Abstract
While memory T-cells represent a hallmark of adaptive immunity, little is known about the genetic mechanisms regulating the longevity of memory CD4 T cells. Here, we studied the dynamics of gene expression in antigen specific CD4 T cells during infection, memory differentiation, and long-term survival up to nearly a year in mice. We observed that differentiation into long lived memory cells is associated with increased expression of genes inhibiting cell proliferation and apoptosis as well as genes promoting DNA repair response, lipid metabolism, and insulin resistance. We identified several transmembrane proteins in long-lived murine memory CD4 T cells, which co-localized exclusively within the responding antigen-specific memory CD4 T cells in human. The unique gene signatures of long-lived memory CD4 T cells, along with the new markers that we have defined, will enable a deeper understanding of memory CD4 T cell biology and allow for designing novel vaccines and therapeutics.
Keywords: Memory T cell, Gene, CD4 T cell, Memory cell markers, Cell longevity, Genetic programs
1. Introduction
Memory T cells are the linchpin of the adaptive immune response. Compared to naïve T cells, popular dogma characterizes memory T cells as present in higher numbers, with greater sensitivity for antigen recognition, and having faster responsiveness with little to no requirement for co-stimulation [48]. However, most of these “memory” characteristics have been determined by studying CD8 memory T cells, and much less is known about CD4 memory T cells. This knowledge deficit is critical to address, as CD4 memory T cells activate dendritic cells, promote B cell antibody production, and help CD8 T cells become robust secondary effectors through cytokine production [7,33,59,61,63]. Some of the barriers to studying CD4 memory T cells are that, unlike specific CD8 memory T cells that have proven relatively straightforward to detect, antigen-specific CD4 memory T are less frequent, more heterogeneous, lack specific surface markers, and require onerous tetramer staining procedures [18]. While many of the findings from the CD8 memory field may inform CD4 memory cell biology, in vivo evidence shows that qualitatively and quantitatively, these responses differ. Memory CD4 T cells have been characterized as having a faster decay than CD8 memory T cells [28], which is in conflict with the observation that in humans CD4 memory T cells can persist for up to 75 years [21]. The maintenance of CD4 memory T cells has been linked to nonspecific tonal TCR signaling in some studies [13,34], and in other cases appears to be independent of cognate antigen [64].
We and others have explored the signals that induce such long-term gene expression changes that allow for memory cell survival, and found that B cells are required for induction of CD4 memory T cells [11,12,40,58,69]. Moreover, we have established that differentiation from CD4 T effector to CD4 memory T states occurs upon the engagement of TCR by suboptimal doses of antigen during the contraction phase of infection [11,58]). When the level of antigen reaches low levels, B cells become the primary antigen presenting cells (APC) to capture and internalize the Ag via BCR-mediated endocytosis. B cells then present the low levels of peptide:MHC complexes (pMHC) to antigen experienced CD4 T cells, which induces differentiation into a state of dormancy by a CTLA-4 associated mechanism[12,45]. It is likely that presentation of low numbers of pMHC II by B cells to antigen experienced CD4 T cells may select the highest affinity memory CD4 precursors to be endowed with a gene expression program that promotes quiescence, and therefore, longevity. Dormant memory T would avoid unnecessary cell division and cytokine production associated with shortening of chromosomes, or autoinflammatory conditions [19,70], while maintaining the capacity to respond polyfunctionally in secondary responses upon challenge with Ag plus TLR ligands [11,12,44,45,58].
To better understand the genetic programs that lead to the development of memory CD4 T cells and their long survival, we examined the gene expression profiles of antigen-specific CD4 T cells from naive state through activation and differentiation from early memory to long-term memory up to near a year in mice. Our data presented here reports specific genetic signatures that denote long-term antigen-specific memory CD4 T cell populations. In addition, the findings that several CD4 memory T cell-surface markers were shared between mice and humans, supports the welcomed notion that the underlying principles and markers found in murine systems may be translatable to the generation of long-term CD4 memory T cells induced by vaccines against the deadly SARS CoV-2 virus.
2. Methods and materials
2.1. Mice.
BALB/c and DO11.10 TCR Tg mice were obtained from Jackson Labs. DR1 (DRB1*0101) Tg mice [56]were backcrossed to MHC class II KO mice for 12–16 generations to eliminate their endogenous class II proteins (I-Af) and were inbred to homozygosity. All mice were housed in the Johns Hopkins University animal facilities under virus-free conditions in accordance with protocols approved by the Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
2.2. Study Participants
Peripheral blood mononuclear cells (PBMCs) for human sort and phenotyping experiments were obtained either from HIV−/HCV− leukapheresis samples (Stem Cell) or whole blood (Johns Hopkins). All participants provided written consent.
2.3. Peptides, Proteins, H5N1 influenza vaccine and 2017–2018 attenuated flu vaccine.
Peptides OVA(323–339) (ISQAVHAAHAEIN-EAGR) (Global-Peptides] and H5N1-HA(259–274) (SNGNFIAPEYAYK-IVK), (Elim Biopharmaceuticals Inc) were at > 90% purity. Chicken OVA protein (Grade VI) was from Sigma-Aldrich. Both Inactivated influenza vaccine, A/H5N1 Influenza Vaccine and 2017–2018 attenuated flu vaccine were obtained from BEI Resources (beiresources.org).
2.4. Antibodies.
Mouse FITC-CD4 and eFluor 605-CD44 [BD-Pharmingen]; KJ1.26 anti-TCR (Caltag Laboratories), Brilliant Violet 570-CD4 and Alexa Fluor 700-CD44 were from [Biolegend], FITC-Klotho,PE-Cy7-CD99, PE-Cy5-VDR, and PE-Cy7-CNR2 were from [Bioss]; APC-CCR10 and APC-ITGA3 were from [R&D Systems]; Fixable viability dye eFluor 780 was from [eBioscience]. Antibodies for human PBMC staining were as follows: from Biolegend, CD99-FITC,CD69-BV421 and CD69-APC , CD45R0-APC and CD45R0-BV421, CD3-AF700, CD4-Pe/Cy7, CD45RA-PE, Itga3 (CD49c)-PE, IL-7R-BV510, CD25-APC, HLA-DR-APC. From BD Biosciences, CCR10-APC and CCR10-PerCP Cy5.5. and viability dye eFluor 780 as above. Relevant mouse anti human isotype antibodies to IgG1k or IgG2ak were utilized for the following fluorophores: BV421, BV510, FITC, PE, and APC for both isotypes (Biolegend).
2.5. Adoptive transfer and cell isolation for cDNA microarray.
An estimated 6,000 Splenocytes of 8 weeks old female DO11.10 mice were transferred into age-matched BALB/c recipients i.v., and injected with 2.5 million pfu of Vaccinia-OVA virus in PBS (no adjuvant) i.p. 24 h later. Draining lymph nodes and spleens were extracted at times 0-, 9 days, 5 weeks, 6- and 10.5 months later. Cells were stained and CD44hi cells positive for CD4 and KJ1.26 Ab were sorted on a high-performance MoFlo sorter. Cells from sex and age-matched old naïve DO11.10 mice served as naive controls. Cells were lysed and sent for the microarray analysis (Miltenyi Biotec).
2.6. Microarray data collection and gene expression analysis.
mRNA was isolated from lysed cells by magnetic bead technology and corresponding cDNA was amplified by PCR. The cDNA was then examined by gel and electropherogram to ensure the integrity. The amplified cDNA was labeled with Cy3 fluorescent dye and hybridized overnight to Agilent Whole Mouse Genome Oligo Microarrays. The fluorescence signal was detected by Agilent’s Microarray Scanner System (Agilent Technologies). The microarray image files were then processed by Agilent Feature Extraction Software (FES) and Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware). Quality assessment was then performed on the microarray data. Spatial images of the foreground and background signals were made for each microarray sample to check any potential image artifacts, using the R/bioconductor package mArray. The distribution of the data was checked with density and box plots. The microarray signal data were normalized with the quantile normalization method to reduce the obscuring variations between microarrays (Bolstad et al., 2003). Between-treatment and between-replicate variations were examined with the pair-wise MvA plots, in which the base 2 log ratios (M) between two samples are plotted against their averaged base 2 log signals (A). Principal Component analysis (PCA) was also performed to assess sample variability. The probes of incomplete functional annotation including “unknown”, “predicted gene”, “hypothetical protein” and “RIKEN cDNA” were filtered out in the downstream analysis. After the probe level data processing above, the differential gene expression between the different conditions was assessed by a statistical linear model analysis using the R/bioconductor package limma (Ritchie et al., 2015), which results in more stable inference and improved power (Smyth, 2004). The lists of differentially expressed genes were obtained by the criteria of nominal p-value < 1% and fold change cutoff > 2 and visualized by volcano plots. The probe sequences of the selected genes were verified by mapping to UCSC genome browser mouse database. The final gene lists were submitted to DAVID Bioinformatics Resources software (https://david.ncifcrf.gov/) for functional enrichment analysis. The gene lists from microarray were also normalized by log2 and uploaded to Expander software to cluster genes based on the dynamics of their expression patterns at different time points with CLICK algorithm. The cutoff homogeneity value for each gene cluster is 0.5 to ensure tight clustering. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [17] and are accessible through GEO Series accession number GSE151583 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE151583)
2.7. Immunization of DR1 Tg mice for memory or activated H5N1 specific T-cells.
6–8 weeks old Tg DR1 mice were immunized with CpG + H5N1 vaccine (memory) or with H5N1 peptide (activated). Draining inguinal lymph nodes (activated), spleen (memory) from each of 5 immunized mice were harvested 10 months (memory) or 8 days (activated) after immunization.
