Abstract
PDCD2 is an evolutionarily conserved protein with previously characterized homologs in Drosophila (zfrp8) and budding yeast (Tsr4). Although mammalian PDCD2 is essential for cell proliferation and embryonic development, the function of PDCD2 that underlies its fundamental cellular role has remained unclear. Here, we used quantitative proteomics approaches to define the protein-protein interaction network of human PDCD2. Our data revealed that PDCD2 specifically interacts with the 40S ribosomal protein uS5 (RPS2) and that the PDCD2-uS5 complex is assembled co-translationally. Loss of PDCD2 expression leads to defects in the synthesis of the small ribosomal subunit that phenocopy a uS5 deficiency. Notably, we show that PDCD2 is important for the accumulation of soluble uS5 protein as well as its incorporation into 40S ribosomal subunit. Our findings support that the essential molecular function of PDCD2 is to act as a dedicated ribosomal protein chaperone that recognizes uS5 co-translationally in the cytoplasm and accompanies uS5 to ribosome assembly sites in the nucleus. As most dedicated ribosomal protein chaperones have been identified in yeast, our study reveals that similar mechanisms exist in human cells to assist ribosomal proteins coordinate their folding, nuclear import and assembly in pre-ribosomal particles.
INTRODUCTION
Ribosomes are evolutionarily conserved molecular structures required for protein synthesis and consist of ribonucleoprotein (RNP) complexes composed of ∼80 ribosomal proteins and four noncoding ribosomal RNAs (rRNAs) in eukaryotes. The synthesis of new ribosomes imposes a major commitment in terms of cellular energy consumption, involving transcription of all three RNA polymerases as well as the spatial and temporal action of >300 ribosome assembly factors (1). Ribosome biogenesis begins in the nucleolus with the RNA polymerase I-dependent synthesis of a precursor rRNA (pre-rRNA) that is co-transcriptionally assembled into a large RNP particle via the recruitment of specific ribosomal proteins and early ribosome maturation factors (2). This large pre-ribosome particle is rapidly converted into precursors of the small (40S) and large (60S) ribosomal subunits via a complex sequence of endonucleolytic and exonucleolytic RNA cleavage events (1). Ultimately, the 40S ribosomal subunit comprises 33 ribosomal proteins assembled around the 18S rRNA, whereas the 60S subunit is constituted of three different rRNAs (28S, 5.8S and 5S) along with 47 ribosomal proteins.
During ribosome biogenesis, millions of ribosomal proteins are produced in the cytoplasm and need to be imported into the nucleus for incorporation into pre-ribosomal particles (3). As most ribosomal protein genes are essential for cell viability, the central mechanisms responsible for recognition of the 80 ribosomal proteins in the cytoplasm and how each ribosomal protein is imported into the cell nucleus for ribosome assembly still remain poorly understood. In the past few years, the idea that dedicated chaperones assist ribosomal proteins to coordinate their folding, nuclear import, and/or assembly in pre-ribosomal particles has gained significant interest (4). To date, most of the evidence supporting the existence of specific ribosomal protein chaperones has been reported in the budding yeast, Saccharomyces cerevisiae. In this species, specific binding partners to a handful of ribosomal proteins have been identified and shown to be important for the accumulation of free ribosomal proteins, the import of ribosomal proteins into the nucleus, and accompanying ribosomal proteins to assembly sites in the nucleolus (5–10). Accordingly, mutations in dedicated ribosomal protein chaperones usually result in ribosome biogenesis defects that resemble those observed upon depletion of their ribosomal protein partners. In humans, the protein BCCIPβ has been proposed to act as a nuclear chaperone for the 60S ribosomal subunit protein uL14/RPL23 (11). However, BCCIPβ does not appear to be required for the synthesis of 60S ribosomal subunits (11). Therefore, the extent to which the requirement for dedicated ribosomal protein chaperones is conserved in mammalian cells remains unclear.
As ribosomal proteins are abundant, ubiquitous, with potential for binding nucleic acids, they are also prime candidates for specialized functions via extra-ribosomal activities. Indeed, specific ribosomal proteins have been shown to contribute to important cellular processes and biological pathways beyond the ribosome (12,13). Furthermore, the wealth of data emerging from protein-protein interaction studies strongly suggests the occurrence of ribosomal proteins involved in extra-ribosomal complexes whose function remains unknown. We previously uncovered that the 40S ribosomal protein uS5 (RPS2) forms an extra-ribosomal complex with the protein arginine methyltransferase PRMT3 in fission yeast (14), and notably, that the uS5-PRMT3 complex is conserved from yeast to humans (15,16). More recently, we disclosed the existence of a new extra-ribosomal complex between uS5 and the zinc finger protein ZNF277, revealing that ZNF277 and PRMT3 compete for association with uS5 by forming mutually exclusive complexes (17). Human uS5 also forms mutually exclusive complexes with the programmed cell death 2 (PDCD2) and PDCD2-like (PDCD2L) proteins (15). Interestingly, PDCD2 and PDCD2L show homology to S. cerevisiae Tsr4, an essential protein that was recently reported to function as a dedicated chaperone for uS5 in budding yeast (5,9). However, in contrast to S. cerevisiae Tsr4, PDCD2L is not essential in human cells and its absence does not result in obvious defects in pre-40S maturation (15), arguing against a role for PDCD2L in chaperoning uS5 in human cells. In contrast, loss of PDCD2 function results in ribosome biogenesis defects (15), but the underlying molecular mechanism remains unknown.
Studies therefore support an intricate network of evolutionarily conserved protein-protein interactions involving extra-ribosomal uS5 (5,9,14,15,17–19). In this study, we investigated the functional significance of the uS5-PDCD2 association (15). PDCD2 was originally identified in a screen for mRNAs upregulated upon apoptosis in rat cells (20). PDCD2 is essential for embryonic development in mice and Drosophila (21,22). Interestingly, PDCD2 is also essential for the maintenance of embryonic stem cells in mice (22), a finding consistent with experiments using hematopoietic stem cells in Drosophila (23). Despite harboring a MYND-type zinc finger, a domain that has been shown to promote protein-protein interactions within transcriptional repressor complexes (24,25), and evidence of interactions with the Host Cell Factor 1 (26), no transcription-related activity has yet been reported for PDCD2. Thus, the molecular mechanism underlying the functional role of PDCD2 in cell growth and self-renewal remains elusive.
Here, we report that PDCD2 functions as a dedicated chaperone for the 40S ribosomal protein uS5 in human cells. We show that PDCD2 associates with uS5 co-translationally and that a loss-of-function of PDCD2 in human cells results in 40S maturation defects that phenocopy a deficiency in uS5. The PDCD2-uS5 association is required for 40S ribosomal subunit biogenesis, as PDCD2 variants impaired for the interaction between PDCD2 and uS5 were not able to restore the ribosome biogenesis defects caused by a PDCD2 deficiency. Importantly, we show that PDCD2 promotes the cellular accumulation of free uS5 and its incorporation into 40S ribosomal subunit precursors. Our findings, together with recent work by others (5,9), indicate that PDCD2 supports an evolutionarily conserved and essential function in chaperoning nascent uS5, sustaining the view that the requirement for dedicated ribosomal protein chaperones is conserved in mammalian cells.
MATERIALS AND METHODS
Human cell culture
HEK293-FT, U2OS, HeLa-FT and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% of tetracycline-free fetal bovine serum. Inducible expression of GFP, GFP-uS5, BirA, and BirA-PDCD2 was achieved by flippase-mediated recombination in HEK293-FT or HeLa-FT cells, as previously described (27). Inducible shRNA-expressing cells were generated using the pTRIPZ lentiviral inducible vector (GE Healthcare). Induction of GFP-tagged proteins was achieved with 500 μg/ml of doxycycline for 24 h, whereas the induction of shRNA cell lines was achieved with 2 × 48 h incubation with 1500 μg/ml doxycycline. Activation of BirA-tagged proteins was achieved with 50 μM biotin for 24 h. siRNAs were transfected with Lipofectamine 2000 at a final concentration of 25 nM (siControl, siPDCD2-1, siPDCD2-2 and siPDCD2L) or 32 nM (siuS5) for 72 h.
Affinity purification (AP) and proximity-dependent biotinylation (PDB) assays
For SILAC-based AP and PDB experiments, proteins were metabolically-labeled with stable isotopes of arginine and lysine in cell culture, as previously described (17). Briefly, HEK 293-FT cells expressing GFP- or BirA-tagged versions of proteins were grown in media containing labeled amino acids. 24–48 h after induction with doxycycline (SILAC) or biotin (BioID) cells were collected in lysis buffer and incubated at 4°C for 20 min. Lysates were centrifuged for 10 min at 13 000 rpm at 4°C and equal amount of proteins were incubated with GFP-trap beads from ChromoTek (Martinsried, Germany) or Dynabeads M-270 Streptavidin (Thermo Fisher) for 3 h at 4°C. Beads were then washed and proteins were subjected to two rounds of elution. SILAC eluates were vaccum-concentrated and resuspended in reducing buffer. Gel electrophoresis, in-gel digestions, LC–MS/MS, and analysis of SILAC ratios were performed as described previously (15,17).
Protein analysis and antibodies
Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by immunoblotting using the following primary antibodies: anti-PDCD2 and anti-LTV1 (ab133324 and ab122100, respectively; Abcam), anti-PNO1, anti-bystin, anti-eL19 and anti-uS3 (sc-133263, sc-271722, sc-100830 and sc-376008, respectively; Santa Cruz Biotechnology), anti-GFP (11814460001; Roche), anti-tubulin (T5168; Sigma-Aldrich) and anti-uS5 (a generous gift from Mark Bedford). Membranes were then probed with either a donkey anti-rabbit antibody conjugated to IRDye 800CW (926-32213; Li-Cor) or a goat anti-mouse antibody conjugated to Alexa Fluor 680 (A-21057; Life Technologies). Protein detection was performed using an Odyssey infrared imaging system (Li-Cor).
Reconstitution assay in E. coli
HA-tagged PDCD2 and Flag-tagged uS5 proteins were co-expressed in E. coli Rosetta 2 cells. For the co-expression of both proteins, the PDCD2 cDNA was cloned in the pCDFDuet-1 vector (Millipore Sigma) in frame with DNA sequences encoding a C-terminal HA tag, while the uS5 cDNA was cloned in the pETDuet-1 vector (Millipore Sigma) in frame with DNA sequences encoding a C-terminal Flag tag. For the control experiment expressing PDCD2-HA alone, E. coli Rosetta 2 cells were co-transformed with the pETDuet-1 vector alone. Bacterial cultures, induction, cell lysis and binding assays were performed as previously described (17).
