Abstract
Background
This study addresses whether existing specific transcriptional profiles can improve and support the current status of the definition of ulcerative colitis (UC) remission apart from the existing endoscopic, histologic, and laboratory scoring systems. For that purpose, a well-stratified UC patient population in remission was compared to active UC and control patients and was investigated by applying the next-generation technology RNA-Seq.
Methods
Mucosal biopsies from patients in remission (n = 14), patients with active UC (n = 14), and healthy control patientss (n = 16) underwent whole-transcriptome RNA-Seq. Principal component analysis, cell deconvolution methods, gene profile enrichment, and pathway enrichment methods were applied to define a specific transcriptional signature of UC in remission.
Results
Analyses revealed specific transcriptional signatures for UC in remission with increased expression of genes involved in O-glycosylation (MUC17, MUC3A, MUC5AC, MUC12, SPON1, B3GNT3), ephrin-mediated repulsion of cells (EFNB2E, EFNA3, EPHA10, EPHA1), GAP junction trafficking (TUBA1C, TUBA4A, TUBB4B, GJB3, CLTB), and decreased expression of several toll-like receptors (TLR1, TLR3, TLR5, TLR6).
Conclusions
This study reveals specific transcriptional signatures for remission. Partial restoration and improvement of homeostasis in the epithelial mucus layer and revival of immunological functions were observed. A clear role for bacterial gut flora composition can be implied. The results can be useful for the development of treatment strategies for UC in remission and may be useful targets for further investigations aiming to predict the outcome of UC in the future.
Keywords: ulcerative colitis, molecular signatures, remission
Specific transcriptional signatures for ulcerative colitis in remission reveal that partial restoration and improvement of the epithelial mucus layer and revival of immunological functions in the mucus layer are endorsed by the expression of protective genes. In addition, a clear role for bacterial gut flora composition can be implied.
INTRODUCTION
Ulcerative colitis (UC) is a chronic inflammatory disorder that requires long-term treatment to achieve remission.1 The inflammation status of UC is usually determined by endoscopic, histologic, and laboratory parameters.2 Current management programs aim for induction and maintenance of clinical remission to prevent disease-related and treatment-induced complications.3, 4 Different scoring systems for UC activity are used to evaluate endoscopic disease activity and activity status, but none have yet had all properties fully assessed.5, 6 However, there is no consensus on how to define clinical remission as no validated definition exists. The last guidelines from the European Colitis and Crohn’s Organization included clinical and endoscopic parameters where no mucosal lesions should appear (Mayo endoscopic grade 0).7 Despite this instruction, in most clinical studies Mayo grade 1 is included in the definition of clinical remission.8, 9 It is well known that clinical and endoscopic remission as defined above indeed may involve persisting microscopic inflammatory activity even in the absence of gastrointestinal symptoms.10-13 This activity can result in progressive accumulation of bowel damage, such as dysmotility, fibrosis, and increased risk of colorectal neoplasm.10, 14-17 There is a need for standardization of the assessment of remission and validation that gives a prognostic value.18 Therefore, an urgent need to characterize the complex pathogenic and healing mechanisms in UC still exists.
It is believed that the elucidation of key inflammatory pathways and perturbations involved by integrated clinical and genomic analyses should provide insight into the molecular events and pathways that are involved during remission. Recently, attempts have been made to describe transcriptional levels in UC remission by using microarrays.19-21 However, hybridization-based methods are restricted to predefined and often well-annotated species and RNA probe sets. Next-generation sequencing techniques have no such restrictions, which may lead to the discovery of potential new transcripts with a meaning for UC in remission. A transcriptomic study using RNA-Seq technology has been recently reported for the pediatric UC cohort PROTECT.22, 23
This study is the first to describe the entire transcriptomic landscape of adult UC in remission using next-generation sequencing technology with a focus on a clearly defined and representative group of UC patients in remission.
The establishment of robust and more specific biomarker groups and genomic signatures of UC remission may also lead to novel therapeutic approaches and is believed to pave the way for the development of personalized treatment options for UC patients in the future.
METHODS
Patient Material
A standardized sampling method was used to collect mucosal biopsies (N = 44) from patients in remission (n = 14), newly diagnosed treatment-naïve UC patients with mild to moderate disease (n = 14), and control patients (n = 16). Normal biopsies were taken from an earlier study.24 Patients’ UC was diagnosed based upon established clinical endoscopic and histological criteria as defined by the European Colitis and Crohn’s Organization guidelines. The grade of inflammation was assessed during colonoscopy using the UC disease activity index endoscopic subscore of 3-10 for mild to moderate disease.7 Tumor necrosis factor-alpha mRNA expression levels were measured by real-time quantitative polymerase chain reaction (qPCR), thereby indicating the grade of UC activity.20 Fecal calprotectin was measured in all patients representing remission and in 8 patients representing active UC with the Calprest ELISA kit (Eurospital). A total Geboes score was assessed for all biopsies representing remission and for 9 samples representing active UC.25 Patients in the remission group remained in remission for more than 1.5 years after discontinuation of treatment. The samples were taken from an established biobank approved by the Norwegian Board of Health. Patient characteristics are depicted in Table 1.
TABLE 1.
Patient Characteristics
| Characteristics | Control Patients (n = 16) | Active UC (n = 14) | UC in Remission (n = 14) |
|---|---|---|---|
| Gender (male/female) | 11/5 | 9/5 | 9/5 |
| Age, y, mean ± SD | 52.9 ± 16.9 | 40.7 ± 13.9 | 46.5 ± 16.0 |
| Clinical score ± SD | 0 | 7.78 ± 1.52 | 0.44 ± 1.01* |
| Endoscopy score mean ± SD | 0 | 1.79 ± 0.43 | 0 |
| Geboes score (total) | n.d. | 6.53 ± 2.93 | 1.07 ± 1.73 |
| TNF-α copies/µg RNA ± SD | 3,663 ± 1,973 | 15,907 ± 9,623 | 4,645 ± 1,830 |
| Calprotectin (mg/kg) mean ± SD | n.d. | 587.5 ± 483.8† | 53.9 ± 41.1 |
| Extension of disease‡ | _ | 2/9/3 | 2/8/4 |
| Medication§ | _ | _ | 14/4/5 |
aAverage score of 9 patients.
†Average calprotectin levels in 11 patients.
‡Proctitis/left-sided colitis/pancolitis.
§5-aminosalicylic acids/steroids/immunosuppressives.
n.d. indicates not determined; TNF-α, tumor necrosis factor-alpha.
