Graphical abstract
Abbreviations: PHD finger, Plant Homeodomain finger; NSD1, Nuclear receptor-binding SET (Su(var)3–9, Enhancer of zeste, Trithorax) domain protein 1; Nizp1, (NSD1-interacting Zn-finger protein); PHDVC5HCHNSD1, Fifth PHD and C5HCH tandem domain of NSD1; C2HRNizp1, C2HR finger domain of Nizp1; NMR, Nuclear Magnetic Resonance; VS, Virtual Screening; STD, saturation transfer difference
Keywords: PHD finger, NSD1, Nizp1, Protein-protein interactions, Virtual screening, NMR
Abstract
PHD fingers are small chromatin binding domains, that alone or in tandem work as versatile interaction platforms for diversified activities, ranging from the decoding of the modification status of histone tails to the specific recognition of non-histone proteins. They play a crucial role in their host protein as mutations thereof cause several human malignancies. Thus, PHD fingers are starting to be considered as valuable pharmacological targets. While inhibitors or chemical probes of the histone binding activity of PHD fingers are emerging, their druggability as non-histone interaction platform is still unexplored. In the current study, using a computational and experimental pipeline, we provide proof of concept that the tandem PHD finger of Nuclear receptor-binding SET (Su(var)3–9, Enhancer of zeste, Trithorax) domain protein 1 (PHDVC5HCHNSD1) is ligandable. Combining virtual screening of a small subset of the ZINC database (Zinc Drug Database, ZDD, 2924 molecules) to NMR binding assays and ITC measurements, we have identified Mitoxantrone dihydrochloride, Quinacrine dihydrochloride and Chloroquine diphosphate as the first molecules able to bind to PHDVC5HCHNSD1 and to reduce its documented interaction with the Zinc finger domain (C2HRNizp1) of the transcriptional repressor Nizp1 (NSD1-interacting Zn-finger protein). These results pave the way for the design of small molecules with improved effectiveness in inhibiting this finger-finger interaction.
1. Introduction
PHD fingers are small and abundant non-catalytic Zn2+ binding domains annotated in more than 170 human chromatin-related proteins [1]. They are often present in single or multiple copies or in tandem with other epigenetic reader domains in so called erasers or writers, i.e. enzymes harboring catalytic domains able to change the chromatin modification status [2], [3], [4]. They are typically involved in the recognition of specific histone marks (H3K4me0, H3K4me3, H3K9me3) to modulate the transcriptional activity of the hosting protein [5], [6]. Mutations of PHD fingers can lead to aberrant protein functions that on turn can trigger different pathological conditions, including neurological and developmental diseases, cancer and immunological disorders [7]. Hence, similarly to other chromatin binding domains such as BRD [8] and PWWP [9], PHD fingers are becoming appealing epigenetic drug targets, thus representing a promising pharmacological alternative to classical enzymatic inhibition strategies of their host protein [10], [11], [12], [13], [14]. Importantly, the versatile structural scaffold of PHD fingers, alone or in tandem with other chromatin binding domains, results in diversified functions that go beyond the perceiving of the epigenetic landscape [15]. In particular, they have emerged as multifaceted interaction platforms, well-suited for bridging their host proteins with other subunits of bigger macromolecular chromatin complexes [12], [16], [17], herewith offering additional pharmacological intervention opportunities. A paradigmatic example of PHD fingers serving as hub for protein–protein interactions is represented by the tandem PHD finger of Nuclear receptor-binding SET (Su(var)3–9, Enhancer of zeste, Trithorax) domain protein 1 (NSD1). It is composed by a canonical (PHDV) and a degenerate (C5HCH) PHD finger domain that form together an indivisible structure (PHDVC5HCHNSD1) [18] (Fig. 1). NSD1 is a histone methyl transferase whose translocation or mutations lead respectively to pediatric acute myeloid leukemia (AML) [19] and to the inherited overgrowth disease Sotos Syndrome [20]. Herein PHDVC5HCHNSD1 appears to play a pathophysiological role, as it contributes to oncogenesis and inappropriate Hox genes activation in AML condition [19] and is hotspot for Sotos Syndrome point mutations [18]. Moreover, it mediates the recruitment of its host protein to the transcriptional repressor Nizp1 (NSD1 interaction zinc finger protein) [21], [22] via a direct finger-finger interaction with the Zinc-finger domain of Nizp1 (C2HRNizp1), herewith reinforcing its repressive activity [22] (Fig. 1). The pathophysiological role of this repressive complex is still elusive, therefore the identification of chemical probes able to interfere with the PHDVC5HCHNSD1/C2HRNizp1 interaction would be a desirable tool to dissect its biological outcomes. Modulators of this finger-finger interaction might create new opportunities for the development of inhibitors of aberrant NSD1 function(s) and complement the classical blockade of its histone methyl transferase activity [23]. In this framework we have previously solved the NMR structure of PHDVC5HCHNSD1, and shown that it does not recognize Histone H3 tail peptides, but it binds to C2HRNizp1. We have generated a three-dimensional model of the complex and shown that the interaction (Kd = 4 μM) is highly specific. In particular, an exposed evolutionary conserved RWR loop of C2HRNizp1 accommodates into the PHDVC5HCHNSD1 interdomain groove and creates hydrophobic and electrostatic intermolecular interactions [18] (Fig. 2A). The biophysical assessment of this finger-finger interaction has created the premises for studies aiming at the identification of small molecules and/or molecular probes able to interfere with NSD1/Nizp1 complex and modulate NSD1 activity. Identification of protein–protein interactions (PPIs) modulators offers a wide range of opportunities in terms of specific inhibition. However, the targeting of PPIs with small molecules poses formidable challenges, mainly because of the large multifaceted binding surfaces of PPIs that make their druggability quite difficult [24], [25]. Here, coupling the virtual screening of a small subset of the ZINC database (Zinc Drug Database, ZDD) to NMR binding assays and ITC measurements, we have identified 3 compounds able to bind to PHDVC5HCHNSD1 and to reduce the interaction with C2HRNizp1. These results pave the way for the design of more efficient inhibitors of this finger-finger interaction.
Fig. 1.
