Abstract
Introduction: The receptor-binding domain (RBD) in SARS-CoV-2 binds strongly to angiotensin-converting enzyme 2 (ACE2) receptors and causes coronavirus disease 2019 (COVID-19). Antimicrobial photodynamic therapy (aPDT) is a well-established treatment option for the treatment of several viral infections. This in silico study was conducted to target the RBD of SARS-CoV-2 as a target site for aPDT.
Methods: SARS-CoV-2-RBD was selected as a novel target for indocyanine green (ICG) as a photosensitizer during aPDT to exploit its molecular modeling, hierarchical nature of protein structure, and physico-chemical properties using several bioinformatic tools. The binding mode of the RBD to ICG was assessed via protein-ligand docking.
Results: The results of a computational biology analysis revealed that SARS-CoV-2-RBD has 223 amino acids with a molecular weight of 25098.40 Da. RBD is most similar to 6W41 with an E-value of 4e-167, identity of 100%, and query cover of 100%. The aliphatic index of the RBD protein sequences was 71.61, suggesting that the protein is stable in a broad spectrum of temperatures. The predicted structure of RBD showed that it is a protein with a positive charge and a random coil structure (69.51%). Four ligands were modeled in this entry, including one N-acetyl-D-glucosamine (NAG), one glycerol (GOL), and two sulfate ions (SO4 ), to which ICG desires to bind in the molecular docking analysis.
Conclusion: Molecular modeling and simulation analysis showed that SARS-CoV-2-RBD could be a substrate for binding to ICG during aPDT to control the spread of COVID-19.
Keywords: Antimicrobial photodynamic therapy, Bioinformatics tools, Indocyanine green, In silico, SARS-CoV-2
Introduction
The novel and highly pathogenic coronavirus disease 2019 (COVID-19) from Wuhan, China is a critical global public health emergency causing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 Receptor recognition by SARS-CoV-2 is one of the main factors involved in viral infectivity and pathogenesis, which can be used as a major target for vaccination and antiviral strategies.2
The entry of SARS-CoV-2 into host cells is mediated by a virus-surface spike protein.3 This protein contains a receptor-binding domain (RBD) that recognizes its host receptor, angiotensin-converting enzyme 2 (ACE2).4 Inhibition of RBD binding to its receptor not only prevents the virus from entering the host cells but also makes it possible to reduce the proliferation and pathogenicity of the virus.5
No specific treatments, drugs, or biologics are available for the prevention, management, and/or treatment of COVID-19. Light-based technology has been used to prevent and treat different viral infections without causing harm to the host tissue.6 The light-based approach can increase these effects through combination with a photosensitizer as a photosensitizing agent, which is known as antimicrobial photodynamic therapy (aPDT).7
The clinical applications of aPDT as a potentially effective and safe treatment modality for infectious diseases were initially directed against superficial viral diseases including herpes genitalis.8,9 In addition to in vitro studies investigating different viruses, several clinical trials have evaluated the use of systemic and topical aPDT as a treatment approach and adjuvant therapy with surgery, cryotherapy or chemotherapy against viral infections in recent years.10-14 Several studies have revealed that lipid-enveloped viruses are more susceptible to inactivation using aPDT compared to non-enveloped viruses.15 Although there is no evidence specifically supporting the effect of aPDT on viral lipids and/ or proteins, some studies have investigated the effect of ROS on viral lipids.16-18 Since there are viral lipids in the envelope of the COVID-19, it should probably be sensitive to the effects of aPDT. Nevertheless, it is important to identify the relevant viral molecular targets with appropriate sensitivity to aPDT. According to previous studies,19-21 in silico analysis of proteins helps to identify the appropriate substrate and interaction with its receptor to select an efficient photosensitizer in order to enhance the aPDT function. In the present study, the molecular modeling and physico-chemical characteristics of RBD were investigated as a substrate for binding to indocyanine green (ICG) photosensitizer using several biological databases and bioinformatics tools.
Materials and Methods
Molecular Modeling and Validation
Based on the newly released sequence of COVID-19, the amino acid sequence of SARS-CoV-2-RBD was obtained from National Center for Biological Information (NCBI; http://www.ncbi.nlm.nih.gov) and the obtained sequence was verified by Uniprot KB (https://www.uniprot.org/help/uniprotkb). The Protein-Basic Local Alignment Search Tool (BLAST; https://www.ncbi.nlm.nih.gov/blast/) was performed in Protein Data Bank (PDB; www.rcsb.org) to find similar sequences to the query sequence.
Physical and Chemical Characteristics of SARS-CoV-2-RBD
The physical and chemical characteristics of SARS-CoV-2-RBD such as molecular weight (Mw), composition of amino acid, atomic composition, theoretical isoelectric point (PI), estimated half-life, extinction coefficient, aliphatic index, instability index and Grand Average of Hydropathicity (GRAVY) were computed by the ProtParam tool (http://www.expasy.org/cgi-bin/protpraram).
Secondary Structure Predictions of SARS-CoV-2-RBD
PSI-blast based secondary structure prediction (PSIPRED; http://bioinf.cs.ucl.ac.uk/psipred/) and Garnier-Osguthorpe-Robson (GOR) IV (http://cib.cf.ocha.ac.jp/bitool/GOR/) were used to predict the secondary structure of SARS-CoV-2-RBD.