2.8. Staining for MHC-II Tetramers.
PE-conjugated DR1/H5N1-HA(259–274) tetramers were produced in our laboratory. Biotinylated DR1 monomers, PE-conjugated CLIP tetramers, and the conjugation protocol were provided by the NIAID Tetramer Core Facility. Cells were first cultured for 6 days with or without Ag stimulation and stained with either DR1/H5N1-HA(259–274) or DR1/CLIP tetramers before staining with antibodies.
2.9. Selection of surface markers for memory CD4 T cells.
The 4 gene lists that were either up-regulated or down-regulated at 9 days, 5 weeks, 6 months and 10.5 months compared to day 0 were submitted to online custom Venn diagram (http://bioinfogp.cnb.csic.es/tools/venny/index.html) for comparison. The genes that were only shared between 6 months and 10.5 months but not in 9 days or 5 weeks were called “Late-memory specific” and chosen for further comparison with two other gene lists (genes that were differentially expressed between 10.5 months and age-matched old naïve mice, or genes that were differentially expressed between old naïve mice and day 0) (Supplemental Figure 8). Genes that were shared among up-regulated “Late-memory specific”, down-regulated “Late-memory specific” and “10.5 months versus old naïve” but not in “old naïve versus young naïve” group were selected as “true Late-memory specific genes” (Supplemental Table I)
These “true Late-memory specific genes” were submitted into DAVID software to identify genes that are associated with gene ontology terms “plasma membrane” or “membrane”. The fold changes and the availability of fluorophore-conjugated commercial antibodies for these genes were used as criteria for selecting the final targets of flow cytometry. Although Cnr2 and Vdr were found only differentially expressed > 2-fold between 10.5 month post-immunization and age-matched old naïve mice, they were still included for consideration because Cnr2 plays important roles in regulation of metabolism or cell survival and Vdr is important in regulation of metabolism/cell survival.
2.10. Sorting human PBMC.
Peripheral blood mononuclear cells (PBMCs) from HIV- and HCV- healthy donors were isolated via Ficoll-paque density centrifugation. CD4 T cells were then isolated by negative selection (Stem Cell, Easy Sep) and resting CD4 T cells were obtained from this population by depleting cells expressing HLA-DR, CD25 and CD69 (Miltenyi). Resting CD4 T cells were stained in bulk for 1) CD99-FITC (Biolegend), CCR10-APC (BD Biosciences), and Propridium Iodide to distinguish viable cells; 2) CD4-PeCy7 only (Biolegend); or 3) CD45RA-PE (Biolegend) and CD45RO-APC (Biolegend). These three populations of cells were sorted independently on the MoFlo.
2.11. High-Dimensional Flow Analysis (ExCYT).
In order to perform a holistic analysis of the flow cytometry data, we utilized ExCYT, a software package for high-dimensional cytometry analysis to conduct dimensionality reduction via t-SNE, clustering via Gaussian Mixture Models, and visualization with the provided tools within the package [62]. Preprocessing of the data within the software included doing conventional lymphocyte and L/D gating followed by an optional CD4 + gate. At this point, 1000 cells were randomly sampled from each sample and were used for downstream analyses. First, a t-SNE analysis was conducted to reduce the data from its original high-dimensional space to two dimensions for visualization purposes. This allowed us to compare populations that may be differentially used by cohorts of our samples. We then applied a Gaussian mixture model (as implemented in ExCYT) to cluster our data. From these clusters, we used the software to identify clusters of data that were differentially utilized between cohorts. Finally, using the high-dimensional box plots that ExCYT produces, we could characterize these differentially utilized clusters via their high-dimensional parameters.
2.12. In vitro recall of human cells with 2017–2018 flu vaccine.
From the CCR10/CD99 co-stained sample, all four populations were isolated (CD99hiCCR10+, CD99loCCR10+, CD99loCCR10−, CD99hiCCR10−), although for some donors the CCR10+CD99lo population was too limited for subsequent stimulation assays. Sorted cells were collected in RPMI containing 20% FBS, washed once with PBS, counted and then resuspended at 10e6 cells/ml. About 400–500,000 sorted CD4 T cells were plated at a 1:2 ratio with CD4-depleted autologous PBMCs, such that the CD4 T cells represented in the well solely derived from the sorted fraction. This combined culture was subject to stimulation with: media only, 1 ul of 2017–2018 strain of flu vaccine (BEI resources), or 10 ug/ml HIV lysate (PepMix, JPT). After 17–19 hrs, cells were stained with CD69-BV421 (Biolegend), CD3-AF700 (Biolegend), CD4-PeCy7 (Biolegend), and eBio780 Fixable Viability Dye (eBio), fixed with 4% formaldehyde, and run on an LSRII.
3. Results
3.1. Tracking gene expression changes of antigen specific CD4 T cells undergoing differentiation to long-lived memory.
To examine the distinct genetic programs utilized by antigen-specific CD4 T cells differentiating into long-lived memory cells, we monitored gene expression changes in CD4 T cells undergoing various stages of differentiation post-immunization for up to 10.5 months. DO11.10 transgenic (Tg) CD4 T cells specific for the OVA(323–339) epitope in complex with I-Ad were chosen for adoptive transfer experiments into naïve BALB/c recipient mice. Supplementary Fig. 1a depicts characterization of the naïve DO11.10 Tg T cells that were adoptively transferred to the recipient hosts. To ensure that the transferred DO11.10 cells were within physiological endogenous clonal frequencies, we transferred 6000 DO11.10 Tg CD4 T cells into each BALB/c host. As the engraftment efficiency for transferred cells is about 10–15%, it is expected that only 600–900 DO11.10 cells would survive in each recipient, which is considered to be within physiological ranges [25]). To evaluate memory cell development at different stages post immunization in an acute infection, a recombinant Vaccinia-OVA virus was used as the immunogen a day after cell transfer. Of note, this recombinant strain clears within 42 days post immunization in mice [9]. Clearance of the immunogen was an important aspect of the experimental design, as we sought to elucidate long-term gene expression changes that occurred upon acute rather than chronic infection, in order to avoid T cell exhaustion. Four groups of mice (15 per group, pooled into 3 biological replicates) were immunized on the same day, but were sacrificed at different time points after immunization; day 9, Week\ 5 (1.5 month), months 6 and 10.5 (Fig. 1a), which were selected to represent activated, early memory, late memory, and long-term memory stages’ respectively. Lymphocytes from these four groups of mice that stained positive for CD44, DO11.10, and CD4 were FACS-sorted as antigen-experienced CD4 T cells. Two additional control groups were also included for gene analyses: (1) young naïve mice sacrificed on day 0 and, (2) old naïve mice age-matched with mice immunized for 10.5 months and sacrificed at the same time. The latter control group was included to distinguish the effects of genes involved in aging from those involved in memory T cell persistence in our analysis. CD44lo DO11.10 CD4 T cells were FACS sorted from these two groups (Fig. 1a). The mRNA from the lysed DO11.10 CD4 T cells were isolated and amplified for microarray analysis. Because of the expected low numbers of CD4 memory T cells in 6 and 10.5 months, we used the Miltenyi μMACS™ SuperAmp™ technology established for RNA amplification of rare cells [2,57]. We used the criteria of nominal p-value < 0.01 and fold change cutoff > 2 to obtain a list of differentially expressed genes from the microarray data. The probe sequences of the selected genes were verified by mapping to UCSC genome browser mouse database before further functional analyses.