Schematic representation of NSD1 and Nizp1 domain architecture and of the interaction between PHDVC5HCHNSD1 and C2HRNizp1. C-terminal residue numbers of NSD1 and Nizp1 and domain boundaries of PHDVC5HCHNSD1 and C2HRNizp1 are indicated. The representation is not drawn to scale.
Fig. 2.
Ligandability assessment of PHDVC5HCHNSD1. A) Data driven docking model of PHDVC5HCHNSD1/C2HRNizp1 complex [18]. PHDVC5HCHNSD1 and C2HRNizp1 are represented in surface and cartoon, respectively. B) FTmap consensus sites (CSs) identified on the PHDVC5HCHNSD1 structure. The protein and the CSs clusters, ranked from the highest to the lowest populated, are shown in solid and mesh surface, respectively.
2. Materials and methods
2.1. FTMap analysis
Hot spots identification was performed using the FTMap computational map server (www.ftmap.bu.edu). The lowest energy structure of PHDVC5HCHNSD1, as determined by NMR, (PDB code PDB 2NAA) was uploaded into the FTMap server and ran according to instructions [26]. The results were visually inspected using PyMol [27].
2.2. Virtual screening
The in silico screening of the ZINC Drug Database (ZDD) against PHDVC5HCHNSD1 was performed using the virtual screening workflow of the Schrödinger software suite 2019-3 (Schrödinger, LLC, New York, NY, 2019). The lowest energy structure of PHDVC5HCHNSD1 (PDB 2NAA) [18] used for virtual screening was prepared with the Protein Preparation Wizard tool of Maestro [28]. The orientation of the solvent exposed hydroxyl groups of Serine, Threonine and Tyrosine were optimized. A restrained minimization was run using the OPLS3 force field [29] with a root mean square deviation (RMSD) tolerance on heavy atoms of 0.3 Å. The receptor grid was generated defining the centroid of active site residues E2204 and S2123, which are directly involved in PHDVC5HCHNSD1/C2HRNizp1 interaction [18] with a box dimension of 12 Å × 15 Å × 18 Å. As S2123 plays a fundamental role in complex formation [18], we allowed rotation of its hydroxyl group. LigPrep [30] was used to prepare the 2924 compounds of the ZDD with Epik [31], [32] at 7 ± 2.0 pH units using standard parameters.
Virtual screening was carried out in three steps including high-throughput virtual screening (HTVS), Standard Precision (SP), and extra precision (XP) docking stage, after each step, the best 30, 50 and 20% compounds in terms of docking score were kept, respectively. To obtain a post-processing ligand binding energy a molecular mechanics generalized Born surface area (MM-GBSA) calculation was performed on the final 172 compounds with Prime [33]. The best 50 molecules in terms of ΔG of binding were selected for visual inspection. Among them 19 unique compounds, commercially available and showing good solubility in water were purchased for experimental validation. ADME-related properties of the selected compounds were evaluated using QikProp [30] program running in normal mode. QikProp generates physically relevant descriptors, and uses them to perform ADME predictions, number of violations of Lipinski’s rule of five [34] and of Jorgensen’s rule of three [35].
2.3. Reagents
The following molecules were purchased from Sigma-Merck: ZINC08101116 (Gentamicine sulfate), ZINC03872123 (Clozapine), ZINC00001547 (Hydroxystilbamidine bis(methanesulfonate)), ZINC08214681 (Streptomycin), ZINC08214483 (Amikacin), ZINC08214692 (Tobramycin), ZINC04213094 (Isoemetite), ZINC18098320 (Chlorhexidine), ZINC01530861 (Chloroquine diphosphate), ZINC00601274 (Astemizole), ZINC03830246 (Quinacrine dihydrochloride), ZINC03794794 (Mitoxantrone dihydrochloride), ZINC08214590 (Kanamycin). The following molecules were purchased from Mcule ZINC22443609 (plerixafor), ZINC3830583 (Clindamycin hydrochloride), ZINC52981502 (Netilmicin), ZINC03830924 (Idabucirin HCl), ZINC04097283 (Lyncomicyn hydrochloride) and ZINC14880004 (Rolitetracycline).
2.4. Sample preparation for NMR and binding assays
Murine PHDvC5HCHNSD1 (Glu2117–Asp2207, NCBI Reference Sequence: NM 008739.3) and murine C2HRNizp1 (residues Glu397–Lys434, corresponding to Glu398–Lys435 in the human sequence, NCBI Reference Sequence: NM 032752.3) were expressed in E. coli and purified as described in [18]. As the mouse PHDVC5HCHNSD1 sequence shares 99% identity with the human one (residues Glu2116–Asp2206 in the human sequence NM 022455.4), we adopted the human sequence numbering scheme to map ligand interactions. Uniformly 15N-labeled PHDvC5HCHNSD1 was expressed by growing E. coli BL21 (DE3) cells in minimal bacterial medium containing 15NH4Cl, as sole nitrogen source. The NMR buffer of both PHDvC5HCHNSD1 and C2HRNizp1 contained D2O 10% (v/v) 20 mM sodium phosphate pH 6.3, 0.15 M NaCl, 2 mM dithiothreitol (DTT), 50 μM ZnCl2 with 0.15 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). Stock solution of ligands (10 mM) contained 20 mM phosphate buffer, pH 6.3, 0.15 M NaCl.
2.5. NMR measurements
NMR spectra were recorded at 295 K on a Bruker Avance 600 MHz spectrometer (Karlsruhe, Germany) equipped with a triple-resonance TCI cryoprobe with an x, y, z-shielded pulsed-field gradient coil. Spectra were processed with Topspin™ 3.2 (Bruker) and analyzed with CcpNmr Analysis 2.3 [36]. 1H-15N-HSQC assignments of PHDvC5HCHNSD1 were taken from the BMRB databank (accession number: 25933). 1H assignment of the 1–3 has been performed acquiring and analyzing classical 2D 1H–1H TOCSY (TOtal Correlation SpectroscopY, tmix = 60 ms), 2D 1H1H NOESY (Nuclear Overhauser Effect SpectroscopY, tmix = 200 ms, T = 295 K) in 20 mM phosphate buffer, pH 6.3, 0.15 M NaCl.
