Abstract
SUMOylation is an important post-translational modification that participates in a variety of cellular physiological and pathological processes in eukaryotic cells. Sirt2, a NAD+-dependent deacetylase, usually exerts a tumor-suppressor function. However, the role of SUMOylation in cancer cells is not fully known. In this study, we found that SUMOylation can occur in the Sirt2 protein at both lysine 183 and lysine 340 sites. SUMOylation did not affect Sirt2 localization or stability but was involved in P38-mTORC2-AKT cellular signal transduction via direct deacetylation on a new substrate MAPK/P38. SUMOylation-deficient Sirt2 lost the capability of suppressing tumor processes and showed resistance to the Sirt2-specific inhibitor AK-7 in neuroblastoma cells. Here, we revealed the important function of Sirt2-SUMOylation, which is closely associated with cellular signal transduction and is essential for suppressing tumorigenesis in neuroblastoma.
Keywords: Sirt2, SUMOylation, Acetylation, Cell Signaling, Neuroblastoma
Abbreviations
- ADP
Adenosine Diphosphate
- ATP
Adenosine Triphosphate
- CDS
Coding Sequence
- CHX
Cycloheximide
- CPT
Cisplatin
- CQ
chloroquine
- EGF
Epidermal Growth Factor
- FBS
Fetal Bovine Serum
- GST
Glutathione S transferase
- HDAC
Histone Deacetylase
- IC50
50% Inhibiting Concentration
- NAD
Nicotinamide Adenine Dinucleotide
- NAM
Nicotinamide
- PTM
Post-Translational Modification
- SAE
SUMO1 Activating Enzyme
- SENP
SUMO1/Sentrin Specific Peptidase
- SUMO
Small Ubiquitin-like Modifier
- TSA
Trichostatin A
- UBC9
Ubiquitin Carrier Protein 9
- UTR
Untranslated Region
- WT
Wide Type
Introduction
The human Sirt2 protein, a homolog of the budding yeast silent information regulator 2 (Sir2), is a member of the Sirtuin family of NAD+-dependent deacetylases and ADP-ribosyltransferases [1, 2]. Accumulating evidence has revealed that Sirt2 plays an important role in multiple cell events [3], [4], [5], which involve a variety of biological and pathological conditions, including development, neural activity, obesity, and carcinogenesis [2, 6, 7]. Accordingly, various Sirtuin inhibitors have been developed and assessed for their effects on different diseases by targeting specific protein structures, biological activities, or signaling pathways [8, 9].
As an important deacetylation enzyme, Sirt2 requires the inevitable NAD-nicotinamide exchange reaction to deacetylate the specific lysine on its substrates, such as those found in histone H4, α-tubulin, p65, and p53 [10], [11], [12], [13]. Traditionally, Sirt2 maintains genome integrity and suppresses tumorigenesis depending on its deacetylation activity [14]. In addition, it acts as a substrate and can be affected by other enzymes or regulators after post-translational modification (PTM). The histone acetyltransferase p300 is able to acetylate Sirt2 and attenuate its deacetylase activity to enhance the acetylation level of its specific substrates [15]. Sirt2 phosphorylation at Ser331, triggered by cyclin-dependent kinases, inhibits its catalytic activity and decreases the cell motility [16, 17]. However, ERK1/2 interacts with Sirt2 and enhances the stability of Sirt2 protein as well as deacetylase activity, which is probably due to Sirt2 phosphorylation at other sites [18].
SUMOylation is a reversible PTM involved in protein subcellular localization [19], stress response [20], transcriptional repression [21], DNA repair [22], and many other cellular processes [23, 24]. Small ubiquitin-like modifier (SUMO) can be covalently attached to proteins as a monomer or a lysine-linked polymer via the sequential action of E1 complex SAE1/SAE2, E2 (UBE2I/UBC9), and E3 enzymes (PIAS, RANBP2, or CBX4), and can be removed by SUMO1/Sentrin-specific proteases (SENPs). In the Sirtuin family, Sirt1, Sirt3, and Sirt6 have been shown to be SUMOylated and show significant biological and physiological functions in cellular activity or tumor processes. SUMOylation of Sirt1 increases its deacetylase activity and inactivates apoptotic proteins in response to genotoxic stress [25]. Similarly, Sirt6 SUMOylation can also increase its deacetylation activity and exert a tumor suppressive function [26]. However, mitochondrial deacetylase Sirt3 activity can be retrained by SUMOylation, but SENP1 translocated into mitochondria upon metabolic stress activates Sirt3 to reduce fat mass and antagonize high-fat diet-induced obesity [27].
Here, we reported that SUMOylation also occurred within the Sirt2 protein and that it ensured its deacetylase activity to suppress the tumor processes in neuroblastoma. In addition, de novo evidence revealed that Sirt2 was capable of directly deacetylating P38/MAPK and inhibited P38-mTORC2-AKT signaling, which was also closely associated with its SUMOylation function.
Experimental procedures
Cell cultures
Human embryonic kidney 293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Hyclone, Logan, UT, USA) with 10% FBS (BioSun, Shanghai, China) and 1% penicillin-streptomycin (Hyclone). SH-SY5Y cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone) with 10% FBS and 1% penicillin-streptomycin. All cells were cultured at 37°C in a 5% CO2 humidified incubator. Transfection of cells was performed using Hiff Trans Liposomal Transfection Reagent (Yeasen, Shanghai, China). DMEM, RPMI 1640, and penicillin-streptomysin solution were purchased from Hyclone. Polybrene and Cell Counting Kit reagents were purchased from Yeasen.
Antibodies and reagents
Antibodies against HA-Tag Rabbit (C29F4, #3724), HA-Tag mouse (6E2, #2367), acetyl-lysine (#9441), p-P38 (Thr180/Tyr182) (D3F9, #4511), P38(#9212), p-ERK (E-4, #sc-7383), ERK (#4696), p-AKT (Ser473, 193H12, #4058), AKT (#9272), p-mTOR (Ser2448, #2971), mTOR (#2983), p-p70 S6K (Thr421/Ser424, #9204), S6K (#9202), and UBC9 (D26F2, #4786) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against SENP1 (A1260) were purchased from ABclonal (Woburn, MA, USA). Antibodies against Flag-tag (F1804), MG132, and cycloheximide were purchased from Sigma-Aldrich (St. Louis, MO, USA). Antibodies against Sirt2 (#ab67299) and SUMO1 (#Y299) were purchased from Abcam (Cambridge, UK). Puromycin (P8230) and rapamycin (R8140) were purchased from Solarbio (Beijing, China). Regents of SB202190 (#S1077), LY294002 (#S1105), NAD+ (#S2518), trichostatin A (TSA) (#S1045), nicotinamide (NAM) (#S1899), and protease inhibitor cocktail (EDTA-Free,100 × in DMSO) were obtained from Selleck (Houston, TX, USA). The KOD-plus-mutagenesis kit was purchased from Toyobo (Osaka, Japan). Protein A/G magnetic beads were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Ni-NTA beads were purchased from Qiagen (Hilden, Germany).
