Abstract
Metabolic labeling of RNAs with noncanonical nucleosides that are chemically active, followed by chemoselective conjugation with imaging probes or enrichment tags, has emerged as a powerful method for studying RNA transcription and degradation in eukaryotes. However, metabolic RNA labeling is not applicable for prokaryotes, in which the complexity and distinctness of gene regulation largely remain to be explored. Here, we report 2′-deoxy-2′-azidoguanosine (AzG) as a noncanonical nucleoside compatible with metabolic labeling of bacterial RNAs. With AzG, we develop AIR-seq (azidonucleoside-incorporated RNA sequencing), which enables genome-wide analysis of transcription upon heat stress in Escherichia coli. Furthermore, AIR-seq coupled with pulse-chase labeling allows for global analysis of bacterial RNA degradation. Finally, we demonstrate that RNAs of mouse gut microbiotas can be metabolically labeled with AzG in living animals. The AzG-enabled metabolic RNA labeling should find broad applications in studying RNA biology in various bacterial species.
INTRODUCTION
Cellular RNA levels result from the interplay of RNA transcription, processing and degradation (1–3). To dissect these tightly regulated processes, it is desirable to selectively analyze nascent RNAs in addition to measurements on total RNAs. Several complementary methods with nascent RNA-specificity have been developed and have greatly facilitated the understanding of gene regulation networks (4). One of the methods exploits chemically active nucleoside analogs (i.e. noncanonical nucleosides) that can serve as surrogates of natural nucleosides and be used for RNA synthesis in living cells (5). The noncanonical nucleoside-incorporated RNAs are then chemically conjugated with fluorophores for imaging or affinity tags for enrichment and sequencing. Since the noncanonical nucleosides can only be incorporated into newly transcribed RNAs, this method allows for studying transcription by separation of nascent RNAs from the pre-existing populations. Furthermore, by applying noncanonical nucleosides in pulse-chase labeling experiments, RNA degradation can be quantified and profiled by monitoring the decay of pulse-labeled RNAs (6–8). A variety of nucleoside analogs have been developed for metabolic RNA labeling in various eukaryotic cells (9–16). Among them, 4-thiouridine (4SU) and 5-ethynyluridine (EU) are two most widely used noncanonical nucleosides that can be conjugated via thiol coupling chemistry and click chemistry, respectively (9,10). Although the metabolic RNA labeling technique has been instrumental for studying RNA dynamics in eukaryotes, it is not applicable to bacteria.
For a long time, prokaryotic transcriptomes were generally considered to be much simpler. As a result, whole-transcriptome studies in bacteria lagged behind eukaryotes until recently (17). For the past two decades, transcriptomics has re-shaped our view on the complexity, dynamics, and regulatory mechanisms of bacterial transcriptomes (18). For example, bacterial mRNAs are now known to be generally regulated by hundreds of small noncoding RNAs (ncRNAs), which rivals the scope of microRNA regulation in eukaryotic cells (19,20). Furthermore, mRNA expression in bacteria dynamically changes in responses to cellular and environmental stimuli by regulation of transcription initiation and regulatory elements such as riboswitches and RNA thermometers (21–23). However, methods for analyzing prokaryotic nascent RNAs remain limited. It is therefore of great interest to develop a metabolic RNA labeling strategy for studying bacterial RNA biology.
Here, we report 2′-deoxy-2′-azidoguanosine (AzG) as a noncanonical nucleoside for bacterial labeling. AzG enables the development of AIR-seq (azidonucleoside-incorporated RNA sequencing), in which bacterial RNAs are metabolically labeled with AzG, click-labeled, and selectively enriched for analysis. We first apply AIR-seq for a genome-wide analysis of transcription upon heat stress in E. coli. AIR-seq reveals heat shock-induced transcripts that were not apparent by RNA-sequencing (RNA-seq) analysis of total RNAs. Furthermore, AIR-seq coupled with the pulse-chase labeling strategy allows for global analysis of mRNA degradation in E. coli, with no need of transcription inhibition. The results categorize bacterial transcripts with different decay rates. Finally, we demonstrate that metabolic RNA labeling with AzG can be applied to various bacterial species as well as the gut microbiotas in living mice.
MATERIALS AND METHODS
Bacterial strain and culture condition
Escherichia coli K12 (HfrH) was purchased from China Center of Industrial Culture Collection (Beijing, China). Acinetobacter baumannii ATCC 19606 and Staphylococcus aureus ATCC 6538 were kindly provided by Xiaoguang Lei's lab at Peking University. Salmonella typhimurium SL1344 was kindly provided by Xiaoyun Liu's lab at Peking University. Rhodococcus erythropolis ATCC 4277 and Bacillus subtilis W800 were kindly provided by Xiaolei Wu's lab at Peking University. Klebsiella pneumoniae was clinically isolated and kindly provided by Xiaoguang Lei's lab at Peking University. Bacteria were grown in nucleoside-free medium [M9 + 0.4% (wt/vol) glucose + 10 g/l tryptone] at 37°C with shaking at 220 rpm.
Metabolic labeling of RNA with nucleoside analogs
For bacterial labeling, overnight cultured bacteria were diluted 1:100 in fresh medium and grown to an OD600 of ∼0.3. The bacteria were incubated with nucleoside analogs including EU, 4SU, AzA, AzU, AzC or AzG with indicated conditions. For mammalian cell labeling, HeLa cells were grown to ∼80% confluence and incubated with EU at the indicated concentration for 12 h.
Rifampicin inhibition, RNase treatment, and guanosine competition
The bacteria were incubated with 100 μM AzG and 500 ng/μl rifampicin. The DBCO-Cy5-reacted RNA was treated with 0.5 μg/μl RNase A and 1 U/μl RNase I for 1 h at 37°C before loading to the agarose gel. The bacteria were treated with 100 μM AzG in the presence of guanosine at varied concentrations for 2 h.
Cell viability and proliferation assay
For proliferation analysis, the bacteria were incubated with indicated nucleoside analogs, during which the OD600 values were measured every 2 h. For cell viability analysis, bacteria cultures were centrifuged, washed by PBS, and resuspended in PBS by the same volume. Then, the bacteria were incubated with 50 μM propidium iodide at room temperature for 10 min, washed by PBS for three times, and analysed by flow cytometry. Live and dead bacteria cells showed different fluorescence intensity using a 488-nm laser and a 670-LP filter. The cell viability was calculated as the ratio of live cells to all cells.