Titrations. For NMR titrations, at each titration point a 2D water-flip-back 1H-15N-edited HSQC spectrum was acquired with 2048 (160) complex points for 1H (15N), respectively, apodized by 90° shifted squared (sine) window functions and zero filled to 256 points for indirect dimension. Assignment of the labelled proteins in the presence of the ligands was obtained following individual cross-peaks through the titration series. For each residue the weighted average of the 1H and 15N chemical shift perturbation (CSP) was calculated as CSP = [(Δδ2HN + Δδ2N/25)/2]1/2 [37].
Ligand titrations have been performed on 15N-PHDvC5HCHNSD1 adding 0.5, 1, 2, 3, 6, 10, 20 equivalents of ligands to the labelled protein. In order to minimize dilution and NMR signal loss, titrations were carried out by adding small aliquots of concentrated ligands (typically 10 mM, dissolved in 20 mM phosphate buffer, pH 6.3, 0.15 M NaCl) to the 15N labelled protein samples (0.13 mM, dissolved in NMR buffer).
Dissociation constant estimation. The apparent dissociation constants of the ligands-15N- PHDvC5HCHNSD1 interactions were estimated from least-squares fitting of CSPs as a function of total ligand concentration according to the following equation:
with a = 8(Ka/δb)∙[Pt], b = 1 + Ka∙([Lti] + [Pt]), and c = δb∙Ka∙[Lti], [Lti] is the total ligand concentration at each titration point, [Pt] is the total protein concentration, Ka = 1/Kd is the association constant, and δb is the chemical shift of the resonance in the complex, δi is the absolute change in chemical shift for each titration point. The Kd were obtained from the average plus the standard deviation of the fitting values of 7 residues, whose resonances were well resolved in the HSQC spectra and had CSP > avg + sd. Kd and δb were used as fitting parameters using the Xmgrace program (http://plasma-gate.weizmann.ac.il/Grace/).
Saturation Transfer Difference (STD) experiments. STD experiments have been performed on 2 mM ligands in the presence of 0.1 mM PHDvC5HCHNSD1 in NMR buffer. STD experiments were acquired using a pulse scheme (Bruker pulse sequence: stddiffesgp.3) with excitation sculpting with gradients for water suppression and spin-lock field to suppress protein signals. The spectra were acquired using 128 scans, a spectral width of 9600 Hz, 64 K data points for acquisition. For protein saturation, a train of 60 Gaussian shaped pulses of 50 ms was applied, for a total saturation times of 3 s, with relaxation delays of 3 s. On- and off-resonance irradiations were set at 0 ppm and at 107 ppm, respectively. STD spectra were obtained by internal subtraction of the on-resonance spectrum from the off-resonance spectrum. To analyze the STD effect we used the amplification factor (AFSTD) [38]. The AFSTD at a given ligand concentration ([L]T) was obtained by multiplying the relative STD effect of a given ligand hydrogen (ISTD/I0) (where ISTD and I0 correspond to the peak integral in the STD spectrum and in the off-resonance spectrum, respectively, and ISTD = I0-ISAT, where ISAT is the intensity of the saturated peak) with the molar ratio of ligand in excess relative to the protein ([L]T/[P]):
To obtain an epitope mapping we quantitatively expressed the differences in ASTD for the different hydrogens calculating the relative STD effects (STD%), whereby for each ligand the hydrogen with highest ASTD was set to 100%, and used as a reference to calculate the relative STD effects for the other protons.
2.6. Isothermal titration calorimetry competition experiments
ITC titrations were performed at 23 °C using a VP-ITC isothermal titration calorimeter (MicroCal LLC, Northampton, MA, USA). Recombinant proteins and ligands were dissolved in the same buffer (20 mM NaH2PO4 /Na2HPO4 pH 7.2, 0.15 M NaCl, 2 mM beta-mercaptoethanol, 10 μM ZnCl2). The ΔG of binding between C2HRNizp1 and PHDVC5HCHNSD1 was previously determined in [18]. To monitor the inhibition activity of 1–3, we stepwise titrated 1.5–3 mM C2HRNizp1 into a cell containing a pre-formed protein–ligand complex (formed by 0.15 mM PHDVC5HCHNSD1 with a 20-fold excess of either molecule 1, 2 or 3) up to a three or five-fold molar excess. The quantity of heat absorbed or released in the process was measured. Control experiments were performed under identical conditions to determine the dilution heat of the titrant C2HRNizp1 into buffer and of the buffer into protein–ligand samples. Data were analyzed with the software ORIGIN 7.0®.
3. Results
3.1. Assessment of PHDVC5HCHNSD1 ligandability
PPIs are traditionally classified as difficult pharmacological targets because of their ‘poor druggability’ reputation, mainly due to the structural and dynamic complexity of the interfacial features [24], [39]. However, recent successful studies have challenged this assumption demonstrating that the targeting of protein–protein interfaces is feasible, thus increasing the possibilities of pharmacological intervenes [40]. In particular, PPIs can be mediated by so-called “hot spots”, where specific hydrophobic interactions and, to a lesser extent, polar interactions drive most of the affinity [41]. This is the case for PHDVC5HCHNSD1/C2HRNizp1 interaction where hydrophobic and polar contacts formed by the R415W416R417 (RWR) loop of C2HRNizp1 and the PHDVC5HCHNSD1 interdomain groove represent the major driving force for complex formation, as assessed by mutagenesis studies [18]. Prompted by these results, we applied FTMap [26] to assess in silico the ligandability of the surface of PHDVC5HCHNSD1. Herein, the druggability of a target protein was defined based on two criteria: i) the number of probe clusters within a consensus site (CS) higher than 16 and ii) the existence of at least one additional weaker hot spot within 8 Å from the CS itself. FTMap identified 7 CSs (Fig. 2B), with the first two being the largest one that fulfilled the druggability requirements. In particular, the highest populated site (CS1) was composed by 20 probe clusters and was close to one weaker hot spot (CS7, consisting of 4 probes). The second one (CS2) contained 19 probes and was within 8 Å from two weaker hot spots (CS3, and CS4 consisting of 16 and 13 probes, respectively). Interestingly, CS2 was located at the interface between PHDvNSD1 and C5HCHNSD1, in the C2HRNizp1 interaction surface (Fig. 2), supporting the notion that this region could be druggable and appropriate for a virtual screening campaign.
