Abstract
Background
Cholera epidemics continue to challenge disease control, particularly in fragile and conflict-affected states. Rapid detection and response to small cholera clusters is key for efficient control before an epidemic propagates. To understand the capacity for early response in fragile states, we investigated delays in outbreak detection, investigation, response, and laboratory confirmation, and we estimated epidemic sizes. We assessed predictors of delays, and annual changes in response time.
Methods
We compiled a list of cholera outbreaks in fragile and conflict-affected states from 2008 to 2019. We searched for peer-reviewed articles and epidemiological reports. We evaluated delays from the dates of symptom onset of the primary case, and the earliest dates of outbreak detection, investigation, response, and confirmation. Information on how the outbreak was alerted was summarized. A branching process model was used to estimate epidemic size at each delay. Regression models were used to investigate the association between predictors and delays to response.
Results
Seventy-six outbreaks from 34 countries were included. Median delays spanned 1–2 weeks: from symptom onset of the primary case to presentation at the health facility (5 days, IQR 5–5), detection (5 days, IQR 5–6), investigation (7 days, IQR 5.8–13.3), response (10 days, IQR 7–18), and confirmation (11 days, IQR 7–16). In the model simulation, the median delay to response (10 days) with 3 seed cases led to a median epidemic size of 12 cases (upper range, 47) and 8% of outbreaks ≥ 20 cases (increasing to 32% with a 30-day delay to response). Increased outbreak size at detection (10 seed cases) and a 10-day median delay to response resulted in an epidemic size of 34 cases (upper range 67 cases) and < 1% of outbreaks < 20 cases. We estimated an annual global decrease in delay to response of 5.2% (95% CI 0.5–9.6, p = 0.03). Outbreaks signaled by immediate alerts were associated with a reduction in delay to response of 39.3% (95% CI 5.7–61.0, p = 0.03).
Conclusions
From 2008 to 2019, median delays from symptom onset of the primary case to case presentation and to response were 5 days and 10 days, respectively. Our model simulations suggest that depending on the outbreak size (3 versus 10 seed cases), in 8 to 99% of scenarios, a 10-day delay to response would result in large clusters that would be difficult to contain. Improving the delay to response involves rethinking the integration at local levels of event-based detection, rapid diagnostic testing for cluster validation, and integrated alert, investigation, and response.
Keywords: Armed conflict, Cholera, Communicable disease control, Epidemics, Outbreaks, Refugees, Surveillance
Background
Cholera transmission was reported in 34 countries in 2018 and 55 countries in 2019 [1, 2]. The disease is estimated to be substantially under-recorded [3]. Large cholera epidemics frequently coincide with armed conflict and humanitarian crises, including those in Democratic Republic of the Congo, Iraq, Somalia, South Sudan, and Yemen [4–8]. At the start of a cholera outbreak, transmission is driven by the low capacity to detect and isolate the first identified cases. Inadequate preparedness and poor access to case management drives increased mortality. The rapid detection and control of small outbreaks is therefore key for efficient control before an epidemic propagates [9].
In 2017, the Global Task Force on Cholera Control (GTFCC) recommended that countries increase their capacity to contain small outbreaks, using rapid response teams, to aid efforts to substantially reduce global transmission by 2030 [10]. However, little is known about the global capacity for rapid detection of, and response to, cholera outbreaks. In a review of the detection of all-pathogen outbreaks in Africa reported in the World Health Organization’s (WHO) Disease Outbreak News from 1996 to 2014, the median time from onset of symptoms of the first identified case (or health facility visit, if unavailable) to discovery of the outbreak (defined, for example, as the declaration of the outbreak or appearance in an official report) was 27 days [95% CI 20–31.5] [11]. A review of all-pathogen outbreaks in fragile and conflict-affected states from 2000 to 2010 found a similar median delay of 29 days [range 7–80] from symptom onset of the first identified case to detection of the outbreak and a median delay of 7 days [range, 0–30] from detection to investigation [12]. For cholera, a month-long delay in detection represents approximately 6 median serial intervals and, thus, a high potential for uncontrolled transmission [13].
To understand the potential for early detection and rapid response for cholera outbreaks in fragile and conflict-affected states, we examined temporal trends in cholera epidemics to evaluate with what delays the first case or cluster presented, was detected, investigated, responded to, and was confirmed by laboratory culture. We modeled epidemic sizes corresponding to these delays. To explain these delays, we investigated the mechanisms for early warning of these outbreaks, predictors of delays, and global improvements in reducing delays.
Methods
Compilation of cholera outbreaks
The period of 2008 to 2019 was chosen to reflect recent experience with cholera response. A list of countries that appeared ≥ 2 times during this period on the World Bank’s Harmonized List of Fragile Situations, and had a documented cholera burden as per the GTFCC’s 2017 list of cholera-affected countries, was compiled (Additional file 1) [10, 14]. Small-island states affected mainly by climate change rather than conflict were excluded. A list of countries meeting the fragility criteria but not included in the GTFCC list were included if they were documented using other sources as having had cholera outbreaks from 2008 to 2019 (i.e., Iraq, Myanmar, and Syria). Cholera-affected countries that did not meet the fragility criteria but either (a) hosted refugees (i.e., Kenya, Tanzania, and Uganda) and/or (b) border fragile and conflict-affected states with cholera outbreaks (i.e., Benin, Ethiopia, Niger, Nigeria, Tanzania, and Zambia) were included. Given that no annual list of annual cholera outbreaks exists, a list of outbreaks was compiled using a two-step process. We first reviewed the WHO’s annual cholera reports to identify which countries reported transmission during the study period [15]. Countries that do not routinely report cholera to WHO but are known from other sources to have had cholera outbreaks were included (e.g., Ethiopia and Myanmar). We then sought details on the occurrence of sub-national outbreaks from the WHO’s Disease Outbreak News and UNICEF’s Cholera Outbreaks in Central and West Africa Bulletin (2015–9) [16, 17]. The GTFCC definition of a cholera outbreak was applied (cholera-free region: ≥ 1 culture-confirmed case and evidence of local transmission, or year-round transmission: unexpected increase in magnitude or timing of suspect cases over 2 weeks with laboratory confirmation) [18]. As stool sampling and transport is often unfeasible in insecure settings, we included instances where cholera alerts were identified (e.g., one case of acute watery diarrhea (AWD) testing positive for cholera by rapid diagnostic test (RDT)) [19]. Finally, we included cholera alerts that triggered the cholera investigation mechanism, but where testing detected another pathogen, in order to explore detection and investigation mechanisms.
Compilation of reports on cholera outbreaks
We searched the peer-reviewed literature for further identification and reporting on cholera outbreaks. Peer-reviewed articles were sourced from PubMed/MEDLINE using a date-specific keyword search (“country AND cholera”). Given that only a small number of outbreaks are reported in the scientific literature, we searched the gray literature, including epidemiological summaries, national cholera preparedness and response plans, and non-peer-reviewed studies. The sources included the following: (1) Reliefweb (https://reliefweb.int/, a repository of documents and data from humanitarian crises) using a date-specific keyword search (“country AND cholera”; “UNHCR AND cholera”) and (2) regional outbreak bulletins and journals including the WHO EMRO Weekly Epidemiological Monitor (2008 to 2019), WHO AFRO Outbreaks and Emergencies Bulletin (2017–9), WHO SEARO Journal of Public Health, WHO WPRO Western Pacific Surveillance and Response Journal, and UNICEF Cholera Outbreaks in Central and West Africa Bulletin (2015 to 2019) [16, 20–23]. The Program for Monitoring Emerging Diseases (ProMED) database of disease observations from media sources was used to fill in missing information on dates, but was not used as the primary source of information [24]. When little information was available from the sources above, websites of ministries of health and crisis-specific surveillance systems (e.g., early warning alert and response systems or networks (EWARS/EWARN) or disease early warning systems (DEWS)) were searched. An example includes the EWARN of the Syrian Assistance Coordination Unit for Northern Syria [25].
Inclusion criteria and data extraction
Outbreaks were included if at least two of the following dates were available: (1) dates of symptom onset of the primary case, and/or case presentation, and/or outbreak detection, and (2) dates of investigation and/or response. If the date of symptom onset for the primary case was missing, it was estimated as 5 days before the date of case presentation (equal to the median delay for outbreaks with available date of symptom onset), or date of outbreak detection if date of case presentation was unavailable. If the date of case presentation was unavailable, it was replaced by the date of outbreak detection. The earliest dates of (1) symptom onset for the first identified primary case, (2) case presentation to a health facility, (3) detection of outbreak/alert raised, (4) investigation by local health authorities, (5) response, and (6) laboratory confirmation by culture were extracted (Table 1). We defined the date of response as the earliest date by which a cholera-specific control measure was applied to the outbreak-affected area (e.g., water, sanitation, and hygiene (WASH) activities, setup of case management, active case-finding, community-based activities, and delivery of cholera kits). The starting month and year of the outbreak, geographical context (i.e., urban, rural, or displacement camp), type of crisis or fragility (i.e., armed conflict, fragile state, natural disaster, refugee setting, non-fragile state bordering a fragile state), and the WHO region were extracted. Any additional information on factors that may have contributed to the observed delay, including presence of an early warning function, was extracted. Details on the signal type for outbreak detection were recorded, if available, as a (1) formal alert detected by health workers reported immediately within the surveillance system, (2) informal alert from community members or a non-governmental organization (NGO) reported immediately, or (3) weekly data analysis of surveillance trends.
Table 1.
Dates used to estimate delays in detection, investigation, and response for cholera outbreaks
| Date | Defined as earliest date (by priority) |
|---|---|
| Date of start of outbreak |
1. Symptom onset for first identified case 2. Case presentation to health facility (less 1 day) |
| Date of alert/outbreak detection | Alert issued from health facility, community health worker, community member, local public health office, or laboratory |
| Date of investigation | Investigation by local authorities |
| Date of earliest response | Any cholera-specific response activity (case-finding, control measures by health facility or public health office, household/community WASH, case management) |
| Date of laboratory confirmation | First documented culture confirmation |
Analysis of delays and their predictors
For each outbreak, median delays and their interquartile ranges (IQR) were calculated by subtracting the date of symptom onset of the primary case from the dates of (1) case presentation, (2) outbreak detection, (3) investigation, (4) earliest response, and (5) laboratory confirmation. For each outbreak, the dates were graphed on a timeline.
To investigate the association between the observed delay from symptom onset of the primary case to response and potential predictor variables, a multivariate ordinary least-squares regression model was used. Delay to response was log-transformed to produce a normalized distribution. Extreme values in delay to response were judged to represent meaningful delays rather than data errors and were retained in the dataset. Predictor variables included signal type, context, crisis, WHO region, and year of outbreak onset (to detect any secular trend). Akaike information criterion (AIC) and a step-wise selection process was used to assess model fit and complexity. In separate regressions, year of outbreak onset was used as a predictor variable to investigate secular trends for delays to case presentation, outbreak detection, investigation, and confirmation. Loess curves were used to visualize the temporal trends using a smoothed trend line that down-weighted extreme values [26]. Percent change and 95% confidence intervals were presented for each regression.
Branching process model
A preliminary review of retrieved reports demonstrated that the early epidemic sizes at the dates that the outbreak was detected and responded to were rarely documented. Instead, to estimate the potential early epidemic sizes at each delay, a branching process model was used to estimate the median and range of epidemic sizes at the time points indicated by the median delays to case presentation, investigation, response, and confirmation [27–29]. We simulated multiple outbreaks using 10,000 runs and calculated the proportion of these outbreaks with early epidemic sizes below the threshold of 20 cases in a 5 to 30 day period. We selected this threshold arbitrarily as the outbreak size with potential to be contained. Transmission started with a seed case(s) which generated secondary cases from a negative binomial distribution Z ~ NegB(RE, k) with a mean equivalent to the reproduction number (RE, 2.5 [13, 30], reflecting early and high transmission potential among an unvaccinated population) and heterogeneity introduced by a dispersion parameter (k, 4.5, reflecting low overdispersion in RE) [31]. Each new infection drew a time of infection from a serial interval distribution S ~ gamma(shape = 0.5, rate = 0.1) with a median of 5 days [4, 13, 32]. We assumed that at the time of outbreak detection there were 3 seed cases and that all resulting infectious persons were symptomatic. Simulations would end by chance when either the cases did not produce additional secondary cases, or they reached 1000 cases (representing a large outbreak). In a sensitivity analysis, we considered outbreaks of larger size at detection (i.e., 10 and 20 seed cases).