Fig. 1. Dynamics of gene expression patterns in DO11.10 CD4 T cells.
a) Schematics of the experimental design. 6000 DO11.10 Tg cells, specific to the OVA (323–339) epitope in complex with H-2d, were adoptively transferred into 8-week old BALB/c recipient mice 24 h before immunization with Vaccinia-OVA virus. Dynamics of gene expression were studied in FACS-sorted CD4 KJ1.26+ CD44+ T cells from lymph nodes and spleens of the recipient mice at day 9, 5 weeks (35 days), 6 months (180 days) and 10.5 months (320 days) post immunization. FACS-sorted CD4 KJ1.26+ CD44− T cells from age-matched naïve mice served as age control for day 0 and 10.5 months in microarray experiments. For each time point, cell samples were pooled from 5 mice for each cage with 3 cages in total, thus 3 biological replicates were used for statistics in microarray. b). Selection of gene clusters based on expression dynamics. The microarray data was submitted into Expander software, which grouped all the genes into 16 clusters based on their expression patterns on day 0, day 9, week 5, month 6, and month 10.5 post immunization. The y-axis shown in each cluster is the fold changes of the relative intensities of genes normalized by log2.
Genes were analyzed using the Expander software [65], which grouped the genes into 16 gene clusters based on the dynamics of their expression patterns across the five time points (Fig. 1b, and Methods). To find genes specific for the development and longevity of CD4 memory T cells, we defined gene clusters as memory-specific if their expression patterns met one of the following three conditions: a) gene expression continuously went up after 5 weeks, b) gene expression continuously went down after 5 weeks, or c) gene expression did not change from month 6 to month 10.5, but the gene expression in month 6 was either higher or lower than week 5. Separating differential gene expression using these criteria showed a remarkable pattern of memory-specific gene signatures at the 10.5-month time point, often appearing many months after the initial priming event. Per the above criteria, six gene clusters (1, 2, 3, 5, 7 and 8) were selected as being “memory-specific” and were submitted to the DAVID Bioinformatics Resources software for gene ontology (GO) Biological Process analysis. Five GO terms among the entire top enriched GO Biological Process terms were found: regulation of apoptosis, regulation of proliferation, response to DNA damage, lipid metabolism, and carbohydrate metabolism (Supplementary Fig. 1b).
To find out how the genes associated with these five GO terms might contribute to the longevity of CD4 memory T cells, we examined the dynamics of differentially expressed genes between day 0 and 10.5 months in each GO term category separately (Figs. 2, 3 & Supplementary Figs. 2-6). Interestingly, the heatmaps of global genetic changes revealed that there were two major gene expression patterns. One set of genes maintained low expression levels at the activation phase but higher expression levels in the memory phase; while the other set of genes displayed higher expression levels in the activation phase but were downregulated during the memory phase. A PubMed literature search on every gene in these five GO term heatmaps provided some understanding for potential functions of these genes, linking their expression patterns to CD4 memory T cell development.
Fig. 2. Expression dynamics and functions of genes in three gene ontology (GO) terms.
Genes that were differentially expressed between 10.5 months post immunization and day 0 were submitted to DAVID database to identify those associated with the five GO terms enriched in “memory specific clusters” in Fig. 1b. Global expression heatmaps of genes associated with GO terms “cell proliferation” (a), “DNA repair” (b) or “apoptosis” (c) were created with a row Z-Score ranges from −4 to 4(left panel). A few representative genes from each GO term gene group were selected with fold changes in their expression levels at different time points comparing to day 0 (middle panel). The potential function and related pathways of these selected genes were depicted (right panel). Genes were differentially (fold change > 2, p-value < 0.05) upregulated (pink box), downregulated (blue box) or not differentially expressed (white box) in 10.5 months post immunization compared to day 0. Genes in red circle have been shown in literature to be able to form complexes. Arrows between genes/pathways indicate positive regulation and lines with blunt end between genes/pathways indicate negative regulation by literature references.
Fig. 3. Expression dynamics and functions of genes in two gene ontology (GO) terms.
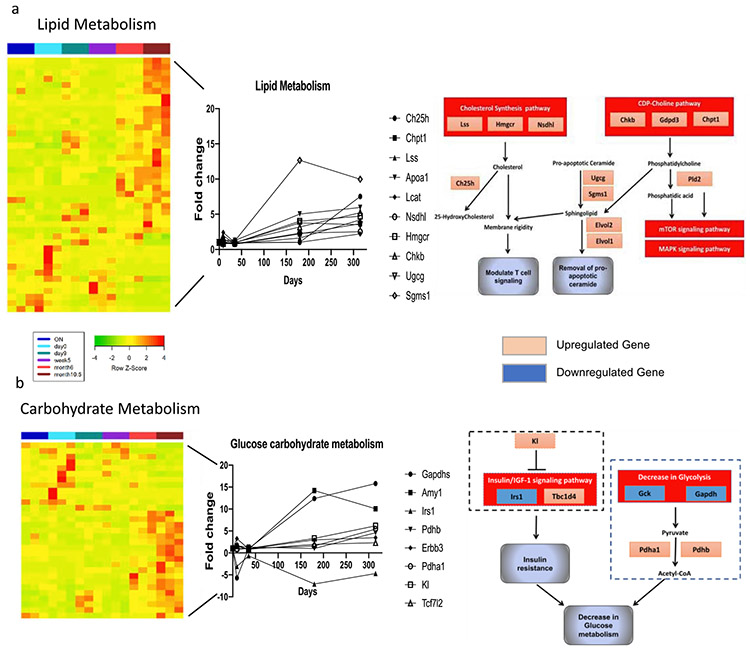
Genes that were differentially expressed between 10.5 months post immunization and day 0 were submitted to DAVID database to identify those associated with the five GO terms enriched in “memory specific clusters” in Fig. 1b. Global expression heatmaps of genes associated with GO terms “lipid metabolism”(a) or “carbohydrate metabolism”(b) were created with a row Z-Score ranges from −4 to 4 (left panel). A few representative genes from each GO term gene group were selected with fold changes in their expression levels at different time points comparing to day 0 (middle panel). The potential function and related pathways of these selected genes were depicted (right panel). Genes were differentially (fold change > 2, p-value < 0.05) upregulated (pink box), downregulated (blue box) in 10.5 months post immunization compared to day 0. Genes in dotted square are viewed as a whole mechanism impacting other pathways. Arrows between genes/pathways indicate positive regulation and lines with blunt end between genes/pathways indicate negative regulation by literature references.
3.2. Genes regulating cell proliferation, DNA damage, and survival are enriched in long-term CD4 memory T cells
Using this approach, we formulated pathways by which these genes differentially expressed at 10.5 months could help in supporting the longevity and survival of CD4 memory T cells. For example, we found most of the differentially-expressed genes in GO terms linked to cell proliferation and DNA repair response were associated with the tumor suppressor p53, which has been known for both negatively regulating cell proliferation and enhancing DNA repair response under oxidative stress [26,68]. Fig. 2a & 2b depict two representative sets of genes with proposed roles in the p53 pathway, or downstream in the DNA base excision repair pathway. Most of these representative p53 associated genes (e.g. Tial1, Msx1, Ing5, Vdr, Rxra, Ncoa6, Gadd45a, Alkbh2), when compared to young naïve mice, were found to be expressed at significantly higher levels at 6 months and 10.5 months post immunization (Fig. 2a & b), which highlights the importance of DNA repair response and reduced proliferation in long-lived memory cells. It is noteworthy that we also identified several genes that can limit access to DNA for replication and transcription. One such example is the histone gene Hist1h2ai, which encodes the nucleosome component histone H2A1. Because the nucleosome regulates access to certain regions during DNA replication in cell division, it can serve as a gene repressor [22,42]. Down-regulation of Hist1h2ai during CD4 activation may, therefore, facilitate an increase in DNA replication and cell proliferation. The 7-fold up-regulation of Hist1h2ai at 6 months and 10.5-month compared to day 0 post immunization suggests that late-stage long-lived CD4 memory T cells may have restricted DNA replication or transcriptional capabilities. This is consistent with a state of quiescence important for the longevity of CD4 memory T that has been shown experimentally [11,12,37,44,45].
In addition to observing a tightly-regulated proliferation and DNA repair program, long-lived CD4 memory T cells require the activity of pathways involved in prevention of cell apoptosis (Fig. 2c). Such programs have been found to be involved in maintaining long-lived CD8+ memory T cells [15,20]. Consistent with this notion, we observed that genes associated with the apoptosis GO term were mostly upregulated at 6 months and 10.5 months (Fig. 2c). The PI3K/Akt and Ras-ERK pathways, which regulate responses to apoptosis are among those genes and are highlighted. In general, most upregulated genes at these later time points have a role in the activation of pro-survival/anti-apoptotic proteins and inhibition of pro-apoptotic proteins. An example is Pou4fi (also known as Brn-3a), which was upregulated 18–20 fold at both 6 month- and 10.5-month time points, activates pro-survival proteins Bcl-2 and Bcl-xL, and cooperates with p53 to induce cell cycle arrest [8,31]. Altogether, these dynamic gene expression changes suggest that during CD4 memory differentiation, a combination of programs that limit proliferation, reduce DNA damage, and prevent apoptosis help to maintain long-term memory T cell longevity.
