Abstract
Purpose:
Genitopatellar syndrome and Say-Barber-Biesecker-Young-Simpson syndrome are caused by variants in the KAT6B gene and are part of a broad clinical spectrum, called KAT6B disorders, whose variable expressivity is increasingly being recognized.
Methods:
We herein present the phenotypes of 32 previously unreported individuals with a molecularly confirmed diagnosis of a KAT6B disorder, report 23 new pathogenic KAT6B variants, and review phenotypic information available on all published individuals with this condition. We also suggest a classification of clinical subtypes within the KAT6B disorder spectrum.
Results:
We demonstrate that cerebral anomalies, optic nerve hypoplasia, neurobehavioral difficulties and distal limb anomalies other than long thumbs and great toes, such as polydactyly, are more frequently observed than initially reported. Intestinal malrotation and its serious consequences can be present in affected individuals. Additionally, we identified four children with Pierre-Robin sequence, four individuals who had increased nuchal translucency/cystic hygroma prenatally, and two fetuses with severe renal anomalies leading to renal failure. We also report an individual in which a pathogenic variant was inherited from a midly affected parent.
Conclusion:
Our work provides a comprehensive review and expansion of the genotypic and phenotypic spectrum of KAT6B disorders that will assist clinicians in the assessment, counseling and management of affected individuals.
Keywords: KAT6B, KAT6B-related disorders, KAT6B disorders, Genitopatellar Syndrome, Say-Barber-Biesecker-Young-Simpson syndrome, SBBYSS
INTRODUCTION
Epigenetic regulation through histone acetylation is essential for proper growth and development, and its role in human genetic diseases is increasingly recognized. KAT6B (formerly known as MYST4 and MORF) encodes a highly conserved histone acetyltransferase that is part of the MYST family 1 and regulates the expression of multiple genes. 2 Previous work has demonstrated that KAT6B preferentially acetylates lysine 14 of histone H3. 2,3 KAT6B functions in a multi-subunit complex with other proteins including KAT6A, BRPF1 and ING5. 4 Exome sequencing has enabled identification of de novo heterozygous variants in KAT6B as the etiology of both genitopatellar syndrome (GPS) (OMIM 606170) and Say-Barber-Biesecker-Young-Simpson syndrome (SBBYSS) (OMIM 603736), a variant of Ohdo syndrome. 5–7
GPS is a skeletal dysplasia characterized by hypoplastic or absent patellae, flexion contractures of the hips and knees, agenesis of the corpus callosum, microcephaly, craniofacial dysmorphisms, and genitourinary anomalies.8 Ohdo syndrome was first described as a genetic condition characterized by intellectual disability in association with congenital heart disease and dysmorphisms. 9 Subsequently, Young and Simpson reported a more severe phenotype later referred to as the SBBYS variant of Ohdo syndrome, or SBBYS syndrome. 10–12 SBBYSS is characterized by blepharophimosis, dacryostenosis, ptosis, a mask-like facial appearance and long thumbs/great toes. 5 GPS and SBBYSS have been historically described as distinct disorders with respect to clinical findings but with several overlapping features. Indeed, both the GPS and SBBYSS phenotypes include significant global developmental delay/intellectual disability, hypotonia, genital abnormalities, patellar hypoplasia/agenesis, congenital heart defects, dental anomalies, hearing loss, and thyroid anomalies. 13 To date, about 90 individuals with KAT6B disorders have been reported, including 18 with GPS, 58 with SBBYSS, and 13 described as having an intermediate phenotype.