All analyses were carried out in R statistical software version 4.0.3 [33].
Results
Description of outbreaks
Seventy-six outbreaks from 34 countries met the inclusion criteria. Overall, 1970 documents were reviewed, and 138 documents were retained (1–4 documents per outbreak including 28 peer-reviewed articles and 110 gray literature sources as listed in Additional file 2) [34–171]. Where countries reported acute watery or severe diarrhea as a proxy for cholera (e.g., Ethiopia, Myanmar) [172, 173] or where surveillance was poor due to conflict in remote areas (Myanmar, Northern Nigeria, and Syria as documented by Sparrow and colleagues) [19], few or no reports were located. Few reports from endemic countries with ongoing transmission (e.g., Cameroon, Democratic Republic of the Congo, Somalia) were found, likely due to the difficulty in ascertaining start dates. Three false alerts resulting in the exclusion of cholera were identified in Cameroon (2) and Syria (1); we described these outbreaks qualitatively and left them out of the quantitative analysis [100, 101, 108]. One alert of an RDT-positive case where culture could not be obtained due to ongoing conflict was identified in Syria and kept in the quantitative analysis (noting that confirmation was not possible) [83, 84]. Fifty-one (67%) of the 76 outbreaks were missing the date of onset of symptoms for the primary case.
Narrative descriptions and sources of information for outbreaks are compiled in Additional file 2. Most reports were from Africa (80.3%, mainly Chad, South Sudan, Burundi, and Uganda) and the Eastern Mediterranean region (13.2%, mainly Yemen, Iraq, and Syria) (Table 2). Outbreaks occurred during armed conflicts (e.g., Afghanistan, South Sudan, Yemen), after natural disasters (e.g., cyclones in Mozambique, post-earthquake in Nepal), in fragile situations (e.g., Angola, Chad), in refugee settlements (e.g., camps in Kenya and Tanzania), and in countries bordering cholera-affected fragile states (e.g., Benin, Tanzania). Most reports (56.6%) were from urban sites. Where the information was available (55/76 outbreaks), most (83.6%) were detected through formal and informal alerts compared with weekly data analysis (16.4%).
Table 2.
Characteristics of outbreaks, 2008–2019
| Characteristic | N (%) |
|---|---|
| WHO region | |
| Africa (AFRO) | 61 (80.3) |
| Eastern Mediterranean (EMRO) | 10 (13.2) |
| South-East Asia (SEARO) | 3 (4.0) |
| Americas (PAHO) | 1 (1.3) |
| Western Pacific (WPRO) | 1 (1.3) |
| Context | |
| Urban | 43 (56.6) |
| Rural | 27 (35.5) |
| Refugee or displacement camp | 6 (7.9) |
| Crisis | |
| Fragile situation | 31 (40.8) |
| Armed conflict | 25 (32.9) |
| Country bordering FCAS | 10 (13.2) |
| Natural disaster | 5 (6.6) |
| Refugee setting | 5 (6.6) |
| Surveillance system N = 47 (not reported for 29 outbreaks) | |
| Early warning function | 36/47 (76.6) |
| Through routine surveillance | 22/36 (61.1) |
| EWARS/DEWS | 14/36 (38.9) |
| Routine surveillance | 11/47 (23.4) |
| Signal N = 55 (not reported for 21 outbreaks) | |
| Alert | 46/55 (83.6) |
| Formal alert | 37/46 (80.4) |
| Informal alert | 9/46 (19.6) |
| Weekly data analysis | 9/55 (16.4) |
Delays and potential epidemic sizes
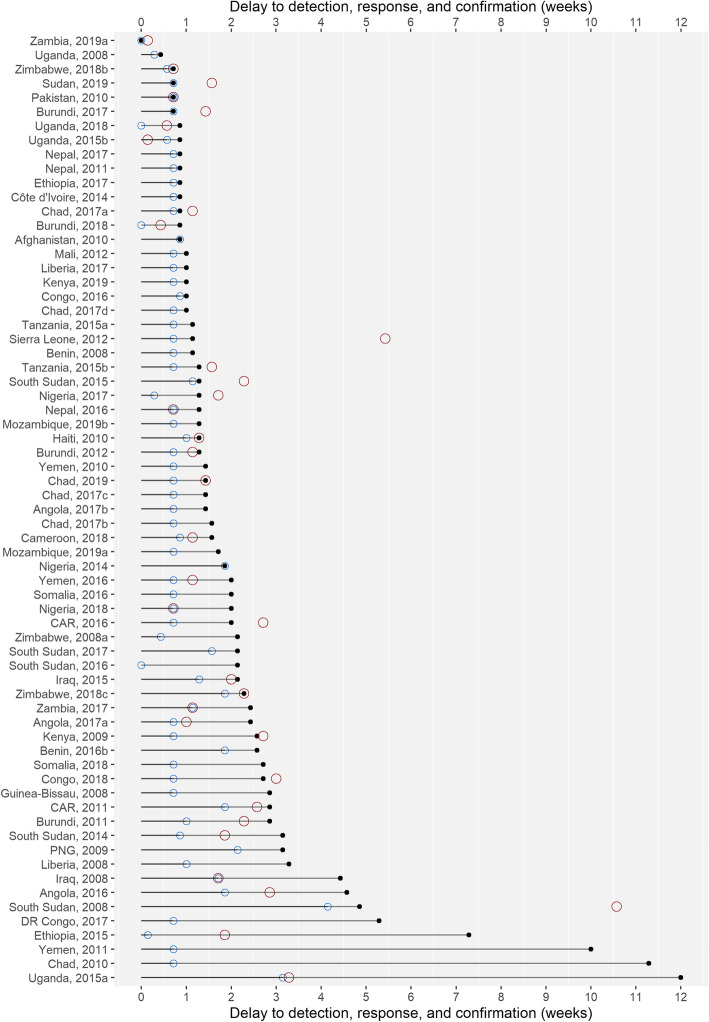
Median delays are listed in Table 3, and histograms of the delays are listed in Additional file 3. The timelines of the individual outbreaks are visualized in Fig. 1. The median delay from date of the first identified case’s symptom onset to case presentation at the health facility was 5 days (IQR 5–5). The median delays between symptom onset of the primary case and detection (5 days, IQR 5–6), investigation (7 days, IQR 5.8–13.3), and response (10 days, IQR 7–18) spanned 1 to 2 weeks. Across countries, these delays varied; investigations and responses were routinely launched on the same or next day in Cameroon and Nepal, while long delays of 70 days in Uganda in 2015, 79 days in Chad in 2010, and 84 days in Yemen in 2011 were reported. The median delay to laboratory confirmation, for the 41/76 outbreaks for which the information was available, was 11 days (IQR 7–16), similar to the delay to response. Countries affected by conflict frequently had delays from symptom onset of the primary case to response greater or equal to 2 weeks (Fig. 1).
Table 3.
Median delays (with interquartile range (IQR) and range)
| Delay | Median delay (days) (IQR) | Range (days) |
|---|---|---|
| Delay to case presentation (n = 76) | ||
| Symptom onset to case presentation | 5 (5–5) | 0–22 |
| Delay to detection (n = 76) | ||
| Symptom onset to outbreak detection | 5 (5–6) | 0–29 |
| Case presentation to outbreak detection | 0 (0–0.3) | 0–24 |
| Delay to investigation (n = 48) | ||
| Symptom onset to investigation | 7 (5.8–13.3) | 0–84 |
| Case presentation to investigation | 2 (1–8) | 0–62 |
| Delay to response (n = 67) | ||
| Symptom onset to response | 10 (7–18) | 0–84 |
| Case presentation to response | 6 (2.5–13.5) | 0–74 |
| Delay to confirmation (n = 41) | ||
| Symptom onset to confirmation | 11 (7–16) | 0–74 |
Fig. 1.
Delay in weeks from date of onset of symptoms to outbreak detection (blue circle), response (black circle), and confirmation (red circle), by outbreak, 2008–2019 (excluding outbreaks missing response date)
Several outbreaks were detected when already-large, challenging containment (e.g., Afghanistan, 2011, 255 cases, ID 2; Chad, 2017, 50 cases and 13 deaths, ID 19; Ethiopia, 2015, 268 cases, ID 28; Haiti, 2010, > 1000 cases, ID 32). Table 4 summarizes the model-simulated early epidemic sizes that the outbreaks could have reached by the date of different delays and different initial outbreak sizes. With 3 seed cases at detection, a median delay to case presentation of 5 days resulted in a median epidemic size of 9 cases (upper range, 29 cases), with nearly all outbreaks < 20 cases (98.6%). A median delay to response of 10 days resulted in a median epidemic size of 12 cases (upper range, 47 cases), with a comparable proportion of outbreaks < 20 cases (92.6%). Lengthening the delay to response to 30 days resulted in an upper range of 72 cases, with 67.7% of outbreaks remaining < 20 cases. Using 10 seed cases to simulate outbreaks of larger size at detection, a median delay to case presentation of 5 days resulted in a median epidemic size of 28 cases (upper range, 55 cases), with a minority of outbreaks < 20 cases (5.7%). With a median delay to response of 10 days delay, there was a median epidemic size of 34 cases (upper range, 67 cases), with < 1% of outbreaks < 20 cases. At 30 days, the upper range was 100 cases, with < 1% of outbreaks remaining < 20 cases. With 20 seed cases at detection, outbreaks enlarged quickly, reaching a median of 55 cases (range 30–89) with a median delay to response of 5 days and median of 65 cases (range 40–110) with a median delay to response of 10 days.
Table 4.
