Splicing | ||||||||
| ENSG00000162944 | RFTN2 | protein coding | 2 | 198 332 948 | 198 540 769 | Yes | No | No | No | ||
| ENSG00000234270 | RPL36P20 | lincRNA | 3 | 41 996 937 | 41 997 255 | Yes | No | No | No | ||
| ENSG00000114268 | PFKFB4 | protein coding | 3 | 48 455 117 | 48 699 448 | Yes | No | No | No | ||
| ENSG00000106536 | POU6F2 | protein coding | 7 | 39 017 598 | 39 532 694 | No | No | Yes | No | ||
| ENSG00000255147 | RP11-655M14.4 | pseudogene | 11 | 67 516 514 | 67 524 439 | Yes | No | No | No | ||
| ENSG00000110057 | UNC93B1 | protein coding | 11 | 67 758 575 | 67 772 452 | Yes | No | No | No | ||
| ENSG00000260492 | RP11-686F15.3 | protein coding | 12 | 54 144 231 | 54 145 619 | Yes | No | No | No | ||
| ENSG00000260105 | AOC4P | pseudogene | 17 | 40 917 939 | 41 126 386 | No | No | Yes | No | ||
| ENSG00000188825 | LINC00910 | lincRNA | 17 | 41 447 213 | 41 466 567 | No | No | Yes | No | ||
| ENSG00000186566 | GPATCH8 | protein coding | 17 | 42 472 652 | 42 580 798 | No | No | Yes | No | ||
| ENSG00000161692 | DBF4B | pseudogene | 17 | 42 829 632 | 42 785 976 | No | No | No | Yes | ||
| ENSG00000030582 | GRN | pseudogene | 17 | 42 422 614 | 42 430 470 | No | No | Yes | No | ||
| ENSG00000214425 | LRRC37A4P | pseudogene | 17 | 43 627 701 | 43 578 685 | No | No | No | Yes | ||
| ENSG00000168591 | TMUB2 | protein coding | 17 | 42 264 338 | 42 269 099 | No | No | Yes | No | ||
Yes/No represents whether the gene was associated with the trait through gene expression or RNA splicing.
Abbreviation: BMD, bone mineral density; CHR, chromosome.
Functional annotation of the significant genes
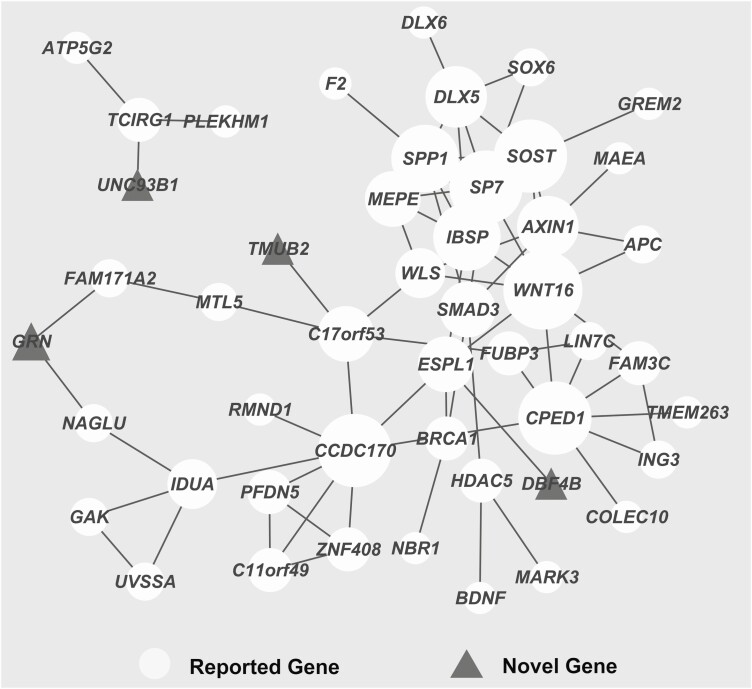
We next sought to identify the interactions between the novel genes in our results and known genes in previous GWAS studies. As expected, these genes formed a significant PPI network (Fig. 3) (PPI enrichment P-value < 1), and 4 novel genes (DBF4B, GRN, TMUB2, and UNC93B1) detected in our study were directly connected to the known genes, such as ESPL1, NAGLU, and TCIRG1. The top 5 genes with the highest degree of connectivity, namely, WNT16, CPED1, CCD170, SOST, and SP1, have been associated with BMD or fracture in previous GWAS.