3.2. Virtual screening: targeting the PHDVC5HCHNSD1/C2HRNizp1 interaction surface
We used the Schrödinger Suite 2019-3 (Schrödinger Inc., LLC) to dock into PHDVC5HCHNSD1 (pdb code: 2NAA) the ZINC Drug Database, composed by 2924 commercially available compounds, including worldwide commercially available approved drugs as pure compounds, Drugbank approved molecules and nutraceuticals, and Food and Drug Administration approved drugs. The screening workflow consisted in three sequential docking steps comprising a high-throughput virtual screening (HTVS), a Standard Precision (SP), and an extra precision (XP) docking stage. 172 compounds emerging from the XP step were subsequently re-ranked according to molecular mechanics generalized Born surface area (MM-GBSA) calculations (Fig. 3). The best 50 molecules in terms of binding energy were selected for visual inspection. Finally, 19 unique compounds, commercially available and showing good water solubility were selected for further experimental validation (Supplementary Table S1). The “drug-likeness”, the bioavailability and the pharmacokinetic profiles of the candidates were assessed on the basis of Lipinski’s “Rule of Five” (Ro5) [34], Jorgensen’s “Rule of three” (Ro3) [35] and ADME properties (absorption, distribution, metabolism and excretion), respectively (Supplementary Table S1). As proposed by Jorgensen and Duffy [35] to assess the pharmacokinetic profiles of the selected compounds we used an overall ADME-compliance score (#stars), indicating the number of property descriptors computed by QikProp that fall outside the 95% range of similar values for known drugs. Herein, we observed that 21.1% and 26.3% of the compounds showed no Lipinski and Jorgensen violations, respectively, while the 85% and 100% showed ≤2 Ro5 and Ro3 violations, respectively (Supplementary Fig. S1). Moreover, the 79% of the selected candidates showed suitable ADME properties with up to a maximum of 5 molecular descriptors and predicted properties falling outside the normal range of known drugs, thus indicating that the selected molecules are expected to have good pharmacokinetic properties (Supplementary Fig. S2).
Fig. 3.

Virtual screening workflow.
3.3. Hits validation
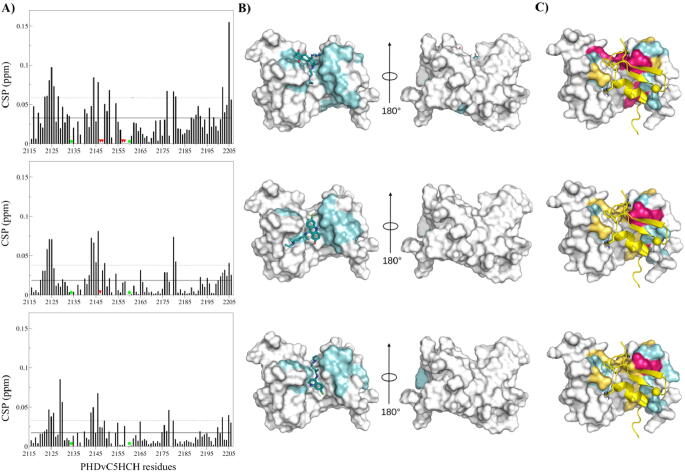
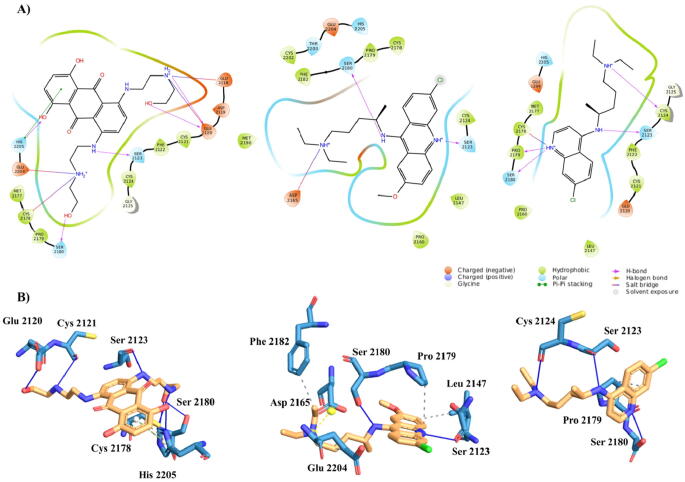
The water-soluble and commercially available hits identified in the virtual screening (19 molecules) (Table 1) were subsequently tested for their binding to PHDVC5HCHNSD1 by NMR spectroscopy using protein-based methods [42] to assess possible interactions and mapping thereof on PHDVC5HCHNSD1 structure. We stepwise titrated purified 15N-labeled PHDVC5HCHNSD1 (0.13 mM) with increasing concentrations of ligand (up to 2.5 mM), whereby for each titration point we recorded heteronuclear single-quantum coherence (HSQC) spectra. 3 molecules turned out to be negative for binding, as judged by their 15N HSQC spectra that superimposed perfectly on a reference experiment (Supplementary Fig. S3A), and 7 molecules induced protein aggregation. 6 molecules caused very weak and scattered chemical shift perturbations (CSPs) on the protein surface, suggestive of aspecific interactions (Supplementary Fig. S4). Only 3 compounds (1, ZINC03794794 – Mitoxantrone dihydrochloride; 2, ZINC03830246 – Quinacrine dihydrochloride; 3, ZINC01530861 – Chloroquine diphosphate) (Fig. 4A) proved to be positive, and induced several small but significant CSPs, indicative of weak interactions occurring in the fast exchange regime on the NMR time scale (Fig. 4B). By fitting the shifts of selected NMR peaks upon addition of increasing ligands concentrations we estimated a Kd of 1.2 ± 0.4 mM for 1, 1.4 ± 0.3 mM for 2 and 4.7 ± 1.4 mM for 3 (Fig. 4C, Table 1). Overall, the pattern of significant CSPs (CSP > avg + sd) was similar for the titrated molecules, with resonances of C2124NSD1, G2125NSD1, A2144NSD1, C2146NSD1 S2180NSD1, H2205NSD1 shifting upon addition of 1–3 (Fig. 5A, Supplementary Table S2). Here, projection onto the docking poses of the residues significantly shifting during ligands titrations allowed to generate a mapping of the interactions, which indicates that the three hits targeted one face of the inter-domain groove between PHDv and C5HCH (Fig. 