SUMOylation assays
Three methods were used to determine SIRT2 SUMOylation. (1) Ni-NTA pull-down assay: Briefly, 293T cells were co-transfected with plasmid combination for 48h and were lysed in imidazole containing Buffer 1 (6 M guanidine-HCl, 0.1M NaH2PO4, 0.01M Tris-HCl, 10-mM β-mercaptoethanol, and 5 mM imidazole, pH 8.0). The cell lysates were incubated with Ni-NTA beads at 4°C overnight, and then the beads were successively washed by buffer 2 (8 M Urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, 10mM β-mercaptoethanol, pH8.0), buffer 3 (8 M Urea, 0.1 M NaH2PO4, 0.01M Tris-HCl, 10 mM β-mercaptoethanol, 0.2% TritonX-100, pH6.3), and buffer 4 (8M Urea, 0.1M NaH2PO4, 0.01M Tris-HCl, 10 mM β-mercaptoethanol, 0.1% TritonX-100, pH 6.3). Purified protein was eluted with elution buffer (200 mM imidazole, 0.15 M Tris-HCl, 30% glycerol, 0.72 M β-mercaptoethanol, and 5% SDS, pH 6.7) as previously described [28] and determined by immunoblotting with anti-HA antibody. (2) Immunoprecipitation assay: 293T cells transfected with plasmid combination were harvested and lysed at 48 h in 1% SDS lysis buffer (containing 5 mM EDTA, 5 mM EGTA, 20 mM N-ethylmaleimide, and a complete protease inhibitor cocktail). The cell lysate was diluted in pre-RIPA (20 mM NaH2PO4/Na2HPO4, 150 mM NaCl, 1% Triton-100, 0.5% deoxycholic acid sodium salt) at a ratio of 1:10, and was incubated with 8 μL of Protein A/G Beads and 2 μL of anti-Flag antibody overnight at 4°C. After washing 3 times, the protein samples were subjected to western blotting analysis. (3) GST-pull-down assay: Escherichia coli BL21 (DE3) harboring plasmid pGEX-6p-1/Sirt2 with or without pE1E2S1 construct was stimulated by 0.2 mM IPTG for 10 h at 16°C and was lysed according to the instruction of B-PER Protein Extraction Reagent (Thermo Fisher Scientific). Lysate was then incubated with Glutathione Hicap Matrix (Qiagen) overnight at 4°C. After washing three times with lysis buffer, western blotting was performed to determine the Sirt2 SUMOylation in vitro.
Immunoprecipitation and acetylation assays
HEK293T cells were transfected with HA-tagged P38 with or without Flag-tagged Sirt2 for 48 h and lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.1% SDS, 1 mM EDTA, and a complete protease inhibitor cocktail). For the acetylation assay, HEK293T cells were cultured with 2 μM TSA for 16 h before harvest. Cell lysates were incubated with Protein A/G Mix Magnetic Beads and proper antibodies at 4°C overnight followed by immunoblotting to determine the interaction between P38 and Sirt2 protein, or the acetylation level of P38 in vitro and in vivo.
Cell proliferation assay
The CCK8 kit (Biotool, Shanghai, China) was used to measure the cell proliferation of the constructed stable SH-SY5Y cell lines according to the manufacturer’ s instructions. Briefly, 1000 cells/well were resuspended in 100 μL of medium and seeded into a 96-well plate. After culturing at 24 h intervals for 3 days, the cells were treated with 10% CCK8 solution and incubated at 37°C for one hour. The absorbance at 450 nm was measured with a microplate reader (Thermo Fisher Scientific). The experiments were performed independently in triplicate, and the data are presented as the mean ± S.E.M.
Migration and invasion assays
The migration assay was performed as previously described [29]. Briefly, SH-SY5Y stable cells harboring wild HA-tagged Sirt2 or its mutants were resuspended in 200-μL serum-free RPIM medium. Then, the cell suspension was added into the upper uncoated (4 × 104 cells/well for migration) or Matrigel matrix-coated (8 × 104 cells/well for invasion) chambers (Corning, Corning, NY, USA). The complete medium containing 20% FBS was added to the lower chambers as a chemoattractant at 37°C in humidified 5% CO2. The cells that migrated or invaded to the lower side of the upper chambers were fixed and stained with 0.1% crystal violet solution for counting and analysis. The experiment was performed independently in triplicate, and the data are presented as the mean ± S.E.M.
Soft agar colony formation assay
The designated cells were seeded into soft agar to determine the effect of anchorage-independent growth as described previously [29]. Briefly, 2 mL medium containing 0.6% base agar gel (Ameresco, Framingham, PA, USA) was solidified as the lower layer in 6-well plates. A total of 1000 cells were seeded into 2 mL medium containing 0.35% base agar gel and 5% FBS and layered onto the base in 6-well plates. Cell colonies developed in soft agar were stained with 0.01% crystal violet after 2 wk. The colonies were photographed under an inverted microscope and the number of colonies was counted using ImageJ software.
Xenograft tumor model
The SH-SY5Y cells xenograft tumor model was developed in the back of nude mice as described previously [29]. Five-wk-old male BALB/c nude mice were subcutaneously injected with 100μL of PBS containing 2 × 106 SH-SY5Y cells stably expressing Sirt2-shRNA, HA-Sirt2WT, HA-Sirt2K183R, HA-Sirt2K340R, or HA-Sirt2K183/340R. The wild-type SH-SY5Y cell was used as a mock control. The operated mice were sacrificed at 2 wk after xenograft, and the tumor weight was measured. All animal operations were conducted with the approval and guidance of the Shanghai Jiao Tong University Medical Animal Ethics Committee.
Statistical analysis
All data were analyzed as means ± S.D. for western blotting, or means ± S.E.M for migration, invasion, soft agar, and mouse xenograft experiments. Statistical calculations were performed using SPSS and the graphs were constructed using the GraphPad Prism software. Differences between individual groups were analyzed using a two-tailed and unpaired t test. A P-value <0.05(*), <0.01(**), or <0.001(***) was considered statistically significant.
Results
Human Sirt2 can be SUMOylated in vitro and in vivo
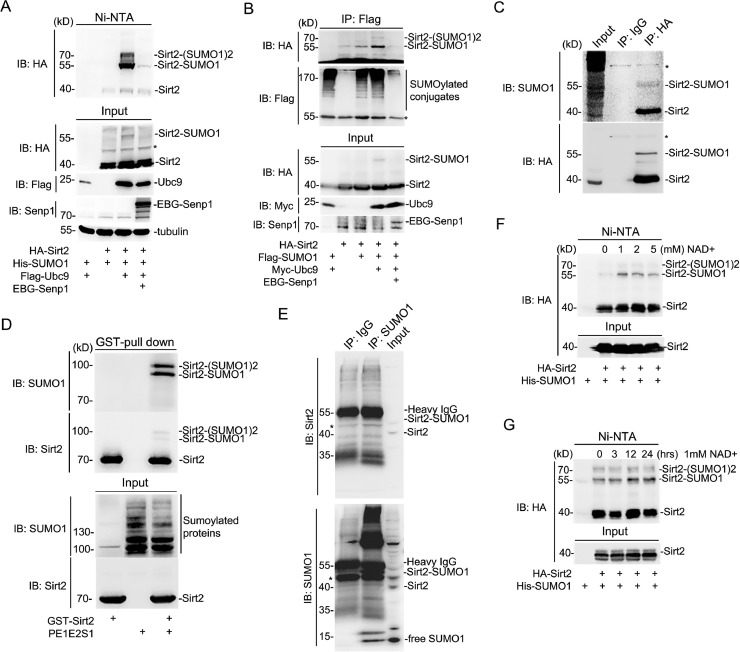
To determine whether Sirt2 can be SUMOylated, HEK293T cells were used to overexpress HA-tagged Sirt2 with or without His-tagged SUMO1 and FLAG-tagged UBC9, cell lysis after transfection was subjected to Ni-NTA pull-down assay and western blot analysis. Two major shifted Sirt2 protein bands were observed and their levels were enhanced or attenuated by the E2 enzyme UBC9 or deSUMOylation enzyme SENP1, respectively (Fig. 1A). Consistent with this, the immunoprecipitation assay also showed the same outcome for Sirt2 SUMOylation events in HEK293T cells (Fig. 1B). To identify the endogenous SUMOylation of the Sirt2 protein, SH-SY5Y cells stably expressing HA-Sirt2 were harvested and subjected to immunoprecipitation with an HA antibody, which showed that Sirt2 SUMOylation could indeed occur in vivo according to the detection by SUMO1 antibody and HA antibody (Fig. 1C). In the prokaryotic expression system, an in vitro GST-pull down assay also proved that GST-tagged SIRT2 could be SUMOylated when co-transfected with SUMO1, E1, and E2 enzymes in the BL21 E. coli host (Fig. 1D). Endogenous SUMOylation on Sirt2 was also identified in SH-SY5Y cells through immunoprecipitation assay with SUMO1 and Sirt2 antibodies (Fig. 1E). In addition to SUMO1 modification, SUMO2 and SUMO3 modifications were also positively identified in HEK293T cells (Fig. S1A).
