Abstract
The advent of highly effective treatments targeting the disease biology of chronic lymphocytic leukemia (CLL) has transformed the therapeutic field tremendously. However, transformation into an aggressive B-cell lymphoma, called Richter syndrome (RS), remains highly challenging since the treatment options for this condition are still insufficient. Exploratory drug testing and experimental studies are restricted by the lack of satisfactory models. We have established U-RT1, a cell line derived from a highly proliferating RS clonally related to the patient's underlying CLL. The cell line shows morphological features and an immunophenotype of RS-DLBCL (non-GCB). Molecular analysis revealed a complex karyotype with driver aberrations characteristic for RS such as loss of TP53 and CDKN2A. Furthermore, U-RT1 displays a chromosomal gain of the NOTCH1 gene locus and strong immunoreactivity for BCL-2. These features suggest that U-RT1 is the first eligible model system for investigations on the pathogenesis of RS and novel treatment options.
Keywords: Richter transformation, Richter syndrome, In vitro model, CLL, DLBCL
Abbreviations: aCGH, Array-based comparative genomic hybridization; CHOP, Cyclophosphamide, doxorubicin, vincristine and prednisone; CLL, Chronic lymphocytic leukemia; DLBCL, Diffuse large B-cell lymphoma; EBV, Epstein Barr virus; FCS, Fetal calf serum; FISH, Fluorescence in situ hybridization; FR, Framework region; GCB, Germinal center B-cell like; RS, Richter syndrome
Introduction
The development of an aggressive B-cell lymphoma in patients with a previous or concomitant diagnosis of chronic lymphatic leukemia (CLL) is defined as Richter syndrome (RS) or Richter transformation (RT), first described by Maurice Richter in 1928 [1]. Around 10% of patients with CLL undergo RT [2] and suffer from increasing lymphadenopathy, deteriorating general condition, fever, weight loss, and night sweats [3]. Despite intensive therapy, the prognosis of RS is poor and most patients die within the first year of diagnosis [4].
In most cases, patients show the diffuse large B-cell lymphoma (DLBCL) variant of RS (RS-DLBCL), but in 10% of cases the CLL evolves into Hodgkin lymphoma, termed Hodgkin variant of RS (HvRS) [5]. The diagnosis of RS is generally made on histology. The RS-DLBCL cells are morphologically distinct from small monomorphic CLL cells, appearing as large blast-like neoplastic B cells with small nucleoli. RS-DLBCL shows a diffuse growth pattern and a highly proliferative potential [6]. Immunohistologically, most cases of RS-DLBCL no longer express CD5 and CD23, giving a different immunophenotype from the underlying CLL [7]. Using the Hans algorithm, 90% to 95% of RS-DLBCLs are the more aggressive activated non-germinal centre B-cell subtype (non-GCB DLBCL), lacking immunoreactivity for CD10 but showing a variable expression of BCL6 and positivity for MUM1 [8].
In about 80% of cases, CLL progresses along a linear model of evolution leading to a clonally related aggressive B-cell lymphoma, the other 20% of cases are clonally unrelated to the underlying CLL [4,7,9]. Clonally unrelated lymphomas are thought to be CLL-independent neoplasms resulting from the patients’ treatment, with a compromised immune system and possibly Epstein Barr virus (EBV) infection. The survival of patients with this type of secondary lymphoma is comparable to that of patients with de novo DLBCL, while clonally related RS shows an inferior response to current treatment options [4].
Clinical risk factors for developing RS include an advanced stage of CLL, a large tumor mass (bulky disease), diffuse bone marrow involvement, and quantity and type of previous therapy (especially treatment with purine nucleoside analogues such as fludarabine in combination with alkylating agents such as chlorambucil) [10], [11], [12]. Biological risk factors for RS in CLL have been found to be NOTCH1 mutation (especially in exon 34) [8], stereotyped B-cell receptor (“subset 8”) [9] expression of CD38 or ZAP70, and the absence of del13q14 and unmutated immunoglobulin heavy chain variable gene [13], [14], [15]. CLL cells harboring inactivated TP53 or disruption of 17p13 have also been shown to be at higher risk of CLL progression and RT [11,[15], [16], [17]].
While the proliferation of large B-cell lymphoma often depends on deregulation of specific B-cell signalling pathways, the detailed pathogenesis of RS remains unclear. Nevertheless, 90% of cases of clonally related RS show aberrations in TP53, NOTCH1, MYC or CDKN2A, thus affecting regulators of apoptosis and proliferation [8,18,19]. This biology partly explains the poor response of patients to the chemotherapy regimen currently used in diffuse large B-cell lymphoma (R-CHOP). In particular, RS harbouring mutations of TP53 or del17p13 is often refractory and carries a significantly higher mortality than RS with TP53 wildtype [4]. Interestingly, RS lacks many of the frequently described genetic defects found in de novo DLBCL such as mutation or gene rearrangement of BCL-2 or BCL6 [4,20], EZH2 or GNA13 mutations (GCB DLBCL), or perturbation of the NFκB pathway due to mutations in CD79a/b, MYD88 or TNFAIP (non-GCB DLBCL) [4,18].
Despite the multitude of tested combinations of chemotherapy and novel immunotherapies, the results are still unsatisfactory and the overall survival of patients with clonally related RS-DLBCL remains poor [6,21]. Traditionally, therapeutic regimens for RS resemble those of de novo DLBCL [10] with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), usually in combination with rituximab (R-CHOP) [22], and autologous or allogeneic stem cell transplantation as postremission or salvage therapy for patients without limiting comorbidities [23].
New approaches in the management of relapsed and refractory CLL include traditional chemotherapy regimens in combination with novel agents like the BTK inhibitor Ibrutinib, which has shown remarkable efficacy [6,24,25] and was tested in a small case series of RS [25,26]. Recent studies also surprisingly revealed high expression of PD-1 on neoplastic B cells of clonally related RS, while most cases of CLL and de novo DLBCL lacked immunoreactivity for PD-1 [27]. The first trials in patients with RS have shown an overall response of 44% to the PD-1 small molecule inhibitor pembrolizumab, with an overall survival of 11 mo [28].