RNA Isolation
Total RNA was isolated using the Allprep DNA/RNA Mini Kit from Qiagen (catalog number 80204) and the QIAcube instrument (Qiagen), according to the manufacturer’s protocol. The RNA quantity and purity were assessed by using the NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE). The Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA) and the RNA StdSens Analysis Kit (Bio-Rad, catalog number 700–7103) were used to evaluate RNA integrity, according to the instruction manual. The RNA samples were kept at −70°C until further use. All RNA samples used for analyses had a RNA integrity number value of between 8.0 and 10.0.
qPCR
The tumor necrosis factor-alpha levels in biopsies were measured by using qPCR. The RNA quantity was assessed with NanoVue Plus (GE Healthcare, United Kingdom). Synthesis of cDNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen, catalog number 205314) and the QuantiNova Probe PCR Kit (Qiagen, catalog number 208256). Beta-actin was used as the housekeeping gene. For the detection, a CFX Connect Real Time PCR Detection System (Bio-Rad) was used. The results were measured in copies/µg. Tissue samples with values <7,000 copies/µg RNA were considered noninflamed tissues, and values >7,000 copies/µg RNA were considered inflamed tissues as depicted in Table 1.26
Library Preparation and Next-Generation Sequencing
Whole transcriptome libraries of UC in remission samples were prepared with the TruSeq Stranded Total RNA LT Sample Prep Kit from Illumina (catalog number RS-122–2203). The amount of input material was 1 µg of total RNA. The Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and the Agilent DNA 1000 kit (cat number 5067-1504) were used to assess RNA library quality, according to the instruction manual. The RNA libraries comprised fragments with an average size of 307 base-pairs. The libraries were normalized to 10 nM and subsequently sequenced with the NextSeq 550 instrument (Illumina) according to the manufacturer’s instructions. The average number of uniquely mapped reads per sequencing run were 88 million reads per sample.
Data Analysis
Base calling and quality scoring were performed on the NextSeq 550 Illumina sequencer. Initial quality checks were performed by the NextSeq 550 on-board computer. The STAR-2.5.2b assembly algorithm and htseq-count were used for downstream analysis. Transcripts were aligned to human genome assembly GRCH38p.11 (https://www.ncbi.nlm.nih.gov/grc/human/data), and DESeq2 was used to compile and normalize from the individual raw htseq count matrixes. Transcripts with an average read count of <30 after DESeq2 normalization were excluded from further analysis. The comparisons between the UC, remission (R), and normal (N) samples were made using the LIMMA package with EBayes to shrink probewise sample variances on the DESeq2 normalized data. Only transcripts with a log2 fold change >0.5 and a Benjamini-Hochberg adjusted P < 0.05 in either UC-R or R-N comparisons were kept for further analysis.27
Profile analysis was conducted on the basis of the LIMMA results. The profile N < R > UC designated a situation where the log2 (R)-log2 (N) fold change was >0.5 and where the log2 (R)-log2 (UC) fold change was greater than 0.5. Note that the comparison UC-N was not considered in the profiles, as this study focused on remission-specific profiles. Both comparisons had a P adjusted value <0.05. The profile N > R = UC designated a situation where log2 (N) > log2 (R) by more than 0.5 and was significant (adjusted P < 0.05), but the absolute value of log2 (UC)-log2 (R) was <abs (0.5) and/or the comparison was not significant after the P value adjustment. Profiles included were N < R < UC, N > R > UC, N < R > UC, N > R < UC, N >R = UC, N < R = UC, N = R > UC, and N = R < UC. The gene set overrepresentation test was conducted using the enrichPathway algorithm of the R Bioconductor ReactomePA package (https://bioconductor.org/packages/release/bioc/html/ReactomePA.html).28, 29 Genes for each profile were entered into the enrichPathway algorithm. Reactome pathways that had a significant (adjusted P < 0.05) overrepresentation of genes were reported for all profiles.
For the estimation of specific cell populations in patient samples, an analysis was performed using the R/Bioconductor CellMix manual (http://web.cbio.uct.ac.za/~renaud/CRAN/web/CellMix/) with the immune response in silico (IRIS) weighted marker list characteristic for the different cell types.30 The epithelial marker cadherin 1, the phosphatidylinositol glycan anchor biosynthesis class F, the epithelial cell adhesion molecule, the L1 cell adhesion molecule, and the laminin subunit alpha 1 were added to the IRIS marker list and weighted strongly to give an estimate of the presence of epithelial fractions in the patient samples.
Heat maps were generated using the pHeatmap R package (https://CRAN.R-project.org/package=pheatmap). Only genes that were present in one of the reactome overrepresented pathways were considered. Heat maps were drawn for genes found in an overrepresented reactome pathway for the following profiles: N < R < UC, N > R > UC, N < R > UC, and N > R < UC. Processed RNA-Seq data were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE128682.
Ethical Considerations
The participants signed an informed and written consent form. The study was approved by the Regional Ethics Committee of North Norway and the Norwegian Social Science Data Services (REK Nord 2012/1349).
RESULTS
Whole Transcriptional Profile Characterization of UC in Remission
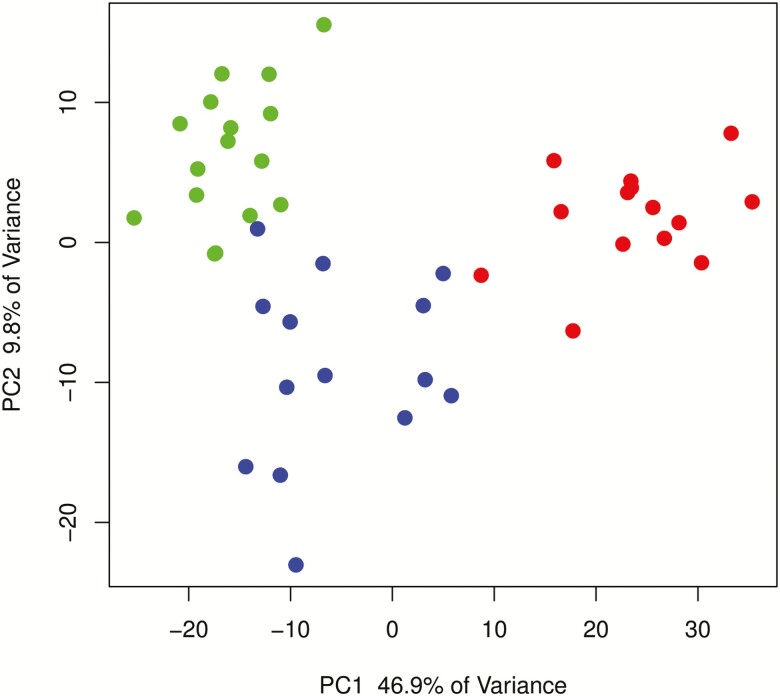
The entire transcriptome representing UC in remission was established by RNA-Seq, which revealed 13,927 differentially expressed genes (DEGs). Initial principal component analysis (PCA) of the entire DESeq2 log-normalized count matrix revealed a clear distinction between UC in remission, normal, and active UC samples along the first principal component with 46.9% and the second principal component with 9.8% of the total variance (Fig. 1). There were no differences in the UC remission sample profile that indicated any gender separation.
