Abstract
Fluorescent ligands are versatile tools for the study of G protein-coupled receptors. Depending on the fluorophore, they can be used for a range of different applications, including fluorescence microscopy and bioluminescence or fluorescence resonance energy transfer (BRET or FRET) assays. Starting from phenylpiperazines and indanylamines, privileged scaffolds for dopamine D2-like receptors, we developed dansyl-labeled fluorescent ligands that are well accommodated in the binding pockets of D2 and D3 receptors. These receptors are the target proteins for the therapy for several neurologic and psychiatric disorders, including Parkinson’s disease and schizophrenia. The dansyl-labeled ligands exhibit binding affinities up to 0.44 nM and 0.29 nM at D2R and D3R, respectively. When the dansyl label was exchanged for sterically more demanding xanthene or cyanine dyes, fluorescent ligands 10a-c retained excellent binding properties and, as expected from their indanylamine pharmacophore, acted as agonists at D2R. While the Cy3B-labeled ligand 10b was used to visualize D2R and D3R on the surface of living cells by total internal reflection microscopy, ligand 10a comprising a rhodamine label showed excellent properties in a NanoBRET binding assay at D3R.
Subject terms: G protein-coupled receptors, Medicinal chemistry, Chemical tools
Introduction
Dopamine receptors belong to the large family of G protein-coupled receptors (GPCRs). With their transmembrane architecture, these proteins are responsible for signal transduction from the extracellular environment to intracellular compartments. Dopamine receptors respond to the binding of the neurotransmitter/hormone dopamine and are divided into two sub-groups, the D1-like (D1R, D5R) and the D2-like (D2R, D3R, D4R) family, depending on their G protein coupling preference1. D2-like receptors represent the main targets for the therapy of severe neurological and psychiatric disorders including Parkinson’s disease, schizophrenia, restless legs syndrome, and addiction1. Over the past years, high-resolution X-ray crystal structures or cryo-electron microscopy maps have become available for the entire subfamily (D2R: 6LUQ2, 6CM43, 6VMS4; D3R: 3PBL5; D4R: 6IQL6, 5WIV7, 5WIU7), enabling structure-based drug design7 as an alternative to ligand-based drug development. Independent from the employed design strategy, drug discovery campaigns require fast and reliable in vitro assays to determine target affinity, selectivity, kinetics and functional activity of novel lead structures. Whereas numerous assay technologies, such as enzyme fragment complementation, fluorescence or bioluminescence resonance energy transfer (FRET or BRET), proximity-based assays and even label-free methods have been employed for the determination of functional activity at GPCRs8, the characterization of a ligand’s affinity towards a given receptor is mostly based on radioligand competition. This binding assay has been proven useful due to the high chemical similarity of unlabeled and labeled probe molecules, the highly specific detection of radioactive labels, and the relatively easy assay format9. Nevertheless, radioligand-based methods have several disadvantages. Besides the costs and problems associated with the disposal of radioactive waste and regulatory requirements, high-affinity radioactive probe molecules have to be available for the receptor of interest9.
Over recent years, fluorescence-based technologies have emerged as exciting alternative to study ligand affinity10–12. Similar to radioactive probes, fluorescent molecules can be detected at very low concentration with high specificity. Although the synthesis of small-molecule fluorescent ligands is far from trivial, it requires lower level regulatory and safety precautions compared to the preparation of radioligands. Moreover, several fluorescence-based technologies like resonance energy transfer (RET) and fluorescence anisotropy (also known as fluorescence polarization) do not require separation of unbound ligands and assays can thus be run in a homogenous, fast “mix-and-measure” setup10,13. In combination with sophisticated techniques like total internal reflection fluorescence (TIRF) microscopy, fluorescent ligands allow to detect and track GPCRs with single molecule resolution14–16. Fluorescent ligands have also been used to monitor receptor internalization17, and to study receptor oligomerization in native tissue18. Very recently, the development of a small and bright luciferase variant (nanoluciferase, Nluc)19 has greatly facilitated the implementation of BRET-based ligand binding assays11. In contrast to Renilla luciferase, which is frequently used to study intracellular protein–protein interactions, N-terminal Nluc fusion proteins can be easily targeted to the cell surface11. NanoBRET between fluorescently labeled ligands and Nluc-receptor fusion proteins has already been used to study ligand binding at a number of therapeutically relevant GRCRs, including β-adrenergic11, adenosine11, histamine20, free fatty acid21, and chemokine22 receptors. Despite the high therapeutic relevance of the D2-like receptor family, only a small number of fluorescent ligands targeting these receptors have been reported so far. Whereas a fluorescently labeled agonist (2-(N-phenethyl-N-propyl)amino-5-hydroxytetralin scaffold, PPHT-red)23,24 has been employed in FRET ligand binding studies at D2R (Tag-lite, Cisbio), NanoBRET binding assays have not yet been established for this receptor family. Besides the agonistic PPHT derivatives, different fluorescent ligands comprising an antagonistic N-(p-aminophenethyl)spiperone (NAPS) scaffold have also been described25–28. For example, a Cy3B-labeled NAPS derivative has been used for microscopic analyses of ligand binding at D3R29. In our group, we have developed Cy3B-labeled fluorescent ligands with an N-propylamino-5-hydroxytetraline or phenylpiperazine substructure for TIRF microscopy studies of D2R and D3R homodimerization in living cells14.
Taking advantage of this expertise, we describe the design, molecular docking, chemical synthesis and pharmacological characterization of novel fluorescent ligands targeting the D2-like receptor subfamily. Starting from well-known dopamine receptor recognition elements, we developed dansyl-labeled fluorescent ligands that possess excellent binding affinity at D2R and D3R receptors. We demonstrate that our design strategy is suitable for accommodating larger xanthene and cyanine fluorophores, while maintaining high-affinity dopamine receptor binding. Depending on the fluorophore, our novel ligands can be used to label D2R and D3R in living cells (TIRF microscopy) or in a newly established D3R-NanoBRET ligand binding assay that will open up the path for future drug discovery campaigns.
Results
Ligand design and synthesis
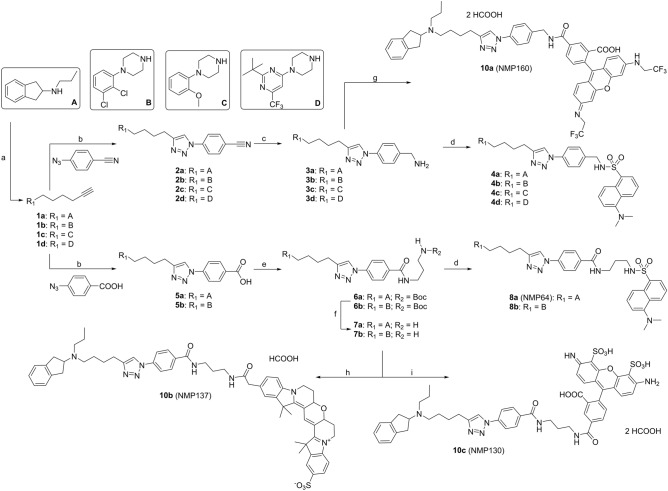
For the development of fluorescent probes targeting the D2-like receptor family, we made use of four different privileged scaffolds that are frequently found in dopamine receptor ligands30. The N-propyl substituted 2-amino-dihydro-1H-indene (building block A, Fig. 1), is a well-known dopamine-isostere and exhibits agonistic properties31,32. In contrast, 1,4-disubstituted phenyl- or pyrimidyl-piperazines (1,4-DAPs, building blocks B–D, Fig. 1) with different substituents attached to the aromatic core represent the main receptor recognition element of atypical antipsychotics such as aripiprazole and cariprazine30. While the 2-methoxy or 2,3-dichloro substituted phenylpiperazines are known to bind to all D2-like receptor subtypes, the pyrimidylpiperazine (building block D) selectively targets D3R33. Previous studies have revealed the importance of a second lipophilic moiety for high-affinity binding to dopamine receptors30,34. Thus, we envisioned connection of the primary pharmacophores to a triazole-benzylamine or triazole-benzoic acid moiety through a flexible four-carbon aliphatic linker. This linker size was chosen so that ligands should possess high affinity for both D2R and D3R, while a considerably shorter linker would have been beneficial for the D4R subtype30. For an initial set of fluorescent ligands, we focused on the incorporation of a dansyl label, a naphthalene-derived fluorophore emitting green light. The dansyl fluorophore has a relatively low molecular weight and is commercially available as reactive sulfonyl chloride (dansyl chloride). The dansyl dye is widely used to label proteins for fluorescence polarization measurements due to its favorable lifetime or as an acceptor for resonance energy transfer from protein tryptophan residues. It is also known for its large Stokes shift, resulting in a good signal-to-noise ratio35. Since we have previously found that the binding pockets of D2R and D3R can accommodate flexible linkers14 but also sterically more demanding substituents in their extended binding pocket36,37, installation of the dansyl fluorophore was envisioned through sulfonamide-formation either directly with the primary benzylamine (ligands 4a-d) or through addition of a short 1,3-diaminopropane spacer to the benzoic acid precursors (ligands 8a,b). The latter strategy has already been successful for the development of Cy3B-labeled dopamine receptor ligands in the context of TIRF microscopy14,17.
