Highlights
-
•
It is important to develop biomarkers for the early detection of lung cancer.
-
•
By probing a protein microarray containing more than 9000 antigens with sputum of lung cancer patients and cancer-free smokers, we identified 28 lung tumor antigens (TAs).
-
•
A panel of three TA-associated autoantibody (TAAb) biomarkers was further developed that had 81% sensitivity and 83% specificity for diagnosis of lung cancer.
-
•
This study provides the first evidence for the potential of sputum TAAbs as biomarkers for the early detection of lung cancer.
Keywords: Lung cancer, Sputum, Autoantibodies, Biomarkers, Diagnosis
Abstract
Tumor antigens (TAs) can initiate host immune responses and produce TA-associated autoantibody (TAAbs), potential cancer biomarkers. Sputum is directly generated from the upper and lower airways, and thus can be used as a surrogate sample for the diagnosis of lung cancer based on molecular analysis. To develop sputum TAAb biomarkers for the early detection of lung cancer, the leading cause of cancer death, we probed a protein microarray containing more than 9,000 antigens with sputum supernatants of a discovery set of 30 lung cancer patients and 30 cancer-free smokers. Twenty-eight TAs with higher reactivity in sputum of lung cancer cases vs. controls were identified. The diagnostic significance of TAAbs against the TAs was determined by enzyme-linked immunosorbent assays (ELISAs) in sputum of the discovery set and additional 166 lung cancer patients and 213 cancer-free smokers (validation set). Three sputum TAAbs against DDX6, ENO1, and 14–3-3ζ were developed as a biomarker panel with 81% sensitivity and 83% specificity for diagnosis of lung cancer, regardless of stages, locations, and histological types of lung tumors. This study provides the first evidence that sputum TAAbs could be used as biomarkers for the early detection of lung cancer.
Introduction
Lung cancer is the leading cause of cancer-related deaths, accounting for 28% of all cancer deaths [1]. 85% of lung tumors are non-small cell lung cancers (NSCLCs) [1], which comprise two major histological subtypes: squamous cell carcinoma (SCC) and adenocarcinoma (AC). Given that the 5-year survival rate depends on the stage of lung cancer, the early detection of NSCLC is clinically important [1, 2]. Efforts have been made to develop tumor cells-derived proteins as circulating cell-free (e.g., serum or plasma) biomarkers for lung cancer. For instance, analysis of CEA, CYFRA-21-2, SFTBP57, EGFR, ProGRP58, and Ciz159 proteins in plasma showed promise for the diagnosis of lung cancer [3, 4]. However, since the circulating tumor proteins that are released from the cancer cells are few and diluted in a large serum or plasma volume, the sensitivity of the circulating cell-free molecular biomarkers in blood is very low, particularly for the early stages of NSCLC [5].
Self-proteins (antigens) of tumors can be affected by specific point mutations, misfolding, overexpression, aberrant glycosylation, truncation, or aberrant degradation [6,7]. The tumor antigens (TAs) elicit immune responses and stimulate TA-associated autoantibodies (TAAbs) before or during tumor formation [8], which may provide biomarkers for the diagnosis of cancer [9]. Compared with the TAs, TAAbs have several benefits as biomarkers. First, although the TAs are usually at an undetectable level in body fluids, the TAAbs response to the TAs are often amplified in the immune responses, and hence more easily detectable [7]. Second, as opposed to TAs that are frequently transiently elevated in body fluids, the corresponding TAAbs are relatively lasting, and thus are more stable and readily detected [8]. Third, TAAbs are highly specific and handily detectable in small volumes of specimens with well-established secondary reagents. Indeed, the potential use of determination of TAAbs in serum or plasma for the diagnosis of lung cancer was demonstrated [10]. For example, a serum based-earlyCDT-lung assay that consisted of six TAAbs against p53, NY-ESO-1, CAGE, GBU4–5, Annexin 1, and SOX2 could detect lung cancer with 38% sensitivity and 88% specificity [11]. However, the sensitivity of the serum TAAb biomarkers was not sufficient for the early diagnosis of lung cancer in the clinical settings.
Sputum is coughed up from the lower airways and deep lungs, and hence could act as a mirror to pulmonary diseases [12,13]. Sputum essentially consists of cellular and liquid fractions [14,15]. The cellular portion contains exfoliated airway epithelial cells from the bronchial tree [12, 13]. We and others have shown that genetic and epigenetic analyses of exfoliated respiratory epithelial cells could provide an approach for the early detection of lung cancer [[16], [17], [18], [19], [20],12,13,[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]]. The liquid fraction of sputum is complex dilute aqueous solution of lipids, glycoconjugates, and plasma-derived mediators and proteins [12,13]. In contrast with the epithelial cells exfoliated from local respiratory tract sites, sputum supernatant comprises the liquid portion that is produced by the cells lining the bronchi and airways where NSCLCs are immersed in the bio-fluid [12,13]. Furthermore, sputum supernatant may comprise local- and systemic-derivative lung TAs and the corresponding TAAbs, which raises the rationale for using sputum as a diagnostic medium for detection of NSCLC with TAAb biomarkers. Therefore, we hypothesized that the analysis of TAAbs in sputum may provide an alternative means for the noninvasive diagnosis of lung cancer. The objective of this study was to validate the hypothesis by identifying and developing TAAb biomarkers in sputum for the early detection of NSCLC.