Sucrose gradient analysis
To analyze the sedimentation pattern of PDCD2, ultracentrifugation using sucrose gradients was performed as previously described (15,17). Briefly, cycloheximide-treated cells were washed with PBS, and lysis buffer (supplemented with 50 μg/mL cycloheximide) was added directly to a 15-cm dish. Cells were scraped, incubated 15 min at 4°C, and centrifuged for 10 min at 4°C to remove cell debris. Approximately 5–10 mg of total protein was loaded onto a 5–50% sucrose gradient and centrifuged for 3 h at 40 000 rpm in a SW41 rotor (Beckman Coulter). The gradient was then fractionated by upward displacement with 55% (w/v) sucrose using a gradient fractionator (Brandel Inc.) connected to a UA-6 UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm. 0.6-ml fractions were collected, proteins were precipitated using a chloroform-methanol method, and analyzed by western blotting. For ribosome distribution profiles, 5% of the cleared lysate was kept for western blotting analysis. Ribosome subunits curves were reproduced using a DI-1100 instrument and the WinDaq software (DATAQ Instruments).
RNA analysis
Total RNA was prepared by using TRIzol (Life Technologies) and pre-rRNA species were analyzed by Northern blotting as previously described (15) with the following modifications: 5 μg/well of total RNA were mixed with 0.6 volume of loading dye (75% formamide, 45 mM tricine, 45 mM triethanolamine, 0.75 mM EDTA, 0.03% bromophenol blue, 0.6 M formaldehyde) and loaded onto a 0.8% denaturing agarose gel (0.8% agarose, 30 mM tricine, 30 mM triethanolamine, 0.4 M formaldehyde) in running buffer (30 mM tricine, 30 mM triethanolamine) at 200 V. RNAs were then transferred onto nylon membranes and hybridized using DNA probes. Oligo probes used in this study were ITS1 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′), 5′ETS (5′-AGACGAGAACGCCTGACACGCACGGCAC-3′), ITS2 (5′-GGGGCGATTGATCGGCAAGCGACGCTC-3′), 18S (5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′) and 28S (5′-CCCGTTCCCTTGGCTGTGGTTTCGCT-3′). Signals were detected using a Typhoon Trio instrument and quantified by ImageQuant TL (GE Healthcare).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described (28) using antibodies specific for RNA polymerase I subunits, RPA116 and RPA194 (generous gift from Ingrid Grummt) as well as a IgG control antibody (abcam ab46540). Briefly, cells were washed and cross-linked for 10 min at 37°C by addition of formaldehyde at a final concentration of 1%. Cells were washed and sonicated with a Branson Sonifier 450 at 20% amplitude with 10-s pulses at 3 min cycle. The chromatin extracts were diluted 10-fold in ChIP dilution and precleared by incubating with 40 μl salmon sperm DNA/protein A-agarose 50% gel slurry for 2 h at 4°C. Proteins were immunoprecipitated at 4°C overnight and washed extensively. DNA-protein cross-links were reversed by incubation at 65°C overnight followed by proteinase K treatment. DNA was recovered by purification with the Qiaquik PCR purification column (QIAGEN).
RNA-seq analysis
Libraries for control and PDCD2-depleted HeLa cells were prepared using total RNA from two independent knockdown experiments and analyzed using the Illumina HiSeq technology. Reads containing adapter sequences (>11 nt) were removed from the dataset, and low quality bases at read termini were trimmed using the fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html), discarding short reads (<32 nt) after trimming. Tophat was used to perform a spliced alignment to the human reference genome build hg19. After mapping, CoCo (29) was used to assign reads to features while correcting read counts for nested genes and multiple mapping reads. Differential gene expression analysis was performed using the DESeq2 R package (v1.24) (30). The list of genes annotated as ribosome biogenesis genes was downloaded from the amigo website (http://amigo.geneontology.org/amigo/term/GO:0042254).
Complementation assay
To generate an shRNA-resistant version of PDCD2, the sequence 5′-ca/gat/cat/ctg/gac/cat/at-3′ in the PDCD2 cDNA was mutated to 5′-cc/gac/cac/ctc/—/—/– -3′ to prevent siRNA-mediated targeting of the transgenic PDCD2 mRNA. This shRNA-resistant version of PDCD2 is deleted for three nonconserved amino acids (D179/H180/I181) and reconstituted a stable complex with uS5 similar to the full-length version of PDCD2 (data not shown). Functional complementation assays were performed using HeLa cells conditionally expressing PDCD2-specific or nontarget control shRNAs. Briefly, shCtrl and shPDCD2 cells were induced with 1500 ng/mL doxycycline for 72 h and then transferred into 60-mm cell culture dishes. Twenty-four hours later, cells were transfected with the empty pgLAP1 vector, the pgLAP1 vector containing the shRNA-resistant version of PDCD2, or pgLAP1 vectors containing the shRNA-resistant version of PDCD2 with additional mutations in the PDCD2 TYPP and MYND domains (shown in Figure 4A). Induction of GFP and the various versions of GFP-tagged PDCD2 was allowed for 48 h using 1500 ng/mL doxycycline. Total cell extracts were prepared for analysis of protein levels and pre-RNA processing was analyzed by Northern blotting using total RNA.
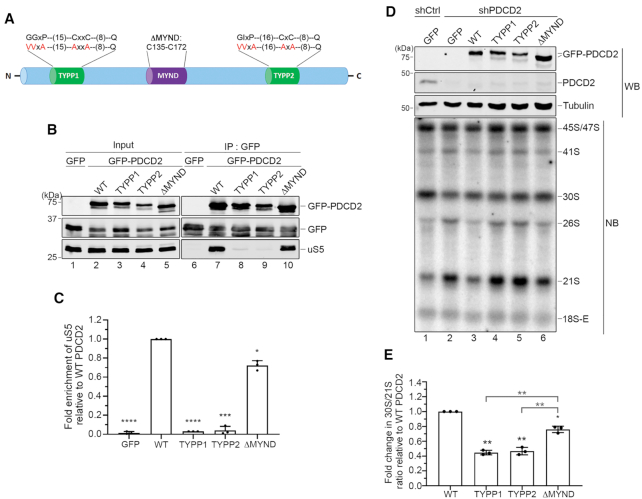
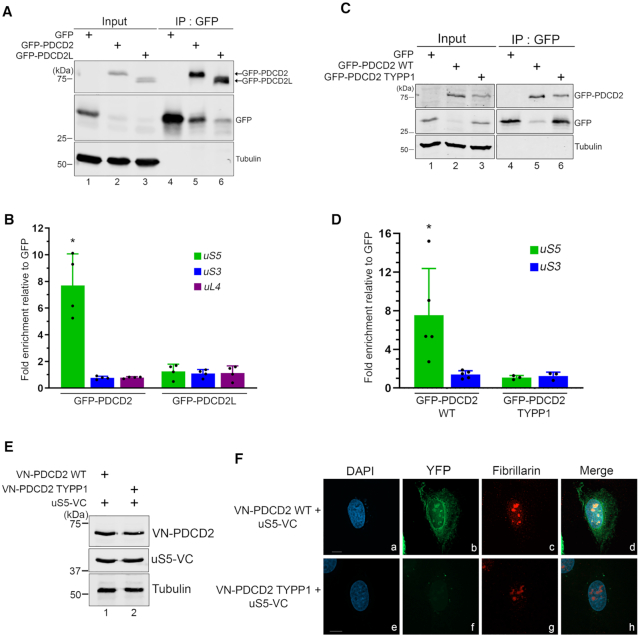
Figure 4.
The capacity of PDCD2 to form a complex with uS5 is required for the function of PDCD2 in 40S ribosomal subunit biogenesis. (A) Schematic representation of human PDCD2 and modifications introduced in this study. The evolutionarily conserved TYPP domain and the MYND type zinc finger are shown. Valine and alanine residues shown in red represent the various substitutions introduced into the N- and C-terminal repeats (TYPP1 and TYPP2, respectively) of the TYPP domain. The ΔMYND version of PDCD2 harbors a 38-aa deletion from residue 135 to 172. (B) Western analysis of total extracts (lanes 1–5) and anti-GFP precipitates (lanes 6–10) prepared from HeLa cells that were transiently transfected with the indicated versions of PDCD2 (lanes 2–5 and 7–10) or the with a GFP control plasmid (lanes 1 and 6). (C) Quantification of uS5 levels recovered in GFP immunoprecipitates normalized to the levels of GFP-PDCD2. Values were expressed relative to the wild-type (WT) version of PDCD2, which were set to 1.0. The data and error bars represent the average and standard deviation from three independent experiments. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001; Student's t-test. (D) Western blot (WB) and northern blot (NB) analysis from HeLa cells induced to express a nontarget control (lane 1) and PDCD2-specific (lanes 2–6) shRNA before transfection with plasmids expressing either GFP alone (lanes 1–2) or the indicated shRNA-resistant versions of GFP-PDCD2 (lanes 3–6). Pre-rRNAs were detected by using a probe complementary to sequences in the ITS1 region. (E) Quantification analysis of 30S/21S pre-rRNA ratios as determined by northern blot analysis (as D) from three independent experiments. The fold change in 30S/21S pre-rRNA ratio in PDCD2-depleted cells is presented relative to cells expressing the wild-type shRNA-resistant version of PDCD2 (WT). *P-value < 0.05; **P-value <0.01 , as determined by Student's t test.
RNA co-immunoprecipitation assay
The RNA co-immunoprecipitation assays were performed as previously described (17). 15-cm dishes of HEK293-FT cells conditionally expressing GFP, GFP-PDCD2L, GFP-PDCD2, and GFP-PDCD2 TYPP mutant were induced using doxycycline for 48 h. Cells were washed twice with PBS and 5% of cells were kept for total RNA extraction (input fraction). Cells were then resuspended in lysis buffer and incubated 15 min at 4°C. Lysate was centrifuged at 13 000 rpm for 10 min at 4°C and 5% of the lysate was kept for protein analysis. The remainder of the lysate was incubated with GFP-Trap beads for 3 h at 4°C. The beads were washed 5 times with lysis buffer and 10% of the beads were kept for protein analysis. RNA was extracted from the remainder of the beads using TRIzol reagent (Life Technologies) and analyzed by RT-qPCR. Total and copurified RNAs were analyzed by RT-qPCR using the following gene-specific primers: uS5 (5′-TATGCCAGTGCAGAAGCAGACC-3′/5′-CCTCCTTGGAGCACTTAACAC-3′), uS3 (5′-CAAGAAGAGGAAGTTTGTCGC-3′/5′-GAACATTCTGTGTTCTGGTGG-3′), uL4 (5′-GCAGGTCATCAGACTAGTGC-3′/5′-GGTTTTGGTTGGTGCAAAC-3′), GAPDH (5′-GTCAGCCGCATCTTCTTTTG-3′/5′-GCGCCCAATACGACCAAATC-3′).