Figure 1.
Synthesis of fluorescent ligands 4a-d, 8a,b and 10a-c. Reagents and conditions: (a) 6-chlorohex-1-yne, K2CO3, KI, CH3CN, reflux, 68–85%; (b) 4-azido-benzonitrile or 4-azidobenzoic acid, 5 mol% CuSO4·5H2O, 10 mol % Na-ascorbate, 1:1:1 mixture of tert.-butanol:H2O:CH2Cl2, rt, 76–92%; (c) LiAlH4, THF, 0 °C to rt, crude; (d) dansyl chloride, triethylamine, CH2Cl2; 0 °C, 53–88%; (e) tert.-butyl (3-aminopropyl)carbamate, DIPEA, TBTU, DMF/CH2Cl2, 0 °C, 85–88%; (f) 4 M HCl, dioxane, rt, 98%, (g) 5′-carboxy-N,N′-bis(2,2,2-trifluoroethyl)rhodamine (9)38, DIPEA, TBTU, CH2Cl2/DMF, 0 °C, 24 h, 23%; (h) Cy3B NHS ester, DIPEA, DMF, rt, 24 h, 66%; (i) Alexa488 TFP ester, DIPEA, DMF, rt, 24 h, 58%.
Synthesis of the dansyl-labeled ligands started from the secondary amines N-propyl-2,3-dihydro-1H-inden-2-amine, 2,3-dichlorophenylpiperazine, 2-methoxyphenylpiperazine or 2-(tert-butyl)-4-(piperazin-1-yl)-6-(trifluoromethyl)pyrimidine (Fig. 1, building blocks A–D), that were reacted with 6-chlorohex-1-yne in a nucleophilic displacement reaction in analogy to previously described protocols39,40. The resulting terminal alkynes 1a-d were subjected to a copper(I)-catalyzed azide-alkyne cycloaddition with 4-azido-benzonitrile14,41, giving access to the 1,4-disubstituted triazoles 2a-d in excellent yield. Subsequent reduction of the benzonitriles with LiAlH4 afforded the primary amines 3a-d, which were used in the next step without further purification. In the final step, dansyl-labeled title compounds 4a-d were obtained by reaction with dansyl chloride in presence of triethylamine. Fluorescent ligands 8a,b comprising an additional aminopropane spacer were prepared in a very similar reaction sequence. To this end, the terminal alkynes 1a,b were first reacted with 4-azido-benzoic acid42 in a copper(I)-catalyzed azide-alkyne cycloaddition14,41 yielding the triazoles 5a,b. The benzoic acid moiety was then coupled to mono Boc-protected 1,3-diaminopropane43 using TBTU as the coupling reagent, yielding intermediates 6a,b. Boc-deprotection was carried out using hydrochloric acid in dioxane, before the resulting primary amines 7a,b were reacted with dansyl chloride to afford the title compounds 8a and 8b.
Despite the advantages of the dansyl label, it is not ideally suited for all popular fluorescence-based technologies, for example bioluminescence resonance energy transfer. For instance, its fluorescence properties are highly sensitive to solvent polarity and the fluorescence excitation maximum (λmax ≈ 350 nm)35 prevents usage in NanoBRET assays. In recent years, many organic fluorophores with emission maxima in the red spectral range and enhanced photochemical properties have been developed. Chemically, these fluorophores often belong to the xanthene or cyanine dye families, but differ in their substituents, leading to a variety of net charges and different degrees of hydrophilicity44. We selected three different fluorophores, the commercially available dyes Alexa488 and Cy3B and a recently described bis-trifluoroethyl substituted rhodamine38 for the synthesis of our fluorescent dopamine receptor probes. Alexa488 and Cy3B, but also tetramethylrhodamine dyes (TAMRA), have already been shown to be suitable for characterizing ligand binding to GPCRs11,22,29,45, and especially Cy3B-labeled ligands have proven useful for fluorescence microscopy14,17,29. For the synthesis of these ligands, we focused on the N-propyl substituted 2-amino-dihydro-1H-indene scaffold. 5 ´-Carboxy-N,N ´-bis(2,2,2-trifluoroethyl)rhodamine (9) was prepared as described previously38 and connected directly to the benzylamine 3a using TBTU as the coupling reagent to give the desired fluorescent ligand 10a in a yield of 28%. Alexa488 and Cy3B were purchased as tetrafluorophenyl (TFP) or N-hydroxysuccinimid (NHS) esters, respectively, and reacted with the 1,3-dipropylamino-substituted benzoic acid 7a under basic conditions in DMF, affording the fluorescent ligands 10b (Cy3B) and 10c (Alexa488).
Molecular docking
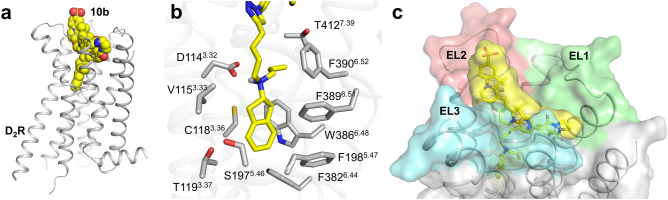
To explore whether addition of the fluorescent moieties would be tolerated by the dopamine receptors and to explore their location upon binding of the probes to D2R and D3R, docking studies were performed with the representative ligands 8a and 10b, comprising a dansyl and a Cy3B fluorophore, respectively, and the crystal structures of D2R (complex with risperidone, PDB-ID: 6CM43) or D3R (complex with eticlopride, PDB-ID: 3PBL5). As expected from the ligand design, typical receptor-ligand interactions in the orthosteric binding pocket3,5 including the salt bridge formation with Asp3.32 were observed for the indanylamine moiety of 8a and 10b in both receptors (Fig. 2, Supplementary Fig. S1). These results indicate that the presence of the fluorescent moiety does not hinder the pharmacophore from adopting its canonical binding pose. Thus, the addition of long and flexible aliphatic spacers, which were previously employed in fluorescent probes targeting D2R or D3R14,17, is not strictly required. According to the docking results, the fluorescent moieties of both ligands lay between the extracellular loops of the receptors, mainly interacting with hydrophobic amino acid sidechains. Between the two receptor subtypes, binding poses of the ligands only differed marginally (Fig. 2, Supplementary Fig. S1).
Figure 2.
Docking pose of 10b at D2R. (a) Overview of the 10b pose in the D2R. (b) The stick representation of the indanylamine pharmacophore in the orthosteric binding pocket shows an ionic interaction of the protonated nitrogen with D1143.32, while the aromatic ringsystem is accommodated in a hydrophobic pocket formed by V1153.33, C1183.36, T1193.37, S1975.46, F1985.47, F3826.44, W3866.48 and F3896.51. The propyl substituent of the protonated amine points into a hydrophobic cleft formed by W3866.48, F3906.52 and T4127.39. (c) Surface representation of the extracellular loop region. 10b is colored yellow, while EL1, EL2 and EL3 are colored in green, red and cyan, respectively. The polar sulfonate group of 10b is directed outwards towards the solvent.
