Abstract
Photolyases are ubiquitously occurring flavoproteins for catalyzing photo repair of UV-induced DNA damages. All photolyases described so far have a bilobal architecture with a C-terminal domain comprising flavin adenine dinucleotide (FAD) as catalytic cofactor and an N-terminal domain capable of harboring an additional antenna chromophore. Using sequence-similarity network analysis we discovered a novel subgroup of the photolyase/cryptochrome superfamily (PCSf), the NewPHLs. NewPHL occur in bacteria and have an inverted topology with an N-terminal catalytic domain and a C-terminal domain for sealing the FAD binding site from solvent access. By characterizing two NewPHL we show a photochemistry characteristic of other PCSf members as well as light-dependent repair of CPD lesions. Given their common specificity towards single-stranded DNA many bacterial species use NewPHL as a substitute for DASH-type photolyases. Given their simplified architecture and function we suggest that NewPHL are close to the evolutionary origin of the PCSf.
INTRODUCTION
Sunlight is the predominant source of energy for life on Earth. However, the ultraviolet (UV) range of the solar electromagnetic spectrum is harmful by damaging DNA and thus acts as genotoxic environmental factor. UV-irradiation of deoxyribonucleic acid (DNA) can directly cause mainly two types of UV-lesions, the cyclobutane pyrimidine dimers (CPDs) with ∼80% and the pyrimidine–pyrimidone (6–4) photoproducts ((6–4)-PPs) (1) with ∼20% occurrence. Beside these major lesions other, indirectly UV-induced DNA modifications are found (2). Photochemical [2+2] cycloaddition of two adjacent pyrimidines along the C5–C6 bonds results in the cis–syn CPD stereoisomer, whereas (6–4)PP are generated by [2+2] cycloaddition that is followed by a Paternò-Büchi rearrangement of the oxetane intermediate (3).
To avoid mutagenic and genotoxic effects of UV-lesions of DNA, different protective mechanisms have evolved in nature. Beside nucleotide excision repair, the main pathway for repair of UV-lesions in higher mammals (4,5), light-dependent repair is catalysed by DNA photolyases (PHLs). These enzymes revert either CPD or (6–4)PP lesions by light-driven and transient electron transfer (ET) from their fully reduced, catalytic flavin cofactor, FADH−, to the lesion site. Together with the evolutionary related cryptochromes (CRY), which mediate light-driven signalling, photolyases form the ancient photolyase/cryptochrome protein superfamily (PCSf). Based on substrate specificity photolyases are separated in CPD and (6–4) photolyases. On a phylogenetic level, five groups of photolyases have been identified: class I-III CPD photolyases, eukaryotic and bacterial (6–4) photolyases (6). The latter group, also designated as FeS-BCP or CryPro, is phylogenetically only distantly related to other PCSf subgroups. Apart the catalytic FAD chromophore, bacterial (6–4) PHLs bind additionally an [Fe4S4]-cluster and 6,7-dimethyl-8-ribityllumazine (DLZ) as cofactors (7,8). In contrast, most cryptochromes are incapable to repair UV-damaged DNA, but act as light-dependent photoreceptors for regulating growth, development, circadian rhythm and even as magnetoreceptors in some animals (9–12). Exceptions are DASH-like (Drosophila, Arabidopsis, Synechocystis, Homo) CRYs, which repair CPD lesions within single-stranded DNA (ssDNA) (13), or bifunctional cryptochromes like aCRY from Chlamydomonas reinhardtii that catalyse (6–4)PP repair as well (14,15).
Independently of their divergent functions, CRYs and photolyases have highly similar structures, i.e. a conserved bilobal domain architecture with the FAD chromophore bound to the C-terminal all-α helical domain. An antenna chromophore for light-harvesting is often bound by the N-terminal domain with its characteristic Rossmann-like fold. Several antennae types are found in photolyases and cryptochromes: 5,10-methenyltetrahydrofolate (MTHF, pterin type) (16–18), 8-hydroxy-7,8-didemethyl-5-deaza-riboflavin (8-HDF, deazaflavin type) (19,20), flavin mono-nucleotide (FMN) (21), DLZ (8,22) or a second FAD molecule (23). Given that FADH− is indispensable for enzymatic function of photolyases, photoreduction of oxidized flavin (FADox) and semiquinone radical state (FADHo) species by blue-light is crucial for maintenance of catalytic activity. Photoreduction aka photoactivation depends on an intra-protein electron transfer chain involving mostly a tryptophan triad as common structural motif within the PCSf. Apart from the classical tryptophan triad as found in class I photolyases like EcCPDI from Escherichia coli (24), several alternative or extended electron transfer pathways have been described within different subfamilies of the PCSf (17,25–29).
The evolutionary origin of the PCSf is still under debate. One proposal states that the bacterial (6–4) PHL with their [Fe4S4]-cluster represent the ancestors of the PCSf (6,30,31). Our study shows a novel subfamily of the PCSf with CPD photolyase activity that is restricted to several clades of aquatic bacteria. With an average length of ∼370 aa members of this so-called NewPHL subfamily are small compared to other PCSf orthologs (>450 aa). Most importantly, NewPHLs have an inversed topology with the catalytic FAD-domain at the N-terminus and a second domain of unknown function at the C-terminus. Biochemical characterization of NewPHLs from Dinoroseobacter shibae (DsNewPHL) and Methylobacterium mesophilicum (MmNewPHL) showed a lack of any bound antenna chromophores, but similar photochemical properties and in vitro repair activities against CPD lesions as known from other CPD photolyase classes. Interestingly, NewPHLs are specific for single stranded DNA (ssDNA) like their bacterial relatives, the DASH cryptochromes. Given the simplified architecture and size of NewPHL this type of single-strand specific photolyases may represent the ancestor of the photolyase-cryptochrome superfamily.
MATERIALS AND METHODS
Plasmid constructs
NewPHL-coding plasmids were obtained as synthetic genes from BioCat (Heidelberg, Germany). The constructs are based on the pET28a(+)-vector system (Novagen, Germany) and codon-optimized for heterologous gene expression in E. coli. Sequence information of DsNewPHL and MmNewPHL are deposited under UniProt entries A8LJA9 (gene_ID: Dshi_1389) and M7YZC8 (gene_ID: MmSR116_2909), respectively. DsNewPHL and MmNewPHL are each fused to an N-terminal His6-tag for purification via immobilized metal ion affinity chromatography (IMAC).
Recombinant expression and purification of DsNewPHL and MmNewPHL
The expression vector pET28a-DsNewPHL was transformed into E. coli BL21(DE3) Gold cells (Stratagene). For protein expression, bacteria were grown in 2 l LB-medium supplemented with kanamycin at 37°C to an OD600 of 0.6–0.8. Thereafter, expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mm. Cells were shaken at 18°C for 20 h, collected by centrifugation and resuspended in buffer A (50 mm NaH2PO4, 100 mm NaCl, 20% (v/v) glycerol, pH 8). Cell disruption was performed with a French pressure cell (Aminco) with a constant pressure of 1000 psi; cell debris was pelleted by centrifugation at 4°C and 15 000 rpm for 60 min. After cell disruption, DsNewPHL was purified with IMAC and heparin affinity chromatography using a NGC system (Bio-Rad). The supernatant was sterile-filtered and applied to a buffer A-equilibrated column packed with Ni2+-NTA agarose matrix (Qiagen). The column was washed by 4 column volumes (CVs) of buffer A and afterwards by 4 CVs of 4% buffer B (50 mm NaH2PO4, 100 mm NaCl, 500 mm imidazole, 20% glycerol (v/v), pH 8). DsNewPHL was eluted with 35% buffer B. For heparin affinity chromatography IMAC fractions were pooled and diluted 1:4 with buffer A and applied to the heparin column (HiTrap heparin, GE Healthcare). The column was washed with buffer A for 4 CVs and eluted with a linear gradient towards 50% buffer C (Heparin elution buffer: (50 mm NaH2PO4, 2000 mm NaCl, 20% Glycerol (v/v), pH 8). The result of in vitro purification was monitored by SDS-PAGE analysis. The same protocol was used for MmNewPHL. After the second purification step, both NewPHL enzymes were obtained with overall yields of 20–40 mg/l expression culture with a purity of ≥95%. Overexpression and purification of MmCPDII was done as described before (20).
Co-expression of DsNewPHL or MmNewPHL with FO synthase was performed like described above with the additional plasmid pCDF-His6Fbic that encodes the bifunctional FO synthase (7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase) from Streptomyces coelicolor (15,20).