The KAT6B pathogenic variant spectrum from previously published individuals includes 56 variants: 22 substitutions, 22 small intragenic deletions, 10 small intragenic duplications and two small intragenic deletions associated with an insertion. The types of variants at the protein level include 32 frameshift, 20 nonsense, two missense and two splicing defects. Pathogenic variants are most often located in exon 18, the last exon of the gene. More proximal pathogenic variants have typically been associated with milder phenotypes and are thought to lead to nonsense-mediated decay and KAT6B haploinsufficiency. 5 Premature termination codons in the last exon or the last 50 nucleotides of the penultimate exon typically cause the mRNAs to escape the NMD pathway and be translated into aberrant proteins with either loss- or gain-of-function effects. 14,15 Interstitial 10q21.3q22.2 deletions encompassing KAT6B have been reported in eight individuals who presented with some features overlapping with KAT6B disorders, such as hypotonia, developmental delay, feeding difficulties and craniofacial dysmorphisms. 16 Of note, Preiksaitiene et al reported a 5 Mb 10q22.1q22.3 deletion encompassing KAT6B in an individual with blepharophimosis, minor dysmorphisms and developmental delay, compatible with SBBYSS. 17 Through genotype-phenotype correlation studies, we observed that pathogenic variants causing the more severe GPS phenotype were located proximally in exon 18 and could lead to the expression of a truncated protein lacking the C-terminal domain. A gain-of-function effect was hypothesized to occur from altered binding affinity or dysregulated interactions of KAT6B with interacting proteins leading to clinical findings more specific to GPS, 18 whereas the SBBYSS phenotype would result from a loss of KAT6B functions. This has not yet been tested experimentally. Recently, more proximal pathogenic variants in KAT6B exons 3, 7, 11, and 14–17 (for GPS, only in exons 17 and 18) were identified in individuals with GPS, SBBYSS and the intermediate phenotype. The increasing identification of individuals with an intermediate phenotype having a variant previously identified in individuals with GPS or SBBYSS make phenotype predictions based on genotype imprecise.
KAT6B pathogenic variants almost always occurred de novo when parental testing has been performed. Recently, Kim et al. reported a family with an inherited pathogenic variant in exon 11 causing relatively mild disease in three individuals with SBBYSS. 19 Another inherited pathogenic splice variant in intron 5 has been found in six related individuals with mild SBBYSS phenotypes. 20 GPS and SBBYSS are thus part of a broad phenotypic spectrum and the variable expressivity of KAT6B disorders is being increasingly recognized, motivating research and delineation of an expanded allelic series.
Here, we review the clinical phenotypes of all reported individuals with molecularly confirmed KAT6B pathogenic variants in the literature and compare them with 32 newly identified individuals with GPS, SBBYSS and intermediate phenotypes. We aim to better define the phenotypic spectrum of KAT6B disorders. We present new KAT6B pathogenic variants and update a publicly accessible LOVD (Leiden Open Variation Database) KAT6B variant database (https://databases.lovd.nl/shared/genes/kat6b), which catalogs all known KAT6B pathogenic variants.
MATERIAL AND METHODS
Individuals with molecularly proven KAT6B disorders from different clinical centers were included in this series. Some individuals presented with clinical features suggestive of a KAT6B disorder and underwent targeted KAT6B sequencing. Molecular analysis via Sanger sequencing in these cases was performed at Baylor College of Medicine in Houston, USA or at Sainte-Justine Hospital Research Center in Montreal, Canada. 6 Other individuals underwent exome sequencing (ES) as part of research projects involving patients with suspected rare Mendelian genetic conditions. However, KAT6B variants for most individuals were identified by clinical testing performed in commercial laboratories (panel or ES). These individuals have been enrolled in the present study following the discovery of a KAT6B pathogenic variant, through communication between clinical teams. Others have been recruited through MatchMaker Exchange 21 and GeneMatcher. 22 Details about the sequencing technique used for each individual is presented in Table S1. Information was uniformly obtained from each clinical team using a structured phenotypic table (Table S2). Each individual from this cohort was evaluated regarding the presence of major features suggestive of GPS or SBBYSS (Table 1) and the patients were subdivided in the following four different clinical subtypes (Table S3):
Table 1.
Major clinical features suggestive of GPS and SBBYSS
| Major features suggestive of GPS | Major features suggestive of SBBYSS |
|---|---|
| • Genital anomalies • Patellar hypoplasia/agenesis • Contractures at the hips, knees and/or clubfoot • Agenesis of the corpus callosum • Renal anomalies (hydronephrosis or multiple renal cysts) |
• Long thumbs/great toes • Immobile mask-like face • Blepharophimosis and/or ptosis • Lacrimal duct anomalies • Patellar hypoplasia/agenesis |
GPS: Genitopatellar syndrome/SBBYSS: Say-Barber-Biesecker-Young-Simpson syndrome
Adapted from Lemire G, Campeau PM, Lee BH. KAT6B Disorders. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews((R)). Seattle (WA): University of Washington, Seattle University of Washington, Seattle.