Figure 3.
PPI network of significant genes associated with TB-BMD or fracture in our analysis. Gene pairs with interaction scores larger than 0.4 were retained in the PPI network. Four novel genes (DBF4B, GRN, TMUB2, and UNC93B1) detected in our study were directly connected to the genes reported in previous GWASs or TWASs. CHR, chromosome; GWAS, genome wide association study; PPI, protein-protein interaction; TB-BMD: total body BMD; TWAS, transcriptome-wide association study.
To find the interactions between novel and known genes in our findings and prioritize genes that may play more important roles in osteoporosis, we then performed the GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The Gene Ontology (GO) analysis showed that these genes prioritized by our analyses were significantly enriched () in 17 biological processes (Table 4), such as Wnt signaling pathway (), osteoblast differentiation (), and skeletal system development (). The KEGG analysis showed that these genes were enriched in 5 pathways (Table 4), such as signaling pathways regulating the pluripotency of stem cells (), Wnt signaling pathway (mathgraphic>), and Hippo signaling pathway ().
Table 4.
Significantly Enriched GO Terms and KEGG Pathways
| Data Source | Term | Count | % | P | Genes |
|---|---|---|---|---|---|
| GO-BP | GO:0 001 501: Skeletal system development | 5 | 6.76 | 1.74E-03 | DLX6, DLX5, FGFRL1, PKDCC, MEPE |
| GO:0 032 332: Positive regulation of chondrocyte differentiation | 3 | 4.05 | 2.27E-03 | SMAD3, PKDCC, SOX6 | |
| GO:0 060 021: Palate development | 4 | 5.41 | 2.91E-03 | DLX6, DLX5, TIPARP, PKDCC | |
| GO:0 016 055: Wnt signaling pathway | 5 | 6.76 | 5.32E-03 | WNT16, SOST, WLS, AXIN1, APC | |
| GO:0 001 649: Esteoblast differentiation | 4 | 5.41 | 7.00E-03 | IBSP, DLX5, SP7, SPP1 | |
| GO:0 030 501: Positive regulation of bone mineralization | 3 | 4.05 | 7.61E-03 | SLC8A1, SMAD3, PKDCC | |
| GO:0 045 893: Positive regulation of transcription, DNA-templated | 7 | 9.46 | 1.25E-02 | FUBP3, SOST, DLX5, SMAD3, BRCA1, AXIN1, NR1H3 | |
| GO:1 904 885: Beta-catenin destruction complex assembly | 2 | 2.70 | 1.86E-02 | AXIN1, APC | |
| GO:0 045 732: Positive regulation of protein catabolic process | 3 | 4.05 | 2.13E-02 | TIPARP, AXIN1, APC | |
| GO:0 090 090: Negative regulation of canonical Wnt signaling pathway | 4 | 5.41 | 2.33E-02 | SOST, PFDN5, AXIN1, APC | |
| GO:0 007 155: Cell adhesion | 6 | 8.11 | 2.87E-02 | IBSP, MAEA, CNTNAP1, SPON2, SPP1, APC | |
| GO:0 001 503: Ossification | 3 | 4.05 | 3.63E-02 | IBSP, SOST, SPP1 | |
| GO:0 051 481: Negative regulation of cytosolic calcium ion concentration | 2 | 2.70 | 3.69E-02 | SLC8A1, SMAD3 | |
| GO:0 060 070: Canonical Wnt signaling pathway | 3 | 4.05 | 3.88E-02 | SMAD3, AXIN1, APC | |
| GO:0 048 646: Anatomical structure formation involved in morphogenesis | 2 | 2.70 | 4.05E-02 | DLX6, DLX5 | |