5B). Interestingly, the resonances of residues mostly affected by ligands binding coincided or were nearby those significantly affected by C2HRNzip1 interaction (Fig. 5C, Supplementary Table S2). We next performed saturation transfer difference (STD) experiments as orthogonal NMR methods to further validate the binding of molecules 1–3. STD is a well-recognized ligand-based NMR method, commonly used to probe ligand–protein interactions of medium-weak affinity (high nM to mM) [43]. The experiment is based on the intermolecular saturation transfer from the protein to the binding part of the ligand. Thus, the part of the ligand in direct proximity to the protein shows the most intense NMR signals, herewith allowing for the identification of those ligand hydrogens which are closest to the receptor [43]. Reference and STD spectra performed on a solution containing 2 mM ligand and 0.1 mM PHDVC5HCHNSD1 exhibited narrow and well resolved lines. Significant STD effects were observed for ligands 1–3 unequivocally demonstrating their binding to PHDVC5HCHNSD1 (Fig. 6A,B). To obtain group ligand epitopes [38], i.e. to identify those hydrogens in close contact with PHDVC5HCHNSD1, for each molecule we calculated the relative STD effect (STD%) at 2 s of saturation time. We observed that for the three molecules the aromatic protons displayed the highest relative STD effect (with 70–100% saturation), implying their close proximity to the protein. Conversely, the aliphatic protons contributed to a lesser extent suggesting only a modest involvement in the interaction (Fig. 6A,B). Overall, MM-GBSA refined docking poses of the three ligands were consistent with NMR chemical shift perturbations and in part with STD epitope mapping (Fig. 5, Fig. 7). Ligand Interaction Diagrams of the binding site showed that the aromatic moieties of all the compounds bound to the hydrophobic groove at the interface of PHDv and C5HCH establishing critical apolar interactions with residues L2147NSD1, C2178NSD1, P2179NSD1, F2182NSD1, E2204NSD1 and H2205NSD1 (Fig. 7A,B). In the docking poses all the compounds created also polar interactions with residues around the hydrophobic interface groove, whereby each compound established at least two hydrogen bonds with S2180NSD1 and the carbonyl of S2123NSD1. Molecule 1 and molecule 3 established additional polar contacts with E2120NSD1, C2121NSD1, C2178NSD1, H2205NSD1, and with the carbonyl of C2124NSD1, respectively (Fig. 7). However, the relatively low percentage of saturation in STD experiments observed for the methylene nearby the hydroxyl groups of molecule 1 and the N-ethyl groups of molecules 2 and 3 suggests that these aliphatic chains do not stably interact with PHDvC5HCHNSD1. Collectively, validation of the virtual screening hits through ligand based and protein based NMR experiments allowed to identify three aromatic ligands that specifically target the inter-domain region of PHDvC5HCHNSD1 that is recognized by the RWR motif of C2HRNizp1.
Table 1.
Molecules emerging from VS selected for experimental validation. For positive hits the dissociation constant (Kd) are reported. For negative hits the type of interaction (aspecific, aggregation (*) or no binding (–) is reported.
| # | ZINC code | Commercial Name | Interaction/Kd |
|---|---|---|---|
| 1 | ZINC03794794 | Mitoxantrone dihydrochloride | 1.2 ± 0.4 mM |
| 2 | ZINC03830246 | Quinacrine dihydrochloride | 1.4 ± 0.3 mM |
| 3 | ZINC01530861 | Chloroquine diphosphate | 4.7 ± 1.4 mM |
| 4 | ZINC18098320 | Chlorhexidine | * |
| 5 | ZINC08101116 | Gentamicin sulfate | Aspecific |
| 6 | ZINC22443609 | Plerixafor | * |
| 7 | ZINC08214590 | Kanamycin | – |
| 8 | ZINC14880004 | Rolitetracycline | * |
| 9 | ZINC03872123 | Clozapine | Aspecific |
| 10 | ZINC03830583 | Clindamicin hydrochloride | – |
| 11 | ZINC00001547 | Hydroxystilbamidine bismethanesulfonate | * |
| 12 | ZINC52981502 | Netilmicin | Aspecific |
| 13 | ZINC08214681 | Streptomycin | Aspecific |
| 14 | ZINC08214483 | Amikacin | * |
| 15 | ZINC08214692 | Tobramycin | Aspecific |
| 16 | ZINC00601274 | Astemizole | * |
| 17 | ZINC03830924 | Idabucirin hydrochloride | * |
| 18 | ZINC04097283 | Lyncomicyn hydrochloride | – |
| 19 | ZINC04213094 | Isoemetite | Aspecific |
Fig. 4.