Anaerobic IMAC purification of NewPHLs
Cells were harvested by centrifugation at 17 000 × g and 4°C for 25 min and stored at -80°C. All further steps were performed under anoxic conditions in an anaerobic chamber (COY-System) including cell disruption, purification with Ni-NTA column and UV/Vis spectroscopy. The COY-System provides a strict anaerobic atmosphere of 0–5 ppm oxygen using a palladium catalyst and hydrogen gas mix of 5%. The detection is carried out by an anaerobic monitor (CAM-12) that shows oxygen and hydrogen concentrations. The samples are cooled by a dry bath (ALB6400 Cool Block) and precooled buffers were used at 4°C. Buffers were previously degassed by alternately purging with argon and applying vacuum for 3 min each, for a total of 15 cycles. All solutions and materials were brought into the COY system through an air lock and evacuated for at least four cycles. After purification via IMAC the protein samples were rebuffered via PD-10 columns and UV/Vis spectroscopy was performed under strictly anaerobic conditions in buffer A.
Analytical size exclusion chromatography (SEC)
The oligomeric state of DsNewPHL in solution was determined via analytical size exclusion chromatography. A Superdex 75 increase 10/300 (GE Healthcare) column and a protein concentration of 1 mg/l was used and SEC was performed in a HEPES buffer system (20 mm HEPES, 100 mm NaCl, 5% glycerol, pH 8) at 4°C. The oligomeric state was determined with a previous calibration curve (y = –0,182·x +3567, with y = lg10(MM) and x = elution volume) according to manufacturer manual.
Analysis of protein-bound Cofactor
In addition to UV–vis spectroscopy, determination of the FAD cofactor was performed with thin layer chromatography (TLC). Therefore, an established protocol was used without further modifications (32).
Characterization of NewPHLs by UV–Vis spectroscopy
All spectrophotometric measurements and the in vitro activity assay of recombinant NewPHLs were performed in a V-660 photometer (JASCO) at 10°C in buffer A. For the in vitro photoreduction of the protein-bound FAD cofactor 50 mm dithiothreitol (DTT) was added to the protein solution. The sample was incubated for 5 min in darkness and afterwards illuminated with blue light with a maximum emission wavelength of 455 nm (ILS – Intelligent LED Solutions) and a light intensity of 5.9 mW·cm−2 (224 μmol·m−2·s−1) at 4 cm distance. Spectra were taken at different time points ranging from 1 min to 90 min.
In order to obtain the fully oxidized cofactor, DsNewPHL or MmNewPHL were incubated for 40 h in darkness with 20 mm potassium ferricyanide K3[Fe(CN)6]. Afterwards the oxidant was removed via a PD10 desalting column (GE Healthcare) and an absorption spectra was measured to determine the cofactor/protein ratio. For occupation calculation, absorption values at 280 and 450 nm with the known extinction coefficients for FADOx and the respective proteins were used (18,33). In vitro photoreduction of NewPHLs harboring only FADox were performed like described above with time points ranging from 15 to 180 min.
Fluorescence spectroscopic analysis of MmNewPHL
MmNewPHL with only FADox after in vitro oxidation with K3[Fe(CN)6] in buffer A was used for fluorescence spectroscopy with a final concentration of 0.8 mg/ml. The fluorescence was measured with a RF-5301PC fluorescence spectrometer (Shimadzu, Japan). For detection of FAD cofactor an emission spectrum was measured at 420 nm excitation wavelength and an excitation spectrum at 520 nm emission wavelength with a total volume of 100 μl each sample. Background fluorescence, determined for a negative control reaction without protein, was subtracted from each measured value.
In vitro activity assay for ssDNA with CPD lesion
Single-stranded oligonucleotide dT18 (Thermo scientific) was used as a DNA-template for in vitro repair assays. A simplified protocol of Pokorny et al. (34) was used for the introduction of the CPD lesion. 20 μM oligo(dT)18 in 10 mm Tris·HCl, 1mm EDTA, pH 7.5 was placed in a Quartz Suprasil cell (Hellma) and was made anaerobic by bubbling with a stream of argon for 20 min. The following DNA-damaging was performed with an UV transilluminator (6 × 15 W, TF-20M, Vilber Lourmat). Ten cycles of 3 min UV-irradiation followed by 3 min of interruption were applied, and the sample was cooled on ice throughout the interruptions. UV induced damage was monitored by absorption spectroscopy after each second cycle; the decrease in absorbance at 265 nm indicates the introduction of CPD lesions. Formation of (6–4)PP lesions did not exceed 5% of UV-photoproducts as delineated from absorbance increase at 325 nm (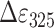 = 6000 M−1·cm−1). The average CPD lesion per oligonucleotide was calculated using difference absorptions at 265 nm with
= 6000 M−1·cm−1). The average CPD lesion per oligonucleotide was calculated using difference absorptions at 265 nm with 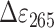 = 19 000 M−1·cm−1 (35). For photometric activity assay, 1 μm fully reduced NewPHL was added to 5 μm CPD-damaged DNA, incubated for 5 min in darkness and illuminated with blue light for 90 min. Enzymatic repair activity was detected by monitoring the increase of the absorption at 265 nm with absorption spectroscopy. MmCPDII was used as a positive control for repair activity under the same experimental conditions. Final concentrations for the additional in vitro activity assay of DsNewPHL are 100 nmDsNewPHL, 5 μm oligo(dT)18 with T<>T lesions, 0.175 mm DTT.
= 19 000 M−1·cm−1 (35). For photometric activity assay, 1 μm fully reduced NewPHL was added to 5 μm CPD-damaged DNA, incubated for 5 min in darkness and illuminated with blue light for 90 min. Enzymatic repair activity was detected by monitoring the increase of the absorption at 265 nm with absorption spectroscopy. MmCPDII was used as a positive control for repair activity under the same experimental conditions. Final concentrations for the additional in vitro activity assay of DsNewPHL are 100 nmDsNewPHL, 5 μm oligo(dT)18 with T<>T lesions, 0.175 mm DTT.
Restriction-site restoration assay
A fluorophore-labeled DNA strand (50 nt) with a single centrally-located cyclobutane thymine dimer (T<>T) was prepared as described before (34) except that the label was DY782 (Eurofins). The lesion containing DNA strand was heated together with a complementary 40mer strand (1.25 molar excess in annealing reactions) to 95°C for 5 min and the temperature was then gradually lowered by 1.2°C a minute to a final temperature of 11°C. DNA probes were kept at –20°C.
The MmNewPHL, DsNewPHL and MmCPDII proteins were diluted with buffer A. All assays contained proteins in a final concentration of 500 nm. Proteins were prepared freshly and pre-irradiated with a blue light LED source (445 ± 5 nm, 50 μmol m−2 s−1) for 120 min on ice in presence of 50 mm DTT as external reductant. For the final assay, duplex DNA probe was added in a final concentration of 50 nm. The assay reaction also contained 10% glycerol and 1× buffer O (50 mm Tris–HCl pH 7.5, 100 mm NaCl, 10 mm MgCl2, 100 μg/ml BSA; Thermo Scientific) for the following digestion with the restriction enzyme VspI (Thermo Scientific). Repair assays were incubated in the dark on ice for 10 min to allow binding of proteins to the probe and irradiated afterwards for 60 min with wavebands centered at 385 ± 5 nm (25 μmol·m−2·s−1) using a LED light source. Some probes were kept in darkness for control. All reactions were kept on ice. After irradiation, 10 units of VspI were added to each probe to accomplish digestion at 37°C for 120 min. VspI was inactivated at 65°C for 20 min. For denaturation, reactions were mixed with formamide loading buffer in a 21:4 ratio (60% formamide, 20 mm EDTA, pH 7.5, 10% glycerol) heated to 95°C for 15 min and separated on acrylamide gels (4–15%) containing 7 m urea (Bio-Rad Laboratories) for separation of DNA. Gels were analyzed as described before (34).
HDX-MS analysis of intrinsic protein order
HDX-MS was essentially carried out as described previously (36) aided by a robotic two-arm autosampler (LEAP Technologies). 7.5 μl (50 μM) DsNewPHL were diluted with 67.5 μl of D2O-containing buffer (50 mm sodium phosphate, pH 8.0, 100 mm NaCl) and incubated for 10, 95, 1000 or 10 000 s at 25°C. H/D exchange was stopped by mixing 55 μl of the reaction with an equal volume of quench buffer (400 mm KH2PO4/H3PO4, 2 m guanidine–HCl, pH 2.2) kept at 1°C and immediately injected into an ACQUITY UPLC M-class system with HDX technology (Waters) (37). DsNewPHL was digested with immobilized pepsin at 12°C in water +0.1% (v/v) formic acid at a flow rate of 100 μl/min and the resulting peptides trapped on a C18 column at 0.5 °C. After 3 minutes, the C18 trap column was placed in line with an ACQUITY UPLC BEH C18 1.7 μm 1.0 × 100 mm column (Waters) and the peptides separated at 0.5 °C with a gradient of water +0.1% (v/v) formic acid (eluent A) and acetonitrile +0.1% (v/v) formic acid (eluent B) at 30 μl/min flow rate as follows: 0–7 min/95–65% A, 7–8 min/65–15% A, 8–10 min/15% A, 10–11 min/5% A, 11–16 min/95% A. Mass spectra were recorded on a G2-Si HDMS mass spectrometer (Waters) in High Definition MS (HDMS) positive ion mode. [Glu1]-fibrinopeptide B (Waters) was used for lock-mass correction. Non-deuterated samples of DsNewPHL were prepared similarly employing non-deuterated buffer (50 mm sodium phosphate pH 8.0, 100 mm NaCl). Here, mass spectra were acquired in Enhanced High Definition MS (HDMS) positive ion mode (38,39) Between samples, the immobilized pepsin was washed three times with 80 μl of 4% (v/v) acetonitrile and 0.5 m guanidine hydrochloride. All measurements were performed in triplicates. Peptide identification and assignment of deuterium incorporation was done using the PLGS and DynamX 3.0 software (Waters), respectively, as described elsewhere (36).