3.3. Long-lived CD4 memory T cells express a unique lipid metabolism gene signature
Recent studies have described that during the shift from effector to memory states, CD8 T cells undergo changes in their energy metabolism from glucose to lipid metabolism[3,53,66]. This shift in metabolism is thought to promote CD8 memory T cell longevity and function [53]. In light of these studies, we compared the expression levels of genes related to regulation of metabolism in naïve versus memory T cells over time. Fig. 3a & b and Supplemental Figs. 1, 5 & 6 show that a lipid metabolism program, rather than glucose metabolism, predominates in CD4 memory T cells.
A detailed assessment of this lipid metabolism program suggested that choline and ceramide metabolism as well as cholesterol synthesis and metabolism, were important for long-lived CD4 memory T cells. As shown in Fig 3a, a list of genes involved in regulating these metabolism pathways had increased expression levels at 6 and 10.5 months compared to young naive (Fig. 3a and Supplementary Fig. 5). Although the exact role of cholesterol in CD4 memory T cells remains unclear, the levels of cellular cholesterol could impact the rigidity of T cell membranes. Increased cholesterol in the membrane can influence T cell signaling by affecting TCR clustering and T cell proliferation, both of which have clear implications for CD4 memory T cell function [5].
In addition to cholesterol metabolism, ceramide metabolism plays a role in cellular persistence, as ceramide is associated with mediating apoptosis and antagonizing insulin-stimulated glucose uptake [6]. Consistent with this, we observed an upregulation of the genes involved in reducing ceramide levels. For example, Sgms1 converts ceramide to sphingomyelin via the addition of phosphatidylcholine, reduces reactive oxygen species, and facilitates normal mitochondrial function. Ugcg, which encodes an enzyme catalyzing glycosylation of ceramide, has anti-apoptotic and pro-survival effects ([14,24,32,36,41,46,60,71]). A central protective role of these genes may be in limiting the overall ceramide levels in long-lived CD4 memory T cells, which may be similar to a specific strategy often co-opted by various tumors to enhance cell longevity [47]. For example, we observed an upregulation of genes mediating synthesis of phosphatidylcholine in the CDP-choline pathway (Fig. 3a), which could facilitate the generation of sphingolipid from ceramide, or be converted to phosphatidic acid by Pld2 to activate mTOR and MAPK signaling pathway [29,55]. Taken together, this unique genetic program involving lipid metabolism helps to maintain the longevity of CD4 memory T, primarily by affecting cellular cholesterol and ceramide levels, and allowing for more efficient lipid oxidation to drive cellular metabolic needs.
In parallel to the finding of upregulated lipid metabolism, we observed a downregulation of glucose/carbohydrate metabolism in long-lived CD4 memory T cell, which was exemplified by the decreased expression of genes mediating glycolysis and increased expression levels of genes inhibiting the insulin signaling pathway in 10.5 months post immunization. A few representative genes in these two pathways were shown as examples in Fig. 3b. Among these genes, the expression of the Irs1 gene, was sharply downregulated > 6-fold in CD4 memory T cells at 6 months and 10.5-months post immunization (Fig. 3b). This gene encodes for insulin receptor substrate 1, which can regulate insulin signaling [27]. Downregulation of this gene, together with other genes (e.g. Kl) involved in insulin signaling, indicates an increased level of insulin resistance in the CD4 memory T cells, which would potentially decrease the uptake of glucose into the cells and allow those cells to shift from using predominantly glycolysis to lipid oxidation for maintaining cellular energy requirements. This shift in cellular metabolism, which has been demonstrated to be a major hallmark of CD8 + memory T cell biology [49,52].
In summary, this specific genetic program mediating the upregulation of lipid metabolism and downregulation of glucose metabolism in long-lived CD4 memory T cells highlight a unique immunometabolic pathway of regulating ceramide metabolism that may be augmented therapeutically.
3.4. Verification of microarray-described gene expression provides novel markers of long-term CD4 memory T cells
Our microarray data was obtained from analyses of adoptively transferred DO11.10 Tg T cells into naïve recipient mice. To validate these results in a polyclonal context, we took advantage of the availability of HLA-DR1 Tg mice and an in-house prepared tetramer reagent against the immunodominant epitope of H5N1 influenza HA protein that we have previously identified [23,35]. HLA-DR1 Tg mice have a full T cell repertoire [56], therefore represent more physiological conditions for T memory formation. We immunized HLA-DR1 Tg mice with an attenuated H5N1 influenza vaccine in CpG and examined the development and responsiveness of the CD4 memory T cell specific to the immunodominant epitope, H5N1-HA(259–274) peptide challenge. Specific CD4 memory T cells were tracked using HA(259–274)-specific MHC-II tetramers. To enrich HA(259–274) specific resting CD4 memory T cells, and validate their quiescent phenotype, T cells from 10.5 month immunized mice were placed in culture for 7 days with or without antigen. By doing so, non-specific or irrelevant CD4 T cells die off during the lengthy in vitro culture, while quiescent memory T cells survive and are enriched. The results (Fig 4a) showed that these CD44hi HA(259–274)/DR1 tetramer positive CD4 memory T cells did not proliferate upon in vitro challenge with either H5N1-HA1 protein or H5N1 vaccine, unlike the robust increase in the number of HA(259–274)-specific CD4 T cells from recently immunized effector CD4 T to the in vitro challenge with either H5N1-HA1 protein or attenuated vaccine (Supplementary Fig. 7). The finding that long-lived H5N1-HA(259–274) specific CD4 memory T cells developed in DR1 Tg mice are quiescent, revealed generality of our findings and justified further evaluation of the microarray data.
Fig. 4. Expression of selected membrane associated proteins in quiescent H5N1 specific memory CD4 T cells in DR1 mice.
a) DR1 mice were injected with H5N1 vaccine and CpG i.p., and 4 months later, draining lymph nodes were extracted and cells incubated for 7 days without any stimulation, or stimulated by either H5N1 HA1 protein or H5N1 vaccine in the media. The cells were then stained with H5N1 (Left top panels) or CLIP tetramers (Left bottom panels), as well as CD44 and other antibodies for flow cytometry analysis. Dots show the CD44hi Tetramer+ lymphocytes after exclusion of macrophages, B cells and CD8+ T cells. The flow data from 4 repeated experiments is summarized on the right panel, p = 0.88. b) Expression dynamics of several selected membrane associated genes during memory CD4 T cell development (c) Expression of different selected membrane/surface markers on CD4 CD44hi H5N1 tetramer+ memory cells in spleen (orange), CD4 CD44hi H5N1 tetramer+ activated cells (red) and CD4 CD44lo naïve cells (blue). The cells are from the CD4 CD44hi Tetramer+ population (for activated CD4 T cells and CD4 memory T cells) or CD4CD44lo population (for naïve CD4 T cells) shown in Supplemental figure 9. d) Summary of MFI levels in protein expression among memory CD4 T cells, activated CD4 T cells and naïve T cells from 3 repeated experiments. Data are presented as mean ± standard deviation. Asterisks denote statistical significance: **** P < .0001, *** P < .001, ** P < .01, * P < .05, Student’s two-tailed paired ratio T-test.
We chose to examine the protein expression of a few selected genes encoding proteins expressed on cell membranes and expression increased during long-term CD4 memory T establishment (i.e. by 6- and 10.5-months post-immunization) (Fig. 4b, Supplemental Figure 8, Supplemental table I and Methods). We reasoned that this strategy could also yield potential long-term memory markers that could easily be detected by fluorescence staining. Seven proteins were chosen including IL-7R, a well-established marker for CD8 and CD4 memory T precursor cells (Fig. 4b).
To evaluate the protein expression of the putative memory markers we used H5N1-HA(259–274)/DR1 specific CD4 memory T cells ten months post immunization with H5N1 influenza vaccine in CpG. Similar to the setup described in Fig. 4a, H5N1-HA(259–274)/DR1 specific CD4 memory T cells were enriched by in vitro culture for 7 days in the absence of antigen stimulation to allow dying of nonspecific cells. Enriched cells were then co-stained with antibodies against CD4, CD44, and others listed in the Methods. For comparison, we also assessed these markers on “activated” H5N1-specific CD4 T cells from H5N1-HA(259–274) peptide immunized DR1 Tg mice. Lymphocytes from the draining lymph nodes harvested 9 days post-immunization and cultured with the same peptide for 7 days in vitro served as positive controls. Supplementary Fig. 9 shows a high percentage of HA(259–274)/DR1 tetramer+ effector CD4 cells (~9%) in recently immunized mice versus memory populations that expectedly were only ~ 1.6% tetramer+.