| GO:0 033 147: Negative regulation of intracellular estrogen receptor signaling pathway | 2 | 2.70 | 4.41E-02 | CRIPAK, BRCA1 | |
| GO:0 007 049: Cell cycle | 4 | 5.41 | 4.80E-02 | DBF4B, MAEA, BRCA1, GAK | |
| KEGG | hsa04550: Signaling pathways regulating pluripotency of stem cells | 5 | 6.76 | 1.27E-03 | WNT16, DLX5, SMAD3, AXIN1, APC |
| hsa04310: Wnt signaling pathway | 4 | 5.41 | 1.17E-02 | WNT16, SOST, AXIN1, APC | |
| hsa05217: Basal cell carcinoma | 3 | 4.05 | 1.49E-02 | WNT16, AXIN1, APC | |
| hsa04390: Hippo signaling pathway | 4 | 5.41 | 1.50E-02 | WNT16, SMAD3, AXIN1, APC | |
| hsa05210: Colorectal cancer | 3 | 4.05 | 1.94E-02 | SMAD3, AXIN1, APC |
Abbreviations: GO-BP, biological process in gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; Wnt, wingless-related integration site.
Common genes shared by TWAS and published mRNA expression profiles
To partially validate our TWAS results (based on predicted gene expression) in the true transcriptional profiles, we compared our TWAS results with 3 published mRNA expression profile analysis results of osteoporosis (including bone cells (27), MSCs (28), and B cells (29)). Among the 88 genes prioritized in our results, 18 genes showed significantly different expression signals between osteoporotic patients and controls in MSCs (such as SMAD3, IBSP, and SOST), 2 genes (KLHL8 and MEPE) showed a significant signal in bone cells, and no genes showed significant signals in B cells (Table 5). All of the partially validated 20 genes have also been reported in previous GWASs in the GWAS catalog or precious TWASs in the TWAS hub and UKBBeta.
Table 5.
The Common Genes Shared by TWAS and the Differently Expressed Genes Identified by mRNA Expression Profiles
| Ensemble ID | Gene Name | CHR | FC (OP/CTR) | Cell type |
|---|---|---|---|---|
| ENSG00000145332 | KLHL8 | 4 | 1.14 | Bone |
| ENSG00000152595 | MEPE | 4 | 0.6 | Bone |
| ENSG00000166949 | SMAD3 | 15 | 43.27 | MSC |
| ENSG00000029559 | IBSP | 4 | 9.41 | MSC |
| ENSG00000167941 | SOST | 17 | 4.6 | MSC |
| ENSG00000141349 | G6PC3 | 17 | 4.52 | MSC |
| ENSG00000127418 | FGFRL1 | 4 | 3.69 | MSC |
| ENSG00000127415 | IDUA | 4 | 2.84 | MSC |
| ENSG00000188554 | NBR1 | 17 | 2.77 | MSC |
| ENSG00000183023 | SLC8A1 | 2 | 2.59 | MSC |
| ENSG00000118785 | SPP1 | 4 | 2.53 | MSC |
| ENSG00000149091 | DGKZ | 11 | 2.47 | MSC |
| ENSG00000033050 | ABCF2 | 7 | 2.42 | MSC |
| ENSG00000108797 | CNTNAP1 | 17 | 2.13 | MSC |
| ENSG00000139055 | ERP27 | 12 | 2.12 | MSC |
| ENSG00000148943 | LIN7C | 11 | 2.04 | MSC |
| ENSG00000107164 | FUBP3 | 9 | 0.46 | MSC |
| ENSG00000090686 | USP48 | 1 | 0.45 | MSC |
| ENSG00000196937 | FAM3C | 7 | 0.44 | MSC |
| ENSG00000071243 | ING3 | 7 | 0.38 | MSC |
Abbreviations: CHR, chromosome; CTR, control; FC, fold change; MSC, mesenchymal stem cell; OP, osteoporosis; TWAS, transcriptome-wide association study.