Bioinformatics-based detection and analysis of the NewPHL subfamily
For identification of the novel subgroup of the PCSf we used the sequence similarity network (SSN) tools (40). The SSN analysis was based on the InterPro database entry IPR005101 and PFAM entries PF000875 and PF004141, which cover the whole photolyase-cryptochrome superfamily. A threshold of ≤10−70 for the E-value for pairwise sequence alignment was chosen and all sequences with an identity ≥90% are summarized as one node. Further processing of the resulted clusters for graphical representation was performed with Cytoscape (41). All sequence information from the SSN analysis are summarized in Supplementary Table S1 (Excel data sheet).
The homology model of DsNewPHL was built with Chimera and Modeller v9.21 (42). The search for acceptable template structures was performed with the Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) (43) using the PDB as selected sequence database. The template with the highest sequence similarity is given by the structure of the S. elongatus class I CPD photolyase (PDB entry 1TEZ). Visualization and analysis of the model was done with PyMOL v2.1. For secondary structure predictions the PSIPRED (44) webserver was used (http://bioinf.cs.ucl.ac.uk/psipred).
The multiple sequence alignment was performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (45) with NewPHL orthologs DsNewPHL (Uniprot ID: A8LJA9), MmNewPHL (Uniprot ID: M7YZC8), ElNewPHL from Erythrobacter litoralis (Uniprot ID: Q2N8F4), PmNewPHL from Prochlorococcus marinus (Uniprot ID: A0A0A2A9H3), RcNewPHL from Rhodospirillum centenum (Uniprot ID: B6IPR5), the DASH-Crys AtCry3 (Uniprot ID: Q84KJ5) from Arabidopsis thaliana and SynCry from Synechocystis (Uniprot ID: P77967) and the class I CPD photolyases SynCPDI (Uniprot ID: P05327) and EcCPDI from E. coli (Uniprot ID: P00914). Results were edited with the MView online tool (https://www.ebi.ac.uk/Tools/msa/mview/) (45). For a detailed alignment of the FAD-binding site corresponding sequence fragments were taken from the Pfam Database PF03441: DsNewPHL (78–212 aa), MmNewPHL (76–212 aa), ElNewPHL (73–201 aa), PmNewPHL (71–196 aa), RcNewPHL (137–270 aa), AtCry3 (368–526 aa), SynCry (288–478 aa), SynCPDI (280–469 aa) and EcCPDI (272–466 aa).
For the WebLogo analysis of NewPHLs a multiple sequence alignment of 200 NewPHL orthologs was performed with Clustal Omega and the WebLogo was created with Version 3.7 (http://weblogo.threeplusone.com/) (46).
Sequence covariation analysis
Analysis of the NewPHL cluster were used for covariation analysis with the GREMLIN webserver (www.gremlin.bakerlab.org/index.php) (47). For that we retrieved all NewPHL sequences from the previous SSN analysis, removed all fragments and redundant sequences and subjected after multiple-sequence alignment with Clustal Omega the remaining 587 sequences to GREMLIN (0 iterations, E-value cut-off 10−10, >75% coverage). The resulting covariance analysis with 577 remaining sequences and a sequence/length ratio of 1.43 was analysed. The 14 statistically most significant covariations for residues within the catalytic and Z-domains (80% < score probability < 99.9%; average 92%) belong to the rim region for sealing the FAD chromophore as shown by mapping on the homology model of DsNewPHL.
RESULTS
Bioinformatics analysis of the PCSf and identification of NewPHL and NewCRY subfamilies
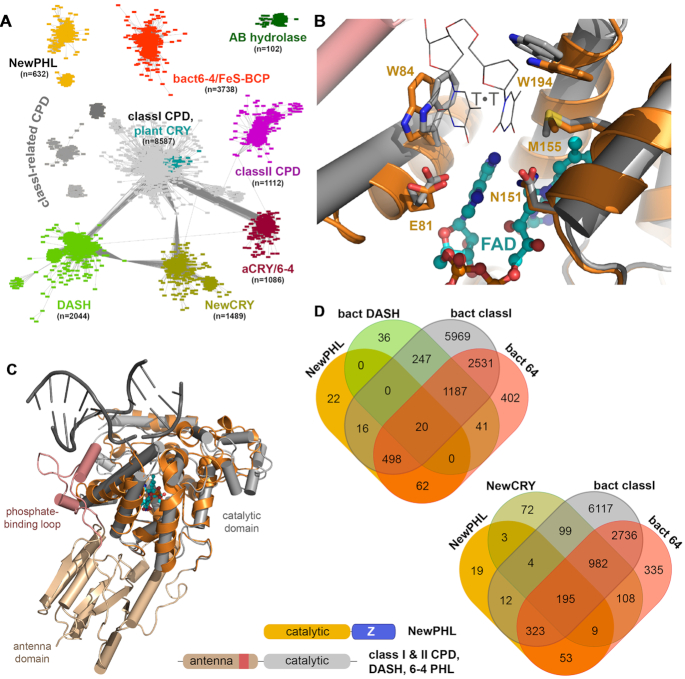
Previous bioinformatics analyses of the PCSf and its families relied on the characteristic two-domain architectures of photolyases and cryptochromes. We wondered whether there existed ancestral photolyases, which still lacked the N-terminal domain with its ancillary antenna function. To chart the entire PCSf for this purpose we combined the protein families PF00875, PF04244 from the PFAM database with IPR005101 from INTERPRO for sequence-similarity network (SSN) analysis (40). Whereas the PF00875 and PF04244 families cover both domains of members of the PCSf, i.e. the N-terminal antenna domain and catalytic C-terminal domain, the family definition of IPR005101 covers only the C-terminal domain. The resulting SSN for this dataset (Figure 1A and Supplementary Table S1 for sequence information) consists of 19 524 non-redundant sequences and can be subdivided into 11 clusters when applying a pairwise E-value cut-off of < 10−70 for edge definition. As expected, the largest part of the PCSf SSN is covered by class I CPD photolyases (8587; 44%), whose four subclusters include not only the class III photolyases with their distinct mode of MTHF antenna binding (17) but also plant cryptochromes. The second largest subfamily is represented by the bacterial (6–4) photolyases (FeS-BCP, CryPro) with 3738 members (19%), followed by DASH cryptochromes (10%) and a new subfamily, here designated as NewCRY (8%). The latter family, formerly annotated as CPD photolyases, is only found in bacteria and haloarchaea. However, NewCRY members lack conserved residues within the active site, which are essential for light-driven DNA repair, such as the conserved glutamate for interaction with the 5′ pyrimidine (EcCPDI: E274) or the asparagine (EcCPDI: N341) for interaction with the 3′ pyrimidine of the CPD lesion. Accordingly, only a function for light-dependent signalling appears currently feasible for this subfamily given that one of its representatives, VcCry2 from Vibrio cholera, has been found to bind FAD (18).
Figure 1.
Bioinformatics analysis of the whole photolyase-cryptochrome superfamily. (A) Sequence-similarity network of the PCSf including a combined set of sequences from PFAM families PF00875, PF04244 and INTERPRO family IPR005101. Each of the 9136 nodes represents sequences of a pairwise sequence of >50% (overall 19524 sequences), connecting edges represent BLAST E-value scores of <10−70. Two novel, exclusively bacterial PCF families have been identified by this SSN, NewCRY and NewPHL. Notably, class III CPD photolyases are an unresolved subset within the cluster for class I CPD photolyases and plant cryptochromes. A small cluster designated as AB hydrolases represents plant gene products, which harbour an N-terminal domain similar to the antenna domain of the PCSf and a C-terminal α/β-hydrolase domain. (B) Homology model of the NewPHL ortholog from Dinoroseobacter shibae (orange) that is superimposed to the template structure of the class I CPD photolyase of Synechococcus elongatus (PDB code: 1TEZ) in complex with substrate CPD-DNA (grey, sequence identity 30%). Its predicted active site structure of DsNewPHL (orange) harbours all residues required for CPD repair activity by SynCPDI (gray). Furthermore, the C4-carbonyl groups of the 5′- and 3′-thymines are predicted to form hydrogen bonds with the N6 amino group of the FADH− cofactor. (C) Overall view of the DsNewPHL catalytic domain in comparison to CPD-DNA bound state of SynCPDI. (D) Venn diagrams showing species (co)distribution between PCSf subfamilies with members of bacterial origin. A schematic representation of the domain architecture of the NewPHLs in comparison to canonical photolyases is shown in the lower part of the figure.