Simulated epidemic sizes (with standard deviation (SD) and range), and proportion of outbreaks < 20 cases for outbreaks of 3, 10, and 20 seed cases at detection
| 3 seed cases | 10 seed cases | 20 seed cases | ||||
|---|---|---|---|---|---|---|
| Delay from onset of symptoms (primary case) | Median delay (days) | Median epidemic size (SD, range) | % outbreaks < 20 cases | Median epidemic size (SD, range) | % outbreaks < 20 cases | Median epidemic size (SD, range) |
| Case presentation or outbreak detection | 5 | 9 (3.7, 3–29) | 98.6 | 28 (5.7, 12–55) | 5.7 | 55 (7.4, 30–89) |
| Investigation | 7 | 10 (4.4, 3–40) | 96.9 | 31 (6.1, 11–61) | 1.6 | 60 (8.2, 34–99) |
| Response | 10 | 12 (5.1, 3–47) | 92.6 | 34 (7.0, 16–67) | < 1 | 65 (8.8, 40–110) |
| Confirmation | 11 | 12 (5.4, 3–50) | 91.9 | 35 (7.1, 13–69) | 67 (9.1, 41–105) | |
| Delay from onset of symptoms (primary case) | Counterfactual delay (days) | Median epidemic size (SD, range) | % outbreaks < 20 cases | Median epidemic size (SD, range) | % outbreaks < 20 cases | Median epidemic size (SD, range) |
| 14-day delay | 14 | 14 (6.0, 3–51) | 85.8 | 37 (7.8, 17–79) | < 1 | 70 (9.7, 41–113) |
| 21-day delay | 21 | 16 (7.4, 3–63) | 76.6 | 40 (9.0, 16–87) | 74 (10.8, 43–124) | |
| 30-day delay | 30 | 18 (8.9, 3–72) | 67.7 | 43 (10.3, 18–100) | 78 (12.2, 43–131) | |
Legend: DRC Democratic Republic of the Congo, CAR Central African Republic, PNG Papua New Guinea
Factors associated with delays
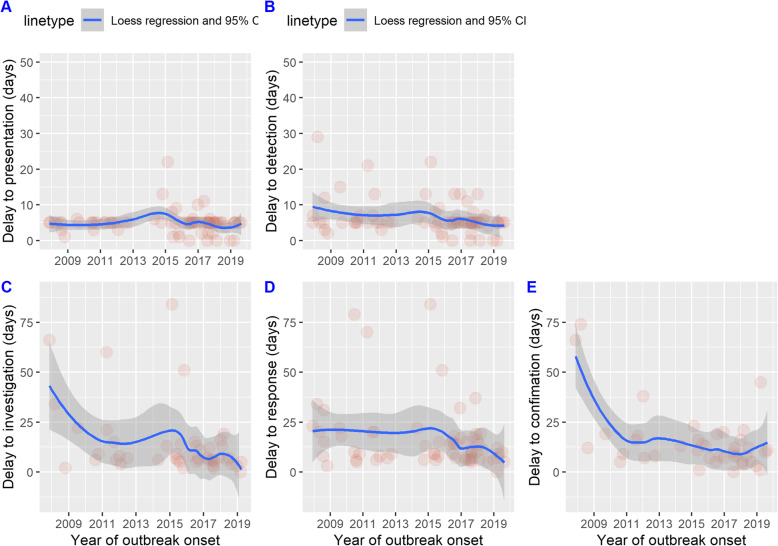
Given that the signal type was complete for only 55/76 observations, two models were implemented: a multivariable adjusted model (including year of outbreak onset, WHO region, context, and crisis type), and a bivariate model for signal type only (informal/formal alert versus weekly data analysis). Using AIC for the multivariable model, including only year of outbreak onset returned the lowest AIC score (Additional file 4). A weak crude association between year of outbreak onset and delay to response, with an annual decrease in response time of 5.2% (95% CI 0.5–9.6, p = 0.03), was found (visualized in Fig. 2). This model met the assumptions for linearity and homogeneous variance and explained 6% of the variance. Similar decreases in delay to detection, investigation, and confirmation were found (Fig. 2 and Additional file 5). In the second model, alerts (versus data analysis) were associated with a reduction in response time of 39.3% (95% CI 5.7–61.0, p = 0.03) (boxplot displayed in Fig. 3). The model met the assumptions for ordinary least-squares regression but one extreme value in delay affected the leverage. The model explained 8% of the variance.
Fig. 2.
Scatterplot of cholera outbreaks by delay between date of symptom onset of the primary case and dates of a presentation, b detection, c investigation, d response, and e confirmation, and Loess curves, as a function of outbreak start date, 2008 to 2019. Red dots represent individual outbreaks over time and gray shading indicates 95% CI of Loess regression
Fig. 3.
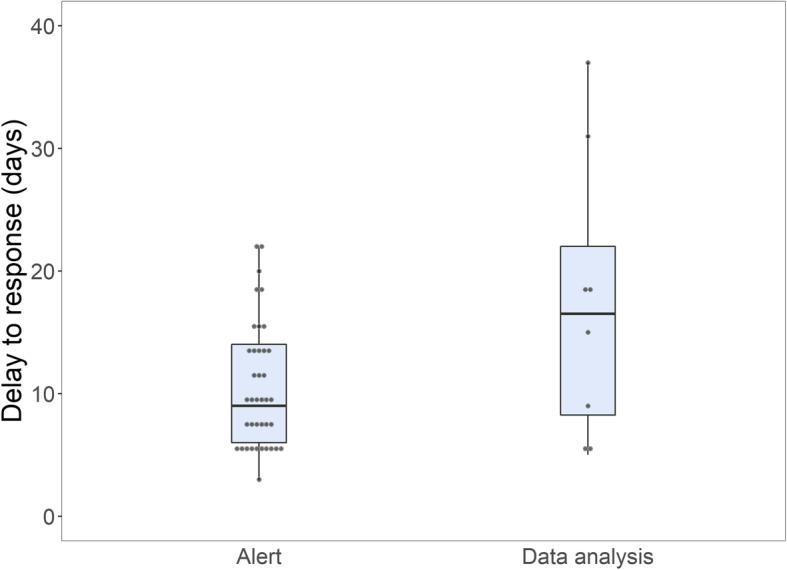
Delay from onset of symptoms of the primary case to response, by signal type (immediately-notified alert compared with weekly data analysis), 2008–2019 (N = 49/76 outbreaks with information on signal type available). Gray dots represent individual outbreaks
More information from the examination of outbreaks is illustrative of the use of alerts. Of the 83.6% of outbreaks detected through alerts, 37/46 (80.4%) were through alerts by a health worker or community health worker and 9/46 (19.6%) through informal alerts by community members. For example, in 2015, in Aleppo, Syria, an alert was issued through the EWARN via phone after RDT testing of a suspect, and an investigation initiated based on the positive result (2-day delay to investigation, ID 63). In 2017, in a displacement camp in Northern Nigeria, an alert of a suspect case was issued by MSF by phone through the EWARS on the same day of case presentation (2-day delay to investigation, ID 48), demonstrating the rapid recognition of a suspect case by health workers [171]. Comparisons of outbreaks within countries are instructive. In Benin, of two outbreaks in rural areas detected in 2016, one outbreak was detected through an immediate call to public health authorities (5-day delay to detection, ID 7) while for the other outbreak, issuance of the alert on the weekly set day of routine surveillance data transmission resulted in a 13-day delay to detection (ID 8). In Central African Republic in 2011, an alert from the community of multiple suspect cases was issued late (13-day delay, ID 16) compared with 2016, when an alert from Red Cross volunteers was issued in half the time (5-day delay, ID 17).
In several instances, early warning systems further benefited from rapid investigation and response. For example, in Afghanistan, in 2010, the DEWS provided a response mechanism to link the detection of a large cluster of 60 suspect cases in a remote and insecure village by a local NGO with rapid action which reportedly led to containment within a month (6-day delay to investigation, ID 1). In 2011, the DEWS in Afghanistan detected an already-large outbreak of 255 suspect cases in multiple clusters but with a rapidly administered response (21-day delay to investigation, ID 2). Reduced transmission within 3 months followed. In Liberia in 2017, a suspect case that died en route to the health facility was detected based on symptoms, triggering a rapid response to isolate additional cases in the index case’s village (7-day delay to investigation, ID 38). In Chad in 2017, two suspect cases among children which resulted in rapid progression to death were reported to the local health facility, who investigated the source village and found a larger cluster of 50 cases and 13 deaths (6-day delay to investigation, ID 19). Though already a large outbreak, this led to a response on the following day.
Information in reports suggested improvements in surveillance, investigation, and response over time. In Cameroon in 2016, two false alerts for cholera later attributed to food poisoning and rotavirus were made by health workers and community members respectively and led to rapid investigation upon detection, testing by RDT and culture, and ongoing control activities during the investigation period (ID 13, 14). In Somalia, faster response in insecure urban areas using EWARS in 2016 and 2018 can be compared to a lack of a comprehensive early response during ongoing transmission over 2 months in 2008 (14- and 19-day delays versus 2-month delay, ID 54–56). Nepal’s EWARS facilitated rapid detection and response to clusters from 2011 onwards (total delays 6–9 days, ID 42–44) [75, 130, 162]. In 2016, RDT capacity was added at health facilities to enable better discrimination between alerts of cholera or diarrhea due to other pathogens [161].
Potential factors related to long delays to response
Long delays from symptom onset of the primary case to response (~ 2 weeks) were observed in 29/67 (43.2%) outbreaks for which a response date was available. These appeared to be related to poor sensitivity of the formal surveillance system due to the remote locations of outbreaks [63] (Papua New Guinea, ID 52); insecurity posed by armed conflict (Somalia, 2008, ID 54; South Sudan, 2008, ID 57; Yemen, 2011, ID 72); reliance on laboratory confirmation to declare an outbreak before initiating a comprehensive response (Iraq, 2008, 2015, ID 33, 34, South Sudan, 2014, ID 58); assuring government declaration and mobilization of non-governmental actors (CAR, 2011, ID 16); a less effective local response which required reinforcement by capacity from the national level or other partners (Congo, 2018, ID 25; Ethiopia, 2015, ID 28; Guinea-Bissau, 2008, ID 31; South Sudan, 2017, ID 61; Uganda, 2015, ID 68; Zimbabwe, 2018, ID 79) [113]; and missed superspreading events (e.g., a funeral in Zimbabwe, 2018, ID 79) [136].
Discussion
In an era of large-scale cholera epidemics in conflict settings like Yemen and previously cholera-free settings like Haiti, improving and sustaining early detection and response to small outbreaks remains critical for averting large-scale epidemics. Reducing delays in the timelines of patients presenting to health facilities, increasing capacity of health workers to recognize suspect cases of cholera, and reinforcing local investigation and response therefore remain as important as vaccination and other emerging tools. Some of the largest outbreaks in recent years in South Sudan (2014–6), Ethiopia (2015 onwards), and Zambia (2017–8) have suffered from late detection and/or response, which has led to surges of cases that have overwhelmed health systems [131, 140, 174–176].
Our findings indicate that from 2008 to 2019, median delays from symptom onset of the primary case to case presentation at the health facility and to response were approximately 5 days and 10 days, respectively. Longer delays to response were documented across the whole time period. Evaluations from Nigeria, Yemen, and other settings have shown that reasons for delays to detection include poor population access to health services due to disrupted health systems and/or insecurity, difficulty in discerning diarrhea and dehydration due to cholera from other causes without rapid diagnostics, reliance on laboratory confirmation before initiating response, and less effective local response [171, 177, 178]. Epidemic control more than 2 weeks post-onset carries a strong risk of epidemic propagation, particularly where the population is highly mobile. Our simple model simulations and sensitivity analyses suggest that with 3 seed cases, in 2 to 33% of scenarios such delays could result in clusters of 20 or more cases that would be difficult to contain. Comparatively, a field investigation and preliminary response to contain transmission done at the time of case presentation (~ 1 week) could potentially reduce the probability of reaching these epidemic sizes to 2 to 4% of scenarios. If larger outbreaks of 10 seed cases are detected, even within ~ 1 week, 95% of outbreaks could accumulate 20 or more cases, and thus would be difficult to contain.
Early detection and response are major aims of the Ending Cholera roadmap. There are two reasons to believe that policy and practice have somewhat narrowed the gap between detection and response. First, we found a global improvement in time to response that corroborates a previous analysis of improvements for detection of all-pathogen outbreaks in low- and middle-income countries from 1996 to 2014 [11]. This may be related to more attention and investment by governments and the GTFCC to the impacts of cholera epidemics in fragile states, given a decade of large and devastating cholera outbreaks in Haiti and across West and Central Africa, the Horn of Africa, and the Gulf of Aden [10]. Some countries appear to have documented improved capacity for detection and response, as shown in this analysis (e.g., Chad, Nepal, Somalia). This may be reflected by investments into strategies like the Joint External Evaluation process which have specified critical gaps for improvement [179–181].