Materials and methods
Study population
The study was performed under a research protocol (HP-40666) approved by the Institutional Review Board of University of Maryland Baltimore. Our enrollment criteria were individuals between the ages of 50–75 who had more than a 30 pack-year smoking history. The clinical diagnosis of lung cancer was made with histopathologic examinations of specimens obtained by CT-guided transthoracic needle biopsy, transbronchial biopsy, videotape-assisted thoracoscopic surgery, or surgical resection. The surgical pathologic staging was determined according to the TNM classification of the International Association for the Study of Union Against Cancer with the American Joint Committee on Cancer and the International Staging System for Lung Cancer. Histopathological classification was determined according to the World Health Organization classification. When tumors were located in the bronchus (cartilage bearing airway), they were classified as central lung cancers. When tumors did not arise from the bronchus and instead were associated with bronchioles, they were classified as peripheral lung cancer. A total of 196 lung cancer patients and 243 cancer-free smokers were enrolled. The cases and controls were divided into two sets. The first one consisted of 30 stage I NSCLC lung cancer patients and 30 cancer-free smokers (Table 1) and used as a discovery set. The second one included 68 stage I NSCLC, 46 stage II, and 52 stage III-IV patients, as well as 213 cancer-free smokers (Table 2) and was used as a validation set. To mitigate the confounding effects, the cases and controls were matched 1:1 by sex, age, cigarette smoking, and chronic obstructive pulmonary disease (COPD) in both sets of specimens. Information of lung tumor stages, locations, and histological types were included in the tables.
Table 1.
Characteristics of NSCLC patients and cancer-free smokers in cohort 1*.
| NSCLC cases (n = 30) | Controls (n = 30) | |
|---|---|---|
| Age | 66.32 (SD 11.37) | 65.79 (SD 11.33) |
| Sex | ||
| Female | 12 | 12 |
| Male | 18 | 18 |
| Race | ||
| White | 10 | 10 |
| African American | 20 | 20 |
| Pack-years (median) | 36.29 | 35.78 |
| Stage | all are stage I | |
| Histological type | ||
| Adenocarcinoma | 16 | |
| Squamous cell carcinoma | 14 | |
| Locations | ||
| Central tumors | 13 | |
| Peripheral tumors | 17 |
Abbreviations: NSCLC, non-small cell lung cancer.
All P>0.05.
Table 2.
Characteristics of NSCLC patients and cancer-free smokers in cohort 2 *.
| NSCLC cases (n = 166) | Controls (n = 213) | |
|---|---|---|
| Age | 67.25 (SD 10.94) | 66.18 (SD 11.15) |
| Sex | ||
| Female | 64 | 82 |
| Male | 102 | 131 |
| Race | ||
| White | 122 | 155 |
| African American | 44 | 58 |
| Pack-years (median) | 36.37 | 35.49 |
| Stage | ||
| Stage I | 68 | |
| Stage II | 46 | |
| Stage III-IV | 52 | |
| Histological type | ||
| Adenocarcinoma | 86 | |
| Squamous cell carcinoma | 80 | |
| Locations | ||
| Central tumors | 78 | |
| Peripheral tumors | 88 |
Abbreviations: NSCLC, non-small cell lung cancer.
All p > 0.05.
Sputum collection and preparation
We collected sputum from the participants as described in our previous reports [12,13,[21], [22], [23],25,27, 28,[34], [35], [36], [37], [38], [39]]. Sputum specimens were mucoid and of lower respiratory origin as indicated by the presence of bronchial epithelial cells [12,13,21,35]. Only sputum with a squamous epithelial cell count of <10 cells/high-power field was used in this study [12,35]. Each of the sputum samples was diluted with 3 ml of phosphate buffered saline (PBS) (Thermo Fisher Scientific, Grand Island, NY) per 1 gram of sputum. The diluted samples were passed through an 18-gage needle for mechanical disruption [12], and then centrifuged for 10 min. The supernatant was collected and further centrifuged at 3500 rpm for 20 min and stored at −80 °C until use.
Protein microarray profiling
The ProtoArray human protein microarrays (Invitrogen, Carlsbad CA) were used for profiling TAs in the sputum samples of discovery set (Table 1). Briefly, the microarray slides were treated with the blocking buffer (Invitrogen) at 4 °C for 1 h. We then probed the arrays with a 1:10 dilution of each sample. We incubated the microarrays at 4 °C for 2 h, which were washed five times with PBS buffer. We diluted Alexa Fluor®647-conjugated goat anti-human IgG antibody into probe buffer (Invitrogen) to a 1 µg/mL final concentration, which was added to each array and incubated for 1.5 h at 4 °C. We removed the secondary antibody, and washed the arrays, which were scanned using an Axon GenePix 4000B fluorescent microarray scanner. We analyzed the results by using the Prospector Imager and Analyzer (Invitrogen). We probed the microarrays with goat anti-GST antibody to check the uniformity of the proteins spotted in the protein microarrays followed by incubation with Alexa Fluor 555-labeled antigoat IgG. We also incubated other control microarrays only with the secondary antibody Alexa Fluor 647-labeled anti-human IgG for background determination.