Binding affinity and functional activity
Affinities for the dopamine receptor subtypes D1–D4 along with the related 5-HT1A, 5-HT2 and α1 receptors were determined by radioligand binding for the synthesized fluorescent ligands and some of the central intermediates (Table 1, Supplementary Table S1). In general, ligand affinities were found to be mostly dependent on the pharmacophore. Ligands comprising an N-propyl-2-aminoindane or phenylpiperazine substructure showed Ki values in the low nanomolar range for all D2-like receptors, with the highest affinities observed at the D3R subtype (0.26–13 nM), while the affinities for the D1R were substantially lower (180 – > 10,000 nM). The direct comparison of the dansyl-labeled ligands 4a-c with their benzonitrile precursors 2a-c clearly demonstrates that the insertion of the naphthyl-derived fluorophore is well tolerated by D2LR, D2SR, D3R and D4R, as highly similar binding affinities were determined in the presence and absence of the fluorescent moiety. As expected, ligands 2d and 4d, containing a pyrimidylpiperazine as primary dopamine receptor recognition element, were found to be selective for the D3R subtype. The introduction of the dansyl moiety further increased the D3R-selectivity, as it had no major influence on the D3R affinity (9.5 vs. 11 nM), but decreased the affinity for the D2LR and D2SR subtypes by a factor of ~ 20. Since 4d is also selective for D3R over the investigated serotonin and α1 receptors, it could potentially be used in fluorescence microscopy for D3R localization studies in tissues. Connection of the dansyl label and the lipophilic moiety through a short 1,3-diaminopropylene-spacer did not substantially improve binding affinities at the D2R and D3R, indicating that this additional spacer is not strictly required for the small dansyl fluorophore. However, with a Ki value of 0.44 nM, ligand 8a displayed the highest D2SR affinity of the entire series. When the small dansyl fluorophore was exchanged for the sterically more demanding xanthene or cyanine dyes, binding affinities slightly decreased for the D2S and D2L receptors (4.8–46 nM). Similar to their dansyl analogs, fluorescent ligands 10a–c displayed the strongest binding affinity for the D3R subtype (0.76–0.97 nM). With the exception of the D3R-selective ligands 2d and 4d, the series of fluorescent ligands and precursors also showed affinity for 5-HT1AR (13–260 nM) and α1-AR (1.1–150 nM). This is not unexpected, since the 2-methoxyphenylpiperazine is a common motif in α1-AR antagonists46. At 5-HT2R, observed affinities ranged from 220 to 2,200 nM, only.
Table 1.
Binding affinities of the test compounds.
| Ki [nM]a | ||||
|---|---|---|---|---|
| [3H]spiperone | ||||
| hD2LR | hD2SR | hD3R | hD4R | |
| 2a | 8.7 ± 2.5 (4) | 6.5 ± 1.7 (4) | 0.41 ± 0.09 (4) | 33 ± 7 (4) |
| 2b | 15 ± 5 (3) | 6.3 ± 0.2 (3) | 1.0 ± 0.2 (3) | 120 ± 61 (3) |
| 2c | 77 ± 17 (3) | 34 ± 8 (3) | 13 ± 4 (3) | 140 ± 10 (3) |
| 2d | 360 ± 60 (4) | 350 ± 30 (4) | 9.5 ± 1.9 (4) | 5,000 ± 300 (4) |
| 4a | 5.2 ± 0.6 (4) | 1.7 ± 0.3 (5) | 0.29 ± 0.02 (5) | 27 ± 3 (5) |
| 4b | 20 ± 8 (3) | 5.1 ± 1.7 (3) | 1.9 ± 0.7(3) | 59 ± 20 (3) |
| 4c | 17 ± 7 (3) | 8.4 ± 3.4 (3) | 6.5 ± 2.3 (3) | 32 ± 6 (3) |
| 4d | 7,600 ± 3,000 (4) | 6,000 ± 1,400 (4) | 11 ± 2 (4) | > 10,000 (4) |
| 7a | 3.5 ± 0.4 (8) | 1.8 ± 0.2 (8) | 0.26 ± 0.03 (8) | 8.5 ± 0.8 (8) |
| 8a (NMP64) | 1.2 ± 0.2 (4) | 0.44 ± 0.12 (4) | 0.32 ± 0.05 (4) | 10 ± 4 (4) |
| 8b | 13 ± 6 (4) | 3.6 ± 1.6 (4) | 1.9 ± 0.9 (4) | 130 ± 10 (4) |
| 10a (NMP160) | 46 ± 7 (4) | 21 ± 4 (4) | 0.97 ± 0.23 (4) | 56 ± 4 (4) |
| 10b (NMP137) | 10 ± 2 (5) | 4.8 ± 1.0 (5) | 0.90 ± 0.26 (5) | 50 ± 7 (4) |
| 10c (NMP130) | 23 ± 5 (4) | 15 ± 4 (4) | 0.76 ± 0.11 (4) | 47 ± 9 (4) |
Binding affinities of the test compounds for human D2LR, D2SR, D3R, D4R were determined by radioligand competition.
aData represent mean ± SEM of (n) individual experiments, each performed in triplicates.
In order to evaluate the impact of the fluorophores on the functional activity of the ligands, we determined their capacity to elicit β-arrestin-2 recruitment to the D2SR employing an assay based on enzyme fragment complementation (Pathhunter, DiscoverX). Since the presence of GRK2 is known to be important for a sensitive detection of ligand effects in HEK293 cells, GRK2 was coexpressed together with the D2SR fused to the enzyme donor. When the cells were incubated with the fluorescent ligands 8a and 10a–c, substantial stimulation of β-arrestin-2 recruitment was observed, confirming the agonistic nature of the N-propyl-2-aminoidane dopamine-isostere (Supplementary Fig. S2). In good agreement with its high binding affinity, the dansyl-labeled ligand 8a acted as a full agonist (Emax 98 ± 2%) and displayed a potency (EC50 51 ± 11 nM) that is only slightly lower compared to the reference agonist quinpirole (EC50 20 ± 3 nM, Emax 100 ± 1%). Incorporation of the larger trifluoroethyl-rhodamine, Cy3B or Alexa488 fluorophores led to a two- to eightfold reduction in ligand potency alongside with slightly reduced ligand efficacy for the fluorescent agonists 10a (EC50 410 ± 70 nM, Emax 89 ± 3%), 10b (EC50 410 ± 60 nM, Emax 79 ± 3%) and 10c (EC50 100 ± 20 nM, Emax 93 ± 3%), respectively. In agreement with our docking studies, these results indicate that D2-like receptors can accommodate the small dansyl- but also larger xanthene or cyanine-derived fluorescent ligands. However, not only the size but also the type of the fluorophore can have an impact on binding affinity, intrinsic activity and potency. This is illustrated in particular by the Cy3B-labeled ligand 10b, which has threefold higher affinity but fourfold lower potency for β-arrestin-2 recruitment at D2SR compared to 10c that only differs from 10b in terms of the fluorophore.
TIRF microscopy
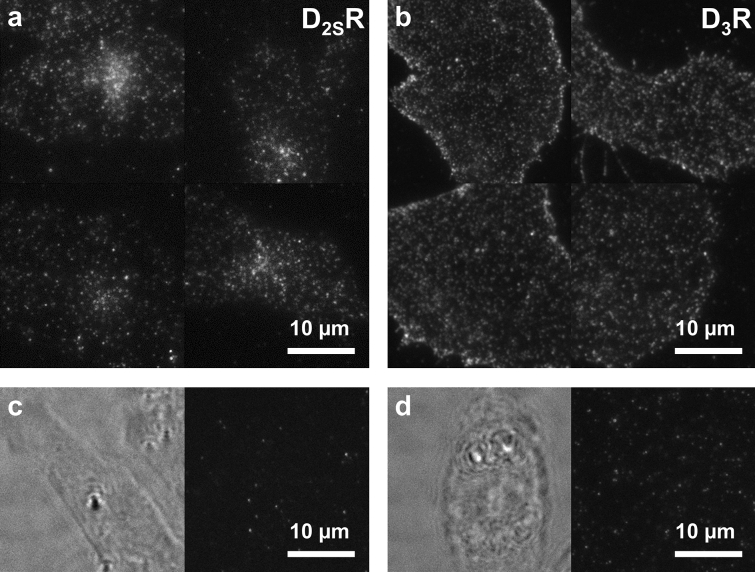
Fluorescence microscopy, in particular TIRF microscopy, is a powerful method to study the expression, distribution and interactions of GPCRs in the cytoplasmic membrane with high spatial and temporal resolution14–17,47,48. We employed our previously developed protocol for the imaging of dopamine receptors14 to verify that our newly developed fluorescent ligands are generally suitable for fluorescence imaging. For TIRF microscopy, we focused on the Cy3B-labeled ligand 10b, as the same fluorophore was previously used for the imaging of D2R and D3R14,17. Indeed, when CHO cells stably expressing D2SR or D3R were labeled with 10b in 10 nM (D2SR) or 1 nM (D3R) concentration, receptors were detected in the cytoplasmic membrane as discrete fluorescent spots (Fig. 3a,b). While D3R was found to be evenly distributed over the entire cell membrane, fluorescently labeled D2SR showed a more inhomogeneous distribution with clusters of fluorescent puncta. Employing a set of different fluorescently labeled D2R ligands, we could previously show that these clusters correspond to internalized receptors17. It is not surprising that we could not observe such clusters for D3R, because D3R is known to hardly undergo agonist-mediated β-arrestin recruitment and internalization49. When cells were pretreated with the D2/3R antagonist spiperone, neither clustered fluorescent puncta nor labeling of the receptors at the cell surface were observed, indicating that non-specific binding and uptake of ligand 10b were negligible (Fig. 3c,d).