FIGURE 1.
PCA showing the difference between active UC (red), UC remission (blue), and normal control (green) samples of the top 5,000 most variable transcripts after DESeq2 rlog normalization. Principal component 1 explained 46.9 % of the total variance, and principal component 2 explained 9.8 % of the total variance.
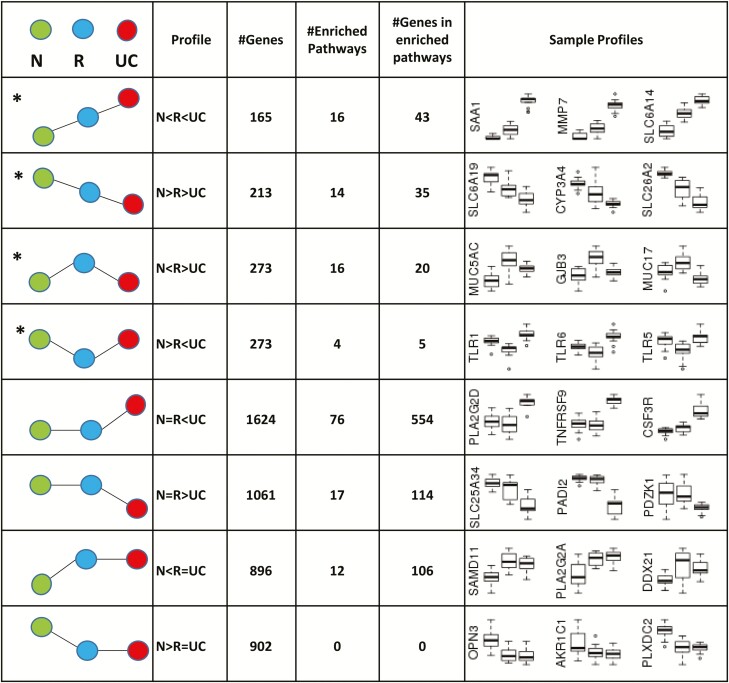
To further describe and analyze the transcriptome, the differentially expressed transcripts were adjusted to P < 0.05 and a log2 fold change > 0.5. The resulting number of significant DEGs (n = 5,407; Supplementary Data 1) was used for further downstream analyses. Fig. 2 depicts the assignment of significantly DEGs of normal (green), UC (red), and remission (blue) groups to clusters specific for all possible matching profiles. The number of genes found in each profile, the number of overrepresented reactome pathways, the total number of genes found in overrepresented reactome pathways, and a sample box plot for 3 genes from each profile are shown. For further interpretation of the results, we describe 4 of the 8 possible profiles, indicated by an asterisk in the figure, in detail. They represent genes (n = 924) in an intermediate state in remission (N < R < UC; N > R > UC) and genes specific for remission (N < R > UC; N > R < UC; Fig. 2). The gene clusters of each profile underwent reactome pathway enrichment as described in detail in “Methods” above, which resulted in a total of 50 annotated pathways representative for these profiles (Supplementary Data 2). Genes in the remaining profiles have been published in previous research.24 All genes and pathways matching any of the profiles can be found in Supplementary Data 1.
FIGURE 2.
Overview of transcriptional profiles found for UC in remission. Assignment of significantly DEGs of normal (green), UC (red), and remission (blue) groups to clusters specific for all possible matching profiles. Transcripts with a minimum fold change difference of 0.5, an adjusted P < 0.05, and a minimum of 30 reads are depicted as # genes. Genes left after pathway enrichment of the different profiles are indicated. The first column is a pictorial representation of profiles. The number of genes found in each profile, the number of enriched reactome pathways, the total number of genes found in enriched reactome pathways, and sample box plots for 3 genes from each profile are shown. Profiles indicated with (*) are considered and discussed in this study.
A comparison of the data obtained for UC vs normal samples with recent published transcriptomic data for the pediatric UC cohort PROTECT showed a strong correlation of 0.94, indicating that the datasets are comparable (Supplementary Data 3).22 Therefore, we used a remission dataset from the PROTECT cohort including a 115 glucocorticoid response gene signature and compared it with a remission dataset containing 4 glucocorticoid patient samples from this study. Four significantly regulated genes— PHLDA3, DSG3, ABCA12, and XKR9—overlapped with the PROTECT glucocorticoid response gene signature. In addition, the 4 patient samples with glucocorticoid treatment did not separate from the remaining remission samples in the PCA (Fig. 1). All the results regarding the comparison with the datasets from the PROTECT study are shown and summarized in Supplementary Data 3.