The results shown in Fig. 4c & d compared the expression levels of the seven selected proteins among memory, activated, or naïve CD4 T cells. Because the number of naïve antigen-specific CD4 T cells from unimmunized mice was too small after culturing for 7 days, naïve CD4 T cells from mice immunized with H5N1 vaccine and gated from the CD4+ CD44lo population served as naïve controls. As shown, all selected genes were expressed robustly in CD4 memory T cells compared to naive and activated CD4 T cells, except for the expression of Itga3, which showed some overlap between memory and activated CD4 T cells.
Overall, these experiments validated our microarray data and revealed genetic signatures specific for long-term murine CD4 memory T cells. From these genetic programs, we observed specific cell-surface markers that were enriched on antigen-specific murine CD4 memory T cells emerged that can now be used for identification of long-lived memory CD4 T cells in mice. We wondered whether these cell-surface markers could not only denote murine CD4 T cell memory populations, but also human ones.
3.5. Evaluation and testing of long-term CD4 T cell memory markers in healthy human donors
We next attempted to examine memory CD4 T cells from healthy donors for expression of the new memory markers. CD45R0 is a commonly-used marker of memory cells in humans, and cells that express CD45RA isoform classically denote naïve or effector cells [43]. To that end, we measured surface expression levels of the selected markers defined in murine memory T cells in 12 healthy individuals by flow cytometry and correlated them with expression of CD45RA and CD45R0. We observed an enrichment in cells expressing CD99, CCR10 and Itga3 in the CD45R0 compartment at comparable levels to that observed with the canonical memory marker, IL-7R (Fig. 5a-c). Importantly, we included activation markers, CD25, CD69 and HLA-DR in our panel to exclude cells that may have been activated in vivo, and therefore would not resemble a quiescent, long-lived memory CD4 T cell phenotype (Supplementary Figure 10). Interestingly, we observed about a 3-fold increase in MFI of CD99 in resting CD4 CD45R0 versus CD45RA + cells, corresponding to a 1-log increase on the flow plots (Fig. 5c), which was twice as high as the expression levels of IL-7R. Furthermore, CCR10 and Itga3 appeared to mainly be expressed on CD45R0 cells (Fig. 5c). Importantly, gating on any of these three markers is over 95% predictive of CD45R0, or “antigen experienced,” status (Fig. 5b), suggesting that while not all CD45R0 cells have these markers, these markers are exclusively present on CD45R0 cells. Fig. 6
Fig. 5. Several putative long-term memory markers from the murine microarray analysis exclusively co-localize with existing human memory marker CD45R0.
a) The percentage of CD99hi, CCR10+, Itga3+ and IL7R+ expression gated on resting CD4 T cells of 12 healthy donors. b) Resting CD4T cells that either expressed high levels of the putative memory markers from our microarray analysis – CD99 (CD99hi), CCR10, or Itga3 – were assayed for the presence or absence of canonical memory marker CD45R0. Notably, nearly 100% of cells expressing these putative markers also expressed CD45R0 (CD45R0+). c) Cells that either express or lack CD45R0, indicated as CD45R0− in the figure (i.e. CD45RA+), were assayed for relative frequencies of CD99hi, CCR10+, Itga3+ and IL7R+. The percentages and MFI of CD99, CCR10 and Itga3 positive populations in either CD45R0+ or CD45R0− compartments reveal an enrichment of these markers in the CD45R0+ compartment, to a degree comparable to that of IL-7Rhi cells. Data are presented as mean or mean ± standard deviation. Asterisks denote statistical significance: **** P < .0001, *** P < .001, Student’s two-tailed paired ratio T-test, n = 12.
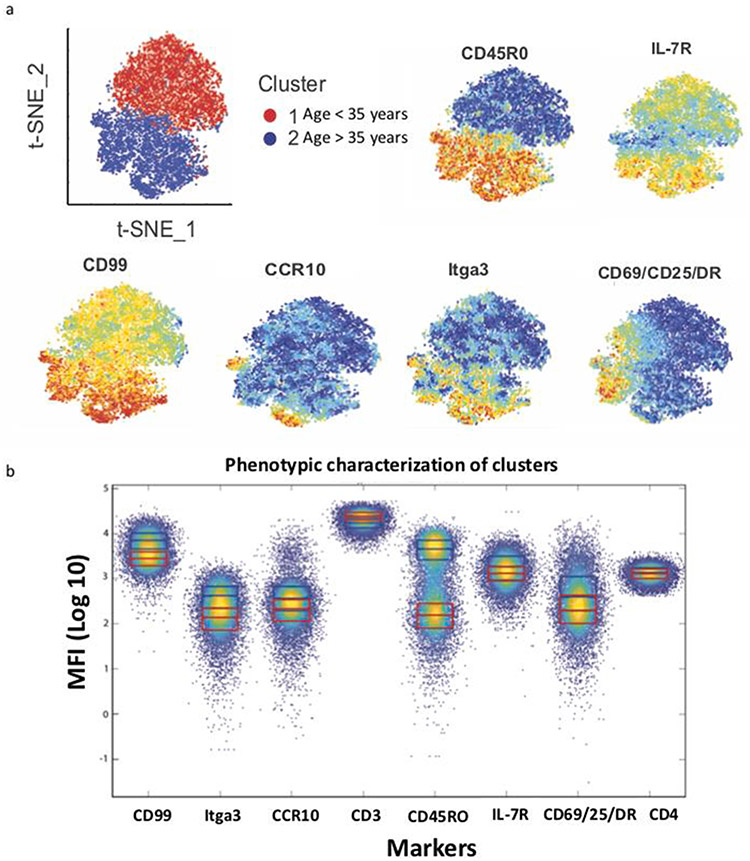
Fig. 6. Expression of memory markers increase with aging in human cohorts.
a) The median expression levels of various protein markers for CD4 memory T cells stained by flow cytometry were summarized for comparison between two cohorts of individuals with different age ranges (> =35 years or < 35 years). Cluster 1 represents protein expression levels in cohorts of healthy donors (age < 35 years) while cluster 2 represents protein expression levels in cohorts of healthy donors (age > = 35 years). The protein expression levels used by ExCyt for t-SNE analysis were based on CD45RO expression from flow cytometry staining of CD4 T cells in two cohorts of healthy human individuals with varied age and gender (N = 12). b) CD99, CCR10, Itga3 and IL7R all have higher expression levels in healthy donors with age > =35 years, and notably all co-localize with cells expressing high levels of CD45R0.
CD99 has been found to be related to optimal co-stimulation of T cells [50] and highly expressed on activated/memory T cells from lymph nodes of patients with gastric cancer or chronic peptic ulcer [51]. It has also recently been reported to be upregulated on human fetal CD4 T memory T cells [39], while CCR10 has been shown to mark memory CD4 T cells that home to the skin [30]. Itga3, or integrin subunit alpha, makes up half of the α3β1 integrin and recently has been shown to be a critical regulator in Th17 cell differentiation [72]. However, none of these features (CD99hi, CCR10+, Itga+) have been used to stain human memory cells in the past, and represent novel markers for potentially long-lived subsets in the human. Of the markers, CD99 and CCR10 appeared to have the most distinguishable staining pattern for detecting memory populations, as Itga3 appears to be expressed at low levels (data not shown). We therefore followed up exclusively with these two markers for functional tests.