Discussion
Genome-wide association studies are a powerful approach to identify genetic variants for complex diseases; however, the biological functions for many of those trait-associated variants are still unclear. A TWAS integrates GWASs and gene expression datasets to identify gene–trait associations, which has garnered substantial interest within the genetics field and have been conducted for many traits and tissues. Compared with GWAS, which is based on the associate between variants and traits, a TWAS can associate genetically predicted gene expression and disease risk, with which we could prioritize candidate causal genes and tissues for the interested traits. Despite this success, there are still some caveats and shortcomings in a TWAS. First, the associations detected by a TWAS do not represent the causal effects from genes to traits, as the direction of effects may be reverse. Second, similar to a GWAS, a TWAS often identified multiple candidate genes in a locus because of sharing GWAS variants in LD. Third, the noncausal genes may be prioritized as potential causal genes because of the correlation of their expression with the true causal genes. Fourth, current TWAS methods commonly use cis eQTLs to predict gene expression. Unfortunately, cis eQTLs can only explain a small component of total expression, which depends on genetic, environmental, and technical components. Fifth, when considering multitissue effects, it is important to determine carefully gene expressions in which tissue or cell types will influence the interested traits (47).
In this study, we performed a powerful multitissue-based TWAS to integrate the information embedded in gene expression data, with GWAS results for osteoporosis-related traits (TB-BMD and fracture) to identify putative novel osteoporosis-associated genes.
In total, we identified 46 genes associated with BMD and 30 genes associated with fracture, which showed significant S-MultiXcan signals in both discovery and validation datasets. Among them, multiple genes have been reported to have an effect on osteoporosis in previous studies, such as WNT16 (48, 49), CPED1 (50, 51), and SOST (52–54), partially validating our novel approach in recovering some of the well-known genes for osteoporosis. However, we only detected 1 common gene (FAM3C), which was known to modulate osteogenic cell differentiation and affect bone volume and cortical BMD (55), with the recent TWAS for SPN BMD, FNK BMD, and FA BMD based on muscle skeleton and peripheral blood (46). Our study was based on the multitissue-based method S-MultiXcan, while their analyses were based on the single-tissue-based method FUSION. The differences between these studies were not only caused by the BMD analyzed from different sites, but also the reference panels for predicting gene expression.
Beyond the analysis on gene expression levels, we also considered the effects of RNA splicing on osteoporosis, which may enable the detection of additional novel genes (37). Several skeletal system diseases are related to RNA splicing (16). Our findings highlighted the importance of RNA splicing as a novel mediator of genetic effects on osteoporosis. For example, we identified that 1 of the intron excisions in BRCA1 is associated with fracture. BRCA1 is a gene involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks (56). Mutations in BRCA1 were associated with increased odds of bone loss in women (57), but the biological process is unclear.
The tissues or cell types that a gene is expressed in could reflect the biological processes and functions of the gene (58). Through SMR analysis, we observed that the effects of some genes varied widely in different tissues, such as UVSSA. For instance, in this study we demonstrated that the expression of UVSSA showed a negative effect to osteoporosis in some tissues, such as adipose visceral omentum. UVSSA encodes the UV-stimulated scaffold protein A (UVSSA), which forms a complex with ubiquitin-specific peptidase 7 (USP7) (59). The interaction of UVSSA and USP7 plays an important role in regulating transcription-coupled nucleotide excision repair in human cells (60). It has been reported that protein deubiquitinase USP7 is an essential player in osteogenic differentiation of human adipose-derived stem cells (hASCs) through its catalytic activity in hASCs (61). In contrast, we detected that UVSSA showed a positive effect to osteoporosis in other tissues, such as the brain anterior cingulate cortex BA24, suggesting that it may have multiple mechanisms to affect the traits in different tissues. Furthermore, we also observed that some genes showed consistent potential causal effects in widespread tissues, such as LRRC37A4P and CRIPAK. The absence of tissue specificity cross tissues suggests that the function of these genes may start early in development and have widespread influence on downstream phenotypic consequences (62). For instance, CRIPAK encodes the cysteine-rich inhibitor of p21-activated protein kinase 1 (Pak1). Pak1 is involved in a variety of cell life activities and plays an important role in several cellular processes, including cytoskeleton reorganization, promotion of cell survival, and estrogen receptor (ER) signaling (63).