Genes in an Intermediate State in UC Remission
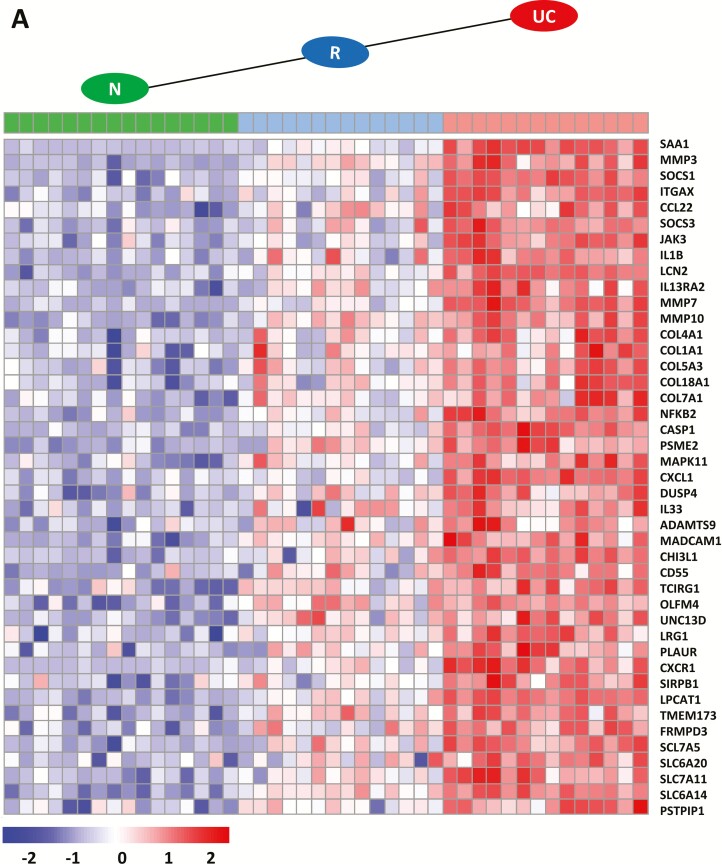
The different expression profile patterns with the number of genes (n = 78) including the 3 groups—N, R, and active UC for the intermediate state of remission (N < R < UC; N > R > UC)—are shown in Fig. 3. Genes that have been previously shown to have an increased expression in active UC24 showed increased expresssion but to a lesser extent during remission (n = 43) (Fig. 3A). Functional annotation (Table 2) revealed involvement in neutrophil degranulation (CHI3L1, CD55, TCIRG1, OLFM4, UNC13D, LRG1, PLAUR, CXCR1, SIRPB1, LPCAT1, TMEM173, FRMPD3, CXCL1) and degradation of the extracellular matrix, represented by members of the collagen family (COL4A1, COL1A1, COL5A3, COL18A1, COL7A1) and several matrix metallopeptidases (MMP3, MMP7, MMP10). Further, genes were annotated for interleukin signaling (NFKB2, CASP1, SAA1, SOCS1 and SOCS3, IGTAX, CCL22, JAK3, IL-1B, LCN2, IL-13RA2) and amino acid transport across the plasma membrane, represented by several members of the solute carrier family (SLC7A5, SLC6A20, SLC7A11, SLC6A14).
FIGURE 3.
Heatmap of genes found in enriched reactome pathway profiles in an intermediate state in remission with indicated profile patterns. A, Transcripts with significantly higher expression in remission (R; blue) than in normal (N; green) samples, and transcripts with significantly higher expression in UC (red) than in R samples (N < R < UC). B, Transcripts with significantly lower expression in R (blue) than in N (green) samples, and transcripts with significantly lower expression in UC (red) than in R samples (N > R > UC). Only transcripts with an absolute fold change >0.5, an adjusted P <0.05, and a minimum of 30 reads are depicted. Transcripts were normalized from raw count to log2 values by using DESeq2. Fold change and P values were calculated with LIMMA.
TABLE 2.
Reactome-Enriched Pathways and Genes of Indicated UC Remission Profiles for N, UC, and R
| Pathway | Gene Symbol |
|---|---|
| Enriched pathways for N < R < UC (adjusted P < 0.05) | |
| Interleukin-4 and interleukin-13 signaling | SAA1, MMP3, SOCS1, ITGAX, CCL22, SOCS3, JAK3, IL-1B, LCN2, IL-12RA2 |
| Collagen degradation | MMP7, MMP10, COL4A1, COL1A1, COL5A3, COL18A1, COL7A1 |
| Signaling by interleukins | PSME2, MAPK11, CXCL1, DUSP4, IL-33 |
| Extracellular matrix organization | ADAMTS9, MADCAM1 |
| Neutrophil degradation | CHI3L3, CD55, TCIRG1, OLFM4, UNC13D, LRG1, PLAUR, CXCR1, SIRPB1, LPCAT1, TMEM173, FRMPD3 |
| Amino acid transport across plasma membrane | SLC7A5, SLC6A20, SLC7A11, SLC6A14 |
| NLRP3 inflammasome | PSTPIP1, CASP1, NFKB2 |
| Enriched pathways for N > R > UC (adjusted P < 0.05) | |
| Glucuronidation | UGT1A10, UGT1A9, UGT1A7, UGT1A6, UGT1A5, UGT1A4, UGT2B7, UGT2B7 |
| Biological oxidations | NR1H4, SULT1A2, CYP27A1, SLC26A2, CYP3A4, MAOA |
| Solute carrier-mediated transport | SLC16A1, SLC3A1, SLC4A4, SLC6A19, SLC22A5, SLC26A3, SLC30A10, SLC22A4, SLC36A1, SRI |
| Bile acids and bile salt metabolism | ABCB11, AMACR |
| Abacavir transport and metabolism | ABCG2, ABCB1, PCK1 |
| Metallothioneins (binding metals) | MT1E, MT1M, MT1F |
| Digestion | GUCA2B, GUCA2A, ALPI |
| Enriched pathways for N < R > UC (adjusted P < 0.05) | |
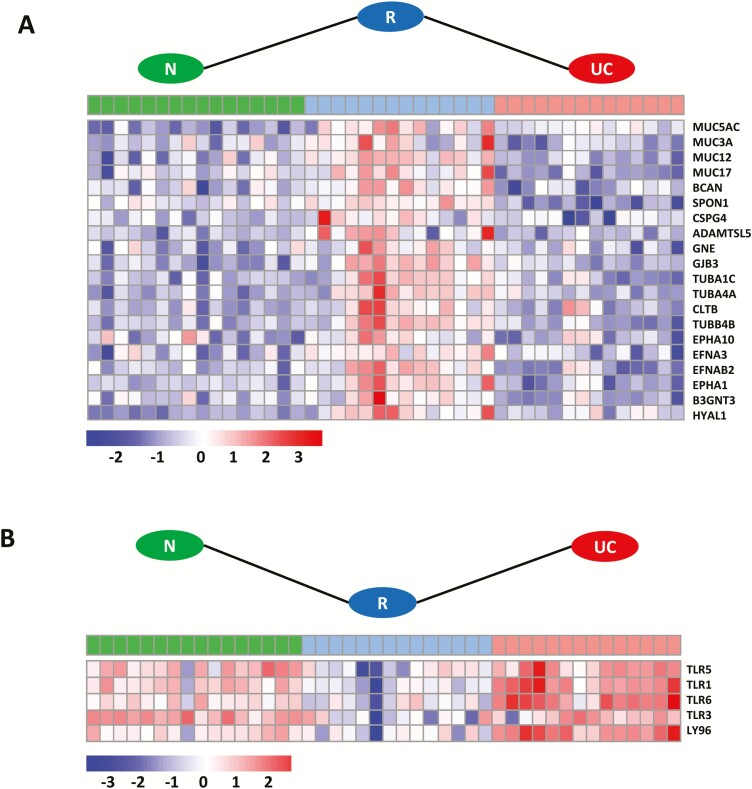
| O-linked glycosylation; O-linked-glycosylation of mucins | MUC5AC, MUC3A, MUC12, MUC17, B3GNT3 |
| O-linked glycosylation | SPON1, ADAMTSL5 |
| Diseases of glycosylation | GNE |
| Chondroitin sulfate and dermatan sulfate degradation | CSPG4, BCAN, HYAL |
| GAP junction trafficking | GJB3, TUBA1C, TUBA4A, CLTB, TUBB4B |
| EPH-ephrin-mediated repulsion of cells | EPHA10, EFNA3, EFNB2, EPHA1 |
| Enriched pathways for N > R < UC (adjusted P < 0.05) | |
| TLR signaling cascade; MyD88 deficiency (TRL2/4); IRAK4 deficiency (TRL2/4) | TLR1, TLR6, LY96 |
| TLR signaling cascade | TLR3, TLR5 |
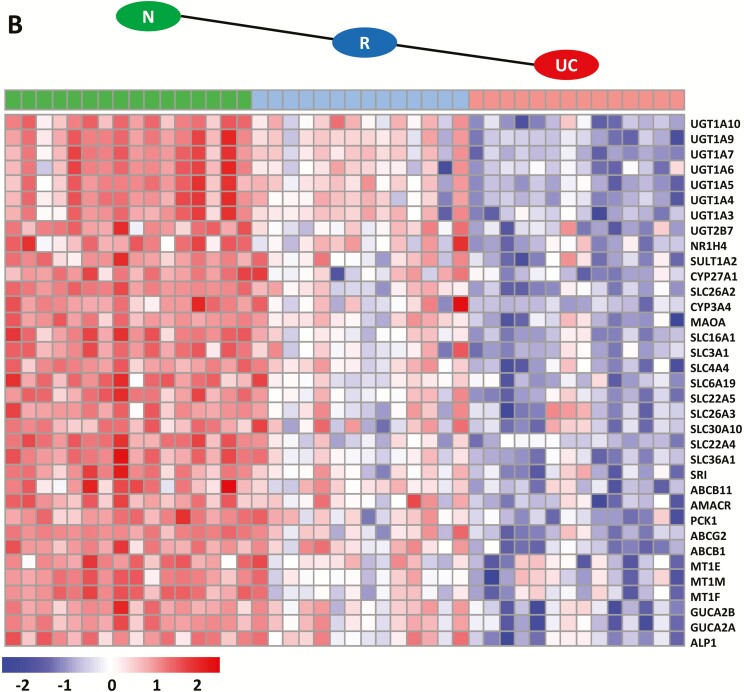