Enzyme-linked immunosorbent assay (ELISA)
We purified recombinant fusion proteins for the selected TAs using UltimateTM ORF Clones (Invitrogen) according to the manufacturer's instructions. We examined expression of recombinant protein in SDS–PAGE and Coomassie blue staining. We used Western blot analysis to confirm if the bands were reactive with reference antibodies [40]. ELISA analysis of TAAbs against the corresponding TAs was carried out using a protocol as previously described [41,42]. Briefly, we coated purified proteins to a 96-well microliter plate (Fisher Scientific) at concentration of 0.5 μg/mL. We diluted the supernatant of sputum at 1:100 and horseradish peroxidase (HRP)—conjugated goat anti-human IgG at 1:4000, which were used as the primary and secondary antibodies, respectively. The reaction was sopped with 5% SDS, and the optical density (OD) was measured in an ELISA reader (Thermo Fisher Scientific) at 405 nm. We calculated the ELISA relative absorbance of each sputum sample–antigen reaction by the OD450 of expressed antigens divided by median OD450 of all antigens tested. We used the median value to normalize the background of each sputum specimen. Each sample was analyzed in triplicate.
Statistical analysis
To analyze microarray data, we compared the signal intensity from each protein with the signal from the negative control features among the array and assigned a CI-p value for each protein. We identified the significant signals and calculated the Z-scores, which reflected the signal strength relative to all protein features. We compared the two groups and identified the proteins that had increased signal values in both groups. We calculated a p value for each protein across the hypothesis that there was no signal increase in one group compared with another. We compared clusters using Multi Experiment Viewer (Dana-Farber Cancer Institute, Boston, MA) based on Pearson's correlation distant metrics using normalized values in log10 to picture the discrimination between the two groups.
To analyze ELISA data, we used a one-tailed Student's t-test to evaluate whether the means of normal and cancer cases were statistically different from each other. We used Pearson's correlation analysis to determine the relationship of each targets with demographic characteristics of the patients and cancer-free controls. We used final clinical and pathologic diagnoses as reference standards to estimate sensitivity and specificity of the analysis of each biomarker. Furthermore, we analyzed the significantly associated TAAbs by using logistic regression models with constrained parameters as least absolute shrinkage and selection operator (LASSO) to eliminate the less relevant variables [33]. We estimated functions of the combined biomarkers by logistic regression with or without adjustment for known risk factors for NSCLC. Receiver-operator characteristic (ROC) curve analysis was undertaken using expression level for each biomarker from cancer patients and cancer-free controls by analyze-it software (analyze-it Software, Leeds, UK). We also generated a 95% confidence interval for the difference in the area under the ROCs (AUCs) by using the bootstrap. We established the optimal cut-off value by using the Youden index. Moreover, to compare different biomarker panels, we computed their AUCs to determine the sensitivity and specificity as previously described [43].
Results
TAs of lung cancer in sputum identified by protein microarrays
The lowest average background in sputum was 482 relative fluorescence units (RFUs), and the highest average background was 517 RFUs. Mean background signal values on the arrays were within the expected range for all sputum samples. Furthermore, signal used values for protein (antigen) features reached the scanner maximum of ∼65,000 RFU, indicating a dynamic range >2 logs. Therefore, the experiments and data acquisition of the microarray analysis were performed under optimal conditions. Altogether, 28 TAs (Table 3) were identified that showed significantly differential signal intensities between lung cancer patients and cancer-free smokers with a p< 0.05 and signal intensity ratio of two groups being greater than 3.0. Among the 28 antigens, 26 or two displayed increased or decreased levels in lung cancer cases vs. cancer-free smokers, respectively.
Table 3.
28 TAs showed significantly differential signal intensities between lung cancer patients and cancer-free smokers.
| GenBank ID | Protein Description | Ratio* | pvalue |
|---|---|---|---|
| BC039826.1 | DEAD (Asp‐Glu‐Ala‐Asp) box polypeptide 6 (DDX6) | 43.6 | 3.36E−03 |
| NP_001419.1 | Enolase 1 (ENO1) | 33.8 | 7.36E−03 |
| BC050424.1 | F‐box protein 38 (FBXO38) | 26.2 | 2.76E−03 |
| NP_001129173.1 | 14–3–3 protein zeta (14–3–3ζ) | 19.7 | 5.60E−03 |
| NP_640,343.1 | Cancer/testis antigen 1 (CTAG1A) | 15.9 | 4.50E−03 |
| PV3665 | Casein kinase 1, delta (CSNK1D) | 12.1 | 7.32E−03 |
| CAA56734.1 | Cell surface associated Mucin 1 (MUC1) | 10.7 | 1.78E−03 |
| BC026100.2 | Testis‐specific transcript, Y‐linked 8 (TTTY8) | 8.4 | 1.80E−03 |
| BC059404.1 | B‐cell CLL/lymphoma 6, member B (BCL6B) | 7.8 | 4.01E−02 |
| NM_002742.1 | Serine/threonine‐protein kinase D1 (PRKD1) | 7.5 | 2.37E−02 |
| NP_000691.1 | Annexin 1 (ANXA1) | 6.9 | 2.86E−02 |
| BC001497.2 | RAN binding protein 5 (RANBP5) | 5.1 | 2.35E−02 |
| NM_005118.2 | tumor necrosis factor superfamily, member 15 (TNFSF15) | 4.6 | 3.21E−03 |
| BC005220.1 | Chaperonin containing TCP1, subunit 8 (theta) (CCT8) | 45.1 | 8.33E−03 |
| AAH04364.1 | Transmembrane protein 39B (TMEM39B) | 3.2 | 3.65E-02 |
| NM_006792.2 | Mortality factor 4 (MORF4) | 43.3 | 2.33E−02 |
| NM_203,454. | Putative C‐>U‐editing enzyme APOBEC‐4 (APOBEC4) | 28.3 | 1.67E−02 |
| XM_936,115.1 | HLA class II histocompatibility antigen, DQ(3) alpha chain (HLA-DQA3) | 22.6 | 2.13E-02 |
| PV3825 | Casein kinase 1, gamma 1 (CSNK1G1) | 21.9 | 1.67E−02 |
| NM_002622.3 | Prefoldin subunit 1 (PFDN1) | 17.3 | 2.63E−02 |
| NM_031966.4 | Cyclin B1 (CCNB1) | 4.7 | 2.67E−02 |
| BC033708.1 | Ral GEF with pH domain and SH3 binding motif 1 (RALGPS1) | 4.8 | 4.50E−03 |
| AAF66640.1 | Heat shock protein HSP60 (HSP60) | 40.3 | 2.38E−02 |
| NM_005517.2 | High‐mobility group nucleosomal binding domain 2 (HMGN2) | 3.5 | 8.54E−03 |
| BC041157.1 | Thromboxane A synthase 1 (TBXAS1) | 3.6 | 9.34E−03 |
| BC096097.2 | Histidine ammonia‐lyase (HAL) | 4 | 6.23E−03 |
| NM_172,160.1 | Methylcrotonoyl‐Coenzyme A carboxylase 1 (alpha) (MCCC1) | 10.6# | 2.53E−03 |
| BC013119.1 | Signal‐induced proliferation‐associated 1 like 2 (SIPA1L2) | 3# | 3.33E−02 |
signal intensity ratio of two groups.