Molecules 1–3 bind to 15N labelled PHDVC5HCHNSD1. A) The chemical structures of the 3 hits. B) Superposition of 15N HSQC spectra of PHDVC5HCHNSD1 (0.13 mM, 20 mM phosphate buffer, pH 6.3, 0.15 M NaCl, T = 295 K) in the absence (in black) and in the presence (red) of twenty-fold excess of 1 (top), 2 (middle), and 3 (bottom). C) Plot of normalized chemical shift change versus concentration for selected residues, when 1 (top), 2 (middle) and 3 (bottom) are titrated into 15N-PHDVC5HCH NSD1. Dissociation constants were measured by fitting average CSP at each concentration of titrant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Mapping of the ligands binding sites. A) Histograms showing chemical shift perturbations (CSPs) of 15N‐labeled PHDVC5HCHNSD1 amides (0.1 mM) upon addition of 20‐fold excess of 1 (top), 2 (middle), 3 (bottom). Missing residues are prolines. Green circles indicate residues whose amides resonances are not present in the free protein because of exchange with the solvent. Red triangles indicate residues whose amide resonances disappear upon ligand addition (C2146NSD1, L2147NSD1, N2148NSD1, W2157NSD1, E2158NSD1 for 1 and L2147NSD1 for 2). Residues with CSPs > avg + sd elicited by 1 (top), 2 (middle), 3 (bottom) (or disappearing upon ligand binding) are projected in cyan on B) their corresponding binding pose; on C) PHDVC5HCHNSD1 in complex with C2HRNizp1 (cartoon representation). Residues whose resonances were perturbed upon addition of C2HRNizp1 are shown in yellow. Residues affected by both the ligands and C2HRNzip1 are highlighted in magenta. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Ligands epitope mapping. A) Aromatic and aliphatic regions of the STD experiment performed on 1 (top), 2 (middle), 3 (bottom). Resonance assignments are indicated according to (B). On- and off-resonance spectra are shown in the lower and upper panels, respectively. B) Low (0% < STD% < 30%), medium (30% ≤ 8STD% < 70%), and high (STD% ≥ 870%) relative STD percentage are mapped with green, yellow, and red circles, respectively on the 2D chemical structure. 1H assignments of 1–3 are summarized in Supplementary Table S3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Binding pose of 1–3. A) Ligand Interaction diagram and B) binding pose of 1 (left), 2 (center) and 3 (right). Molecules and protein interacting residues are shown in orange and blue sticks, respectively. Hydrogen bond, salt bridge, π-stacking and hydrophobic interactions are shown with blue line, yellow, green and grey dot lines, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Molecules 1–3 reduce the interaction between C2HRNizp1 and PHDVC5HCHNSD1
As both the CSPs mapping of the interaction on PHDVC5HCHNSD1 and the docking poses suggested that 1–3 obstructed to a certain extent the C2HRNizp1 binding site (Fig. 1B, Fig. 7), we asked whether these molecules, despite their low binding affinity, were able to interfere at least in part in the interaction between PHDVC5HCHNSD1 and C2HRNizp1. To verify this hypothesis, we performed ITC experiments titrating C2HRNizp1 into PHDVC5HCHNSD1 solutions that were previously saturated with a twenty-fold excess of 1–3 and compared the results with ITC titrations in the absence of ligands. Herein, we observed that all the three molecules reduced the interaction between C2HRNizp1 and PHDVC5HCHNSD1 by one order of magnitude, with the dissociation constant increasing from 4 μM to 40–50 μM in the absence and presence of ligands, respectively (Fig. 8A–D, Table 2). Saturation of PHDVC5HCNSD1 with 1–3 reduced the contributions of both the enthalpic and the entropic terms to the ΔG of binding. These results, in accordance with the observed CSPs in NMR titrations, support the notion that 1–3 target at least in part the C2HRNizp1 binding site herewith weakening the interaction between PHDVC5HCHNSD1 and. C2HRNizp1.
Fig. 8.
Molecules 1–3 reduce the interaction between C2HRNizp1 and PHDVC5HCHNSD1. ITC-binding curves of C2HRNizp1 to PHDVC5HCHNSD1, in the presence of twenty-fold excess of A) 1, B) 2, C) 3 and D) in the absence [18] of 1–3. The upper panel shows the sequential heat pulses for domain-domain binding, and the lower panel shows the integrated data, corrected for heat of dilution and fit to a single-site-binding model using a nonlinear least-squares method (line). The dissociation constants are indicated.
Table 2.
Thermodynamic parameters of the interaction between C2HRNizp1 and PHDvC5HCHNSD1 in the absence and in the presence of a twenty-fold excess of molecules 1–3. ΔG, ΔH, ΔS, stoichiometry (n) and dissociation constants (Kd) as measured by ITC experiments at T = 293.15 K are indicated.
| ΔG (kcal/mol) | ΔH (kcal/mol) | ΔS (cal/mol) | N | Kd (μM) | |
|---|---|---|---|---|---|
| No ligand [18] | −7.3 ± 0.1 | 4.1 ± 0.1 | 38.7 ± 0.1 | 1.1 ± 0.1 | 3.8 ± 0.7 |
| 1 | −5.9 ± 0.1 | 3.6 ± 0.1 | 32.2 ± 0.2 | 0.9 ± 0.1 | 40.5 ± 2.2 |
| 2 | −5.8 ± 0.1 | 2.5 ± 0.2 | 27.8 ± 0.1 | 1.0 ± 0.1 | 54.1 ± 1.1 |
| 3 | −5.9 ± 0.1 | 2.7 ± 0.1 | 29.0 ± 0.2 | 1.0 ± 0.1 |
|
5. Discussion and conclusions
PHD fingers are small epigenetic readers able to decode the modification status of histones [5], [6]. They are involved in several diseases, especially in cancer, thus their pharmacological targeting is starting to be considered as a complementary therapeutic strategy to the classical enzymatic inhibition of their host proteins [44]. These small Zn2+ binding domains are versatile structural platforms well suited for diversified protein–protein interactions. This is the case for the PHD fingers of MLL1 [45] of Pygo2 [46] and of Sp140 [47], interacting respectively with Cyp33, Bcl9 and Pin1, or for the second PHD finger of AIRE, that functions as a hub for multiple protein–protein interactions [16]. While inhibitors or chemical probes of the histone binding activity of PHD fingers are starting to emerge [11], [12], [13], the druggability of PHD fingers (alone or in tandem) as non-histone interaction platform remains completely unexplored. This is possibly due to the limited information on both the biological significance and the molecular details at the basis of these interactions. Hence, identification of chemical probes targeting these complexes would be extremely enlightening in terms of their structural and functional understanding, even though tackling large PPI targets with synthetic molecules still remains a considerable