Abstract
Hydrogen sulfide (H2S) is a gaseous signaling molecule that plays multiple roles in plant development. However, whether endogenous H2S plays a role in fruit ripening in tomato is still unknown. In this study, we show that the H2S-producing enzyme l-cysteine desulfhydrase SlLCD1 localizes to the nucleus. By constructing mutated forms of SlLCD1, we show that the amino acid residue K24 of SlLCD1 is the key amino acid that determines nuclear localization. Silencing of SlLCD1 by TRV-SlLCD1 accelerated fruit ripening and reduced H2S production compared with the control. A SlLCD1 gene-edited mutant obtained through CRISPR/Cas9 modification displayed a slightly dwarfed phenotype and accelerated fruit ripening. This mutant also showed increased cysteine content and produced less H2S, suggesting a role of SlLCD1 in H2S generation. Chlorophyll degradation and carotenoid accumulation were enhanced in the SlLCD1 mutant. Other ripening-related genes that play roles in chlorophyll degradation, carotenoid biosynthesis, cell wall degradation, ethylene biosynthesis, and the ethylene signaling pathway were enhanced at the transcriptional level in the lcd1 mutant. Total RNA was sequenced from unripe tomato fruit treated with exogenous H2S, and transcriptome analysis showed that ripening-related gene expression was suppressed. Based on the results for a SlLCD1 gene-edited mutant and exogenous H2S application, we propose that the nuclear-localized cysteine desulfhydrase SlLCD1 is required for endogenous H2S generation and participates in the regulation of tomato fruit ripening.
Subject terms: Molecular biology, Metabolism, Plant physiology
Introduction
Hydrogen sulfide (H2S) is a gaseous signaling molecule that is widely present in living organisms. Accumulating evidence has confirmed multiple functions of H2S in plant root development, stomatal movement, postharvest senescence, petiole abscission, and response to abiotic stresses1–6. Similar to the role of nitric oxide in the regulation of fruit ripening and senescence, multiple studies have found that H2S can alleviate postharvest ripening and senescence of fruits, such as strawberry, kiwifruit, and banana, by regulating the antioxidant system and ethylene pathway2,7,8. H2S fumigation could reduce the fruit decay index and respiratory intensity of strawberry2. Exogenous H2S could act as a regulator of fruit ripening by antagonizing the effect of ethylene in tomatoes9. Transcriptome analysis indicated that H2S could delay the ripening and senescence of kiwifruit by modulating genes involved in cell wall degradation and in the ethylene signaling pathway10. H2S could also perform a signaling role through persulfidation, which is the posttranslational modification of cysteine residues (R-SHs) in target proteins by covalent addition of thiol groups to form persulfides (R-SSHs)11,12. For instance, H2S was found to inhibit ethylene synthesis by inhibiting the activity of 1-aminocyclopropane-1-carboxylic acid (ACC) oxidases (ACOs) by persulfidation, thereby alleviating tomato ripening and senescence12. Previous research on the role of H2S in alleviating fruit ripening and senescence has been conducted mainly through exogenous H2S fumigation, while whether endogenous H2S generation affects fruit ripening is still unclear.
In plants, endogenous H2S can be produced through the sulfate assimilation pathway in chloroplasts, where the enzyme is sulfite reductase (SiR)13. Endogenous cytosolic H2S could also be generated from l-cysteine by cysteine-degrading enzymes14. l-cysteine desulfhydrase 1 (DES1), belonging to the O-acetylserine thiol lyase (OASTL) family, was characterized as the key enzyme in this process15. LCD (l-cysteine desulfhydrase, with l-Cys as the substrate) has also been shown to catalyze the degradation of cysteine to H2S, ammonia, and pyruvate. In Arabidopsis, the H2S-deficient mutant lcd displayed increased stomatal density and stomatal index values, and H2S was found to act downstream of jasmonic acid signaling to regulate stomatal development in cotyledons16. Furthermore, LCD expression was activated by the Ca2+/calmodulin2-binding transcription factor TGA3 in Arabidopsis to increase H2S production and bolster Cr6+ tolerance3.
Ethylene is a key hormone in the regulation of fruit ripening and senescence, especially in respiratory climacteric fruits such as tomato17. The ethylene synthesis pathway has been characterized in tomato, and increased expression of ACS1A, ACS2, ACS4, ACO1, and ACO3, genes encoding ACC synthase and ACC oxidase, play an important role in tomato fruit ripening18. The transition from green to red is one of the phenotypic characteristics of fruit ripening in tomato. Chlorophyll b reductase (NYC1), pheophytinase (PPH), pheophorbide a oxygenase (PAO), and SGR1 (stay-green protein) are required for chlorophyll degradation19. Carotenoid biosynthesis starts with the condensation of two geranylgeranyl diphosphate (GGPP) molecules by phytoene synthase (PSY) to form phytoene. Phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS) are enzymes that catalyze two symmetric dehydrogenation reactions, converting 15-cis phytoene to tetra-cis-lycopene17. In tomato, PSY1, PDS, and ZDS are key genes in the regulation of carotenoid synthesis. During fruit softening, cell wall components undergo degradation by pectin methylesterase, polygalacturonase (PG), cellulase (CEL), and xyloglucan-degrading enzymes (XTHs)17.
Our previous research showed that exogenous H2S delayed the ripening of postharvest tomato fruits by modulating the antioxidant system and ethylene signaling pathway9. In the present research, transcriptome sequencing was performed in tomato fruit treated with H2S, and the genes involved in amino acid metabolism and ripening-related genes were shown to be enriched. We found two LCD-encoding genes, namely, SlLCD1 and SlLCD2, in tomato, whereas their role in regulating fruit ripening is unknown. In the present research, the subcellular localization of SlLCD1 was studied in tobacco cells, and the role of SlLCD1 in sulfur metabolism and fruit ripening was studied by virus-induced gene silencing (VIGS) and CRIPSR/Cas9-mediated gene editing in tomato fruit to understand the role of H2S in fruit ripening.
Results
Expression profile of the SlLCD genes in tomato plants
The expression profiles of the SlLCD genes in tomato plants were analyzed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Fig. 1). SlLCD1 gene expression increased gradually during fruit development and ripening, whereas that of SlLCD2 did not change too much. The expression of SlLCDs was also explored in different tissues of Micro-Tom by analysis of the public database TomExpress http://tomexpress.toulouse.inra.fr/login. The data showed that increased SlLCD1 expression was observed during fruit ripening, suggesting the possible role of SlLCD1 and H2S in the regulation of fruit ripening (Fig. S1).
Fig. 1. SlLCD expression pattern in different tissues of tomato.
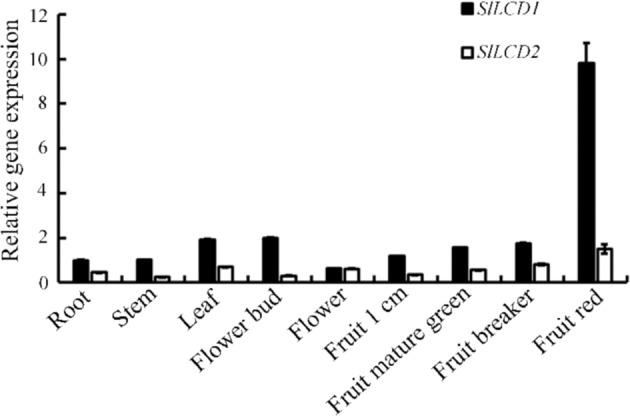
The tissues include root, stem, leaf, flower bud, flower, 1 cm fruit, mature green fruit, breaker fruit, and red fruit. Values are the means ± SDs of three replicates
Subcellular localization of SlLCD1 and SlLCD2
As SlLCD1 showed a potential nuclear localization signal (NLS) at its N-terminus (H18LAKKPKLS), we predicted that SlLCD1-GFP might localize to the nucleus. Confocal and fluorescence microscopy confirmed that SlLCD1 showed strong nuclear localization, as fluorescence from SlLCD1-GFP merged with the nuclear stain Hoechst 33342 in transfected tobacco leaf cells (Fig. 2). Different mutants of SlLCD1 were constructed, and a schematic diagram of the intended mutations in SlLCD1 is shown in Figs. S2 and 2A. To verify whether the NLS segment was essential for the nuclear localization of SlLCD1, a truncation mutant, SlLCD1 (Δ1–26)-GFP, which lacked the first 26 amino acid residues at the N-terminus was constructed, and the truncation mutant failed to localize to the nucleus but was located in the cytoplasm. The basic amino acids lysine (K) and arginine (R) are key factors defining monopartite NLSs; thus, we mutated the native NLS of SlLCD1 to a series of variants by replacing lysine residues with glutamines: SlLCD1 (K21Q K22Q K24Q), SlLCD1 (K21Q), SlLCD1 (K22Q), SlLCD1 (K24Q), and SlLCD1 (K21Q K22Q). Nuclear localization of SlLCD1 was abolished in the SlLCD1 (K21Q K22Q K24Q) mutant, whereas SlLCD1 (K21Q), SlLCD1 (K22Q), and SlLCD1 (K21Q K22Q) retained nuclear localization. Surprisingly, the SlLCD1 (K24Q) mutant failed to localize to the nucleus (Fig. 2B), suggesting that K24 in SlLCD1 was essential for the nuclear localization of SlLCD1. As shown in Fig. 2C, SlLCD2-GFP showed strong colocalization with chloroplast autofluorescence, suggesting that SlLCD2 was mainly localized to the chloroplast. The homologous proteins in 16 other plant species were analyzed to determine whether the potential NLS found in SlLCD1 is conserved in other plant species. As shown in Supplementary Table 1, at least one member of the LCD proteins in the plant contained a potential NLS at the N-terminus.