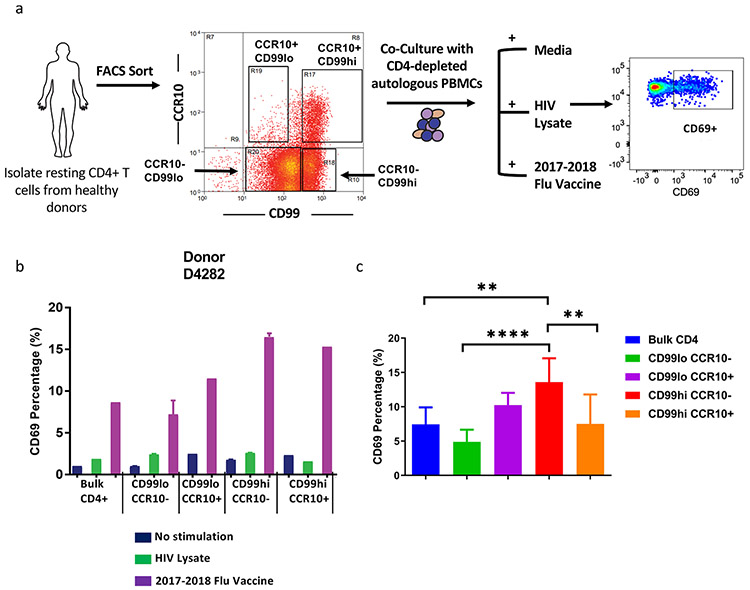
To measure whether cells expressing CD99 and CCR10, either as double, or single positive cells contained more antigen-specific CD4 T cells, we sorted these populations from the peripheral blood of healthy human donors who had been vaccinated with the 2017–2018 attenuated flu vaccine and performed in vitro recall experiments. Resting CD4 T cells from the same human donors were challenged in vitro with the 2017–2018 flu vaccine or HIV lysate as a negative control (Fig 7a). Notably, CD4 T cells expressing one or both of these markers responded more robustly to the flu vaccine than bulk CD4 T cells, suggesting an enrichment of Ag-specific cells in these populations (Fig. 7b-c, Supplemental Fig. 11). CD99hi cells responded most robustly to rechallenge. Interestingly, CD99loCCR10− cells responded less well than bulk CD4 T cells, most prominently observed in donor D4282, suggesting that removing cells expressing CD99, CCR10, or both reduces the flu-specific memory CD4 T cell population. These observations are consistent with the fact that levels of CD99 and the percent of cells expressing CCR10 appear to segregate with CD45R0 + memory T cells (Fig. 7c).
Fig. 7. CD4 T cells that have high expression levels of adhesion molecule CD99 exhibit enhanced responsiveness to in vitro flu vaccine challenge than the bulk CD4 T cell population, and depleting cells expressing high levels of this marker blunts the anti-flu recall response.
a) Four different CD4 T cell subsets (CD99lo CCR10−, CD99lo CCR10+, CD99hi CCR10−, CD99hi CCR10+) were sorted based on the expression of CD99 and CCR10 and cultured with media alone, HIV lysate or the 2017–2018 attenuated flu vaccine in vitro with CD4-depleted autologous PBMCs for 17–19 h. The percentage of CD69+ cells was used as readout for activation. b) The percentages of CD69+ CD4 T cells from different sorted subsets in a representative healthy donor under three different in vitro stimulation conditions are shown. All healthy human donors were vaccinated with 2017–2018 flu vaccine a year ago before donating blood. The error bars represent mean ± SD. c) Summary of responses from stimulated four sorted subsets to 2017–2018 flu vaccine in three healthy donors. Error bars represent mean ± SD. ** P < .01, **** P < .0001. Student’s two-tailed paired ratio T-test.
4. Discussion
A major unanswered question in the field of CD4 T cell memory is understanding what specific genetic/phenotypic characteristics distinguish long-lived memory CD4 T cells from effector and short-lived antigen-specific CD4 T cells. Addressing this knowledge deficit would have profound implications for designing more durable vaccines aimed at inducing CD4 T cell responses that augment humoral and cytotoxic T cell immunity for the lifetime of an individual. Here, we have examined the dynamics of gene expression in antigen-specific CD4 T cell differentiation in an acute infection from naïve to activated and memory states up to nearly a year post immunization using sensitive microarray technology.
Our approach improves on other studies in at least two distinct ways: 1) we have studied antigen-specific clonal populations under physiologically relevant conditions in mice with polyclonal TCR repertoires, and 2) we have studied the kinetics of memory development for over 10.5 months. Following antigen-specific cells enabled us to identify true long-term memory CD4 T cells based on the parameter of time rather than previously-reported memory surface marker expression. Most existing studies often choose to define memory T cells 30 days post antigen stimulation and rely on the expression of certain memory markers (e.g. CD45R0 in human, CD44 in mice) to track “antigen experienced” cells. However, such memory markers may change based on the activation state of the cell, and what one believes to be a memory population may simply be a recently-activated T cell population that inaccurately represents a long-term memory phenotype.
Another particular strength of our study is that we have studied gene expression in two murine antigenic systems: DO11.10 CD4 Tg T-cells in BALB/c mice, H5N1-HA(259–274) specific CD4 T cells in polyclonal DR1 Tg mice, and importantly, in healthy human donors. Studying the dynamics of gene expression in CD4 T cells in an acute infection model from their naïve state to acute activation, contraction, early memory, and late memory transitions under physiological conditions, enabled a systemic evaluation of genes that are highly likely to contribute to establishment of long-lived memory phenotype. Memory CD4 T cells developed for a viral infection may be heterogeneous [67]. Our previous studies on cytokine production from Vaccinia-OVA specific memory CD4 T cells indicated that the long-lived memory CD4 T cells conformed to a Th1-differentiated phenotype [11]. This was also confirmed by our microarray data analysis, which shows that only Th1 signature genes, including T-bet, were differentially upregulated in the activation/memory stages (data not shown). Additionally, a comparison of our microarray data with recently published RNA-seq data from early memory CD4 T cells [10] found some shared genes despite the differences in the antigenic systems and gene detection technologies. Moreover, several of our murine long-lived memory CD4 T cell genes have similar expression patterns to that of human long-lived CD8 memory T cells [1]. These findings highlight the idea that in long-lived memory T cells regardless of species, antigens, or T cell types certain expression patterns are shared.
Two highly significant conclusions can be drawn from our gene analyses. First, there is a gene expression program initiated from the first priming of a CD4 T cell that has long-ranging effects at the memory stage, by reducing the cell’s proliferative capacity and immune activation, while increasing the cell’s ability to respond to DNA damage and prevent apoptosis. This signature is consistent with our prior observations that antigen-experienced CD4 T cells become quiescent upon encounter with low avidity engagement of TCR during the resolution of infection. Importantly, the increased expression of a majority of these genes were first detected at the effector to memory differentiation phase, and continued through 6 months and for some genes through 10.5 months, strongly suggesting that memory T longevity requires genetic programs that continuously and actively work together. In addition, several metabolism-related genes were also dynamically regulated at the memory stage. Notably, we observed gene expression changes consistent with enhanced insulin resistance, which may play an important role in transitioning long-term CD4 memory T cells from a primarily glucose to lipid metabolism profile, similar to CD8+ memory T cells. Most strikingly, however, was the differential expression of genes involved in ceramide metabolism, presumably to limit levels of this biomolecule. Such a program may represent a unique mechanism by memory CD4 T cells to ensure persistence. Moreover, understanding the molecular “switch” that promotes high levels of this pathway may guide efforts to induce enhanced, durable CD4 T cell memory responses.
While it is currently unclear how such a signature may be triggered in Ag-specific CD4 T cells at first priming, we have shown in the past that B cells are the predominant antigen presenting cell at the contraction phase of an immune response [12]. It is possible that B cell presentation of low numbers of pMHC-II complexes selects for high-affinity antigen-experienced CD4 memory T cell precursors, and that the physical and signaling characteristics of this interaction initiates a genetic and epigenetic program that allows for cellular persistence. Further mechanistic studies on the molecular events involving memory CD4T cell fate determination would help to elucidate this critical transition.
To validate the microarray data, we assessed the protein expression of some of the genes whose protein products are known to be expressed on the cell membrane. Such markers should ideally serve as new CD4 T cell markers associated with different phases of memory development and longevity. Indeed, all selected genes were expressed strongly on long-lived (10.5 months) CD4 memory T cells, in contrast to either weak upregulation, or no change on activated CD4 T cells, and lower expression levels at early memory phase (six months). Remarkably, Cnr2 expression was only enhanced by 10.5 months, suggesting that memory longevity requires continuous upregulation of genes even at very late stages of memory CD4 T cell life. The rest of the 5 selected genes showed a gradual increase in expression, reaching their maximum expression by 300 days post immunization, an observation that signifies the likely importance of those proteins to regulating the longevity of long term CD4 memory T cells. While a mechanistic link between expressions of these genes and their connection to CD4 T cell longevity remain to be established, some clues can be found from literature. For example, the Klotho protein has long been associated with anti-aging characteristics through a variety of mechanisms, such as anti-oxidation, anti-senescence, anti-autophagy, and modulation of many signaling pathways, including insulin-like growth factor and Wnt signaling [4,38]. High expression of Cnr2 in developing B cells has also been shown to associate with a stem-cell like characteristic, which has been associated with a memory CD4 phenotype [54]. Therefore, it would be of great interest to explore the molecular roles of these proteins in long-lived memory CD4 T cells in future work.