Fluorescence microscopy
For the visual analysis of YFP-tagged uS5, a stable HeLa-FT cell line that conditionally induces the expression of uS5-YFP were first transfected with various siRNAs (siRNA control, siPDCD2 and siPDCD2L). Forty-eight hours post-transfection, the expression of uS5-YFP was induced using 500 μg/ml of doxycycline for 24 h. Cells were fixed, permeabilized, and analyzed for YFP fluorescence and nucleolar staining using a fibrillarin-specific antibody (Santa Cruz biotechnology). BiFC experiments were performed as previously described using human U2OS cells (17). Briefly, cells were co-transfected with Venus vector N (VN-) or Venus vector C (VC-) fusion proteins and fixed 24 h post-transfection before analysis by fluorescence microscopy. For all microscopy experiments, slides were mounted on a coverslip with Slowfade Gold antifade solution (Life Technologies). Images were captured by a Zeiss Axio Observer microscope using a 63× oil objective. Images used for quantifications of fluorescence pixel intensities come from samples of two independent experiments that were processed and imaged identically. Quantifications of YFP signal intensities were made on 137–154 nucleoli from 63 to 70 cells of each condition in experimental duplicates using ImageJ (31). The Kruskal–Wallis test, the uncorrected Dunn's tests, and the graphic representations were performed using GraphPad Prism™ version 8 (GraphPad Software).
Ribosome fractionation
Preparation of ribosomes and nonribosomal fractions was performed as described previously (32) with minor modifications. Briefly, HeLa-FT cells conditionally expressing GFP or GFP-uS5 were first transfected with nontarget control and PDCD2-specific siRNAs. Forty-eight hours post-transfection, the expression of GFP and GFP-uS5 was induced for 24 h using 500 μg/ml of doxycycline and cells were treated with 100 ug/ml of cycloheximide for 1 minute at 37°C. Cells were then washed with PBS, scraped in PBS, and centrifuged for 3 min at 4°C. Cells were resuspended in lysis buffer (32) and incubated for 20 min on ice. After centrifugation, the cleared lysate was layered over a 20% (w/v) sucrose cushion and centrifuged at 70 000 rpm at 4°C in a TLA-110 ultracentrifuge rotor for 4 h. The cytosolic nonribosomal fraction was collected, and proteins were precipitated with methanol and chloroform. The ribosome pellets were directly resuspended in SDS-PAGE loading buffer. Proteins from ribosomal and nonribosomal fractions were subjected to western blot analysis.
RESULTS
Human PDCD2 interacts with free ribosomal protein uS5
As a first step toward defining the molecular function of PDCD2 in human cells, we screened for PDCD2-associated proteins using two independent, yet complementary approaches: affinity purification coupled to mass spectrometry (AP-MS) and proximity-dependent biotinylation coupled to MS (PDB-MS). Importantly, both AP-MS and PDB-MS were coupled to quantitative proteomics, as assessed by stable isotope labeling by amino acids in cell culture (SILAC) that classifies interactions on the basis of specificity (ratio of peptide intensities between the experimental pulldown and a control purification) and protein abundance, as estimated by the sum of peptide signal intensities of a given protein normalized to its molecular mass (33). The AP-MS analysis of PDCD2 was previously described (15), using a stable cell line that conditionally expresses a GFP-tagged version of PDCD2. For the PDB-MS analysis, we generated a stable cell line that conditionally expresses PDCD2 fused to the BirA* biotin ligase. In total, the AP-MS and PDB-MS approaches identified 64 (15) and 8 (see Supplementary Table S1) PDCD2-associated proteins, respectively, with a SILAC enrichment ratio >2.0, a minimum of two unique peptides, and an amino acid sequence coverage >5% (Figure 1A–C). Notably, four proteins were identified in both the AP-MS and PDB-MS analysis (Figure 1C): PDCD2, the 40S ribosomal protein uS5, the General Transcription Factor II-I (GTF2I), and the alpha subunit of the nascent polypeptide-associated complex (NACA). Noteworthy is the identification of PRMT3 and ZNF277 in the AP-MS analysis of PDCD2 (Figure 1A), as these two proteins are known to form independent extra-ribosomal complexes with uS5 (15–17). Accordingly, this study will focus on the functional significance of the uS5-PDCD2 association.
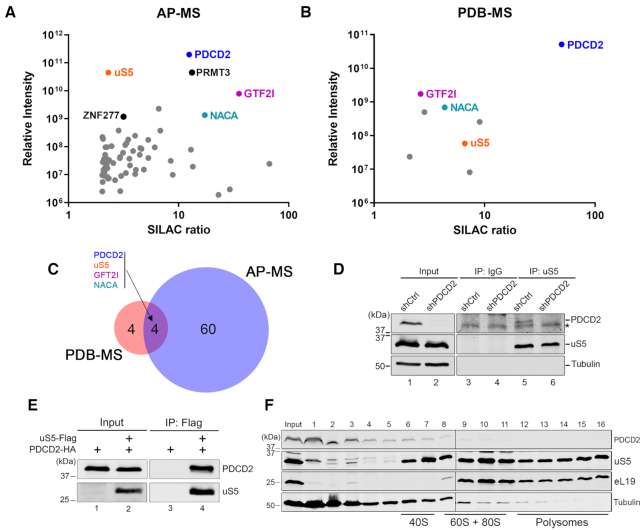
Figure 1.
PDCD2 forms a complex with the free uS5 protein in human cells. (A) Scatterplot showing results from GFP-PDCD2 affinity purification-coupled to mass spectrometry (AP-MS) plotted by SILAC ratio on the x-axis (peptide intensity originating from the GFP-PDCD2 purification versus the GFP control), which reflects specificity; and relative peptide intensity up the y-axis (total peptide intensity for each protein), reflecting the relative abundance of each protein in the purification. (B) Scatterplot showing results from proximity-dependent biotinylation (PDB) assay using BirA-PDCD2 coupled to SILAC-based mass spectrometry (MS) quantifications, as described in (A). (C) Venn diagram showing overlap between PDCD2-associated proteins identified by AP-MS (n = 64) and PDB-MS (n = 8). (A–C) The four proteins common to AP-MS and PDB-MS are shown in color. (D) Western blot analysis of cell extracts (lanes 1–2) as well as uS5 (lanes 5–6) and IgG (lanes 3–4) immunoprecipitates prepared using extracts from HeLa cells that conditionally express control (lanes 1, 3 and 5) and PDCD2-specific (lanes 2, 4 and 6) shRNAs. Antibodies used for Western blotting are indicated on the right. The asterisk (top panel) indicates the presence of a non-specific protein detected using the PDCD2 antibody. (E) Western blot analysis of total extracts (Input, lanes 1–2) and anti-Flag precipitates (IP, lanes 3–4) prepared from E. coli that co-expressed PDCD2 and uS5 (lanes 2 and 4) or that expressed PDCD2 alone (lanes 1 and 3). (F) Western analysis of the indicated proteins (right) using fractions of centrifuged sucrose gradients that were prepared using extracts of HeLa cells. The positions of the 40S, 60S/80S, and polysomes sedimentation are indicated at the bottom.
First, we wanted to validate the uS5-PDCD2 association by testing whether a complex is formed between the endogenous proteins in human cells. We therefore performed a reciprocal coimmunoprecipation (coIP) assay where we affinity purified endogenous uS5 and examined for copurification of endogenous PDCD2 by Western blotting. As shown in Figure 1D, a band revealed using a PDCD2-specific antibody was detected in the uS5 precipitate (lane 5, top panel), but not in the control immunoprecipitation (lane 3). As an additional control, immunoprecipitation of endogenous uS5 using extracts prepared from PDCD2-deficient cells (using a PDCD2-specific shRNA [see Materials and Methods]) did not result in the detection of PDCD2 in uS5 precipitates (Figure 1D, compare lanes 5–6). We thus conclude that endogenous versions uS5 and PDCD2 form a protein complex that can be biochemically isolated from human cells.
Next, we addressed whether uS5 directly interacts with PDCD2 by testing if a uS5-PDCD2 complex could be reconstituted in vitro using recombinant proteins. Notably, co-expression of uS5 and PDCD2 in Escherichia coli was able to reconstitute a complex as demonstrated by the detection of PDCD2 after affinity purification of uS5-Flag under stringent conditions (Figure 1E, lane 4). In contrast, a control purification using extracts of E. coli that did not express uS5-Flag did not recover PDCD2 (Figure 1E, lane 3), despite the presence of PDCD2 in the extract (Figure 1E, lane 1). These data using a heterologous expression system support a direct physical interaction between PDCD2 and uS5.
As uS5 was the only ribosomal protein detected in both AP-MS and PDB-MS analyses of PDCD2, this suggested that PDCD2 likely associates with free, non-ribosomal uS5. Accordingly, assessment of PDCD2 distribution after velocity sedimentation on sucrose gradients showed that the majority of PDCD2 was distributed in the low-density fractions (Figure 1F, see fractions 1–3), with the remaining low level of PDCD2 signal gradually decreasing into higher density fractions (fractions 4–11). This fractionation analysis thus supports the view that PDCD2 primarily associates with free uS5, which is consistent with the protein-protein interaction assays of PDCD2 (Figure 1A and B).
PDCD2 is required for small ribosomal subunit production in human cells
We previously reported that siRNA-mediated depletion of PDCD2 results in a 40S ribosomal subunit deficit that leads to the accumulation of free 60S ribosomal subunits (15). To further characterize the ribosome biogenesis defects associated with a deficiency in PDCD2, we attempted to generate PDCD2-null cell lines by using genome editing via CRISPR/Cas9. However, we could not get clones lacking complete expression of PDCD2, in contrast to its paralog PDCD2L (15). This is consistent with results from genome-scale loss-of-function screenings in cancer cell lines indicating that PDCD2, but not PDCD2L, is essential for cell viability (34). We therefore generated conditional human cell lines that induce PDCD2-specific shRNAs after addition of doxycycline to the culture media. Using this system, we depleted more than 90% of the total cellular levels of PDCD2 in a doxycycline-dependent manner (see Figure 1D, lanes 1–2). Importantly, conditional depletion of PDCD2 caused a marked accumulation of free 60S subunit, resulting in a 2.5-fold increase in the free 60S:40S ratio relative to control cells (Figure 2A, yellow profile). The altered stoichiometry between 40S and 60S ribosomal subunits in PDCD2-deficient cells also caused a marked reduction in the levels of 80S monosomes (Figure 2A).