Homology Modeling of SARS-CoV-2-RBD
The modeling of the three-dimensional structure of the protein was performed by SWISS-Model Workspace (https://swissmodel.expasy.org/interactive). After optimization, the three-dimensional model was verified via ERRAT, VERIFY3D, and PROCHECK that are available at the Structure Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/) and ProsA programs (https://prosa.services.came.sbg.ac.at/prosa.php).
Global Model Quality Estimation and Qualitative Model Energy Analysis (QMEAN) scores were used to analyze SARS-CoV-2-RBD. The global quality of the entire model and the local per-residue analysis of different regions within a model were determined by QMEAN.
Phosphorylation Site Prediction
The NetPhos 3.1 server (http://www.cbs.dtu.dk/services/NetPhos/) was used to predict threonine, serine, or tyrosine phosphorylation sites in SARS-CoV-2-RBD using the ensembles of neural networks. Predictions were made for the following 17 kinases: CKI, CKII, EGFR, INSR, ATM, p38MAPK, DNAPK, PKA, PKB, SRC, GSK3, cdk5, PKC, cdc2, RSK, and CaM-II.
Functional Analysis of SARS-CoV-2-RBD
The Protter server (http://wlab.ethz.ch/protter/) was then used to determine the interactive integration and embodiment of annotated and predicted protein sequence features together with experimental proteomic evidence. Moreover, the query sequence was also assessed to find the family of protein using the motif finder server (http://www.genome.jp/tools/motif/). The sites cleaved by chemical agents and/or proteases in SARS-CoV-2-RBD were predicted by the ExPASy peptide cutter (https://web.expasy.org/peptide_cutter/). Also, Scanprosite (https://prosite.expasy.org/scanprosite/) was used to scan SARS-CoV-2-RBD for matches against the PROSITE collection of motifs as well as against its own patterns.
Ligand Geometry for Docking Simulations
The chemical structure of SARS-CoV-2-RBD ligands was obtained from Drugbank. The PDBQT files of these ligands were prepared. Eventually, molecular docking simulations were carried out to examine interactions between SARS-CoV-2-RBD and ICG as a photosensitizer during aPDT using molecular docking (SYBYL-X 2.1 program).
Results
Sequence Retrieval Analysis
From the BLAST results, PDB ID: 6W41 was found with maximum similarity. The total score and E-value for protein alignment between SARS-CoV-2-RBD and 6W41 were 460 and 4e-167, respectively, as well as the identity and query cover between them were 100%. It has been found that SARS-CoV-2-RBD has 223 amino acids with a molecular weight of 25098.40 Da.
Predicted Primary Protein Sequence Characterization of SARS-CoV-2-RBD
The primary structure of SARS-CoV-2-RBD was analyzed using the ExPasy ProtParam tool. The results suggested that the total number of negatively (Glu + Asp) and positively (Lys + Arg) charged residues with the total number of 3482 atoms are 16 and 23 respectively.
The compositions and percentage of amino acids present in SARS-CoV-2-RBD estimated by UniProt software are: Ala (A) 5.4%; Arg (R) 4.9%; Asn (N) 9.4%; Asp (D) 4.0%; Cys (C) 4.0%; Gln (Q) 3.1%; Glu (E) 3.1%; Gly (G) 6.7%; His (H) 0.4%; Ile (I) 4.0%; Leu (L) 6.3%; Lys (K) 5.4%; Met (M) 0.0%; Phe (F) 7.2%; Pro (P) 5.8%; Ser (S) 7.6%; Thr (T) 5.8%; Trp (W) 0.9%; Tyr (Y) 6.7%; Val (V) 9.0%; Pyl (O) and Sec (U): 0.0%.
The amount of light absorbed by the protein at a given wavelength was determined by the extinction coefficient. The extinction coefficient of the SARS-CoV-2-RBD in water was found to be 33850 M-1 cm-1 at 280 nm. In addition, its PI was 8.91.
GRAVY for SARS-CoV-2-RBD was computed to be -0.259. This estimated low range value of the GRAVY index indicates that it is hydrophilic, the possibility of better interaction with water. According to the results, the aliphatic index for the RBD protein sequences was 71.61, which suggested the protein is constant for a broad range of temperatures. The estimated half-life of the RBD protein sequence of SARS-CoV-2 was found to be 1 h (mammalian reticulocytes, in vitro), 2 min (yeast, in vivo), and 2 min (Escherichia coli, in vivo). As a result, the SARS-CoV-2-RBD can be a suitable candidate for aPDT procedure.