Detailed case studies of cholera outbreaks provide practical observations on the mechanisms of surveillance, diagnosis, and response which can reduce delays. Early detection with high-quality epidemiological data has been augmented with the use of sentinel site surveillance at hospitals equipped with RDTs and trained and vigilant health workers in Kathmandu, Nepal [162]; community-based surveillance using existing community health worker or Red Cross volunteer networks to enable early warning of clusters in the community before patients appear at health facilities in Central African Republic and Haiti [71, 152, 182, 183]; and other event-based surveillance mechanisms, including phone hotlines and mobile phone fleets, to enable immediate notification of suspect events in public, private, and NGO clinics and in the community, as seen in Northern Nigeria and Cameroon [171, 184, 185]. Response should not be delayed by poor laboratory capacity. A potentially stronger role for health workers in local facilities exists in their use of enriched, high-specificity RDTs [186] and aligned probable case definitions to validate clusters of suspected cholera cases that can trigger an immediate investigation and response [162, 171]. This is directly applicable in remote districts and insecure areas where laboratory confirmation will be slow. Timely field investigation and preliminary response remains promising as most outbreak reports cited the use of an early warning alert system, with several examples of integrated investigation and response capacity. We consider that an integrated alert and at least a preliminary response to an outbreak within 1 week of onset should be possible in fragile settings [187]. However, despite the presence of EWARS in nearly 80% of the outbreaks examined, the median delay to response was 10 days. Where EWARS was used successfully to link early detection with a preliminary and robust response, for example in Afghanistan (2010–1), Nepal (2011–6), and Northern Nigeria (2018), a timely response was judged to be dependent on adequate and trained human resources (e.g., district-level rapid response teams or local health facility staff capable of multidisciplinary investigation and a generic response [63, 162]), and the ability to mount at least a preliminary response moving forward independent of laboratory confirmation [52, 75, 100, 101, 139, 161, 171]. For example, investigation and response were integrated in Afghanistan (2010) where a local NGO was trained rapidly to carry out a comprehensive community response, as they had more access to the area than health authorities in an insecure area [52]. In Chad (2017), investigation was carried out by the staff of a local health facility, who also initiated the preliminary community response [137].
It is important to consider the limitations inherent to a retrospective review of data from secondary sources. First, as no global registry of cholera outbreaks exists, we relied on the manual compilation of available situation reports and articles. The most comprehensive source, the WHO’s current compilation of annual cholera data, does not provide detailed information on outbreaks and misses non-reporting countries. The small annual numbers of outbreaks pre-2015 documented here may reflect the few global data sources available. As well, larger outbreaks are more likely to be detected, responded to, and therefore documented and included here. Second, the delays are estimates of reality; dates from situation reports are likely inaccurate to an unquantifiable degree as the exact dates of local investigation and response may be subjective and are documented infrequently. The identification of the primary case(s) depends on the depth of the field investigation, and with a multi-pathway pathogen like Vibrio cholerae that causes a range of disease severity, transmission chains may be missed. Fifty-one (67%) of the 76 outbreaks were missing the date of onset of symptoms for the primary case, which then had to be estimated, limiting accuracy. The dates of response were based on the judgment of the timing of the first transmission-reducing intervention and thus may represent variable intensity of response across outbreaks. Of note, the longest delays to response noted during outbreaks in Chad, Ethiopia, Somalia, and Uganda were related to the first viable response after an inadequate local response. To address these inconsistences, we sourced multiple reports per outbreak to triangulate the information and obtain a clear timeline, and excluded a large number of outbreaks where reports lacked detailed dates. Second, we note that while outbreaks are likely to occur during conflict, they are difficult to detect amidst violence where surveillance coverage is poor [19]. These outbreaks, with potentially high mortality, may have gone undetected, unless they occurred in urban areas and/or propagated to a point of being overwhelming, as in South Sudan in 2014 and Yemen in 2016 [4, 131]. Third, the simple branching process models used here for demonstrating early epidemic growth are for illustration purposes only. These models did not take into account key sources of uncertainty including initial susceptibility to infection (influenced by prior cholera infection or vaccination), heterogeneity in contact and transmission routes, depletion of susceptible persons, and the time-varying Rt value.
The documentation of the occurrence and features of cholera outbreaks is currently very heterogeneous. A real-time global database using standard outbreak event reports maintained by WHO regional offices to prospectively log data and metrics from outbreaks of cholera, as well as other epidemic-prone diseases, would yield superior accuracy for the annual evaluation of timeliness. WHO AFRO’s Weekly Bulletin on Outbreaks and Emergencies provides an existing template which can feed into such a global database [20]. WHO AFRO has used this tool to provide annual metrics of timeliness in outbreak response for epidemic-prone diseases from 2017 to 2019, demonstrating reduced time from symptom onset of the primary case to outbreak detection (defined as alerting national authorities) from 14 (IQR 6–37) days in 2017 to 4 (IQR 1–11) days in 2019 [188].
Conclusions
Cholera epidemics will continue to appear unpredictably and cause serious morbidity and mortality in countries affected by armed conflict and fragility. Cholera surveillance and response is dependent on rethinking the timely detection, investigation, and response to primary cases at the local level. This includes reinforcing outbreak detection through event-based surveillance, consistent weekly reporting using standard case definitions, systematic use of enriched RDTs, and integrating early investigation with preliminary local response. These measures should increasingly underpin the detection and containment of emerging epidemics.
Supplementary Information
Additional file 1. Countries investigated.
Additional file 2. Compilation of outbreaks by country, date of onset, delays (detection, investigation, response), signal, source, and description of investigation and response.
Additional file 3. Histograms of delays from symptom onset to (A) case presentation, (B) outbreak detection, (C) investigation, (D) response, and (E) confirmation.
Additional file 4. Overview of alternative models for the main analyses.
Additional file 5. Model parameters for additional delay analyses.
Acknowledgements
We would like to thank Abdihamid Warsame, Lauren D'Mello-Guyett, Kevin Van Zandvoort, and Alicia Rosello for advice on the methodology.
Abbreviations
- AIC
Akaike information criterion
- AWD
Acute watery diarrhea
- CI
Confidence interval
- DEWS
Disease early warning system
- EWARN
Early warning alert and response network
- EWARS
Early warning alert and response system
- FCAS
Fragile and conflict-affected situation
- IQR
Interquartile range
- GTFCC
Global Task Force for Cholera Control
- NGO
Non-governmental organization
- OCV
Oral cholera vaccination
- RE
Effective reproduction number
- RDT
Rapid diagnostic test
- SD
Standard deviation
- WASH
Water, sanitation, and hygiene
- WHO
World Health Organization
Authors’ contributions
RR, FF, FC, and JE contributed to the study design. RR contributed to the data retrieval and compilation and analysis. RR, FF, FC, and JE contributed to the interpretation. RR, FC, JE, and FF contributed to the manuscript writing. All authors read and approved the final manuscript.
Funding
RR is funded by a Doctoral Foreign Study Award from the Canadian Institutes of Health Research (Award no. DFS-164266). FC and WJE are funded by UK Research and Innovation as part of the Global Challenges Research Fund (Grant No. ES/P010873/1).
Availability of data and materials
The dataset supporting the conclusions of this article is available in the Github repository, [https://github.com/ruwanepi/Detection-analysis].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-020-01865-7.
References
- 1.WHO Cholera, 2018. Weekly Epidemiol Rec. 2019;48(94):561–580. [Google Scholar]
- 2.WHO Cholera, 2019. Weekly Epidemiol Rec. 2020;37(95):441–448. [Google Scholar]
- 3.Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis. 2015;9(6):e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camacho A, Bouhenia M, Alyusfi R, Alkohlani A, Naji MAM, de Radigues X, Abubakar AM, Almoalmi A, Seguin C, Sagrado MJ, et al. Cholera epidemic in Yemen, 2016-18: an analysis of surveillance data. Lancet Glob Health. 2018;6(6):e680–e690. doi: 10.1016/S2214-109X(18)30230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingelbeen B, Hendrickx D, Miwanda B, van der Sande MAB, Mossoko M, Vochten H, Riems B, Nyakio JP, Vanlerberghe V, Lunguya O, et al. Recurrent cholera outbreaks, Democratic Republic of the Congo, 2008-2017. Emerg Infect Dis. 2019;25(5):856–864. doi: 10.3201/eid2505.181141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubogo M, Mohamed AM, Ali AH, Ali AH, Popal GR, Kiongo D, Bile KM, Malik M, Abubakar A. Oral cholera vaccination coverage in an acute emergency setting in Somalia, 2017. Vaccine. 2020;38(Suppl 1):A141–A147. doi: 10.1016/j.vaccine.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Lam E, Al-Tamimi W, Russell SP, Islam Butt MOU, Blanton C, Musani AS, Date K. First oral cholera vaccination campaign in Iraq during an outbreak and humanitarian crisis: findings from the coverage survey, 2015. Am J Trop Med Hyg. 2016;95(5):567. [Google Scholar]
- 8.Connolly MA, Gayer M, Ryan MJ, Salama P, Spiegel P, Heymann DL. Communicable diseases in complex emergencies: impact and challenges. Lancet. 2004;364(9449):1974–1983. doi: 10.1016/S0140-6736(04)17481-3. [DOI] [PubMed] [Google Scholar]
- 9.Ratnayake R, Finger F, Azman AS, Lantagne D, Funk S, Edmunds WJ, Checchi F. Highly targeted spatiotemporal interventions against cholera epidemics, 2000-19: a scoping review. Lancet Infect Dis. 2020. 10.1016/S1473-3099(20)30479-5. [DOI] [PubMed]
- 10.Global Task Force on Cholera Control . Ending cholera. A global roadmap to 2030. Geneva: WHO; 2017. [Google Scholar]
- 11.Kluberg SA, Mekaru SR, McIver DJ, Madoff LC, Crawley AW, Smolinski MS, Brownstein JS. Global capacity for emerging infectious disease detection, 1996-2014. Emerg Infect Dis. 2016;22(10):E1–E6. doi: 10.3201/eid2210.151956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruckner C, Checchi F. Detection of infectious disease outbreaks in twenty-two fragile states, 2000-2010: a systematic review. Confl Health. 2011;5:13. doi: 10.1186/1752-1505-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azman AS, Rumunu J, Abubakar A, West H, Ciglenecki I, Helderman T, Wamala JF, Vazquez Ode L, Perea W, Sack DA, et al. Population-level effect of cholera vaccine on displaced populations, South Sudan, 2014. Emerg Infect Dis. 2016;22(6):1067–1070. doi: 10.3201/eid2206.151592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Bank . Fragile and conflict-affected situations lists from FY06 to FY20. 2020. [Google Scholar]
- 15.WHO . Weekly epidemiological record: cholera articles. 2020. [Google Scholar]
- 16.UNICEF . UNICEF West and Central Africa cholera platform. UNICEF regional office for West and Central Africa. 2020. [Google Scholar]
- 17.WHO: Disease outbreak news. WHO. 2020: https://www.who.int/csr/don/en/ Accessed 16 Nov 2020.
- 18.Global Task Force on Cholera Control (Surveillance Working Group) Interim guidance document on cholera surveillance. Global Task Force on Cholera Control. 2017. [Google Scholar]
- 19.Sparrow A, Almilaji K, Tajaldin B, Teodoro N, Langton P. Cholera in the time of war: implications of weak surveillance in Syria for the WHO's preparedness-a comparison of two monitoring systems. BMJ Glob Health. 2016;1(3):e000029. doi: 10.1136/bmjgh-2016-000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO Regional Office for Africa . Weekly bulletins on outbreaks and other emergencies. Brazzaville: WHO Regional Office for Africa; 2020. [Google Scholar]
- 21.WHO Regional Office for the Western Pacific . Western Pacific surveillance and response journal. Manilla: WHO Regional Office for the Western Pacific; 2020. [Google Scholar]
- 22.WHO Regional Office for the Eastern Mediterranean. Weekly epidemiological monitor. Cairo: WHO Regional Office for the Eastern Mediterranean; 2020. http://www.emro.who.int/pandemic-epidemic-diseases/information-resources/weekly-epidemiological-monitor.html. Accessed 2 Dec 2020.
- 23.WHO Regional Office for South-East Asia . WHO SEARO journal of public health. New Delhi: WHO Regional Office for South-East Asia; 2020. [Google Scholar]
- 24.ProMED-mail. http://www.promedmail.org. Accessed 12 December 2020.