Figure 3.
TIRF microscopy imaging with fluorescent ligand 10b. TIRF images of CHO cells stably expressing (a) D2SR or (b) D3R labeled with 10b (10 nM for D2SR, 1 nM for D3R) visualize the receptor distribution on the cell surface and demonstrate the suitability of the fluorescent ligand for high resolution fluorescence microscopy. (c, d) Validation of specific labeling: representative brightfield (left) and TIRF images (right) of a CHO cell stably expressing (c) D2SR or (d) D3R, preincubated with the D2/3R antagonist spiperone (10 μM) and treated with 10b (10 nM) show only a few non-specifically adhered, immobile fluorescent spots and no cell-specific labeling.
NanoBRET
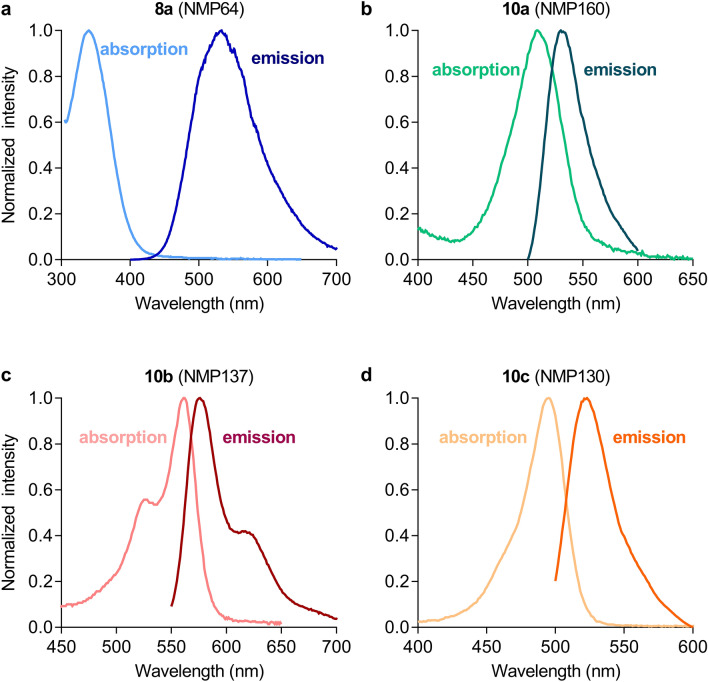
To further take advantage of the newly developed ligands as fluorescent probes, we planned to establish a NanoBRET assay for the detection of ligand binding at dopaminergic receptors. In this assay, ligand binding is detected through RET between the bioluminescent Nluc19 enzyme fused to the receptor N-terminus and the fluorescent ligand, which only occurs if the two molecules are in sufficient proximity to each other11. As a first step, we obtained absorbance and emission spectra of ligands 8a and 10a-c to identify most suitable candidates. As shown in Fig. 4, ligands 8a, 10a and 10c possess a maximum emission in the range of 540–550 nm, while the emission maximum of the Cy3B fluorophore in 10b is red-shifted (emission maximum at ~ 600 nm). Analysis of the absorption spectra confirmed the expected spectral properties of the employed fluorophores. While absorption occurred only in the UV range for the dansyl-labeled ligand 8a, compounds 10a-c showed significant absorption in the area of 450–550 nm (up to 600 nm for 10b). Thus, ligands 10a-c could serve as BRET acceptors in combination with the Nluc enzyme as BRET donor, which shows luminescence in the range of 400–550 nm (maximum at 460 nm)19.
Figure 4.
Spectral properties of fluorescent ligands 8a and 10a-c. Absorption and fluorescence emission spectra of the fluorescent ligands 8a and 10a-c were obtained on a microplate reader and normalized to the respective maximum signal of each sample.
For the development of the assay, we focused on the D3R, since our fluorescent ligands showed the highest affinity for this receptor subtype. To this end, a membrane targeted Nluc enzyme11 (secNluc, Promega) and D3R were fused in frame by polymerase chain reaction and cloned into pcDNA3.1 for mammalian expression. A second construct carrying an N-terminal HA export sequence and a FLAG-tag50 in front of the Nluc was generated, which allowed the detection of cell surface expression by ELISA17. Upon transient transfection into HEK293T cells, both variants of the Nluc-D3R fusion protein were well expressed, as determined by radioligand saturation (Bmax 2,500–16,200 fmol·mg−1 protein for secNluc-D3R and 5,000–5,700 fmol·mg−1 protein for FLAG-Nluc-D3R, compared to 2,200–4,800 fmol·mg−1 protein for wild type receptors (wtD3R), respectively), and both constructs showed the expected Nluc emission spectra in the presence of the substrate furimazine (Supplementary Fig. S3). A direct comparison of Flag-D3R and FLAG-Nluc-D3R in an ELISA with an antibody directed against the N-terminal FLAG-tag (Supplementary Fig. S3) showed that the presence of the N-terminal enzyme even improved cell surface expression. On the other hand, the N-terminal Nluc had no influence on the receptor-ligand recognition properties, as binding affinities for the reference ligands haloperidol, cariprazine, aripiprazole and fluspirilene were found to be highly similar to those obtained with wtD3R (Supplementary Table S2). In a G protein activation assay (IPOne, Cisbio) with the reference agonist quinpirole, even slightly better potencies were observed for the Nluc-D3R fusion constructs compared to unmodified receptors (Supplementary Fig. S3, EC50 ± SEM: 14 ± 4 nM for wtD3R, 3.7 ± 0.4 nM for secNluc-D3R and 4.6 ± 0.8 nM for FLAG-Nluc-D3R, respectively), which are likely a result of their higher expression level.
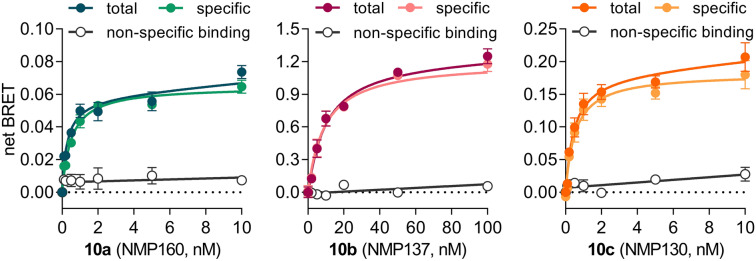
To find out whether fluorescent ligands 10a-c are indeed suitable for BRET-based ligand binding assays, we performed saturation binding assays with live HEK293T cells expressing secNluc-D3R. For all three ligands, typical saturation hyperbolas were observed (Fig. 5). Application of 10 µM haloperidol efficiently prevented binding of the fluorescent ligands and demonstrated a low contribution of non-specific binding to the detected netBRET signal. Analysis of the KD values revealed identical affinities for the trifluoroethyl-rhodamine and Alexa488-labeled ligands 10a and 10c (KD ± SEM 0.72 ± 0.07 nM for 10a, n = 5; 0.72 ± 0.08 nM, n = 4 for 10c, respectively), that were in good agreement with their Ki values obtained by radioligand competition (Table 1). In contrast, the determined affinity of the Cy3B-derivative 10b was substantially lower (KD ± SEM 12.1 ± 1.7 nM, n = 4), which was surprising, given that its radioligand Ki value (0.90 ± 0.26 nM) was similar to those of 10a and 10c. Almost identical results were obtained, when additional saturation experiments with 10a (KD ± SEM: 0.90 ± 0.14 nM, n = 5) and 10b (KD ± SEM: 11.3 ± 1.6 nM, n = 4) were performed with membranes from HEK293T cells expressing secNluc-D3R instead of whole live-cells (Supplementary Fig. S4). This indicates that the higher KD value of 10b observed by NanoBRET is not a result from general differences between membrane and whole-cell assays.