Fluorescence microscopy
The AzG-treated bacteria were harvested by centrifugation and washed for three times by PBS, fixed with 4% PFA for 15 min at r.t. After washing for three times with PBS, the bacteria were permeabilized with 0.1% Triton X-100 for 15 min at r.t., and washed three times with PBS. The bacteria were reacted with 200 μM CuSO4, 800 μM THPTA, 50 μM alkyne-Cy5, and 2.5 mM freshly prepared sodium ascorbate in PBS for 1 h at r.t., followed by washing three times with PBS containing 1% Tween-20 and imaged by fluorescence microscopy on a Zeiss LSM 700 laser scanning confocal microscope under 63X magnification.
Extraction of RNA and DNA
The bacteria were harvested by centrifugation. The total RNA was extracted by TRIzol (Ambion) following the manufacturer's instructions. Extraction of DNA was performed following the manufacturer's instructions of EasyPure Bacteria Genomic DNA kit (Transgen Biotech).
In-gel fluorescence scanning of RNA
For SPAAC coupling, 30 μg RNA in 20 μl DEPC-treated water was reacted with 50 μM DBCO-Cy5 (stock solution in RNase-free DMSO) for 1 h at r.t. For CuAAC, the RNA was reacted with 100 μM alkyne-Cy5 (or azide-Cy5), 500 μM CuSO4, 2 mM THPTA, and 5 mM freshly prepared sodium ascorbate for 1 h at r.t. After extraction by TRIzol to remove the excess dye and re-suspension in 10 μl DEPC-treated water, the reacted RNA was mixed with 5 μl 50% glycerol, followed by separation by 1% agarose gel with or without GelRed and scanning on a ChemDoc MP imaging system (Bio-Rad) and a Typhoon FLA 9500 laser scanner (GE Healthcare).
In-gel fluorescence scanning of DNA
For SPAAC coupling, 2 μg DNA in 10 μl water was reacted with 50 μM DBCO-Cy5 for 1 h at r.t. For CuAAC, the DNA was reacted with 100 μM azide-Cy5, 500 μM CuSO4, 2 mM THPTA, and 5 mM freshly prepared sodium ascorbate for 1 h at r.t. The reacted DNA was extracted with 1 vol. of PCI. After centrifugation at 20 000 g for 10 min, the upper aqueous phase was washed with 1 vol. of chloroform, precipitated with 1 vol. of isopropanol and 0.1 vol. of 3 M NaCl aqueous solution at r.t. for 10 min. The precipitation was washed with 1 ml 75% ethanol, air dried, and resuspended in 10 μl DEPC-treated water. The DNA was mixed with 5 μl 50% glycerol, followed by separation by 1% agarose gel with or without GelRed and scanning on a ChemDoc MP imaging system (Bio-Rad) and a Typhoon FLA 9500 laser scanner (GE Healthcare).
Total RNA-seq
Total RNA extracted from AzG or vehicle-treated bacteria were sequenced with rRNA depleted on an Illumina HiSeq X Ten platform in Novegene Co. Reads with 150 bp paired-end were generated for each sample. RNA-seq was performed on biological replicates.
AIR-seq
For heat shock experiments, the bacteria were incubated with 100 μM AzG for 10 min at 37°C or 42°C, followed by extraction of total RNA with TRIzol. For the pulse-chase experiments, the bacteria were incubated with 100 μM AzG for 2 h, harvested, cultured with 100 μM guanosine for 0, 5 or 10 min, one-tenth volume of a ‘stop solution’ composed of 10% buffer-saturated phenol in ethanol were added and then chilled rapidly, followed by extraction of total RNA with TRIzol. The extracted total RNA was reacted with 50 μM DBCO-PEG4-biotin at r.t. for 1 h. After extraction with 1 vol. of PCI and centrifugation at 20 000 g for 10 min, the upper aqueous phase was washed with 1 vol. of chloroform, precipitated by 3 vol. of ethanol and 0.1 vol. of 3 M NaCl aqueous solution at −80°C overnight. The precipitation was washed with 75% ethanol, air dried, and resuspended in DEPC-treated water. After washing twice with PBS containing 0.1% Tween-20, Dynabeads Myone streptavidin C1 beads were added and incubated on an orbital shaker at 1200 rpm for 1 h at r.t. After washing three times with high-salt wash buffer [100 mM Tris–HCl (pH 7.5), 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl, 0.02% Na-deoxycholate] for 10 min, the beads were resuspended in biotin elution buffer [12.5 mM biotin, 75 mM NaCl, 7.5 mM Tris–HCl (pH 7.5), 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl, 0.02% Na-deoxycholate] and incubated on an orbital shaker at 1500 rpm for 20 min at r.t., followed by heating at 65°C for 10 min on a thermo shaker. The solution was collected and the beads were eluted one more time. Then, the RNA was precipitated by 3 vol. of ethanol, 0.1 vol. of 3 M NaCl aqueous solution and 150 ng/μl glycogen at –80°C overnight. After centrifugation at 20 000 g for 50 min at 4°C, the RNA was washed with 1 ml of 75% ethanol, air dried, and resuspended in DEPC treated water. The samples were then fragmented and subjected to RNA-seq along with the input RNA. The samples were sequenced on an Illumina HiSeq X Ten platform and 150 bp paired-end reads were generated for each sample. In this study, rRNA was not depleted for AIR-seq. AIR-seq was performed in duplicates.
RNA metabolic labeling of mouse gut microbiotas
C57BL/6 mice were treated with 50 μl of 20 mM AzG by oral gavage for three times with an interval of 1.5 h. After three times of gavage, the mice were sacrificed and the gut microbiotas were collected according to a published procedure (24). The microbiotas were resuspended in RNase-free PBS, and adjusted to an OD600 of approximately 1.0. 500 μl of microbiotas were used for RNA extraction and in-gel fluorescence scanning. 500 μl of microbiotas were used for fluorescence imaging.
RNA dot blot
60 μg RNA in 20 μl DEPC-treated water was reacted with 50 μM DBCO-PEG4-biotin (stock solution in RNase-free DMSO) for 1 h at r.t. After extraction by TRIzol to remove the excess reagent, the resulted RNA was dissolved in 10 μl DEPC-treated water. 2 μl of RNA was loaded to a nylon membrane (hybond−N+) and crosslinked by irradiation with 254-nm UV light at 0.15 J/cm2 twice using a UV crosslinker (CL-1000, UVP). The membrane was blocked by 10% SDS for 1 h and washed by TBST for three times. Then, the membrane was stained with streptavidin-HRP (0.5 μg/ml) for 1 h and followed by washing with 10% SDS, 5% SDS, 1% SDS, and three times of TBST for 5 min each. The blot was acquired using HRP substrate and peroxide solution (Millipore) on Tanon-5200 Multi.