the proteins’ signal intensity ratio is calculated by mean of signal intensity of cancer-free smokers divided by that lung cancer patients, suggesting the proteins has a low level in lung cancer patients vs. cancer-free smokers.
The proteins in bold are chosen for subsequent ELISA validation based on criteria: 1) having differential signals between two the populations with a p ≤ 0.01 and, 2) having signal intensity ratio between the two populations being >10.0.
Validation of the identified lung TAs by ELISA in sputum
From the 28 antigens, 8 antigens (Tables 3 and 4) were chosen for subsequent ELISA validation based on criteria: 1) having differential signals between the two populations with a p ≤ 0.01 and, 2) having signal intensity ratio between the two populations being >10.0. ELISA was first performed in sputum samples of the discovery set that was used in the protein microarray analysis. Of the 8 TAs tested, seven or one had augmented or reduced interaction levels with TAAbs in sputum of lung cancer cases vs. cancer-free smokers, respectively (All p < 0.01). The tested TAs by ELISA displayed the same significant direction as by protein array (All p < 0.01) (Table 4). To further validate the TAs, we analyzed TAAbs against the 8 TAs in a validation set with a relatively large sample size that consisted of 166 patients diagnosed with lung cancer in different stages, locations, and histological types and 213 cancer-free smokers. Consistent with data observed in discovery set, the TAAbs in validation set displayed a considerably different level in sputum of lung cancer patients vs. control subjects (All p < 0.01).
Table 4.
ELISA validation of eihgt lung cancer-antigens in cohort 1.
| Protein Description | Relative OD in cancer-free smokers | Relative OD in cancer patients | pvalues |
|---|---|---|---|
| DDX6 | 0.2125 (0.4131) | 1.112 (1.1276) | 6.92E−08 |
| ENO1 | 0.0046 (0.0186) | 0.15655 (0.3968) | 0.00399 |
| FBXO38 | 0.03077 (0.0496) | 0.3220 (0.5037) | 0.00002 |
| 14–3–3ζ | 0.00652 (0.0168) | 0.09751 (0.2493) | 0.006 |
| ESO-1 | 0.00831 (0.0303) | 0.2315 (0.6001) | 0.0052 |
| CSNK1D | 0.0967 (0.2643) | 0.4955 (0.8259) | 0.0005 |
| MUC1 | 0.05822 (0.1446) | 0.76187 (1.5741) | 0.00095 |
| MCCC1 | 4.2286 (0.6328) | 1.7976 (1.6222) | 9.41E−09 |
Development of sputum TAAb biomarkers for lung cancer
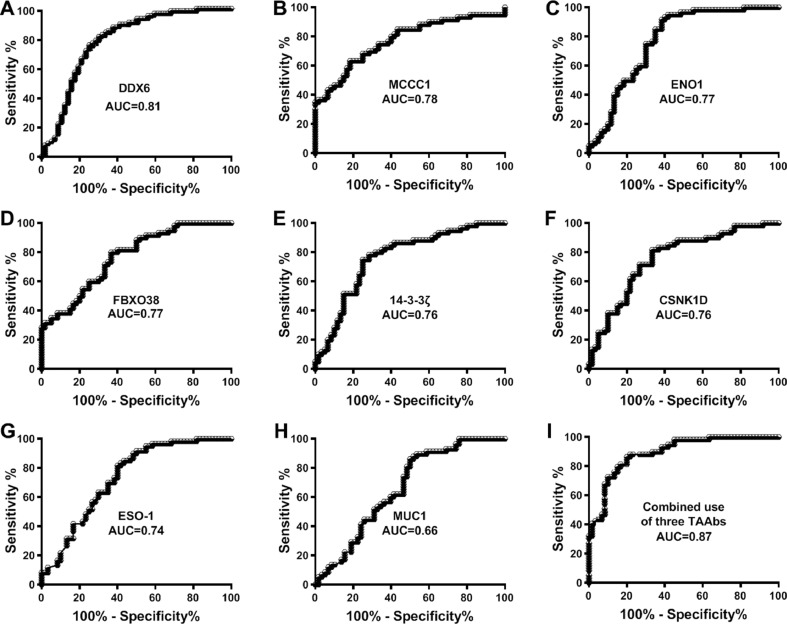
We evaluated the potential of using TAAbs against the 8 TAs for diagnosis of NSCLC in the 166 cases and 213 controls. The TAAbs exhibited AUC values of 0.66–0.81 in distinguishing NSCLC patients from cancer-free smokers with 60–74% sensitivities and 60–78% specificities (Fig. 1 and Table 5). We used a stepwise logistic regression model to select the optimal panel of biomarkers. Three TAAbs, which were against DDX6, ENO1, and 14-3-3ζ respectively, were selected as the best biomarkers in a panel. The optimal cutoff for the combined biomarkers was U = 0.3628, where U=−6.2573+1.3529*log (DDX6)-1.9647*log (ENO1)+2.3165*14-3-3ζ. Any subject with U≥0.3628 was classified as a NSCLC case. The combined use of the three TAAbs produced 0.88 AUC in distinguishing NSCLC patients from the controls, which was significantly higher than that of any single one (p < 0.05) (Fig. 1). Furthermore, the analysis of the three biomarkers produced 81% sensitivity and 83% specificity for diagnosis of NSCLC (Table 5). Interestingly, combined analysis of the three TAAbs did not display a statistical difference between stages, locations, and histological types of lung cancer (All p > 0.05). Furthermore, the prevalence of the biomarkers was related with pack-years of smoking (p < 0.05), however, not associated with patient age and gender.