Further therapeutic options are still urgently needed for RS. Cell culture model systems for clonally related RS are therefore needed to explore novel targeted agents and immunotherapies as potential treatment options. We describe the first stable RS cell line, U-RT1, established from a cervical lymph node of a patient with CLL who developed multiple aggressive B-cell lymphomas. The diffuse large B-cell lymphoma from which the U-RT1 cell line was established turned out to be clonally related to the previously diagnosed CLL.
Material and methods
Patient data and patient's tissue
This study was triggered by encountering a 60 year-old male Caucasian patient with CLL and B symptoms. Initially, the patient was treated with fludarabine and cyclophosphamide in combination with the CD20 monoclonal antibody rituximab. Partial remission was achieved even though the rituximab had to be discontinued due to intolerance. Relapse occurred about 4.5 y after diagnosis, when the patient was initially treated with bendamustine and alemtuzumab (CD52 monoclonal antibody) and later on with COP (cycloposphamide, vincristine sulfate, and prednisone). The disease remained stable for 11 mo, followed by progression. At this time, genetic analyses revealed that the CLL harboured a 13q14.3 deletion and a loss of one copy of TP53 (del17p13.1). The remaining TP53 copy had mutated (c.342-343del2bpins1bp). IGH rearrangement was identified to be VH3 family specific (V3-30-3 homology 100%). The patient was treated with the BCL-2 inhibitor venetoclax for about 5 wk. During this time, he developed an EBV-positive aggressive B-cell lymphoma in the stomach, diagnosed on gastric biopsy. A cervical lymph node was excised and showed an aggressive DLBCL. No TP53 loss could be detected here, but a small subset of cells showed a 13q14.3 deletion. The patient gave his written informed consent for the scientific use of his tissues. Directly after surgery, about 1 cm³ of the lymph node was transferred into Iscove's Modified Dulbecco's Medium (IMDM) containing 10% fetal calf serum (FCS) and antibiotics and used to establish the cell line. The patient died from progressive disease 2 wk after the excision of the lymph node.
Primary cell culture
The lymph node tissue was minced and vortexed. Further tissue disintegration was performed with collagenase IV treatment under standard conditions. Sequential centrifuging and resuspending resulted in 4 primary cell cultures. The primary cultures were kept in IMDM containing 10% FCS and antibiotics, together with insulin, transferrin, and selenium (ITS medium) for the first 6 wk of culture. Two of the 4 primary cultures showed a time-dependent increase of large B-cell-like cells. A single cell separation of the growing cells was performed using a fluorescence-activated cell sorter (BD FACSAria Ilu, BD bioscience, San Jose, CA, USA). A total of 576 single cells were seeded into wells of 96-well plates (Nunc), each containing 100 µL of ITS medium. The cells were incubated at 37°C and 5% CO2. Cell growth was monitored in five wells. At a stage of 100 to 500 cells, the subclones were transferred to 24-well plates (Nunc) and subsequently into 25 cm² cell culture flasks (Nunc). Karyograms and immunocytochemistry panels of the subclones were compared and revealed no striking differences. One of the clones was therefore further expanded and the resulting cell line was named U-RT1 (Ulm-Richter Transformation 1).
Immunohistochemistry and immunocytochemistry
Cell blocks were made for protein analysis in U-RT1 cells. A volume of 10 mL of the cell suspension was centrifuged at 130 g for 5 min. The cell pellet was then resuspended in 5 mL of 100% ethanol and spun down again. The pellet was resuspended in 1 mL of 100% ethanol and one drop of hematoxylin and transferred into a 1.5 mL reaction tube (Eppendorff, Hamburg, Germany). The tube was placed upside down on a filter paper to remove residual ethanol. The pellet so obtained was fixed in buffered formalin for 4 h, then embedded in paraffin.
Immunohistochemistry and immunocytochemistry was performed on 2 µm tissue sections. Depending on the antibody used, different antigen retrieval procedures were required. Supplementary Table 1 gives a summary of the antibodies used, the antigen retrieval techniques, dilutions, and incubation times. The RED kit (DAKO, Glostrup, Denmark) was used to detect antigen-bound antibodies. Counterstaining of the cell nuclei was achieved by staining with hematoxylin for 5 min and incubating in H2O for 10 min at 20°C.
EBV detection
EBV DNA was detected using a nested touchdown PCR approach. Annealing temperatures were subsequently reduced, starting at 68°C and going down to 58°C (final annealing temperature was used for 30 cycles). Primer sequences are given in Supplementary Table 2. Additionally, an EBV-encoded RNA chromogenic in situ hybridization (EBER CISH) was performed on sections of the pelleted cells to detect EBV-specific mRNA. The sections were rinsed in Xylol, ethanol (100%), ethanol (70%), and H2O. A digestion step using pepsin (20 min, 37°C) was subsequently performed. After incubation in distilled water and ethanol (100%), the sections were air dried. The sections were then hybridized with EBER-specific probes (2 h, 56°C). They were then washed in CISH washing buffer and EBERs were detected using the ZytoFast Plus CISH Implementation Kit (ZytoVision, Bremerhaven, Germany) as described in the manufacturer's handbook.
Clonality testing (heavy chain rearrangement)
The rearrangement of the immunoglobulin heavy chain gene was determined by multiplex PCR using framework region 1, 2, and 3 (FR1, FR2, and FR3) specific primers, as described by van Dongen et al. [29]. In short, the FR1, FR2, and FR3 segments were amplified using a mixture of family-specific primers (primer sequences and PCR programs are given in Supplementary Table 2A and B). The amplicons were separated in a capillary sequencer (GenomeLabGeXP, Beckman Coulter, USA) to determine the exact length (GeneMarker V2.70; SoftGenetics, State College, PA, USA), resulting in a defined amplicon triplet representing a specific IGH rearrangement.