Fig. 1.
Human Sirt2 can be SUMOylated in vitro and in vivo. (A) His-SUMO1 conjugates were isolated with Ni-NTA agarose under denaturing conditions and immunoblotted with antibodies against mouse-HA, mouse-Flag or rabbit-SENP1. (B) Cell extracts were immunoprecipitated with mouse-Flag antibody and immunoblotted with antibodies against mouse-HA, mouse-Flag, mouse-Myc or rabbit-SENP1. (C) SH-SY5Y cells overexpressing HA tagged Sirt2 were extracted and cell lysis was immunoprecipitated with mouse-HA or normal mouse IgG antibodies and immunoblotted with antibodies against rabbit-SUMO1 or mouse-HA. The light-chain-specific HRP-conjugated anti-mouse/rabbit IgG was used for the second antibody for immunoprecipitation assay. (D) SUMOylation reaction was carried out in DE3 bacterial cells expressing GST tagged Sirt2. Bacterial extracts were immunoblotted with antibodies against rabbit-SUMO1 or rabbit-Sirt2 after isolation with GST agarose. (E) SH-SY5Y cell extracts were used for immunoprecipitated with antibodies against rabbit-SUMO1 and immunoblotted with rabbit-Sirt2 antibody. (F and G) 293T cells expressing HA-Sirt2 and His-SUMO1 were treated with gradient concentration NAD+ (0, 1, 2, or 5 mM) for 24 h or treated with 1 mM NAD+ for different time periods (0, 3, 12 or 24 h). Cell extracts were immunoblotted with antibody against mouse-HA after the method of Ni-NTA pull down. The asterisks represent nonspecific bands. GST, Glutathione S transferase.
As Sirt2 is a NAD+-dependent deacetylase, we wondered whether Sirt2 SUMOylation could be stimulated by NAD+ in cells. Surprisingly, 1 mM NAD+ obviously enhanced the Sirt2 SUMOylation level, but higher NAD+ concentration would not help further improvement (Fig. 1F). However, it had a significant and positive time effect on Sirt2 SUMOylation under 1mM NAD+ stimulation (Fig. 1G). In contrast, EGF, H2O2, or even cisplatin (CPT) did not affect the SUMOylation level of Sirt2 (Fig. S1B–D). Together, these evidences indicate that Sirt2 can be SUMOylated in vitro and in vivo, which can be specifically stimulated by NAD+.
Sirt2-SUMOylation does not affect the localization and stability of Sirt2 in cells
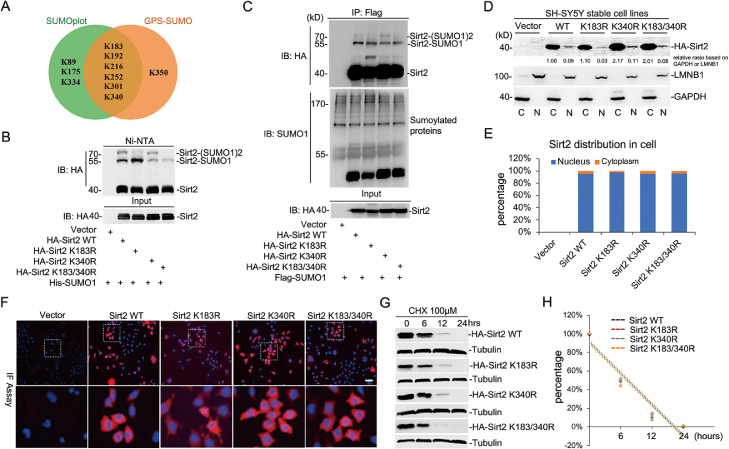
To determine which lysine was conjugated by the SUMO1 molecule, SUMOplot Analysis Program (http://www.abgent.com.cn/tools/sumoplot) and GPS-SUMO 2.0 Online Service (http://sumosp.biocuckoo.org/online.php) were applied to predict the probability of SUMOylation sites within Sirt2 protein (Fig S2A). There were six potential SUMOylation sites predicted by both softwares with different system approaches [30], including Lys183, Lys192, Lys216, Lys252, Lys301, and Lys340 (Fig. 2A and Fig. S2A). Sequentially, substitution of this lysine (K) to arginine (R) revealed that both K183R and K340R significantly attenuated the SUMOylation level of Sirt2 (Fig. S2B and Fig. 2B). Intriguingly, the K183R mutation within the Sirt2 protein notably removed a shifted protein band but enhanced the level of another one; however, the K183R and K340R double mutations caused a significant decline in the total Sirt2 SUMOylation level according to both the Ni-NTA pull-down and immunoprecipitation assays (Fig. 2B and C).
Fig. 2.
Sirt2-SUMOylation does not affect the localization and stability of Sirt2 in cells. (A) Putative SUMOylation sites of human Sirt2 predicted by SUMOplot and GPS-SUMO software were shown in the pie chart. (B and C) 293T cells were co-transfected with His-SUMO1/Flag-SUMO1 and HA-tagged Sirt2 or its mutants at K183 and K340. SUMOylated Sirt2 was isolated by Ni-NTA agarose or immunoprecipitated with mouse-Flag antibody and detected with antibodies against mouse-HA or -Flag using the method of western-blot. (D and E) SH-SY5Y cell lines stably expressing HA-tagged Sirt2 or its mutants were extracted by the Nuclear/Cytosol Fractionation kit. Cell extracts were subjected to immunoblot. Sirt2 distribution in cells was calculated by Image J software. (F) The localization of HA-Sirt2K183R, HA-Sirt2K340R and HA-Sirt2K183/340R in SH-SY5Y cells were shown by immunofluorescence staining with the anti-HA antibody. Nuclear DNA was stained with DAPI. (G) SH-SY5Y cells expressing HA-Sirt2K183R, HA-Sirt2K340R and HA-Sirt2K183/340R were treated with 100 μM CHX for different time periods (0, 6,12 or 24 h). Mixture extracts were immunoblotted by antibodies against HA and Tubulin. (H) Its half-life period is shown as the line graph according to the quantification of protein level by using Image J software. CHX, Cycloheximide.
To assess whether SUMOylation can affect Sirt2 localization and stability in the cell, HA-tagged Sirt2 wide type or mutants were cloned into the pGreenPuro-Dual vector (see description in Fig. 3G) and transferred into SH-SY5Y cells through lentiviral infection, respectively, and the expression level was assessed by western blotting (Fig S2C). The nucleus/cytosol extraction assay revealed that SUMOylation did not alter the localization of Sirt2, regardless of K183, K340, or K183+340 (Fig. 2D and E). Moreover, the immunofluorescence assay also showed a consistent outcome (Fig. 2F). Sirt2 protein stability was controlled by the proteasome system, but not the lysosomal pathway, according to the protein level detection in cells after MG132 or chloroquine treatment (Fig. S2D). However, SUMOylation on Sirt2 did not significantly affect its stability by detecting the protein half-life after cell treatment with cycloheximide (Fig. 2G). Taken together, these data indicate that both K183 and K340 are the main SUMOylation sites in Sirt2, but the SUMOylation on Sirt2 impacts neither of its localization nor stability in neuroblastoma cells.