Prediction and Characterization of Secondary Structures of SARS-CoV-2-RBD
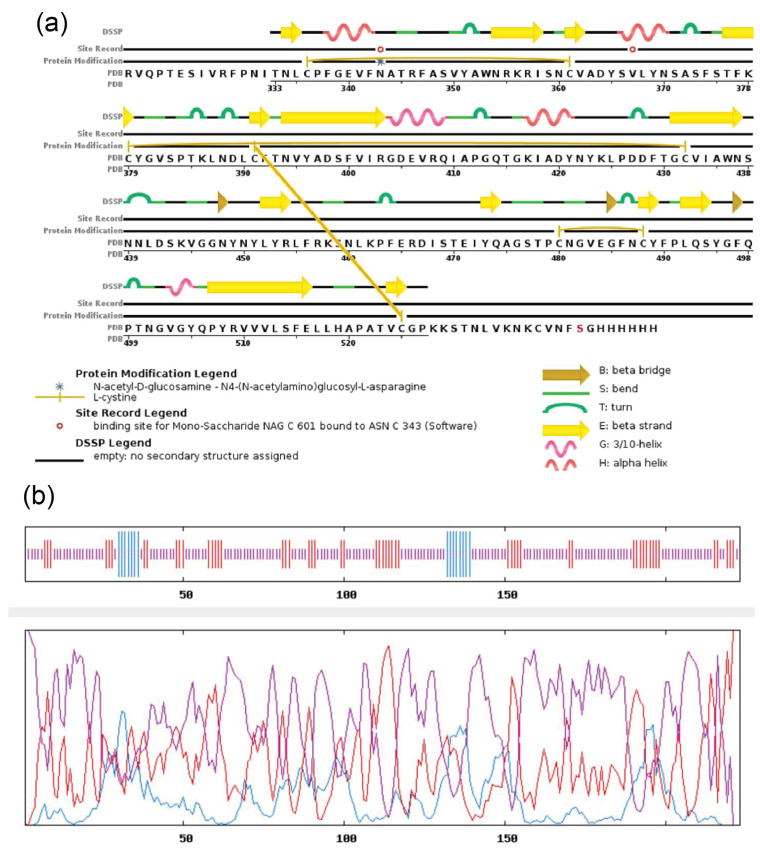
Secondary structure prediction showed the spatial arrangement and location of each amino acid in SARS-CoV-2- RBD (Figure 1a-b). In our designed secondary structure of SARS-CoV-2-RBD, the random coil, extended strand, and alpha helice were 155, 53, and 15 residues, respectively. As shown, random coils in protein structure have the main roles in conformational changes, flexibility, and enzymatic turnover.
Figure 1.
Secondary Structure Prediction of SARS-CoV-2-RBD Using: (a) PSIPRED, (b) GOR IV.
Protein Three-Dimensional Modeling
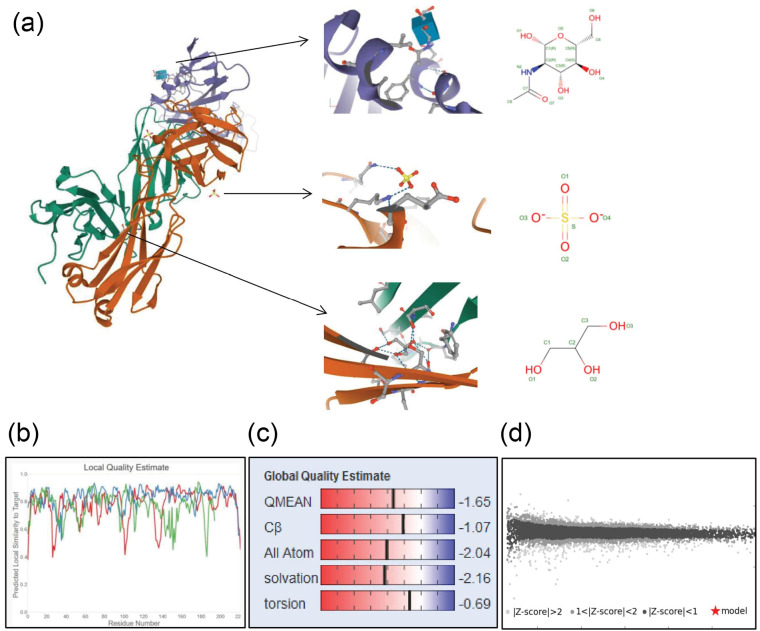
Homology modeling of SARS-CoV-2-RBD was done by a template sequence whose structure has been solved by X-ray diffraction and/or NMR spectroscopy by the program called Swiss model. The selected template sequence for three-dimensional modeling of SARS-CoV-2-RBD was 6W41 with a query sequence of 100% and the resolution of 3.08 angstrom (Ǻ), R-value observed of 0.224, and the oligo-State of homo-trimer (Figure 2). Also, the quality of the structures produced through homology modeling was confirmed using QMEAN. The QMEAN and global scores were -1.65 and 0.86 respectively (Figure 2).
Figure 2.
Bioinformatics Analysis and Computer Simulation Molecular Modeling of SARS-CoV-2-RBD; a) Three-dimensional structures, b) Local model quality overall model quality, c) QMEAN value, d) Comparison with a non-redundant set of SARS-CoV-2-RBD structure.