Genes that were downregulated in remission to a lesser extent than in active UC (n = 35) are depicted in Fig. 3B. Those genes could be functionally annotated to pathways (Table 2) including biological oxidation, in particular glucuronidation (represented by 8 different UDP glucuronosyltransferase family members: UGT1A3, UGT1A4, UGT1A 5, UGT1A 6, UGT1A7, UGT1A9, UGT1A10, UGT2B7; and 2 cytochrome members: CYP27A1 and CYP3A4), solute carrier-mediated transmembrane transport and related disorders (represented by 10 solute carrier family members), synthesis of bile acids (ABCB11 and AMACR), metallothioneins (MT1E, MT1M, MT1F), and digestion (GUCA2B and GUCA2A; ALPI). All genes representing these regulatory profiles can be found in Supplementary Data 2.
Genes Specific for UC Remission
Specific expression profile patterns with the number of genes (n = 25) including samples for the N, R, and active UC (N < R > UC; N > R < UC) groups are shown in Fig. 4. Overrepresented pathways of genes (Table 2 and Supplementary Data 4) with increased expression (n = 20) during remission (Fig. 4A) are mainly involved in diseases of glycosylation, represented by different mucins (MUC5AC, MUC3A, MUC12, MUC17), BCAN, SPON1, CSPG4, ADAMTSL5, GNE, and B3GNT3. Others could be annotated to GAP junction trafficking, such as tubulins (TUBA1C, TUBA4A, TUBB4B), CLTB, and GJB3, and in the ephrin-mediated repulsion of cells represented by ephrins (EFNA3 and EFNB2), and ephrin type A receptors (EPHA10 and EPHA1) (Fig. 4A). Only 5 genes with decreased expression during remission could be enriched and annotated to overrepresented pathways for diseases of the immune system, in particular diseases associated with the toll-like receptor (TLR) signaling cascade including TLR1, TLR3, TLR5, and TLR6 and the innate immune signal transduction adaptor MyD88 and interleukin-1 IRAK4 deficiency (Fig. 4B). All genes representing these regulatory profiles can be found in Supplementary Data 2, and all remaining genes after gene set enrichment representing these regulatory profiles can be found in Table 2 and Supplementary Data 3.
FIGURE 4.
Heatmap of specific genes for UC in remission with indicated profile patterns. A, Transcripts with significantly higher expression in remission (R; blue) than in normal (N; green) and UC (red) samples; N < R > UC. B, Transcripts with significantly lower expression in R (blue) than in N (green) and UC (red) samples; N > R < UC. Transcripts with a minimum fold change difference of 0.5, an adjusted P < 0.05, and a minimum of 30 reads are depicted. Transcripts were normalized from raw count to log2 values by using DESeq2. Fold change and P values were calculated with LIMMA.
Changes in Fractions of Cell Populations
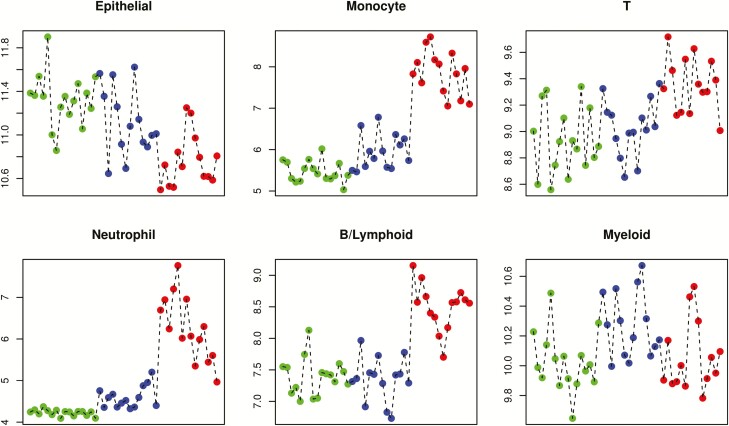
To estimate specific cell populations in the UC remission samples compared with the control and active UC tissue samples, a cell deconvolution method was applied as described in “Methods” above. The deconvolutions were restricted to the following cell types: epithelial cells, monocytes, T-cells, neutrophils, B cells/lymphoid cells, and myeloid cells. The results showed a clear difference of cell fractions present in the UC remission samples when they were compared with the active UC and control samples (Fig. 5). The epithelial cell fraction showed intermediate levels in the remission samples when they were compared with the normal samples and active UC samples. A decrease in fractions of monocytes, T-cells, and B cells/lymphoid cells was observed for the remission samples when they were compared with the active UC samples. High fractions of neutrophils observed for active UC samples were almost reduced to levels of normal samples in remission. The fractions of myeloid cells remained unchanged in the remission samples compared with those in the normal samples and active UC samples. The results of the deconvolution experiments are summarized in Fig. 5.