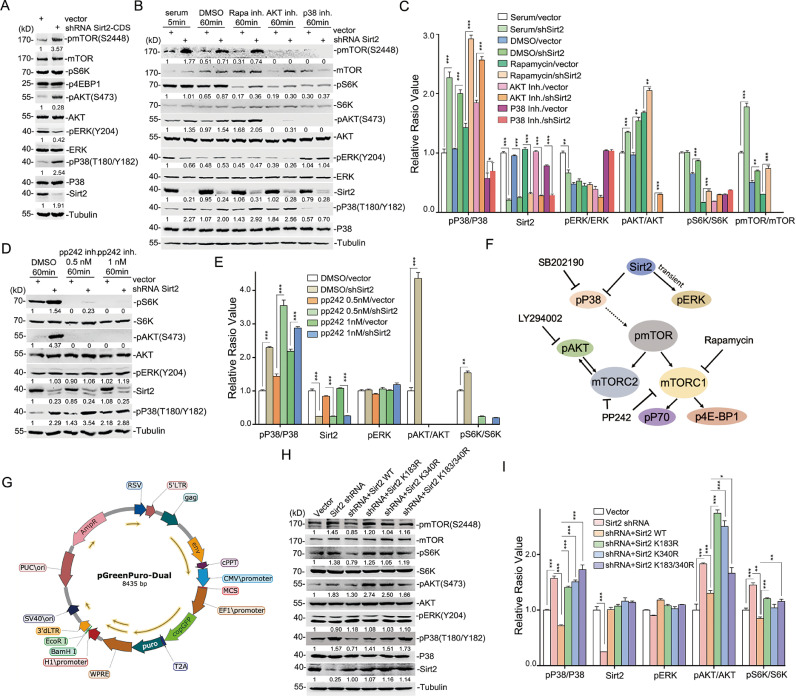
Fig. 3.
Sirt2-SUMOylation mainly suppresses P38-mTORC2-AKT signaling in neuroblastoma cells. (A) SH-SY5Y cells infected with lentiviral expressing shRNA targeting to Sirt2 3’-UTR or control were starved for 2 h and then stimulated by complete culture medium for 5 min. Cell lysis was collected and resolved by SDS-PAGE for western-blot analysis. (B–E) SH-SY5Y cells infected with lentiviral expressing Sirt2 shRNA or control were treated with mTORC1 inhibitor Rapamycin (20 ug/mL), AKT inhibitor LY294002 (50 μM), P38 inhibitor SB202190 (20 μM), pp242 (0.5 or 1 μM) or dimethylsulfoxide (DMSO) as a control for 60 min. Cell lysis was performed and visualized by SDS-PAGE for western-blot analysis (B and D). Protein levels were quantified by Image J software and showed as a graph (C and E). (F) A snapshot of the relationship of P38-mTOR-AKT signal mediated by Sirt2. (G) The homemade lentivector contained CMV promoter for overexpressing foreign gene, H1 promoter for expressing shRNA and EF1a promoter for Puromycin and EGFP expression. The vector map was drawn by using DNAMAN software. (H and I) Cell lines of SH-SY5Y harboring silenced endogenous Sirt2 with or without expressing exogenous wild-type Sirt2 or its mutants were harvested for western blot analysis (H). Protein levels were quantified by Image J software and are shown as a graph (I). All the western-blot analysis was repeated at least 3 times. *P < 0.05, **P < 0.01, ***P < 0.001.
Sirt2-SUMOylation mainly inhibits P38-mTORC2-AKT signaling in neuroblastoma cells
As an important deacetylation enzyme, Sirt2 participates in various cell activities such as aging, cell signal transduction, and carcinogenesis [31]; however, its signaling mechanisms in cells are still not fully understood. Considering this, we silenced the expression of Sirt2 in SH-SY5Y cells through the pGreenPuro-dual lentiviral system with the specific shRNA. pP38, pAKT, and pmTOR were significantly enhanced, but pERK was short-term attenuated when Sirt2 expression was abrogated after starvation following complete culture medium stimulation (Fig. 3A left two lanes in 3B, Fig. S3A). mTORC1 and mTORC2 are the two main sensors of mTOR signaling, while the activation of pS6K (pP70)/p4E-BP1 or pAKT reflects the mTORC1 or mTORC2 signaling pathway, respectively. Here, Sirt2 silencing did not significantly enhance the activation of p4E-BP1, but only slightly affected pS6K (Fig. 3A, Supplemental Fig. S3A). These data indicated that Sirt2 was mainly involved in P38, AKT, and mTORC2 signal transduction in neuroblastoma cells. To clarify the regulation role between P38 and mTORC2/AKT caused by Sirt2 abrogation, several inhibitors were applied for further analysis (Fig. 3B–E). Rapamycin, a mTOR-specific inhibitor, inhibited mTORC1 activation but had no effect on mTORC2, and it did not affect the levels of pP38, pmTOR, and pAKT under the Sirt2 silencing conditions (Fig. 3B middle 4, 5 lanes and C). In addition, AKT inhibitor LY294002 did not affect P38 activation, but significantly abolished the pmTOR and pS6K levels (Fig. 3B right, 3,4 lanes, and C). Intriguingly, both pAKT and pmTOR were suppressed when the P38 signal was blocked by SB202190 (Fig. 3B right 1, 2 lanes and C). In addition, pp242, an inhibitor of both mTORC1 and mTORC2, was used to confirm that the mTOR signal did not affect the Sirt2-associated P38 activity (Fig. 3D and E). These data indicated that Sirt2 in neuroblastoma mainly suppressed P38-mTORC2-AKT signaling (Fig. 3A–E). The strategy of cell signal transduction mediated by specific inhibitors showed that Sirt2 could only slightly or temporarily affect mTORC1 or ERK signaling, but was mainly responsible for suppressing the P38-mTORC2-AKT signaling in neuroblastoma (Fig. 3F).
To rescue Sirt2 expression in SH-SY5Y cells harboring silenced endogenous Sirt2 for reobserving signal transduction, we constructed a homemade lentivector based on pGreenPuro shRNA lentivector (System Biosciences, Heidelberg, Switzerland), termed pGreenPuro-Dual. This construct contained two independent expression cassettes that generated noncoding RNA transcripts (i.e., shRNA, miRNA, or lncRNA) by the H1 promoter, and produced mRNA for translation by the CMV promoter (Fig. 3G). This all-in-one lentiviral vector can be used for recombinant lentivirus packaging to silence endogenous genes and express exogenous genes (i.e., wild-type or mutants) with only one infection force on the target cell, which avoids the uncontrollable changes in cellular behavior caused by multiple viral infections and drug selections.
Then, the SH-SY5Y cells were subjected to infection with recombinant lentivirus harboring shRNA targeting the endogenous Sirt2-3’UTR and rescued exogenous HA-Sirt2-WT, -K183R, -K340R or -K183/340R, and cells infected with virus generated by pGreenPuro-Dual vector or pGreenPuro-Dual/shRNA were set as the system control. After puromycin selection, the endo- or exo-Sirt2 protein expression levels in these cell lines were quantified by western blotting (Fig. S3B). Surprisingly, reintroduced Sirt2-WT suppressed the activation of P38, mTOR, and AKT resulting from endo-Sirt2 ablation, while SUMOylation-deficient Sirt2 (K183R, K340R, or K183/340R) did not resume normal signal transduction in SH-SY5Y cells (Fig. 3H and I). Taken together, these results indicated that Sirt2-SUMOylation was critical in inhibiting the P38-mTORC2-AKT signaling pathway in neuroblastoma.