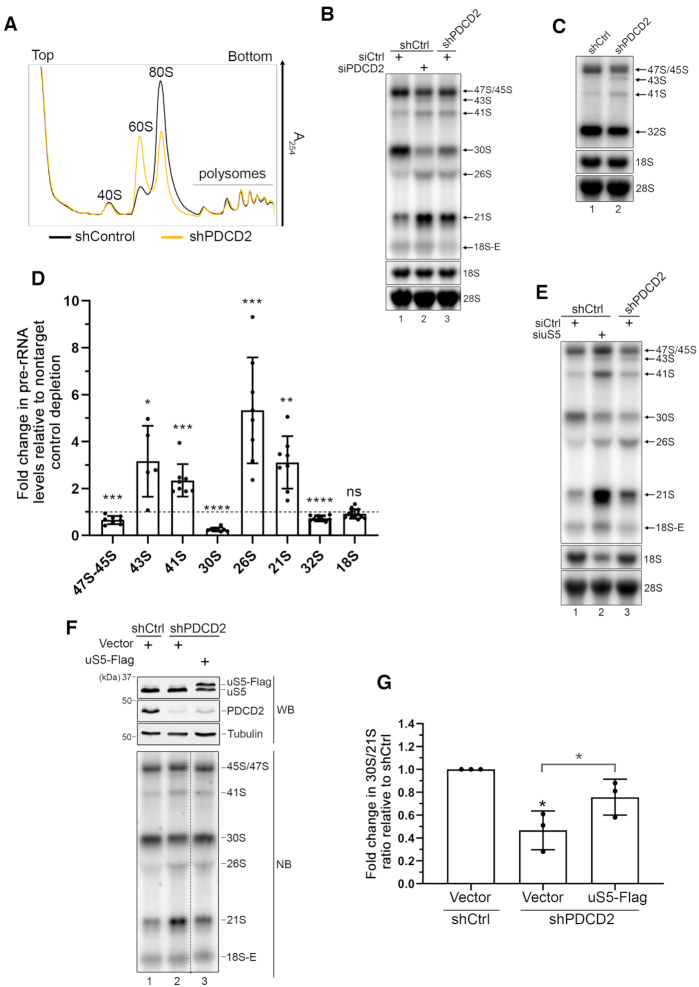
Figure 2.
PDCD2 contributes to 40S ribosomal subunit biogenesis. (A) Sucrose gradient analysis of total extracts prepared from HeLa cells that were previously induced to express a control nontarget (black profile) or a PDCD2-specific (yellow profile) shRNA for 96 h. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated. (B) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1–2) or a PDCD2-specific (lane 3) shRNA and that were transfected with nontarget (lanes 1 and 3) or PDCD2-specific (lane 2) siRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the ITS1 region. Pre-rRNA species are indicated on the right. (C) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1) or a PDCD2-specific (lane 2) shRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the ITS2 region. Pre-rRNA species are indicated on the right. (D) Levels of the indicated pre-rRNA were normalized to the mature 28S rRNA and are expressed relative to values for cells treated with a nontarget control depletion. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value > 0.05; *P-value <0.05; **P-value <0.01; ***P-value <0.001; ****P value <0.0001, as determined by Student's t test. (E) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lanes 1–2) or a PDCD2-specific (lane 3) shRNA and that were transfected with nontarget (lanes 1 and 3) or uS5-specific (lane 2) siRNA. Pre-rRNAs were detected by using a probe complementary to sequences in the 5′ ITS1 region. Pre-rRNA species are indicated on the right. (F) Western blot (WB, Top) and Northern blot (NB, Bottom) analysis from HeLa cells induced to express a nontarget control (lane 1) and PDCD2-specific (lanes 2–3) shRNA before transfection with a control plasmid (lanes 1–2) or a DNA construct expressing a Flag-tagged version of uS5 (lane 3). Pre-rRNAs were detected by using a probe complementary to sequences in the ITS1 region. Bottom panel: the data in lanes 1 to 3 were from the same blot, with the dashed line indicating some intervening lanes that were cropped out. (G) Quantification analysis of 30S/21S pre-rRNA ratios as determined by northern blot analysis (as F) from three independent experiments. The fold change in 30S/21S pre-rRNA ratio in PDCD2-depleted cells is presented relative to cells expressing a control nontarget shRNA (shCtrl). *P-value <0.05, as determined by Student's t test.
The accumulation of free 60S ribosomal subunits is generally the consequence of a deficiency in 40S subunit production and/or availability (35). We therefore examined rRNA precursors associated with 40S ribosomal subunit production by northern blotting using a probe complementary to internal transcribed spacer 1 (ITS1) sequences (see Supplementary Figure S1 for position of ITS1 probe). As can be seen in Figure 2B, depletion of PDCD2 in HeLa cells using independent methods and targeting sequences (siRNA and shRNA) resulted in similar changes in pre-rRNA levels relative to control cells (compare lanes 2–3 to lane 1). Although the steady-state level of mature 18S rRNA was not visibly affected by knocking down PDCD2, cells deficient for PDCD2 showed robust accumulation of the 21S pre-rRNA (Figure 2B and D), which is consistent with defective maturation of pre-40S particles (35). Because steady-state levels of the 18S-E precursor were low in our cell line (Figure 2B), it was difficult to accurately measure the impact of PDCD2 depletion on this pre-rRNA species. Levels of 43S, 41S and 26S rRNA precursors were also increased after PDCD2 knockdown. Conversely, a marked reduction in 30S pre-rRNA levels was noted in cells deficient for PDCD2 (Figure 2B and D). The accumulation of 43S and 41S pre-rRNA was also seen using a probe complementary to ITS2 sequences (see Supplementary Figure S1 for details about ITS2 probe), together with reduced levels of 47S/45S precursors (Figure 2C and D). In addition, a small reduction in the levels of 32S pre-rRNA was also detected with the ITS2 probe (Figure 2C and D). The greater effect of the PDCD2 knockdown on the levels of the 30S pre-rRNA relative to 32S precursors (Figure 2D) is consistent with a more important contribution of PDCD2 to 40S ribosomal subunit biosynthesis compared to the 60S ribosomal subunit (see Supplementary Figure S1). Collectively, the effects of a PDCD2 deficiency on ribosome biogenesis are consistent with altered ITS1 processing (reduced cleavage at site 2), uncoupling of cleavage at sites A0 and 1 (accumulation of 43S and 26S), and a reduction in 3′ end processing of the 21S pre-rRNA (see Supplementary Figure S1).
Since PDCD2 mainly associated with the uS5 ribosomal protein in human cells (Figure 1), we next compared the effects of depleting PDCD2 and uS5 on pre-rRNA processing. The effects of knocking down uS5 on pre-rRNA processing were generally consistent with a previous analysis of pre-rRNA profiles resulting from the individual depletion of 40S ribosomal proteins (36). Although the pre-rRNA processing defects associated with the knockdown of uS5 were generally more robust than for PDCD2, the depletion of uS5 largely phenocopied the pre-rRNA processing defects associated with PDCD2 loss-of-function: 21S and 26S accumulation, 30S reduction, as well as 41S and 43S accumulation (Figure 2E, compare lanes 2–3). The fact that depletion of either uS5 or PDCD2 resulted in virtually the same pre‐rRNA processing defects suggests that their cooperation is indispensable for proper ribosome biogenesis. In S. cerevisiae, increased dosage of a client ribosomal protein can partially compensate for the absence of the dedicated ribosomal protein chaperone (5,9). To assess whether increased dosage of uS5 can rescue the pre-rRNA processing defects associated with a PDCD2 loss-of-function, we transfected PDCD2-deficient cells with a construct that expresses a Flag-tagged version of uS5 or a vector control. As shown in Figure 2F–G, increased expression of uS5 significantly restored the pre-rRNA defects detected in PDCD2-depleted cells relative to the control vector (compare lane 3 to lane 2; quantifications in Figure 2G). Taken together, these data support the conclusion that PDCD2 is required for optimal production of 40S ribosomal subunit via a mechanism that involves interaction with uS5.
rDNA transcription and expression of ribosome biogenesis genes are not affected in PDCD2-deficient cells
PDCD2 contains a MYND-type zinc finger, a protein–protein interaction domain that has been linked to chromatin remodeling and other transcription-related processes (37). Accordingly, the reduction in 47S/45S precursors detected by both ITS1- and ITS2-specific probes in cells deficient for PDCD2 (Figure 2B–D) could be explained by a role of PDCD2 in regulating rDNA transcription by RNA polymerase I (RNAPI). Yet, as ITS1 and ITS2 probes cannot readily distinguish between the primary 47S precursor and its processed 45S intermediate, we used a specific probe to assess the levels of 47S primary transcript (see Supplementary Figure S1 for the position of the 5′ETS probe). As shown in Figure 3A and B, the use of a 47S-specific probe did not reveal changes in the level of the primary rRNA transcript in PDCD2-depleted cells. Yet, as RNA detection is an indirect measure of transcription activity, we used chromatin immunoprecipitation (ChIP) assays to assess RNAPI occupancy along the human rDNA locus. Antibodies specific to two independent subunits of the RNAPI complex, namely RPA194 and RPA116, were used for the immunoprecipitation procedure using chromatin prepared from PDCD2-depleted and control human cells. ChIP assays of RPA194 and RPA116 at the rDNA locus showed similar density profiles in both PDCD2-deficient and control cells (Figure 3C). Together with the 47S-specific Northern blot analysis, the RNAPI ChIP assays indicate that human PDCD2 is not required for normal rDNA transcription. Accordingly, these data argue that the reduced levels of 47S/45S precursors detected using ITS1- and ITS2-specific probes in cells deficient for PDCD2 can be largely attributed to decreased levels of the 45S pre-rRNA intermediate.
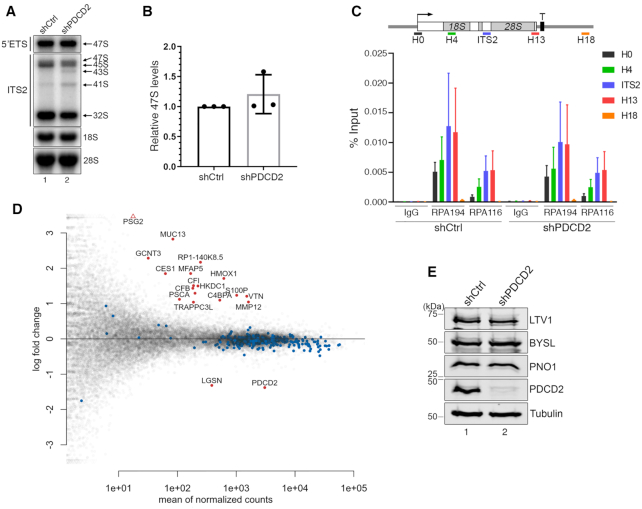
Figure 3.