Data processing
The nucleotide sequences of all transcripts on E. coli reference genome were downloaded from Ensemble (ASM584v2) with transcript types marked. The raw data were filtered by TrimGalore v0.6.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads with a mapping quality score Q < 30 were filtered out. The cleaned data were mapped to the whole genome using HISAT2 v2.0.5 with default parameters and no spliced alignment (25). The resulting SAM files were converted to BAM format by SAMtools v1.3 (26). The mapped reads were then counted to different genes by featureCounts v1.6.3 with multiple overlap allowed (27). FPKM values were calculated by StringTie v2.1.4 (28). The replication analysis was performed with those genes with an FPKM > 1. The differential expression analysis was performed using DESeq2 v1.20.0 (29). The Gene Ontology analyses were performed using DAVID (30). For RNA stability analysis, the highly expressed rRNAs including rrlG, rrsD, rrlC, rrlD, rrlH, rrsC, rrlA, rrsG, rrsH and rrlB were selected for normalization. The FPKM of each transcript at different time points was adjusted by normalizing the total FPKM of the 10 rRNAs. The relative abundance (RA) of each transcript at 5 min or 10 min was calculated as the ratio of FPKM to that at 0 min. Then, we set up a high-confidence filter to exclude outliers by considering: (i) the standard derivation (STD) of RA should be < 0.4 at both 5 and 10 min; (ii) the RA at 5 min and the ratio of RA at 10 min to RA at 5 min should be < 1.2.
RESULTS
Design and synthesis of bacteria-compatible noncanonical nucleosides
Aiming to develop nucleoside analogs compatible with bacterial RNA labeling, we first set out to evaluate the two nucleoside analogs widely used in eukaryotic cells. E. coli cells were incubated with 4SU or EU at varied concentrations for 2 h, which resulted in no detectable labeling even at high concentrations (Supplementary Figure S1A, B). Moreover, severe suppression of bacterial growth was observed with both 4SU and EU (Supplementary Figure S1C, D), which was in agreement with previous observations that 4SU impaired cellular metabolism in E. coli (31). Cell viability was significantly impaired with 4SU at concentrations higher than 100 μM, while EU did not induce significant decrease in cell viability (Supplementary Figure S1E, F). In addition, tRNAs of bacteria contain endogenous 4SU generated via thiolation of uracil (32), which may interfere with metabolic labeling if exogenous 4SU would be used. Of note, although EU does not incorporate into DNA in mammalian cells (10), significant labeling of DNA was observed in EU-treated E. coli, presumably due to metabolic conversion of EU to EdUTP (Supplementary Figure S1G). Together, these results demonstrate that 4SU and EU are not applicable for bacteria.
Based on previous reports on 2′-modified nucleosides (33–36), we then turned our attentions to 2′-deoxy-2′-azidonucleosides, including 2′-deoxy-2′-azidoadenosine (AzA), 2′-deoxy-2′-azidouridine (AzU), 2′-deoxy-2′-azidocytidine (AzC), and AzG, in which the hydroxyl group at the C2′ position of ribose was substituted with an azide (Figure 1A). We reasoned that the 2′-azido group (i) could mimic 2′-hydroxyl for RNA incorporation in bacteria based on the recently reported metabolic incorporation of AzA and AzC in mammalian cells (13,16), (ii) should block conversion to deoxynucleotides, (iii) might alleviate the cytotoxicity in bacteria and (iv) would enable conjugation via strain-promoted azide-alkyne cycloaddition (SPAAC or copper-free click chemistry) (37) to avoid RNA degradation by copper-induced radicals (38).
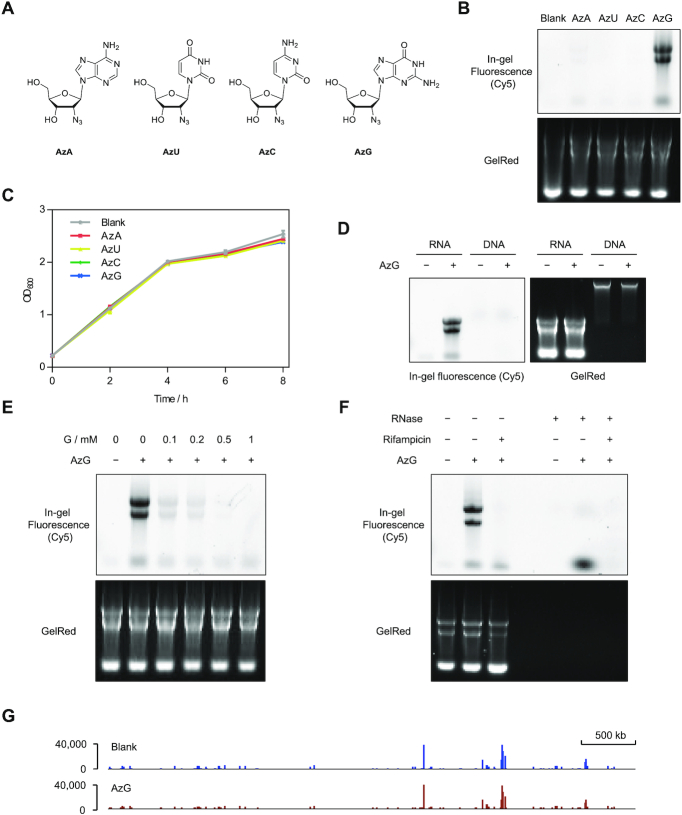
Figure 1.