Sirt2 is a promised deacetylase of MAPK/P38 and its enzyme activity relates to SUMOylation
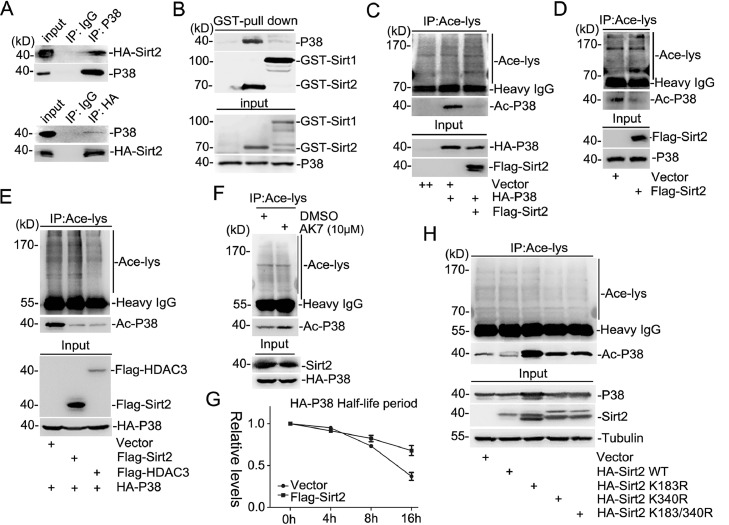
Acetylation of lysine residues within proteins is relevant to the physiological activities of the targeted proteins. P38 is reversibly acetylated by PCAF/P300 and deacetylated by HDAC3, which regulates its affinity for ATP binding and P38 kinase activity [32]. However, there is no evidence yet to illuminate the relationship between Sirt2 and P38. To investigate whether Sirt2 could directly regulate P38, the immunoprecipitation assay revealed that Sirt2 could interact with P38 in cells (Fig. 4A). Consistent with this, the GST-pull down assay also proved that GST-Sirt2 recombinant protein could specifically bind to endogenous P38 in SH-SY5Y cells, but GST-Sirt1 protein could not (Fig. 4B). As one of the deacetylation enzymes, we wondered whether Sirt2 could regulate the acetylation level of P38. The immunoprecipitation assay showed that Sirt2 could obviously decrease both exogenous and endogenous P38 acetylation levels (Fig. 4C and D). Compared with the known deacetylase HDAC3, Sirt2 showed a similar deacetylation ability of P38 protein (Fig. 4E). Moreover, the P38 acetylation level was obviously enhanced by treatment with the Sirt2-specific inhibitor AK-7 (Fig. 4F). Traditionally, acetylated proteins are more stable compared to their unacetylated counterparts because unacetylated lysines are usually targeted for ubiquitination-mediated proteasomal degradation [33]. Considering this, we compared the half-life of P38 with or without Sirt2 expression. The results showed that deacetylated P38 was more stable than acetylated one (Fig. 4G and Fig. S3C), which was consistent with the observation that Sirt2 deficiency can stabilize NFATc2 to enhance its transcription activity [34]. To assess whether Sirt2-SUMOylation could affect its deacetylation activity, SH-SY5Y stable cell lines harboring Sirt2-WT or -mutants were used for immunoprecipitation with acetyl-lysine antibody and the acetylated-P38 level indicated that SUMOylation significantly impaired the deacetylation ability of Sirt2 (Fig. 4H). Thus, these results together suggested that Sirt2 is a promised deacetylase of P38, and the enzyme activity is closely associated with its SUMOylation.
Fig. 4.
Sirt2 is a promising deacetylase of MAPK/P38 and its enzyme activity relates to SUMOylation. (A) HEK293T cells were transfected with HA-Sirt2. Cell extracts were immunoprecipitated for western blot analysis. (B) GST-Sirt1 or -Sirt2 was expressed in DE3 bacterial cells. Purified recombinants by GST-pull down and was immunoblotted with GST or P38 antibodies. (C) Exogenous Flag-Sirt2 and HA-P38 were co-expressed in HEK293T cells. Cell lysis was subjected to immunoprecipitation with antibody against Ace-lysine and immunoblotted with HA, Flag, or Ace-lysine antibodies. (D) HEK293T cell extracts with or without overexpressed Flag-Sirt2 were immunoprecipitated with Ace-lysine or normal IgG antibody and immunoblotted with Flag, P38, or Ace-lysine antibodies. (E) HEK293T cells co-transfected with HA-P38 and Flag-Sirt2/Flag-HDAC3 or its vector were harvested, immunoprecipitated with Ace-lysine antibody and immunoblotted with antibodies against HA, Flag, or Ace-lysine. (F) HEK293T cells expressing HA-tagged P38 were treated with AK7 (10 μM) or DMSO for 24 h. Cell extracts were immunoprecipitated with Ace-lysine antibody and immunoblotted with antibodies against HA, Sirt2, or Ace-lysine. (G) HEK293T cells expressing HA-P38 with or without exogenous Flag-Sirt2 were treated with 100 μM CHX for different time periods, and then were harvested for western blot analysis. The degradation rate of P38 is quantified as the line diagram. (H) HEK293T cells were transfected with or without wild-type HA-Sirt2 or its mutants were harvested and extracted for immunoprecipitation with Ace-lysine antibody and immunoblotted with P38, HA, tubulin, or Ace-lysine antibodies. CHX, Cycloheximide; GST, Glutathione S transferase.
Sirt2-SUMOylation is critical in suppressing the phenotypes of neuroblastoma cells
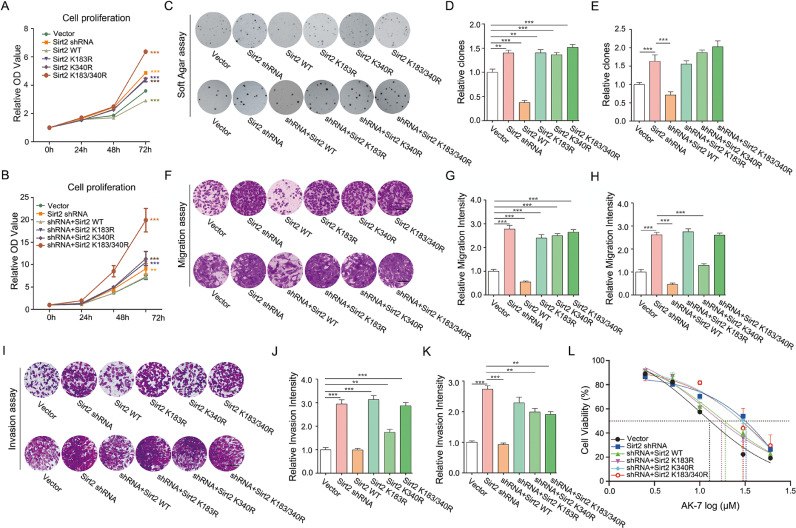
To assess the phenotypes induced by Sirt2-SUMOylation in neuroblastoma cells, we first detected the proliferation ability of SH-SY5Y cell lines with silenced Sirt2 or directly overexpressed Sirt2 variants (i.e., WT, K183R, K340R, or K183/340R), which showed Sirt2 ablation or Sirt2-SUMOylation deficiency significantly facilitated the proliferation capability, but Sirt2-WT showed an opposite effect (Fig. 5A). Meanwhile, reapplied Sirt2-WT, but not Sirt2-K183R, -K340R, or -K183/340R, re-established the proliferation in Sirt2-silenced cells (Fig. 5B). According to the soft-agar colony-forming assay, knockdown of Sirt2 or overexpression of Sirt2 mutants (K183R, K340R, or K183/340R) enhanced clone formation, yet overexpressed Sirt2-WT had significantly suppressed clone formation (Fig. 5C upper panel and D). Similarly, re-applied Sirt2 variants in Sirt2-deficient cells showed almost the same outcomes (Fig. 5C lower panel and E). To inspect the cell mobility and invasiveness resulting from Sirt2-SUMOylation, migration and invasion assays were conducted using the cell chamber in 24-well plates. The results revealed that Sirt2 was able to significantly suppress the migration and invasion abilities of SH-SY5Y cells (Fig. 5F-K). However, SUMOylation-deficient Sirt2, whatever overexpression (Fig. 5F, I upper panel and G, J) or rescued expression (Fig. 5F, I lower panel and H, K), intensified the cell capabilities of migration and invasion in vitro. In addition, we wondered whether Sirt2-SUMOylation affected drug susceptibility, and the IC50 of CPT and AK-7 were detected in the cell lines mentioned above. The results showed that CPT, a frequently used chemical drug for neuroblastoma, did not have any effect on SH-SY5Y cells with or without Sirt2 or its mutants compared to the control (Fig S3D). While AK-7 proved to be able to inhibit Sirt2 activity and enhance the phosphorylation of P38 in SH-SY5Y according to our previous report [35], there was a significant difference in drug-effect between SH-SY5Y cells harboring Sirt2-WT and those harboring Sirt2-mutants (K340 mainly) (Fig. 5L). Together, these data revealed that Sirt2-SUMOylation played an important role in inhibiting the malignant phenotypes in neuroblastoma cells and contributed to the cell sensitivity to the Sirt2-specific inhibitor AK-7 in vitro.