Fig. 1.
Receiver-operator characteristic (ROC) curves of 8 TAAbs in sputum of 166 patients diagnosed with NSCLC and 213 cancer-free smokers.
Table 5.
diagnostic performance of eight lung cancer-antigens in cohort 2.
| Protein Description | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|
| DDX6 | 0.8062 (0.7367 to 0.8905) | 73.68% (62.32% to 83.13%) | 75.44 (62.24% to 85.87%) |
| MCCC1 | 0.7762 (0.6912 to 0.8593) | 70.00% (56.79% to 81.15%) | 71.67% (58.56% to 82.55%) |
| ENO1 | 0.7747 (0.6883 to 0.8611) | 75.00% (62.14% to 85.28%) | 70.00% (56.79% to 81.15%) |
| FBXO38 | 0.7686 (0.6864 to 0.8508) | 61.67% (48.21% to 73.93%) | 70.00% (56.79% to 81.15%) |
| 14–3–3ζ | 0.7656 (0.6789 to 0.8522) | 58.33% (44.88% to 70.93%) | 78.62% (67.67% to 89.22%) |
| CSNK1D | 0.7615 (0.6744 to 0.8473) | 71.67% (58.56% to 82.55%) | 73.33% (60.34% to 83.93%) |
| ESO-1 | 0.7358 (0.6457 to 0.8259) | 70.00% (56.79% to 81.15%) | 65.00% (51.60% to 76.87%) |
| MUC1 | 0.6636 (0.5634 to 0.7639) | 60.34% (46.64% to 72.95%) | 60.34% (46.64% to 72.95%) |
| Combined use of three TAAbs | 0.8667 (0.7935 to 0.9499) | 81.36% (69.09% to 90.31)% | 83.33% (71.48% to 91.71%) |
Abbreviations: AUC, the area under receiver operating characteristic curve; CI, confidence interval.
Discussion
Since sputum is directly secreted from the lower airways and deep lungs where lung tumors exist, it has the advantages as surrogate material over serum or plasma for specifically diagnosing lung cancer. First, sampling of sputum is simple and does not require trained health care professionals. Second, sputum is microbiologically stabilized after the collection. Third, the collection can be done anywhere, including at home. Fourth, large numbers of sputum samples can be collected rapidly, and therefore, are particularly useful for screening a large number of high-risk individuals or for usage in decentralized regions. Finally, and most importantly, since sputum originates from the deep lungs where lung tumors are immersed in the biofluid, sputum may constitute a direct source of TAAb biomarkers for the early detection of NSCLC.
In this study, we first comprehensively profiled proteins in sputum using microarray and identified TAs in sputum of lung cancer patients. Sputum TAAbs against TAs (DDX6, ENO1, and 14-3-3ζ) were further developed as a biomarker panel that had 81% sensitivity and 83% specificity for diagnosis of lung cancer. In addition, the observed TAAb responses were independent of patients’ characteristics, including race, gender, and tumor stage and type, suggesting that the sputum TAAb biomarkers could be used for equally detecting the major types of NSCLC at the early stages, regardless of locations of the primary tumors.
ENO1 exists on the surface of cells and has important functions in cell migration and tissue invasion. Activated ENO1 has a direct coupling to glycolytic activity. Furthermore, ENO1 is upregulated by HIF1α in response to hypoxia [44]. We have found that ENO1 gene has increased levels in both genomic copy number and transcript in lung cancer cells compared with normal cells, and its dysregulation is involved in lung tumorigenesis [45]. Furthermore, we developed a panel of six genomic probes including one targeting ENO1 gene that could be analyzed in the exfoliated airway epithelial cells of sputum for lung cancer detection [22]. Moreover, our previous study revealed that ELISA analysis of ENO1 protein in sputum supernatant had a diagnostic value in distinguishing patients with stage I lung cancer from cancer-free individuals [12].