AzG is compatible with metabolic RNA labeling in E. coli. (A) Chemical structures of four 2′-deoxy-2′-azidonucleosides. (B) In-gel fluorescence scanning showing total RNA that were isolated from E. coli treated with 100 μM individual 2′-deoxy-2′-azidonucleosides for 2 h, and reacted with DBCO-Cy5. (C) Growth curves of E. coli treated with vehicle or 100 μM individual 2′-deoxy-2′-azidonucleosides. Error bars represent mean ± s.d. Results are from three independent experiments. (D) In-gel fluorescence scanning showing total RNA and DNA that were isolated from E. coli treated with 100 μM AzG for 2 h, and reacted with DBCO-Cy5. (E) In-gel fluorescence scanning showing total RNA that were isolated from E. coli treated with 100 μM AzG in the presence of guanosine at varied concentrations, and reacted with DBCO-Cy5. (F) E. coli were cultured with 100 μM AzG in the presence or absence of the transcription inhibitor rifampicin. Total RNA were extracted and reacted with DBCO-Cy5 followed by treatment with or without RNase and detected by in-gel fluorescence scanning. In (B)–(F), GelRed-stained gels demonstrate equal loading. (G) Whole-genome alignment of the RNA-seq data from E. coli treated with or without AzG.
The chemical synthesis of the four 2′-deoxy-2′-azidonucleosides has been previously reported (39–41). AzA, AzU and AzC were commercially obtained and the purity was confirmed by NMR analysis. For AzG, we synthesized by adapting a synthetic route for 2′-deoxy-2′-chloroguanosine (42). Using guanosine as the starting material, AzG was chemically synthesized in eight steps with an overall yield of 1.8% (Supplementary Scheme S1).
To evaluate 2′-deoxy-2′-azidonucleosides for labeling bacterial RNAs, E. coli were treated with 100 μM 2′-deoxy-2′-azidonucleosides for 2 h, followed by isolation of the total RNA and reaction with the aza-dibenzocyclooctyne-Cy5 conjugate (DBCO-Cy5) via copper-free click chemistry. In-gel fluorescence scanning showed significant RNA labeling with AzG, but not with the other three analogs (Figure 1B). The labeling was observable with the reaction time of 10 min and the fluorescence intensity increased gradually by extending the reaction time to 1 h (Supplementary Figure S2A). Importantly, copper-free click chemistry maintained the RNA integrity (Supplementary Figure S2B). In contrast, CuAAC labeling of AzG-incorporated E. coli RNA resulted in significant RNA degradation (Supplementary Figure S2C). Furthermore, AzG as well as the other 2′-deoxy-2′-azidonucleosides did not affect E. coli growth (Figure 1C). As expected, AzG did not label DNA, indicating that the 2′-azido group of AzG could effectively block its conversion to deoxyguanoside (Figure 1D). The AzG-labeling of RNAs was abolished by competition with guanosine (G), transcription inhibition, or RNase digestion, further confirming the RNA labeling specificity of AzG (Figure 1E, F). Interestingly, G was able to compete off the AzG labeling with high efficiency, which might be attributed to weakened base paring of AzG with C (33). Another possibility might be that the enzymes converting guanosine to guanosine triphosphate do not well tolerate the 2′-azide modification. Moreover, metabolic incorporation of AzG did not affect transcription at the transcriptome level as analyzed by RNA-seq (Figure 1G and Supplementary Figure S3). These results demonstrate that AzG can serve as a surrogate for guanosine and be efficiently and specifically incorporated into newly transcribed E. coli RNAs.
Optimization of metabolic RNA labeling using AzG in E. coli
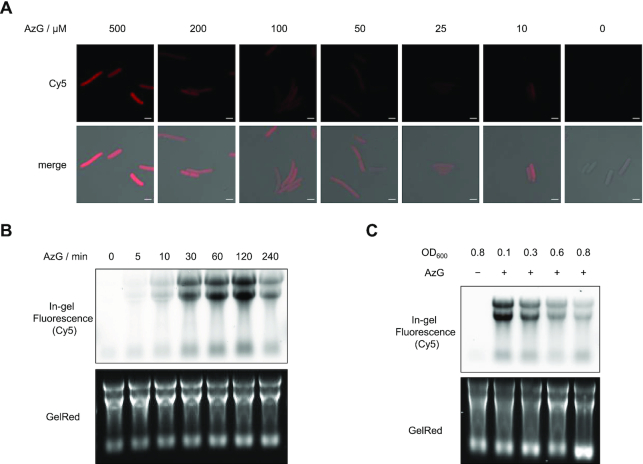
A series of experiments were performed to optimize the AzG-labeling procedure for E. coli. After the reaction of AzG-incubated E. coli cells with alkyne-Cy5 via click chemistry, the metabolic labeling of RNAs with AzG were visualized by confocal fluorescence microscopy. Fluorescence signals distributed in the cells were observed with the AzG concentration as low as 10 μM and gradually increased with increasing the AzG concentration (Figure 2A). The AzG labeling could be detected with labeling time as short as 5 min, which allows for detection of RNA dynamics on a time scale of minutes (Figure 2B). The labeling increased upon prolonging the incubation time up to 2 h. Further extending the incubation time to 4 h decreased the labeling intensity and adding AzG again at 2 h did not result in higher labeling (Supplementary Figure S4). This could be attributed to the effects of bacterial density on AzG labeling. Interestingly, lower bacterial density resulted in higher labeling efficiency (Figure 2C). Importantly, AzG did not induce cytotoxicity as showed by the permeability and proliferation assay (Supplementary Figure S5).
Figure 2.
Optimization of AzG-labeling conditions. (A) Confocal fluorescence microscopy images showing E. coli treated with AzG at varied concentrations for 2 h, followed by click reaction with alkyne-Cy5. Scale bars, 2 μm. (B) In-gel fluorescence showing total RNA from E. coli treated with 100 μM AzG for varied durations of time. (C) In-gel fluorescence scanning showing total RNA that were isolated from E. coli treated with 100 μM AzG at varied starting cell concentrations, and reacted with DBCO-Cy5. In (B) and (C), GelRed-stained gels demonstrate equal loading.
We then sought to determine the incorporation efficiency of AzG. The incorporation ratio of nucleoside analogs can be determined by nuclease digestion of the incorporated RNA into single nucleosides, followed by UPLC-MS/MS quantification. However, the 2′-azide of AzG was found to impair nuclease digestion to single nucleosides (33). To circumvent this issue, we compared AzG-incorporated E. coli RNA with EU-incorporated HeLa RNA by in-gel fluorescence scanning (Supplementary Figure S6). In agreement with previous reports (10), the incorporation efficiency of EU was determined by UPLC-MS/MS as approximately 1.0%, based on which the incorporation efficiency of AzG at 100 μM for 2 h was estimated to be 0.2%. For RNA transcripts with an average length of 1,000 nts, approximately one molecule of AzG was incorporated into every two transcripts. Considering that 0.2% substitution of G labels a sufficient amount of transcripts for detection while inducing minimum perturbation to RNA structure, we therefore chose treating E. coli at OD600 of 0.3 with 100 μM AzG for the following experiments.