FIGURE 5.
Estimation of cell fractions in patient samples using the Bioconductor CellMix package and the IRIS marker set as described in “Methods.” The blue dots indicate normal control samples, the red dots indicate active UC samples from a former study,24 and the green dots indicate UC in remission. The y axis depicts the mean weighted expression of IRIS marker genes within each cell type set.
DISCUSSION
This study provides a unique, comprehensive, and quantitative record of high-resolution gene expression in UC patients in remission using next-generation RNA-Seq technology and provides for the first time well-defined transcriptional signature patterns for UC in remission obtained by pathway enrichments. A clearly defined patient sample group representing UC in remission was used to characterize the representative transcriptional signature of UC in remission (Table 1). Initial PCA revealed a quite complex pathology of remission (Fig. 1). Some remission samples clustered close to UC and others close to N, demonstrating the variability within remission and the challenge of defining remission-specific profiles statistically. The assignment of genes to different expression profiles (Fig. 2) and gene set enrichment (Table 2) made it possible to show that genes found in an intermediate state in remission showed reduced but not completely diminished levels of inflammatory signatures when compared with active UC (Fig. 3). This decrease may represent the remaining quiescent microscopic disease activities during remission as suggested by others.7, 10, 11, 13 Examples of considerable downregulated transcripts in remission for previously reported top upregulated transcripts in active UC are genes involved in the degradation of the extracellular matrix, collagen degradation, neutrophil degranulation, and signaling by interleukins (Table 2 and Supplementary Data 3). The downregulation of the 2 inflammatory bowel disease (IBD) susceptibility genes CXCL1 and CXCR1 (Fig. 3) involved in neutrophil degranulation was in agreement with almost absent fractions of neutrophils present during remission (Fig. 5). These results concurred with a recent transcriptomic cohort study including pediatric UC patients.22 Note also that a downregulation of transcripts for MMP3, MMP10, CH13L1, and CXCL1 during remission has been reported by using real-time qPCR.21
The involvement of the microflora and their importance in the onset, development, and preservation of UC has been implied.31–34 During remission, several genes involved in patient antibacterial response showed reduced expression, like the transporter SLC6A14; the IBD disease marker LCNC2; SAA1, which represents an important link between mucosal T-cells and microbial communities; and CHI3L1, which lessens bacterial adhesion and invasion on/into colonic epithelial cells.35-38 These observations are in concordance with the results obtained by cell deconvolutions, which revealed an improvement of the mucus layer in the remission samples when compared with the active UC samples by showing decreased fractions of monocytes, T-cells, and B cells/lymphoid cells and enhanced fractions of epithelial cells (Fig. 5).
However, the main focus of this study was to determine more specific transcriptional signatures involved in remission. By assigning transcripts to different expression profiles and using gene set enrichment methods, we were able to uncover transcripts in the remission samples that were differentially up- or downregulated as compared to the UC and N samples (Fig. 4). Surprisingly, enriched pathways for upregulated genes in the remission samples were found for pathways such as O-glycosylation, GAP junction trafficking, and ephrin-mediated repulsion of cells (Table 2). It is well acknowledged that gastrointestinal O-glycosylated mucins are constituents of luminal barrier function and are the first line of host defense against invading pathogens.39 The observed increased expression of the mucin MUC17 and its paralog MUC3A during remission (Fig. 4A) may prevent bacterial invasion through barrier function by maintaining the integrity of the surface epithelial layer and homeostasis on the mucus surface.39-41 In addition, MUC17 has been shown to be instrumental in limiting the epithelial adhesion and invasion of enteroinvasive Escherichia coli.42, 43 In concordance with these results was the observation of a partial restoration of the epithelial mucus layer (Fig. 5). However, epithelial cell fractions did not reach levels as seen in the normal samples, which indicates that mucosal healing in UC remission was not fully accomplished. Epithelial cells are tightly coupled together through specialized intercellular junctions, including tight junctions and gap junctions,44 and the increased expression of genes annotated for GAP junctions in this study (TUBA1C, TUBA4A, TUBB4BCLTB, GNE, GJB3) confirm this notion. In addition, the observed increased expression of MUC12 may support protection of the epithelial cells in the mucosa: previous findings showed a significant downregulation of MUC12 in the colon and ileum in patients with Crohn disease.45, 46
In addition to the mucins, other genes involved in O-linked glycosylation such as B3GNT3 (lymphocyte homing and lymphocyte trafficking), SPON1, and ADAMTSL5 (metalloendopeptidase activity) showed increased expression (Fig. 4A). In particular, SPON1 has been shown to regulate macrophage microbicidal activity by effectively phagocytizing the bacteria Salmonella typhimurium and E. coli.47-50
An increasing body of evidence suggests that epithelial cells also directly exchange information at cell-cell contacts via the ephrin family of receptor tyrosine kinases and their membrane-associated ephrin ligands.51, 52 It has been reported that impaired intestinal epithelial barrier and abnormal T-cell responses are evident during IBD.53, 54 This implies that the observed increased levels of EFNB2 in the remission samples may be directly involved in the activation of T-cell development,54, 55 in the organization of stem cell compartments, and in the ordered migration of epithelial cells along the intestinal villus axis.44 Increased expression of EFNB2 has been also found to enhance intestinal wound healing in patients with Crohn disease and may play a potential role in the EphB/ephrin-B system as a pharmacological target in intestinal inflammatory disorders.56, 57
In addition, some of the differentially regulated genes have been reported to be associated with the development of colorectal cancer such as MUC5AC, the ephrin receptors EPHA10 and EPHA10, and OLFM4.58-60 However, the value of these genes as reliable predictors for disease outcome remains to be elucidated.