Sequencing (FR1 IGH)
To determine the sequence of the FR1 segment of the rearranged IGH gene, the FR1 segment was amplified using a multiplex PCR with a primer mix of all family primers (VH1-VH6 FR1; Supplementary Table 2A). The amplicons were separated on a 3% agarose gel, cut out and purified using the PeqGold Microspin Gel extraction kit (Peqlab Biotechnologie GmbH; Erlangen, Germany). The purified amplicon was used as a template in a second PCR round using single family-specific primer pairs. The resulting amplicons were gel separated, purified, and Sanger sequenced (Eurofins Genomics). Sequence comparison and phylogenetic trees were performed using the Clustal Omega Multi Sequence Alignment online software (http://www.ebi.ac.uk/Tools/msa/clustalo/; 25.11.2019).
Fluorescence in situ hybridization
One-μm thick sections of formalin-fixed paraffin-embedded tissue were deparaffinized, hydrated, and incubated in 10 mM citrate buffer (pH 6.0) in a preheated steamer (98°C) for 15 min. Protein breakdown was achieved by incubating the slides with 0.025% pepsin solution (dissolved in 1% hydrochloric acid) at 37°C, gently shaking. After 20 min, the same amount of pepsin was added to the solution for another 10 min of incubation.
Afterwards, the slides were incubated twice for 5 min in saline-sodium citrate buffer before rinsing in distilled water, dehydrating in 70%, 80%, and 96% ethanol, and air drying.
For denaturation and hybridization, the DNA-Fluorescence in situ hybridization (FISH) probe was mixed with appropriate amounts of hybridization buffer, applied to the target area on the tissue slide (0.2 µL probe per slide), and covered with a coverslip (18 × 18 mm). The edges of the coverslip were sealed thoroughly with Fixogum glue and the denaturation was achieved through 5 mi at 75°C on a temperature-controlled hot plate. For hybridization, the slides were incubated overnight in a dark, humidified incubator at 37°C.
After the removal of glue and coverslip, the slides were washed 3 times for 5 min in a formamide-containing solution at 42°C and 3 times for 5 min in washing solution in a 60°C water bath. Counterstaining was performed with DAPI (60 µg/mL) in an antifade solution for 30 s. Signals from 100 nuclei were analysed. For translocation probes, the sample was considered positive for rearrangement if more than 15% of the signals were separated (at least 4 signals’ distance). The detection limit for amplifications and deletions was determined by testing tonsil tissue. Means and standard deviations (SD) were calculated. The cut-off for deletion/amplification detection was set to 3 × SD.
Multicolor fluorescence in situ hybridization
Metaphase spreads were prepared with standard colcemid treatment for 3 h, hypotonic lysis of the cells and methanol-acetic acid fixation. For karyogram analyses, the slides were incubated in 0.025% trypsin solution (in Hanks balanced salt solution/phosphate buffered saline 1:1) and then stained with Giemsa solution for 2 to 3 min. After rinsing in water, the target area of the wet slides was covered with a coverslip (24 × 50 mm) and analysed using Ikarus software (MetaSystems, Altlussheim, Germany). Multicolor FISH was similar to the FISH method described above but with some differences. The prepared metaphase slides were incubated in acidic pepsin solution for only 30 s. Five-µL 24XCyte-probe was applied to the target area and 10 metaphases per slide were documented and analyzed using ISIS software (MetaSystems).
Array comparative genomic hybridization
DNA from U-RT1 was isolated using the DNeasy Kit from Qiagen. A total of 500 ng DNA was used for the SurePrint G3 Human CGH Array 8 × 60K (Agilent Technologies, Inc., Santa Clara, USA). Labelling and hybridization was done following the manufacturer's protocol. The Sure Scan Microarray Scanner (Agilent) was used for measurement. Data analysis was performed with Genomic Workbench software from Agilent.
BCL-2 inhibitor testing and apoptosis/necrosis detection
U-RT1 cells were seeded 10,000 to a well in 96-well plates (Nunc) and incubated in IMDM containing 10% FCS and varying concentrations of the BCL-2 inhibitors venetoclax (ABT-199), ABT-737, HA14-1, or the CDK4/6 inhibitor palbociclib (Selleckchem, Munich, Germany). After 48 h’ incubation at 37°C and 5% CO2, cell viability was detected using a 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. Detection was performed using standard protocols for MTT viability assays. In short, inhibitor-treated and untreated U-RT1 cells were incubated with MTT by adding 10 µL MTT stock solution (12 mM) to 100 µL cell suspension. After a 3-h incubation period (37°C, 5% CO2), 100 µL of a cell lysis solution (10% sodium dodecyl sulfate in 0.01 M HCL – phosphate buffered saline) were added. During the following 3 h, the suspension was mixed by pipetting up and down 3 to 5 times to ensure a uniform solution. The absorption of the lysates was then measured at 570 and 690 nm using a multiplate reader (Biotek, Bad Friedrichshall, Germany). After background subtraction, the mean absorptions were normalized to the absorption of the lysates of untreated cells. Dose-response curves and IC50 concentrations were calculated using GraphPad Prism6 software.
Results
Characterization of the RS model cell line U-RT1
Morphology and immunohistochemistry
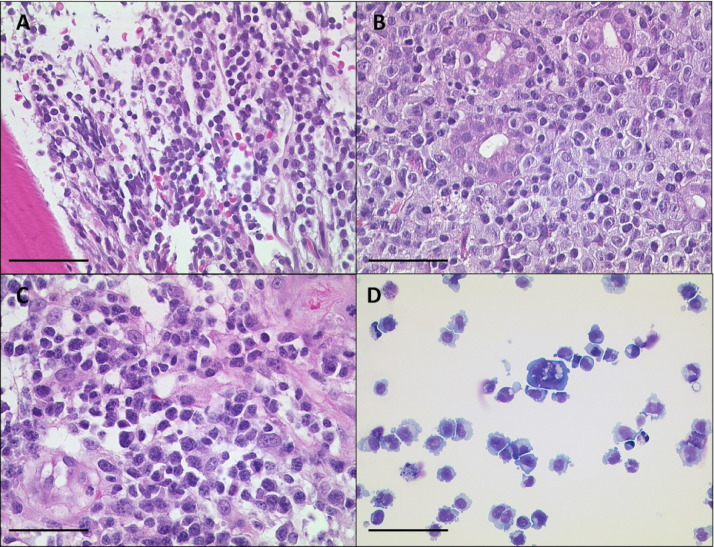
U-RT1 consists of large polymorphic B cells with scant cytoplasm and oval to round nuclei containing fine chromatin and a few membrane-bound nucleoli. This morphology resembles the centroblastic variant of diffuse large B cell lymphoma. In addition, 5% to 10% of U-RT1 cells appear to be multinucleated cells. Figure 1 shows haematoxylin and eosin (H&E) staining of the patient's bone marrow biopsy displaying small, monomorphic CLL cells, the aggressive lymphoma which the patient developed in the stomach, an aggressive lymphoma of the lymph node, and the U-RT1 cell line.