Fig. 2. Subcellular localization of the SlLCD1 protein in tobacco cells.
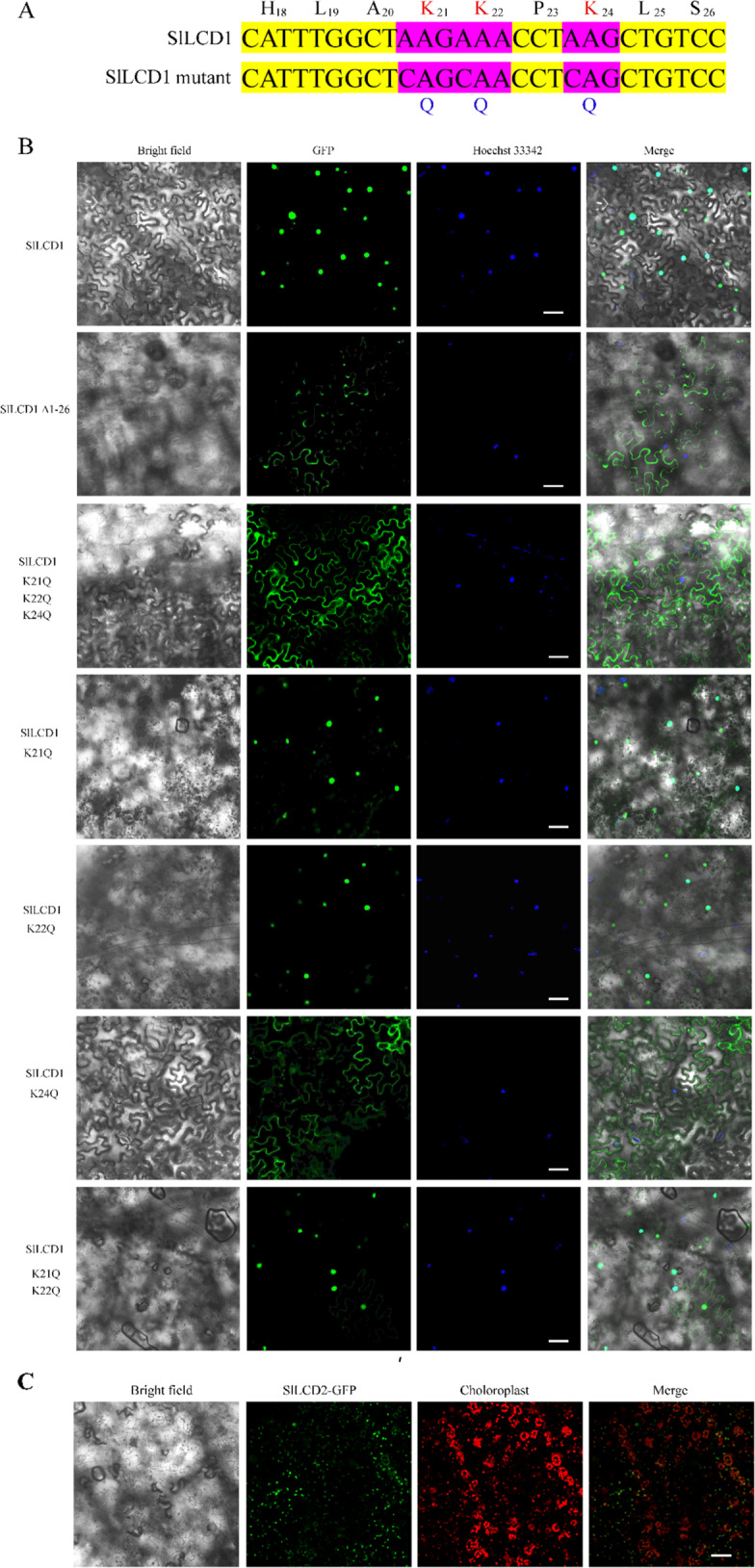
A The K (lysine) residues in SlLCD1 were substituted with Q (glutamine). B Confocal image of tobacco leaf cells expressing SlLCD1-GFP or a truncation (Δ1–26) or the following site-directed mutant GFP fusion proteins: SlLCD1 (K21Q K22Q K24Q)-GFP, SlLCD1 (K21Q)-GFP, SlLCD1 (K22Q)-GFP, SlLCD1 (K24Q)-GFP, and SlLCD1 (K21Q K22Q)-GFP. Hoechst 33342 represents the nucleus. The red fluorescence signal in C (SlLCD2-GFP) represents the autofluorescence of chloroplasts. Scale bars represent 50 μm
VIGS of SlLCD1 accelerates tomato fruit ripening
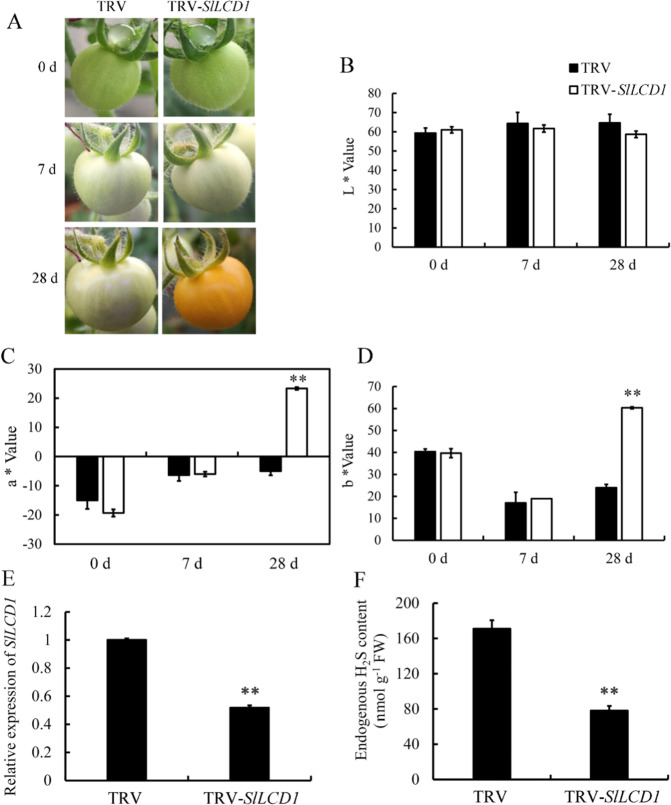
VIGS constructs containing the tobacco rattle virus (TRV)-LCD1 were used to silence the expression of LCD1. As shown in Fig. 3A, fruit infected with TRV-LCD1 showed pink coloration on day 28 after infection, while control fruit infected with the empty vector remained white-green at day 28. Chromaticity data showed that LCD1-silenced fruit exhibited a higher a value on the green (−) to red (+) axis compared with control fruit and a higher b* value on the blue (−) to yellow (+) axis (Fig. 3C, D), whereas the value of lightness (L) was not significantly changed (Fig. 3B). The silencing effect of VIGS-LCD1 was confirmed by RT-qPCR, as shown in Fig. 3E. The expression of LCD1 was downregulated significantly by VIGS and reached approximately half of that in control fruit. LCD catalyzes the generation of H2S with cysteine as its substrate; thus, in fruit where LCD1 is silenced, a decrease in the H2S content in fruit might be expected. Accordingly, Fig. 3F shows that tomato fruit harboring the VIGS-LCD1 construct contained a significantly lower level of H2S than the control.