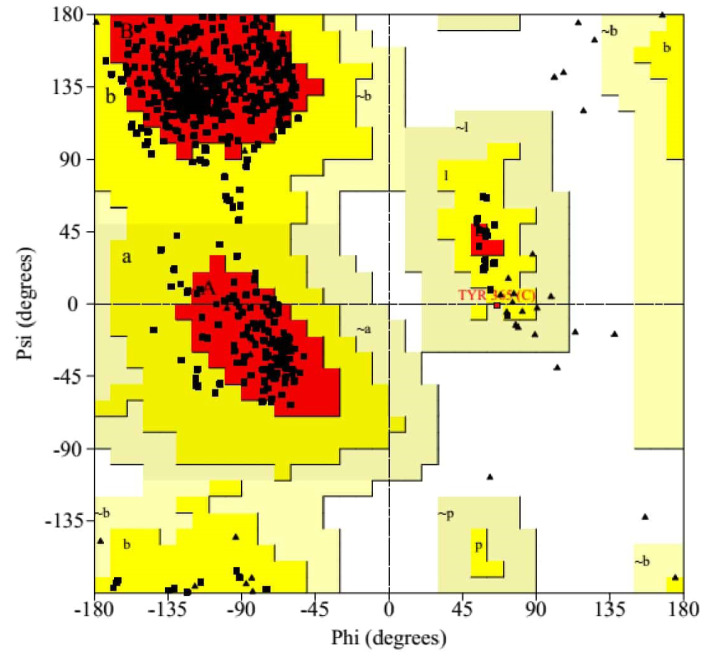
The residues of SARS-CoV-2-RBD were classified according to their regions in the quadrangle in the Ramachandran plot analysis. According to the Ramachandran plot analysis (Figure 3), 86.6% (473/638) of residues were placed in the most favored region [A, B, L]; among them, 72 (13.2%) and 1 (0.2%) residues were present in the additional allowed region [a, b, l, p] and in generously allowed regions [∼a,∼b,∼l,∼p] respectively. There was no residue (0.0%) in disallowed regions. The numbers of end-residues (excl. Gly and Pro), proline, and glycine residues were 5, 36, and 51 respectively.
Figure 3.
Ramachandran Plot of SARS-CoV-2-RBD by the PROCHECK Server.
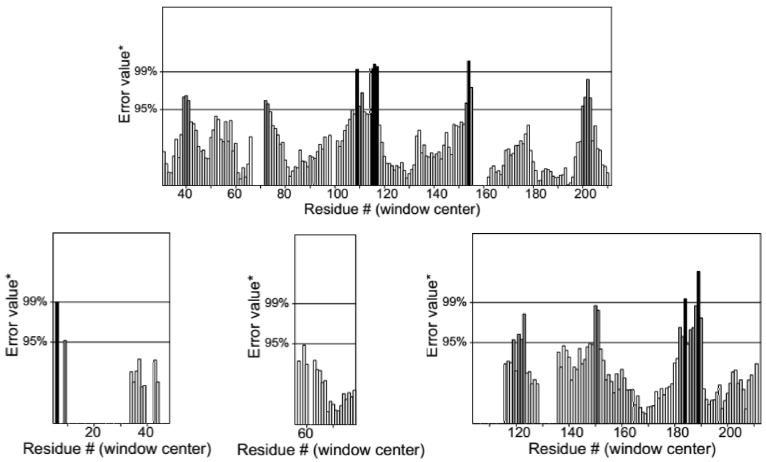
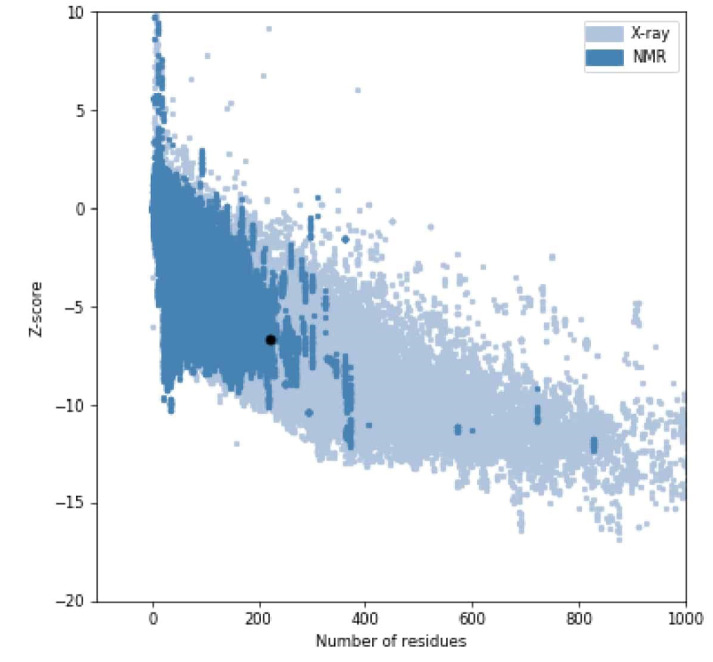
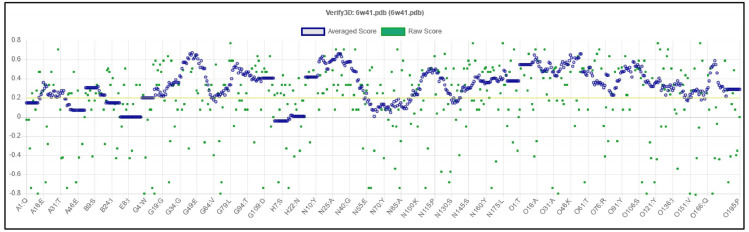
The mean of the overall quality factor of the predicted model was 88.85, which displayed that there were no bonded atomic interactions of the current model in the normal range (Figure 4). Also, the energy profile of the predicted model was consistent with the reliable conformation with the Z-Score of -6.62 based on the results of overall model quality (Figure 5). The compatibility of the three-dimensional structure model with each amino acid sequence was scored by VERIFY3D. The finding in Figure 6 verified the correctness of a protein model by the compatibility of its three-dimensional profile with its sequence.
Figure 4.
Evaluation of the Protein Model of SARS-CoV-2-RBD by ERRAT.
Figure 5.