Fig. 5.
Sirt2-SUMOylation is critical in suppressing the phenotypes of neuroblastoma cells. (A–H) SH-SY5Y cells lines stably over-expressing HA-tagged Sirt2-WT, -mutants or shRNA targeting to Sirt2 3’-UTR were constructed with the pGreenPuro-Dual lentiviral system. The cell line harboring vector was set as the negative control. SH-SY5Y cells lines stably expressing shRNA targeting to Sirt2 3’-UTR or co-expressing shRNA and Sirt2-WT or -mutants were also generated by pGreenPuro-Dual lentiviral system. The cell line harboring vector or shRNA was set as system control. Cell proliferation was detected with CCK-8 proliferation assay (A and B). 1000 SH-SY5Y cells were seeded in the top layer with 0.35% agar for clone formation assay. The photos were taken at 10 days after seeding. The number of colonies was counted by Image J and analyzed by SPSS software (C–E). 4 × 104 SH-SY5Y cells were seeded in the upper uncoated chamber. The photos were taken after 0.1% crystal violet staining at 24 h post cell seeding. The number of cells was assessed by Image J and analyzed by SPSS software (F–H). A total of 8 × 104 SH-SY5Y cells were seeded in the upper matrix-coated chamber. The photos were taken after 0.1% crystal violet staining at 48 h post cell seeding. The number of cells was assessed by Image J and analyzed by SPSS software (I–K). (L) Cell viability was detected by treatment with a gradient concentration of AK7 for 48 hours. All the experiments were repeated at least 3 times. *P < 0.05, **P < 0.01, ***P < 0.001.
Sirt2-SUMOylation exerts tumor-suppressor property in mice and Sirt2 expression is closely related to patient prognosis
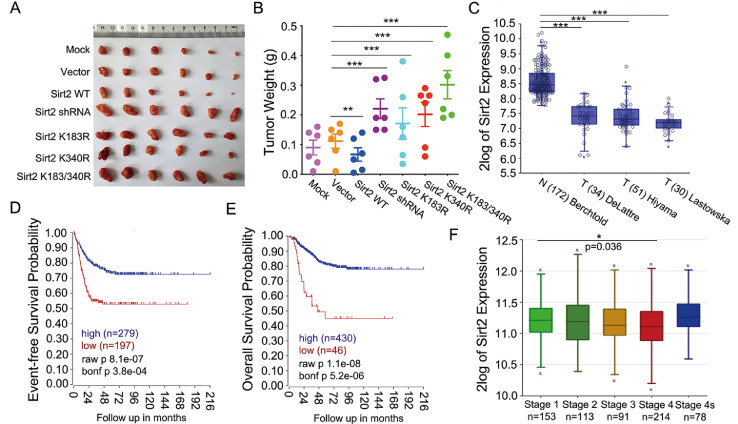
In the xenograft mouse model, the tumor size or weight was measured at 2 wk after subcutaneous injection. Compared with the vector or mock group, both the weight and size of the tumors from the Sirt2-deficient or -mutant cell injection group were much greater. However, overexpression of Sirt2-WT significantly suppressed tumor growth (Fig. 6A and B). To trace expression changes of Sirt2 in the development and progression of neuroblastoma, a series of statistical analyses were performed using the R2 online database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Compared to normal brain tissue, Sirt2 expression was significantly suppressed in tumors from three different datasets (Fig. 6C). The survival curve also revealed that lower Sirt2 expression predicted a poor survival probability, both event-free and overall (Fig. 6D and E). Moreover, with the increase in tumor pathological stage, the expression of Sirt2 showed a downward trend according to the R2 dataset analysis, particularly in stage 4 (Fig. 6F). Moreover, neuroblastoma from stage 4s, which is similar to those from stages 1 and 2 in molecular characteristics [36], showed the same Sirt2-expression pattern (Fig. 6F). Taken together, these results indicated that Sirt2 expression was closely associated with the development of neuroblastoma and patients’ prognosis. SUMOylation on Sirt2 seemed essential for suppressing the tumorigenesis of neuroblastoma in mice.
Fig. 6.
Sirt2-SUMOylation exerts tumor-suppressor effects in mice and Sirt2 expression is closely related to patients’ prognosis. (A and B) Five-wk old nude mice were subcutaneously injected with 2 × 106 SH-SY5Y cells stably expressing Sirt2-shRNA, -WT, or mutants. Mice were injected with naked or lentivector infected SH-SY5Y cell line was set as the mock or control group, respectively. All mice were sacrificed and tumors were dissected, photographed, and weighted at 2 wk (A). The weight of tumors was analyzed by SPSS software (B). (C) R2 online database was used for comparison among normal brain tissue (Berchtold 172 cases) and tumor tissues (DeLattre 34 cases; Hiyama 51 cases; Lastowska 30 cases). (D and E) Kaplan curve representing event-free survival probability (D) or overall survival probability (E) associated with Sirt2 gene expression was generated by R2 online software (Accession: GSE45547, 649 cases). A total of 173 samples lacking survival data were omitted from the analysis. (F) Sirt2 expression levels were compared among patients with different pathological stages (Accession: GSE45547, 649 cases). One way analysis of variance (ANOVA) was used for the statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Neuroblastoma, an embryonal tumor of the autonomic nervous system, is the most common extracranial tumor in children and infants. Nearly half of all neuroblastomas occur in children younger than 2 years of age. Neuroblastoma accounts for approximately 6% to 10% of childhood tumors and 15% of childhood tumor mortality. Neuroblastoma is a neurosecretory tumor that may originate in any ridge of the sympathetic nervous system, typically in the adrenal medulla or paraspinal ganglia [37]. Recent advances in neuroblastoma research have focused on inhibition of the cell signaling pathway, tyrosine kinase activity, differentiation, or immunotherapy, and increasing evidence has revealed that AKT is a clinically relevant target in neuroblastoma [38, 39].
Human deacetylase Sirt2, which is closely related to yeast Hst2p, localizes in the cytoplasm in mammalian cells [10] and regulates various cellular processes by promoting deacetylation of multitudinous substrates in diverse cellular compartments. In cancer, Sirt2 frequently displays tumor suppressor function [31, 40]; however, this intrinsic mechanism is still not clearly understood. PTM is an important process to ensure that proteins exert further cellular functions in eukaryotes. To date, it has been reported that Sirt2 can be phosphorylated or acetylated by altering the deacetylation activity, which directly or indirectly affects the biological function of its substrates [16, 18]. In this work, we identified a novel PTM-SUMOylation on Sirt2 at both K183 and K340 sites, which makes a great contribution to its tumor-suppressor function as well as the cell signal transduction. Because Sirt2 is a NAD+-dependent deacetylase [41, 42], we also revealed that Sirt2-SUMOylation can be specifically and positively regulated by NAD+ in the cell, which logically highlights the importance of SUMOylation to Sirt2 protein function (Fig 1F and G). SUMOylation is called a small ubiquitin-like modifier because of its partial similarity to ubiquitination [43]. It often influences protein stability and subcellular localization [44]. However, we did not observe significant changes in either its localization or stability following a series of point-mutation assays (Fig. 2), which prompted us to speculate the other biological functions of Sirt2-SUMOylation.