Figure 5.
NanoBRET saturation curves for the fluorescent ligands 10a-c. Saturation binding experiments were performed with live HEK293T cells expressing secNluc-D3R and the fluorescent ligands 10a, 10b and 10c, comprising a trifluorethyl-rhodamine derivative, a Cy3B and an Alexa488 fluorophore, respectively. Non-specific binding was determined in the presence of 10 µM haloperidol. Data points show mean ± SEM of one representative out of n = 5 (for 10a) or n = 4 (for 10b,c) experiments, with each condition carried out in triplicate. The netBRET signal was calculated as the difference between total BRET and the signal obtained in the absence of a fluorescent ligand.
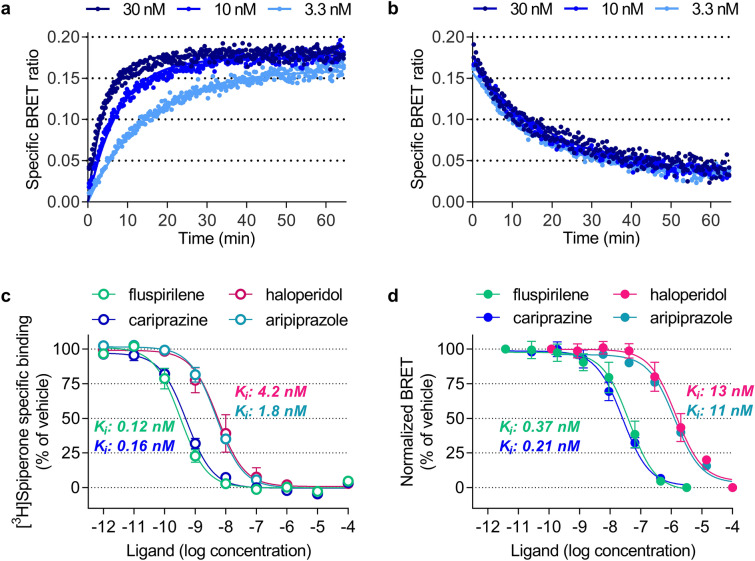
Association and dissociation experiments performed with fluorescent ligand 10a at room temperature confirmed its excellent properties and showed a concentration dependent association profile (Fig. 6a), with an association rate constant of 1.02 ± 0.30 × 107 min−1 × M−1 (Kon ± SEM, n = 10, Supplementary Table S3). As expected, dissociation of 10a was independent from the employed concentration (Fig. 6b), resulting in a mean residence time of 19 ± 1 min (mean ± SEM, n = 3). This is about 1.5-fold faster than previously observed for the radioligand [3H]spiperone at D3R51.
Figure 6.
Kinetics of fluorescent ligand 10a at D3R and competition binding experiments. (a) Association of 10a at room temperature was measured by NanoBRET using secNluc-D3R membrane preparations and threefold serial dilutions of 10a (3.3–30 nM). (b) Dissociation of 10a was initiated by addition of 10 μM haloperidol after equilibrium had been reached. Data show duplicates of an individual representative experiment. (c,d) Competition binding curves for reference antipsychotics obtained with (c) the radioligand [3H]spiperone and membranes from HEK293T cells expressing wtD3R receptors or (d) 100 nM fluorescent ligand 10a and secNluc-D3R membranes. Obtained IC50 values were transformed into Ki values applying the equation of Cheng and Prusoff52. Data show mean ± SEM of n = 4 independent experiments.
Taking advantage of the high-affinity binding and the excellent signal to noise ratio of 10a in the membrane-based BRET assay, we sought to determine the affinity of the antipsychotics haloperidol, fluspirilene, cariprazine and aripirazole for a direct comparison with the data from radioligand binding experiments (Fig. 6c,d). Independent from the employed concentration of 10a (10 nM or 100 nM), obtained binding affinities of the four antipsychotics showed nearly the same rank order and were only slightly lower (Supplementary Table S4, Fig. 6d) than those obtained by classical radioligand binding (Supplementary Table S2, Fig. 6c). These results demonstrate that 10a can be successfully employed in membrane-based NanoBRET assays to determine the Ki values of non-labeled ligands.
Discussion
Fluorescent ligands represent versatile tools for the investigation of diverse biological questions. Similar to radioligands, they can be detected in very low concentration and with high specificity. In the context of GPCR research, fluorescent ligands have been successfully employed to study receptor internalization17, receptor-receptor interactions within the cellular membrane14–16,18, and recently also ligand binding11,22–24,29,45,53, by techniques like fluorescence microscopy, fluorescence polarization and resonance energy transfer. Starting from phenylpiperazine and indanylamine scaffolds, known dopamine-isosteres with antagonistic or agonistic properties, respectively, we have designed and synthesized a small library of fluorescent ligands for D2-like receptors. Our initial efforts were directed towards the synthesis of dansyl-labeled probe molecules, as this fluorophore is cost efficient and readily available as reactive sulfonyl chloride. The obtained fluorescent ligands exhibited binding affinities for D2R and D3R in the subnanomolar range and ligand 8a also acted as highly potent D2R agonist. Despite these favorable characteristics, the application of the ligands is hampered by the fluorescence properties of the dansyl dye, that are in principle amenable to fluorescence microscopy, but not optimally suited for modern RET applications. Encouraged by the binding properties of ligands 4a and 8a, we exchanged the dansyl fluorophore by cyanine or xanthene moieties, both of which have been frequently used in fluorescence microscopy or NanoBRET binding assays11,14,17,22,29,45. While this exchange had almost no influence for ligand recognition properties at D3R, the fluorophore slightly reduced binding affinity at D2R.The fluorescent labels also slightly affected the intrinsic activity of the ligands when tested in a β-arrestin-2 recruitment assay at D2R. It should be noted that the employed fluorophores not only differ in their molecular weight and steric demand, but also their overall lipophilicity and net charge, which may influence not only receptor recognition and activation, but also the tendency of non-specific binding44.
Taking advantage of our newly developed fluorescent ligands, we sought to establish a fluorescent ligand binding assay with the D3R subtype serving as an example case. In principle, ligands 10a-c comprising a bis-trifluoroethylrhodamine, a Cy3B or an Alexa488 fluorophores should be well suited for both, FRET and BRET-based technologies. In FRET-based ligand binding assays, the receptor of interest is labeled with a fluorophore, either through the binding of a fluorescently labeled antibody or a self-labeling tag, that can be linked to a small molecule fluorophore12. In contrast, NanoBRET binding assays11 make use of a small and bright luciferase variant (Nluc)19, that is fused to the extracellular part of the investigated GPCR. In both cases, binding of the fluorescent ligand is detected based on RET between the receptor as light emitting donor and the fluorescent ligand serving as the acceptor10. Both RET-assays have been proven extremely useful for the characterization of ligand binding at GPCRs, especially if high affinity radioligands are not available or if binding kinetics10,23,24 are central to the investigated research question. Typical saturation hyperbolas were obtained for all three ligands when we performed NanoBRET assays with living HEK293T cells expressing an Nluc-D3R fusion protein. For two of the ligands, 10a and 10c, observed binding affinities were almost identical to those derived from classical radioligand binding studies. This was also true, when NanoBRET experiments were performed with cell membranes instead of whole cells. For the Cy3B-labled ligand 10b, the NanoBRET KD was approximately tenfold higher. On the other hand, 10b showed and excellent behavior in TIRF microscopy studies with CHO cells expressing D2R and D3R. This illustrates that despite similar absorption and emission spectra, the choice of the fluorophores may be critical not only for ligand binding to the target but has to be tailored to the desired application of the fluorescent ligand.
In summary, we have developed a set of high affinity fluorescent ligands for D2R and D3R receptors, which are the main targets in the treatment of severe neurological and psychiatric diseases. Depending on the employed fluorophore, the ligands can be used in high-resolution TIRF microscopy or to study ligand binding by resonance energy transfer. The established D3R-NanoBRET assay can be equally performed in whole cells and membranes and can be used for ligand binding screenings and characterization of novel ligands for D3R in the future.
Methods
General procedures (GP) for chemical synthesis
Synthesis of the fluorescent ligands was achieved employing the following general procedures. Detailed protocols and analytical data for the individual compounds are provided as Supplementary Methods and Supplementary Figs. S5 and S6.