Among the candidate genes in our TWAS, we detected 18 genes significantly differently expressed between osteoporotic patients and controls in MSCs (such as SMAD3, IBSP, and SOST), and 2 genes (KLHL8 and MEPE) showed significant signals in bone cells through published mRNA expression profiles. All of these genes were reported to be associated with bone-in in previous GWASs in the GWAS catalog (64) or TWASs in the TWAS hub (45) and UKBBeta (36). For instance, SMAD3 encodes the SMAD 3 protein, a member of SMAD protein family. The SMAD 3 protein is involved in the regulation of producing of RANKL, which is crucial for the differentiation of osteoclast (65, 66). The partial validation in results of differently expression gene analysis with mRNA profiles in an adequate tissue could reflect the robust of our results, but mRNA profiles in single tissues cannot reveal all of the associations between gene expression and osteoporosis. Bone homeostasis is related to multiple biological processes, such as immune system and hormone levels, so osteoporosis is affected by the interaction of gene expression in multitissues or cells as well as environmental factors (21, 22, 54). We did not detect any common genes shared in a TWAS and results of differently expressed gene analysis in B cells. The analysis of mRNA profiles in B cells only contained 20 unrelated postmenopausal white women and detected 29 significant genes totally. So, we assume that the small sample size of mRNA profiles limited the detecting power, which supports an enormous advantage of TWAS methods that can be applied to large samples.
Through PPI network analysis, we detected that 4 novel genes (GRN, DBF4B, TMUB2, and UNC93B1) have a direct connection with known osteoporosis-related genes. There was some evidence to support the potential roles of the 4 genes in bone metabolism:
GRN. Granulin precursor is best known as a glial protein for which deficiency leads to the most common inherited form of frontotemporal dementia (67). GRN is an adipokine associated with diet-induced obesity and insulin resistance (68, 69). GRN can promote bone resorption and suppress bone formation in female mice (70).
DBF4B. DBF4 zinc finger B encodes a regulator of the cell division cycle 7 homolog (Cdc7) protein, and the complex of Dbf4b protein with Cdc7 is required for the DNA replication (71). The pre-mRNA splicing of DBF4B is regulated by SRSF1 (72), which was identified as a hub gene of PPI and co-expression networks for osteoporosis in postmenopausal women (73).
UNC93B1. Unc-93 homolog B1 encodes a protein that is involved in innate and adaptive immune response by regulating toll-like receptor (TLR) signaling (74). Toll-like receptors have critical roles in osteoarthritis and fracture healing (75, 76).
TMUB2. The function of transmembrane and ubiquitin-like domain containing 2 is unclear. Interestingly, the expressions of TMUB2 and C17orf53 were both regulated by a cis-eQTL rs227584 (77), which was significantly associated with BMD and fracture and may alter C17orf53 protein interaction with NIMA-related kinase 2 (Nek2) and subsequently influence osteoblast growth and activity (78). Therefore, the association signals we observed in TMUB2 may be related to the association between C17orf53 expression and fracture, which is also significant in our analyses.
In light of the novel findings and the validation of well-known genes/pathways, our study may also have some limitations: First, there are no bone tissues/cells in the available references, such as osteoclast and osteoblast panels. Therefore, tissue-specific associations in the bone tissues/cells may not be detected. Second, the sample size of reference panels in GTEx are relatively small at present, and thus some gene expression levels may not be accurately predicted (47). Third, RNA expression/splicing was predicted through linear models; thus, the associations between variants, genes, and traits through nonlinear interaction (if any) cannot be identified. Fourth, we only included lymphoblastoid cells in the RNA splicing level analysis. RNA splicing is highly tissue-specific and a TWAS analysis in a single tissue/cell cannot reveal the complete list of the genes associated with osteoporosis. Transcriptome-wide association study analyses including more tissues/cells for osteoporosis is needed in future studies.