Fig. 3. VIGS of SlLCD1 accelerates tomato fruit ripening.
A Phenotype of SlLCD1-silenced fruits of the Micro-Tom cultivar. Left: tobacco rattle virus (TRV)-infected fruit in which ripening was not affected by TRV. Right: TRV-SlLCD1-infected fruit showed accelerated color transition from green to red. Photos were taken at 0, 7, and 28 days after infection with Agrobacterium tumefaciens strain GV3101 containing the vectors. B L* indicates lightness. C The a* value stands for chromaticity on a green (−) to red (+) axis. D b* Indicates chromaticity on the blue (−) to yellow (+) axis. Values are the means ± SDs of three replicates. The symbol ** stands for P < 0.01. The gene expression of SlLCD1 (E) and the content of H2S (F) in SlLCD1-silenced fruits (silenced by VIGS). Values are the means ± SDs of three replicates. The symbol ** stands for P < 0.01
Generation of the LCD1 mutation by CRISPR/Cas9
For stable transformation of SlLCD1, the CRISPR/Cas9 vector containing two sgRNA targets of LCD1 was generated and introduced into Agrobacterium tumefaciens strain GV3101 for tomato transformation. For the positive T1 plants, the gene fragment of SlLCD1 was amplified with genomic DNA as a template with primers that flanked both sgRNA targets. As shown in Fig. 4A, sequencing showed that lcd1-7 contained a T residue inserted near the PAM sequence, and lcd1-9 had a deletion of G near the PAM. The sequence surrounding the first sgRNA is shown, while the second sgRNA is not shown, as the nearby sequence was not edited. Both the insertion and deletion of a single base led to the formation of a premature stop codon, eliminating the cysteine desulfhydrase domain of LCD1. Specifically, the T insertion in lcd1-7 caused a severe frameshift mutation, and the translation stopped after the 53rd amino acid residue. In lcd1-9, the one-base deletion caused a mutated peptide sequence after the 46th amino acid residue, and the translation stopped at the 48th amino acid due to a premature stop codon. To confirm that this mutation affected LCD1 function, we measured the endogenous H2S content of wild-type and lcd1-7/9 mutant plants. Figure 4B shows that mutation of both lcd1-7 and lcd1-9 resulted in significantly lower levels of H2S in comparison to the wild type, suggesting that the mutations altered the enzymatic function of LCD1 as a cysteine desulfhydrase. The enzymatic activity of SlLCDs was also confirmed by ectopic expression in Escherichia coli, and the result in Fig. S3 shows that both SlLCDs could catalyze the production of H2S with l-cysteine as the substrate.
Fig. 4. Identification of SlLCD1 gene-edited plants.
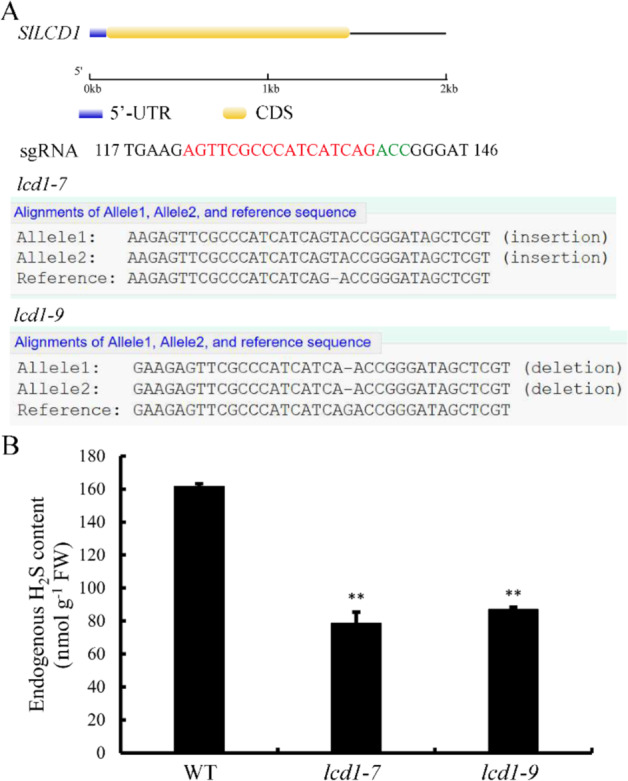
A Gene structure of SlLCD1, which is indicated as the 5’-UTR (untranslated region) and CDS (coding sequence). Generation of SlLCD1 mutations by CRISPR/Cas9 using single-guide RNAs (sgRNAs). Sequences of the SlLCD1 mutants lcd1-7 and lcd1-9 are shown. sgRNA target and a protospacer-adjacent motif (PAM) are indicated in red and green, respectively. Deletions and insertions are indicated by dashes. B Endogenous H2S content in leaves of the wild type (WT) and the lcd1-7 and lcd1-9 mutants of tomato. The values are the means ± SDs of three replicates. The symbol ** stands for P < 0.01
Role of LCD1 in tomato fruit ripening
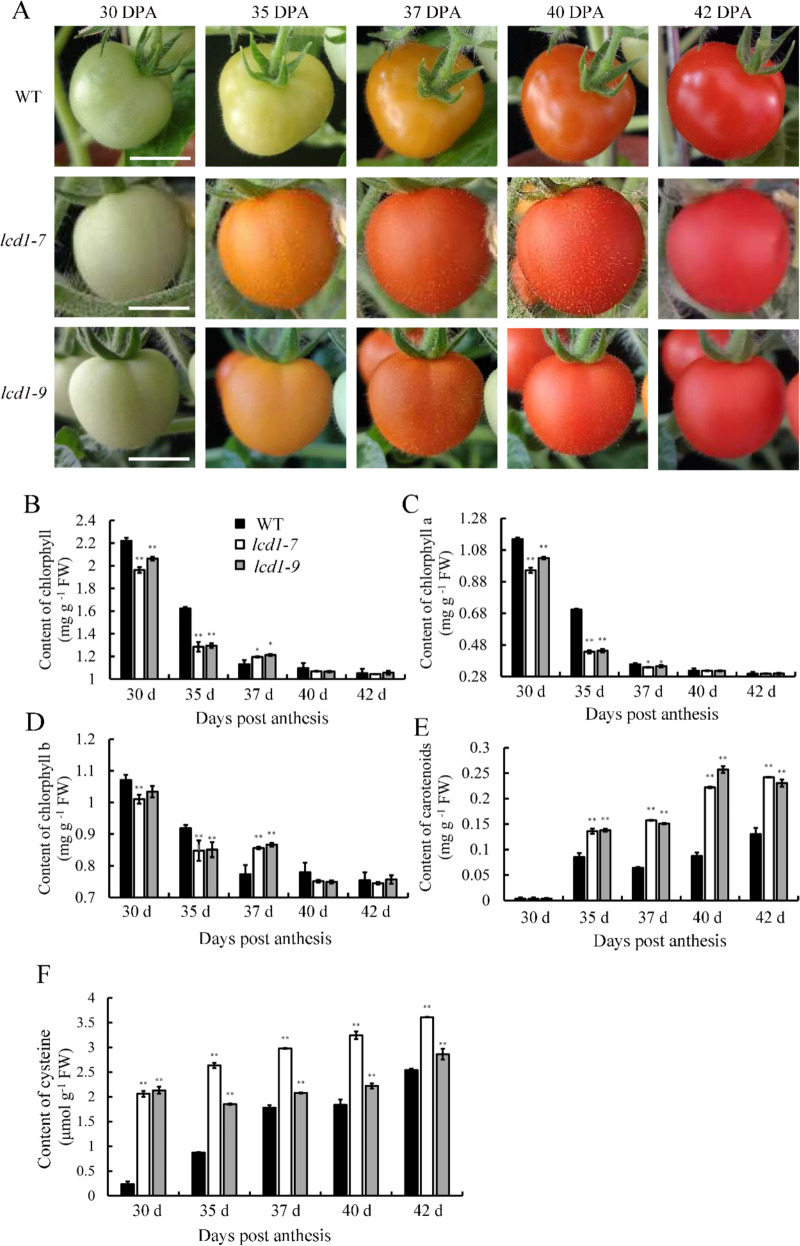
We characterized the phenotypes of the WT and lcd1 mutant tomato plants by measuring plant heights and fruit coloration. As shown in Fig. S4, both lcd1-7 and lcd1-9 led to shorter plant heights, suggesting that mutation of LCD1 affected the growth of tomato plants. The role of the LCD1 mutation in fruit ripening was observed in the first flowering branch. As shown in Fig. 5A, tomato fruit turned pink at 37 days post anthesis (DPA) and became fully red at 40 DPA. In contrast, the lcd1-7 and lcd1-9 mutants turned orange at 35 DPA, suggesting that LCD1 played a negative role in fruit ripening. We also measured tomato fruit ripening by determining the levels of chlorophylls and carotenoids. As shown in Fig. 5B, the total chlorophyll content decreased gradually from 30 to 42 DPA in the wild-type and lcd1 mutants. However, the LCD1 mutation caused significantly lower chlorophyll content at 30 and 35 DPA compared with the wild type, and similar trends were observed when chlorophylls a and b were measured (Fig. 5C, D). Figure 5E shows changes in carotenoids during fruit ripening in the wild type and lcd1 mutants. The carotenoid content increased gradually from 30 to 42 DPA in the wild type, while much higher levels were observed in the lcd1 mutants. At 37 DPA and 42 DPA, the carotenoid content in the lcd1 mutants was approximately two times that in wild-type fruit. Overall, the metabolism of pigments further suggested the role of LCD1 in the regulation of fruit ripening.