AIR-seq for profiling transcription upon heat stress in E. coli
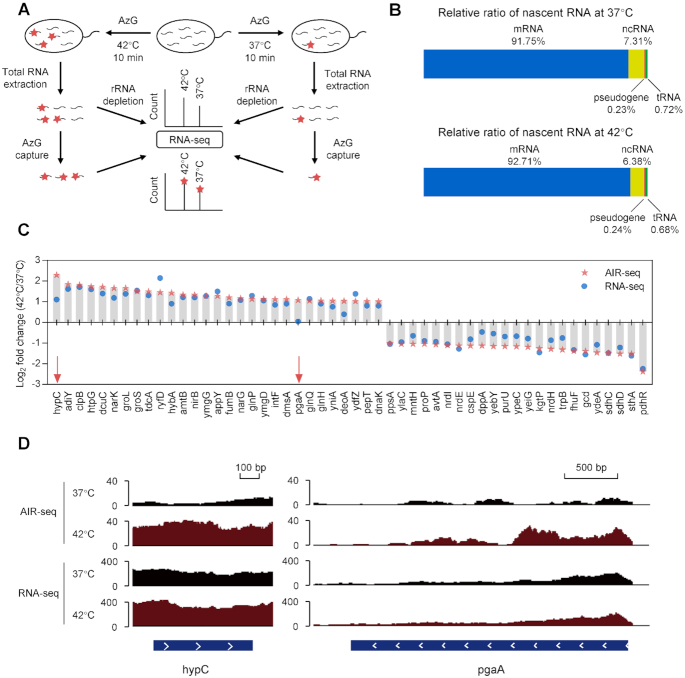
Based on metabolic labeling of RNAs with AzG, we developed AIR-seq (azidonucleoside-incorporated RNA sequencing) for probing transcription in E. coli upon heat stress (Figure 3A). E. coli grown under normal condition at 37°C or under mildly heat shocked condition at 42°C were metabolically labeled with AzG for 10 min, followed by extraction of total RNA. After reacting with DBCO-biotin, the newly transcribed RNAs were isolated by streptavidin beads and subjected to RNA-seq. In parallel, the total RNA samples were analyzed by RNA-seq for comparison. The sequencing results exhibited high correlation between two biological replicates, indicating overall good reproducibility of AIR-seq (Supplementary Figure S7A). We next tested whether the number of guanosines in a transcript influenced the extent of enrichment, given that transcripts with more guanosines might have a better chance to be labeled. The number of guanosines was plotted against the enrichment ratio of individual transcripts, which showed no correlation (Supplementary Figure S7B).
Figure 3.
AIR-seq for profiling transcription upon heat shock in E. coli. (A) Workflow of AIR-seq and RNA-seq for profiling RNA transcription in E. coli upon heat shock (42°C, 10 min). (B) Relative level of different types of RNAs in AIR-seq. rRNAs were not included. (C) Log2 fold change of RNA counts of individual genes upon heat shock by AIR-seq and RNA-seq. Examples in (D) were indicated with red arrows. (D) hypC (left) and pgaA (right) tracks in total RNA seq and AIR-seq upon heat shock.
As expected, both AIR-seq and RNA-seq identified various types of RNAs, including mRNAs, tRNAs, pseudogenes, and other ncRNAs (Figure 3B and Supplementary Figure S7C). The relative abundance of individual genes were then compared between the heat shock and normal samples. Both RNA-seq and AIR-seq identified heat shock-induced and suppressed genes (Supplementary Figure S8 and Supplementary Table S1). Gene Ontology (GO) analysis indicated the enrichment of genes involved in ‘response to heat’ (Supplementary Figure S9). The enrichment ratios for individual genes were generally higher in AIR-seq, demonstrating the superior sensitivity of AIR-seq owing to selective enrichment of the newly synthesized transcripts (Figure 3C). Importantly, AIR-seq revealed heat shock-induced transcripts that were not apparent in RNA-seq. This distinction between two methods could result from transcripts already abundant before heat shock. For example, hypC and pgaA exhibited 2.2- and 1.0-fold increase in RNA-seq of the total transcript pool, while AIR-seq revealed 4.9- and 2.1-fold increase in the new transcript pool (Figure 3C, D and Supplementary Figure S10).
Genome-wide profiling of RNA degradation in E. coli
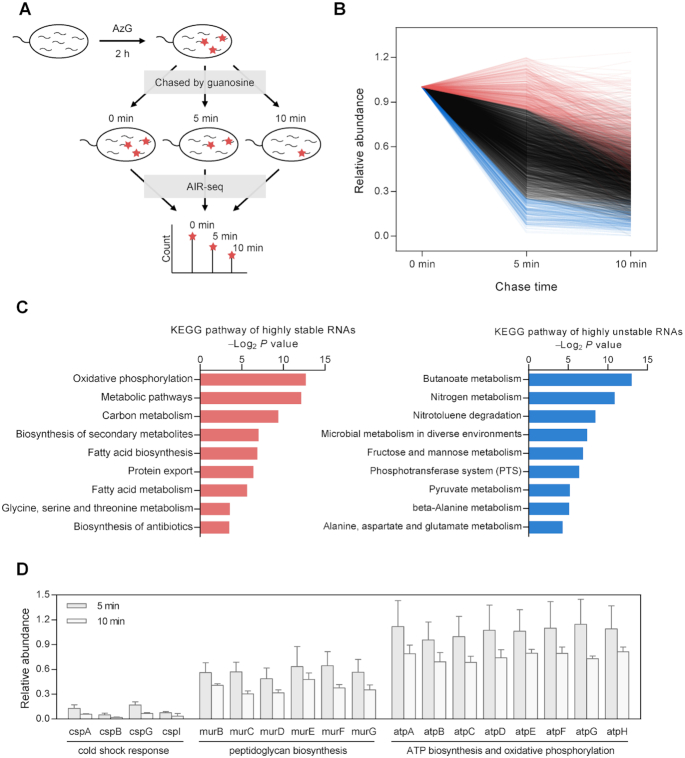
We next applied AIR-seq to study RNA degradation. Dissecting RNA degradation from RNA synthesis is essential for understanding the dynamics of gene expression. To study RNA degradation by RNA-seq, global inhibition of transcription has to be performed in order to measure RNA decay without interference from RNA synthesis (43,44). However, transcription inhibitors are generally toxic, can affect the abundance of many transcripts, and cause delay in RNA decay by residual RNA synthesis (44,45). We envisioned that AIR-seq, in conjunction with the pulse-chase labeling, would enable genome-wide analysis of RNA degradation in E. coli with no need of transcription inhibition (Figure 4A). After pulse labeling with AzG for 2 h, the E. coli cells were chased with guanosine for 0, 5 and 10 min, respectively, followed by AIR-seq analysis. Decay profiles of individual transcripts during the chase phase were determined by normalizing to rRNAs (Figure 4B and Supplementary Figure S11). Among 3052 transcripts, rRNAs, and tRNAs exhibited a relative longer half-life comparing to the mRNAs, pseudogenes, and other types of ncRNAs (Supplementary Figure S12). The list of 3052 transcripts were classified into three groups: highly stable transcripts with >85% remained at 5 min, highly unstable transcripts with <25% remained at 5 min, and the rests which degraded between 25% and 85% (Figure 4B and Supplementary Table S2).