The expression of ribosome biogenesis genes is not affected in cells deficient for PDCD2. (A) Northern blot analysis of mature and precursor rRNAs using total RNA prepared from HeLa cells that were previously induced to express a control nontarget (lane 1) or a PDCD2-specific (lane 2) shRNA. Pre-rRNAs were detected by using probes complementary to sequences in the 5′ ETS (47S-specific, top panel) and in the ITS2 region (middle panel). Pre-rRNA species are indicated on the right. (B) Levels of the 47S pre-rRNA were normalized to the mature 28S rRNA and are expressed relative to values for cells treated with control shRNAs. Data and error bars represent the means and standard deviations, respectively, from three independent experiments. (C) ChIP analysis along the rDNA gene using chromatin prepared from HeLa cells that were previously induced to express a control nontarget (shCtrl) or a PDCD2-specific (shPDCD2) shRNA. (Top) Bars below the rDNA gene show the positions of PCR products used for ChIP analyses. (Bottom) Antibodies specific to RNA polymerase I subunits (RPA194 and RPA116) and a rabbit IgG control were used for the ChIP assays. Error bars indicate SD. N = 3 biological replicates from independent cell cultures. (D) MAplot showing the log2 fold change of each gene (y-axis) as a function of the mean gene expression (x-axis) from an RNA-seq experiment on two independent PDCD2-depleted and control HeLa cells. Significantly differentially expressed genes in PDCD2-deficient cells are shown in red (|log2FC| > 1, FDR<0.01), including PDCD2. None of the 330 ribosome biogenesis genes (blue dots) are among the significantly differentially expressed genes. (E) Western blot analysis of the indicated proteins using extracts prepared from HeLa cells that were previously induced to express a control nontarget (lane 1) or a PDCD2-specific (lane 2) shRNAs.
Next, we examined whether PDCD2 controls the expression of ribosomal protein genes and/or genes encoding ribosome biogenesis factors as an explanation for the defects in 40S ribosomal subunit production caused by knocking down PDCD2 in human cells. We therefore analyzed total RNA prepared from two independent PDCD2 knockdown experiments in HeLa cells by strand-specific RNA-seq. In total, few genes were mis-regulated in PDCD2-deficient cells: 17 genes were up-regulated and 2 were down-regulated (Figure 3D; Supplementary Table S2), including PDCD2 that was one of the two significantly down-regulated genes. Importantly, expression analysis of genes annotated with a biological function related to ribosome biogenesis, including ribosomal protein genes, did not reveal changes in mRNA abundance based on RNA-seq (see Figure 3D, blue dots). We also monitored the protein levels of specific 40S subunit maturation factors, including LTV1, BYSL, and PNO1, which were not affected by a deficiency in PDCD2 (Figure 3E). We therefore conclude that the defects in 40S ribosomal subunit production caused by the depletion of human PDCD2 are unlikely to be the consequence of a role of PDCD2 in controlling the expression of ribosome biogenesis genes.
The PDCD2-uS5 interaction is required for 40S ribosomal subunit biogenesis
PDCD2 belongs to a family of proteins containing TYPP (Tsr4, YwqG, PDCD2L, PDCD2) domains (38). The TYPP domain is structured around two similar repeats, each consisting of GGxP and CxxC-like motifs as well as a highly conserved glutamine (Q) residue (see Figure 4A). In PDCD2, the two repeats of the TYPP domain are separated by a MYND-type zinc finger (Figure 4A). To determine whether the association between PDCD2 and uS5 is required for the PDCD2-dependent defects in pre-40S maturation (Figure 2), variants of human PDCD2 that can no longer bind to uS5 were needed. We therefore generated a set of PDCD2 alleles that express variants with amino acid substitutions or deletion to characterize the functional domains of PDCD2. Substitutions were introduced at evolutionarily conserved residues within various domains of human PDCD2, including the GGxP and CxxC-like motifs in the N-terminal and C-terminal repeat of the TYPP domain (Figure 4A). We also generated a version of PDCD2 that displays a 38-aa deletion, removing the entire MYND zinc finger domain (Figure 4A). These PDCD2 alleles were transfected into HeLa cells and expressed as GFP fusion proteins to define regions of PDCD2 necessary for uS5 association using a copurification assay. Immunoprecipitates were prepared to isolate PDCD2 using GFP-based affinity purification and analyzed for the ability of the different PDCD2 variants to copurify with uS5 by Western blotting. As shown in Figure 4B, endogenous uS5 was recovered in GFP immunoprecipitates prepared from extracts of cells that expressed wild-type GFP-PDCD2 (lane 7), but not from extracts of control cells that expressed GFP alone (lane 6), demonstrating the specificity of the copurification assay. Notably, amino acid substitutions of conserved residues within the individual repeats of the TYPP domain of PDCD2 abolished the interaction between uS5 and PDCD2 (Figure 4B, lanes 8–9; quantifications in Figure 4C). In contrast, the PDCD2-uS5 association was only reduced by 30% when the MYND zinc finger domain of PDCD2 was deleted (Figure 4B, lane 10 and Figure 4C). These results indicate that the TYPP domain is a critical determinant of PDCD2 required for the interaction with uS5.
To test whether the ribosome biogenesis defects associated with the loss-of-function of PDCD2 was dependent on the interaction with uS5, we established a functional complementation assay to assess the ability of the different PDCD2 mutants to rescue the altered pre-rRNA profile caused by a PDCD2 deficiency (Figure 2). For this, we engineered shRNA-resistant versions of wild-type and mutant PDCD2 alleles (see Experimental Procedure for details) to allow transient expression of PDCD2 in HeLa cells expressing a PDCD2-specific shRNA. Expression of a wild-type, shRNA-resistant version of GFP-tagged PDCD2 significantly restored the perturbed pre-rRNA levels observed in cells deficient for endogenous PDCD2 compared to the expression of GFP alone (Figure 4D, compare lane 3 to lane 2, and see Supplementary Figure S2). We next examined the extent to which the TYPP mutants and the MYND deletion complemented a PDCD2 deficiency. As shown in Figure 4D, PDCD2 mutants in the N- and C-terminal repeats of the TYPP domain showed altered pre-rRNA levels similar to PDCD2-depleted cells that expressed GFP alone (compare lanes 4–5 to lane 2), whereas wild-type PDCD2 reinstated a pre-rRNA profile similar to human cells expressing a control nontarget shRNA (compare lane 3 to lane 1). Interestingly, a version of PDCD2 with a deletion of the MYND zinc finger domain was able to partially suppress the aberrant pre-rRNA profile detected upon knockdown of PDCD2 compared to wild-type PDCD2 (Figure 4D, compare lane 6 to lane 3). Notably, quantification analysis of multiple independent experiments revealed that the MYND deletion was significantly more effective than the TYPP mutants at complementing a PDCD2 loss-of-function (Figure 4E), similar to analysis of PDCD2-uS5 interactions (Figure 4C). Together, these results indicate that the ability of PDCD2 to form a complex with uS5 depends on a functional TYPP domain and is important for the role of PDCD2 in small ribosomal subunit synthesis.
PDCD2 associates with uS5 co-translationally
Because uS5 was identified as the main PDCD2-associated ribosomal protein as determined by independent proteomics approaches (Figure 1) and since the uS5-PDCD2 interaction is important for 40S ribosome biogenesis (Figure 4), we explored the possibility that PDCD2 could play a chaperoning role to nascent uS5. Accordingly, the S. cerevisiae homolog of PDCD2, Tsr4, was recently shown to act as a dedicated chaperone for uS5/Rps2 (5,9). One frequent observation of dedicated ribosomal protein chaperone is a co-translational recruitment via the nascent ribosomal protein (4,7). To test whether PDCD2 is recruited to uS5 in a co-translational manner, we affinity-purified GFP-PDCD2 from human cell extracts (Figure 5A, lane 5) that were previously treated with cycloheximide to block translation elongation, thereby maintaining ribosome-mRNA associations. In addition, we also affinity-purified the paralog of human PDCD2, PDCD2L (Figure 5A, lane 6) as well as GFP alone (Figure 5A, lane 4). RNA was subsequently isolated from the GFP-Trap beads and analyzed by RT-qPCR for the specific enrichment of uS5 mRNA. The data were normalized to the GAPDH mRNA to control for experimental variations, and the values were set to 1.0 for the control GFP purification. As shown in Figure 5B, we observed a significant enrichment of uS5 mRNA in GFP-PDCD2 pull-downs, whereas the uS5 mRNA did not copurify with GFP-PDCD2L. The enrichment of uS5 transcripts in GFP-PDCD2 was specific, as mRNAs encoding uS3 (RPS3) and uL4 (RPL4) were not found to be selectively enriched with GFP-PDCD2 (Figure 5B). We next examined whether a TYPP mutant of PDCD2, which fails to associate with uS5 (Figure 4), can copurify the uS5 mRNA. Notably, affinity purification of PDCD2 with substitutions in the N-terminal TYPP repeat (TYPP1) did not show any enrichment of uS5 transcripts (Figure 5C-D), suggesting that a PDCD2-uS5 protein-protein interaction is required for the copurification of the uS5 mRNA with PDCD2. We therefore conclude that PDCD2 recognizes uS5 in a co-translational manner.
Figure 5.
PDCD2 binds to uS5 in a cotranslational manner. (A) Western blot analysis of total extracts (lanes 1–3) and anti-GFP precipitates (lanes 4–6) prepared from HeLa cells that stably express GFP (lanes 1 and 4), GFP-PDCD2 (lanes 2 and 5), and GFP-PDCD2L (lanes 3 and 6). (B) Fold mRNA enrichment (IP/input ratio) for the indicated mRNAs in GFP, GFP-PDCD2, and GFP-PDCD2L immunoprecipitates was analyzed by RT-qPCR and normalized to a control housekeeping mRNA (GAPDH). Values were then set to 1.0 for the control GFP purification. *P ≤ 0.05; Student's t-test. (C) Western blot analysis of total extracts (lanes 1–3) and anti-GFP precipitates (lanes 4–6) prepared from HeLa cells that stably express GFP (lanes 1 and 4), GFP-PDCD2 wild-type (lanes 2 and 5), and GFP-PDCD2 TYPP mutant (lanes 3 and 6). (D) Analysis of mRNA enrichment as described in panel B, but using GFP, GFP-PDCD2 WT, and GFP-PDCD2 TYPP immunoprecipitates. *P ≤ 0.05, Student's t-test. (E) Western blot analysis of total extracts prepared from U2OS cells that were previously transfected with the indicated DNA constructs for 24 h. The blots were analyzed using antibodies specific for PDCD2 (top), uS5 (middle), and Tubulin (bottom). (F) Representative BiFC images showing interaction between VN-PDCD2 and uS5-VC in living human cells. U2OS cells that co-expressed uS5-VC with either the wild-type VN-PDCD2 (a–d) or the VN-PDCD2 TYPP mutant (e-h) were fixed and simultaneously analysed by direct fluorescence (b and f) and immunostaining for the nucleolar marker fibrillarin (c and g). DNA staining with DAPI shows the nucleus of each cell (a and e).