GP I: Synthesis of terminal alkynes39,40
To a mixture of a secondary amine, K2CO3 (2 eq) and KI (1 eq) in CH3CN was added 6-chlorohex-1-yne at room temperature and the reaction mixture was heated to reflux overnight. After addition of H2O, the aqueous phase was extracted with CH2Cl2. The combined organic layers were dried over Na2SO4 and evaporated under reduced pressure to give the crude product.
GP II: Cu(I)-catalyzed 1,3-cycloaddition41
To a mixture of corresponding alkynes and aromatic azides in a solvent system of tert.-BuOH-H2O-CH2Cl2 (1:1:1) was added CuSO4·5H2O (5 mol %) and sodium ascorbate (10 mol %) and the suspension was stirred at room temperature. After the completion of the reaction, the suspension was diluted by the addition of H2O, extracted with CH2Cl2, and the combined organic layers were dried over Na2SO4. After evaporation, the crude residue was purified by silica-gel column chromatography with CH2Cl2 followed by 95:5 CH2Cl2-MeOH.
GP III: N-dansylation54
To a mixture of the crude primary amine and triethylamine (1:1) in CH2Cl2 was added dansyl chloride at 0 °C. The reaction mixture was stirred overnight at room temperature and the product was extracted with CH2Cl2. The combined organic layers were dried over Na2SO4 and concentrated in vacuo to obtain the crude product. Target compounds were isolated by silica-gel column chromatography with CH2Cl2 followed by 98:2 CH2Cl2-MeOH.
GP IV: Amide coupling
To a solution of the benzoic acid derivatives and DIPEA in CH2Cl2 at 0 °C was added TBTU in anhydrous DMF and the mixture was stirred for 30 min before a solution of tert.-butyl 3-aminopropylcarbamate in CH2Cl2 was added. After stirring for 2–3 h, saturated NaHCO3 solution was added and the mixture was extracted with CH2Cl2. The combined organic layers were dried over Na2SO4 and concentrated in vacuo to obtain the crude product. The compounds were isolated by silica-gel column chromatography with 95:5 CH2Cl2-MeOH.
Molecular docking
Docking of 8a and 10b was performed analogously to previously described protocols55. Ligands were geometry optimized by means of Gaussian1656 at the B3LYP/6–31 (d,p) level of theory (attributing a formal charge of + 1) and subsequently docked into the crystal structures of the D2R (PDB-ID: 6CM4) and the D3R (PDB-ID: 3PBL) using AutoDock Vina57. We applied a search space of 40 Å × 40 Å × 40 Å due to the large Cy3B moiety of 10b and to ensure a complete coverage of the binding pocket. Based on the scoring function and experimental data, four ligand-receptor complexes were selected, which were subsequently submitted to energy minimization using the PMEMD module of the AMBER 18 program package58. The all-atom force field ff14SB and the general AMBER force field (GAFF) were used for the receptors and ligands, respectively. Parameters for 8a and 10b were assigned using antechamber, and charges were calculated using Gaussian16 at the HF/6–31 (d,p) level of theory and the RESP procedure according to the literature59. A formal charge of + 1 was defined for the ligands.
Radioligand competition experiments
Affinities of the fluorescent ligands and precursors towards the human D2LR, D2SR, D3R, D4R60, and porcine D1R, α1-AR, 5-HT1AR and 5-HT2R61 were determined as described previously. For D2LR, D2SR, D3R or D4R, membranes from CHO cells stably expressing these receptors and the radioligand [3H]spiperone were used. Binding studies with D1R were carried out with homogenates obtained from porcine striatum and [3H]SCH23390. For α1R, 5-HT1AR and 5-HT2R homogenates of porcine cerebral cortex and the radioligands [3H]prazosin for α1R, [3H]WAY100,635 for 5-HT1AR, and [3H]ketanserin in the presence of 10 µM prazosin for 5-HT2R were used. Detailed concentrations, KD values, etc. are summarized in Supplementary Table S5. Non-specific binding was determined in the presence of 10 µM haloperidol (D1R-D4R), WAY100,635 (5-HT1AR), ketanserin (5-HT2R) or prazosin (α1R). Data analysis was performed using the algorithms for non-linear regression in PRISM6.0 to provide an IC50 value which was transformed into the Ki value employing the equation of Cheng and Prusoff52. Radioligand competition with Nluc-D3R membranes was performed in an analogous manner.
TIRF microscopy
TIRF microscopy was performed as previously described14. In brief, CHO-K1 cells stably expressing D2SR or D3R were seeded on 18 mm glass slides coated with fibronectin in phenol red-free DMEM/F12 supplemented with 10% FBS and were allowed to adhere over night at 37 °C, 5% CO2. Cells were washed twice with phenol red-free DMEM/F12 containing 10% FBS and incubated with 10b (1 nM for D3R, 10 nM for D2SR) for 1 h at 37 °C, before they were washed another three times. Specific labeling was confirmed by preincubation with 10 μM spiperone for 2 h, followed by incubation with the fluorescent ligand 10b (10 nM). Glass slides were mounted into a custom-built imaging chamber (500 µL) and washed with imaging buffer (137 mM NaCl, 5.4 mM KCl, 2.0 mM CaCl2, 1.0 mM MgCl2, and 10 mM, HEPES, pH 7.4) twice, before the imaging chamber was placed on the microscope stage. TIRF imaging was performed at 24.0 ± 0.3 °C on a Nikon TI-Eclipse inverted microscope equipped with a 100x, 1.49 NA oil-immersion objective. For excitation, a Nikon D-Eclipse C1 laser box (561 nm), a 561/14 nm excitation filter and a dichroic long-pass mirror (561 nm) were used. Emitted light was passed through an emission filter 609/54 nm (Semrock Rochester) and projected onto a water-cooled EM-CCD camera(Polar Series Accel 250 LC, Thermo Scientific) at -98 °C (512 × 512 FT, DU-897, Andor). To ensure homogenous illumination, only the central quarter of the chip (300 × 300 pixel) was used for analysis. Microscope control and image acquisition were performed with the NIS Elements software (Nikon Instruments).
Measurement of absorption and emission spectra
Absorption spectra were recorded on a CLARIOstar (BMG Labtech, Ortenberg, Germany) microplate reader. 1 mM solutions of the ligands were prepared in DMSO and measured either directly (8a) or after dilution to 10 µM in PBS (10a-c). Emission spectra were collected employing an excitation wavelength of 335/10, 470/10 or 520/10 nm, respectively. Spectra were background corrected and normalized to the maximum absorbance/emission of each sample.
cDNA constructs
For the generation of Nluc-D3R fusion constructs in pcDNA3.1, sequences of the Nluc enzyme19 (pNLF1-N or pNLF1-secN, Promega) and D3R (DRD3, cdna.org) were amplified by polymerase chain reaction and fused in frame with a 4 AA linker (GSSG) by Gibson Assembly62 (New England Biolabs). To achieve surface expression, the fusion protein was either N-terminally tagged with an HA-signal sequence and a FLAG-tag50 or the secretory version of the enzyme19 (pNLF1-secN) was used. Sequence integrity was verified by DNA sequencing (Eurofins Genomics).
Radioligand saturation binding
HEK293T cells were grown to a confluence of 70–80% and transfected with the Nluc-D3R plasmids using polyethyleneimine (PEI) as the transfection reagent (PEI/DNA ratio 3:1). After incubation in DMEM/F12 with 10% FBS at 37 °C, 5% CO2 for 48 h, cells were harvested and cell membranes were prepared as described previously34. The protein concentration was determined using the method of Lowry and bovine serum albumin as standard63. Membranes (protein concentration 5–20 µg·mL−1) were incubated with the radioligand [3H]spiperone (0.05 nM–2.00 nM, Perkin Elmer) for 1 h at 37 °C in binding buffer (50 mM Tris, 1 mM EDTA, 5 mM MgCl2, 100 µg·mL−1 bacitracin, 5 µg·mL−1 soybean trypsin inhibitor, pH 7.4). Non-specific binding was determined in the presence of 10 µM haloperidol. Reactions were terminated by filtration through GF/B filters soaked with 0.3% PEI solution. Dried filters were sealed with scintillation wax and bound radioactivity was determined with a Microbeta Counter (Perkin Elmer). Data were analyzed with the one-site saturation binding model implemented in PRISM8.0 (GraphPad Software Inc., San Diego, CA) to determine the equilibrium dissociation constant (KD) and the receptor expression level (Bmax).