Figure 1.
H&E staining of CLL, the lymph node, and the cell line U-RT1 from the same patient. (A) The bone marrow biopsy shows small neoplastic lymphatic cells of the CLL. (B) Gastric diffuse large B-cell lymphoma and (C) diffuse large B-cell lymphoma in the lymph node of the same patient. (D) May-Grunwald-Giemsa (MGG) staining of cytospins of the U-RT1 cell line shows polymorphic B cells with scant cytoplasm and oval to round nuclei and scattered multinucleated cells. The black line indicates 50 µm. CLL, chronic lymphocytic leukemia.
We also performed immunohistochemistry to detect the expression of DLBCL and RS-related antigens in the patient's lymphoma tissues. Comparing the expression profiles of the CLL with the expression profiles seen in the lymphoma tissues, we found clear differences in the expression of CD5, CD10, CD23, BCL-2 and PAX-5. Immunocytochemistry staining of U-RT1 cells revealed expression of CD20, CD23, BCL-2, and PAX-5. The cells have in part lost CD5, are negative for CD10, CD38, BCL6, and Cyclin D1, and show a high proliferation rate of approximately 80% as determined by KI-67 antigen detection. The full immunohistochemistry panel is shown in Table 1. Using the Hans algorithm, U-RT1 and the corresponding lymphoma can be assigned as non-GCB DLBCL. The population doubling time is about 36 h.
Table 1.
Protein detection in patient tissue and U-RT1 cell block sections by immunohistochemistry and immunocytochemistry.
| Antigen | CLL Bone Marrow | Lymphoma Stomach | Lymphoma Lymph Node | U-RT1 |
|---|---|---|---|---|
| BCL-2 | +++ | +++ | +++ | +++ |
| BCL6 | + | − | − | − |
| CD5 | +++ | − | ++ | + |
| CD10 | − | − | + | − |
| CD20 | +++ | +++ | +++ | ++ |
| CD23 | +++ | + | − | +++ |
| CD30 | n.a. | − | − | −/+ |
| CD38 | ++ | +++ | +++ | − |
| CDK4 | n.a. | − | − | ++ |
| CDK6 | n.a. | − | − | − |
| MYC | + | ++ | + | + |
| Cyclin D1 | − | − | − | − |
| IgD | +++ | − | − | − |
| IgM | +++ | − | − | + |
| Ki-67 | ++ | +++ | +++ | +++ |
| MUM1 | n.a. | − | ++ | ++ |
| P16 | n.a. | − | − | − |
| P53 | n.a. | n.a. | n.a. | − |
| PAX-5 | +++ | +++ | +++ | +++ |
| PD-L1 | n.a. | - | + | + |
| Rb | n.a. | n.a. | n.a. | +++ |
| ZAP-70 | n.a. | − | +++ | − |
Immunohistochemistry panels for U-RT1, gastric lymphoma, lymphoma found in the lymph node, and the underlying chronic lymphatic leukemia all from the same patient. Standard CLL markers were used. The stains for P53 and Rb missing in the lymphomas are due to the amount of tumor tissue available. Results were evaluated as follows: no reaction (−), 30% cells positive (+), 30% to 70% cells positive (++), more than 70% cells positive (+++).
CLL = chronic lymphocytic leukemia.
Short tandem repeat analysis
To confirm the origin of the cell line, we compared the short tandem repeat signature of the cell line and patient lymph node tissue. In all of the 12 markers tested, the cell line's short tandem repeat profile fitted with the original lymph node tissue (Supplementary Table 3).
To prove the clonal relation of the cell line and the underlying CLL, B cell-specific IGH gene rearrangement analyses were performed, resulting in a single triplet signal for FR1-, FR2-, and FR3-specific amplicons. In the patient's bone marrow sample, we detected a specific clonal event representing the CLL (Figure 2). The subclones of U-RT1 exhibited the FR1, FR2, and FR3 amplicon lengths, confirming that the cell line represents an aggressive DLBCL transformed from an underlying CLL. IGH rearrangement was shown to be the same as initially detected in the CLL (V3-30-3 homology 100%). Furthermore, the TP53 mutation found in the CLL (c.342-343del2bpins1bp) was also found in U-RT1. Surprisingly, standard clonal analyses did not detect this clone in either the gastric lymphoma or the lymph node. At least 2 clonally independent lymphomas were detected at these sites. Further analysis was performed by reamplifying and Sanger sequencing the FR1-specific amplicons using family-specific primers. A cluster analysis of the resulting sequences showed that the clonally dependent RS was detectable in addition to the predominant clonally independent DLBCL clones in the lymphomas of the stomach and lymph node. To summarise, we were able to identify at least two clonally independent and one clonally related RS in the patient. The U-RT1 cell line represents the latter.
Figure 2.
Clonality analyses in DNA of patient tissues and the U-RT1 cell line established. (A) The cell line represents the clone detected in the bone marrow specimen of the CLL. The gastric lymphoma and the lymph node tissue show differing clonal events. (B) Phylogram of the sequence comparison of amplicons produced by family-specific FR1 primer to amplify parts of the rearranged IGH locus. Products using VH2, VH3, VH4, and VH6 FR1 family primers were amplifiable and compared, indicating at least three clonal events that were independent of each other. CLL, chronic lymphocytic leukemia.