We also used bimolecular fluorescence complementation (BiFC) assays to address the subcellular localization of the uS5–PDCD2 interaction in living cells. BiFC consists of fusing two non-fluorescent fragments of the yellow fluorescent protein (YFP) to two proteins from a stable complex leading to restoration of fluorescence within a cell by reconstituting the split YFP, thereby providing indication about the cellular localization of a complex (39). We used an improved version of YFP named Venus (40) to create fusions with uS5 and PDCD2: the N-terminal fragment of Venus (VN) was fused to the N-terminus of PDCD2, whereas the C-terminal fragment of Venus (VC) was fused to the C-terminus of uS5 (Figure 5E, lane 1). The VN-PDCD2 and uS5-VC constructs were co-transfected in U2OS cells, and 24 h post-transfection, the cells were fixed and visualized by fluorescence microscopy. As shown in Figure 5F, the uS5-PDCD2 interaction resulted in a reconstituted Venus signal that was found to localize to the nucleolus, the nuclear periphery, and the cytoplasm (see panel b). This signal was specifically triggered by the PDCD2-uS5 interaction, as combining fusions of uS5 and a TYPP mutant version of PDCD2 produced background signal (Figure 5F, panel f), consistent with biochemical data indicating that a functional TYPP domain is required for PDCD2 to interact with uS5 (Figure 4). As controls, we confirmed that each fusion protein was properly expressed (Figure 5E, lanes 1–2) and that the BiFC localization pattern of the uS5-PDCD2 interaction was observed in multiple cells (Supplementary Figure S3). These results reveal that the uS5-PDCD2 complex is found in the cytoplasm, at the nuclear periphery, and in the nucleolus.
PDCD2 promotes the accumulation of soluble uS5 protein and its incorporation into 40S ribosomal subunits
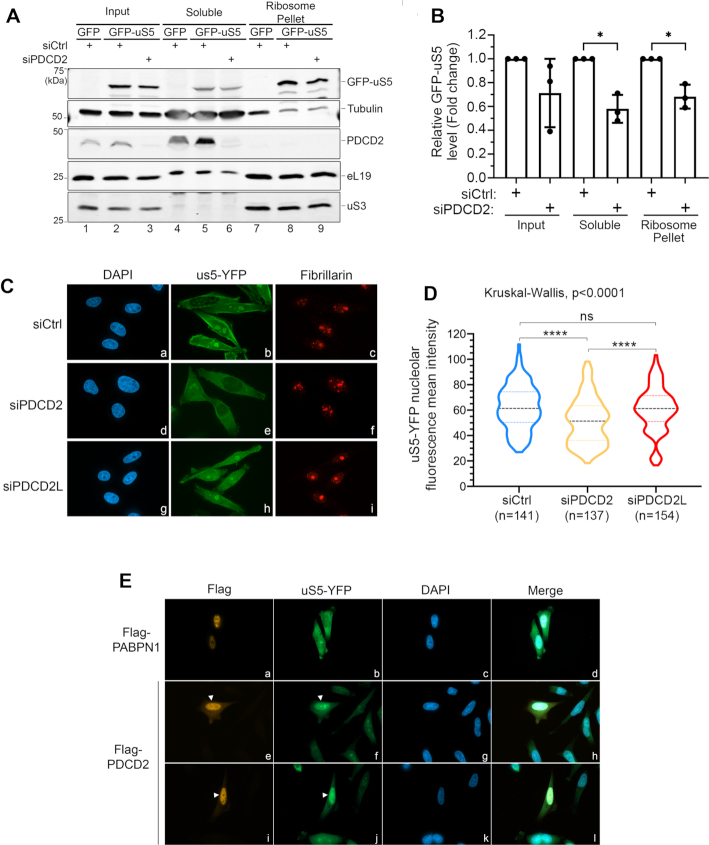
Despite the fact that PDCD2 is not a stable component of 40S ribosomal particles (Figure 1), knockdown of PDCD2 in human cells results in 40S maturation defects that phenocopy a deficiency in uS5 (Figure 2), indicating that the effect of PDCD2 on 40S production must be exerted through uS5. Accordingly, the co-translational recognition of nascent uS5 by PDCD2 (Figure 5) is consistent with a chaperoning function for PDCD2. Normally, chaperones dedicated to ribosomal proteins are recruited co-translationally in the cytoplasm to coordinate the folding, nuclear import, and/or incorporation of nascent ribosomal proteins into ribosomal subunit precursors (5,7–9,41). To assess whether PDCD2 acted on newly-synthesized uS5, we designed an experimental strategy to analyze de novo synthesized uS5 in conditions of PDCD2 deficiency. Specifically, we used PDCD2-specific and control nontarget siRNAs to knockdown PDCD2 expression for 48 h. PDCD2-deficient and control cells were subsequently induced to express a GFP-tagged version of uS5 or GFP alone for 24 h. Extracts were prepared and subjected to ultracentrifugation through a sucrose cushion to separate ribosomal complexes from the ribosome-free cytosol. Notably, these experiments revealed that the levels of GFP-uS5 in the soluble and pellet fractions were weakly, but significantly reduced in PDCD2-deficient cells relative to cells transfected with control siRNAs (Figure 6A, lanes 5–6 and 8–9; Supplementary Figure S4A and B; Quantifications in Figure 6B). We also quantified the levels of GFP-uS5 relative to independent ribosomal proteins: eL19 and uS3. This quantification analysis showed reduced levels of GFP-uS5 in the ribosome pellet (Supplementary Figure S4C), but not in the soluble fraction (Supplementary Figure S4D) of PDCD2-deficient cells. It should be noted, however, that only a small proportion of ribosomal proteins are recovered in the soluble fraction (between 0–4% of total), thereby resulting in increased variation in the quantification analysis of soluble ribosomal proteins. Taken together, these data suggested that less uS5 reaches pre-40S ribosomal subunits assembly sites in PDCD2-deficient cells. To get further evidence that loss of PDCD2 function impairs the accumulation of uS5 into ribosome assembly sites, we analyzed the subcellular localization of YFP-tagged uS5 in PDCD2-deficient cells. uS5-YFP was used for the fluorescence microscopy as it gave a better signal-to-noise ratio than GFP-uS5-expressing cells. Importantly, both uS5-YFP and GFP-uS5 copurified similar levels of endogenous PDCD2 (Supplementary Figure S4E). As shown in Figure 6C and Supplementary Figure S4F, the overall nucleolar signal of uS5-YFP was reduced in PDCD2-depleted cells compared to control cells (compare panel e to panel b). As an additional control, no such reduction of uS5-YFP nucleolar signal was observed in cells deficient for the paralog of human PDCD2, PDCD2L (Figure 6C, panel h). Quantification of uS5-YFP nucleolar fluorescence intensity revealed a significant reduction in the mean nucleolar signal in PDCD2-depleted cells compared to control cells and PDCD2L-depleted cells (Figure 6D). We also examined the impact of increased dosage of PDCD2 on the subcellular localization of uS5-YFP. Notably, we found that uS5-YFP accumulated in the nucleoplasm of cells that overexpressed a Flag-tagged version of PDCD2 (Figure 6E, panels e–f and i–j; see arrowheads), whereas adjacent non-transfected cells showed mainly cytoplasmic and nucleolar uS5-YFP signal. As an additional control, the overexpression of a different nuclear protein, Flag-PABPN1, did not result in the accumulation of uS5-YFP in the nucleoplasm (Figure 6E, panels a-b). Collectively, the results presented in Figure 6 indicate that PDCD2 is important for the accumulation of nascent uS5 protein and its incorporation into nucleolar pre-40S ribosomal subunits.
Figure 6.
PDCD2 contributes to uS5 accumulation and pre-40S incorporation. (A) HeLa cells that were previously depleted of PDCD2 using sequence-specific siRNAs (lanes 3, 6, and 9) or treated with a control nontarget siRNA (lanes 1–2, 4–5 and 7–8) were induced to express GFP alone (lanes 1, 4 and 7) or GFP-tagged uS5 (lanes 2–3, 5–6, and 8–9) for 24 h. Cell extracts (lanes lanes 1–3) were fractioned on a sucrose cushion to separate the ribosomal fraction (lanes 7–9) from the ribosome-free cytosol (lanes 4–6) and both fractions were subjected to Western blot analysis. (B) Levels of GFP-uS5 were normalized to Tubulin levels and are expressed relative to values for cells treated with control siRNAs. Data and error bars represent the means and standard deviations, respectively, from three independent experiments. *P-value < 0.05, as determined by Student's t test. (C) HeLa cells that conditionally express uS5-YFP were transfected with a nontarget control siRNA (panels a-c), siRNAs that target PDCD2 (panels d–f), and siRNAs targeting PDCD2L (panels g–i). 48 h post-transfection, uS5-YFP expression was induced with doxycycline for 24 h, cells were then fixed and simultaneously analysed by direct fluorescence (b, e and h) and immunostaining for the nucleolar marker fibrillarin (c, f and i). DNA staining with DAPI shows the nucleus of each cell (a, d and g). (D) Violin plots with nucleolar YFP fluorescence mean intensity scores for cells transfected with the indicated siRNAs from two independent replicates. Statistical differences were calculated using a Kruskal–Wallis test among the three groups (siCtrl, siPDCD2, and siPDCD2L), and Dunn's uncorrected tests for pairwise comparison. P-values are indicated as follows: ****P < 0.0001; ns, not significant. (E) HeLa cells that conditionally express uS5-YFP were transfected with DNA constructs expressing Flag-PABPN1 (panels a–d) or Flag-PDCD2 (panels e–l) and simultaneously induced to express uS5-YFP with doxycycline. 24 h post-transfection/induction, cells were then fixed and simultaneously analysed by direct fluorescence (b, f and j) and immunostaining using a Flag antibody (a, e and i). DNA staining with DAPI shows the nucleus of each cell (c, g and k). Arrowheads (e, f, i and j) show Flag-PDCD2-transfected cells that accumulate uS5-YFP in the nucleus.