- 25.Assistance Coordination Unit: Early warning alert and response network . Assistance coordination unit. 2020. [Google Scholar]
- 26.Harrell FE. Regression modeling strategies : with applications to linear models, logistic regression, and survival analysis. New York: Springer; 2001. [Google Scholar]
- 27.Althaus CL. Ebola superspreading. Lancet Infect Dis. 2015;15(5):507–508. doi: 10.1016/S1473-3099(15)70135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucharski AJ, Eggo RM, Watson CH, Camacho A, Funk S, Edmunds WJ. Effectiveness of ring vaccination as control strategy for Ebola virus disease. Emerg Infect Dis. 2016;22(1):105–108. doi: 10.3201/eid2201.151410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukandavire Z, Morris JG: Modeling the epidemiology of cholera to prevent disease transmission in developing countries. Microbiol Spectr. 2015;3(3):1–6. [DOI] [PMC free article] [PubMed]
- 31.Moore SM, Shannon KL, Zelaya CE, Azman AS, Lessler J. Epidemic risk from cholera introductions into Mexico. Plos Curr. 2014;6. 10.1371/currents.outbreaks.c04478c7fbd9854ef6ba923cc81eb799. [DOI] [PMC free article] [PubMed]
- 32.Kahn R, Peak CM, Fernandez-Gracia J, Hill A, Jambai A, Ganda L, Castro MC, Buckee CO: Incubation periods impact the spatial predictability of cholera and Ebola outbreaks in Sierra Leone. Proc Natl Acad Sci U S A. 2020;117(9):5067–73. [DOI] [PMC free article] [PubMed]
- 33.R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for statistical Computing; 2020. [Google Scholar]
- 34.UN Mission in Liberia: UNMIL Humanitarian Situation Report. UN Mission in Liberia. 2007, No. 131: https://reliefweb.int/report/liberia/liberia-unmil-humanitarian-situation-report-no-131. Accessed 16 Nov 2020.
- 35.International Federation of the Red Cross and Red Crescent: Sudan: Cholera Final report, Emergency appeal n° MDRSD001. International Federation of the Red Cross and Red Crescent. 2008: https://reliefweb.int/sites/reliefweb.int/files/resources/F1F7E51BB603CAA7852574AC00580E32-Full_Report.pdf. Accessed 16 Nov 2020.
- 36.Kpayli M: Southeasterners suffer another cholera outbreak. Liberian Times 2008: https://reliefweb.int/report/liberia/liberia-southeasterners-suffer-another-cholera-outbreak. Accessed 16 Nov 2020.
- 37.Medair: Sudan: Cholera intervention in Yei. Medair. 2008: https://reliefweb.int/report/sudan/sudan-cholera-intervention-yei. Accessed 16 Nov 2020.
- 38.Médecins Sans Frontières: Sewage problems cause cholera outbreak in Zimbabwe. Méd Sans Front 2008: https://www.msf.org/sewage-problems-cause-cholera-outbreak-zimbabwe. Accessed 16 Nov 2020.
- 39.Ministry of Health Iraq, WHO Representative’s Office Iraq: Summary: Week 34 ending 24-August 2008. Ministry of Health Iraq. 2008, Sitrep no 74: https://reliefweb.int/sites/reliefweb.int/files/resources/329E5325F2C5F87C852574C10063268A-Full_Report.pdf. Accessed 16 Nov 2020.
- 40.ProMED-mail.: Cholera, diarrhea & dysentery update 2008 (34). 2008, Archive Number: 20080904.2765: https://promedmail.org/promed-post/?id=108549. Accessed 16 Nov 2020.
- 41.Unknown: Guinea-Bissau: Cholera epidemic reaches capital, Bissau. The New Humanitarian. 2008: https://reliefweb.int/report/guinea-bissau/guinea-bissau-cholera-epidemic-reaches-capital-bissau. Accessed 16 Nov 2020.
- 42.Unknown: Guinea-Bissau: WHO confirms cholera outbreak. The New Humanitarian. 2008: https://reliefweb.int/report/guinea-bissau/guinea-bissau-who-confirms-cholera-outbreak. Accessed 16 Nov 2020.
- 43.WHO Cholera, Iraq. Weekly Epidemiol Rec. 2008;39(83):349–356. [Google Scholar]
- 44.WHO . Cholera in Zimbabwe: epidemiological bulletin number 1. Harare: WHO; 2008. [Google Scholar]
- 45.WHO . Cholera in Zimbabwe: epidemiological bulletin number 2. Harare: WHO; 2008. [Google Scholar]
- 46.WHO EMRO: Cholera in Iraq. WHO Regional Office for the Mediterranean 2008, 1(36): https://applications.emro.who.int/dsaf/epi/2008/Epi_Monitor_2008_1_36.pdf?ua=1&ua=1. Accessed 16 Nov 2020.
- 47.WHO EMRO: Cholera in Somalia. WHO Regional Office for the Eastern Mediterranean. 2008, 1(10): http://applications.emro.who.int/dsaf/epi/2008/Epi_Monitor_2008_1_10.pdf?ua=1. Accessed 16 Nov 2020.
- 48.WHO EMRO: Cholera outbreak in southern Sudan. 2008, 1(22 & 23). http://applications.emro.who.int/dsaf/epi/2008/Epi_Monitor_2008_1_22-23.pdf?ua=1. Accessed 2 Dec 2020.
- 49.International Federation of the Red Cross and Red Crescent: Disaster relief emergency fund (DREF) Operation, Papua New Guinea: Cholera, dysentery and influenza outbreaks, DREF operation n° MDRPG004. International Federation of the Red Cross and Red Crescent. 2009(Glide No. EP-2009-000185-PNG): https://reliefweb.int/sites/reliefweb.int/files/resources/EFAA8C5B1D718088C125762B00415B26-Full_Report.pdf. Accessed 16 Nov 2020.
- 50.Centers for Disease Control and Prevention Update: cholera outbreak --- Haiti, 2010. MMWR Morb Mortal Wkly Rep. 2010;59(45):1473–1479. [PubMed] [Google Scholar]
- 51.Disasters Emergency Committee: Cholera Hits Pakistan Three Months After Floods Starts. Disasters Emergency Committee. 2010: https://www.dec.org.uk/press-release/cholera-hits-pakistan-three-months-after-floods-start. Accessed 16 Nov 2020.
- 52.Government of Afghanistan, WHO: Afghanistan: Cholera outbreak under control, say MoPH, WHO. Government of Afghanistan and World Health organization. 2010(July 7, 2020): https://reliefweb.int/report/afghanistan/afghanistan-cholera-outbreak-under-control-say-moph-who. Accessed 28 Aug 2010.
- 53.WHO EMRO: Suspected cholera in Afghanistan. WHO Regional Office for the Mediterranean. 2010, 3(18): http://applications.emro.who.int/dsaf/epi/2010/Epi_Monitor_2010_3_38.pdf?ua=1. Accessed 16 Nov 2020.
- 54.WHO EMRO: Cholera in Pakistan. WHO Regional Office for the Eastern Mediterranean. 2010, 3(43): https://applications.emro.who.int/dsaf/epi/2010/Epi_Monitor_2010_3_43.pdf?ua=1&ua=1. Accessed 16 Nov 2020.
- 55.WHO EMRO: Cholera in Yemen. WHO Regional Office for the Eastern Mediterranean. 2010, 3(27): http://applications.emro.who.int/dsaf/epi/2010/Epi_Monitor_2010_3_27.pdf?ua=1. Accessed 16 Nov 2020.
- 56.WHO Global Alert and Response: Cholera in Pakistan. WHO. 2010: https://www.who.int/csr/don/2010_10_25/en/. Accessed 16 Nov 2020.
- 57.Dunoyer J, Sudre B: Rapport de capitalisation au sujet de l'épidémie de choléra au Tchad, 2010. Ministere de la Sante Publique 2011: https://reliefweb.int/sites/reliefweb.int/files/resources/rapport-de-capitalisation-epid%C3%A9mie-de-chol%C3%A9ra-au-tchad-2010-acf.pdf. Accessed 16 Nov 2020.
- 58.Gbary AR, Dossou JP, Sossou RA, Mongbo V, Massougbodji A. Epidemiologic and medico-clinical aspects of the cholera outbreak in the Littoral department of Benin in 2008. Med Trop (Mars) 2011;71(2):157–161. [PubMed] [Google Scholar]
- 59.International Federation of the Red Cross and Red Crescent: Disaster relief emergency fund (DREF). Burundi: Cholera. International Federation of the Red Cross and Red Crescent. 2011(GLIDE n° EP-2011-000126-BDI): https://reliefweb.int/sites/reliefweb.int/files/resources/Full_Report_2277.pdf. Accessed 16 Nov 2020.
- 60.Unknown: UNICEF responds to new cholera outbreak claiming lives in Central African Republic. UNICEF 2011: https://www.unicef.org/media/media_60000.html. Accessed 16 Nov 2020.
- 61.Luquero FJ, Banga CN, Remartínez D, Palma PP, Baron E, Grais RF. Cholera epidemic in Guinea-Bissau (2008): the importance of “place”. PLoS One. 2011;6(5):e19005. doi: 10.1371/journal.pone.0019005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Penguele A., Djeintote M., Balekouzou A., Tembeti J., Feilema P., Kazambu D., D. D: Cholera outbreak investigation in the Central African Republic, October – November 2011. Central African Republic Field Epidemiology and Laboratory Training Program 2011: https://www.cdcfoundation.org/sites/default/files/upload/image/2011CholeraOutbreakReport.pdf. Accessed 16 Nov 2020.
- 63.Rosewell A, Dagina R, Murhekar M, Ropa B, Posanai E, Dutta SR, Jennison A, Smith H, Mola G, Zwi A, et al. Vibrio cholerae O1 in 2 coastal villages, Papua New Guinea. Emerg Infect Dis. 2011;17(1):154–156. doi: 10.3201/eid1701.100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Unknown: A Dozen killed in cholera outbreak. The New Humanitarian. 2011: https://www.thenewhumanitarian.org/news/2011/09/14/dozen-killed-cholera-outbreak. Accessed 16 Nov 2020.
- 65.WHO EMRO: Cholera in Afghanistan. WHO Regional Office for the Eastern Mediterranean. 2011, 4(18 & 19): http://applications.emro.who.int/dsaf/epi/2011/Epi_Monitor_2011_4_18-19.pdf?ua=1. Accessed 16 Nov 2020.
- 66.WHO EMRO: Suspected cholera outbreaks in Afghanistan. WHO Regional Office for the Eastern Mediterranean. 2011, 4(26 & 27): http://applications.emro.who.int/dsaf/epi/2011/Epi_Monitor_2011_4_26_27.pdf?ua=1. Accessed 16 Nov 2020.
- 67.WHO EMRO: Cholera in Yemen. WHO Regional Office for the Eastern Mediterranean. 2011, 4(42): http://applications.emro.who.int/dsaf/epi/2011/Epi_Monitor_2011_4_42.pdf?ua=1. Accessed 16 Nov 2020.
- 68.WHO EMRO: Humanitarian health action: growing needs for emergency life-saving health care in Yemen, monthly highlights. WHO Regional Office for the Eastern Mediterranean 2011: https://www.who.int/hac/crises/yem/highlights/june2011/en/. Accessed 16 Nov 2020.
- 69.Croix-Rouge du Burundi: La Croix-rouge du Burundi dans la luttre contre l'épidémie de choléra. Croix-Rouge du Burundi 2012: https://reliefweb.int/report/burundi/la-croix-rouge-du-burundi-dans-la-lutte-contre-l%E2%80%99%C3%A9pid%C3%A9mie-de-chol%C3%A9ra. Accessed 16 Nov 2020.
- 70.International Federation of the Red Cross and Red Crescent: Mali: preventing the spread of cholera near Gao. International Committee of the Red Cross. 2012: https://www.icrc.org/en/doc/resources/documents/news-release/2012/mali-news-2012-07-05.htm. Accessed 16 Nov 2020.
- 71.International Federation of the Red Cross and Red Crescent: Disaster Relief Emergency Fund (DREF) final report, Central African Republic: Cholera outbreak, DREF operation n° MDRCF009. International Federation of the Red Cross and Red Crescent. 2012(GLIDE n° EP-2011-000153-CAF): https://reliefweb.int/sites/reliefweb.int/files/resources/MDRCF009finrep.pdf. Accessed 16 Nov 2020.