Genetic features of U-RT1
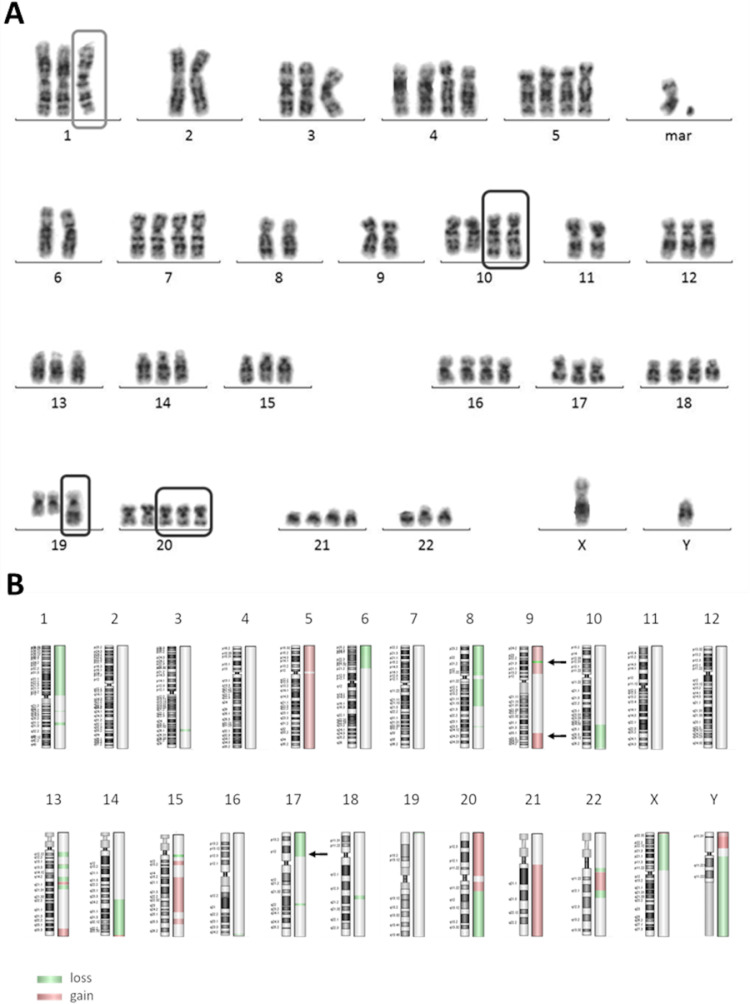
U-RT1 cells have a complex karyotype with >50 chromosomes. In a classical karyogram, aberrations in chromosomes 1, 2, 10, 19, and 20 were observed (Figure 3). The aberration in chromosome 1 was identified as isochromosome 1q. Multicolour FISH analyses revealed the following translocations t(1;11), t(2;19), t(8;17), t(9;22), t(10;15), and t(11;20).
Figure 3.
(A) Karyogram of the U-RT1 cell line. The cell line displays a pseudo-triploid set of chromosomes with recurring abnormalities in chromosomes 1, 10, 19, and 20. (B) Overview of chromosomal aberrations in U-RT1 detected by aCGH. Green marking indicates loss of genetic material in the labelled region, red areas harbour a gain of genetic material in the corresponding chromosomal segment. Aberrations were determined using the ADM-2 algorithm of the Agilent Genomic Workbench. Arrows indicate the location of TP53 (17p13.1), CDKN2A (9p21.3), and NOTCH1 (9q34.3). CLL, chronic lymphocytic leukemia.
Array-based comparative genomic hybridization (aCGH) of U-RT1 also shows a complex pattern of aberrations, with chromosomes 1p, 5, 8, 9, 15q, 20, 21, X and Y being the most affected. These findings confirm the results of the karyogram and mFISH analysis. Interestingly, aberrations that are described as hallmarks of de novo DLBCL development such as BCL-2 and c-Myc rearrangement (Supplementary Figure 1A and B) or driver mutations in the NFkB pathway regulating genes MYD88 or CD79 [4,18,20] are not seen in U-RT1. Instead, aberrations were found in TP53, CDKN2A and NOTCH1, which are 3 of the 4 most frequently affected genes associated with the development of RS [8,18,19]. Figure 3B gives an overview of the chromosomal aberrations found in U-RT1 using aCGH.
FISH analyses using TP53-specific FISH probes showed a loss of one copy of TP53, which was previously seen in the underlying CLL. In addition, U-RT1 lacks both CDKN2A copies. PCRs with CDKN2A-specific primers resulted in an absence of any amplicon, thus verifying the loss of CDKN2A detected in the FISH experiment (Supplementary Figure 1).
BCL-2 inhibitors induce cell death in U-RT1 and DLBCL cell lines
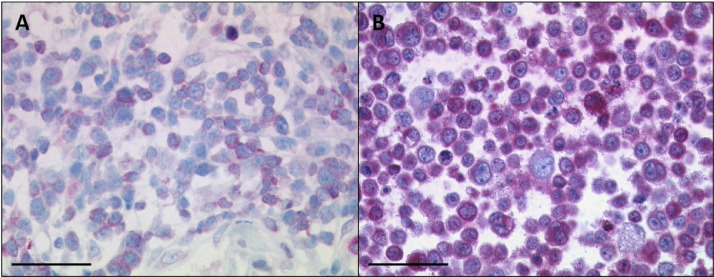
BCL-2 inhibitors are currently in focus for treating CLL [6,30,31]. Immunohistochemistry revealed a strong positivity of BCL-2 in U-RT1 and the lymph node from which the cell line was established (Table 1, Figure 4). Therefore, we tested whether the BCL-2 family inhibitors venetoclax (ABT-199), ABT-737, and HA14-1 were capable of inducing cell death in the U-RT1 cell line (Figure 5 A–C).
Figure 4.
BCL-2 staining of the U-RT1 cell line (B) and the lymph node with the diffuse large B-cell lymphoma from which the cell line was established (A). Both the cell line and the lymphoma show immunoreactivity for the BCL-2 antibody. The black line indicates 50 µm.