DISCUSSION
Ribosomal proteins are highly abundant, ubiquitous, and basically charged proteins with potential for binding nucleic acids. Accordingly, ribosomal proteins are prime candidates for non-specific interactions before they are stably incorporated into ribosomal subunits. As ribosomal proteins are synthesized in the cytoplasm and need to be transported into the nucleus for ribosome assembly, completing this process becomes a demanding assignment given their abundance and susceptibility for non-specific interactions. Interestingly, the last few years have seen the emergence of factors, termed ‘dedicated chaperones’, that recognize specific ribosomal proteins to facilitate their solubility, traffic, and/or assembly, but that are not stable constituents of the mature ribosome (4). To date, most of these specific factors have been identified in budding yeast and it has remained unclear if the function of dedicated chaperones is conserved between yeast and humans. In this study, we report that human PDCD2 functions as an evolutionarily conserved chaperone dedicated to the 40S ribosomal protein uS5 (RPS2). This conclusion is supported by several observations: (i) PDCD2 specifically and directly interacts with uS5 (Figure 1), (ii) PDCD2 and uS5 form an endogenous protein complex in human cells (Figure 1), (iii) a deficiency in PDCD2 results in 40S ribosome biogenesis defects that phenocopy a depletion of human uS5 (Figure 2), (iv) increased uS5 dosage restored the ribosome biogenesis defects associated with a PDCD2 deficiency (Figure 2), (v) PDCD2 recognizes nascent uS5 in a co-translational manner (Figure 5), (vi) PDCD2 is important for the accumulation of soluble uS5 and its incorporation into 40S particles (Figure 6) and (vii) the yeast homolog of PDCD2, Tsr4, was recently shown to function as a dedicated chaperone for S. cerevisiae uS5 (5,9). The role of PDCD2 in chaperoning free uS5 is also likely conserved in invertebrates, as data supporting a physical interaction between uS5 and PDCD2/Zfrp8 was reported in Drosophila (18). Collectively, these findings support that the need for a dedicated chaperone to promote the accumulation of soluble uS5 protein is conserved from yeast to humans.
Depletion of PDCD2 and uS5 from human cells led to the accumulation of various rRNA processing intermediates related to 40S subunit production (Figure 2). Some of those intermediates are associated with inefficient processing of the 5′-ETS (45S, 43S, 41S, 30S and 26S), which occurs in the context of the Small Subunit (SSU) processome (1,2) before uS5 is incorporated into pre-40S particles (42). Accordingly, the accumulation of these large rRNA processing intermediates is likely to be an indirect consequence of a deficiency in uS5 and PDCD2, as certain factors that act in the SSU processome may become limiting as they are inefficiently recycled from late pre-40S particles (such as 21S-containing precursors). Indeed, as 21S and 18S-E rRNA processing intermediates exist in the context of pre-40S particles, the robust accumulation of 21S (Figure 2) in cells depleted for PDCD2 and uS5 is more consistent with a direct effect of a uS5 deficiency.
Mammalian PDCD2 localizes to the cytoplasm and the nucleus at steady state (Supplementary Figure S5), consistent with previous data from the Human Protein Atlas (43). Specifically, our BiFC analysis revealed the presence of uS5-PDCD2 complexes in the nucleolus and the cytoplasm of human cells (Figure 5). These observations suggest that PDCD2 recognizes uS5 co-translationally in the cytoplasm and accompanies uS5 to ribosome assembly sites in the nucleus. Consistent with a model in which PDCD2 escorts uS5 into the nucleus of human cells, uS5-YFP nucleolar signal was significantly reduced in PDCD2-deficient cells (Figure 6D). We also found that overexpression of PDCD2 results in nuclear accumulation of uS5-YFP (Figure 6E), supporting the view that PDCD2 accompanies uS5 into the nucleus. A model in which PDCD2 escorts uS5 into the nucleus contrasts with the chaperoning function of Tsr4 in S. cerevisiae where Tsr4 appears to be restricted to the cytoplasm (5). Surprisingly, uS5-GFP accumulates in the nucleus of TSR4-deleted cells (9), raising the possibility that Tsr4 could contribute to the nuclear pool of uS5 in S. cerevisiae. However, as TSR4-null cells show several morphological defects (9), the nuclear accumulation of uS5-GFP in the tsr4Δ mutant could also result from indirect consequences of ribosome biogenesis defects. As for many ribosomal proteins, the nuclear import pathway of uS5 remains unknown. Although ensemble algorithms (44) predict weak nuclear localization signals (NLSs) in the N-terminal region of human PDCD2, it remains to be determined whether PDCD2 functions as an import adaptor to promote uS5 nuclear import. An active role in the nuclear import of uL5 (Rpl11) and uL18 (Rpl5) was previously shown for the Syo1 chaperone protein in S. cerevisiae (45). Alternatively, the association of PDCD2 with uS5 could induce a conformational change in uS5 that exposes a cryptic NLS.
Our study established a critical role for the conserved TYPP domain of PDCD2 in chaperoning uS5. Remarkably, a detailed computational analysis of this evolutionarily conserved domain superfamily had predicted a general role for the TYPP domain in chaperone-like activities by mediating protein-protein interactions (38). Based on the structure of the B. subtilis YwqG protein (pdb id: 1PV5), the two-repeat structure of the TYPP domain stack against each other by forming five-stranded β-sheets into a single globular domain (38). Consistent with a similar two-region globular fold in human PDCD2, substitutions in conserved residues in either the N- or C-terminal repeat region of the TYPP domain abrogated the interaction with uS5 and the ability of PDCD2 to complement a PDCD2 loss-of-function (Figure 4). Together with data indicating that S. cerevisiae Tsr4 recognizes an unstructured region in the N-terminus of uS5 (5,9), our findings suggest that the TYPP domain could function in binding intrinsically disordered regions or unstructured peptides. Given that the positively-charged N-terminal region of uS5 is specific to eukaryotes, it is likely that bacterial proteins encompassing a TYPP domain, such as the uncharacterized YwqG proteins of B. subtilis and E. coli, will chaperone client proteins other than the prokaryotic homologue of uS5. Interestingly, many ribosomal protein chaperones appear to recognize eukaryotic-specific regions in their client ribosomal proteins (5,7–9,46). Whether the acquisition of eukaryotic-specific extension/domains and dedicated chaperones are linked to the emergence of a nuclear/nucleolar compartment for ribosome biogenesis is an intriguing question in evolutionary biology.
In humans, two different genes encode homologs of S. cerevisiae Tsr4: PDCD2 and PDCD2L that share the presence of a TYPP domain and protein-protein interactions with uS5. In contrast to PDCD2, however, PDCD2L is not essential for cellular viability and pre-rRNA processing defects are not readily detectable in PDCD2L-null cells (15). Furthermore, our results revealed that whereas PDCD2 binds to uS5 co-translationally, PDCD2L does not (Figure 5). Thus, despite sharing sequence similarity with Tsr4 and the ability to interact with uS5, it appears that the ribosomal protein chaperone-like function in humans is specific to PDCD2, while PDCD2L was likely co-opted for new functions in ribosome biogenesis. Indeed, PDCD2L physically associates with pre-40S ribosomal subunits and contains a strong leucine-rich nuclear export signal that is required for CRM1 binding (15), which makes it a potential candidate as an adaptor protein to facilitate CRM1-mediated nuclear export of 40S precursors, a function that is likely undertaken by a redundant set of adaptor proteins. In addition, the N- and C-terminal repeat regions of the PDCD2 TYPP domain are separated by a MYND domain, a specialized zinc finger that promotes protein-protein interactions (47) that is not found in human PDCD2L and S. cerevisiae Tsr4. Our results indicated that the MYND type zinc finger of PDCD2 was not essential for its interaction with uS5 (Figure 4), consistent with the fact that PDCD2L and Tsr4 efficiently bind to uS5 without the presence of a MYND domain. Future studies will therefore be required to define the functional role of the PDCD2 MYND domain.
Our AP-MS analysis of GFP-PDCD2 recovered PRMT3 and ZNF277, two proteins that we previously reported to compete for binding to free uS5 (17). Accordingly, this suggests that PDCD2 can form two different higher-order complexes with uS5, which either additionally contain ZNF277 or PRMT3. The functional relevance of these mutually exclusive complexes (PDCD2-uS5-PRMT3 vs PDCD2-uS5-ZNF277) remains to be determined. Interestingly, these complexes appear to be evolutionarily conserved (48), suggesting that they play important biological roles. However, neither PRMT3 nor ZNF277 appear to be required for ribosome biogenesis, as siRNA-mediated knockdowns show no sign of altered ribosome production (15,17). As ZNF277 was shown to associate with RNA (49) and chromatin (50), it will be interesting to see whether these higher-order complexes contribute to gene regulation.
PDCD2 is essential for embryonic development in mice (22), Zebrafish (51) and Drosophila (21). In mouse embryonic stem cells, loss of PDCD2 results in loss of proliferation and failure of S phase entry (52). By virtue of associations between PDCD2 and chromatin remodeling proteins HCFC1 and NCOR1 (26), together with the fact that MYND domain-containing proteins are known to interact with nuclear factors that function in transcription regulation (24,53,54), it was speculated that PDCD2 functions as a transcriptional regulator required for cellular proliferation and differentiation. As yet, however, direct evidence to support that PDCD2 functions in transcription regulation is still lacking. Our RNA-seq analysis in fact revealed only a few genes that were misregulated in PDCD2-deficient HeLa cells (Figure 3). Of potential interest is the observation that four (CFI, CFB, C4BPA, VTN) of the 17 significantly upregulated genes appear to function in the regulation of complement activation, a system that plays key roles in the defense against pathogens and the clearance of apoptotic cells (55). It remains possible that PDCD2 may contribute to gene regulation in a cell type-specific manner; however, PDCD2 is ubiquitously expressed across all tissues and cell types (56). Thus, given the data presented in this study, it is likely that the molecular function of PDCD2 essential for cellular proliferation and embryonic development is related to ribosome biogenesis via its critical role as a ribosomal protein chaperone. Consistent with this model, the conditional inactivation of PDCD2 results in p53 activation and cell cycle arrest (52), which are hallmarks of ribosome biogenesis defects as a result of a ribosomal protein deficiency such as in ribosomopathies (57,58).
In summary, our study reveals that the evolutionarily conserved PDCD2 protein functions as a specific ribosomal protein chaperone in human cells. The connection between altered ribosome biogenesis and cancer is now well established (3), supporting the idea that inhibition of ribosome biogenesis is a promising strategy in cancer chemotherapy. Because PDCD2 is critical for ribosome biogenesis, essential for cell proliferation and down-regulated in differentiated cells, PDCD2 represents a potentially attractive target for cancer treatment. Targeting PDCD2 could be particularly valuable for specific cancers, such as liver cancer, where elevated PDCD2 expression is associated with a significantly reduced probability of patient survival (59).