Fig. 5. SlLCD1 mutation caused accelerated fruit ripening in tomato.
A Fruit phenotypes of lcd1-7 and lcd1-9 mutant plants during 42 days post anthesis (DPA). B–F The levels of total chlorophyll, chlorophyll a, chlorophyll b, carotenoids, and cysteine in fruit of wild-type and mutant plants during 42 days post anthesis (DPA). The values are the means ± SDs of three replicates. The symbol ** stands for P < 0.01. Bar 1 cm
Due to the role of LCD in catalyzing cysteine synthesis3,14, we speculated that cysteine might accumulate in the lcd1 mutant, which was confirmed by the data shown in Fig. 5F. Cysteine levels accumulated gradually in the wild type during fruit ripening. However, significantly higher amounts of cysteine were observed in lcd1-7/9 at 30 DPA, where the cysteine content in lcd1 mutant fruit was approximately ten times that in wild-type fruit. At 35–42 DPA, the cysteine content of lcd1-9 was lower than that of lcd1-7 but still significantly higher than that of the wild type. The increased cysteine content in lcd1 mutant fruit suggested that the LCD1 mutation failed to decompose cysteine, which resulted in accumulated cysteine.
Effects of the LCD1 mutation on the expression of ripening-related genes
To explore the molecular mechanism underlying the difference in chlorophyll content during the ripening process of wild-type and lcd1-7 tomato, the relative expression levels of the key genes involved in the chlorophyll degradation pathway, i.e., NYC1, PAO, PPH, and SGR1, were analyzed. As shown in the heatmap (Fig. 6) and bar chart in Fig. S5, the expression levels of the NYC1, PAO, PPH, and SGR1 genes were significantly increased in the fruit with the LCD1 mutation during ripening. Although increased gene expression of NYC1, PAO, PPH, and SGR1 was observed in the wild type, the levels were lower than those in lcd1 mutant fruit. Among these genes, the expression levels of the PPH and SGR1 genes in the fruit of the lcd1 mutant at 35 DPA were nearly ten times that in wild-type fruit. Thus, it could be concluded that increased gene expression in the chlorophyll degradation pathway may contribute to accelerated chlorophyll degradation in lcd1 mutant fruit.
Fig. 6. SlLCD1 mutation increases the expressions of ripening related genes during tomato fruit ripening.
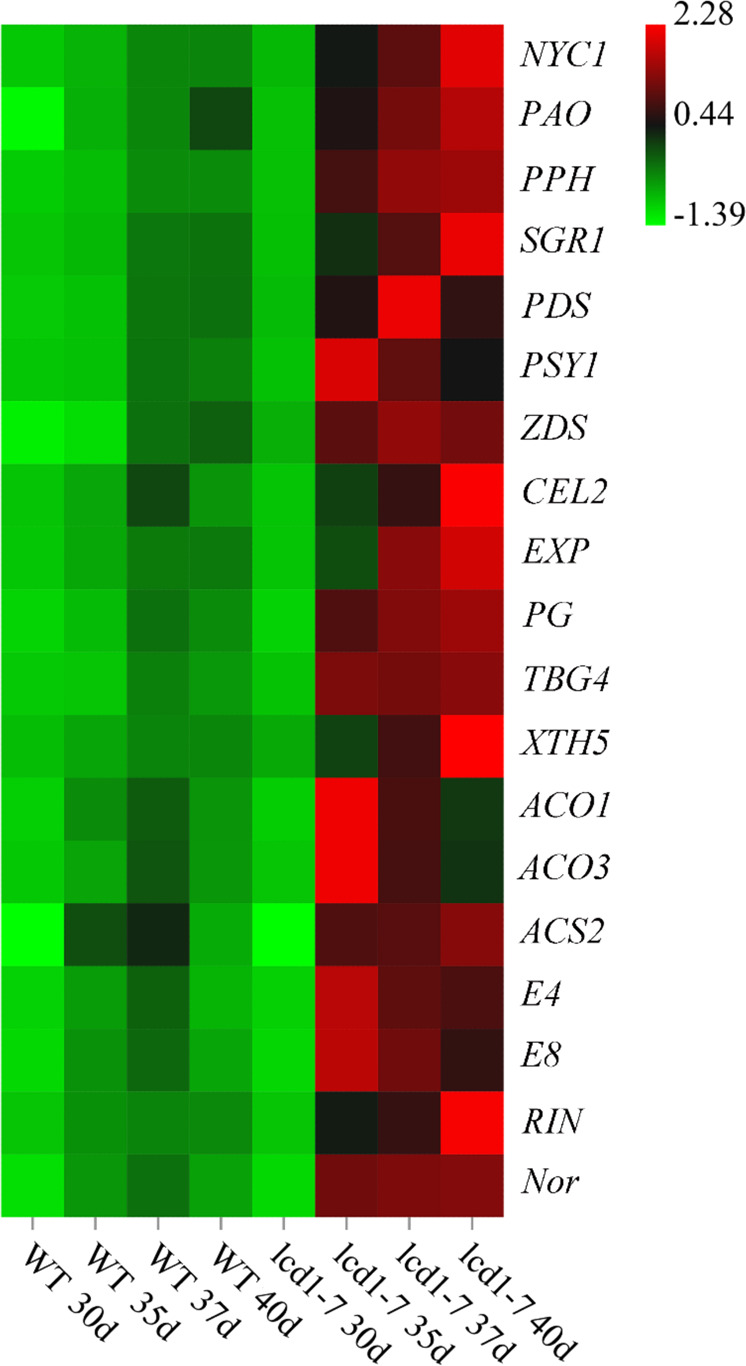
Heatmap of the relative gene expression of NYC1, PAO, PPH, SGR1, PDS, PSY1, ZDS, CEL2, EXP, PG, TBG4, XTH5 ACO1, ACO3, ACS2, E4, E8, RIN, and Nor in fruit of wild-type and lcd1-7 mutant plants at 40 days post anthesis (DPA)
Because the LCD1 mutation caused carotenoid accumulation in fruit, the expression of PSY1, PDS, and ZDS involved in carotenoid biosynthesis was analyzed in the wild type and in the lcd1 mutant. Figure 6 shows that the expression of the PDS, PSY1, and ZDS genes was significantly increased in lcd1 mutant fruit compared with that in the wild type. For example, the expression of the PSY1 gene in fruit of the lcd1 mutant was 74 times that in fruit of the wild type at 35 DPA (Fig. S5).