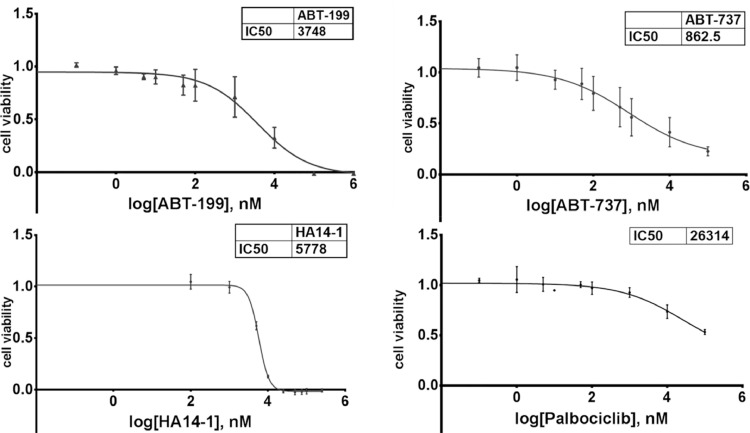
Figure 5.
Inhibition of proliferation, i.e. induction of cell death in U-RT1 cells by treating with various reagents. LD50 was reached using 3748 nM of ABT-199 (A) and 5778 nM HA14-1 (C). 862.5 nM ABT-737 led to the half-maximum inhibition of the cells (IC50; B). No specific inhibition of proliferation was induced with palbociclib (D).
Incubation of U-RT1 cells with increasing amounts of venetoclax led to a dose-dependent reduction of viable cells reaching half-maximum effects with 3.75 µM of the reagent, indicating a nonspecific toxicity.
Treating the cells with ABT-737 led to a maximum reduction in viability to about 16% using high concentrations of 100 µM. Nonetheless, the IC50 of ABT-737 was about 860 nM, pointing to a specific growth inhibition effect even at lower concentrations. Finally, a third BCL-2 inhibitor (HA 14-1) was used to inhibit cellular growth of U-RT1 in vitro. HA14-1 displayed a very small range of action between no effect (up to about 2 µM), half-maximum effect (IC50 = LD50 = 5.8 µM), and 100% dead cells when treated with about 15 µM.
We showed that U-RT1 displays a total genomic loss of the CDKN2A gene (Supplementary Figure 1D) and the corresponding lack of p16 at protein levels whereas CDK4 and Rb were detectable in the cell line (Table 1). This implies a CDK4/6-mediated uncontrolled proliferation and is in line with a responder phenotype of the CDK4/6-inhibitor palbociclib [32]. Interestingly, treating U-RT1 with varying concentrations of this inhibitor did not lead to reduced cellular proliferation (Figure 5D).
Discussion
At the present time, RS is not completely understood. In particular, the prognosis of RS clonally related to the underlying CLL is extremely unfavorable as it shows a very poor response to standard DLBCL treatments. Chigrinova et al. showed that 2 main genetic pathways lead to the transformation of CLL into RS [19]. Inactivation of TP53 and CDKN2A covers about 50% of the cases, with trisomy 12 in another 30%. We showed that one copy of TP53 (with the remaining copy being mutated) and both copies of CDKN2A were lost in U-RT1. Furthermore, aCGH revealed a gain of the NOTCH1 gene locus. This RS cell line can therefore be used as a model system for the majority of RS cases. Differences and commonalities of CLL-related RT and de novo DLBCL play a profound role in establishing new therapies. In our U-RT1 model, we showed that cell survival can be affected by BCL-2 family inhibitors. The patient himself represents a case with clinical and biological features of a high-risk CLL treated with several different regimens. The transformation of the CLL into an aggressive RS fits well with its characterisation as high-risk CLL displaying TP53 and CDKN2A loss. The interesting fact that the patient developed at least two different secondary lymphomas may be explained by the multiple chemotherapies.
The fact that the RS clone which gave rise to the U-RT1 cell line was initially not detectable in the lymph node tissue from which the line was established is interesting. It points to a lower proportion of cellular material of this clone compared with the other lymphomas. This is especially surprising considering the high proliferation capability of U-RT1 as measured by the KI-67 index of >80%. The secondary lymphomas were clearly detectable in the primary cell cultures of the lymph node tissue, but single cell separation resulted in regrowth of the RS clone that was represented in clearly smaller quantities. Subcloning from a single cell stage is evidence for the aggressiveness of a lymphoma. A second subcloning of the U-RT1 cell line revealed a clonal efficacy of about 6% (data not shown) whereas only 0.9% of the primary culture regrew from single cells. This may also be an indication for a reduced percentage of the RS clone in the lymph node.
BCL-2 inhibition is currently being used as a treatment option in CLL. But little is known about the efficacy of BCL-2 family inhibitors in RS. Pham et al. recently tested venetoclax on 26 DLBCL and 10 MCL cell lines [33]. These lines responded in a wide range of inhibitor concentrations ranging from nM to high µM. The authors defined the responder cut-off as 100 nM. Although a reduction of viable cells could be achieved in U-RT1, the amounts of venetoclax (IC50 = 3.75 µM) needed define U-RT1 as a nonresponder to this BCL-2 inhibitor. ABT-737 was developed by Oltersdorf et al. in 2005 as a BH3 mimetic with high affinity binding to BCL-2, BCL-xL and BCL-w [34]. This small molecule displayed cytotoxic effects to B-cell tumor cell lines, cells from CLLs, and small lung cancer cell lines (reviewed by Cory and Adams [35]). Treating multiple myeloma cell lines revealed half-maximum induced cell death in concentrations ranging from 5 to 15 µM, which were 10 times higher than the concentrations needed in U-RT1 [36]. HA14-1 was used to treat follicular lymphoma cell lines [37]. Here, the cell line HF4.9 was the line with the lowest lethal dose of 50% of the cells (LD50) of 4.5 µM and HF1A3 showed the highest tolerance (LD50 = 12.6 µM), positioning U-RT1 (LD50 = 5.8 µM) as a cell-line sensitive to BCL-2 inhibition with HA14-1. Wang et al. described H14-1 to be a competitive BCL-2 inhibitor that reaches its half-maximum binding competition with Bak-BH3 at concentrations about 9 µM and inducing apoptosis in the human myeloid leukaemia cell line HL-60 with an LD50 of about 20 µM [38]. These published data point to a vulnerability of the U-RT1 cells to BCL-2 inhibition. In vitro, the proliferation of U-RT1 was drastically reduced by treating the cells with all 3 BCL-2 family inhibitors. However, the cells had to be classified as nonresponders to the inhibitor with highest specificity to BCL-2 (venetoclax). Induced cell death in the U-RT1 cells may, therefore be due mainly to inhibitory effects on BCL-xL and BCL-w but not to BCL-2.