Live-cell NanoBRET
For whole cell NanoBRET saturation assays, HEK293T cells were transfected with secNluc-D3R employing Mirus TransIT-293 (3:1 reagent to DNA ratio). After 24 h at 37 °C, 5% CO2, cells were detached using Versene and transferred to white, F-bottom 384-well plates at a density of 2,500 cells/well and incubated for another 24 h at 37 °C, 5% CO2. Cells were washed with PBS and incubated for 30 min at 37 °C with 5% FBS in DMEM without phenol red, before the fluorescent ligands (5 µL, diluted into DMEM without phenol red with 5% FBS from a 1 mM DMSO-stock) were added. Non-specific binding was determined in the presence of 10 µM haloperidol. After 1 h incubation with the ligands at 37 °C, furimazine (Promega, final dilution 1:500 to 1:2,500) was added, followed by a 5 min incubation at room temperature in the dark. BRET was measured as the ratio of acceptor fluorescence and donor luminescence employing a CLARIOstar microplate reader equipped with 475/30 nm and 535/30 or 580/30 emission filters, respectively. Total, non-specific and specific binding were analyzed employing the algorithms for one-site saturation binding implemented in PRISM6.0 or 8.0.
Membrane-based NanoBRET
SecNluc-D3R membranes were resuspended in assay buffer20 (50 mM Na2HPO4, 50 mM KH2PO4, 1 mg·mL−1 saponin, 5% FBS, pH 7.4), diluted to a total protein concentration of 0.5–3 μg/well in 384-well plates and incubated with the fluorescent ligands 10a or 10b (0–100 nM) at 37 °C for 90 min. Non-specific binding was determined in the presence of 10 μM haloperidol. After addition of furimazine (1:5,000, final volume 35 µL), and 5 min incubation at room temperature in the dark, BRET readings were obtained using a CLARIOstar plate reader equipped with a 475/30 nm donor together with a 535/30 nm (for 10a) or 620/30 nm (for 10b) acceptor emission filter. Data were analyzed as described for live-cell NanoBRET. For association kinetics, secNluc-D3R membranes were added to various concentrations of 10a (1–100 nM) in the presence or absence of 10 μM haloperidol and furimazine (1:2,500). For dissociation, membranes were preincubated with 10a for 1 h, before furimazine was added and dissociation was initiated with 10 μM haloperidol. Kinetic assays were monitored at room temperature and minimal possible cycle time of the CLARIOstar was used for each measurement. Data was fitted to one phase association and dissociation equations in PRISM6.0 to determine the kinetic constants. In competition binding experiments, membranes were incubated with serial dilutions of unlabeled ligands and the fluorescent ligand 10a (10 or 100 nM) for 90 min at 37 °C. Addition of furimazine (1:5,000) and BRET measurements were carried out as described above. Data analysis was performed using the one site—fit Ki equation in PRISM6.0 to determine the inhibition constants (Ki) of the unlabeled ligands.
Figure preparation
Figures were prepared using ChemDraw, version 18.0.0 (Fig. 1, Perkin Elmer Informatics, www.informatics.perkinelmer.com), PyMOL Molecular Graphics System, version 2.2.1 (Fig. 2, Supplementary Fig. S1, Schrödinger, LLC, www.pymol.org), NIS-Elements AR, version 3.22.13 (Fig. 3, Nikon Instruments, https://www.microscope.healthcare.nikon.com/de_EU/products/software), and GraphPad Prism version 8.0.0 for Windows, (Fig. 4–6, Supplementary Figs. S2–S4, GraphPad Software, San Diego, California USA, www.graphpad.com).
Supplementary Information
Acknowledgements
We thank Prof. Michel Bouvier, IRIC, University of Montreal, for providing the plasmid encoding GRK2 and Dr. Christopher Lang, FAU Erlangen-Nürnberg, for the synthesis of 5′-carboxy-N,N′-bis(2,2,2-trifluoroethyl)rhodamine.
Author contributions
A.A. designed, conducted and analyzed membrane NanoBRET experiments. K.F. designed Nluc constructs, performed and analyzed live-cell NanoBRET and radioligand saturation assays with supervision from H.H. and D.W. N.P. designed and synthesized fluorescent ligands under the supervision of P.G. M.F.S. performed and analyzed molecular docking. A.T. performed and analyzed TIRF microscopy. H.H. performed and analyzed radioligand competition and β-arrestin-2 recruitment assays. D.W. has been responsible for the overall project strategy and provided project supervision. A.A. and D.W. wrote the manuscript with contributions from all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL. D.W. and P.G. receive funding from the German Research Foundation (DFG, GRK1910).
Data availability
The data that support the findings of this study are available within the Supplementary Information files and/or from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78827-9.
References
- 1.Beaulieu J-M, Espinoza S, Gainetdinov RR. Dopamine receptors—IUPHAR review 13. Br. J. Pharmacol. 2015;172:1–23. doi: 10.1111/bph.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan L, et al. Haloperidol bound D2 dopamine receptor structure inspired the discovery of subtype selective ligands. Nat. Commun. 2020;11:1074. doi: 10.1038/s41467-020-14884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S, et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018;555:269. doi: 10.1038/nature25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin J, et al. Structure of a D2 dopamine receptor–G-protein complex in a lipid membrane. Nature. 2020;584:125–129. doi: 10.1038/s41586-020-2379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Cao C, He L, Wang X, Zhang XC. Crystal structure of dopamine receptor D4 bound to the subtype selective ligand L745870. eLife. 2019;8:e48822. doi: 10.7554/eLife.48822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017;358:381–386. doi: 10.1126/science.aan5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R, Xie X. Tools for GPCR drug discovery. Acta Pharmacol. Sin. 2012;33:372–384. doi: 10.1038/aps.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan CA. Chapter 10: GPCR-radioligand binding assays. In: Shukla AK, editor. Methods in Cell Biology. Cambridge: Academic Press; 2016. pp. 191–215. [DOI] [PubMed] [Google Scholar]
- 10.Soave M, Briddon SJ, Hill SJ, Stoddart LA. Fluorescent ligands: bringing light to emerging GPCR paradigms. Br. J. Pharmacol. 2020;177:978–991. doi: 10.1111/bph.14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoddart LA, et al. Application of BRET to monitor ligand binding to GPCRs. Nat. Methods. 2015;12:661–663. doi: 10.1038/nmeth.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emami-Nemini A, et al. Time-resolved fluorescence ligand binding for G protein–coupled receptors. Nat. Protoc. 2013;8:1307–1320. doi: 10.1038/nprot.2013.073. [DOI] [PubMed] [Google Scholar]
- 13.Sridharan R, Zuber J, Connelly SM, Mathew E, Dumont ME. Fluorescent approaches for understanding interactions of ligands with G protein coupled receptors. Biochim. Biophys. Acta. 2014;1838:15–33. doi: 10.1016/j.bbamem.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabor A, et al. Visualization and ligand-induced modulation of dopamine receptor dimerization at the single molecule level. Sci. Rep. 2016;6:33233. doi: 10.1038/srep33233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hern JA, et al. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasai RS, et al. Full characterization of GPCR monomer–dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabor A, Möller D, Hübner H, Kornhuber J, Gmeiner P. Visualization of ligand-induced dopamine D2S and D2L receptor internalization by TIRF microscopy. Sci. Rep. 2017;7:10894. doi: 10.1038/s41598-017-11436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albizu L, et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat. Chem. Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall MP, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoddart LA, et al. Development of novel fluorescent histamine H1-receptor antagonists to study ligand-binding kinetics in living cells. Sci. Rep. 2018;8:1572. doi: 10.1038/s41598-018-19714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiansen E, Hudson BD, Hansen AH, Milligan G, Ulven T. Development and characterization of a potent free fatty acid receptor 1 (FFA1) fluorescent tracer. J. Med. Chem. 2016;59:4849–4858. doi: 10.1021/acs.jmedchem.6b00202. [DOI] [PubMed] [Google Scholar]