DATA AVAILABILITY
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE151094.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mark Bedford for the antibody specific to human uS5; Ingrid Grummt for antibodies to RPA116 and RPA194; and Ulrike Kutay for the uS5-YFP DNA construct. Jennifer Lafontaine is thanked for the generation of the GFP-uS5 HeLa conditional cell line and Yann Vanrobaeys is acknowledge for the initial analysis of the RNA-seq data.
Contributor Information
Anne-Marie Landry-Voyer, Department of Biochemistry & Functional Genomics, Université de Sherbrooke, Sherbrooke, QC J1E 4K8, Canada.
Danny Bergeron, Department of Biochemistry & Functional Genomics, Université de Sherbrooke, Sherbrooke, QC J1E 4K8, Canada.
Carlo Yague-Sanz, Department of Biochemistry & Functional Genomics, Université de Sherbrooke, Sherbrooke, QC J1E 4K8, Canada.
Breac Baker, Department of Biochemistry & Functional Genomics, Université de Sherbrooke, Sherbrooke, QC J1E 4K8, Canada.
Francois Bachand, Department of Biochemistry & Functional Genomics, Université de Sherbrooke, Sherbrooke, QC J1E 4K8, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research Grant [MOP-273292 to F.B.]; C.Y.S. is supported by a postdoctoral fellowship from the Fonds de recherche du Québec – Santé (FRQS); F.B. holds a Canada Research Chair in Quality Control of Gene Expression. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bassler J., Hurt E.. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2019; 88:281–306. [DOI] [PubMed] [Google Scholar]
- 2. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscipl. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pelletier J., Thomas G., Volarevic S.. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018; 18:51–63. [DOI] [PubMed] [Google Scholar]
- 4. Pillet B., Mitterer V., Kressler D., Pertschy B.. Hold on to your friends: dedicated chaperones of ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays. 2017; 39:1–12. [DOI] [PubMed] [Google Scholar]
- 5. Black J.J., Musalgaonkar S., Johnson A.W.. Tsr4 is a cytoplasmic chaperone for the ribosomal protein Rps2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2019; 39:e00094-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koch B., Mitterer V., Niederhauser J., Stanborough T., Murat G., Rechberger G., Bergler H., Kressler D., Pertschy B.. Yar1 protects the ribosomal protein Rps3 from aggregation. J. Biol. Chem. 2012; 287:21806–21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pausch P., Singh U., Ahmed Y.L., Pillet B., Murat G., Altegoer F., Stier G., Thoms M., Hurt E., Sinning I. et al.. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 2015; 6:7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pillet B., Garcia-Gomez J.J., Pausch P., Falquet L., Bange G., de la Cruz J., Kressler D.. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear Pre-60S assembly site. PLoS Genet. 2015; 11:e1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rossler I., Embacher J., Pillet B., Murat G., Liesinger L., Hafner J., Unterluggauer J.J., Birner-Gruenberger R., Kressler D., Pertschy B.. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic. Acids. Res. 2019; 47:6984–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ting Y.H., Lu T.J., Johnson A.W., Shie J.T., Chen B.R., Kumar S.S., Lo K.Y.. Bcp1 is the nuclear chaperone of Rpl23 in Saccharomyces cerevisiae. J. Biol. Chem. 2017; 292:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wyler E., Wandrey F., Badertscher L., Montellese C., Alper D., Kutay U.. The beta-isoform of the BRCA2 and CDKN1A(p21)-interacting protein (BCCIP) stabilizes nuclear RPL23/uL14. FEBS Lett. 2014; 588:3685–3691. [DOI] [PubMed] [Google Scholar]
- 12. Lu H., Zhu Y.F., Xiong J., Wang R., Jia Z.. Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol. Res. 2015; 177:28–33. [DOI] [PubMed] [Google Scholar]
- 13. Molavi G., Samadi N., Hosseingholi E.Z.. The roles of moonlight ribosomal proteins in the development of human cancers. J. Cell. Physiol. 2019; 234:8327–8341. [DOI] [PubMed] [Google Scholar]
- 14. Bachand F., Silver P.A.. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004; 23:2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Landry-Voyer A.M., Bilodeau S., Bergeron D., Dionne K.L., Port S.A., Rouleau C., Boisvert F.M., Kehlenbach R.H., Bachand F.. Human PDCD2L is an export substrate of CRM1 that associates with 40S ribosomal subunit precursors. Mol. Cell. Biol. 2016; 36:3019–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swiercz R., Person M.D., Bedford M.T.. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005; 386:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dionne K.L., Bergeron D., Landry-Voyer A.M., Bachand F.. The 40S ribosomal protein uS5 (RPS2) assembles into an extra-ribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction. J. Biol. Chem. 2019; 294:1944–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Minakhina S., Naryshkina T., Changela N., Tan W., Steward R.. Zfrp8/PDCD2 interacts with RpS2 connecting ribosome maturation and gene-specific translation. PLoS One. 2016; 11:e0147631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perreault A., Gascon S., D’Amours A., Aletta J.M., Bachand F.. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 2009; 284:15026–15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Owens G.P., Hahn W.E., Cohen J.J.. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol. Cell. Biol. 1991; 11:4177–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minakhina S., Druzhinina M., Steward R.. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development. 2007; 134:2387–2396. [DOI] [PubMed] [Google Scholar]
- 22. Mu W., Munroe R.J., Barker A.K., Schimenti J.C.. PDCD2 is essential for inner cell mass development and embryonic stem cell maintenance. Dev. Biol. 2010; 347:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minakhina S., Steward R.. Hematopoietic stem cells in Drosophila. Development. 2010; 137:27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lutterbach B., Sun D., Schuetz J., Hiebert S.W.. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol. 1998; 18:3604–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masselink H., Bernards R.. The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene. 2000; 19:1538–1546. [DOI] [PubMed] [Google Scholar]
- 26. Scarr R.B., Sharp P.A.. PDCD2 is a negative regulator of HCF-1 (C1). Oncogene. 2002; 21:5245–5254. [DOI] [PubMed] [Google Scholar]
- 27. Bergeron D., Pal G., Beaulieu Y.B., Chabot B., Bachand F.. Regulated intron retention and nuclear Pre-mRNA decay contribute to PABPN1 autoregulation. Mol. Cell. Biol. 2015; 35:2503–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beaulieu Y.B., Kleinman C.L., Landry-Voyer A.M., Majewski J., Bachand F.. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLos Genet. 2012; 8:e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deschamps-Francoeur G., Boivin V., Abou Elela S., Scott M.S.. CoCo: RNA-seq read assignment correction for nested genes and multimapped reads. Bioinformatics. 2019; 35:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Xue S., Ishijima J., Shiroishi T., Barna M.. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011; 145:383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M.. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002; 1:376–386. [DOI] [PubMed] [Google Scholar]
- 34. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M. et al.. Defining a cancer dependency map. Cell. 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cerezo E., Plisson-Chastang C., Henras A.K., Lebaron S., Gleizes P.E., O’Donohue M.F., Romeo Y., Henry Y.. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscipl. Rev. RNA. 2019; 10:e1516. [DOI] [PubMed] [Google Scholar]
- 36. O’Donohue M.F., Choesmel V., Faubladier M., Fichant G., Gleizes P.E.. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010; 190:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spellmon N., Holcomb J., Trescott L., Sirinupong N., Yang Z.. Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 2015; 16:1406–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burroughs A.M., Aravind L.. Analysis of two domains with novel RNA-processing activities throws light on the complex evolution of ribosomal RNA biogenesis. Frontiers in genetics. 2014; 5:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kerppola T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006; 1:1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A.. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002; 20:87–90. [DOI] [PubMed] [Google Scholar]
- 41. Mitterer V., Gantenbein N., Birner-Gruenberger R., Murat G., Bergler H., Kressler D., Pertschy B.. Nuclear import of dimerized ribosomal protein Rps3 in complex with its chaperone Yar1. Sci. Rep. 2016; 6:36714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ameismeier M., Cheng J., Berninghausen O., Beckmann R.. Visualizing late states of human 40S ribosomal subunit maturation. Nature. 2018; 558:249–253. [DOI] [PubMed] [Google Scholar]
- 43. Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M. et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 44. Lin J.R., Mondal A.M., Liu R., Hu J.. Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinformatics. 2012; 13:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauss D., Yoneda Y., Katahira J. et al.. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012; 338:666–671. [DOI] [PubMed] [Google Scholar]
- 46. Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E.. Coordinated ribosomal L4 protein assembly into the Pre-Ribosome is regulated by its Eukaryote-Specific extension. Mol. Cell. 2015; 58:854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matthews J.M., Bhati M., Lehtomaki E., Mansfield R.E., Cubeddu L., Mackay J.P.. It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr. Pharm. Des. 2009; 15:3681–3696. [DOI] [PubMed] [Google Scholar]
- 48. Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E. et al.. A Protein Interaction Map of Drosophila melanogaster. Science. 2003; 302:1727–1736. [DOI] [PubMed] [Google Scholar]
- 49. Ramanathan M., Majzoub K., Rao D.S., Neela P.H., Zarnegar B.J., Mondal S., Roth J.G., Gai H., Kovalski J.R., Siprashvili Z. et al.. RNA-protein interaction detection in living cells. Nat. Methods. 2018; 15:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Negishi M., Saraya A., Mochizuki S., Helin K., Koseki H., Iwama A.. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One. 2010; 5:e12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kramer J., Granier C.J., Davis S., Piso K., Hand J., Rabson A.B., Sabaawy H.E.. PDCD2 controls hematopoietic stem cell differentiation during development. Stem Cells Dev. 2013; 22:58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Granier C.J., Wang W., Tsang T., Steward R., Sabaawy H.E., Bhaumik M., Rabson A.B.. Conditional inactivation of PDCD2 induces p53 activation and cell cycle arrest. Biol Open. 2014; 3:821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lutterbach B., Westendorf J.J., Linggi B., Patten A., Moniwa M., Davie J.R., Huynh K.D., Bardwell V.J., Lavinsky R.M., Rosenfeld M.G. et al.. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 1998; 18:7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Melnick A.M., Westendorf J.J., Polinger A., Carlile G.W., Arai S., Ball H.J., Lutterbach B., Hiebert S.W., Licht J.D.. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 2000; 20:2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bajic G., Degn S.E., Thiel S., Andersen G.R.. Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 2015; 34:2735–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 57. Golomb L., Volarevic S., Oren M.. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014; 588:2571–2579. [DOI] [PubMed] [Google Scholar]
- 58. Mills E.W., Green R.. Ribosomopathies: there's strength in numbers. Science. 2017; 358:eaan2755. [DOI] [PubMed] [Google Scholar]
- 59. Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. et al.. A pathology atlas of the human cancer transcriptome. Science. 2017; 357:eaan2507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE151094.