The second important message from our studies is that the identified murine memory markers were applicable to human memory CD4 T cells. When resting CD4 T cells from the peripheral blood of healthy donors were tested for expression of these proteins, CD99, Ccr10 and Itga proteins were expressed on human memory cells exclusively with other conventional memory markers such as CD45R0. However, human memory cells varied in gene expression of Cnr2 and Kl, which might relate to the lack of information about the timing of prior exposure to the antigens in human samples. This difference may also reflect an evolutionarily divergent transcriptional landscape in long-lived memory cells between mice and humans, where the functions of Klotho and Cnr2 in mice are fulfilled by other protein products in the human. Alternatively, it is possible that human memory CD4T cells in lymphoid organs may express higher transcript and protein levels of these markers, but that those in peripheral blood – our source of PBMCs – may not. Overall, we observed more robust immune responses from sorted human CD4 T cells expressing CD99, and/or CCR10, than bulk CD4 T cells to influenza vaccine challenge in vitro. These data suggest that CD99 and CCR10 molecules on resting CD4 T cells, independent of any other “memory” marker, can designate memory CD4 T cells in humans.
In conclusion, our study has tackled the dynamics of gene expression in specific CD4 T cells generated in vivo under physiological conditions following CD4 T cells during the transition from naïve to activated, memory, and long-term memory stages. We have identified several genetic programs that together reduce cell proliferation and immune activation, protect cells from apoptosis, promote survival, and regulate metabolism in favor of maintaining the longevity of the memory CD4 T cells. These programs appear to be dynamically regulated in antigen-specific memory CD4 T cells at the initial priming event, often only materializing gradually or after 10.5 months (equivalent to > 32 years in the human) [16].
Apart from defining gene expression signatures critical to CD4 T memory persistence, we identified several cell-surface proteins specific to memory CD4 T cells. These markers can now serve as true markers of a long-lived memory CD4 T cell phenotype in mice and even in humans, as they are present on memory CD4 T cells that yield most robust responses to a vaccine challenge. Thus, our studies have important implications for preventative and therapeutic vaccine designs, as well as the study of infectious agents that persist long-term in CD4 T memory cells, including HIV-1 and sars-cov 2.
Supplementary Material
Acknowledgements
We thank Drs. Jonathan Powell, Kevin Shenderov and Im Hong Sun for critical reading of the manuscript, NIAID Tetramer Core Facility for DR1 monomers and CLIP/DR1 tetramers, Bei Resources for influenza vaccine and HA proteins, and Dr. Dennis Zaller for the original DR1 Tg mice. Supported by grants from NIAID, R01AI063764, R21AI101987, and R01AI120634, to SS-N, Howard Hughes Medical Institution to RFS, NIAID F30 AI136704 to SS, and AAI Careers in Immunology Fellowship Award to N.S. and SS-N.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellimm.2020.104210.
References
- [1].Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. , Origin and differentiation of human memory CD8 T cells after vaccination, Nature 552 (2017) 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Appay V, Bosio A, Lokan S, Wiencek Y, Biervert C, Kusters D, Devevre E, Speiser D, Romero P, Rufer N, Leyvraz S, Sensitive gene expression profiling of human T cell subsets reveals parallel post-thymic differentiation for CD4 + and CD8+ lineages, J Immunol 179 (2007) 7406–7414. [DOI] [PubMed] [Google Scholar]
- [3].Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R, mTOR regulates memory CD8 T-cell differentiation, Nature 460 (2009) 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bian A, Neyra JA, Zhan M, Hu MC, Klotho, stem cells, and aging, Clin Interv Aging 10 (2015) 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bietz A, Zhu H, Xue M, Xu C, Cholesterol Metabolism in T Cells, Front Immunol 8 (2017) 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bikman BT, Summers SA, Ceramides as modulators of cellular and whole-body metabolism, J Clinic Investig 121 (2011) 4222–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bourgeois C, Veiga-Fernandes H, Joret AM, Rocha B, Tanchot C, CD8 lethargy in the absence of CD4 help, Eur J Immunol 32 (2002) 2199–2207. [DOI] [PubMed] [Google Scholar]
- [8].Budhram-Mahadeo V, Morris P, Ndisang D, Irshad S, Lozano G, Pedley B, Latchman DS, The Brn-3a POU family transcription factor stimulates p53 gene expression in human and mouse tumour cells, Neurosci Lett 334 (2002) 1–4. [DOI] [PubMed] [Google Scholar]
- [9].Buller RM, Smith GL, Cremer K, Notkins AL, Moss B, Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype, Nature 317 (1985) 813–815. [DOI] [PubMed] [Google Scholar]
- [10].Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. , The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok, Immunity 50 (91–105) (2019) e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dalai SK, Khoruzhenko S, Drake CG, Jie CC, Sadegh-Nasseri S, Resolution of infection promotes a state of dormancy and long survival of CD4 memory T cells, Immunol Cell Biol (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dalai SK, Mirshahidi S, Morrot A, Zavala F, Sadegh-Nasseri S, Anergy in memory CD4+ T cells is induced by B cells, J Immunol 181 (2008) 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].De Riva A, Bourgeois C, Kassiotis G, Stockinger B, Noncognate interaction with MHC class II molecules is essential for maintenance of T cell metabolism to establish optimal memory CD4 T cell function, J Immunol 178 (2007) 5488–5495. [DOI] [PubMed] [Google Scholar]
- [14].Deevska GM, PP D, AA K, G I, M W, SB K Jr, M A, and MN N-K (2017). Novel Interconnections in Lipid Metabolism Revealed by Overexpression of Sphingomyelin Synthase-1. . J Biol Chem. 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ, Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory, Immunity 37 (2012) 1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dutta S, Sengupta P, Men and mice: Relating their ages, Life Sci 152 (2016) 244–248. [DOI] [PubMed] [Google Scholar]
- [17].Edgar R, Domrachev M, Lash AE, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository, Nucleic Acids Res 30 (2002) 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Farber DL, Yudanin NA, Restifo NP, Human memory T cells: generation, compartmentalization and homeostasis, Nat. Rev. Immunol 14 (2014) 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Greider CW, Telomerase discovery: the excitement of putting together pieces of the puzzle (Nobel lecture), Angew Chem Int Ed Engl 49 (2010) 7422–7439. [DOI] [PubMed] [Google Scholar]
- [20].Gupta S, Gollapudi S, Effector memory CD8+ T cells are resistant to apoptosis, Ann N Y Acad Sci 1109 (2007) 145–150. [DOI] [PubMed] [Google Scholar]
- [21].Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK, Duration of antiviral immunity after smallpox vaccination, Nature medicine 9 (2003) 1131–1137. [DOI] [PubMed] [Google Scholar]
- [22].Han M, Grunstein M, Nucleosome loss activates yeast downstream promoters in vivo, Cell 55 (1988) 1137–1145. [DOI] [PubMed] [Google Scholar]
- [23].Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, et al. (2010). A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat Med 16, ppl333–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hartmann D, Wegner MS, Wanger RA, Ferreiros N, Schreiber Y, Lucks J, Schiffmann S, Geisslinger G, Grosch S, The equilibrium between long and very long chain ceramides is important for the fate of the cell and can be influenced by co-expression of CerS, Int J Biochem Cell Biol 45 (2013) 1195–1203. [DOI] [PubMed] [Google Scholar]
- [25].Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK, Naive and memory CD4+ T cell survival controlled by clonal abundance, Science 312 (2006) 114–116. [DOI] [PubMed] [Google Scholar]
- [26].Helton ES, Chen X, p53 modulation of the DNA damage response, J Cell Biochem 100 (2007) 883–896. [DOI] [PubMed] [Google Scholar]
- [27].Hemi R, Paz K, Wertheim N, Karasik A, Zick Y, Kanety H, Transactivation of ErbB2 and ErbB3 by tumor necrosis factor-alpha and anisomycin leads to impaired insulin signaling through serine/threonine phosphorylation of IRS proteins, J Biol Chem 277 (2002) 8961–8969. [DOI] [PubMed] [Google Scholar]
- [28].Homann D, Teyton L, Oldstone MB, Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory, Nat Med 7 (2001) 913–919. [DOI] [PubMed] [Google Scholar]
- [29].Hornberger TA, Sukhija KB, Chien S, Regulation of mTOR by mechanically induced signaling events in skeletal muscle, Cell Cycle 5 (2006) 1391–1396. [DOI] [PubMed] [Google Scholar]