- 72.Ivers LC, Walton DA. The “first” case of cholera in Haiti: lessons for global health. Am J Trop Med Hyg. 2012;86(1):36–38. doi: 10.4269/ajtmh.2012.11-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahamud AS, Ahmed JA, Nyoka R, Auko E, Kahi V, Ndirangu J, Nguhi M, Burton JW, Muhindo BZ, Breiman RF, et al. Epidemic cholera in Kakuma refugee camp, Kenya, 2009: the importance of sanitation and soap. J Infect Dev Ctries. 2012;6(3):234–241. doi: 10.3855/jidc.1966. [DOI] [PubMed] [Google Scholar]
- 74.WHO Outbreak news. Cholera, Sierra Leone. Weekly Epidemiol Rec. 2012;87(36):337–338. [PubMed] [Google Scholar]
- 75.Yadav DK, Tamrakar D, Baral R, Jha P, Gautam S, Pokharel PK. Outbreak of cholera in Tilathi VDC Saptari Nepal. Kathmandu Univ Med J (KUMJ) 2012;10(40):36–39. doi: 10.3126/kumj.v10i4.10992. [DOI] [PubMed] [Google Scholar]
- 76.Dunoyer J, Sudre B, Rebolledo J, Cottavoz P, Rossi M: Le choléra transfrontalier en Sierra Leone et Guinée en 2012 et les stratégies d'intervention associées. Paris: Action Contre la Faim - France, UMR 6249 Laboratoire Chrono-Environnement, Université de France-Comté; 2013.
- 77.International Federation of the Red Cross and Red Crescent: Disaster relief emergency fund (DREF) final report, Burundi: Cholera, DREF operation n° MDRBI009. International Federation of the Red Cross and Red Crescent. 2013(GLIDE n° EP-2012-000187-BDI): https://reliefweb.int/sites/reliefweb.int/files/resources/Burundi%20Cholera%20DREF%20Operation%20Final%20Report.pdf. Accessed 16 Nov 2020.
- 78.Qasem MA, Al-Abhar N, Jumaan A: The Hazard of conflict: cholera outbreak in Abyan, Yemen, 2011. EMPHNET. 2014: http://www.yfetp.com/OPSHandler/Resources/Attachment/RoxyFileman//First%20National%20YFETP%20Conference%20Book%20Feb%202014.pdf. Accessed 16 Nov 2020.
- 79.Republic of South Sudan, Ministry of Health: Cholera Outbreak in Juba, Republic of South Sudan. Ministry of Health of South Suda. 2014: https://reliefweb.int/sites/reliefweb.int/files/resources/Juba-Cholera-15-May-2014.pdf. Accessed 16 Nov 2020.
- 80.UNICEF: Ivory Coast: Cholera and fishermen movement along the coast. UNICEF West and Central Africa Regional Office. 2014, Week 2: https://reliefweb.int/sites/reliefweb.int/files/resources/Cholera_W2_2015%20West%20and%20Central%20Africa.pdf. Accessed 16 Nov 2020.
- 81.UNICEF: Cote d'Ivoire: Confirmation of a cholera outbreak in Abidjan. UNICEF West and Central Africa Regional Office. 2014, Week 41: https://reliefweb.int/sites/reliefweb.int/files/resources/Cholera%20regional%20update_W41_2014%20WCA.pdf. Accessed 16 Nov 2020.
- 82.WHO. Emergencies preparedness and response: cholera outbreak, South Sudan. Juba: WHO; 2014. https://www.who.int/csr/don/2014_05_30/en/. Accessed 16 Nov 20.
- 83.Assistance Coordination Unit: Report on suspected case of Cholera in N Syria: EWARN Report. Assistance Coordination Unit. 2015: https://www.acu-sy.org/en/suspected-case-of-cholera-in-n-syria/. Accessed 16 Nov 2020.
- 84.Assistance Coordination Unit: Epidemiological Weekly Bulletin Syria - Early Warning Alert and Response Network Assistance Coordination Unit. 2015, Epiweek No. 42: https://www.acu-sy.org/en/epi-reports/?b5-file=1954&b5-folder=606. Accessed 16 Nov 2020.
- 85.Bwire G, Malimbo M, Kagirita A, Makumbi I, Mintz E, Mengel MA, Orach CG. Nosocomial cholera outbreak in a mental hospital: challenges and lessons learnt from Butabika National Referral Mental Hospital, Uganda. Am J Trop Med Hyg. 2015;93(3):534–538. doi: 10.4269/ajtmh.14-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cholera Task Force-Iraq: update on current cholera outbreak in Iraq. Cholera Task Force-Iraq. 2015, Sitrep No 1, Epi Week 37: https://reliefweb.int/sites/reliefweb.int/files/resources/SITREP__1__13_Sept_15EXT.pdf. Accessed 16 Nov 2020.
- 87.Ministry of Health South Sudan, WHO: Cholera in Juba, Central Equatoria State, Republic of South Sudan. Ministry of Health of South Sudan. 2015, Situation Report # 1: https://www.who.int/hac/crises/ssd/sitreps/south_sudan_cholera_22june2015.pdf?ua=1. Accessed 16 Nov 2020.
- 88.UNHCR: Burundi situation, UNHCR regional update, No. 0013. UNHCR. 2015, No. 0013: https://reliefweb.int/sites/reliefweb.int/files/resources/UNHCR_BurundiSituation_No0013.pdf. Accessed 16 Nov 2020.
- 89.UNHCR: Burundi situation, UNHCR regional update, No. 0011. UNHCR. 2015, No. 0011: https://data2.unhcr.org/en/documents/download/48620. Accessed 16 Nov 2020.
- 90.UNICEF: Ethiopia: Humanitarian Situation Report. UNICEF. SitRep #7. Addis Ababa. 2015. https://reliefweb.int/sites/reliefweb.int/files/resources/UNICEF%20Ethiopia%20Humanitarian%20Sitrep%20No.%207%2C%20November%20-%20December%202015.pdf. Accessed 16 Nov 2020.
- 91.UNICEF, OCHA: brief preliminary report of UNICEF and OCHA AWD/suspected cholera fact finding mission to Balet Xaawo (as of 12 June 2015). UNICEF. 2015: https://reliefweb.int/sites/reliefweb.int/files/resources/160615_brief_report_of_the_balet_xaawo_unicef_and_ocha_mission.pdf. Accessed 16 Nov 2020.
- 92.WHO: WHO notified of new cholera outbreaks in Tanzania. WHO. 2015: https://reliefweb.int/report/united-republic-tanzania/who-notified-new-cholera-outbreaks-tanzania. Accessed 16 Nov 2020.
- 93.Ethiopian Public Health Institute: Suspected AWD cases. Ethiopian Public Health Institute. 2016, 2(10): https://www.ephi.gov.et/images/pictures/Weekly-Epidemiological-Bulletin-2016-10%20(1).pdf. Accessed 16 Nov 2020.
- 94.International Federation of the Red Cross and Red Crescent: Emergency Plan of Action (EPoA), Central Africa Republic: Cholera Epidemic Outbreak. International Federation of the Red Cross and Red Crescent. 2016: https://reliefweb.int/sites/reliefweb.int/files/resources/MDRCF021.pdf. Accessed 12 Nov 2020.
- 95.Republic of South Sudan, Ministry of Health: Suspect cholera. Ministry of Health of South Sudan. 2016, W26, 27 June-3 July, 2016: https://www.who.int/hac/crises/ssd/south_sudan_epi_3july2016.pdf. Accessed 16 Nov 2020.
- 96.UNICEF: Cholera Outbreaks in Central and West Africa: 2016 Regional Update. UNICEF West and Central Africa. 2016, Week 04: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W4.pdf. Accessed 16 Nov 2020.
- 97.UNICEF: Cholera outbreaks in the West and Central Africa: 2016 Regional Update. UNICEF West and Central Africa. 2016, Week 6: https://reliefweb.int/sites/reliefweb.int/files/resources/snapshot_wca_unicef_cholera_update_w6.pdf. Accessed 16 Nov 2020.
- 98.UNICEF: Cholera Outbreaks in Central and West Africa: 2016 Regional Update. UNICEF West and Central Africa Regional Office. 2016, Week 10: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W10.pdf. Accessed 16 Nov 2020.
- 99.UNICEF: Benin: update Choléra. 2016: https://reliefweb.int/sites/reliefweb.int/files/resources/UNICEF%20Be%C3%ACnin%20Cholera%20Update%2022%20September%202016.pdf. Accessed 16 Nov 2020.
- 100.UNICEF: Cholera alert lifted in Mora, Mayo-Sava Department, Extreme North Cameroon. UNICEF’s West and Central Africa Regional Office. 2016, Week 18: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W18.pdf. Accessed 16 Nov 2020.
- 101.UNICEF: Early warning and early actions in Cameroon, Littoral Region. UNICEF West and Central Africa Regional Office. 2016, Week 26: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W26_-_V1.pdf. Accessed 16 Nov 2020.
- 102.UNICEF: Highlights: Central African Republic. UNICEF West and Central Africa Regional Office. 2016, Week 30: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W30.pdf. Accessed 16 Nov 2020.
- 103.UNICEF: Highlights: Focus on cholera alert in Congo Brazzaville (22/09/2016). UNICEF West and Central Africa Regional Office. 2016, Week 36: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W36.pdf. Accessed 16 Nov 2020.
- 104.UNICEF: Alert: Niger health district Dosso. UNICEF West and Central Africa Regional Office. 2016, Week 40: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA%20Cholera_Update_W40.pdf. Accessed 16 Nov 2020.
- 105.UNICEF: Niger - Dosso. UNICEF West and Central Africa Regional Office. 2016, Week 42: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA%20Cholera_Update_W42.pdf. Accessed 16 Nov 2020.
- 106.WHO AFRO: Ethiopia - response for acute watery diarrhea outbreak in Moyale town. WHO Regional Office for Africa 2016: https://www.afro.who.int/news/ethiopia-response-acute-watery-diarrhea-outbreak-moyale-town. Accessed 16 Nov 2020.
- 107.WHO EMRO: The Ministry of Public Health and population announces cholera cases in Yemen. WHO Regional Office for the Eastern Mediterranean. 2016: http://www.emro.who.int/media/news/the-ministry-of-health-announces-cholera-cases-in-yemen.html. Accessed 16 Nov 2020.
- 108.Assistance Coordination Unit: Epidemiological Weekly Bulletin - Syria Early Warning Alert and Response Network (EWARN). Assistance Coordination Unit. 2017, Epiweek 42: https://www.acu-sy.org/en/epi-reports/?b5-file=4540&b5-folder=606. Accessed 16 Nov , 2020.
- 109.Government of Liberia, WHO: Severe Acute Watery Diarrhea (Cholera). WHO Liberia. 2017, Epi Week 13: https://reliefweb.int/sites/reliefweb.int/files/resources/liberia-early-warning-disease-surveillance-bulletin-week-13.doc.pdf. Accessed 16 Nov 2020.
- 110.Hassan A, Mohammed K, Alaka A, Balami K, Njidda A, Malgwi A, Eteng W, Lawal B, Dalhat M, Balogun M, et al. Outbreak investigation of cholera in an internally displaced persons camp in. Borno state, northeastern Nigeria –. August 2017. In: International conference on (re-)emerging infectious diseases (ICREID). Addis Ababa: Africa Centres for Disease Control and Prevention; 2017.
- 111.Joint Cholera Initiative for Southern Africa (JCISA): Sub-regional up-date on Cholera: Week 4. Joint Cholera Initiative for Southern Africa (JCISA. 2017: https://reliefweb.int/sites/reliefweb.int/files/resources/jcisa_bulletin_8_week_4jan2017.pdf. Accessed 12 Nov 2020.