Treating U-RT1 with the CDK4/6 inhibitor palbociclib surprisingly did not result in a substantial reduction of proliferation although the cells do not express CDKN2A due to complete loss of the gene. Although the loss of the p16 protein was shown on immunohistochemistry and immunocytochemistry, the in vitro proliferation of U-RT1 was only affected at very high inhibitor dosages, known to be cytotoxic in several cell lines tested. Therefore, further cell cycle regulatory mechanisms seem to be dysregulated in U-RT1, giving rise to new studies on cell cycle checkpoint-related aberrations in Richter syndrome.
Although it is obvious that further RS cell lines need to be established, we present the possibility of testing new treatment conditions and searching for novel molecular mechanisms in Richter syndrome using this first cellular model system.
Author contributions
TS, JM, AT, MM and KM performed experiments. JB and ET provided patient data. TS, JM, KM and JB analysed and interpreted the data. SS, PM and KM contributed in design and conception of the work. JM and KM wrote the manuscript. SS, PM, TFEB and JB substantively revised the manuscript. All authors approved the final version of the manuscript.
Footnotes
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest: All authors declare no competing interests.
References
- 1.Richter MN. Generalized reticular cell sarcoma of lymph nodes associated with lymphatic leukemia. Am J Pathol. 1928;4:285–292. 287. [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi D, Gaidano G. Richter syndrome: molecular insights and clinical perspectives. Hematol Oncol. 2009;27:1–10. doi: 10.1002/hon.880. [DOI] [PubMed] [Google Scholar]
- 3.Tsimberidou AM, Keating MJ. Richter syndrome: biology, incidence, and therapeutic strategies. Cancer. 2005;103:216–228. doi: 10.1002/cncr.20773. [DOI] [PubMed] [Google Scholar]
- 4.Rossi D, Spina V, Deambrogi C, Rasi S, Laurenti L, Stamatopoulos K, Arcaini L, Lucioni M, Rocque GB, Xu-Monette ZY. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117:3391–3401. doi: 10.1182/blood-2010-09-302174. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pula B, Salomon-Perzynski A, Prochorec-Sobieszek M, Jamroziak K. Immunochemotherapy for Richter syndrome: current insights. ImmunoTargets Ther. 2019;8:1–14. doi: 10.2147/ITT.S167456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao Z, Quintanilla-Martinez L, Raffeld M, Richter M, Krugmann J, Burek C, Hartmann E, Rudiger T, Jaffe ES, Muller-Hermelink HK. IgVH mutational status and clonality analysis of Richter's transformation: diffuse large B-cell lymphoma and Hodgkin lymphoma in association with B-cell chronic lymphocytic leukemia (B-CLL) represent 2 different pathways of disease evolution. Am J Surg Pathol. 2007;31:1605–1614. doi: 10.1097/PAS.0b013e31804bdaf8. [DOI] [PubMed] [Google Scholar]
- 8.Rossi D, Gaidano G. Richter syndrome: pathogenesis and management. Semin Oncol. 2016;43:311–319. doi: 10.1053/j.seminoncol.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Rossi D, Spina V, Cerri M, Rasi S, Deambrogi C, De Paoli L, Laurenti L, Maffei R, Forconi F, Bertoni F. Stereotyped B-cell receptor is an independent risk factor of chronic lymphocytic leukemia transformation to Richter syndrome. Clin Cancer Res. 2009;15:4415–4422. doi: 10.1158/1078-0432.CCR-08-3266. [DOI] [PubMed] [Google Scholar]
- 10.Parikh SA, Kay NE, Shanafelt TD. How we treat Richter syndrome. Blood. 2014;123:1647–1657. doi: 10.1182/blood-2013-11-516229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parikh SA, Rabe KG, Call TG, Zent CS, Habermann TM, Ding W, Leis JF, Schwager SM, Hanson CA, Macon WR. Diffuse large B-cell lymphoma (Richter syndrome) in patients with chronic lymphocytic leukaemia (CLL): a cohort study of newly diagnosed patients. British journal of haematology. 2013;162:774–782. doi: 10.1111/bjh.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maddocks-Christianson K, Slager SL, Zent CS, Reinalda M, Call TG, Habermann TM, Bowen DA, Hoyer JD, Schwager S, Jelinek DF. Risk factors for development of a second lymphoid malignancy in patients with chronic lymphocytic leukaemia. British journal of haematology. 2007;139:398–404. doi: 10.1111/j.1365-2141.2007.06801.x. [DOI] [PubMed] [Google Scholar]
- 13.Fangazio M, De Paoli L, Rossi D, Gaidano G. Predictive markers and driving factors behind Richter syndrome development. Expert Rev Anticancer Ther. 2011;11:433–442. doi: 10.1586/era.10.237. [DOI] [PubMed] [Google Scholar]
- 14.Eyre TA, Schuh A. An update for Richter syndrome - new directions and developments. Br J Haematol. 2017;178:508–520. doi: 10.1111/bjh.14700. [DOI] [PubMed] [Google Scholar]
- 15.Rossi D, Cerri M, Capello D, Deambrogi C, Rossi FM, Zucchetto A, De Paoli L, Cresta S, Rasi S, Spina V. Biological and clinical risk factors of chronic lymphocytic leukaemia transformation to Richter syndrome. British journal of haematology. 2008;142:202–215. doi: 10.1111/j.1365-2141.2008.07166.x. [DOI] [PubMed] [Google Scholar]
- 16.Hleuhel MH, Ben-Dali Y, Da Cunha-Bang C, Brieghel C, Clasen-Linde E, Niemann CU, Andersen MA. Risk factors associated with Richter's transformation in patients with chronic lymphocytic leukaemia: protocol for a retrospective population-based cohort study. BMJ open. 2019;9 doi: 10.1136/bmjopen-2018-023566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikh SA, Shanafelt TD. Risk factors for Richter syndrome in chronic lymphocytic leukemia. Curr Hematol Malig Repo. 2014;9:294–299. doi: 10.1007/s11899-014-0223-4. [DOI] [PubMed] [Google Scholar]