14–3–3ζ is a family of cellular proteins and could interact with other crucial cellular proteins involved in the tumor development and progression [38]. We previously found that increased 14-3-3ζ protein level was positively associated with stage and grading of NSCLC and inversely related to poor outcomes of the patients [38]. Furthermore, lung cancer cells with down-regulation of 14-3-3ζ were more sensitive to cisplatin in vitro that was associated with the inhibition of cell proliferation, additive G2-M cell cycle arrest, and increased apoptosis [38]. In addition, 14-3-3ζ might act as a proto-oncogene and have oncogenic function in the development of NSCLC tumorigenesis [38]. Interestingly, Qiu et al. previously showed that the occurrence serum TAAb to 14-3-3ζ was one of the three biomarkers that could precede onset of symptoms and diagnosis of lung cancer [46].
We are interested to find that TAAb against DDX6 (also named RCK/p54) is one of the biomarkers in the panel. DDX6 is a member of the family of DEAD/H-box RNA helicases and associated with t(11;14)(q23;q32) chromosomal translocation in B-cell lymphoma cell line RC-K8 [47], [48], [49]. DDX6 has numerous functions, including translation initiation, pre-mRNA splicing, ribosome assembly by acting as an RNA-binding protein. It also contributes to the proliferation and differentiation of the stem and progenitor cells. However, functions of DDX6 in lung carcinogenesis are largely unknown. Herein we show that DDX6 might be a novel TA and detection of the corresponding TAAb in sputum could help diagnose NSCLC at the early stage. Furthermore, the TAAbs are not only noble candidates for the early detection of NSCLC but potential targets for therapeutic intervention of the malignancy.
Although showing promise, the TAAb biomarkers developed in this study are still limited in their sensitivity (81%) and specificity (83%) for diagnosis of lung cancer. We are extensively evaluating all the 28 TAs defined by the protein microarray in a large case-control study to identify additional TAAb biomarkers that can be added to the three so that the diagnostic efficacy of the approach could be improved. Furthermore, it will be useful in the future to combine the TAAb biomarkers with other types of molecular biomarkers, such as DNA and microRNAs, in sputum to create a combined panel with better diagnostic characteristics. In addition, given that the TAs in sputum and blood might have diverse roles in the carcinogenesis of NSCLC through various mechanisms, further comparing sputum and serum ATTb biomarkers for the diagnostic values and determining if their integration would have a synergistic influence on detection of NSCLC is warranted.
The overall 5-year survival rate for stage I NSCLC patients who are typically treated with surgery remains up to 83%. In contrast, only 5–15% and less than 2% of patients with stage III and IV NSCLC are alive after five years [1,2]. These statistics provide the primary rationale to develop biomarker that can improve NSCLC early detection [1,2]. Furthermore, the National Lung Screening Trial (NLST) shows that low-dose CT (LDCT) could detect NSCLC at the early stage and significantly reduce the mortality [2]. However, 25% of smokers screened by LDCT have indeterminate pulmonary nodules (PNs), of which 96% are benign growths. LDCT screening for lung cancer often produces a high false-positive rate and over-diagnosis. Therefore, many patients with benign PNs will undergo harmful diagnostic and therapeutic procedures, presenting a major clinical challenge. The ability to immediately and precisely distinguish malignant from benign PNs at baseline LDCT screening is an unmet need. The TAAb biomarkers might have two important applications. First, they could be used for the early detection of lung cancer patients in high-risk populations (e.g., heavy smokers). Second, the use of the biomarkers in LDCT-based lung cancer screening would i), spare individuals with benign growths from radiation exposure and unnecessary surgical resections or biopsies, and ii), lead to more personalized therapy by enabling effective treatments to be immediately initiated for lung cancer, and hence reduce lung cancer-associated mortality.
In conclusion, using protein microarray together with ELISA, we developed three TAAbs as a small panel of sputum biomarkers for the early detection of lung cancer. The findings from this study may lay the basis to perform a next step to a new diagnostic approach that might have future clinical utility.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgments
We thank the University of Maryland Marlene and Stewart Greenebaum Comprehensive Cancer Center Biostatistics Shared Service to provide statistical analysis services.
Ethical approval and consent to participate
All work was approved by the Ethics Committee of University of Maryland Baltimore
Consent for publication
We obtained consent to publish this paper from all the participants of this study.
Authors' contributions
NL and VKH conducted the experiments and participated in data interpretation. VKH, JD, and NWT participated in coordination and acquisition of data. VKH, NWT and FJ participated in study design, coordination, and data analysis and interpretation, and prepared the manuscript. All authors read and approved the final manuscript.
Footnotes
This work was supported in part by National Cancer Institute grant 1UH2CA251139-01, 5R21CA240556-02, VA merit Award I01 CX000512, and University of Maryland Marlene & Stewart Greenebaum Comprehensive Cancer Center Pilot Grant Program (F. J.).