- 112.Joint Cholera Initiative for Southern Africa (JCISA): Sub-regional up-date on Cholera; Week 8. Joint Cholera Initiative for Southern Africa (JCISA),. 2017: https://reliefweb.int/sites/reliefweb.int/files/resources/jcisa_bulletin_10_week_8feb2017.pdf. Accessed 12 Nov 2020.
- 113.Kwesiga B, Pande G, Ario AR, Tumwesigye NM, Matovu JKB, Zhu BP. A prolonged, community-wide cholera outbreak associated with drinking water contaminated by sewage in Kasese District, western Uganda. BMC Public Health. 2017;18(1):30. doi: 10.1186/s12889-017-4589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ministere de la sante de la Repulique du Benin: Plan D'Endiguement du Cholera, 2017–2021. 2017: http://plateformecholera.info/attachments/article/430/Plan%20d'Endiguement%20Cholera%20du%20Benin.pdf. Accessed 16 Nov 2020.
- 115.Ministry of Health Somalia, WHO: Situation Report for Acute Watery Diarrhoea/Cholera. Ministry of Health Somalia. 2017, Epidemiological Week 50 (11th to 17th Dec 2017): https://reliefweb.int/sites/reliefweb.int/files/resources/week_50_awd_cholera_sitrep_final_0.pdf. Accessed 16 Nov 2020.
- 116.Ministry of Health Zambia: Cholera Update. Ministry of Health, Zambia. 2017: https://www.moh.gov.zm/?p=5394. Accessed 16 Nov 2020.
- 117.Narra R, Maeda JM, Temba H, Mghamba J, Nyanga A, Greiner AL, Bakari M, Beer KD, Chae SR, Curran KG, et al. Notes from the field: ongoing cholera epidemic - Tanzania, 2015-2016. MMWR Morb Mortal Wkly Rep. 2017;66(6):177–178. doi: 10.15585/mmwr.mm6606a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.ProMED-mail: Cholera, diarrhea & dysentery update (122): Asia (Syria). ProMED-mail. 2017, Archive Number: 20171021.5394037: https://promedmail.org/promed-post/?id=5394037. Accessed 16 Nov 2020.
- 119.ProMED-mail.: Cholera, diarrhea & dysentery update (02): Africa. ProMED-mail. 2017, 13 Jan: 20170113.4751889: http://www.promedmail.org. Accessed 12 Nov 2020.
- 120.UNICEF: Alert of suspected cholera outbreak - Eastern Chad - Sila Region - Koukou Angarana (close of Goz Beida). UNICEF West and Central Africa Regional Office. 2017, Week 32: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA_Cholera_Update_W32_-_V2.pdf. Accessed 16 Nov 2020.
- 121.UNICEF: Zoom on Mano River Basin. UNICEF West and Central Africa Regional Office. 2017, Week 16: https://reliefweb.int/sites/reliefweb.int/files/resources/WCA%20Cholera_Update_W16.pdf. Accessed 16 Nov 2020.
- 122.WHO: Cholera – Zambia. WHO. 2017: https://www.who.int/csr/don/11-december-2017-cholera-zambia/en/. Accessed 16 Nov 2020.
- 123.WHO AFRO: Cholera, Burundi. WHO regional Office for Africa. 2017, Week 34: 19–25 August 2017: https://apps.who.int/iris/bitstream/handle/10665/258794/OEW34-192582017.pdf;jsessionid=93BF65989F3FC960D06F2813029628B7?sequence=1. Accessed 16 Nov 2020.
- 124.WHO AFRO: Flambee des cas de cholera au Burundi. WHO Regional Office for Africa. 2017: https://reliefweb.int/sites/reliefweb.int/files/resources/whe-burundi_cholera_update_002_21082017_0.pdf. Accessed 16 Nov 2020.
- 125.WHO AFRO: Cholera, Chad. WHO regional Office for Africa. 2017, Week 34: 19–25 August 2017: http://apps.who.int/iris/bitstream/10665/258794/1/OEW34-192582017.pdf. Accessed 16 Nov 2020.
- 126.WHO AFRO: WHO responds to new acute watery diarrhea outbreak rumour in a remote zone in Somali region of Ethiopia. WHO Regional Office for Africa. 2017: https://www.afro.who.int/news/who-responds-new-acute-watery-diarrhea-outbreak-rumour-remote-zone-somali-region-ethiopia. Accessed 16 Nov 2020.
- 127.WHO AFRO: Cholera in Borno state, Nigeria. WHO Regional Office for Africa. 2017, week 36: 2-8 September 2017. https://apps.who.int/iris/bitstream/handle/10665/258922/OEW36-2892017.pdf?sequence=1. Accessed 2 Dec 2020.
- 128.WHO AFRO: Cholera in Zambia. WHO Regional Office for Africa. 2017, Week 41: 7–13 October 2017: https://apps.who.int/iris/bitstream/handle/10665/259263/OEW41-713102017.pdf?sequence=1. Accessed 16 Nov 2020.
- 129.WHO Hub/Est (Abéché): Situation des cas de cholera dans la delegation sanitaire regionale du Sila. WHO Chad. 2017: https://reliefweb.int/sites/reliefweb.int/files/resources/sitrep_cholera_sila_17_sept_2017.pdf. Accessed 16 Nov 2020.
- 130.Yadav S. Cholera outbreak in Gaidataar: a lesson for further strengthening the task force for epidemic Management in Nepal. JNMA J Nepal Med Assoc. 2017;56(207):374–376. doi: 10.31729/jnma.3284. [DOI] [PubMed] [Google Scholar]
- 131.Abubakar A, Bwire G, Azman AS, Bouhenia M, Deng LL, Wamala JF, Rumunu J, Kagirita A, Rauzier J, Grout L, et al. Cholera epidemic in South Sudan and Uganda and need for international collaboration in cholera control. Emerg Infect Dis. 2018;24(5):883–887. doi: 10.3201/eid2405.171651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Akram K. Investigation of cholera outbreak at Rawalpindi, Pakistan - August 2017. Iproceedings. 2018;4(1):e10579.
- 133.Chimusoro A, Maphosa S, Manangazira P, Phiri I, Nhende T, Danda S, Tapfumanei O, Munyaradzi Midzi S, Nabyonga-Orem J. Responding to cholera outbreaks in Zimbabwe: building resilience over time. In: Claborn D, editor. Current Issues in Global Health. 2018. [Google Scholar]
- 134.Dunoyer J, Moore S, Valingot C, Rebaudet S, Gaudart J, Piarroux R, Sudre B: Epidemiological study of cholera hotspots and epidemiological basins in east and southern Africa: in-depth cholera epidemiological report for South Sudan. Prospective Cooperation and UNICEF 2018: https://plateformecholera.info/attachments/article/639/Cholera%20epidemiology%20in%20South%20Sudan_UNICEF_April%202018_FINAL.pdf. Accessed 16 Nov 2020.
- 135.Government of Burundi, WHO: Rapport de situation n°01 sur la flambee de cholera au Burundi. WHO. 2018: https://reliefweb.int/sites/reliefweb.int/files/resources/30122018_OMS%20Burundi%20Flamb%C3%A9e%20CHOLERA_RAPSIT.pdf. Accessed 16 Nov 2020.
- 136.McAteer JB, Danda S, Nhende T, Manamike P, Parayiwa T, Tarupihwa A, Tapfumanei O, Manangazira P, Mhlanga G, Garone DB, et al. Notes from the field: outbreak of Vibrio cholerae associated with attending a funeral - Chegutu District, Zimbabwe, 2018. MMWR Morb Mortal Wkly Rep. 2018;67(19):560–561. doi: 10.15585/mmwr.mm6719a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monnard G: Reponse à l'épidémie de cholera au Sila et au Salamat, Tchad, 2017: rapport de synthese. UNICEF Chad 2018: https://plateformecholera.info/attachments/article/556/UNICEF%202018%20Rapport%20synth%C3%A8se%20r%C3%A9ponse%20%C3%A9pid%C3%A9mie%20chol%C3%A9ra%20Sila%20-%20Salamat%202017%20FINAL.pdf. Accessed 16 Nov 2020.
- 138.Moore S, Dunoyer J, Sudre B, Valingot C, Rebaudet S, Piarroux R: Epidemiological study of cholera hotspots and epidemiological basins in east and southern Africa. Prospective and Cooperation 2018: https://plateformecholera.info/attachments/article/639/Cholera%20epidemiology%20in%20South%20Sudan_UNICEF_April%202018_FINAL.pdf. Accessed 16 Nov 2020.
- 139.Pande G, Kwesiga B, Bwire G, Kalyebi P, Riolexus A, Matovu JKB, Makumbi F, Mugerwa S, Musinguzi J, Wanyenze RK, et al. Cholera outbreak caused by drinking contaminated water from a lakeshore water-collection site, Kasese District, South-Western Uganda, June-July 2015. PLoS One. 2018;13(6):e0198431. doi: 10.1371/journal.pone.0198431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sinyange N, Brunkard JM, Kapata N, Mazaba ML, Musonda KG, Hamoonga R, Kapina M, Kapaya F, Mutale L, Kateule E, et al. Cholera epidemic - Lusaka, Zambia, October 2017-may 2018. MMWR Morb Mortal Wkly Rep. 2018;67(19):556–559. doi: 10.15585/mmwr.mm6719a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tull K. Knowledge, evidence and learning for development (K4D) Brighton: Institute of Development Studies; 2018. Humanitarian interventions in Ethiopia responding to acute watery diarrhoea. [Google Scholar]
- 142.Evaluation of the UNICEF Level 3 Response to the Cholera Epidemic in Yemen: A Crisis within a Crisis. New York: Evaluation Office, United Nations Children's Fund; 2018.
- 143.UNICEF: Zimbabwe: Cholera outbreak report. UNICEF Zimbabwe. 2018, Sitrep 1: https://reliefweb.int/sites/reliefweb.int/files/resources/UNICEF%20Zimbabwe%20Cholera%20Situation%20Report%20-13%20September%202018.pdf. Accessed 16 Nov 2020.
- 144.WHO: Cholera in Uganda. WHO Regional Office for Africa. 2018, Week 8: 17-23 February 2018: https://apps.who.int/iris/bitstream/handle/10665/260335/OEW8-1723022018.pdf?sequence=1. Accessed 16 Nov 2020.
- 145.WHO AFRO: Cholera in Angola. WHO Regional Office for Africa. 2018, Week 3: 13–19 January 2018: https://apps.who.int/iris/bitstream/handle/10665/259885/OEW03-1319012018.pdf;jsessionid=90D67FBFDC659F22E19940CA3380679E?sequence=1. Accessed 16 Nov 2020.
- 146.WHO AFRO: Cholera in Angola. WHO Regional Office for Africa. 2018, Week 6: 3–9 February 2018: https://apps.who.int/iris/bitstream/handle/10665/260157/OEW6-030922018.pdf?sequence=1. Accessed 16 Nov 2020.
- 147.WHO AFRO: Cholera in Congo. WHO Regional Office for Africa. 2018, Week 16: 14–20 April 2018: https://apps.who.int/iris/bitstream/handle/10665/272386/OEW16-142042018.pdf. Accessed 16 Nov 2020.
- 148.WHO AFRO: Cholera outbreak in Niger. WHO Regional Office for Africa. 2018, Week 29: 14 July-20 July 2018: https://apps.who.int/iris/bitstream/handle/10665/273214/OEW29-1420072018.pdf. Accessed 16 Nov 2020.
- 149.WHO AFRO: Cholera in north-East Nigeria. WHO Regional Office for Africa. 2018, Week 11: 10–16 March 2018: https://apps.who.int/iris/bitstream/handle/10665/260520/OEW11-101632018.pdf?sequence=1. Accessed 16 Nov 2020.
- 150.WHO AFRO: Cholera in Zimbabwe. WHO Regional Office for Africa. 2018, Week 36: 1–7 September 2018: https://apps.who.int/iris/bitstream/handle/10665/274335/OEW36-15092018.pdf. Accessed 16 Nov 2020.