Gene pathway enrichment revealed downregulation of several TLRs in the remission samples (Fig. 4B). It has been suggested that UC may be associated with specific alterations in selective TLR expression in the intestinal epithelium.61, 62 A recent report also implies a role for mucin and TLRs in patient defense against intestinal parasites.42 In this study, TLRs were expressed higher in normal mucosa and active UC samples than in the remission samples. These observations are most likely a result of different treatment strategies applied for this patient group and may be used to establish therapeutic options for UC to obtain better clinical outcomes.63-65
A limitation of this work is the amount of patient samples, with n = 14. The low sample size made it difficult to define molecular remission patterns caused by different treatments. However the number of patients included in this study is still larger than reported for other studies, and PCA (Fig. 1) revealed sufficient separation of the different patient groups.21 It is also possible that the remission signature seen here reflects a response to treatment or natural recovery after initial disease presentation. Such a response could have been evaluated with follow-up for active UC samples, which was not available during this study.
CONCLUSIONS
The data demonstrate that remission is a permanently altered state of UC characterized by a unique transcriptional signature in the mucosa, which is different from that in UC and normal samples. The analysis revealed new and more specific molecular signatures for UC in remission compared with earlier studies conducted with microarrays.19-21 Partial restoration of the epithelial mucus layer and revival of immunological functions in the mucus layer were supported by the expression of protective genes. In addition, a clear role for bacterial gut flora composition can be implied. Some expressed genes have been associated with the development of colorectal cancer, but a role for these genes as reliable predictors for disease outcome remains uncertain. Further studies will tell whether transcriptomic, epigenetic, metagenomic, and meta-transcriptomic signatures together can reveal biomarkers and/or biomarker groups of valuable character to be used for the prognosis of relapse and for the definition of new criteria for de-escalating treatment.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Ingrid Christiansen for the technical help performing the tumor necrosis factor-alpha level measurements and Renate W. Meyer for administrating patient samples.
Supported by: This work was supported by the Northern Norway Regional Health Authority Helse-Nord (SPF-1209–14).
Conflicts of interest: The authors declare no conflict of interest regarding the publication of this article.
REFERENCES
- 1. Peyrin-Biroulet L, Bressenot A, Kampman W. Histologic remission: the ultimate therapeutic goal in ulcerative colitis? Clin Gastroenterol Hepatol 2014;12:929–934.e2. [DOI] [PubMed] [Google Scholar]
- 2. Rogler G, Vavricka S, Schoepfer A, et al. Mucosal healing and deep remission: what does it mean? World J Gastroenterol. 2013;19:7552–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryant RV, Winer S, Travis SP, et al. Systematic review: histological remission in inflammatory bowel disease. Is “complete” remission the new treatment paradigm? An IOIBD initiative. J Crohns Colitis. 2014;8:1582–1597. [DOI] [PubMed] [Google Scholar]
- 4. D’Haens G. Risks and benefits of biologic therapy for inflammatory bowel diseases. Gut. 2007;56:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Travis SP, Higgins PD, Orchard T, et al. Review article: defining remission in ulcerative colitis. Aliment Pharmacol Ther. 2011;34:113–124. [DOI] [PubMed] [Google Scholar]
- 6. Rutter M, Bernstein C, Matsumoto T, et al. Endoscopic appearance of dysplasia in ulcerative colitis and the role of staining. Endoscopy. 2004;36:1109–1114. [DOI] [PubMed] [Google Scholar]
- 7. Magro F, Gionchetti P, Eliakim R, et al. ; European Crohn’s and Colitis Organisation (ECCO) . Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J Crohns Colitis. 2017;11:649–670. [DOI] [PubMed] [Google Scholar]
- 8. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625–1629. [DOI] [PubMed] [Google Scholar]
- 9. Rutgeerts P, Sandborn WJ, Feagan BG, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462–2476. [DOI] [PubMed] [Google Scholar]
- 10. Riley SA, Mani V, Goodman MJ, et al. Microscopic activity in ulcerative colitis: what does it mean? Gut. 1991;32:174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korelitz BI. Mucosal healing as an index of colitis activity: back to histological healing for future indices. Inflamm Bowel Dis. 2010;16:1628–1630. [DOI] [PubMed] [Google Scholar]
- 12. Magro F, Lopes J, Borralho P, et al. Comparison of different histological indexes in the assessment of uc activity and their accuracy regarding endoscopic outcomes and faecal calprotectin levels. Gut. 2018; . doi: 10.1136/gutjnl-2017-315545. [DOI] [PubMed] [Google Scholar]
- 13. DeRoche TC, Xiao SY, Liu X. Histological evaluation in ulcerative colitis. Gastroenterol Rep (Oxf). 2014;2:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bitton A, Peppercorn MA, Antonioli DA, et al. Clinical, biological, and histologic parameters as predictors of relapse in ulcerative colitis. Gastroenterology. 2001;120:13–20. [DOI] [PubMed] [Google Scholar]
- 15. Gupta RB, Harpaz N, Itzkowitz S, et al. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: a cohort study. Gastroenterology. 2007;133:1099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Riley SA, Mani V, Goodman MJ. Why do patients with ulcerative colitis relapse? Gut. 1990;31:179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magro F, Langner C, Driessen A, et al. ; European Society of Pathology (ESP); European Crohn’s and Colitis Organisation (ECCO) . European consensus on the histopathology of inflammatory bowel disease. J Crohns Colitis. 2013;7:827–851. [DOI] [PubMed] [Google Scholar]
- 18. Florholmen J. Mucosal healing in the era of biologic agents in treatment of inflammatory bowel disease. Scand J Gastroenterol. 2015;50:43–52. [DOI] [PubMed] [Google Scholar]
- 19. Bjerrum JT, Hansen M, Olsen J, et al. Genome-wide gene expression analysis of mucosal colonic biopsies and isolated colonocytes suggests a continuous inflammatory state in the lamina propria of patients with quiescent ulcerative colitis. Inflamm Bowel Dis. 2010;16:999–1007. [DOI] [PubMed] [Google Scholar]
- 20. Olsen J, Gerds TA, Seidelin JB, et al. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15:1032–1038. [DOI] [PubMed] [Google Scholar]
- 21. Planell N, Lozano JJ, Mora-Buch R, et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–976. [DOI] [PubMed] [Google Scholar]