In summary, our studies validated some known osteoporosis-associated genes such as WNT16 and CPED1, and also identified some novel genes, which have not been reported in previous GWASs or TWASs, such as GRN and DBF4B. We propose that these findings could provide substantiation of known genes/pathways and also, in particular, furnish novel clues for revealing the pathogenesis of osteoporosis and may contribute to the future development of gene-based intervention/prevention/prediction for osteoporosis.
Acknowledgments
This research was supported in part by using the high-performance computing (HPC) resources and services provided by Technology Services at Tulane University, New Orleans, Louisiana.
Financial Support: This work benefited from the support of grants from the National Institutes of Health [R01MH107354, R01MH104680, R01GM109068, R01AR069055, U19AG055373, R01DK115679, R01AG061917].
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References
- 1. Nih Consensus Development Panel on Osteoporosis Prevention D, Therapy. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285(6):785–795. [DOI] [PubMed] [Google Scholar]
- 2. Foundation NO. America’s bone health: the state of osteoporosis and low bone mass in our nation. http://www.osteoporosisnews.org/advocacy/prevalence/index.htm. Published 2002. Accessed December 12, 2019.
- 3. Becker DJ, Kilgore ML, Morrisey MA. The societal burden of osteoporosis. Curr Rheumatol Rep. 2010;12(3):186–191. [DOI] [PubMed] [Google Scholar]
- 4. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang L, Choi HJ, Estrada K, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23(7):1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng HF, Forgetta V, Hsu YH, et al. ; AOGC Consortium; UK10K Consortium . Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526(7571):112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Medina-Gomez C, Kemp JP, Trajanoska K, et al. Life-Course Genome-wide Association Study Meta-analysis of Total Body BMD and Assessment of Age-Specific Effects. Am J Hum Genet. 2018;102(1):88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morris JA, Kemp JP, Youlten SE, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lappalainen T, Sammeth M, Friedländer MR, et al. ; Geuvadis Consortium . Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X, Joehanes R, Chen BH, et al. Identification of common genetic variants controlling transcript isoform variation in human whole blood. Nat Genet. 2015;47(4):345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212. [DOI] [PubMed] [Google Scholar]
- 13. Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749–761. [DOI] [PubMed] [Google Scholar]
- 14. Solis AS, Shariat N, Patton JG. Splicing fidelity, enhancers, and disease. Front Biosci. 2008;13:1926–1942. [DOI] [PubMed] [Google Scholar]
- 15. Cáceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18(4):186–193. [DOI] [PubMed] [Google Scholar]
- 16. Fan X, Tang L. Aberrant and alternative splicing in skeletal system disease. Gene. 2013;528(1):21–26. [DOI] [PubMed] [Google Scholar]
- 17. Peng H, Zhang Y, Long Z, et al. A novel splicing mutation in COL1A1 gene caused type I osteogenesis imperfecta in a Chinese family. Gene. 2012;502(2):168–171. [DOI] [PubMed] [Google Scholar]
- 18. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamazon ER, Wheeler HE, Shah KP, et al. ; GTEx Consortium . A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barbeira AN, Pividori M, Zheng J, Wheeler HE, Nicolae DL, Im HK. Integrating predicted transcriptome from multiple tissues improves association detection. Plos Genet. 2019;15(1):e1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dar HY, Azam Z, Anupam R, Mondal RK, Srivastava RK. Osteoimmunology: the nexus between bone and immune system. Front Biosci (Landmark Ed). 2018;23:464–492. [DOI] [PubMed] [Google Scholar]
- 22. Bijelic R, Milicevic S, Balaban J. Risk factors for osteoporosis in postmenopausal women. Med Arch. 2017;71(1):25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tenne M, McGuigan F, Besjakov J, Gerdhem P, Åkesson K. Degenerative changes at the lumbar spine–implications for bone mineral density measurement in elderly women. Osteoporos Int. 2013;24(4):1419–1428. [DOI] [PubMed] [Google Scholar]
- 24. Franck H, Munz M. Total body and regional bone mineral densitometry (BMD) and soft tissue measurements: correlations of BMD parameter to lumbar spine and hip. Calcif Tissue Int. 2000;67(2):111–115. [DOI] [PubMed] [Google Scholar]