To establish a role for SlLCD1 in the metabolism of cell wall components in tomato fruit, the expression levels of genes encoding enzymes that play a role in cell wall biosynthesis and degradation were measured in wild-type and lcd1 mutant fruit. Figures 6 and S6 show that the expression of CEL2 (endoglucanase), EXP (expansin), XTH5 (xyloglucan endotransglucosylase), PG (polygalacturonase), and TBG4 (beta-galactosidase) was upregulated in lcd1 tomato fruit compared to wild-type fruit, though the gene expression levels increased gradually during fruit ripening in both the wild type and lcd1-7. Expression of CEL2 in lcd1-7 mutant fruit was upregulated approximately 9-fold at 40 DPA compared to its expression in the wild type. PG and TBG4 were upregulated 10-fold and 47-fold at 35 DPA in lcd1-7 mutant fruit relative to the wild type (Figs. 6 and S6).
Tomato releases ethylene during fruit ripening by upregulating the expression of genes of the ethylene biosynthesis pathway; therefore, we analyzed these genes in wild-type and lcd1-7 tomato fruit. Figure 6 shows the expression of the ethylene biosynthesis genes ACO1, ACO3, and ACS2. Their expression increased gradually in wild-type fruit, suggesting that they were the marker genes of fruit ripening, but the expression levels of ACO1, ACO3, and ACS2 in lcd1-7 were higher than those in wild-type fruit. Among these genes, the expression of ACO1 and ACO3 in lcd1-7 was upregulated by 6-fold and 10-fold, respectively, at 35 DPA compared with the expression in the wild type (Fig. S7).
E4 (encoding the peptide methionine sulfoxide reductase msrA) and E8 (1-aminocyclopropane-1-carboxylate oxidase-like protein) are important ethylene-responsive marker genes in tomato16. As shown in Fig. 6, the relative expression levels of E4 and E8 were upregulated in tomato fruit. The lcd1 mutant showed upregulation of E4 by approximately 7-fold and 8-fold at 35 and 40 DPA, respectively, compared with the wild type (Fig. S7). RIN, a MADS-box family transcription factor, is a positive regulator of tomato fruit ripening and plays a role in the ethylene synthesis pathway. Nor, an NAC family transcription factor, also functions upstream of ethylene synthesis to control fruit ripening. Figure 6 shows that the expression levels of RIN and Nor increased at the beginning of fruit ripening; however, the lcd1 mutation led to increased gene expression. RIN and Nor gene expression in lcd1 tomato fruit was approximately 5- and 6.7-fold that in the wild type at 40 DPA (Fig. S7).
Exogenous H2S treatment delayed tomato fruit ripening and attenuated the expression of ripening-related genes
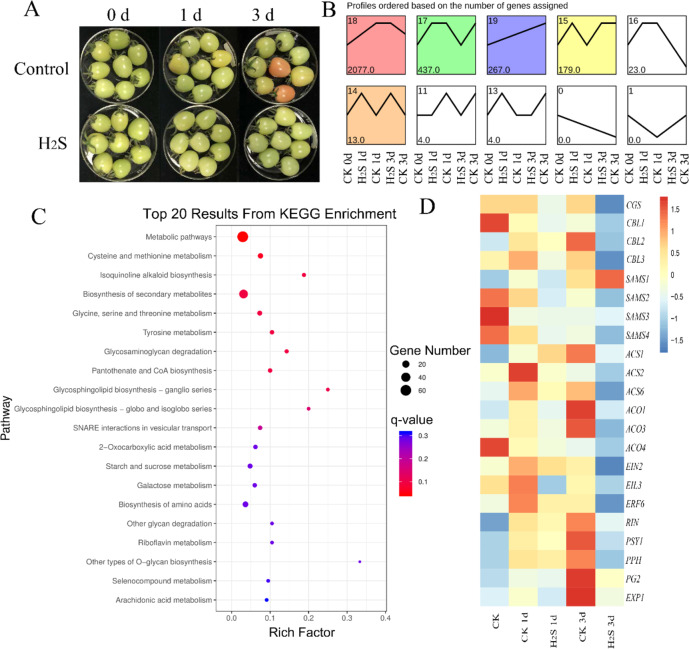
Tomato fruits at the white mature stage that were fumigated with H2S showed markedly delayed fruit ripening (Fig. 7A). To explore the role of H2S in this process, fruit were sampled for transcriptomic analysis at 0, 1, and 3 days post storage, and the expression of ripening-related genes was analyzed. The data in Fig. S8A show that H2S treatment for 1 day induced the expression of 4188 genes and reduced the expression of 4911 genes in comparison to control fruit. On day 3, 9988 genes were upregulated and 6232 genes were downregulated in H2S-treated fruit compared with the control, suggesting that H2S induced profound changes in gene expression. Gene expression patterns were analyzed by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database as shown in Fig. 7B. Profile 17 was enriched in genes that showed upregulation on day 3 for control fruit. Figures 7C and S8B–E show that the genes in profile 17 are involved in the cysteine and methionine metabolism pathways, selenocompound metabolism, porphyrin and chlorophyll metabolism, and plant hormone signal transduction. Then, the genes that showed upregulation upon H2S treatment in profiles 14 and 15 were also analyzed, and the results in Fig. S9 show that H2S treatment affects many metabolic pathways, including primary and secondary metabolism. In addition, the differentially expressed genes in H2S-treated fruit compared with the control on days 1 and 3 were analyzed through KEGG pathway analysis (Figs. S10–13). Generally, H2S treatment upregulated the pathways of ribosome biogenesis, plant hormone signal transduction, glutathione metabolism, ABC transporters, and plant–pathogen interaction compared with the control. In contrast, the pathway of carotenoid biosynthesis was inhibited by H2S.
Fig. 7. Transcriptomic analysis of tomato fruit treated with H2S (H2S) or the control (CK) for 0, 1, and 3 days.
A Images of tomato fruit with H2S (H2S) or the control (CK) for 0, 1, and 3 days. B Differential gene expression pattern analysis based on transcriptomic data. C KEGG analysis based on the genes in profile 17 as shown in B. D Heatmap of gene expression data of ripening-related genes in tomato fruit treated with H2S (H2S) or the control (CK) for 0, 1, and 3 days
We also asked whether the ripening-related genes in tomato fruit were modulated by H2S. The heatmap in Fig. 7D shows that many genes in the methionine pathway, including CGS (cystathionine gamma synthase), CBL1 (cystathionine beta-lyase), CBL2, CBL3, SAM2 (S-adenosylmethionine synthase), SAM3, and SAM4, were repressed in H2S-treated tomato fruit. The ethylene biosynthesis genes ACS1, ACS2, ACS6, ACO1, ACO3, and ACO4 showed increased expression during postharvest storage of tomato, whereas the increase was greatly attenuated by H2S treatment. Many genes involved in ethylene signaling, including EIN2, EIL3, ERF6, and RIN, were upregulated in control fruit at day 3, and H2S inhibited expression. PSY1, PPH, PG2, and EXP1 (encoding an expansin precursor) are ripening-related genes for chlorophyll degradation and fruit softening, and their expression is also repressed by H2S.
Discussion
The research presented in this paper shows that the cysteine desulfhydrase SlLCD1, a key enzyme in sulfur metabolism in plants, that we identified in tomato is localized to the nucleus, and this localization is mediated by a potential NLS. It had previously been proposed that the cysteine desulfhydrase of Arabidopsis (L-CDesI, At3g62130) is a nuclear localized enzyme14,20, but this was not supported by direct experimental evidence. Our work provides evidence that amino acid residues at the N-terminus of SILCD1 do indeed encode a functional NLS. We show that the H18LAKKPKLS domain of SlLCD1 located at the N-terminus is essential for nuclear localization and that K24 is the key amino acid. Mutation of the other lysine residues, namely, K21Q and K22Q, in the putative NLS domain abolished the nuclear localization, and only those sequences containing K24 were located in the nucleus. The detailed mechanism underlying SlLCD1’s nuclear localization is still unknown, but it is likely that importins may mediate its subcellular localization. A major process for the active import of proteins into the nucleus is initiated by the binding of cargo proteins containing NLSs to α‐importins in the cytosol21. For instance, Arabidopsis phosphatidylinositol 4-phosphate 5-kinase 2 was reported to contain a functional nuclear localization sequence and interacts with alpha-importins, which mediate its nuclear localization22. As SlLCD1 is located in the nucleus, we propose that its substrate cysteine is also present in the nucleus. Recently, it was reported that the O-acetylserine(thiol)lyases SlOAS2, 4, and 6 localized in the membrane, cytosol, and nucleus, suggesting that cysteine could be synthesized in the nucleus20. Sulfur-related metabolism is dynamic in the nucleus. For instance, methionine (Met) adenosyltransferase 4 (MAT4)/S-adenosyl-Met synthetase 3, which catalyzes the synthesis of S-adenosyl-Met (SAM), is located in the nucleus in the one-carbon metabolism cycle23.