It has been reported that Sirt2 is closely associated with the activities of P38 and AKT in a variety of physiological conditions or diseases [45], [46], [47]. Hence, we investigated the regulatory relationship between Sirt2, P38, AKT, and even mTOR by using specific chemical inhibitors in neuroblastoma cells, which showed that mTOR-AKT activation resulting from Sirt2 deficiency mainly relied on P38 activation because the mTOR or AKT signal almost vanished after the blocking of P38 activity (Fig. 3B, right lanes 1 and 2). Similarly, we also demonstrated that Sirt2-mediated AKT activity was completely dependent on mTORC2 signaling in SH-SY5Y cells (Fig. 3B, right 3, 4 lanes & E). However, whether P38 can directly regulate mTOR is still unclear, although there is some evidence about the crosstalk between the P38/MAPK and mTOR signaling pathways in human glioblastoma cells [48]. More interestingly, SUMOylation-Sirt2 significantly affected Sirt2-mediated P38-mTORC2-AKT signaling (Fig. 3H and I), which was important for SUMOylation to Sirt2 function. In addition, we have provided evidence that Sirt2 is a promising deacetylase of substrate P38 and directly impaired the phosphokinase activity of P38 in vitro and in vivo, which was consistent with earlier reports that acetylation of P38 enhances its kinase activity [32]. Likewise, the deacetylation activity of Sirt2 has also been shown to be related to its SUMOylation status (Fig. 4). Based on these considerations, we propose that Sirt2 is involved in regulating the P38-mTORC2-AKT signaling axis through the direct deacetylation of P38, which is largely associated with its SUMOylation function.
Although Sirt2 usually functions as a tumor suppressor in cancer, most of the studies focus on seeking its substrates to reflect the regulation mechanism in the process of tumor development, such as deacetylation of P73 [49] and HIF-1α [50]. However, little research has been conducted on the regulatory mechanism of the intrinsic functions of Sirt2. According to our phenotype analysis in vitro and in vivo, we revealed that Sirt2-SUMOylation played a critical role in promoting the processes of neuroblastoma as well as drug sensitivity to AK-7 in SH-SY5Y cells (Fig. 5, Fig. 6A and B). In the clinical setting, we showed that lower Sirt2 expression was closely associated with tumor occurrence or progression and poor prognosis in patients (Fig. 6C–F). Nevertheless, there are still a number of patients harboring high-Sirt2 expression that present rapid progress and short-survival time. This could be due to the Sirt2-SUMOylation deficiency, which causes the loss of Sirt2 activity and the aberrant activation of AKT. Unfortunately, we did not have enough clinical samples and commercial Sirt2-SUMOylation specific antibodies to detect the association between Sirt2-SUMOylation status and patient survival outcomes.
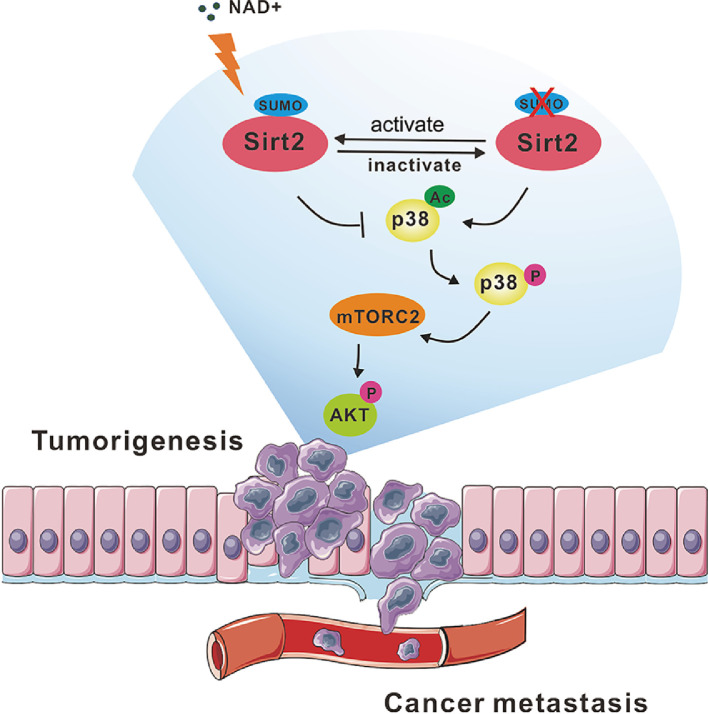
In summary, our findings shed light on a novel biological insight that SUMOylation of Sirt2 is capable of inhibiting the aberrant transduction of the P38-mTORC2-AKT signal through the direct deacetylation effect on a new substrate, MAPK/P38. Sirt2-SUMOylation seemed to function as a ‘brake’ to control P38 acetylation level, helped avoid the excessive activation of P38-mTORC2-AKT, and is essential for its tumor-suppressor function in neuroblastoma (Fig. 7), which may represent an innovative strategy for cancer prognosis or therapy.
Fig. 7.
Graphical abstract. Sirt2 is a NAD+-dependent deacetylase and targets both histone and non-histone protein for deacetylation. NAD+ induced SUMOylation was important for Sirt2 to activate its deacetylase activity. Meanwhile, nonhistone protein MAPK/P38 was revealed as a new substrate of Sirt2 and its acetylation ensured the phosphokinase activity which induced the AKT activation through mTORC2 complex. In neuroblastoma cells, Sirt2-SUMOylation seemed to function as a ‘brake’ to control P38 acetylation level and to avoid the excessive activation of P38-mTORC2-AKT. However, SUMOylation-deficient Sirt2 decreased the capability of deacetylation on P38 and resulted in the aberrant activation of AKT, which ultimately promoted the tumorigenesis or cancer cell metastasis in the development of neuroblastoma.
Conflicts of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare no conflict of interest.
Funding
We acknowledge financial support from the National Natural Science Foundation of China grants 81672708 (to M.X.), 81802563 (to H.Y.), Fundamental Research Program Funding of Ninth People's Hospital affiliated to Shanghai Jiao Tong University School of Medicine to Q.W. (JYZZ045), C.X. (JYZZ074), and H.Y. (JYZZ008G). Fundamental research program funding of Fifth People's Hospital affiliated to Fudan University to W.L. (2019WYFY01).
Author contributions
MX and DW conceptualized, designed and guided the whole work. WL and QW performed most of the experiments, data curation and formal analysis; HY, BC, QF, and RC helped with investigation and methodology; QW and CX collected and analyzed clinical data with software; MX supervised and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.11.013.
Contributor Information
Danhong Wu, Email: danhongwu@fudan.edu.cn.
Ming Xu, Email: mingxu@shsmu.edu.cn, mingxu.msu@gmail.com.