- 18.Fabbri G, Khiabanian H, Holmes AB, Wang J, Messina M, Mullighan CG, Pasqualucci L, Rabadan R, Dalla-Favera R. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. The Journal of experimental medicine. 2013;210:2273–2288. doi: 10.1084/jem.20131448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chigrinova E, Rinaldi A, Kwee I, Rossi D, Rancoita PM, Strefford JC, Oscier D, Stamatopoulos K, Papadaki T, Berger F. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122:2673–2682. doi: 10.1182/blood-2013-03-489518. [DOI] [PubMed] [Google Scholar]
- 20.Matolcsy A, Chadburn A, Knowles DM. De novo CD5-positive and Richter's syndrome-associated diffuse large B cell lymphomas are genotypically distinct. The American journal of pathology. 1995;147:207–216. [PMC free article] [PubMed] [Google Scholar]
- 21.Allan JN, Furman RR. Current trends in the management of Richter's syndrome. International journal of hematologic oncology. 2018;7:IJH09. doi: 10.2217/ijh-2018-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, Morel P, Van Den Neste E, Salles G, Gaulard P. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346:235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 23.El-Asmar J, Kharfan-Dabaja MA. Hematopoietic Cell Transplantation for Richter Syndrome. Biology of blood and marrow transplantation: journal of the American Society for Blood and Marrow Transplantation. 2016;22:1938–1944. doi: 10.1016/j.bbmt.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang M, Shanafelt TD, Call TG, Ding W, Chanan-Khan A, Leis JF, Nowakowski GS, Bowen D, Conte M, Schwager SM. The efficacy of ibrutinib in the treatment of Richter syndrome. Blood. 2015;125:1676–1678. doi: 10.1182/blood-2014-12-610782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamar Z, Kennedy L, Kennedy B, Lynch M, Goad A, Hurd D, McIver Z. Ibrutinib and rituximab induced rapid response in refractory Richter syndrome. Clinical case reports. 2015;3:615–617. doi: 10.1002/ccr3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He R, Ding W, Viswanatha DS, Chen D, Shi M, Van Dyke D, Tian S, Dao LN, Parikh SA, Shanafelt TD. PD-1 Expression in Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma (CLL/SLL) and Large B-cell Richter Transformation (DLBCL-RT): A Characteristic Feature of DLBCL-RT and Potential Surrogate Marker for Clonal Relatedness. The American journal of surgical pathology. 2018;42:843–854. doi: 10.1097/PAS.0000000000001077. [DOI] [PubMed] [Google Scholar]
- 28.Ding W, LaPlant BR, Call TG, Parikh SA, Leis JF, He R, Shanafelt TD, Sinha S, Le-Rademacher J, Feldman AL. Pembrolizumab in patients with CLL and Richter transformation or with relapsed CLL. Blood. 2017;129:3419–3427. doi: 10.1182/blood-2017-02-765685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 30.Fischer K, Al-Sawaf O, Bahlo J, Fink AM, Tandon M, Dixon M, Robrecht S, Warburton S, Humphrey K, Samoylova O. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. The New England journal of medicine. 2019;380:2225–2236. doi: 10.1056/NEJMoa1815281. [DOI] [PubMed] [Google Scholar]
- 31.Seymour JF, Kipps TJ, Eichhorst B, Hillmen P, D'Rozario J, Assouline S, Owen C, Gerecitano J, Robak T, De la Serna J. Venetoclax-Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. The New England journal of medicine. 2018;378:1107–1120. doi: 10.1056/NEJMoa1713976. [DOI] [PubMed] [Google Scholar]
- 32.von Witzleben A, Goerttler LT, Marienfeld R, Barth H, Lechel A, Mellert K, Bohm M, Kornmann M, Mayer-Steinacker R, von Baer A. Preclinical Characterization of Novel Chordoma Cell Systems and Their Targeting by Pharmocological Inhibitors of the CDK4/6 Cell-Cycle Pathway. Cancer research. 2015;75:3823–3831. doi: 10.1158/0008-5472.CAN-14-3270. [DOI] [PubMed] [Google Scholar]
- 33.Pham LV, Huang S, Zhang H, Zhang J, Bell T, Zhou S, Pogue E, Ding Z, Lam L, Westin J. Strategic Therapeutic Targeting to Overcome Venetoclax Resistance in Aggressive B-cell Lymphomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2018;24:3967–3980. doi: 10.1158/1078-0432.CCR-17-3004. [DOI] [PubMed] [Google Scholar]
- 34.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 35.Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer cell. 2005;8:5–6. doi: 10.1016/j.ccr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Kline MP, Rajkumar SV, Timm MM, Kimlinger TK, Haug JL, Lust JA, Greipp PR, Kumar S. ABT-737, an inhibitor of Bcl-2 family proteins, is a potent inducer of apoptosis in multiple myeloma cells. Leukemia. 2007;21:1549–1560. doi: 10.1038/sj.leu.2404719. [DOI] [PubMed] [Google Scholar]
- 37.Skommer J, Wlodkowic D, Matto M, Eray M, Pelkonen J. HA14-1, a small molecule Bcl-2 antagonist, induces apoptosis and modulates action of selected anticancer drugs in follicular lymphoma B cells. Leukemia research. 2006;30:322–331. doi: 10.1016/j.leukres.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 38.Wang JL, Liu D, Zhang ZJ, Shan S, Han X, Srinivasula SM, Croce CM, Alnemri ES, Huang Z. Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7124–7129. doi: 10.1073/pnas.97.13.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]