- [30].Hudak S, Hagen M, Liu Y, Catron D, Oldham E, McEvoy LM, Bowman EP, Immune surveillance and effector functions of CCR10(+) skin homing T cells, J Immunol 169 (2002) 1189–1196. [DOI] [PubMed] [Google Scholar]
- [31].Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS, Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate, J. Biol. Chem 280 (2005) 11851–11858. [DOI] [PubMed] [Google Scholar]
- [32].Ichikawa S, Ozawa K, Hirabayashi Y, Molecular cloning and characterization of the mouse ceramide glucosyltransferase gene, Biochem Biophys Res Commun 253 (1998) 707–711. [DOI] [PubMed] [Google Scholar]
- [33].Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP, CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes, Nature 421 (2003) 852–856. [DOI] [PubMed] [Google Scholar]
- [34].Kassiotis G, Zamoyska R, Stockinger B, Involvement of avidity for major histocompatibility complex in homeostasis of naive and memory T cells, J Exp Med 197 (2003) 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim A, Hartman IZ, Poore B, Boronina T, Cole RN, Song N, Ciudad MT, Caspi RR, Jaraquemada D, Sadegh-Nasseri S, Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources, Nature Commun. 5 (2014) 5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kono M, Dreier JL, Ellis JM, Allende ML, Kalkofen DN, Sanders KM, Bielawski J, Bielawska A, Hannun YA, Proia RL, Neutral ceramidase encoded by the Asah2 gene is essential for the intestinal degradation of sphingolipids, J Biol Chem 281 (2006) 7324–7331. [DOI] [PubMed] [Google Scholar]
- [37].Korb LC, Mirshahidi S, Ramyar K, Sadighi Akha AA, Sadegh-Nasseri S, Induction of T cell anergy by low numbers of agonist ligands, J Immunol 162 (1999) 6401–6409. [PubMed] [Google Scholar]
- [38].Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et al. , Mutation of the mouse klotho gene leads to a syndrome resembling ageing, Nature 390 (1997) 45–51. [DOI] [PubMed] [Google Scholar]
- [39].Li N, van Unen V, Abdelaal T, Guo N, Kasatskaya SA, Ladell K, McLaren JE, Egorov ES, Izraelson M, Chuva de Sousa SM Lopes, et al. , Memory CD4(+) T cells are generated in the human fetal intestine, Nat Immunol 20 (2019) 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Linton PJ, Harbertson J, Bradley LM, A critical role for B cells in the development of memory CD4 cells, J Immunol 165 (2000) 5558–5565. [DOI] [PubMed] [Google Scholar]
- [41].Liu YY, Patwardhan GA, Bhinge K, Gupta V, Gu X, Jazwinski SM, Suppression of glucosylceramide synthase restores p53-dependent apoptosis in mutant p53 cancer cells, Cancer Res 71 (2011) 2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lorch Y, LaPointe JW, Kornberg RD, Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones, Cell 49 (1987) 203–210. [DOI] [PubMed] [Google Scholar]
- [43].Michie CA, McLean A, Alcock C, Beverley PC, Lifespan of human lymphocyte subsets defined by CD45 isoforms, Nature 360 (1992) 264–265. [DOI] [PubMed] [Google Scholar]
- [44].Mirshahidi S, Ferris LC, Sadegh-Nasseri S, The magnitude of TCR engagement is a critical predictor of T cell anergy or activation, J Immunol 172 (2004) 5346–5355. [DOI] [PubMed] [Google Scholar]
- [45].Mirshahidi S, Huang CT, Sadegh-Nasseri S, Anergy in peripheral memory CD4(+) T cells induced by low avidity engagement of T cell receptor, J Exp Med 194 (2001) 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Molano A, Huang Z, Marko MG, Azzi A, Wu D, Wang E, Kelly SL, Merrill AH Jr., Bunnell SC, Meydani SN, Age-dependent changes in the sphingolipid composition of mouse CD4+ T cell membranes and immune synapses implicate glucosylceramides in age-related T cell dysfunction, PLoS One 7 (2012) e47650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Morad SA, Cabot MC, Ceramide-orchestrated signalling in cancer cells, Nat Rev Cancer 13 (2013) 51–65. [DOI] [PubMed] [Google Scholar]
- [48].Murphy K, Weaver C, Janeway's Immunobiology (New York &, Garland Science; ), London, 2016. [Google Scholar]
- [49].O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. , Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development, Immunity 41 (2014) 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Oh KI, Kim BK, Ban YL, Choi EY, Jung KC, Lee IS, Park SH, CD99 activates T cells via a costimulatory function that promotes raft association of TCR complex and tyrosine phosphorylation of TCR zeta, Exp Mol Med 39 (2007) 176–184. [DOI] [PubMed] [Google Scholar]
- [51].Park CK, Shin YK, Kim TJ, Park SH, Ahn GH, High CD99 expression in memory T and B cells in reactive lymph nodes, J Korean Med Sci 14 (1999) 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Patel CH, Powell JD, Targeting T cell metabolism to regulate T cell activation, differentiation and function in disease, Curr Opin Immunol 46 (2017) 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y, Enhancing CD8 T-cell memory by modulating fatty acid metabolism, Nature 460 (2009) 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pereira JP, An J, Xu Y, Huang Y, Cyster JG, Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids, Nat Immunol 10 (2009) 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, Watkins SC, Romero G, Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway, J Biol Chem 274 (1999) 1131–1139. [DOI] [PubMed] [Google Scholar]
- [56].Rosloniec EF, Brand DD, Myers LK, Whittington KB, Gumanovskaya M, Zaller DM, Woods A, Altmann DM, Stuart JM, Kang AH, An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen, J Exp Med 185 (1997) 1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rothwell DG, Li Y, Ayub M, Tate C, Newton G, Hey Y, Carter L, Faulkner S, Moro M, Pepper S, et al. , Evaluation and validation of a robust single cell RNA-amplification protocol through transcriptional profiling of enriched lung cancer initiating cells, BMC Genomics 15 (2014) 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sadegh-Nasseri S, Dalai SK, Korb Ferris LC, Mirshahidi S, Suboptimal engagement of the T-cell receptor by a variety of peptide-MHC ligands triggers T-cell anergy, Immunology 129 (2010) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Seder RA, Ahmed R, Similarities and differences in CD4+ and CD8+ effector and memory T cell generation, Nat Immunol 4 (2003) 835–842. [DOI] [PubMed] [Google Scholar]
- [60].Separovic D, Hanada K, Maitah MY, Nagy B, Hang I, Tainsky MA, Kraniak JM, Bielawski J, Sphingomyelin synthase 1 suppresses ceramide production and apoptosis post-photodamage, Biochem Biophys Res Commun 358 (2007) 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shedlock DJ, Whitmire JK, Tan J, MacDonald AS, Ahmed R, Shen H, Role of CD4 T cell help and costimulation in CD8 T cell responses during Listeria monocytogenes infection, J Immunol 170 (2003) 2053–2063. [DOI] [PubMed] [Google Scholar]
- [62].Sidhom JW, Theodros D, Murter B, Zarif JC, Ganguly S, Pardoll DM, Baras A, ExCYT: A Graphical User Interface for Streamlining Analysis of High-Dimensional Cytometry Data, J Vis Exp. (2019). [DOI] [PubMed] [Google Scholar]
- [63].Sun JC, Bevan MJ, Defective CD8 T cell memory following acute infection without CD4 T cell help, Science 300 (2003) 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Swain SL, Hu H, Huston G, Class II-independent generation of CD4 memory T cells from effectors, Science 286 (1999) 1381–1383. [DOI] [PubMed] [Google Scholar]
- [65].Ulitsky I, Maron-Katz A, Shavit S, Sagir D, Linhart C, Elkon R, Tanay A, Sharan R, Shiloh Y, Shamir R, Expander: from expression microarrays to networks and functions, Nat Protoc 5 (2010) 303–322. [DOI] [PubMed] [Google Scholar]
- [66].Verbist KC, Wang R, Green DR, T cell metabolism and the immune response, Sem. Immunol 24 (2012) 399–404. [DOI] [PubMed] [Google Scholar]
- [67].Verhoeven D, Teijaro JR, Farber DL, Heterogeneous memory T cells in antiviral immunity and immunopathology, Viral Immunol. 21 (2008) 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Vousden KH, Lane DP, p53 in health and disease, Nat Rev Mol Cell Biol 8 (2007) 275–283. [DOI] [PubMed] [Google Scholar]
- [69].Whitmire JK, Asano MS, Kaech SM, Sarkar S, Hannum LG, Shlomchik MJ, Ahmed R, Requirement of B cells for generating CD4+ T cell memory, J Immunol 182 (2009) 1868–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wright WE, Shay JW, Cellular senescence as a tumor-protection mechanism: the essential role of counting, Curr Opin Gene Dev 11 (2001) 98–103. [DOI] [PubMed] [Google Scholar]
- [71].Yano M, Watanabe K, Yamamoto T, Ikeda K, Senokuchi T, Lu M, Kadomatsu T, Tsukano H, Ikawa M, Okabe M, et al. , Mitochondrial dysfunction and increased reactive oxygen species impair insulin secretion in sphingomyelin synthase 1-null mice, J Biol Chem 286 (2011) 3992–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. , Dynamic regulatory network controlling TH17 cell differentiation, Nature 496 (2013) 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.