After class II CPD photolyases and animal(-like) cryptochromes/(6–4) photolyases (each 6%) the second new PCSf subfamily is represented by a cluster, designated as NewPHL (3%), whose members exclusively occur in bacteria. A major feature of NewPHL members, which are among the smallest PCSf representatives with sizes down to ∼300 aa, is the absence of an N-terminal antenna domain found in all other PCSf subfamilies. Homology modelling of their all-α helical FAD-binding domain, e.g. from the D. shibae ortholog (DsNewPHL; M1-S255), with the closest structurally characterized ortholog, the class I CPD photolyase from Synechococcus elongatus, SynCPDI (sequence identity 30%) and multiple sequence alignments of NewPHLs in comparison to class I CPD photolyases and DASH-Crys (see Supplementary Figure S1A) show strict conservation of active site residues required for CPD repair (Figure 1B, C) thus making a function of NewPHL members as CPD photolyases highly likely. Another common feature of NewPHL is their C-terminal domain, called Z-domain (DsNewPHL: W256-S405 aa), with a yet unknown fold and function and a size of about ∼150 amino acids (Figure 1C). Further in silico analysis of NewPHLs with the WebLogo software reveal that the Z-domain is less conserved in contrast to the all-α helical FAD-binding domain (see Supplementary Figure S1B). Only one tryptophan and a GFF motif are highly conserved within the Z-domain, which may indicate a relevant biological function of these residues.
An analysis of species codistribution of bacterial photolyase subfamilies (Figure 1D) shows that NewPHL members almost always accompany a bacterial class I or (6–4) photolyase (596 species from 618 in total, 96%). Interestingly, the same kind of codistribution can be found for bacterial DASH cryptochromes (1495% versus 153, 98%), whereas almost no overlap exists in terms of co-occurrence between NewPHL members and DASH cryptochromes with 3.2% and 1.3% respectively. For comparison, NewCRY members are not only generally found besides bacterial CPDI and (6–4) photolyases (95%), but also show a considerable overlap with NewPHL members (14%). Overall, the lack of co-occurrence of NewPHL and DASH subfamily members in bacterial species resembles the exclusive usage of either class I or class II PHL for repair of double-stranded DNA and may indicate a common in vivo function.
Cofactor analysis of DsNewPHL and MmNewPHL
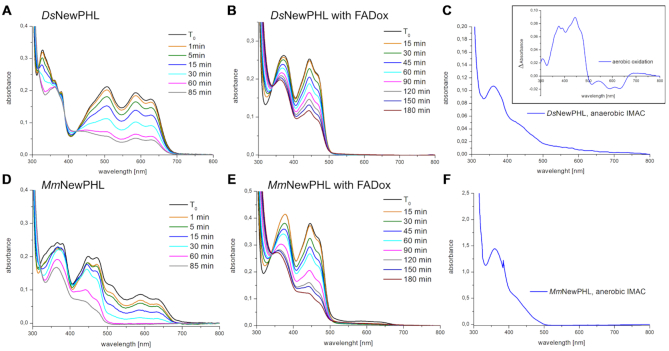
For characterization of the photochemistry and capability of light-driven DNA repair, we established recombinant overproduction for two NewPHL orthologs, DsNewPHL from Dinoroseobacter shibae and MmNewPHL from Methylobacterium mesophilicum. Recombinant DsNewPHL adopts a grey-blue colour after initial purification by immobilized metal ion affinity chromatography (IMAC), whereas MmNewPHL elutes with light green colour. Due to unspecific binding of DNA contaminations to NewPHL further purification via heparin affinity chromatography was required. Analytical size exclusion chromatography shows that His-tagged DsNewPHL and MmNewPHL are monomeric in solution with an apparent molecular mass of ∼47 kDa for DsNewPHL (MMcalc: 46.95 kDa) and ∼39 kDa for MmNewPHL (see Supplementary Figure S2).
Initial absorption spectra after purification (Figure 2A) are characterized by high absorption in the range of 550–700 nm, with peaks at 586 nm and 634 nm. Additional absorption peaks at 507 and 328 nm indicate the presence of FADHo as predominantly bound flavin species in DsNewPHL and MmNewPHL as described before for EcCPDI (48,49). Thin layer chromatography (TLC) confirmed the identity of NewPHL-bound flavins as FAD (Figure 2A, inlay). Given the different colours after purification, MmNewPHL harbours a considerable proportion of FADox besides FADHo as indicated by additional absorption at 445 nm and was accessible to fluorescence spectroscopy due to its readily oxidizable flavin chromophore (Figure 1B). The emission spectrum (λexc = 420 nm) shows a symmetrical shape with a peak at 523 nm, which is comparable to other photolyases harbouring only FAD, for example MmCPDII without the antenna 8-HDF (20). The excitation spectrum (λemit = 520 nm) exhibited a peak at 373 nm and a broad dominant peak around 457 nm.
Figure 2.
Initial spectroscopic characterization and chromophore analysis of NewPHLs. (A) Initial UV–vis spectra after purification for DsNewPHL (solid black line) and after in vitro oxidation of the protein bound FAD with potassium ferricyanide (solid blue line). The spectra for MmNewPHL are shown in dotted lines, after purification (red dotted) and with FADox after oxidation (green dotted). Spectra are normalized at 630 nm for FADH° containing samples and at 375 nm for the NewPHLs with only FADox. (A, inlay) Cofactor analysis via thin layer chromatography (TLC) of heat denatured DsNewPHL (4) and MmNewPHL (5). Flavin adenine dinucleotide (FAD, 1), riboflavin (RF, 2) and flavin mononucleotide (FMN, 3) were used as references. (B) Fluorescence spectrum of MmNewPHL after in vitro oxidation with only FADox bound to the protein. The emission spectrum was recorded with a 420 nm excitation wavelength and the excitation spectrum was recorded with a 520 nm emission wavelength.
Both NewPHLs apparently lack any second antenna chromophore as indicated by the UV–vis spectra of the native proteins and their cofactor extracts (see Supplementary Figure S3). We also excluded a potential interaction between NewPHLs and the 8-HDF antenna, which is not an endogenously available cofactor in E. coli unlike MTHF, DLZ or FAD. By co-expressing the NewPHLs with ScFbiC, a bifunctional enzyme for 8-HDF biosynthesis (15,20), we found that the resulting NewPHLs exhibit unaltered absorption spectra (see Supplementary Figure S4 for MmNewPHL and S5 for DsNewPHL). To determine the protein/FAD ratio, NewPHL were firstly oxidized to the FADox state in vitro using potassium ferricyanide (Figure 2A). After removing the oxidant with a PD10 desalting column the absorption spectrum exhibited peaks at 380, 443, 470 for DsNewPHL (MmNewPHL: 378, 446 and 471 nm) and a weakly pronounced shoulder at 420 nm. Basing the calculation on the absorptions at 280 nm and 455 nm, the known extinction coefficients of FADox and a calculated extinction coefficient of DsNewPHL at 280 nm, we found a nearly 1:1 stoichiometric ratio between protein and cofactor (similar results were obtained for MmNewPHL). This implies that NewPHLs binds no second FAD molecule as antenna chromophore as described for the archaeal photolyase from Sulfolobus tokodaii (23). Overall, these spectroscopic analyses corroborate our notion that NewPHLs only harbour FAD as chromophore.