- 22.Sakyiamah MM, Nomura W, Kobayakawa T, Tamamura H. Development of a NanoBRET-based sensitive screening method for CXCR4 ligands. Bioconjug. Chem. 2019;30:1442–1450. doi: 10.1021/acs.bioconjchem.9b00182. [DOI] [PubMed] [Google Scholar]
- 23.Klein Herenbrink C, et al. The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 2016;7:10842. doi: 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sykes DA, et al. Extrapyramidal side effects of antipsychotics are linked to their association kinetics at dopamine D2 receptors. Nat. Commun. 2017;8:763. doi: 10.1038/s41467-017-00716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monsma FJ, Jr, et al. Characterization of novel fluorescent ligands with high affinity for D1 and D2 dopaminergic receptors. J. Neurochem. 1989;52:1641–1644. doi: 10.1111/j.1471-4159.1989.tb09220.x. [DOI] [PubMed] [Google Scholar]
- 26.Bakthavachalam V, Baindur N, Madras BK, Neumeyer JL. Fluorescent probes for dopamine receptors: synthesis and characterization of fluorescein and 7-nitrobenz-2-oxa-1,3-diazol-4-yl conjugates of D-1 and D-2 receptor ligands. J. Med. Chem. 1991;34:3235–3241. doi: 10.1021/jm00115a012. [DOI] [PubMed] [Google Scholar]
- 27.Madras BK, et al. Fluorescent and biotin probes for dopamine receptors: D1 and D2 receptor affinity and selectivity. Mol. Pharmacol. 1990;37:833–839. [PubMed] [Google Scholar]
- 28.Barton AC, et al. Multiple fluorescent ligands for dopamine receptors. I. Pharmacological characterization and receptor selectivity. Brain Res. 1991;547:199–207. doi: 10.1016/0006-8993(91)90963-V. [DOI] [PubMed] [Google Scholar]
- 29.Allikalt A, Laasfeld T, Ilisson M, Kopanchuk S, Rinken A. Quantitative analysis of fluorescent ligand binding to dopamine D3 receptors using live cell microscopy. FEBS J. 2020 doi: 10.1111/febs.15519. [DOI] [PubMed] [Google Scholar]
- 30.Löber S, Hübner H, Tschammer N, Gmeiner P. Recent advances in the search for D3- and D4-selective drugs: probes, models and candidates. Trends Pharmacol. Sci. 2011;32:148–157. doi: 10.1016/j.tips.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Rusterholz DB, et al. Dopaminergic effects of non-hydroxylated rigid analogs of apomorphine. Eur. J. Pharmacol. 1979;55:73–82. doi: 10.1016/0014-2999(79)90149-3. [DOI] [PubMed] [Google Scholar]
- 32.Tschammer N, Dörfler M, Hübner H, Gmeiner P. Engineering a GPCR−ligand pair that simulates the activation of D2L by dopamine. ACS Chem. Neurosci. 2010;1:25–35. doi: 10.1021/cn900001b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geneste H, et al. Synthesis and SAR of highly potent and selective dopamine D3-receptor antagonists: Variations on the 1H-pyrimidin-2-one theme. Bioorg. Med. Chem. Lett. 2006;16:1934–1937. doi: 10.1016/j.bmcl.2005.12.079. [DOI] [PubMed] [Google Scholar]
- 34.Möller D, et al. Discovery of G protein-biased dopaminergics with a pyrazolo[1,5-a]pyridine substructure. J. Med. Chem. 2017;60:2908–2929. doi: 10.1021/acs.jmedchem.6b01857. [DOI] [PubMed] [Google Scholar]
- 35.Lakowicz JR. Fluorophores. In: Lakowicz JR, editor. Principles of Fluorescence Spectroscopy. Berlin: Springer; 2006. pp. 63–95. [Google Scholar]
- 36.Möller D, et al. Functionally selective Dopamine D2, D3 receptor partial agonists. J. Med. Chem. 2014;57:4861–4875. doi: 10.1021/jm5004039. [DOI] [PubMed] [Google Scholar]
- 37.Weichert D, et al. Molecular determinants of biased agonism at the dopamine D2 receptor. J. Med. Chem. 2015;58:2703–2717. doi: 10.1021/jm501889t. [DOI] [PubMed] [Google Scholar]
- 38.Mitronova GY, et al. New fluorinated rhodamines for optical microscopy and nanoscopy. Chem. Eur. J. 2010;16:4477–4488. doi: 10.1002/chem.200903272. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich K, et al. Dopamine D2, D3, and D4 selective phenylpiperazines as molecular probes to explore the origins of subtype specific receptor binding. J. Med. Chem. 2009;52:4923–4935. doi: 10.1021/jm900690y. [DOI] [PubMed] [Google Scholar]
- 40.Stößel A, et al. Development of molecular tools based on the dopamine D3 receptor ligand FAUC 329 showing inhibiting effects on drug and food maintained behavior. Bioorg. Med. Chem. 2017;25:3491–3499. doi: 10.1016/j.bmc.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 42.Suwal S, Pflum MKH. Phosphorylation-dependent kinase–substrate cross-linking. Angew. Chem. Int. Ed. 2010;49:1627–1630. doi: 10.1002/anie.200905244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrell A, et al. Investigation of the lactam side chain length necessary for optimal indenoisoquinoline topoisomerase I inhibition and cytotoxicity in human cancer cell cultures. J. Med. Chem. 2007;50:2040–2048. doi: 10.1021/jm0613119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes LD, Rawle RJ, Boxer SG. Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers. PLoS ONE. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand F, Klutz AM, Jacobson KA, Fredholm BB, Schulte G. Adenosine A2A receptor dynamics studied with the novel fluorescent agonist Alexa488-APEC. Eur. J. Pharmacol. 2008;590:36–42. doi: 10.1016/j.ejphar.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manetti F, Corelli F, Strappaghetti G, Botta M. Arylpiperazines with affinity toward a1-adrenergic receptors. Curr. Med. Chem. 2002;9:1303–1321. doi: 10.2174/0929867023369961. [DOI] [PubMed] [Google Scholar]
- 47.Calebiro D, et al. Single-molecule analysis of fluorescently labeled G-protein–coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. USA. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasai RS, Ito SV, Awane RM, Fujiwara TK, Kusumi A. The class-A GPCR dopamine D2 receptor forms transient dimers stabilized by agonists: detection by single-molecule tracking. Cell Biochem. Biophys. 2018;76:29–37. doi: 10.1007/s12013-017-0829-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim K-M, et al. Differential regulation of the dopamine D2and D3 receptors by G protein-coupled receptor kinases and β-arrestins. J. Biol. Chem. 2001;276:37409–37414. doi: 10.1074/jbc.M106728200. [DOI] [PubMed] [Google Scholar]
- 50.Guan XM, Kobilka TS, Kobilka BK. Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem. 1992;267:21995–21998. [PubMed] [Google Scholar]
- 51.Frank A, Kiss DJ, Keserű GM, Stark H. Binding kinetics of cariprazine and aripiprazole at the dopamine D3 receptor. Sci. Rep. 2018;8:12509. doi: 10.1038/s41598-018-30794-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y-C, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 53.Rinken A, Lavogina D, Kopanchuk S. Assays with detection of fluorescence anisotropy: challenges and possibilities for characterizing ligand binding to GPCRs. Trends Pharmacol. Sci. 2018;39:187–199. doi: 10.1016/j.tips.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 54.He S-M, Luo Y, Hove-Jensen B, Zechel DL. A fluorescent substrate for carbon–phosphorus lyase: towards the pathway for organophosphonate metabolism in bacteria. Bioorg. Med. Chem. Lett. 2009;19:5954–5957. doi: 10.1016/j.bmcl.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 55.Hiller C, et al. Functionally selective dopamine D2/D3 receptor agonists comprising an enyne moiety. J. Med. Chem. 2013;56:5130–5141. doi: 10.1021/jm400520c. [DOI] [PubMed] [Google Scholar]
- 56.Gaussian 09, Revision B.01 (Gaussian, Inc.: Wallingford, CT, 2010).
- 57.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AMBER18 (University of California, San Francisco, 2018).
- 59.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
- 60.Hübner H, Haubmann C, Utz W, Gmeiner P. Conjugated enynes as nonaromatic catechol bioisosteres: Synthesis, binding experiments and computational studies of novel dopamine receptor agonists recognizing preferentially the D3 subtype. J. Med. Chem. 2000;43:756–762. doi: 10.1021/jm991098z. [DOI] [PubMed] [Google Scholar]
- 61.Bettinetti L, Schlotter K, Hübner H, Gmeiner P. Interactive SAR studies: rational discovery of super-potent and highly selective dopamine D3 receptor antagonists and partial agonists. J. Med. Chem. 2002;45:4594–4597. doi: 10.1021/jm025558r. [DOI] [PubMed] [Google Scholar]
- 62.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 63.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the Supplementary Information files and/or from the corresponding authors upon request.