Overall Model Quality of SARS-CoV-2-RBD With the ProsaWeb Server.
Figure 6.
Evaluation of the Protein Model of SARS-CoV-2-RBD by VERIFY3D.
Functional Analysis of SARS-CoV-2-RBD
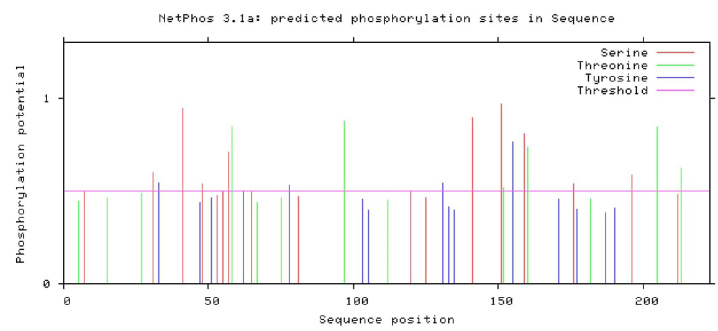
The highest score of phosphorylation site was predicted for serine residue (0.968). After the analysis, the percentage of amino acids composition obtained from the netphosK3.1 server is shown in Figure 7. The server of the ExPASy peptide cutter shows the sequence cleaved by several enzymes (Table 1). The proteinase K cleaved 108 sites of RBD. The positions of cleavage sites were: 2 5 6 8 9 11 14 15 17 20 22 23 24 26 27 29 30 32 33 34 35 40 44 45 47 49 50 51 54 56 58 59 62 64 67 69 72 74 75 77 78 79 82 83 84 88 89 92 93 97 100 101 103 105 107 111 112 115 116 117 118 123 127 131 133 134 135 137 138 143 146 147 150 152 153 154 155 157 160 165 166 168 171 172 174 177 179 182 185 187 190 192 193 194 195 197 198 199 200 202 204 205 206 213 215 216 221 223, while Proline-endopeptidase could cleave only one site in position 145. According to the data, the digestion rate of the protein is good.
Figure 7.
Composition of Amino Acids of SARS-CoV-2-RBD Obtained From the netphosK3.1 Server.
Table 1. The Enzymes That Cleaved the Sequence of SARS-CoV-2-RBD .
| Name of the Enzyme | No. of Cleavages | Positions of Cleavage Sites |
| Arg-C proteinase | 11 | 1 10 28 37 39 85 90 136 139 148 191 |
| Asp-N endopeptidase | 9 | 45 70 79 86 101 108 109 123 148 |
| Asp-N endopeptidase + N-terminal Glu | 16 | 5 21 45 70 79 86 87 101 108 109 123 146 148 152 165 197 |
| BNPS-Skatole | 2 | 35 118 |
| Chymotrypsin-high specificity (C-term to [FYW], not before P) | 31 | 20 24 29 33 35 47 51 56 59 62 74 78 82 103 105 111 118 131 133 135 138 146 155 168 171 177 179 187 190 197 223 |
| Chymotrypsin-low specificity (C-term to [FYWML], not before P) | 45 | 17 20 24 29 33 35 47 50 51 56 59 62 69 72 74 78 82 103 105 111 118 123 131 133 134 135 137 138 143 146 155 168 171 174 177 179 187 190 195 197 199 200 201 215 223 |
| Clostripain | 11 | 1 10 28 37 39 85 90 136 139 148 191 |
| Formic acid | 9 | 46 71 80 87 102 109 110 124 149 |
| Glutamyl endopeptidase | 7 | 6 22 88 147 153 166 198 |
| Hydroxylamine | 2 | 163 183 |
| Iodosobenzoic acid | 2 | 35 118 |
| LysC | 12 | 38 60 68 99 106 126 140 144 210 211 217 219 |
| LysN | 12 | 37 59 67 98 105 125 139 143 209 210 216 218 |
| NTCB (2-nitro-5-thiocyanobenzoic acid) | 9 | 17 42 60 72 113 161 169 206 219 |
| Pepsin (pH1.3) | 45 | 11 16 19 23 24 29 49 50 55 56 58 59 68 69 71 72 73 74 81 82 107 110 111 122 123 133 134 137 145 167 168 172 173 178 194 195 196 197 198 199 200 214 215 222 223 |
| Pepsin (pH>2) | 72 | 11 16 19 23 24 29 32 33 34 35 46 47 49 50 51 55 56 58 59 61 68 69 71 72 73 74 77 78 81 82 102 103 104 105 107 110 111 117 118 122 123 130 131 132 133 134 135 137 145 154 155 167 168 170 172 173 176 177 178 186 189 194 195 196 197 198 199 200 214 215 222 223 |
| Proline-endopeptidase | 1 | 145 |
| Proteinase K | 108 | 2 5 6 8 9 11 14 15 17 20 22 23 24 26 27 29 30 32 33 34 35 40 44 45 47 49 50 51 54 56 58 59 62 64 67 69 72 74 75 77 78 79 82 83 84 88 89 92 93 97 100 101 103 105 107 111 112 115 116 117 118 123 127 131 133 134 135 137 138 143 146 147 150 152 153 154 155 157 160 165 166 168 171 172 174 177 179 182 185 187 190 192 193 194 195 197 198 199 200 202 204 205 206 213 215 216 221 223 |
| Staphylococcal peptidase I | 7 | 6 22 88 147 153 166 198 |
| Thermolysin | 59 | 1 7 8 13 16 19 23 25 28 29 31 33 39 43 44 48 49 53 55 58 63 68 73 76 78 81 82 83 91 99 100 114 115 116 122 126 133 136 137 142 145 156 164 167 173 178 184 191 192 193 194 196 199 203 205 214 215 220 222 |
| Trypsin | 22 | 1 10 28 37 38 39 60 68 85 90 99 106 126 136 139 140 148 191 210 211 217 219 |
As shown in Figure 8, the scanprosite result reveals that there are 223 amino acids and three disulfide bonds with a score of 64.51 are present between 18-43, 61-114, and 73-207. In addition, one functional motif was also detected from the functional study in the case of SARS-CoV-2-RBD. The position and independent E-value of the motif were 12-223 and 2.3e-63 respectively.