Photochemical properties and in vitro photoreduction of NewPHLs
The semiquinone FADHo state of DsNewPHL is unusually stable at 4°C when compared to MmNewPHL and other PHLs or CRYs (20),(36). Oxidation of FADHo was barely detectable within 14 days under aerobic conditions, whereas MmNewPHL shows FAD oxidation within five days under identical conditions. In NewPHLs, the tryptophan triad for ET found in other PCSf subfamilies is apparently incomplete or altered. Only the proximal (DsNewPHL: W192) and medial tryptophans (DsNewPHL: W169) are strictly conserved when compared to the template of homology modelling, SynCPDI (W390, W367). The existence of a third, distal tryptophan at a non-homologous position is feasible in NewPHL, e.g. the conserved W99 in DsNewPHL (Supplementary Figure S1), because distal tryptophans found in other photolyases and cryptochromes anchor a long irregular stretch to their catalytic domain. Nevertheless, both NewPHLs undergo photoreduction from FADHo to FADH− upon blue light illumination and presence of the reducing agent DTT as absorption decreases between 550 and 700 nm and the spectral signature of FADHo between 315 and 400 nm simplifies to a signature characteristic of the FADH− state (49) (Figure 3A, D). Interestingly, when purified under strictly anaerobic conditions from E. coli the FAD chromophore of DsNewPHL adopts an almost pure, fully reduced state (Figure 3C) with a major absorption peak at 362 nm that is comparable to the FADH− state of EcCPDI (35,50). The FADHo → FADH− transition of DsNewPHL proceeds uniformly without formation of other FAD species as indicated by three isosbestic points at 318, 389 and 422 nm. Whereas the end product of photoreduction of DsNewPHL is heterogeneous with a significant contribution of the FADHo state, MmNewPHL can be almost quantitatively reduced to FADH−. Furthermore, we observe no transient accumulation of FADHo during photoreduction of MmNewPHL and DsNewPHL (Figure 3B, E) harbouring only FADox. This suggests that blue-light triggered reduction of FADHo to FADH− is faster than the FADox to FADHo transition under steady-state conditions.
Figure 3.
In vitro photoreduction and anaerobic purification of NewPHLs. (A) Photoreduction of DsNewPHL with 50 mm DTT one day after purification starting with FADHo. All photoreduction experiments were carried out with blue light with a maximum emission wavelength of 455 nm (ILS – Intelligent LED Solutions) and a light intensity of 5.9 mW·cm−2 (224 μmol·m−2·s−1) at 4 cm distance. (B) Photoreduction of DsNewPHL after in vitro oxidation with potassium ferricyanide to FADox. (C) Absorption spectra of DsNewPHL directly after IMAC purification under strictly anaerobic conditions. Difference spectra of aerobic oxidation after anaerobic purification for 300 min, measured with difference between time points T30 min and T300 min is shown within the inlay on the upper right side. (D) Photoreduction of MmNewPHL with 50 mm DTT one day after purification, starting with a mixture of different FAD redox states. (E) The photoreduction of MmNewPHL starting with FADox. (F) The absorption spectrum of MmNewPHL after IMAC under strictly anaerobic conditions.
Unlike other PCSf members studied by us, higher DTT concentrations and longer illumination times are necessary to achieve stoichiometric conversion to FADH− by in vitro photoreduction (25). Comparison with a class II photolyase shows that photoreduction of DsNewPHL and MmNewPHL proceeds about 18–30 times slower under identical conditions (see Supplementary Figure S6). Nevertheless, the prevalence of both NewPHLs for adopting in vivo an FADH− state (Figure 3C, F) indicates that photoreduction is crucial to maintain these photolyases in the catalytically active FADH− state of photolyases as this state oxidizes in the presence of oxygen (inlay, Figure 3C).
NewPHL catalyse CPD lesion repair only in single-stranded DNA
To test whether NewPHL are capable of binding and repairing CPD-damaged DNA, we performed in vitro studies with our NewPHL orthologs. As single-stranded substrate template for CPD repair we used the dT18 oligonucleotide (Thermo Scientific), where we introduced two T<>T dimers on average per template strand by UV-C illumination as tracked and quantified by absorption decrease at 265 nm. Clearly, our CPD substrate DNA is heterogeneous in regard to the number and positions of CPDs along the dT18 strand. Under anaerobic conditions our procedure for CPD formation leads to <5% (6–4)PP lesions within substrate DNA as reflected by almost no absorption increase at 325 nm (51). Catalytic repair of CPD lesions by DsNewPHL or MmNewPHL under blue light illumination was quantified by absorption increase at 265 nm at high enzyme:substrate ratios (1:5) for obtaining maximally achievable CPD-repair (Figure 4A). For comparison and as positive control for repair we used recombinant class II CPD photolyase from Methanosarcina mazei (MmCPDII) without inbuilt antenna chromophore at the same enzyme:substrate ratio (25). Under these conditions, NewPHL were capable to repair twice the number of CPD lesions in the single-stranded DNA substrate (∼70%) than the class II photolyase (35%). To verify light-dependence of repair the assays were carried out under standard conditions as triplicates using low enzyme:substrate ratios of 1:50. As negative controls samples kept in darkness or without added enzyme were used (Figure 4B). As expected, in darkness no significant absorption increase at 265 nm caused by enzyme-catalyzed CPD repair could be observed.
Figure 4.
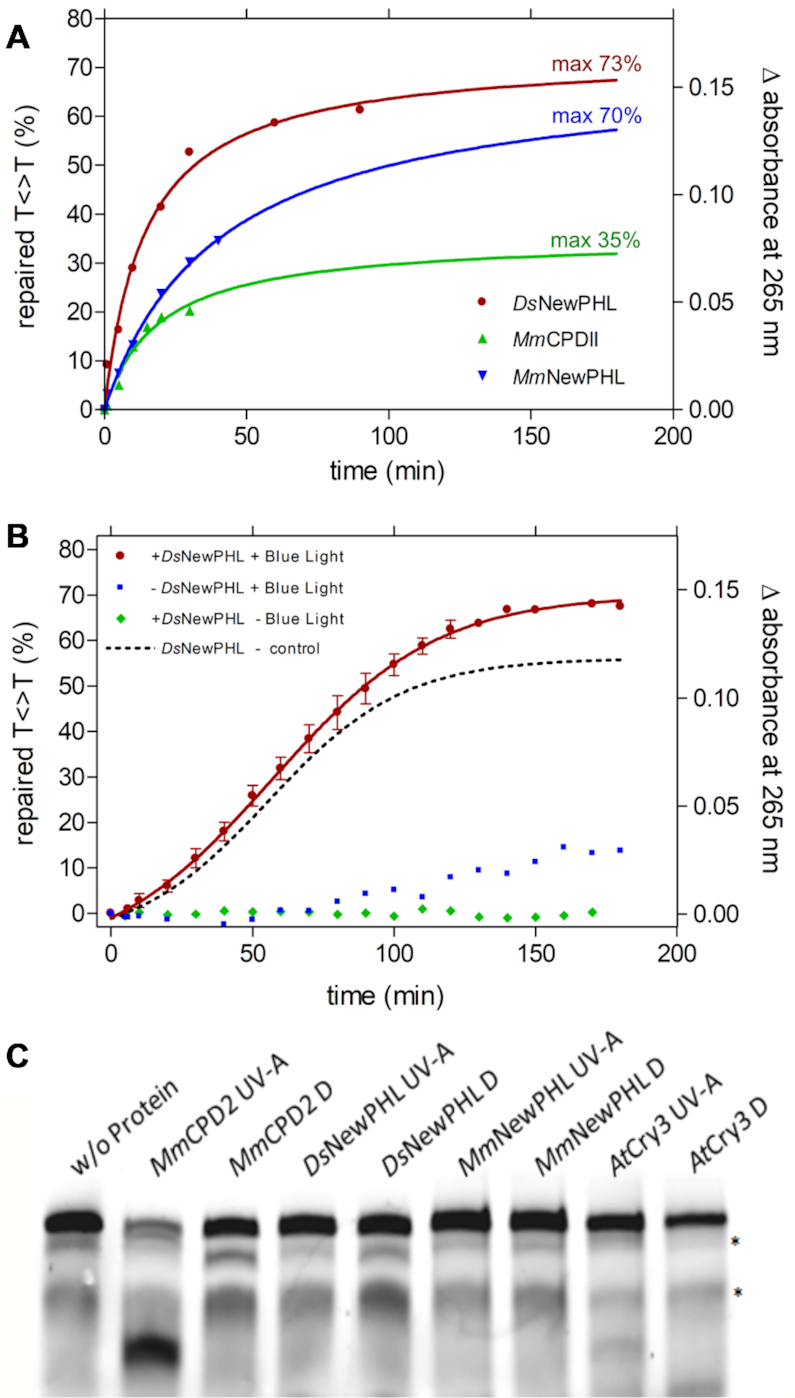
Repair activity and Restriction-site restoration assay of NewPHLs. Repair reactions contained 1 μm (A) or 100 nm (B) previously reduced protein, 5 μm UV-irradiated oligo(dT)18 and 0.175 mm DTT in a volume of 200 μl. Repair of CPD lesions was triggered with blue light (455 nm, 224 μmol·m−1·s−1) at 10°C and followed by UV-Vis spectroscopy. The difference in absorbance at 265 nm was plotted. Reaction-sample without protein and with no illumination served as a control (B). In (A) data points were extrapolated and the maximum repair was determined using an exponential fit. Interestingly, a slight absorption increase under illumination but without added enzyme can be observed after ∼80 min (B) that is caused by onset of DTT oxidation. For comparison the dotted curve shows light-dependent CPD-lesion repair corrected by this background reaction. (C) Restriction-site restoration assay with a single thymine cyclobutane dimer localized in the center of a well formed DNA duplex. The respective DNA probe (50 nm) was incubated with the indicated purified enzymes (each in 500 nm concentration) in the repair reactions in the presence of external reductant DTT (10 mm). Reactions were kept either in the dark (Dark) or under UV-A irradiation (+UV-A, 385 ± 5 nm, 25 μmol·m−2·s−1) for 60 min, and afterwards treated with VspI, denatured and separated on 4–15% polyacrylamide gels containing 7 m urea. The resulting bands were visualized using the Odyssey LiCOR Infrared Imaging system. The upper band shows the uncut substrate, the prominent lower band in the MmCPDII lane represents the repaired substrate cleaved by VspI;* unspecific band.