- 22. Haberman Y, Karns R, Dexheimer PJ, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hyams JS, Davis Thomas S, Gotman N, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. 2019;393:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taman H, Fenton CG, Hensel IV, et al. Transcriptomic landscape of treatment-naive ulcerative colitis. J Crohns Colitis. 2018;12:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geboes K, Riddell R, Ost A, et al. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut. 2000;47:404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olsen T, Goll R, Cui G, et al. Tissue levels of tumor necrosis factor-alpha correlates with grade of inflammation in untreated ulcerative colitis. Scand J Gastroenterol. 2007;42:1312–1320. [DOI] [PubMed] [Google Scholar]
- 27. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 28. Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. [DOI] [PubMed] [Google Scholar]
- 29. Fabregat A, Jupe S, Matthews L, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abbas AR, Baldwin D, Ma Y, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. [DOI] [PubMed] [Google Scholar]
- 31. Barko PC, McMichael MA, Swanson KS, et al. The gastrointestinal microbiome: a review. J Vet Intern Med. 2018;32:9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. Ilar J. 2015;56:192–204. [DOI] [PubMed] [Google Scholar]
- 33. Lane ER, Zisman TL, Suskind DL. The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res. 2017;10:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1–10. [DOI] [PubMed] [Google Scholar]
- 35. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. [DOI] [PubMed] [Google Scholar]
- 36. Stallhofer J, Friedrich M, Konrad-Zerna A, et al. Lipocalin-2 is a disease activity marker in inflammatory bowel disease regulated by IL-17A, IL-22, and TNF-α and modulated by IL23R genotype status. Inflamm Bowel Dis. 2015;21:2327–2340. [DOI] [PubMed] [Google Scholar]
- 37. Tang MS, Bowcutt R, Leung JM, et al. Integrated analysis of biopsies from inflammatory bowel disease patients identifies SAA1 as a link between mucosal microbes with TH17 and TH22 cells. Inflamm Bowel Dis. 2017;23:1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mizoguchi E. Chitinase 3-like-1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology. 2006;130:398–411. [DOI] [PubMed] [Google Scholar]
- 39. Boltin D, Perets TT, Vilkin A, et al. Mucin function in inflammatory bowel disease: an update. J Clin Gastroenterol. 2013;47:106–111. [DOI] [PubMed] [Google Scholar]
- 40. van Putten JPM, Strijbis K. Transmembrane mucins: signaling receptors at the intersection of inflammation and cancer. J Innate Immun. 2017;9:281–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Longman RJ, Poulsom R, Corfield AP, et al. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. J Histochem Cytochem. 2006;54:1335–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moncada DM, Kammanadiminti SJ, Chadee K. Mucin and toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003;19:305–311. [DOI] [PubMed] [Google Scholar]
- 43. Resta-Lenert S, Das S, Batra SK, et al. Muc17 protects intestinal epithelial cells from enteroinvasive E. coli infection by promoting epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2011;300:G1144–G1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez White BE, Getsios S. Eph receptor and ephrin function in breast, gut, and skin epithelia. Cell Adh Migr. 2014;8:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moehle C, Ackermann N, Langmann T, et al. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl). 2006;84:1055–1066. [DOI] [PubMed] [Google Scholar]
- 46. Yamamoto-Furusho JK, Ascaño-Gutiérrez I, Furuzawa-Carballeda J, et al. Differential expression of MUC12, MUC16, and MUC20 in patients with active and remission ulcerative colitis. Mediators Inflamm. 2015;2015:659018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smythies LE, Sellers M, Clements RH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith PD, Smythies LE, Shen R, et al. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Esmann L, Idel C, Sarkar A, et al. Phagocytosis of apoptotic cells by neutrophil granulocytes: diminished proinflammatory neutrophil functions in the presence of apoptotic cells. J Immunol. 2010;184:391–400. [DOI] [PubMed] [Google Scholar]
- 50. Mowat AM, Bain CC. Mucosal macrophages in intestinal homeostasis and inflammation. J Innate Immun. 2011;3:550–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Coulthard MG, Morgan M, Woodruff TM, et al. Eph/ephrin signaling in injury and inflammation. Am J Pathol. 2012;181:1493–1503. [DOI] [PubMed] [Google Scholar]
- 52. Darling TK, Lamb TJ. Emerging roles for eph receptors and ephrin ligands in immunity. Front Immunol. 2019;10:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jin W, Luo H, Wu J. Effect of reduced ephb4 expression in thymic epithelial cells on thymocyte development and peripheral T cell function. Mol Immunol. 2014;58:1–9. [DOI] [PubMed] [Google Scholar]
- 55. Kawano H, Katayama Y, Minagawa K, et al. A novel feedback mechanism by ephrin-b1/b2 in T-cell activation involves a concentration-dependent switch from costimulation to inhibition. Eur J Immunol. 2012;42:1562–1572. [DOI] [PubMed] [Google Scholar]
- 56. Hafner C, Meyer S, Langmann T, et al. Ephrin-b2 is differentially expressed in the intestinal epithelium in Crohn’s disease and contributes to accelerated epithelial wound healing in vitro. World J Gastroenterol. 2005;11:4024–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grandi A, Zini I, Palese S, et al. Targeting the eph/ephrin system as anti-inflammatory strategy in IBD. Front Pharmacol. 2019;10:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walsh MD, Clendenning M, Williamson E, et al. Expression of muc2, muc5ac, muc5b, and muc6 mucins in colorectal cancers and their association with the cpg island methylator phenotype. Mod Pathol. 2013;26:1642–1656. [DOI] [PubMed] [Google Scholar]
- 59. Liu W, Rodgers GP. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016;35:201–212. [DOI] [PubMed] [Google Scholar]
- 60. Herath NI, Spanevello MD, Doecke JD, et al. Complex expression patterns of eph receptor tyrosine kinases and their ephrin ligands in colorectal carcinogenesis. Eur J Cancer. 2012;48:753–762. [DOI] [PubMed] [Google Scholar]
- 61. Kordjazy N, Haj-Mirzaian A, Haj-Mirzaian A, et al. Role of toll-like receptors in inflammatory bowel disease. Pharmacol Res. 2018;129:204–215. [DOI] [PubMed] [Google Scholar]
- 62. Lu Y, Li X, Liu S, et al. Toll-like receptors and inflammatory bowel disease. Front Immunol. 2018;9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baird AC, Mallon D, Radford-Smith G, et al. Dysregulation of innate immunity in ulcerative colitis patients who fail anti-tumor necrosis factor therapy. World J Gastroenterol. 2016;22:9104–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sánchez-Muñoz F, Fonseca-Camarillo G, Villeda-Ramírez MA, et al. Transcript levels of toll-like receptors 5, 8 and 9 correlate with inflammatory activity in ulcerative colitis. BMC Gastroenterol. 2011;11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Atreya R, Bloom S, Scaldaferri F, et al. Clinical effects of a topically applied toll-like receptor 9 agonist in active moderate-to-severe ulcerative colitis. J Crohns Colitis. 2016;10:1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.