endeavor [24]. In the current study we have applied a computational and experimental pipeline to investigate the ligandability of PHDVC5HCHNSD1 as finger-finger interaction platform. To this aim we took advantage of a 3D model of PHDVC5HCHNSD1/C2HRNizp1 interaction [18] and inspected PHDVC5HCHNSD1 structure for possible hotspots suitable for small molecules targeting. These so-called “sticky zones” have emerged as collecting sites of most of the binding energy of inhibitors, thus opening new possibilities for the rational selection of binding molecules [48], [49]. Indeed FTmap analysis [26] predicted the presence of two putative contiguous ligand binding hotspots, that partially coincided with PHDVC5HCHNSD1 interaction surface with C2HRNizp1. To further explore the ligandability of PHDVC5HCHNSD1 we performed a structure-based in silico screening followed by NMR validation of the best hits emerging from our computational protocol. We found three structurally related molecules: the type II topoisomerase inhibitor mitoxantrone (1) used as salvage therapy for AML [50], the antimalaric drugs chloroquine [51] (3) and its acridine analog quinacrine (2), repositioned for treatment of cancer [52]. They all were able to bind with millimolar affinity to PHDVC5HCHNSD1. This value is well in the range of the Kds measured for first hits derived from VS campaigns aiming at the identification of PPI inhibitors. For example, a millimolar affinity has been measured by NMR for Benzimidazoles able to compete with the histone-binding pocket of the Pygo PHD Finger [12]. Similarly, an NMR fragment based screening of the PWWP1 domain of NSD3 yielded hits with mM affinities [9]. Importantly, mapping of the interaction by NMR CSPs methods confirmed that the three molecules target the C2HRNizp1 binding pocket located at the interface between PHDV and C5HCH. This surface is in accordance with the one defined by the Glide docking poses (Supplementary Fig. S5) and with the predicted hotspot region, with 1 accommodating in the CS4 pocket, and 2 and 3 each occupying two adjacent sites, i.e. CS1, CS2 and CS3, CS4, respectively. Orthogonal ligand-based NMR experiments confirmed binding. In particular, epitope mapping via STD experiments suggested that the fused aromatic rings, that are in common to the three molecules, mostly contribute to the interaction. Most likely they create apolar interactions with the small hydrophobic patch at the interdomain interface. Intriguingly, the quinolinyl, acridyl and antraquinone moities of chloroqine (3), quinacrine (2) and mitoxantrone (1), respectively, are reminiscent of the indole moiety of the RWR motif of C2HRNizp1 and might mimic its crucial interaction with PHDVC5HCHNSD1. As a matter of fact, saturation of PHDVC5HCHNSD1 with an excess of the single ligands resulted in a reduction of the finger-finger interaction strength by one order of magnitude, as assessed by ITC measurements. We are aware of the fact that the affinities of the identified compounds are still not appropriate for in vivo applications. PPI inhibitors with in vivo efficacy typically have high nanomolar-low micromolar affinities to their targets. This applies for example for the molecules JQ1 and nutlin, inhibitors of the BET-H4acetylated [53] and of MDM-p53 interactions [54], respectively. Thus, future work should be dedicated to optimize the chemical scaffolds identified in this work in order to generate more potent inhibitors. Our results suggest that the molecules that we have identified mimic only in part the interactions that are crucial for PHDVC5HCHNSD1/C2HRNip1 complex formation. We hypothesize that fragment-based approaches in which the identification and linkage of fragments targeting contiguous hotspots could be a successful strategy. Alternatively, peptidomimetic approaches could be a viable strategy to target non-contiguous hotspots [48], [53]. Design of peptidomimetic of C2HRNizp1 coupled to click-chemistry stabilization are currently ongoing. Nevertheless, the results obtained in this study, show for the first time that a finger-finger interaction, that involves a relatively large interaction surface composed on one side by a RWR signature and by an α-helix, and on the other side by a shallow hydrophobic groove, has proven tractable by small-molecule inhibition. The identified compounds may be thus considered as interesting leads for the development of new and more efficient NSD1/Nizp1 inhibitors. The determination of a high resolution three-dimensional structure of the PHDVC5HCHNSD1/C2HRNizp1 complex will be fundamental for the tailored design of ligands (peptidomimetics, small molecules) targeting the non-histone binding properties of these tandem PHD fingers.
6. Author statement
AB performed the experiments, analyzed the results and wrote the manuscript. MG performed the virtual screening analyzed the results and wrote the manuscript. GQ performed NMR experiments. GM, AB, MG conceived and designed throughout the studies, analyzed and interpreted all the achieved results. GM wrote the manuscript. All authors reviewed and approved the final manuscript.
CRediT authorship contribution statement
Andrea Berardi: Investigation, Formal analysis, Methodology, Validation, Visualization Data curation Writing original draft. Michela Ghitti: Investigation, Formal analysis, Methodology, Validation, Software, Visualization Data curation. Giacomo Quilici: Investigation, Methodology. Giovanna Musco: Conceptualization, Supervision, Writing original drfat, review and editing, funding acquisition.
Acknowledgments
Acknowledgements
Not applicable.
Funding
This work was supported by Italian Ministry of Health (RF-RF-2013-02354880) and by the Italian Association for Cancer Research (AIRC) (IG-21440).
Competing interests
Declarations of interest: none.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.044.
Contributor Information
Andrea Berardi, Email: berardi.andrea@hsr.it.
Giovanna Musco, Email: musco.giovanna@hsr.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu L., Zhen X.T., Denton E., Marsden B.D., Schapira M. ChromoHub: a data hub for navigators of chromatin-mediated signalling. Bioinformatics. 2012;28:2205–2206. doi: 10.1093/bioinformatics/bts340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Zhao S, Patel DJ. Histone Recognition. Cham: Springer International Publishing; 2015. https://doi.org/10.1007/978-3-319-18102-8.