Given the observed repair activity of NewPHL against CPD lesions we wondered whether this subgroup is capable to repair CPD lesions also within double-stranded DNA (dsDNA). For that, we utilized the VspI restriction site restoration assay that was used before by us to assess CPD-repair activity of plantal and fungal DASH cryptochromes (Figure 4C) (34). Again, we used MmCPDII as positive control for activity against dsDNA and cryptochrome 3 from Arabidopsis thaliana, a DASH cryptochromes, whose repair activity is specific for ssDNA, as negative control. Our experiment shows that both NewPHL from D. shibae and M. mesophilicum are incapable to bind and repair CPD lesions within dsDNA. In this way, they mimic the characteristics of bacterial and plantal DASH cryptochromes which act as ssDNA-specific CPD photolyases (34).
The Z-domain seals the FAD-binding site of NewPHLs
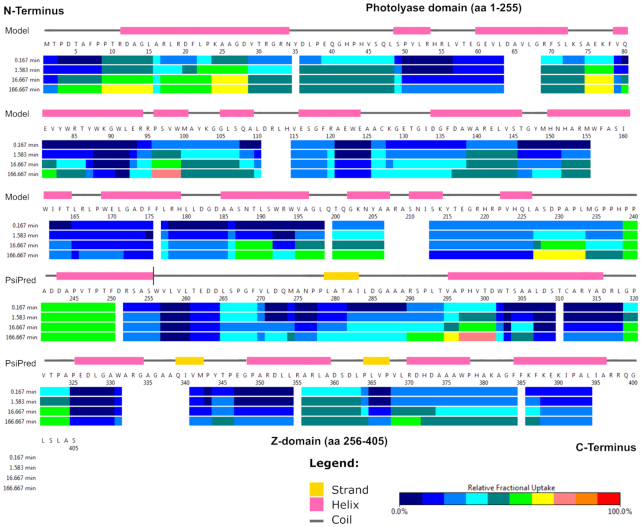
We analysed the folding state and function of the C-terminal Z-domain of NewPHLs by a combination of in silico methods and hydrogen-deuterium exchange mass spectrometry (HDX-MS). Homology modelling of this domain failed due to a lack of suitable template structures. However, an analysis for disordered protein regions with the MoreRONN Webserver (52) predicted that most of the Z-domain is well ordered. Secondary structure predictions with PSIPRED (44) show not only the all-α helical domain that is characteristic of the N-terminal catalytic domain, but also three short β-strands and five α-helices for the Z-domain. The order of the latter's secondary structure elements is not matching the typical motif of a Rossmann fold (53). Accordingly, we suggest that NewPHLs also adopt a bilobal architecture like other PCSf members but with a reversed domain order.
HDX-MS experiments were carried out with recombinant DsNewPHL to gain experimental evidence of putative disordered regions within this NewPHL (54). HDX-MS analysis of DsNewPHL provided 124 peptides, which could be evaluated for deuterium uptake. This corresponds to an overall sequence coverage of 87% with a redundancy of 3.7. Our HDX-MS results support the in silico prediction of a well-ordered Z-domain at the C-terminus. The heat map of DsNewPHL (Figure 6) shows relative deuterium uptake rates for periods of 1 min to 166 min. A short, flexible region at the N-terminus is followed by the mostly well-ordered region of the catalytic FAD-binding domain (Y29-P238). Only the C-terminus of the catalytic domain (P239-F250) exhibits somewhat increased H/D exchange rates. Minor H/D exchange rates within the subsequent region (D251-S405) indicate that the Z-domain is similarly well packed like the catalytic domain.
Figure 6.
HDX-MS analysis of DsNewPHL. Time-resolved hydrogen/deuterium exchange as analysed by mass-spectrometry within the protein is shown in combination with the DsNewPHL sequence and secondary structure prediction. For the N-terminal domain (M1-S255) the secondary structure based on the homology model is shown and for the Z-domain (W256-S405) the secondary structure was predicted with PSIPRED (44).
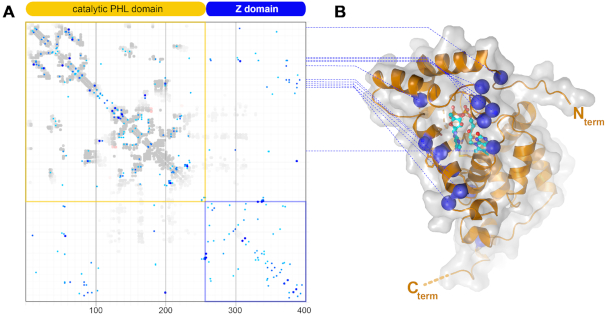
In other photolyases and cryptochromes the N- and catalytic C-terminal domains are almost invariantly packed to each other to seal the pyrimidine edge of the FAD’s isoalloxazine moiety from solvent access, whereas the opposing dimethyl-benzene edge is well-shielded by the catalytic domain itself. To analyse whether the two domains of NewPHLs interact with each other we performed a sequence covariation analysis to predict intramolecular domain-domain contacts. Sequence covariation as performed by the GREMLIN webserver predicts that the Z-domain packs specifically against the catalytic domain (Figure 5) (55). Mapping of covarying residues on the structural model of the catalytic domain unambiguously show their location along a rim-like region close to the pyrimidine edge of the FAD chromophore (DsNewPHL: P9, R53-L56, L63, Y83, T86, E93, R94, G183). As analogous interactions are made by the N-terminal antenna domains in other photolyases and cryptochromes our in silico analysis predicts that catalytic domains of photolyases and cryptochromes generally depend on a second domain or peptide stretching for guaranteeing a well-shielded environment of the FAD chromophore.
Figure 5.
Sequence covariation analysis within the NewPHL family. (A) 577 NewPHL orthologs were subjected to covariation analysis as provided by the GREMLIN webserver (47,55). Whereas the N-terminal catalytic domain (orange) maps to the catalytic domain of photolyases, the C-terminal Z-domain (blue) bears striking similarity for its first 125 amino acids to the antenna-binding domain. (B) Distribution of residues (blue circles) of the catalytic domain, whose covariance strictly depends on residues in the Z-domain (cut-off: scaled score > 0.9).
DISCUSSION
With the present study we show the identification of a novel subgroup of PCSf with sequence similarity network and the initial biochemical characterization of two soluble orthologues, namely DsNewPHL and MmNewPHL, in regard to spectroscopically properties and biological activity within repair of UV-damaged DNA. The NewPHLs harbor only FAD as a chromophore and own an unique domain architecture in comparison to canonical subgroups of PCSf.
Although there are over 50 years of research in the field of photolyases and cryptochromes, the evolutionary origin of these photoreceptors is still under debate. A detailed phylogenetically analysis of PCSfs in comparison to primases presented by Zhang et al. indicates the bacterial (6–4) photolyases as an evolutionary precursor of all other photolyases (8). According to a recent review by Vechtomova et al., the authors pointed out key properties of the cryptochrome/photolyase family including its potential ancestor, which included general the two-domain architecture (56). However, these studies have not yet consider the discovered NewPHLs. In the following the key aspects of cryptochromes/photolyases are briefly described in context of the NewPHLs. The discovery of the first prokaryotic cryptochrome in the cyanobacterium Synechocystis sp. (57) challenged the hypothesis that cryptochromes evolved after the origin of eukaryotic organisms (57,58). It has been shown that DASH cryptochromes and class I CPD photolyases share similarities in DNA recognition, redox activity and DNA binding and may pose a possible evolutionary origin that the photolyases have taken over transcription functions (57),(58). Daiyasue et al. found DASH cryptochromes from vertebrates and fungi and clearly showed that the DASH orthologs are almost ubiquitously distributed and probably the result of horizontal gene transfers. In contrast to cryptochromes, several DASH cryptochromes still show CPD photolyase activity, perhaps as an evolutionary holdover or indicating a special role as being a second CPD photolyase because many organisms in which DASH cryptochromes have been identified so far possess another CPD photolyase gene (Figure 1D) (59). Indeed, Aziz Sancar and coworkers showed that DASH cryptochromes from bacteria, plants and animals are actually cryptochromes/photolyases with a high specificity for cyclobutane pyrimidine dimers in single-stranded DNA (13). Like other photolyases, these DASH photolyases have a second antenna chromophore, MTHF, for efficient energy transfer to FAD (18).