Figure 4.
AIR-seq for profiling RNA degradation. (A) Workflow of pulse-chase labeling with AzG, followed by AIR-seq, for genome-wide measurement of RNA degradation. (B) Distribution of genome-wide decay profiles showing the relative abundance of 3052 transcripts across the time course of the chase phase. Each line represents the decay profile of a specific transcript. Highly stable and unstable genes were plotted in red and blue, respectively. (C) GO analysis of highly stable genes (left) and highly unstable genes (right) showing top enriched terms of KEGG pathway. (D) Representative decay profiles of families of genes involved in cold shock response, peptidoglycan biosynthesis, and ATP biosynthesis and oxidative phosphorylation.
GO analysis indicated that the highly stable mRNAs were enriched in pathways such as oxidative phosphorylation, carbon metabolism, fatty acid biosynthesis, and fatty acid metabolism (Figure 4C and Supplementary Figure S13). Conversely, the highly unstable mRNAs were related to pathways including phosphotransferase system, butanoate metabolism, and fructose and mannose metabolism. Furthermore, we found that transcripts from genes in same pathways tended to have similar degradation profiles (Figure 4D). For example, the cold shock response genes including cspA, cspB, cspG and cspI all degraded quickly in a similar manner. In agreement with our results, cspA was previously shown to have a short half-life at 37°C (23). Genes involved in peptidoglycan biosynthesis, including murB, murC, murD, murE, murF and murG, all degraded at a moderate rate (about 50% remained at 5 min). A group of eight genes including atpA, atpB, atpC, atpD, atpE, atpF, atpG and atpH, which are involved in ATP biosynthesis and oxidative phosphorylation, were highly stable and all had about 90% remained at 5 min. In eukaryotes, including dinoflagellate and mouse embryonic stem cell, transcripts involved in oxidative phosphorylation are very stable (46,47). Taken together, these results indicate that housekeeping genes in various species from prokaryotes to eukaryotes tend to decay at low rates.
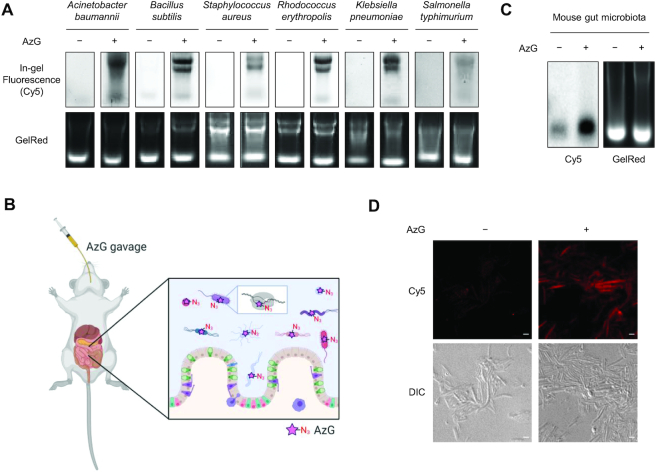
Metabolic RNA labeling in mouse gut microbiotas
The gut microbiotas is a collection of extremely diverse microorganisms, which play essential roles in regulating various physiological and pathological processes (48). In human, this microbial community consists of a comparable number of cells as human cells, with variations between individuals. Remarkably, it is estimated that the gut microbiota contains 100 times more genes than human (49). We envisioned that AzG might be a generic for labeling RNAs in various bacteria species, which should enable labeling of nascent RNAs in gut microbiota (50).
Toward this goal, we first evaluated the generic applicability of AzG by using three representative Gram-negative (Acinetobacter baumannii, Klebsiella pneumoniae, and Salmonella typhimurium) and three representative Gram-positive bacteria (B. subtilis, S. aureus and R. erythropolis). All six bacteria were successfully labeled with 100 μM AzG for 2 h, in a manner similar to E. coli (Figure 5A and Supplementary Figure S14). Interestingly, AzC was also metabolically incorporated into RNAs in B. subtilis (Supplementary Figure S14).
Figure 5.
Metabolic labeling of RNAs in mouse gut microbiotas. (A) In-gel fluorescence scanning showing total RNA that were isolated from Acinetobacter baumannii, Bacillus subtilis, Staphylococcus aureus, Rhodococcus erythropolis, Klebsiella pneumoniae, and Salmonella typhimurium treated with 100 μM AzG for 2 h, followed by reaction with DBCO-Cy5. GelRed-stained gels demonstrate equal loading. (B) Workflow of metabolic RNA labeling of gut microbiotas with AzG in living mice. (C) In-gel fluorescence scanning showing total RNA that were isolated from mouse gut microbiotas treated with 50 μl of 20 mM AzG by oral gavage for three times with an interval of 1.5 h, followed by reaction with DBCO-Cy5. GelRed-stained gels demonstrate equal loading. (D) Confocal fluorescence microscopy images showing gut microbiotas from mice treated with 50 μl of 20 mM AzG by oral gavage for three times with an interval of 1.5 h, followed by click reaction with alkyne-Cy5. Scale bars, 2 μm. For (C) and (D), representative results are shown from three independent experiments.
Next, we sought to demonstrate metabolic RNA labeling of gut microbiotas in living mice (Figure 5B). C57BL/6 mice were administered with AzG by gavage and the intestinal microbiotas were collected after three gavages. In-gel fluorescence scanning, dot blotting, and confocal fluorescence imaging showed that the nascent RNAs in the microbiotas were successfully labeled by AzG (Figure 5C,D and Supplementary Figure S15). These results indicate that AzG is compatible for metabolic labeling of nascent RNAs of gut microbiotas in vivo.