Figure 8.
Position of Disulphide Bonds in SARS-CoV-2-RBD.
Molecular Docking Study
4 ligands were modeled in this entry, including 1 N-acetyl-D-glucosamine (NAG), 1 glycerol (GOL), and 2 sulfate ions (SO4). The number of bonds (or angles) for Mogul statistics and the chemical component dictionary in the model are listed in Table 2. The Z score is the number of standard deviations and |Z| > 2 is considered an outlier worth inspection.
Table 2. Properties of SARS-CoV-2-RBD Ligands .
| Mol | Type | Chain | Bond Lengths | Bond Angles | ||||
| Count | RMSZ | # [Z] > 2 | Count | RMSZ | # [Z] > 2 | |||
| 6 | NAG | C | 14,14,15 | 0.29 | 0 | 17,19,21 | 0.56 | 0 |
| 4 | GOL | L | 5,5,5 | 0.89 | 0 | 5,5,5 | 0.97 | 0 |
| 5 | SO4 | L | 4,4,4 | 0.16 | 0 | 6,6,6 | 0.05 | 0 |
| 5 | SO4 | L | 4,4,4 | 0.16 | 0 | 6,6,6 | 0.06 | 0 |
Three-dimensional plots of the interaction networks of the SARS-CoV-2-RBD ligands with ICG were prepared in Figure 9. Blind docking for the whole protein showed a similar pattern of substrate binding at the exposed binding surface as that of RBD.
Figure 9.
Docking Analysis Between Ligands of the Modeled SARS-CoV-2-RBD Doped With ICG; a) Structure of SARS-CoV-2-RBD; b) Structure of ICG; c) Binding of ICG to sulfate ion ligand; d) Binding of ICG to glycerol ligand; e) Binding of ICG to N-acetyl-D-glucosamine ligand.
The results obtained for all ligands suggest that the RBD interacting with the NAG are Leu123, Phe154, Asn172, Gln186, and Ser194. The residues Leu155, Phe185, and Gln193 of RBD have been reported to interact with GOL, whereas residues Asn187 and Ser194 are described to interact with SO4 (Figure 9c-e).
Discussion
The World Health Organization (WHO) is leading global efforts to minimize the spread of COVID-19 and identify gaps to put an end to this outbreak through developing therapeutics.22 According to the literature, there are many reports on the use of aPDT as an effective methodology against viral infections.10-16 It is confirmed that the type and concentration of the photosensitizer, parameters of the light fluence, and subcellular localization of the photosensitizer in the target site play the main roles in the cell death mechanism during aPDT.23 However, in-vivo use of PDT is limited due to hydrophobicity of photosensitizers, poor target specificity, and limited tissue penetration ability.24
In this study, ICG was used as a photosensitizer. It is soluble in water because of its one or two sulfonate side chains. ICG with one or two sulfonate side chains is an anionic, NIR-absorbing fluorescent contrast agent approved by the US Food and Drug Administration (FDA).23 Moreover, the efficacy of ICG-mediated aPDT against different bacterial infections has been confirmed in several studies.24-27
As commented by Namvar et al,28 ICG with photothermal and photodynamic effects is a good candidate for virus inactivation by aPDT and has proved to be effective against herpes simplex virus. Geralde et al29 evaluated the efficacy of aPDT with ICG (10 µmol/L) as a photosensitizer and extracorporeal illumination using an infrared light source with a 780 nm laser device for the treatment of pneumonia in an experimental mouse model. The findings confirm the potential of aPDT to eliminate Streptococcus pneumoniae and treat infectious diseases by the administration of the ICG and extracorporeal illumination with infrared light. Geralde et al29 also assessed the effect of aPDT on alveolar macrophages, as key lung phagocytic cells in the innate immunity. Notably, the viability of the alveolar macrophage was calculated more than 90% following aPDT with light at 850 nm and ICG suggesting that aPDT does not harm the host immune system and is a safe treatment modality.30
The penetration depth of the light is the main limiting factor in the widespread clinical acceptability of aPDT in the case of deeper and thicker lesions. Among the photosensitizers, the absorption peak of ICG is in near-infrared (810 nm) where penetration into tissues is higher because of low absorption by water, hemoglobin, and melanin, which makes ICG an ideal photosensitizer in extracorporeal aPDT applications for deep-tissue lesions, requiring light penetration through the lung parenchyma.31,32
Because the ACE2 receptor is an initial way for SARS-CoV-2 to enter into target cells, structural analysis of SARS-CoV-2 RBD is essential for ACE2 binding.4 In the present study, RBD was described as a substrate for binding to ICG using molecular modeling and simulation analysis through numerous data banks. It is well established that the prediction of the three-dimensional structure of a protein by bioinformatic research can confirm the data obtained from basic methods such as X-ray crystallography and NMR.19-21
According to the results, RBD is a stable protein in a wide range of temperatures with a molecular weight of 25098.40 Da. ICG can bind to the ligands of SARS-CoV-2-RBD such as NAG, GOL, and SO4 based on the results of docking and penetrate viral cells following light irradiation at a wavelength of 810 nm during aPDT. The generated reactive oxygen species (ROS) can collapse the structure of the viral cells and prevent viral attachment to the host cell surface.23
According to the literature, no study has investigated the use of ICG-aPDT for eradicating COVID-19. This in silico study might contribute to the management of COVID-19 to give an idea about the protein structure and its physical and chemical properties to conduct the extensive tests on the design and manufacture of drugs and vaccines against the COVID-19. As for the use of aPDT for antiviral phototherapy, it is obvious that there is not ‘one’ specific photosensitizer for aPDT against infectious agents including viruses and the choice significantly depends on the type of virus and its application. According to the results of the present in silico study, ICG as a potential candidate showed strong binding to RBD. Given the increased resistance against antiviral agents, aPDT as a minimally invasive treatment modality can be considered valuable support for the treatment of viral diseases.