Appendix. Supplementary materials
References
- 1.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. PNAS. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue T, Nakayama Y, Li Y, Matsumori H, Takahashi H, Kojima H, Wanibuchi H, Katoh M, Oshimura M. SIRT2 knockdown increases basal autophagy and prevents postslippage death by abnormally prolonging the mitotic arrest that is induced by microtubule inhibitors. FEBS J. 2014;281:2623–2637. doi: 10.1111/febs.12810. [DOI] [PubMed] [Google Scholar]
- 4.Newton K, Hildebrand JM, Shen Z, Rodriguez D, Alvarez-Diaz S, Petersen S, Shah S, Dugger DL, Huang C, Auwerx J. Is SIRT2 required for necroptosis. Nature. 2014;506:E4–E6. doi: 10.1038/nature13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.North BJ, Rosenberg MA, Jeganathan KB, Hafner AV, Michan S, Dai J, Baker DJ, Cen Y, Wu LE, Sauve AA. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014;33:1438–1453. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D, Jiang X, He H, Liu D, Yang L, Chen H, Wu L, Geng G, Li Q. SIRT2 functions in aging, autophagy, and apoptosis in post-maturation bovine oocytes. Life Sci. 2019;232 doi: 10.1016/j.lfs.2019.116639. [DOI] [PubMed] [Google Scholar]
- 7.Kim AY, Lee EM, Lee EJ, Kim JH, Suk K, Lee E, Hur K, Hong YJ, Do JT, Park S. SIRT2 is required for efficient reprogramming of mouse embryonic fibroblasts toward pluripotency. Cell Death Dis. 2018;9:893. doi: 10.1038/s41419-018-0920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X, Luo X, Hu J, Wang G. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis. 2018;10:9. doi: 10.1038/s41419-018-1260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Xu Z, Lu Y, Qi B, Bai L, Liu W, Zhang C, Jiang Z. Recent Progress on Discovery of Sirt2 Inhibitors for the Treatment of Various Cancers. Curr Top Med Chem. 2019;19:1051–1058. doi: 10.2174/1568026619666190510103416. [DOI] [PubMed] [Google Scholar]
- 10.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 11.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 13.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 14.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20:487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y, Jin YH, Kim YJ, Kang BY, Choi HJ, Kim DW, Yeo CY, Lee KY. Acetylation of Sirt2 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2008;375:576–580. doi: 10.1016/j.bbrc.2008.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, Luscher-Firzlaff J, Vervoorts J, Lasonder E, Kremmer E, Knoll B. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J Cell Biol. 2008;180:915–929. doi: 10.1083/jcb.200707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhang P, Qi GJ, Jiao FJ, Wang QZ, Yan JG, He F, Zhang Q, Lv ZX, Peng X. CDK5-mediated phosphorylation of Sirt2 contributes to depressive-like behavior induced by social defeat stress. Biochim Biophys Acta Mol Basis Dis. 2018;1864:533–541. doi: 10.1016/j.bbadis.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Choi YH, Kim H, Lee SH, Jin YH, Lee KY. ERK1/2 regulates SIRT2 deacetylase activity. Biochem Biophys Res Commun. 2013;437:245–249. doi: 10.1016/j.bbrc.2013.06.053. [DOI] [PubMed] [Google Scholar]
- 19.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Streich FC, Jr., Lima CD. Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature. 2016;536:304–308. doi: 10.1038/nature19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J, Zuo Y, Wang T, Cao Y, Cai R, Chen FL, Cheng J, Mu J. A crucial role of SUMOylation in modulating Sirt6 deacetylation of H3 at lysine 56 and its tumor suppressive activity. Oncogene. 2016;35:4949–4956. doi: 10.1038/onc.2016.24. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Cao Y, Zheng Q, Tu J, Zhou W, He J, Zhong J, Chen Y, Wang J, Cai R. SENP1-Sirt3 Signaling Controls Mitochondrial Protein Acetylation and Metabolism. Mol Cell. 2019;75:823–834. doi: 10.1016/j.molcel.2019.06.008. e825. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, Ke Y, Baron V, Mercola D, Feng GS. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 2009;28:21–33. doi: 10.1038/emboj.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, Zhu C, Chen C, Liu X, Cheng J. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SH, Zhu Y, Ozden O, Kim HS, Jiang H, Deng CX, Gius D, Vassilopoulos A. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl Cancer Res. 2012;1:15–21. [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai VB, Sundaresan NR, Samant SA, Wolfgeher D, Trivedi CM, Gupta MP. Acetylation of a conserved lysine residue in the ATP binding pocket of P38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol Cell Biol. 2011;31:2349–2363. doi: 10.1128/MCB.01205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation. EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarikhani M, Maity S, Mishra S, Jain A, Tamta AK, Ravi V, Kondapalli MS, Desingu PA, Khan D, Kumar S. SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J Biol Chem. 2018;293:5281–5294. doi: 10.1074/jbc.RA117.000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Lu W, Wei Z, Xu M, Liu X. Neuroprotective effect of Sirt2-specific inhibitor AK-7 against acute cerebral ischemia is P38 activation-dependent in mice. Neuroscience. 2018;374:61–69. doi: 10.1016/j.neuroscience.2018.01.040. [DOI] [PubMed] [Google Scholar]
- 36.van Noesel MM. Neuroblastoma stage 4S: a multifocal stem-cell disease of the developing neural crest. Lancet Oncol. 2012;13:229–230. doi: 10.1016/S1470-2045(12)70012-8. [DOI] [PubMed] [Google Scholar]
- 37.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King D, Yeomanson D, Bryant HE. PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol. 2015;37:245–251. doi: 10.1097/MPH.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 39.Opel D, Poremba C, Simon T, Debatin KM, Fulda S. Activation of Akt predicts poor outcome in neuroblastoma. Cancer Res. 2007;67:735–745. doi: 10.1158/0008-5472.CAN-06-2201. [DOI] [PubMed] [Google Scholar]
- 40.McGlynn LM, Zino S, MacDonald AI, Curle J, Reilly JE, Mohammed ZM, McMillan DC, Mallon E, Payne AP, Edwards J. SIRT2: tumour suppressor or tumour promoter in operable breast cancer. Eur J Cancer. 2014;50:290–301. doi: 10.1016/j.ejca.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Lemos V, de Oliveira RM, Naia L, Szego E, Ramos E, Pinho S, Magro F, Cavadas C, Rego AC, Costa V. The NAD+-dependent deacetylase SIRT2 attenuates oxidative stress and mitochondrial dysfunction and improves insulin sensitivity in hepatocytes. Hum Mol Genet. 2017;26:4105–4117. doi: 10.1093/hmg/ddx298. [DOI] [PubMed] [Google Scholar]
- 42.Katsyuba E, Auwerx J. Modulating NAD(+) metabolism, from bench to bedside. EMBO J. 2017;36:2670–2683. doi: 10.15252/embj.201797135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 44.Ritterhoff T, Das H, Hofhaus G, Schroder RR, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes. Nat Commun. 2016;7:11482. doi: 10.1038/ncomms11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M, Xu M, Zhu C, Wang H, Zhao Q, Zhou F. Sirtuin2 enhances the tumoricidal function of liver natural killer cells in a mouse hepatocellular carcinoma model. Cancer Immunol Immunother. 2019;68:961–971. doi: 10.1007/s00262-019-02337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramakrishnan G, Davaakhuu G, Kaplun L, Chung WC, Rana A, Atfi A, Miele L, Tzivion G. Sirt2 deacetylase is a novel AKT binding partner critical for AKT activation by insulin. J Biol Chem. 2014;289:6054–6066. doi: 10.1074/jbc.M113.537266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turdi S, Li Q, Lopez FL, Ren J. Catalase alleviates cardiomyocyte dysfunction in diabetes: role of Akt, Forkhead transcriptional factor and silent information regulator 2. Life Sci. 2007;81:895–905. doi: 10.1016/j.lfs.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Wang F, Zhou ZW, Xia HC, Wang XY, Yang YX, He ZX, Sun T, Zhou SF. Alisertib induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt/mTOR- and P38 MAPK-mediated pathways in human glioblastoma cells. Am J Transl Res. 2017;9:845–873. [PMC free article] [PubMed] [Google Scholar]
- 49.Funato K, Hayashi T, Echizen K, Negishi L, Shimizu N, Koyama-Nasu R, Nasu-Nishimura Y, Morishita Y, Tabar V, Todo T. SIRT2-mediated inactivation of p73 is required for glioblastoma tumorigenicity. EMBO Rep. 2018;19:e45887. doi: 10.15252/embr.201745587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo KS, Park JH, Heo JY, Jing K, Han J, Min KN, Kim C, Koh GY, Lim K, Kang GY. SIRT2 regulates tumour hypoxia response by promoting HIF-1alpha hydroxylation. Oncogene. 2015;34:1354–1362. doi: 10.1038/onc.2014.76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.