Our results indicate that NewPHLs are for many bacterial organisms an alternative to DASH photolyases by having likewise photolyase activities towards single-stranded nucleic acids. Given up to 30% sequence identity to class I PHL our homology model of the catalytic domain of DsNewPHL shows indeed a considerable degree of structural conservation, e.g. to the class I CPD photolyase of Synechococcus elongatus. Especially, the active site region harbors all known residues required for FAD binding and repair of CPD lesions as described below (60). For example, the L-shaped wedge comprising conserved tryptophans W84 and W194 is predicted to shield the cyclobutane ring by making van-der-Waals interactions with the ring plane of the 5′-thymine and the edge of the CPD-dinucleotide. Furthermore, both thymines form hydrogen bonds via their C4-carbonyl and N3-imide groups to the side chains of E81 and N151. A mutation of the conserved glutamic acid to alanine causes a 60% decrease in the quantum yield for CPD cleavage reaction in the yeast photolyase, indicating a similar role in the NewPHL (61). The intramolecular electron transfer pathway, that is essential for the ability of photoreduction akas photoactivation of photolyases, is predicted to comprise only two (DsNewPHL: W192, W169; MmNewPHL: W190, W167), instead of three tryptophans like in other PCSf subgroups. This may explain our observation that the photoreduction activity of NewPHLs is lower than that of a ‘classical’ photolyase like MmCPDII. A clear difference to other photolyases including DASH is the apparent absence of the phosphate binding loop for interaction with the -1 phosphate of the CPD lesion that is otherwise part of a long linking region between N-terminal antenna and C-terminal catalytic domains. In case of DASH cryptochromes, their incapability to repair CPD lesions in double-stranded DNA is caused by insufficient kinking of bound double-stranded DNA and concomitant flipping of the CPD lesion into the active site (34). This notion can be transferred to NewPHLs with their lack of structural elements such as the phosphate-binding loop. Interestingly, some fungal DASH cryptochromes have been found to repair double-stranded DNA (62), indicating that the loss of dsDNA repair capability may have been a late evolutionary event for providing a class of ssDNA-specific photolyases. In contrast, the simpler architecture of NewPHL may exclude dsDNA repair a priori and makes this class to a prime candidate for an ancestral photolyase only capable of repairing ssDNA.
Another difference to other photolyases is the lack of a known antenna chromophore like MTHF or 8-HDF in our NewPHL, although it cannot be excluded that some other NewPHL orthologs may harbor one. As the catalytically active FAD is N-terminal only the comparably small Z-domain that packs against the catalytic domain for sealing the FAD binding site from solvent may act as a substitute for an antenna-binding domain. However, antenna chromophores are not necessary for DNA binding and repair, since they just broaden the absorption cross section of photolyases by channeling light energy to FADH− (19). The repair activity against single-stranded nucleic acids with CPD lesions is predominantly found in a subset of bacteria, which harbor both CPD and (6–4) photolyases, although its role for bacterial viability has still to be resolved. Accordingly, one may hypothesize that single-strand repair capability is an additional feature for bacterial species anyway capable of withstanding high levels of UV-irradiation.
Clearly, one may postulate two scenarios for the evolutionary position of NewPHL in the PCSf. In the first one, they are closest to ancestral minimal photolyases consisting merely of a catalytic FAD binding domain with a small C-terminal extension for sealing the FAD-binding site. For example, the NewPHL from Halorhodospira halochloris (Uniprot ID: W8KR12) has a length of only 313 amino acids with only 53 residues (V262-R313) in its Z-region. Alternatively, NewPHL could originate from a regular two-domain photolyase as result of an early domain swapping event. According to Occam's razor, the likelihood of NewPHL being close to the evolutionary root of the PCSf is higher than for the highly complex bacterial (6–4) photolyases (FeS-BCP) with their two domains and three different types of cofactors.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge support from the DFG-core facility for interactions, dynamics and macromolecular assembly structure at the Philipps-University Marburg. We thank Sophie Franz-Badur for critical reading of the manuscript.
Author contributions: L.-O.E. designed the research. H.-J.E., M.S., L.S., D.K., L.O.E. performed the research. H.-J.E., M.S., L.-O.E. and A.B. wrote the paper. All authors discussed and edited the manuscript.
Contributor Information
Hans-Joachim Emmerich, Unit for Structural Biochemistry, Department of Chemistry, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany.
Martin Saft, Unit for Structural Biochemistry, Department of Chemistry, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany.
Leonie Schneider, Unit for Structural Biochemistry, Department of Chemistry, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany.
Dennis Kock, Department of Biology, Philipps University Marburg, Karl-von-Frisch-Straße 8, 35032 Marburg, Germany.
Alfred Batschauer, Department of Biology, Philipps University Marburg, Karl-von-Frisch-Straße 8, 35032 Marburg, Germany.
Lars-Oliver Essen, Unit for Structural Biochemistry, Department of Chemistry, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany; Center of Synthetic Microbiology, Philipps University Marburg, Hans-Meerwein Straße 4, 35032 Marburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) within Collaborative Research Center [987 to A.B., L.-O.E., ES152/18 to L.-O.E.]. Funding for open access charge: Deutsche Forschungsgemeinschaft
Conflict of interest statement. None declared.
REFERENCES
- 1. Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003; 103:2203–2237. [DOI] [PubMed] [Google Scholar]
- 2. Schuch A.P., Garcia C.C.M., Makita K., Menck C.F.M.. DNA damage as a biological sensor for environmental sunlight. Photochem. Photobiol. Sci. 2013; 12:1259–1272. [DOI] [PubMed] [Google Scholar]
- 3. Cadet J., Douki T.. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018; 17:1816–1841. [DOI] [PubMed] [Google Scholar]
- 4. Goosen N. Nucleotide excision repair in eukaryotes. Encycl. Biol. Chem. Second Ed. 2013; 10:341–344. [Google Scholar]
- 5. Friedberg E.C. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005; 6:943–953. [DOI] [PubMed] [Google Scholar]
- 6. Ma H., Holub D., Gillet N., Kaeser G., Thoulass K., Elstner M., Krauß N., Lamparter T.. Two aspartate residues close to the lesion binding site of Agrobacterium (6-4) photolyase are required for Mg2+ stimulation of DNA repair. FEBS J. 2019; 286:1765–1779. [DOI] [PubMed] [Google Scholar]
- 7. Oberpichler I., Pierik A.J., Wesslowski J., Pokorny R., Rosen R., Vugman M., Zhang F., Neubauer O., Ron E.Z., Batschauer A. et al.. A photolyase-like protein from agrobacterium tumefaciens with an iron-sulfur cluster. PLoS One. 2011; 6:2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang F., Scheerer P., Oberpichler I., Lamparter T., Krauß N.. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6, 7-dimethyl-8-ribityllumazine antenna chromophore. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.-O., van der Horst G.T.J., Batschauer A., Ahmad M.. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011; 62:335–364. [DOI] [PubMed] [Google Scholar]
- 10. Cashmore A.R. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003; 114:537–543. [PubMed] [Google Scholar]
- 11. Müller P., Ahmad M.. Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception. J. Biol. Chem. 2011; 286:21033–21040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeng Z., Wei J., Liu Y., Zhang W., Mabe T.. Magnetoreception of photoactivated cryptochrome 1 in electrochemistry and electron transfer. ACS Omega. 2018; 3:4752–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Selby C.P., Sancar A.. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17696–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beel B., Prager K., Spexard M., Sasso S., Weiss D., Muller N., Heinnickel M., Dewez D., Ikoma D., Grossman A.R. et al.. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012; 24:2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franz S., Ignatz E., Wenzel S., Zielosko H., Putu E.P.G.N., Maestre-Reyna M., Tsai M.-D., Yamamoto J., Mittag M., Essen L.-O.. Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii. Nucleic Acids Res. 2018; 46:8010–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park H., Kim S., Sancar A., Deisenhofer J.. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995; 268:1866–1872. [DOI] [PubMed] [Google Scholar]