Conclusion
Our findings in the computational biology analysis in the current study warrant detailed examination of the interactions between SARS-CoV-2-RBD and ICG as a photosensitizing agent during aPDT to efficiently inactivate SARS-CoV-2, and can also serve as a model of more accurate prediction of dye-sensitized targets for aPDT in practical applications. This study also provides evidence to speculate that the enhanced sensitizing capacity of SARS-CoV-2 due to successful binding to the ICG may be act to control the spread of SARS-CoV-2 and to reduce disease severity in patients with COVID–19.
Ethical Considerations
This study was approved by the Ethics Committee of Tehran University, Faculty of Medicine (decision No. 48394).
Conflict of Interests
The authors declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgement
This research has been supported by Tehran University of Medical Sciences & Health Services grant No. 99-2-101-48394.
Please cite this article as follows: Pourhajibagher M, Bahador A. Computational biology analysis of COVID-19 receptor-binding domains: a target site for indocyanine green through antimicrobial photodynamic therapy. J Lasers Med Sci. 2020;11(4):433-441. doi:10.34172/ jlms.2020.68.
References
- 1.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020;55(5):105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singhal T. A review of Coronavirus Disease-2019 (COVID-19) Indian J Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M. et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–81. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Miao L, Li JM, Li YY, Zhu QY, Zhou CL. et al. Receptor-binding domain of SARS-Cov spike protein: soluble expression in E coli, purification and functional characterization. World J Gastroenterol. 2005;11(39):6159–64. doi: 10.3748/wjg.v11.i39.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin R, Dai T, Avci P, Jorge AE, de Melo WC, Vecchio D. et al. Light based anti-infectives: ultraviolet C irradiation, photodynamic therapy, blue light, and beyond. Curr Opin Pharmacol. 2013;13(5):731–62. doi: 10.1016/j.coph.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharkwal GB, Sharma SK, Huang YY, Dai T, Hamblin MR. Photodynamic therapy for infections: clinical applications. Lasers Surg Med. 2011;43(7):755–67. doi: 10.1002/lsm.21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang TW, Fiumara N, Weinstein L. Genital herpes: Treatment with methylene blue and light exposure. Int J Dermatol. 1975;14(1):69–71. doi: 10.1111/j.1365-4362.1975.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 9.Roome AP, Tinkler AE, Hilton AL, Montefiore DG, Waller D. Neutral red with photoinactivation in the treatment of herpes genitalis. Br J Vener Dis. 1975;51(2):130–133. doi: 10.1136/sti.51.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shikowitz MJ, Abramson AL, Freeman K, Steinberg BM, Nouri M. Efficacy of DHE photodynamic therapy for respiratory papillomatosis: Immediate and long-term results. Laryngoscope. 1998;108(7):962–967. doi: 10.1097/00005537-199807000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Abramson AL, Shikowitz MJ, Mullooly VM, Steinberg BM, Amella CA, Rothstein HR. Clinical effects of photodynamic therapy on recurrent laryngeal papillomas. Arch Otolaryngol Head Neck Surg. 1992;118(1):25–29. doi: 10.1001/archotol.1992.01880010029011. [DOI] [PubMed] [Google Scholar]
- 12.Mohr H, Lambrecht B, Selz A. Photodynamic virus inactivation of blood components. Immunol Invest. 1995;24(1-2):73–85. doi: 10.3109/08820139509062763. [DOI] [PubMed] [Google Scholar]
- 13.Zhou C, Sun B, Wang F, Dai Z, Han Z, Han J. et al. Coblation plus photodynamic therapy (PDT) for the treatment of juvenile onset laryngeal papillomatosis: case reports. World J Surg Oncol. 2014;12:275. doi: 10.1186/1477-7819-12-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiehe A, O’Brien JM, Senge MO. Trends and targets in antiviral phototherapy. Photochem Photobiol Sci. 2019;18(11):2565–2612. doi: 10.1039/C9PP00211A. [DOI] [PubMed] [Google Scholar]