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., Sicks J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada M., Nishio W., Sakamoto T., Uchino K., Yuki T., Nakagawa A., Tsubota N. Prognostic significance of perioperative serum carcinoembryonic antigen in non-small cell lung cancer: analysis of 1,000 consecutive resections for clinical stage I disease. Ann. Thorac. Surg. 2004;78:216–221. doi: 10.1016/j.athoracsur.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Hanash S.M., Ostrin E.J., Fahrmann J.F. Blood based biomarkers beyond genomics for lung cancer screening. Transl. Lung Cancer Res. 2018;7:327–335. doi: 10.21037/tlcr.2018.05.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belczacka I., Latosinska A., Metzger J., Marx D., Vlahou A., Mischak H., Frantzi M. Proteomics biomarkers for solid tumors: current status and future prospects. Mass Spectrom. Rev. 2019;38:49–78. doi: 10.1002/mas.21572. [DOI] [PubMed] [Google Scholar]
- 6.Desmetz C., Mange A., Maudelonde T., Solassol J. Autoantibody signatures: progress and perspectives for early cancer detection. J. Cell. Mol. Med. 2011;15:2013–2024. doi: 10.1111/j.1582-4934.2011.01355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaenker P., Gray E.S., Ziman M.R. Autoantibody production in cancer–the humoral immune response toward autologous antigens in cancer patients. Autoimmun. Rev. 2016;15:477–483. doi: 10.1016/j.autrev.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Murphy M.A., O'Leary J.J., Cahill D.J. Assessment of the humoral immune response to cancer. J. Proteomics. 2012;75:4573–4579. doi: 10.1016/j.jprot.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Chapman C.J., Murray A., McElveen J.E., Sahin U., Luxemburger U., Tureci O., Wiewrodt R., Barnes A.C., Robertson J.F. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax. 2008;63:228–233. doi: 10.1136/thx.2007.083592. [DOI] [PubMed] [Google Scholar]
- 10.Yang B., Li X., Ren T., Yin Y. Autoantibodies as diagnostic biomarkers for lung cancer: a systematic review. Cell. Death Discov. 2019;5:126. doi: 10.1038/s41420-019-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam S., Boyle P., Healey G.F., Maddison P., Peek L., Murray A., Chapman C.J., Allen J., Wood W.C., Sewell H.F., Robertson J.F. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev. Res. 2011;4:1126–1134. doi: 10.1158/1940-6207.CAPR-10-0328. [DOI] [PubMed] [Google Scholar]
- 12.Yu L., Shen J., Mannoor K., Guarnera M., Jiang F. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin. Lung Cancer. 2014;15:372–378. doi: 10.1016/j.cllc.2014.05.003. e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Q., Todd N.W., Li R., Peng H., Liu Z., Yfantis H.G., Katz R.L., Stass S.A., Jiang F. Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer. 2008;114:275–283. doi: 10.1002/cncr.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishwal S.C., Das M.K., Badireddy V.K., Dabral D., Das A., Mahapatra A.R., Sahu S., Malakar D., Singh I.I., Mazumdar H., Patgiri S.J., Deka T., Kapfo W., Liegise K., Kupa R.U., Debnath S., Bhowmik R., Debnath R., Behera R.K., Pillai M.G., Deuri P., Nath R., Khalo K.P., Sing W.A., Pandit B., Bhattacharya S., Behera D., Saikia L., Khamo V., Nanda R.K. Sputum proteomics reveals a shift in vitamin D-binding protein and antimicrobial protein axis in tuberculosis patients. Sci. Rep. 2019;9:1036. doi: 10.1038/s41598-018-37662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose M.C., Voynow J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 16.Thunnissen F.B. Sputum examination for early detection of lung cancer. J. Clin. Pathol. 2003;56:805–810. doi: 10.1136/jcp.56.11.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belinsky S.A., Palmisano W.A., Gilliland F.D., Crooks L.A., Divine K.K., Winters S.A., Grimes M.J., Harms H.J., Tellez C.S., Smith T.M., Moots P.P., Lechner J.F., Stidley C.A., Crowell R.E. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 18.Belinsky S.A. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat. Rev. Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 19.Varella-Garcia M., Kittelson J., Schulte A.P., Vu K.O., Wolf H.J., Zeng C., Hirsch F.R., Byers T., Kennedy T., Miller Y.E., Keith R.L., Franklin W.A. Multi-target interphase fluorescence in situ hybridization assay increases sensitivity of sputum cytology as a predictor of lung cancer. Cancer Detect. Prev. 2004;28:244–251. doi: 10.1016/j.cdp.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Varella-Garcia M., Schulte A.P., Wolf H.J., Feser W.J., Zeng C., Braudrick S., Yin X., Hirsch F.R., Kennedy T.C., Keith R.L., Baron A.E., Belinsky S.A., Miller Y.E., Byers T., Franklin W.A. The detection of chromosomal aneusomy by fluorescence in situ hybridization in sputum predicts lung cancer incidence. Cancer Prev. Res. 2010;3:447–453. doi: 10.1158/1940-6207.CAPR-09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R., Todd N.W., Qiu Q., Fan T., Zhao R.Y., Rodgers W.H., Fang H.B., Katz R.L., Stass S.A., Jiang F. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin. Cancer Res. 2007;13:482–487. doi: 10.1158/1078-0432.CCR-06-1593. [DOI] [PubMed] [Google Scholar]
- 22.Jiang F., Todd N.W., Li R., Zhang H., Fang H., Stass S.A. A panel of sputum-based genomic marker for early detection of lung cancer. Cancer Prev. Res. 2010;3:1571–1578. doi: 10.1158/1940-6207.CAPR-10-0128. [DOI] [PubMed] [Google Scholar]