- 17. Scheerer P., Zhang F., Kalms J., von Stetten D., Krauß N., Oberpichler I., Lamparter T.. The Class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways. J. Biol. Chem. 2015; 290:11504–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Worthington E.N., Kavakli I.H., Berrocal-Tito G., Bondo B.E., Sancar A.. Purification and characterization of three members of the photolyase/cryptochrome family blue-light photoreceptors from vibrio cholerae. J. Biol. Chem. 2003; 278:39143–39154. [DOI] [PubMed] [Google Scholar]
- 19. Malhotra K., Kim S.T., Walsh C., Sancar A.. Roles of FAD and 8-hydroxy-5-deazaflavin chromophores in photoreactivation by Anacystis nidulans DNA photolyase. J. Biol. Chem. 1992; 267:15406–15411. [PubMed] [Google Scholar]
- 20. Kiontke S., Gnau P., Haselsberger R., Batschauer A., Essen L.-O.. Structural and evolutionary aspects of antenna chromophore usage by Class II photolyases. J. Biol. Chem. 2014; 289:19659–19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klar T., Kaiser G., Hennecke U., Carell T., Batschauer A., Essen L.-O.. Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. ChemBioChem. 2006; 7:1798–1806. [DOI] [PubMed] [Google Scholar]
- 22. Geisselbrecht Y., Frühwirth S., Schroeder C., Pierik A.J.J., Klug G., Essen L.-O.. CryB from Rhodobacter sphaeroides: a unique class of cryptochromes with new cofactors. EMBO Rep. 2012; 13:223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujihashi M., Numoto N., Kobayashi Y., Mizushima A., Tsujimura M., Nakamura A., Kawarabayasi Y., Miki K.. Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J. Mol. Biol. 2007; 365:903–910. [DOI] [PubMed] [Google Scholar]
- 24. Byrdin M., Sartor V., Eker A.P.M., Vos M.H., Aubert C., Brettel K., Mathis P.. Intraprotein electron transfer and proton dynamics during photoactivation of DNA photolyase from E. coli: review and new insights from an ‘inverse’ deuterium isotope effect. Biochim. Biophys. Acta - Bioenerg. 2004; 1655:64–70. [DOI] [PubMed] [Google Scholar]
- 25. Kiontke S., Geisselbrecht Y., Pokorny R., Carell T., Batschauer A., Essen L.O.. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 2011; 30:4437–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nohr D., Franz S., Rodriguez R., Paulus B., Essen L.O., Weber S., Schleicher E.. Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys. J. 2016; 111:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Müller P., Yamamoto J., Martin R., Iwai S., Brettel K.. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6-4) photolyases. Chem. Commun. 2015; 51:15502–15505. [DOI] [PubMed] [Google Scholar]
- 28. Cailliez F., Müller P., Gallois M., De La Lande A.. ATP binding and aspartate protonation enhance photoinduced electron transfer in plant cryptochrome. J. Am. Chem. Soc. 2014; 136:12974–12986. [DOI] [PubMed] [Google Scholar]
- 29. Biskup T., Paulus B., Okafuji A., Hitomi K., Getzoff E.D., Weber S., Schleicher E.. Variable electron transfer pathways in an amphibian cryptochrome: tryptophan versus tyrosine-based radical pairs. J. Biol. Chem. 2013; 288:9249–9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma H., Zhang F., Ignatz E., Suehnel M., Xue P., Scheerer P., Essen L.O., Krauß N., Lamparter T.. Divalent cations increase DNA repair activities of bacterial (6-4) photolyases. Photochem. Photobiol. 2017; 93:323–330. [DOI] [PubMed] [Google Scholar]
- 31. Lamparter T., Zhang F., Oberpichler I., Krauß N., Scheerer P.. Two photolyases in Agrobacterium tumefaciens. FASEB. 2015; 29:879.22. [Google Scholar]
- 32. Deka R.K., Brautigam C.A., Biddy B.A., Liu W.Z., Norgard M.V.. Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. MBio. 2013; 4:e00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Özgür S., Sancar A.. Analysis of autophosphorylating kinase activities of arabidopsis and human cryptochromes. Biochemistry. 2006; 45:13369–13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pokorny R., Klar T., Hennecke U., Carell T., Batschauer A., Essen L.O.. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl Acad. Sci. U.S.A. 2008; 105:21023–21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim S.-T., Sancar A.. Effect of Base, Pentose, and Phosphodiester Backbone Structures on Binding and Repair of Pyrimidine Dimers By Escherichia coli DNA Photolyase. Biochemistry. 1991; 30:8623–8630. [DOI] [PubMed] [Google Scholar]
- 36. Franz-Badur S., Penner A., Straß S., von Horsten S., Linne U., Essen L.O.. Structural changes within the bifunctional cryptochrome/photolyase CraCRY upon blue light excitation. Sci. Rep. 2019; 9:9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wales T.E., Fadgen K.E., Gerhardt G.C., Engen J.R.. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal. Chem. 2008; 80:6815–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geromanos S.J., Vissers J.P.C., Silva J.C., Dorschel C.A., Li G.Z., Gorenstein M.V., Bateman R.H., Langridge J.I.. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009; 9:1683–1695. [DOI] [PubMed] [Google Scholar]
- 39. Li G.Z., Vissers J.P.C., Silva J.C., Golick D., Gorenstein M.V., Geromanos S.J.. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009; 9:1696–1719. [DOI] [PubMed] [Google Scholar]
- 40. Gerlt J.A., Bouvier J.T., Davidson D.B., Imker H.J., Sadkhin B., Slater D.R., Whalen K.L.. Enzyme function initiative-enzyme similarity tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta - Proteins Proteomics. 2015; 1854:1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin D., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Webb B., Sali A.. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016; 2016:doi:10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 44. Buchan D.W.A., Jones D.T.. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019; 47:402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D. et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crooks G., Hon G., Chandonia J., Brenner S.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ovchinnikov S., Kamisetty H., Baker D.. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014; e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Z., Zhang M., Guo X., Tan C., Li J., Wang L., Sancar A., Zhong D.. Dynamic determination of the functional state in photolyase and the implication for cryptochrome. Proc. Natl Acad. Sci. U.S.A. 2013; 110:12972–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu B., Liu H., Zhong D., Lin C.. Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 2010; 13:578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heelis P.F., Sancar A.. Photochemical properties of Escherichia coli DNA photolyase: a flash photolysis study. Biochemistry. 1986; 25:8163–8166. [DOI] [PubMed] [Google Scholar]
- 51. Yamamoto J., Plaza P., Brettel K.. Repair of (6-4) lesions in DNA by (6-4) photolyase: 20 years of quest for the photoreaction mechanism. Photochem. Photobiol. 2017; 93:51–66. [DOI] [PubMed] [Google Scholar]
- 52. Yang Z.R., Thomson R., McNeil P., Esnouf R.M.. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005; 21:3369–3376. [DOI] [PubMed] [Google Scholar]
- 53. Hanukoglu I. Proteopedia: Rossmann fold: a beta-alpha-beta fold at dinucleotide binding sites. Biochem. Mol. Biol. Educ. 2015; 43:206–209. [DOI] [PubMed] [Google Scholar]
- 54. Balasubramaniam D., Komives E.A.. Hydrogen-exchange mass spectrometry for the study of intrinsic disorder in proteins. Biochim. Biophys. Acta - Proteins Proteomics. 2013; 1834:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kamisetty H., Ovchinnikov S., Baker D.. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vechtomova Y.L., Telegina T.A., Kritsky M.S.. Evolution of proteins of the DNA photolyase/cryptochrome family. Biochemistry. (Mosc). 2020; 85:S131–S153. [DOI] [PubMed] [Google Scholar]
- 57. Brudler R., Hitomi K., Daiyasu H., Toh H., Kucho K.I., Ishiura M., Kanehisa M., Roberts V.A., Todo T., Tainer J.A. et al.. Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell. 2003; 11:59–67. [DOI] [PubMed] [Google Scholar]
- 58. Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D.. Cryptochromes: blue light receptors for plants and animals. Science. 1999; 284:760–765. [DOI] [PubMed] [Google Scholar]
- 59. Daiyasu H., Ishikawa T., Kuma K.I., Iwai S., Todo T., Toh H.. Identification of cryptochrome DASH from vertebrates. Genes Cells. 2004; 9:479–495. [DOI] [PubMed] [Google Scholar]
- 60. Mees A., Klar T., Gnau P., Hennecke U., Eker A.P.M., Carell T., Essen L.O.. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004; 306:1789–1793. [DOI] [PubMed] [Google Scholar]
- 61. Vande Berg B.J., Sancar G.B.. Evidence for dinucleotide flipping by DNA photolyase. J. Biol. Chem. 1998; 273:20276–20284. [DOI] [PubMed] [Google Scholar]
- 62. Tagua V.G., Pausch M., Eckel M., Gutiérrez G., Miralles-Durán A., Sanz C., Eslava A.P., Pokorny R., Corrochano L.M., Batschauer A.. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15130–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.