- 3.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49 doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allshire R.C., Madhani H.D. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musselman C.A., Kutateladze T.G. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–9071. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez R., Zhou M.-M. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker L.A., Allis C.D., Wang G.G. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res Mol Mech Mutagen. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez R., Zhou M.-M. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. doi: 10.1016/j.bbi.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böttcher J., Dilworth D., Reiser U., Neumüller R.A., Schleicher M., Petronczki M. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat Chem Biol. 2019;15:822–829. doi: 10.1038/s41589-019-0310-x. [DOI] [PubMed] [Google Scholar]
- 10.Musselman C.A., Kutateladze T.G. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–323. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amato A., Lucas X., Bortoluzzi A., Wright D., Ciulli A. Targeting ligandable pockets on plant homeodomain (PHD) zinc finger domains by a fragment-based approach. ACS Chem Biol. 2018;13:915–921. doi: 10.1021/acschembio.7b01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller T.C.R., Rutherford T.J., Birchall K., Chugh J., Fiedler M., Bienz M. Competitive binding of a benzimidazole to the histone-binding pocket of the pygo PHD finger. ACS Chem Biol. 2014;9:2864–2874. doi: 10.1021/cb500585s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner E.K., Nath N., Flemming R., Feltenberger J.B., Denu J.M. Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry. 2012;51:8293–8306. doi: 10.1021/bi3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrowsmith C.H., Schapira M. Targeting non-bromodomain chromatin readers. Nat Struct Mol Biol. 2019;26:863–869. doi: 10.1038/s41594-019-0290-2. [DOI] [PubMed] [Google Scholar]
- 15.Ali M., Hom R.A., Blakeslee W., Ikenouye L., Kutateladze T.G. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta - Mol Cell Res. 2014;1843:366–371. doi: 10.1016/j.bbamcr.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaetani M., Matafora V., Saare M., Spiliotopoulos D., Mollica L., Quilici G. AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 2012;40:11756–11768. doi: 10.1093/nar/gks933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison E.A., Musselman C.A. Elsevier; 2016. The role of PHD Fingers In Chromatin Signaling. Chromatin Signal. Dis. pp. 127–147. https://doi.org/10.1016/B978-0-12-802389-1.00007-1. [Google Scholar]
- 18.Berardi A., Quilici G., Spiliotopoulos D., Corral-Rodriguez M.A., Martin-Garcia F., Degano M. Structural basis for PHD V C5HCH NSD1 –C2HR Nizp1 interaction: implications for Sotos syndrome. Nucleic Acids Res. 2016;44:3448–3463. doi: 10.1093/nar/gkw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G.G., Cai L., Pasillas M.P., Kamps M.P. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 20.Waggoner D.J., Raca G., Welch K., Dempsey M., Anderes E., Ostrovnaya I. NSD1 analysis for Sotos syndrome: insights and perspectives from the clinical laboratory. Genet Med. 2005;7:524–533. doi: 10.1097/01.GIM.0000178503.15559.d3. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen A.L., Jørgensen P., Lerouge T., Cervin M. Nizp1, a Novel Multitype Zinc Finger Protein That Interacts with the NSD1 Histone Lysine Methyltransferase through a Unique C2HR Motif. Mol Cell Biol. 2004;24:5184–5196. doi: 10.1128/MCB.24.12.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Losson R., Nielsen A.L. Biochimica et Biophysica Acta The NIZP1 KRAB and C2HR domains cross-talk for transcriptional regulation. BBA - Gene Regul Mech. 2010;1799:463–468. doi: 10.1016/j.bbagrm.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Huang H., Howard C.A., Zari S., Cho H.J., Shukla S., Li H. Covalent inhibition of NSD1 histone methyltransferase. Nat Chem Biol. 2020 doi: 10.1038/s41589-020-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayly A.R., Abell C., Skidmore J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat Rev Drug Discovery. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 25.Villoutreix B.O., Kuenemann M.A., Poyet J.-L., Bruzzoni-Giovanelli H., Labbé C., Lagorce D. Drug-like protein-protein interaction modulators: challenges and opportunities for drug discovery and chemical biology. Mol Inform. 2014;33:414–437. doi: 10.1002/minf.201400040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozakov D., Grove L.E., Hall D.R., Bohnuud T., Mottarella S.E., Luo L. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano WL. Pymol: An open-source molecular graphics tool. Graph Syst Version 12r3pre, Schrödinger, LLC n.d.
- 28.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 29.Storer J.W., Giesen D.J., Cramer C.J., Truhlar D.G. Class IV charge models: A new semiempirical approach in quantum chemistry. J Comput Aided Mol Des. 1995;9:87–110. doi: 10.1007/BF00117280. [DOI] [PubMed] [Google Scholar]
- 30.Schrödinger. QikProp,. LCC, New York, NY 2019;3.
- 31.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK a prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 32.Greenwood J.R., Calkins D., Sullivan A.P., Shelley J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 33.Li J., Abel R., Zhu K., Cao Y., Zhao S., Friesner R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinforma. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen W.L., Duffy E.M. Prediction of drug solubility from structure. Adv Drug Deliv Rev. 2002 doi: 10.1016/S0169-409X(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 36.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Genet. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 37.Grzesiek S., Stahl S.J., Wingfield P.T., Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 38.Mayer M., Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 39.Ran X., Gestwicki J.E. Inhibitors of protein–protein interactions (PPIs): an analysis of scaffold choices and buried surface area. Curr Opin Chem Biol. 2018;44:75–86. doi: 10.1016/j.cbpa.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bojadzic D., Buchwald P. Toward small-molecule inhibition of protein-protein interactions: general aspects and recent progress in targeting costimulatory and coinhibitory (immune checkpoint) interactions. Curr Top Med Chem. 2018;18:674–699. doi: 10.2174/1568026618666180531092503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keskin O., Gursoy A., Ma B., Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 42.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Mayer M., Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 44.Dawson M.A., Kouzarides T., Huntly B.J.P. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z., Song J., Milne T.A., Wang G.G., Li H., Allis C.D. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fiedler M., Sannchez-Barrena M.J., Nekrasov M., Mieszczanek J., Rybin V., Mueller J. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zucchelli C., Tamburri S., Quilici G., Palagano E., Berardi A., Saare M. Structure of human Sp140 PHD finger: an atypical fold interacting with Pin1. FEBS J. 2014;281:216–231. doi: 10.1111/febs.12588. [DOI] [PubMed] [Google Scholar]
- 48.London N., Raveh B., Schueler-Furman O. Druggable protein–protein interactions – from hot spots to hot segments. Curr Opin Chem Biol. 2013;17:952–959. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Bakail M., Ochsenbein F. Targeting protein–protein interactions, a wide open field for drug design. C R Chim. 2016;19:19–27. doi: 10.1016/j.crci.2015.12.004. [DOI] [Google Scholar]
- 50.Economides M.P., McCue D., Borthakur G., Pemmaraju N. Topoisomerase II inhibitors in AML: past, present, and future. Expert Opin Pharmacother. 2019;20:1637–1644. doi: 10.1080/14656566.2019.1621292. [DOI] [PubMed] [Google Scholar]
- 51.Plantone D., Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin Drug Invest. 2018;38:653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- 52.Gurova K. New hopes from old drugs: revisiting DNA-binding small molecules as anticancer agents. Futur Oncol. 2009;5:1685–1704. doi: 10.2217/fon.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham A.D., Qvit N., Mochly-Rosen D. Peptides and peptidomimetics as regulators of protein–protein interactions. Curr Opin Struct Biol. 2017;44:59–66. doi: 10.1016/j.sbi.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.