- 15.Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci. 2004;3(5):436–50. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa L, Faustino MAF, Neves MGP, Cunha A, Almeida A. Photodynamic inactivation of mammalian viruses and bacteriophages. Viruses. 2012;4(7):1034–1074. doi: 10.3390/v4071034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baptista MS, Cadet J, Di Mascio P, Ghogare AA, Greer A, Hamblin MR. et al. Type I and type II photosensitized oxidation reactions: Guidelines and mechanistic pathways. Photochem Photobiol. 2017;93(4):912–919. doi: 10.1111/php.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girotti AW. Photosensitized oxidation of membrane lipids: reaction pathways, cytotoxic effects, and cytoprotective mechanisms. J Photochem Photobiol B. 2001;63(1-3):103–113. doi: 10.1016/S1011-1344(01)00207-X. [DOI] [PubMed] [Google Scholar]
- 19.Pourhajibagher M, Bahador A. Evaluation of the crystal structure of a fimbrillin (FimA) from Porphyromonas gingivalis as a therapeutic target for photo-activated disinfection with toluidine blue O. Photodiagnosis Photodyn Ther. 2017;17:98–102. doi: 10.1016/j.pdpdt.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Pourhajibagher M, Bahador A. Outer membrane protein 100 of Aggregatibacter actinomycetemcomitans act as a biopharmaceutical target for photodynamic therapy: An in silico analysis. Photodiagnosis Photodyn Ther. 2016;16:154–160. doi: 10.1016/j.pdpdt.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Pourhajibagher M, Bahador A. Designing and in silico analysis of PorB protein from Chlamydia Trachomatis for developing a vaccine candidate. Drug Res (Stuttg) 2016;66(9):479–483. doi: 10.1055/s-0042-110319. [DOI] [PubMed] [Google Scholar]
- 22.Pramanik K, Ghosh PK, Ray S, Sarkar A, Mitra S, Maiti TK. An in silico structural, functional and phylogenetic analysis with three dimensional protein modeling of alkaline phosphatase enzyme of Pseudomonas aeruginosa. J Genet Eng Biotechnol. 2017;15(2):527–537. doi: 10.1016/j.jgeb.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin C, Yu B, Zhang J, Wu H, Zhou X, Yao H, et al. Methylene blue photochemical treatment as a reliable SARS-CoV-2 plasma virus inactivation method for blood safety and convalescent plasma therapy for the COVID-19 outbreak. [Preprint]. 2020. Available from: 10.21203/rs.3.rs-17718/v1 [DOI] [PMC free article] [PubMed] [Retracted]
- 24.Asnaashari M, Eghbal MJ, Sahba Yaghmayi A, Shokri M, Azari-Marhabi S. Comparison of antibacterial effects of photodynamic therapy, modified triple antibiotic paste and calcium hydroxide on root canals infected with Enterococcus faecalis: an in vitro study. J Lasers Med Sci. 2019;10(Suppl 1):S23–S29. doi: 10.15171/jlms.2019.S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hakimiha N, Khoei F, Bahador A, Fekrazad R. The susceptibility of Streptococcus mutans to antibacterial photodynamic therapy: a comparison of two different photosensitizers and light sources. J Appl Oral Sci. 2014;22(2):80–84. doi: 10.1590/1678-775720130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Laguna V, Pérez-Artiaga L, Lampaya-Pérez V, García-Luque I, Ballesta S, Nonell S. et al. Bactericidal effect of photodynamic therapy, alone or in combination with mupirocin or linezolid, on Staphylococcus aureus. Front Microbiol. 2017;8:1002. doi: 10.3389/fmicb.2017.01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto MC, Prates RA, Kato IT, Núñez SC, Courrol LC, Ribeiro MS. Antimicrobial photodynamic therapy on drug-resistant Pseudomonas aeruginosa-induced infection An in vivo study. Photochem Photobiol. 2012;88(3):590–95. doi: 10.1111/j.1751-1097.2012.01137.x. [DOI] [PubMed] [Google Scholar]
- 28.Namvar MA, Vahedi M, Abdolsamadi HR, Mirzaei A, Mohammadi Y, Azizi Jalilian F. Effect of photodynamic therapy by 810 and 940 nm diode laser on Herpes Simplex Virus 1: An in vitro study. Photodiagnosis Photodyn Ther. 2019;25:87–91. doi: 10.1016/j.pdpdt.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Geralde MC, Leite IS, Inada NM, Salina AC, Medeiros AI, Kuebler WM. et al. Pneumonia treatment by photodynamic therapy with extracorporeal illumination - an experimental model. Physiol Rep. 2017;5(5):13190. doi: 10.14814/phy2.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urbanska K, Romanowska-Dixon B, Matuszak Z, Oszajca J, Nowak-Sliwinska P, Stochel G. Indocyanine green as a prospective sensitizer for photodynamic therapy of melanomas. Acta Biochim Pol. 2002;49(2):387–391. [PubMed] [Google Scholar]
- 31.Crescenzi E, Varriale L, Iovino M, Chiaviello A, Veneziani BM, Palumbo G. Photodynamic therapy with indocyanine green complements and enhances low-dose cisplatin cytotoxicity in MCF-7 breast cancer cells. Mol Cancer Ther. 2004;3(5):537–544. [PubMed] [Google Scholar]
- 32.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]