- 23.Anjuman N., Li N., Guarnera M., Stass S.A., Jiang F. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clin. Transl. Med. 2013;2:15. doi: 10.1186/2001-1326-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta C., Su J., Zhan M., Stass S.A., Jiang F. Sputum long non-coding RNA biomarkers for diagnosis of lung cancer. Cancer Biomark. 2019;26:219–227. doi: 10.3233/CBM-190161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F., Todd N.W., Qiu Q., Liu Z., Katz R.L., Stass S.A. Combined genetic analysis of sputum and computed tomography for noninvasive diagnosis of non-small-cell lung cancer. Lung Cancer. 2009;66:58–63. doi: 10.1016/j.lungcan.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz R.L., Zaidi T.M., Fernandez R.L., Zhang J., He W., Acosta C., Daniely M., Madi L., Vargas M.A., Dong Q., Gao X., Jiang F., Caraway N.P., Vaporciyan A.A., Roth J.A., Spitz M.R. Automated detection of genetic abnormalities combined with cytology in sputum is a sensitive predictor of lung cancer. Mod. Pathol. 2008;21:950–960. doi: 10.1038/modpathol.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N., Ma J., Guarnera M.A., Fang H., Cai L., Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J. Cancer Res. Clin. Oncol. 2014;140:145–150. doi: 10.1007/s00432-013-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen J., Liao J., Guarnera M.A., Fang H., Cai L., Stass S.A., Jiang F. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J. Thorac. Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su J., Anjuman N., Guarnera M.A., Zhang H., Stass S.A., Jiang F. Analysis of lung flute-collected sputum for lung cancer diagnosis. Biomark Insights. 2015;10:55–61. doi: 10.4137/BMI.S26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su J., Leng Q., Lin Y., Ma J., Jiang F., Lee C.J., Fang H. Integrating circulating immunological and sputum biomarkers for the early detection of lung cancer. Biomark Cancer. 2018;10 doi: 10.1177/1179299X18759297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su J., Liao J., Gao L., Shen J., Guarnera M.A., Zhan M., Fang H., Stass S.A., Jiang F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget. 2016;7:5131–5142. doi: 10.18632/oncotarget.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y., Fang H., Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics. 2016;8:109. doi: 10.1186/s13148-016-0275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su Y., Guarnera M.A., Fang H., Jiang F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol. Cancer. 2016;15:36. doi: 10.1186/s12943-016-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Y., Todd N.W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S.A., Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing L., Su J., Guarnera M.A., Zhang H., Cai L., Zhou R., Stass S.A., Jiang F. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin. Cancer Res. 2015;21:484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xing L., Todd N.W., Yu L., Fang H., Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod. Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 37.Yu L., Todd N.W., Xing L., Xie Y., Zhang H., Liu Z., Fang H., Zhang J., Katz R.L., Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int. J. Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan T., Li R., Todd N.W., Qiu Q., Fang H.B., Wang H., Shen J., Zhao R.Y., Caraway N.P., Katz R.L., Stass S.A., Jiang F. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–7906. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 39.Jiang F., Qiu Q., Khanna A., Todd N.W., Deepak J., Xing L., Wang H., Liu Z., Su Y., Stass S.A., Katz R.L. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol. Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai L., Ren P., Liu M., Imai H., Tan E.M., Zhang J.Y. Using immunomic approach to enhance tumor-associated autoantibody detection in diagnosis of hepatocellular carcinoma. Clin. Immunol. 2014;152:127–139. doi: 10.1016/j.clim.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Csorba K., Schmidt S., Florea F., Ishii N., Hashimoto T., Hertl M., Karpati S., Bruckner-Tuderman L., Nishie W., Sitaru C. Development of an ELISA for sensitive and specific detection of IgA autoantibodies against BP180 in pemphigoid diseases. Orphanet. J. Rare. Dis. 2011;6:31. doi: 10.1186/1750-1172-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai L., Tsay J.C., Li J., Yie T.A., Munger J.S., Pass H., Rom W.N., Zhang Y., Tan E.M., Zhang J.Y. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer. 2016;99:172–179. doi: 10.1016/j.lungcan.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Shen J., Liu Z., Todd N.W., Zhang H., Liao J., Yu L., Guarnera M.A., Li R., Cai L., Zhan M., Jiang F. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito T., Funamoto K., Sato N., Nakamura A., Tanabe K., Hoshiai T., Suenaga K., Sugawara J., Nagase S., Okamura K., Yaegashi N., Kimura Y. Maternal undernutrition induces the expression of hypoxia-related genes in the fetal brain. Tohoku J. Exp. Med. 2012;226:37–44. doi: 10.1620/tjem.226.37. [DOI] [PubMed] [Google Scholar]
- 45.Li R., Wang H., Bekele B.N., Yin Z., Caraway N.P., Katz R.L., Stass S.A., Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;25:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 46.Qiu J., Choi G., Li L., Wang H., Pitteri S.J., Pereira-Faca S.R., Krasnoselsky A.L., Randolph T.W., Omenn G.S., Edelstein C., Barnett M.J., Thornquist M.D., Goodman G.E., Brenner D.E., Feng Z., Hanash S.M. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J. Clin. Oncol. 2008;26:5060–5066. doi: 10.1200/JCO.2008.16.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iio A., Takagi T., Miki K., Naoe T., Nakayama A., Akao Y. DDX6 post-transcriptionally down-regulates miR-143/145 expression through host gene NCR143/145 in cancer cells. Biochim. Biophys. Acta. 2013;1829:1102–1110. doi: 10.1016/j.bbagrm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Taniguchi K., Iwatsuki A., Sugito N., Shinohara H., Kuranaga Y., Oshikawa Y., Tajirika T., Futamura M., Yoshida K., Uchiyama K., Akao Y. Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 2018;57:579–589. doi: 10.1002/mc.22781. [DOI] [PubMed] [Google Scholar]
- 49.Tokumaru Y., Tajirika T., Sugito N., Kuranaga Y., Shinohara H., Tsujino T., Matsuhashi N., Futamura M., Akao Y., Yoshida K. Synthetic miR-143 inhibits growth of HER2-positive gastric cancer cells by suppressing KRAS networks including DDX6 RNA helicase. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20071697. [DOI] [PMC free article] [PubMed] [Google Scholar]