- 151.WHO AFRO: Cholera in Zimbabwe. WHO Regional Office for Africa. 2018, Week 4: 20–26 January 2018: http://apps.who.int/iris/bitstream/10665/259942/1/OEW4-202612018.pdf. Accessed 16 Nov 2020.
- 152.WHO Emergencies Preparedness and Response: Cholera – Zimbabwe. WHO. 2018: https://www.who.int/csr/don/20-september-2018-cholera-zimbabwe/en/. Accessed 16 Nov 2020.
- 153.WHO EMRO: New Cholera Outbreak in Somalia. WHO Regional Office for the Eastern Mediterranean. 2018, 11(6): http://applications.emro.who.int/docs/epi/2018/Epi_Monitor_2018_11_06.pdf?ua=1. Accessed 16 Nov 2020.
- 154.Abdullahi KN, Mutindin D, Kabugi W, Mowlid S. Epidemiological description of a protracted cholera outbreak in Hagadera refugee camp and the surrounding host community within Fafi Sub County and Garissa County in Kenya during march-September 2019. Epidemiol Open J. 2019;4(1):31–35. doi: 10.17140/EPOJ-4-116. [DOI] [Google Scholar]
- 155.Cambaza E, Mongo E, Anapakala E, Nhambire R, Singo J, Machava E: Outbreak of Cholera Due to Cyclone Kenneth in Northern Mozambique, 2019. Int J Environ Res Public Health. 2019;16(16):1–9. [DOI] [PMC free article] [PubMed]
- 156.Chen WH, Azman AS. Mitigating cholera in the aftermath of Cyclone Idai. Am J Trop Med Hyg. 2019;101(5):960-2. [DOI] [PMC free article] [PubMed]
- 157.Dan-Nwafor CC, Ogbonna U, Onyiah P, Gidado S, Adebobola B, Nguku P, Nsubuga P. A cholera outbreak in a rural north central Nigerian community: an unmatched case-control study. BMC Public Health. 2019;19(1):112. [DOI] [PMC free article] [PubMed]
- 158.Health Cluster Sudan: Health Cluster Bulletin: the cholera response overview (September – December 2019). Health Cluster Sudan,. 2019: https://reliefweb.int/sites/reliefweb.int/files/resources/hc_bulletin_december_2019-_cholera_outbreak_reposne_overview.pdf. Accessed 16 Nov 2020.
- 159.Kahn R, Mahmud AS, Schroeder A, Aguilar Ramirez LH, Crowley J, Chan J, Buckee CO. Rapid forecasting of cholera risk in Mozambique: translational challenges and opportunities. Prehosp Disaster Med. 2019;34(5):557–562. doi: 10.1017/S1049023X19004783. [DOI] [PubMed] [Google Scholar]
- 160.Nsubuga F, Garang SC, Tut M, Oguttu D, Lubajo R, Lodiongo D, Lasuba M, Mpairwe A. Epidemiological description of a protracted cholera outbreak in Tonj east and Tonj north counties, former Warrap state, South Sudan, may-Oct 2017. BMC Infect Dis. 2019;19(1):4. doi: 10.1186/s12879-018-3640-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roskosky M, Acharya B, Shakya G, Karki K, Sekine K, Bajracharya D, von Seidlein L, Devaux I, Lopez AL, Deen J, et al. Feasibility of a comprehensive targeted cholera intervention in the Kathmandu Valley, Nepal. Am J Trop Med Hyg. 2019;100(5):1088–97. [DOI] [PMC free article] [PubMed]
- 162.Sekine K, Roskosky M. Lessons learned from enhancing sentinel surveillance for cholera in post-earthquake Nepal in 2016. Am J Trop Med Hyg. 2019;100(3):494–496. doi: 10.4269/ajtmh.17-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sudan Humanitarian Country Team: Sudan: humanitarian cholera readiness and response [activities to support national AWD preparedness and response plan 2018-19]. Sudan Humanitarian Country Team 2019: https://reliefweb.int/sites/reliefweb.int/files/resources/191001_Consolidated%20Readiness%20and%20Response%20Plan%20for%20Cholera.pdf. Accessed 16 Nov 2020.
- 164.WHO AFRO: Cholera in Burundi. WHO Regional Office for Africa. 2019, Week 1: 29 December 2018-4 January 2019: https://apps.who.int/iris/bitstream/handle/10665/278952/OEW01-29122018-04012019.pdf?sequence=1&isAllowed=y. Accessed 16 Nov 2020.
- 165.WHO AFRO: Cholera, Chad. WHO Regional Office for Africa. 2019, Week 30: 22–28 July 2019: https://apps.who.int/iris/bitstream/handle/10665/326098/OEW30-2228072019.pdf. Accessed 16 Nov 2020.
- 166.WHO AFRO: Situation Report 1. WHO Regional Office for Africa. 2019, Sitrep 1: https://www.afro.who.int/sites/default/files/2019-05/WHOSitRep1Mozambique06-07-2019.pdf. Accessed 16 Nov 2020.
- 167.WHO Emergencies Preparedness and Response: Cholera – Republic of the Sudan. WHO. 2019: https://www.who.int/csr/don/15-october-2019-cholera-republic-of-the-sudan/en/. Accessed 16 Nov 2020.
- 168.WHO EMRO: OCV campaign Sudan. WHO Regional Office for the Eastern Mediterranean. 2019, 12(44): https://applications.emro.who.int/docs/epi/2019/22244220-2019-12-44.pdf?ua=1&ua=1. Accessed 16 Nov 2020.
- 169.Zambia National Public Health Institute, Ministry of Health Zambia, WHO: Situational Report No. 13. Zambia National Public Health Institute. 2019: https://reliefweb.int/sites/reliefweb.int/files/resources/Cholera%20Sitrep%20as%20at%20%2002_04_19%20%284%29%20sit%20rep%2013_0_0.pdf. Accessed 16 Nov 2020.
- 170.Bompangue D, Moore S, Taty N, Impouma B, Sudre B, Manda R, Balde T, Mboussou F, Vandevelde T. Description of the targeted water supply and hygiene response strategy implemented during the cholera outbreak of 2017-2018 in Kinshasa, DRC. BMC Infect Dis. 2020;20(1):226. doi: 10.1186/s12879-020-4916-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ngwa MC, Wondimagegnehu A, Okudo I, Owili C, Ugochukwu U, Clement P, Devaux I, Pezzoli L, Ihekweazu C, Jimme MA, et al. The multi-sectorial emergency response to a cholera outbreak in internally displaced persons camps in Borno state, Nigeria, 2017. BMJ Glob Health. 2020;5(1):e002000. doi: 10.1136/bmjgh-2019-002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lawpoolsri S, Kaewkungwal J, Khamsiriwatchara A, Sovann L, Sreng B, Phommasack B, Kitthiphong V, Lwin Nyein S, Win Myint N, Dang Vung N, et al. Data quality and timeliness of outbreak reporting system among countries in greater Mekong subregion: challenges for international data sharing. PLoS Negl Trop Dis. 2018;12(4):e0006425. doi: 10.1371/journal.pntd.0006425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.WHO Cholera, 2017. Weekly Epidemiol Rec. 2018;38(93):489–500. [Google Scholar]
- 174.Burki T. End of a cholera epidemic in South Sudan declared. Lancet. 2018;391(10121):647. doi: 10.1016/S0140-6736(18)30261-7. [DOI] [PubMed] [Google Scholar]
- 175.WHO AFRO: Ethiopia - Response for Acute Watery Diarrhea outbreak in Moyale town. https://www.afro.who.int/news/ethiopia-response-acute-watery-diarrhea-outbreak-moyale-town. Accessed 2 Dec 2020.
- 176.Kapata N, Sinyange N, Mazaba ML, Musonda K, Hamoonga R, Kapina M, Zyambo K, Malambo W, Yard E, Riggs M, et al. A Multisectoral emergency response approach to a cholera outbreak in Zambia: October 2017–February 2018. J Infect Dis. 2018;218(suppl_3):S181–S183. doi: 10.1093/infdis/jiy490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Azman AS, Moore SM, Lessler J. Surveillance and the global fight against cholera: setting priorities and tracking progress. Vaccine. 2020;38(Suppl 1):A28–A30. doi: 10.1016/j.vaccine.2019.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Spiegel P, Ratnayake R, Hellman N, Ververs M, Ngwa M, Wise PH, Lantagne D. Responding to epidemics in large-scale humanitarian crises: a case study of the cholera response in Yemen, 2016–2018. BMJ Glob Health. 2019;4(4):e001709. doi: 10.1136/bmjgh-2019-001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Garfield R, Bartee M, Mayigane LN. Validating joint external evaluation reports with the quality of outbreak response in Ethiopia, Nigeria and Madagascar. BMJ Glob Health. 2019;4(6):e001655. doi: 10.1136/bmjgh-2019-001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shahpar C, Lee CT, Wilkason C, Buissonniere M, McClelland A, Frieden TR. Protecting the world from infectious disease threats: now or never. BMJ Glob Health. 2019;4(4):e001885. [DOI] [PMC free article] [PubMed]
- 181.Ojo OE, Dalhat M, Garfield R, Lee C, Oyebanji O, Oyetunji A, Ihekweazu C. Nigeria's joint external evaluation and national action plan for health security. Health Secur. 2020;18(1):16–20. [DOI] [PubMed]
- 182.Kongelf A, Tingberga A, McClelland AL, Jean MC, Dalziel B. International Meeting on Emerging Diseases and Surveillance. Vienna: International Society for Infectious Diseases; 2016. Community-based cholera surveillance by volunteers with mobile phones: a case study from Western area, Haiti. [Google Scholar]
- 183.Ratnayake R, Tammaro M, Tiffany A, Kongelf A, Polonsky JA, McClelland A. People-centred surveillance: a narrative review of community-based surveillance among crisis-affected populations. Lancet Planet Health. 2020;4(10):e483–e495. doi: 10.1016/S2542-5196(20)30221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kuehne A, Keating P, Polonsky J, Haskew C, Schenkel K, Le Polain de Waroux O, Ratnayake R. Event-based surveillance at health facility and community level in low-income and middle-income countries: a systematic review. BMJ Glob Health. 2019;4(6):e001878. doi: 10.1136/bmjgh-2019-001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ngwa MC, Liang S, Mbam LM, Mouhaman A, Teboh A, Brekmo K, Mevoula O, Morris JG., Jr Cholera public health surveillance in the Republic of Cameroon-opportunities and challenges. Pan Afr Med J. 2016;24:222. doi: 10.11604/pamj.2016.24.222.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Debes AK, Ateudjieu J, Guenou E, Ebile W, Sonkoua IT, Njimbia AC, Steinwald P, Ram M, Sack DA. Clinical and environmental surveillance for Vibrio cholerae in resource constrained areas: application during a 1-year surveillance in the far north region of Cameroon. Am J Trop Med Hyg. 2016;94(3):537–543. doi: 10.4269/ajtmh.15-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.WHO. Early warning, alert and response system in emergencies: a field experience of a novel WHO project in north-east Nigeria. Wkly Epidemiol Rec. 2017;92(5):45–5. [PubMed]
- 188.Impouma B, Roelens M, Williams GS, Flahault A, Codeco CT, Moussana F, Farham B, Hamblion EL, Mboussou F, Keiser O. Measuring timeliness of outbreak response in the World Health Organization African region, 2017-2019. Emerg Infect Dis. 2020;26(11):2555–2564. doi: 10.3201/eid2611.191766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Countries investigated.
Additional file 2. Compilation of outbreaks by country, date of onset, delays (detection, investigation, response), signal, source, and description of investigation and response.
Additional file 3. Histograms of delays from symptom onset to (A) case presentation, (B) outbreak detection, (C) investigation, (D) response, and (E) confirmation.
Additional file 4. Overview of alternative models for the main analyses.
Additional file 5. Model parameters for additional delay analyses.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the Github repository, [https://github.com/ruwanepi/Detection-analysis].