DISCUSSION
Metabolic labeling of RNAs with noncanonical nucleosides bearing a reactive group has enabled genome-wide analysis of RNA dynamics in ways that are difficult to achieve by conventional RNA-seq. Although its broad applications in various eukaryotes including vertebrates, insects, and yeasts, no bacterium-compatible noncanonical nucleoside is available until this work. We designed AzG by considering possible differences between eukaryotes and bacteria in the tolerance of unnatural modification on nucleosides by the RNA metabolism enzymes. Using E. coli as a model bacterial system, we have demonstrated that AzG-based AIR-seq can be used for genome-wide analysis of RNA transcription and degradation. Furthermore, AzG is applicable for various kinds of bacteria, even for complex microbial communities such as the gut microbiotas.
It is important to consider whether noncanonical nucleosides may affect RNA function and dynamics. For AzG, it was found to induce resistance to nuclease digestion when incorporated into siRNA (33). It is possible that mRNA and other types of RNA with AzG are more resistant to nucleases. In this work, we minimized the effects of AzG by controlling only one molecule of AzG in a single labeled transcript. Given the typical length of mRNA, AzG should not induce significant perturbation to RNA transcription and degradation.
It is an interesting observation that among the four 2′-deoxy-2′-azidonucleosides only AzG can be metabolically incorporated into RNAs of E. coli and most bacterial species tested. This is probably because nucleoside kinases in these bacteria have varied tolerance toward the 2′-azide modification on the cognate nucleosides. Interestingly, B. subtilis can metabolize both AzG and AzC, which indicates that the tolerance of noncanonical substrates is species-dependent. Moreover, this argue against that only AzG can be transported into bacteria. In fact, recombinant expression of B. subtilis uridine kinase in E. coli resulted in metabolic labeling of E. coli RNA with AzC (Supplementary Figure S16), further supporting this argument.
Engineered nucleoside kinase-noncanonical nucleoside pairs have been exploited for cell-selective metabolic RNA labeling in mammalian cells (16,51). Based on our results in bacteria, it should be possible to engineer bacterial nucleoside kinases for cell-selective AIR-seq, which would facilitate investigation of inter-species communications of bacteria and host-pathogen interactions.
A variety of variants of the metabolic RNA labeling techniques have been developed in eukaryotic systems, which possess desirable characteristics. For example, chemical derivatization of the incorporated 4SU can induce point mutations in RNA-seq, which has been exploited for analysis of nascent RNA with no need of enrichment (47,52,53). It will be of great interest to develop such a chemistry for AzG and AIR-seq of microbiotas.
DATA AVAILABILITY
Data are available in the Gene Expression Omnibus (GEO) under accession number GSE144943.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Guang Ge and Feng Wang in Beijing Okeanos Technology Co. for help on synthesis of AzG, We thank Dr Xiannian Zhang and Prof. Yanyi Huang for helpful advice on data analysis. Part of the analysis was performed on the High Performance Computing Platform of the Center for Life Science.
Contributor Information
Liying Meng, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China.
Yilan Guo, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Beijing National Laboratory for Molecular Sciences, Peking University, Beijing, China.
Qi Tang, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Beijing National Laboratory for Molecular Sciences, Peking University, Beijing, China.
Rongbing Huang, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Beijing National Laboratory for Molecular Sciences, Peking University, Beijing, China.
Yuchen Xie, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China.
Xing Chen, College of Chemistry and Molecular Engineering, Peking University, Beijing, China; Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China; Beijing National Laboratory for Molecular Sciences, Peking University, Beijing, China; Synthetic and Functional Biomolecules Center, Peking University, Beijing, China; Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, Peking University, Beijing, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Projects [2018YFA0507600]; National Natural Science Foundation of China [21672013, 91753206, 21521003]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. A Chinese patent application (application no. 201811348812.X) covering the use of AzG has been filed in which the Peking University is the applicant, and X.C., L.M., R.H. and Y.G. are the inventors.
REFERENCES
- 1. Coulon A., Chow C.C., Singer R.H., Larson D.R.. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat. Rev. Genet. 2013; 14:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Licatalosi D.D., Darnell R.B.. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010; 11:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Houseley J., Tollervey D.. The many pathways of RNA degradation. Cell. 2009; 136:763–776. [DOI] [PubMed] [Google Scholar]
- 4. Wissink E.M., Vihervaara A., Tippens N.D., Lis J.T.. Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet. 2019; 20:705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duffy E.E., Schofield J.A., Simon M.D.. Gaining insight into transcriptome-wide RNA population dynamics through the chemistry of 4-thiouridine. Wiley Interdiscip. Rev. RNA. 2019; 10:e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N. et al.. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011; 29:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Munchel S.E., Shultzaberger R.K., Takizawa N., Weis K.. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell. 2011; 22:2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eisen T.J., Eichhorn S.W., Subtelny A.O., Lin K.S., McGeary S.E., Gupta S., Bartel D.P.. The dynamics of cytoplasmic mRNA metabolism. Mol. Cell. 2020; 77:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melvin W.T., Milne H.B., Slater A.A., Allen H.J., Keir H.M.. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur. J. Biochem. 1978; 92:373–379. [DOI] [PubMed] [Google Scholar]