- 25. Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsuda-Futami E, Shioi A, Jono S, Inaba M, Nishizawa Y, Morii H. Lymphoblastoid interferon-alpha downregulates parathyroid hormone (PTH)/PTH-related peptide (PTHrP) receptor expression in human osteoblastic cells (Saos-2). Bone. 1998;23(3):205–211. [DOI] [PubMed] [Google Scholar]
- 27. Jemtland R, Holden M, Reppe S, et al. Molecular disease map of bone characterizing the postmenopausal osteoporosis phenotype. J Bone Miner Res. 2011;26(8):1793–1801. [DOI] [PubMed] [Google Scholar]
- 28. Benisch P, Schilling T, Klein-Hitpass L, et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. Plos One. 2012;7(9):e45142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao P, Chen Y, Jiang H, et al. In vivo genome-wide expression study on human circulating B cells suggests a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J Bone Miner Res. 2008;23(5):644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis Res Ther. 2007;9(5):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris JA, Kemp JP, Youlten SE, et al. ; 23andMe Research Team . An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51(2):258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bagur A, Vega E, Mautalen C. Discrimination of total body bone mineral density measured by dexa in vertebral osteoporosis. Calcif Tissue Int. 1995;56(4):263–267. [DOI] [PubMed] [Google Scholar]
- 33.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19(1):61–109. [DOI] [PubMed] [Google Scholar]
- 34. Pei YF, Hu WZ, Yan MW, et al. Joint study of two genome-wide association meta-analyses identified 20p12.1 and 20q13.33 for bone mineral density. Bone. 2018;110: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trajanoska K, Morris JA, Oei L, et al. ; GEFOS/GENOMOS consortium and the 23andMe research team . Assessment of the genetic and clinical determinants of fracture risk: genome wide association and mendelian randomisation study. Bmj. 2018;362:k3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barbeira AN, Dickinson SP, Bonazzola R, et al. ; GTEx Consortium . Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li YI, Knowles DA, Humphrey J, et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50(1):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abecasis GR, Auton A, Brooks LD, et al. ; 1000 Genomes Project Consortium . An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 40. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 43. Becker BJ. Combining significance levels. In: The handbook of research synthesis. New York, NY, US: Russell Sage Foundation; 1994:215–230. [Google Scholar]
- 44. Wu Y, Zeng J, Zhang F, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun. 2018;9(1):918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma M, Huang DG, Liang X, et al. Integrating transcriptome-wide association study and mRNA expression profiling identifies novel genes associated with bone mineral density. Osteoporos Int. 2019;30(7):1521–1528. [DOI] [PubMed] [Google Scholar]
- 47. Wainberg M, Sinnott-Armstrong N, Mancuso N, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51(4):592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trajanoska K, Rivadeneira F. The genetic architecture of osteoporosis and fracture risk. Bone. 2019;126:2–10. [DOI] [PubMed] [Google Scholar]
- 49. Movérare-Skrtic S, Henning P, Liu X, et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014;20(11):1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chesi A, Wagley Y, Johnson ME, et al. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun. 2019;10(1):1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kemp JP, Medina-Gomez C, Estrada K, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. Plos Genet. 2014;10(6):e1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sebastian A, Loots GG. Genetics of Sost/SOST in sclerosteosis and van Buchem disease animal models. Metabolism. 2018;80:38–47. [DOI] [PubMed] [Google Scholar]