To understand the role of cysteine desulfhydrases in regulating tomato fruit ripening, we used the BLAST tool in the NCBI database using Arabidopsis LCD as the query and identified two LCD genes in tomato. These tomato LCD sequences were used to examine expression patterns in different tissues at different developmental stages using the TomExpress database24. Among the two LCD genes, SlLCD1 showed increased gene expression during fruit ripening, which was confirmed by RT-qPCR of SlLCD1. When expression of SlLCD1 was silenced with a VIGS construct containing the TRV, accelerated fruit ripening was observed in VIGS-SlLCD1 compared with plants infected with an empty construct. We confirmed the silencing of VIGS-LCD by RT-qPCR, where the expression of LCD1 in silenced fruit was half that in the control fruit, and H2S production was also reduced in silenced plants (Fig. 3E). These observations provide a clear link between reduced H2S content and fruit ripening in tomato when LCD1 is silenced.
These observations linking reduced H2S levels to fruit ripening were confirmed using CRISPR/Cas9 to generate lcd1 mutants. Both lcd1-7 and lcd1-9 showed decreased endogenous H2S content and enhanced fruit ripening compared to control tomato plants. We also analyzed chlorophyll and carotenoid levels in the fruit of CRISPR/Cas9 mutants and showed that the lcd1 mutation resulted in accelerated chlorophyll degradation and enhanced carotenoid accumulation. Surprisingly, we observed increasing cysteine content in both wild-type and lcd1 mutant fruit. We speculate that elevated cysteine levels may increase methionine levels. Methionine is a key intermediate in ethylene synthesis; thus, accelerated fruit ripening in lcd1 could be partially attributed to enhanced C2H4 production via methionine from cysteine25.
The expression of ripening-related genes was analyzed in the lcd1 mutant to study the effect of reduced H2S. Consistent with the observation of accelerated chlorophyll degradation in lcd1, the expression of NYC1, PAO, PPH, and SGR1 was significantly enhanced during fruit ripening. Moreover, the expression of PSY1, PDS, and ZDS, genes involved in carotenoid biosynthesis, was also increased in lcd1 mutant fruit. The modulation of pigment metabolism-related genes may contribute to the degradation of chlorophyll and accumulation of carotenoids in lcd1 mutant fruit.
Tomato fruit ripening has been shown to be accompanied by changes in the expression of genes that play roles in ethylene synthesis and cell wall modification. Our data show that the cell wall metabolism-related enzymes CEL2, EXP, XTH5, PG, and TBG4, the ethylene biosynthesis-related genes ACO1, ACO3, and ACS2, and the ethylene responsive genes E4 and E8 were all expressed at significantly higher levels in lcd1-7 than in wild-type tomato fruit. RIN, a MADS-box family transcription factor, and Nor, a NAC family transcription factor, are TFs that have been shown to be important in regulating ethylene biosynthesis and fruit ripening26–28. We found that the expression levels of RIN and Nor increased significantly at the beginning of fruit ripening in WT tomato, but significantly higher gene expression was found in lcd1. These data implicate the reduction in H2S levels in lcd in accelerated fruit ripening mediated by enhanced ethylene biosynthesis and the expression of ripening-related transcription factors.
Accumulating evidence suggests that exogenous H2S could inhibit the ripening and senescence of multiple fruits and vegetables by antagonizing the effects of ethylene and by regulating ROS metabolism2,7–9. In the present work, tomato fruit were sampled for transcriptome analysis at 0, 1, and 3 days post storage, and the expression of ripening-related genes was analyzed. Genes involved in cysteine and methionine metabolism pathways, selenocompound metabolism, porphyrin and chlorophyll metabolism, and plant hormone signal transduction were enriched in profile 17, and their expression increased in control fruit on day 3 compared to the expression in the H2S treatment group. We infer from our transcriptomic analysis that many genes in the methionine biosynthetic pathway, ethylene biosynthesis, ethylene signaling transduction, chlorophyll degradation, and fruit softening are significantly repressed by H2S.
Overall, we provide evidence that LCD1 in tomato is involved in sulfur metabolism in the nucleus and show that a reduction in endogenous H2S causes accelerated fruit ripening. However, we recognize that the role of endogenous H2S in plant development may be mediated at multiple levels, including by modulation of gene expression, by persulfidation, and by disturbed hormone generation due to changes in sulfur metabolism.
Materials and methods
Potential NLS analysis of LCDs in plants and subcellular localization analysis for SlLCD1 and SlLCD2 in tobacco cells
Putative l-cysteine desulfhydrase proteins were identified by the BLASTP tool in the Phytozome v12.1.6 (https://phytozome.jgi.doe.gov/pz/portal.html#) database and Sweetpotato Genomics Resource (http://sweetpotato.plantbiology.msu.edu/index.shtml) with the l-cysteine desulfhydrase AT3G62130 peptide sequence as a query. The plant species included Arabidopsis thaliana, Brassica oleracea, Citrus sinensis, Cucumis sativus, Fragaria vesca, Glycine max, Ipomoea trifida, Ipomoea triloba, Malus domestica, Medicago truncatula, Oryza sativa, Populus trichocarpa, Solanum lycopersicum, Solanum tuberosum, Sorghum bicolor, Vitis vinifera, and Zea mays. The potential NLS sites were obtained from NLS mapper29 (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) with a criterion score of 5. The protein sequence of SlLCD1 (accession number: LOC101258894) was analyzed and showed a potential NLS at its N-terminus (H18LAKKPKLS). To explore the subcellular localization of the tomato LCD1 and LCD2 (accession number: XM_004238754.4) proteins, the full-length coding sequence for SlLCD1 and SlLCD2 was cloned into p1300-35S-GFP, and a series of SlLCD1 variants fused to GFP were generated: SlLCD1 (Δ1–26)-GFP, SlLCD1 (K21Q K22Q K24Q)-GFP, SlLCD1 (K21Q)-GFP, SlLCD1 (K22Q)-GFP, SlLCD1 (K24Q)-GFP, and SlLCD1 (K21Q K22Q)-GFP. Then, the generated fusion vectors were transformed into Agrobacterium strain GV3101. After overnight culture, the Agrobacterium cells were centrifuged and resuspended in 10 mM MgCl2, 10 mM 2-N-morpholino ethanesulfonic acid (MES), and 200 μM acetosyringone (AS) (pH 5.8) at an OD600 of 0.1–0.2. The mixtures were incubated at room temperature for 2 h and infiltrated into 4-week-old tobacco leaves. After infiltration, the plants were cultivated in the dark for 24 h at 16 °C and then cultivated for 48–72 h under 24 °C/16-h day and 18 °C/8-h night conditions. Hoechst 33342 was used to stain the nucleus30, while the chloroplasts were indicated by their autofluorescence. Cubic tobacco leaves were incubated with Hoechst 33342 (5 μg/mL) for 5 min. The excitation/emission wavelengths were 488 nm/507 nm for GFP, 346 nm/460 nm for Hoechst 33342, and 488 nm/650 nm to 750 nm for chlorophyll autofluorescence. All of the fluorescence signals were detected using a Zeiss LSM710NLO confocal laser scanning microscope. The primers used for the GFP vector are listed in Table S2.