- 10. Jao C.Y., Salic A.. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grammel M., Hang H., Conrad N.K.. Chemical reporters for monitoring RNA synthesis and Poly(A) tail dynamics. ChemBioChem. 2012; 13:1112–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curanovic D., Cohen M., Singh I., Slagle C.E., Leslie C.S., Jaffrey S.R.. Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap. Nat. Chem. Biol. 2013; 9:671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nainar S., Beasley S., Fazio M., Kubota M., Dai N., Corrêa I.R., Spitale R.C.. Metabolic incorporation of azide functionality into cellular RNA. ChemBioChem. 2016; 17:2149–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen K., Aggarwal M.B., Feng C., Balderrama G., Fazio M., Mortazavi A., Spitale R.C.. Spatially restricting bioorthogonal nucleoside biosynthesis enables selective metabolic labeling of the mitochondrial transcriptome. ACS Chem. Biol. 2018; 13:1474–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H.S., Ishizuka T., Kawaguchi M., Nishii R., Kataoka H., Xu Y.. A nucleoside derivative 5-vinyluridine (VrU) for imaging RNA in cells and animals. Bioconjug. Chem. 2019; 30:2958–2966. [DOI] [PubMed] [Google Scholar]
- 16. Nainar S., Cuthbert B.J., Lim N.M., England W.E., Ke K., Sophal K., Quechol R., Mobley D.L., Goulding C.W., Spitale R.C.. An optimized chemical-genetic method for cell-specific metabolic labeling of RNA. Nat. Methods. 2020; 17:311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sorek R., Cossart P.. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010; 11:9–16. [DOI] [PubMed] [Google Scholar]
- 18. Hör J., Gorski S.A., Vogel J.. Bacterial RNA biology on a genome scale. Mol. Cell. 2018; 70:785–799. [DOI] [PubMed] [Google Scholar]
- 19. Barquist L., Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015; 49:367–394. [DOI] [PubMed] [Google Scholar]
- 20. Dutta T., Srivastava S.. Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene. 2018; 656:60–72. [DOI] [PubMed] [Google Scholar]
- 21. Browning D.F., Busby S.J.W.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 22. Sherwood A. V., Henkin T.M.. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 2016; 70:361–374. [DOI] [PubMed] [Google Scholar]
- 23. Kortmann J., Narberhaus F.. Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 2012; 10:255–265. [DOI] [PubMed] [Google Scholar]
- 24. Wang W., Zhu Y., Chen X.. Selective imaging of gram-negative and gram-positive microbiotas in the mouse gut. Biochemistry. 2017; 56:3889–3893. [DOI] [PubMed] [Google Scholar]
- 25. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao Y., Smyth G.K., Shi W.. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 28. Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 31. Bezerra R., Favre A.. In vivo incorporation of the intrinsic photolabel 4-thiouridine into Escherichia coli RNAs. Biochem. Biophys. Res. Commun. 1990; 166:29–37. [DOI] [PubMed] [Google Scholar]
- 32. Machnicka M.A., Olchowik A., Grosjean H., Bujnicki J.M.. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014; 11:1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fauster K., Hartl M., Santner T., Aigner M., Kreutz C., Bister K., Ennifar E., Micura R.. 2′-Azido RNA, a versatile tool for chemical biology: synthesis, X-ray structure, siRNA applications, click labeling. ACS Chem. Biol. 2012; 7:581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hobbs J., Sternbach H., Sprinzl M., Eckstein F.. Polynucleotides containing 2′-amino-2′-deoxyribose and 2′-azido-2′-deoxyribose. Biochemistry. 1973; 12:5138–5145. [DOI] [PubMed] [Google Scholar]
- 35. Torrence P.F., Bobst A.M., Waters J.A., Witkop B.. Synthesis and characterization of potential interferon inducers. Poly(2′-azido-2′-deoxyuridylic acid). Biochemistry. 1973; 12:3962–3972. [DOI] [PubMed] [Google Scholar]
- 36. Padilla R., Sousa R.. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002; 30:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sletten E.M., Bertozzi C.R.. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res. 2011; 44:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang R., Han M., Meng L., Chen X.. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E3879–E3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGee D.P.C., Vargeese C., Zhai Y., Kirschenheuter G., Settle A., Siedem C.R., Pieken W.A.. Efficient synthesis of 2′-amino-2′-deoxypyrimidine 5′-triphosphates. Nucleosides. Nucleotides. 1995; 14:1329–1339. [Google Scholar]
- 40. Klinchan C., Hsu Y.L., Lo L.C., Pluempanupat W., Chuawong P.. Synthesis of non-hydrolyzable substrate analogs for Asp-tRNAAsn/Glu-tRNAGln amidotransferase. Tetrahedron Lett. 2014; 55:6204–6207. [Google Scholar]
- 41. Ikehara M., Maruyama T.. Studies of nucleosides and nucleotides. LXXIX. Purine cyclonucleosides. 37. The total synthesis of an antibiotic 2′-amino-2′-deoxyguanosine. Chem. Pharm. Bull. (Tokyo). 1978; 26:240–244. [Google Scholar]
- 42. Mehta A.P., Abdelwahed S.H., Xu H., Begley T.P.. Molybdopterin biosynthesis: Trapping of intermediates for the MoaA-catalyzed reaction using 2′-deoxyGTP and 2′-chloroGTP as substrate analogues. J. Am. Chem. Soc. 2014; 136:10609–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bernstein J.A., Khodursky A.B., Lin P.H., Lin-Chao S., Cohen S.N.. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen H., Shiroguchi K., Ge H., Xie X.S.. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 2015; 11:808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shaw K.J., Miller N., Liu X., Lerner D., Wan J., Bittner A., Morrow B.J.. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 2003; 5:105–122. [DOI] [PubMed] [Google Scholar]
- 46. Morey J.S., Van Dolah F.M.. Global analysis of mRNA half-lives and de novo transcription in a dinoflagellate, Karenia brevis. PLoS One. 2013; 8:e66347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herzog V.A., Reichholf B., Neumann T., Rescheneder P., Bhat P., Burkard T.R., Wlotzka W., Von Haeseler A., Zuber J., Ameres S.L.. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods. 2017; 14:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lynch S.V., Pedersen O.. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016; 375:2369–2379. [DOI] [PubMed] [Google Scholar]
- 49. Proctor L.M., Creasy H.H., Fettweis J.M., Lloyd-Price J., Mahurkar A., Zhou W., Buck G.A., Snyder M.P., Strauss J.F., Weinstock G.M. et al.. The integrative human microbiome project. Nature. 2019; 569:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franzosa E.A., Hsu T., Sirota-Madi A., Shafquat A., Abu-Ali G., Morgan X.C., Huttenhower C.. Sequencing and beyond: Integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 2015; 13:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Kleiner R.E.. A metabolic engineering approach to incorporate modified pyrimidine nucleosides into cellular RNA. J. Am. Chem. Soc. 2019; 141:3347–3351. [DOI] [PubMed] [Google Scholar]
- 52. Schofield J.A., Duffy E.E., Kiefer L., Sullivan M.C., Simon M.D.. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods. 2018; 15:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riml C., Amort T., Rieder D., Gasser C., Lusser A., Micura R.. Osmium-mediated transformation of 4-thiouridine to cytidine as key to study RNA dynamics by sequencing. Angew. Chem. - Int. Ed. 2017; 56:13479–13483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the Gene Expression Omnibus (GEO) under accession number GSE144943.