- 53. Schwarze UY, Dobsak T, Gruber R, Bookstein FL. Anatomical similarity between the Sost-knockout mouse and sclerosteosis in humans. Anat Rec (Hoboken). 2020;303(9):2295–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Delgado-Calle J, Sato AY, Bellido T. Role and mechanism of action of sclerostin in bone. Bone. 2017;96:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Määttä JA, Bendre A, Laanti M, et al. Fam3c modulates osteogenic cell differentiation and affects bone volume and cortical bone mineral density. Bonekey Rep. 2016;5:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95(11):866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Powell CB, Alabaster A, Stoller N, et al. Bone loss in women with BRCA1 and BRCA2 mutations. Gynecol Oncol. 2018;148(3):535–539. [DOI] [PubMed] [Google Scholar]
- 58. Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. Plos Comput Biol. 2009;5(12):e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Higa M, Tanaka K, Saijo M. Inhibition of UVSSA ubiquitination suppresses transcription-coupled nucleotide excision repair deficiency caused by dissociation from USP7. Febs J. 2018;285(5):965–976. [DOI] [PubMed] [Google Scholar]
- 60. Sarasin A. UVSSA and USP7: new players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med. 2012;4(5):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang Y, Lv L, Li W, et al. Protein deubiquitinase USP7 is required for osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res Ther. 2017;8(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Richardson TG, Hemani G, Gaunt TR, Relton CL, Davey Smith G. A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun. 2020;11(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Talukder AH, Meng Q, Kumar R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006;25(9):1311–1319. [DOI] [PubMed] [Google Scholar]
- 64. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu Z, Greenblatt MB, Yan G, et al. SMURF2 regulates bone homeostasis by disrupting SMAD3 interaction with vitamin D receptor in osteoblasts. Nat Commun. 2017;8:14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen X, Wang Z, Duan N, Zhu G, Schwarz EM, Xie C. Osteoblast–osteoclast interactions. Connect Tissue Res. 2018;59(2):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Olszewska DA, Lonergan R, Fallon EM, Lynch T. Genetics of frontotemporal dementia. Curr Neurol Neurosci Rep. 2016;16(12):107. [DOI] [PubMed] [Google Scholar]
- 68. Kan O, Gorkem U. The effects of serum granulin levels on anthropometric measures and glucose metabolism in infertile women with different ovarian reserve status. Endokrynol Pol. 2019;70(3):255–259. [DOI] [PubMed] [Google Scholar]
- 69. Matsubara T, Mita A, Minami K, et al. PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 2012;15(1):38–50. [DOI] [PubMed] [Google Scholar]
- 70. Wang L, Roth T, Nakamura MC, Nissenson RA. Female-specific role of progranulin to suppress bone formation. Endocrinology. 2019;160(9):2024–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Silva T, Bradley RH, Gao Y, Coue M. Xenopus CDC7/DRF1 complex is required for the initiation of DNA replication. J Biol Chem. 2006;281(17):11569–11576. [DOI] [PubMed] [Google Scholar]
- 72. Chen L, Luo C, Shen L, et al. SRSF1 prevents DNA damage and promotes tumorigenesis through regulation of DBF4B pre-mRNA splicing. Cell Rep. 2017;21(12):3406–3413. [DOI] [PubMed] [Google Scholar]
- 73. Qian GF, Yuan LS, Chen M, et al. PPWD1 is associated with the occurrence of postmenopausal osteoporosis as determined by weighted gene co-expression network analysis. Mol Med Rep. 2019;20(4):3202–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Majer O, Liu B, Woo BJ, Kreuk LSM, Van Dis E, Barton GM. Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature. 2019;575(7782):371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thwaites R, Chamberlain G, Sacre S. Emerging role of endosomal toll-like receptors in rheumatoid arthritis. Front Immunol. 2014;5(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang D, Gilbert JR, Taylor GM, et al. TLR4 inactivation in myeloid cells accelerates bone healing of a calvarial defect model in mice. Plast Reconstr Surg. 2017;140(2):296e–306e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu K, Tan LJ, Wang P, et al. Functional relevance for associations between osteoporosis and genetic variants. Plos One. 2017;12(4):e0174808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhou X, Qiu YH, He P, et al. Why SNP rs227584 is associated with human BMD and fracture risk? A molecular and cellular study in bone cells. J Cell Mol Med. 2019;23(2):898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.