Enzymatic activity determination of SlLCD1 and SlLCD2 expressed in E. coli
The coding sequences of SlLCD1 and SlLCD2 were cloned into the expression vector pJC40 between the NdeI and BamHI restriction sites31. The recombinant plasmids were transformed into DE3 for protein expression. The transformant harboring pJC40-LCD1/2 (DE3) was grown in LB medium containing ampicillin at 37 °C overnight. The culture was then transferred to 100 mL of fresh LB medium containing ampicillin at 37 °C until the OD600 reached approximately 0.6–0.8. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM and incubated overnight at 16 °C and 120 r.p.m. The bacterial cells were harvested by centrifugation at 12,000×g at 4 °C for 10 min and washed twice with 1 M phosphate buffer (pH 7.5 containing 1 mM PMSF) before resuspension. After ultrasound-mediated homogenization, the homogenate was centrifuged at 12,000×g and 4 °C for 20 min and analyzed to determine its ability to catalyze the decomposition of l-cysteine to release H2S according to the method described by Liu et al.20. The experiments were repeated three times, and the results are expressed as the mean ± SD (standard deviation). The related primers are listed in Table S3.
VIGS of SlLCD1 in tomato fruit
The TRV-based vectors pTRV1 and pTRV2 were used for VIGS of SlLCD1 (accession number: LOC101258894). A 387-bp fragment corresponding to nt 29–416 of the SlLCD1 sequence amplified from tomato cDNA by PCR was cloned into pTRV2 to generate pTRV2-SlLCD1. The primers used for pTRV2-SlLCD1 are listed in Table S3. A. tumefaciens strain GV3101 containing the TRV-VIGS vectors was injected on the surface of the fruit petiole as previously described32. A. tumefaciens strain GV3101 containing the vectors was cultured at 28 °C for 16 h in Luria-Bertani medium containing 20 μM AS, 10 mM MES, and 50 μg/mL each of the antibiotics kanamycin, gentamycin, and rifampicin. The Agrobacterium cells were harvested and resuspended in infiltration buffer containing 10 mM MgCl2, 10 mM MES (pH 5.6), and 200 μM AS and finally adjusted to an OD600 of 1.5. Resuspended cells with the pTRV1 and pTRV2 or pTRV2-SlLCD1 vector were then mixed together at a ratio of 1:1 and infected into pedicels of Micro-Tom tomato plants. Tomato petioles infiltrated with pTRV2 without the insert were used as controls. Tomato plants were then stored at 16 °C for 24 h and transferred to normal culture conditions thereafter. The changes in tomato fruit color were measured with a color difference meter (model WSC-100; Konica Minolta, Tokyo, Japan). L* indicates lightness, a* indicates chromaticity on a green (−) to red (+) axis, and b* stands for chromaticity on a blue (−) to yellow (+) axis. Each fruit was measured at four equidistant points around the middle area.
Tomato seeds were removed, and the flesh was sampled in VIGS-SlLCD1 fruit or SlLCD1-gene edited mutants. For tissue expression analysis of SlLCDs, different tissues of tomato were sampled. Total RNA from 0.1 g of frozen samples was extracted using the RNA Extraction Kit (Tiangen, Beijing, China), and cDNA was obtained using a reverse-transcription kit (PrimeScript RT Master Mix; Takara, Kyoto, Japan). The cDNA products were used for gene expression analysis by quantitative polymerase chain reaction (qPCR) performed using a Bio-Rad IQ5 (Hercules, CA). The specific primers used for qPCR were designed based on the coding sequence of the genes as shown in the SGN database (https://solgenomics.net/) or the NCBI database (https://www.ncbi.nlm.nih.gov/ncbisearch) (Table S4). Tubulin gene expression in control tomato plants was used for normalization of data. All analyses were repeated in three technical replicates. The heatmap of gene expression data was prepared by http://www.omicshare.com/tools.
Determination of H2S content in tomato
For the H2S content assay, tomato fruit with VIGS-SlLCD1 or leaves from SlLCD1-gene edited mutants were sampled to analyze the H2S content as described33. The plant samples were homogenized with 1 mL of phosphate buffer solution (pH 7.0, 50 mM) containing 0.1 M ethylene diamine tetraacetic acid (EDTA) and 0.2 M ascorbic acid. Then, the homogenate was mixed with 1 mL of 1 M HCl and placed in a closed vial to release H2S. The released H2S was absorbed by a 1% (w/v) Zn(AC)2 (0.5 mL) trap that was placed in the vial. After incubation for 30 min, 100 μL of 20 mM N,N-dimethyl-p-phenylenediamine and 100 μL of 30 mM FeCl3 were added to the Zn(AC)2 solution. After 15 min in darkness, the amount of H2S released was determined by measuring the absorbance at 670 nm.
CRISPR/Cas9 constructs for SlLCD1 and transformation of S. lycopersicum cv. Micro-Tom
Two SlLCD1 target sites (sgRNA1 and sgRNA2) were designed and selected by the CRISPR-P program (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). The 20-bp oligos of sgRNA were integrated into the AtU3d and AtU3b vectors, and the sgRNA was assembled into the CRISPR/Cas9 binary plasmid by Golden Gate ligation34. A. tumefaciens containing the Cas9-SlLCD1 plasmid was used for stable transformation of tomato35. For confirmation of the SlLCD1 gene-edited plants, genomic DNA was extracted, and the fragment flanking the sgRNA target sequence was amplified from genomic DNA and sent for sequencing. The sequences were decoded on the website http://skl.scau.edu.cn/dsdecode/36. The primer pairs used for vector construction and mutation analyses are listed in Table S4.
Determination of the levels of chlorophyll, carotenoid, and cysteine in tomato fruit
The levels of chlorophyll and carotenoids in tomato fruit were determined according to the methods of Lichtenthaler and Wellburn37 and Nath et al.38, respectively. Tomato flesh (2.0 ± 0.1 g) was sampled from lcd1 mutant fruit at 30, 35, 37, 40, and 42 DPA. Each analysis was repeated three times, and the chlorophyll and carotenoid levels were expressed as mg/g fresh weight (FW).
A cysteine assay kit (Beijing Solarbio Science & Technology Co., Ltd., China) was used to determine the cysteine content. Tomato flesh (0.2 g) as sampled from lcd1 mutant fruit at 30, 35, 37, 40, and 42 DPA and assayed according to the instructions, and the absorbance was measured at 600 nm. The cysteine content was expressed as μmol/mL.
Plant materials and transcriptome analysis
Tomato fruit at the white mature stage were fumigated with H2S released from 150 mL of 0.6 mmol/L sodium hydrosulfide (NaHS) solution or an equal amount of water. The tomato fruit and solutions were stored in 3-L containers at 23 ± 0.5 °C with a relative humidity of 85–90%. For transcriptomic analysis, tomato fruit were harvested at 0, 1, and 3 days post storage. Total RNA was extracted using the Plant RNeasy Extraction Kit (Qiagen, Germany). Sequencing of two biological replicates for each sample was performed using an Illumina GAII platform according to the manufacturer’s instructions. Different gene expression patterns were analyzed using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools). The original gene expression data in FPKM (fragments per kilobase million) in CK 0d, CK 1d, H2S 1d, CK 3d, H2S 3d were compared with CK 0d and then calculated as log2 base values, and the data are shown in Supplementary data 1. The genes in profile 17 (shown in Supplementary data 2) were enriched by the KEGG database, and the related pathways were also exported by the OmicShare tools. The expression of ripening-related marker genes was illustrated in a heatmap by R (v3.3.1).
Statistical analysis
Data were based on three replicates in each experiment, and the experiments were repeated independently three times. Statistical significance was assayed using a one-way analysis of variance with IBM SPSS Statistics (SPSS version 20.0; Armonk, NY), and the results are expressed as the means ± SDs. Significant differences were calculated by a t-test (P < 0.01 or P < 0.05) for significance.
Supplementary information
Supplementary data 1 The original gene expression data in FPKM (Fragments PerKilobase Million) in tomato of CK 0d, CK 1d, H2S 1d, CK 3d, H2S 3d were compared with CK 0d and then calculated in a log2 base
Supplementary data 2 The genes in profile 17 in Fig. 7B
Supplementary data 3 The transcription data of the genes in Fig. 7D
Supplementary data 4 The genes in profile 14 and 15 in Fig. 7B
Supplementary data 5 The differentially expressed genes in H2S treated fruit compared with control
Acknowledgements
We are very grateful for the careful revision from Professor Russell L. Jones at the University of California, Berkeley. This work was supported by the National Natural Science Foundation of China (31970200, 31970312, 31901993, 31670278, 51807046), the Earmarked Fund for the China Agriculture Research System (CARS-10-B1), National Key R&D Program of China (2019YFD1000103, 2019YFD1001303, 2019YFD1001300), the Natural Science Foundations of Anhui Province (1908085MC72), the Key Research and Development Program of Anhui Province (201904a06020031), the Fundamental Research Funds for the Central Universities (JZ2020YYPY0249), and National Undergraduate Training Programs for Innovation of China (No. 202010359054).
Author contributions
K.D.H., X.Y.Z., G.F.Y., and H.Z. conceived and designed the experiments. K.D.H., X.Y.Z., G.F.Y., Y.L.R., C.D., J.T., F.Y., and Z.Q.H. performed the research. K.D.H., X.Y.Z., G.F.Y., X.Y.C., Z.M.X., and L.Y.H. analyzed the data. K.D.H., X.Y.Z., Y.H.L., and H.Z. wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Kang-Di Hu, Xiao-Yue Zhang, Gai-Fang Yao
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00439-1).
References
- 1.Zhang H, et al. Hydrogen sulfide promotes root organogenesis in Ipomoea batatas, Salix matsudana and Glycine max. J. Integr. Plant Biol. 2009;51:1086–1094. doi: 10.1111/j.1744-7909.2009.00885.x. [DOI] [PubMed] [Google Scholar]
- 2.Hu LY, et al. Hydrogen sulfide prolongs postharvest shelf life of strawberry and plays an antioxidative role in fruits. J. Agric. Food Chem. 2012;60:8684–8693. doi: 10.1021/jf300728h. [DOI] [PubMed] [Google Scholar]
- 3.Fang H, et al. The Ca2+/calmodulin2-binding transcription factor TGA3 elevates LCD expression and H2S production to bolster Cr6+ tolerance in Arabidopsis. Plant J. 2017;91:1038–1050. doi: 10.1111/tpj.13627. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, et al. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant. 2020;13:732–744. doi: 10.1016/j.molp.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Shen J, et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell. 2020;32:1000–1017. doi: 10.1105/tpc.19.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, D., Li, J., Li, Z. & Pei, Y. Hydrogen sulfide inhibits ethylene-induced petiole abscission in tomato (Solanum lycopersicum L.). Hortic. Res.7, 14 (2020). [DOI] [PMC free article] [PubMed]
- 7.Li ZR, et al. Hydrogen sulfide alleviates dark-promoted senescence in postharvest broccoli. HortScience. 2015;50:416–420. doi: 10.21273/HORTSCI.50.3.416. [DOI] [Google Scholar]
- 8.Ge Y, et al. Hydrogen sulfide alleviates postharvest ripening and senescence of banana by antagonizing the effect of ethylene. PLoS ONE. 2017;12:e0180113. doi: 10.1371/journal.pone.0180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao GF, et al. Modulation of enhanced antioxidant activity by hydrogen sulfide antagonizing ethylene in tomato fruit ripening. J. Agric. Food Chem. 2018;66:10380–10387. doi: 10.1021/acs.jafc.8b03951. [DOI] [PubMed] [Google Scholar]
- 10.Lin X, et al. Transcriptome analysis reveals delaying of the ripening and cell-wall degradation of kiwifruit by hydrogen sulfide. J. Sci. Food Agric. 2020;100:2280–2287. doi: 10.1002/jsfa.10260. [DOI] [PubMed] [Google Scholar]
- 11.Gotor C, et al. Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 2019;70:4251–4265. doi: 10.1093/jxb/erz225. [DOI] [PubMed] [Google Scholar]
- 12.Jia H, et al. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 2018;9:1517. doi: 10.3389/fpls.2018.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch T, Wachter A. Sulfur metabolism: a versatile platform for launching defence operations. Trends Plant Sci. 2005;10:503–509. doi: 10.1016/j.tplants.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Papenbrock J, Riemenschneider A, Kamp A, Schulz-Vogt HN, Schmidt A. Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants–from the field to the test tube and back. Plant Biol. 2007;9:582–588. doi: 10.1055/s-2007-965424. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez C, Calo L, Romero LC, García I, Gotor C. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010;152:656–669. doi: 10.1104/pp.109.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng G, Zhou L, Wang Y, Zhang G, Chen X. Hydrogen sulfide acts downstream of jasmonic acid to inhibit stomatal development in Arabidopsis. Planta. 2020;251:42. doi: 10.1007/s00425-019-03334-9. [DOI] [PubMed] [Google Scholar]
- 17.Karlova R, et al. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014;65:4527–4541. doi: 10.1093/jxb/eru316. [DOI] [PubMed] [Google Scholar]
- 18.Barry SC, et al. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313X.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- 19.Hörtensteiner S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Lu J, Li H, Wang J, Pei Y. Characterization of the O-acetylserine(thiol)lyase gene family in Solanum lycopersicum L. Plant Mol. Biol. 2019;99:123–134. doi: 10.1007/s11103-018-0807-9. [DOI] [PubMed] [Google Scholar]
- 21.Merkle T. Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep. 2011;30:153–176. doi: 10.1007/s00299-010-0928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerth K, et al. Arabidopsis phosphatidylinositol 4-phosphate 5-kinase 2 contains a functional nuclear localization sequence and interacts with alpha-importins. Plant J. 2017;92:862–878. doi: 10.1111/tpj.13724. [DOI] [PubMed] [Google Scholar]
- 23.Meng J, et al. METHIONINE ADENOSYLTRANSFERASE4 mediates DNA and histone methylation. Plant Physiol. 2018;177:652–670. doi: 10.1104/pp.18.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zouine M, et al. TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J. 2017;92:727–735. doi: 10.1111/tpj.13711. [DOI] [PubMed] [Google Scholar]
- 25.Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP. Amino acid catabolism in plants. Mol. Plant. 2015;8:1563–1579. doi: 10.1016/j.molp.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Vrebalov J, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 27.Gao Y, et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018;5:75. doi: 10.1038/s41438-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, et al. Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 2020;226:460–475. doi: 10.1111/nph.16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl Acad. Sci. USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You F, et al. Genistein protects against Aβ25–35 induced apoptosis of PC12 cells through JNK signaling and modulation of Bcl-2 family messengers. BMC Neurosci. 2017;18:12. doi: 10.1186/s12868-016-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clos J, Brandau S. pJC20 and pJC40–two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr. Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- 32.Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. Virus-induced gene silencing in tomato fruit. Plant J. 2005;43:299–308. doi: 10.1111/j.1365-313X.2005.02441.x. [DOI] [PubMed] [Google Scholar]
- 33.Lai D, et al. Endogenous hydrogen sulfide enhances salt tolerance by coupling the reestablishment of redox homeostasis and preventing salt-induced K+ loss in seedlings of Medicago sativa. Plant Sci. 2014;225:117–129. doi: 10.1016/j.plantsci.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Ma X, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Kimura, S. & Sinha, N. Tomato transformation. Cold Spring Harb. Protoc.10.1101/pdb.prot5084 (2008). [DOI] [PubMed]
- 36.Liu W, et al. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant. 2015;8:1431–1433. doi: 10.1016/j.molp.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983;11:591–592. doi: 10.1042/bst0110591. [DOI] [Google Scholar]
- 38.Nath A, Bagchi B, Misra LK, Deka BC. Changes in post-harvest phytochemical qualities of broccoli florets during ambient and refrigerated storage. Food Chem. 2011;127:1510–1514. doi: 10.1016/j.foodchem.2011.02.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1 The original gene expression data in FPKM (Fragments PerKilobase Million) in tomato of CK 0d, CK 1d, H2S 1d, CK 3d, H2S 3d were compared with CK 0d and then calculated in a log2 base
Supplementary data 2 The genes in profile 17 in Fig. 7B
Supplementary data 3 The transcription data of the genes in Fig. 7D
Supplementary data 4 The genes in profile 14 and 15 in Fig. 7B
Supplementary data 5 The differentially expressed genes in H2S treated fruit compared with control