KAT6B disorder, GPS subtype (at least 2 major features suggesting GPS)
KAT6B disorder, SBBYSS subtype (at least 2 major features suggesting SBBYSS)
KAT6B disorder, intermediate subtype (at least 2 major features of both GPS and SBBYSS, excluding patellar anomalies)
KAT6B disorder, subtype not otherwise specified (cannot be classified in subtype 1, 2 or 3)
Literature search was conducted with PubMed using the key words: KAT6B, SBBYSS and GPS, with the last query on Nov 20th 2019. The identification of all published KAT6B variants was assisted by the LOVD KAT6B database.
Informed consent to publish individuals’ clinical information and photographs was obtained from the parents of the individuals reported in this article.
RESULTS
We present the clinical features and pathogenic variants of 32 previously unreported individuals with KAT6B disorders, including eight individuals with GPS, 15 with SBBYSS, six with the intermediate phenotype, and three with a KAT6B disorder not otherwise specified (Table 2). Representative photographs of some of these individuals are presented in Figure 1. Our literature search identified 89 previously published individuals with a molecularly confirmed KAT6B disorder, and their clinical features are summarized in Table S4. The clinical manifestations are discussed below.
Table 2.
Clinical features of 32 individuals with KAT6B disorders and frequency of these features in previously reported individuals
| Clinical characteristics | GPS - reported in literature (18) | GPS - new cohort | SBBYSS - reported in literature (58) | SBBYSS - new cohort | Intermediate - reported in literature (13) | Intermediate - new cohort | KAT6B disorder NOS - new cohort | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | ||||
| Sex | 56% (10/18) F 44% (8/18) M |
M | M | F | M | M | F | F | F | 53% (31/58) F 47% (27/58) M |
M | F | F | M | M | M | F | F | F | M | F | F | M | F | M | 23% (3/13) F 77% (10/13)M |
F | F | F | F | M | F | F | M | F |
| Increased NT/cystic hygroma or polyhydramnios | 0% (0/18) | 3% (2/58) | + | + | + | + | 23% (3/13) | + | + | + | |||||||||||||||||||||||||
| Developmental delay/intellectual disability | 83% (15/18) | + | + | + | + | 97% (56/58) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 85% (11/13) | + | + | + | + | + | + | + | + | + | ||||
| Microcephaly | 67% (12/18) | + | + | + | + | + | 36% (21/58) | + | 23% (3/13) | + | + | + | + | ||||||||||||||||||||||
| Hypotonia | 22% (4/18) | + | + | 78% (45/58) | + | + | + | + | + | + | + | + | + | + | 62% (8/13) | + | + | + | + | + | + | + | |||||||||||||
| Agenesis/hypoplasia of the corpus callosum | 94% (17/18) | + | + | + | + | + | + | + | + | 10% (6/58) | 46% (6/13) | + | + | + | + | ||||||||||||||||||||
| Seizures | 11% (2/18) | + | 3% (2/58) | 15% (2/13) | + | ||||||||||||||||||||||||||||||
| Optic nerve atrophy/hypoplasia or cortical visual impairment | 17% (3/18) | + | 5% (3/58) | + | + | + | 15% (2/13) | + | + | ||||||||||||||||||||||||||
| Lacrimal duct anomalies | 0% (0/18) | 16% (9/58) | + | + | + | 31% (4/13) | + | + | + | ||||||||||||||||||||||||||
| Hearing loss | 11% (2/18) | 19% (11/58) | + | + | 15% (2/13) | + | + | + | + | + | |||||||||||||||||||||||||
| Blepharophimosis and/or ptosis a | 1% (1/18) | + | 33% (19/58) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 54% (7/13) | + | + | + | + | + | + | + | + | |||||||||
| Mask-like facies a | 1% (1/18) | + | 33% (19/58) | + | + | + | + | + | + | + | + | + | + | + | + | 54% (7/13) | + | + | + | + | + | ||||||||||||||
| Other craniofacial dysmorphisms | 78% (14/18) | + | + | + | + | + | + | + | + | 88% (51/58) | + | + | + | + | + | + | + | + | + | + | + | + | + | 85% (11/13) | + | + | + | + | + | + | + | + | + | ||
| Cleft palate/high-arched palate | 11% (2/18) | + | + | + | + | 26% (15/58) | + | + | + | + | 54% (7/13) | + | + | + | + | + | + | ||||||||||||||||||
| Dental anomalies | 11% (2/18) | + | 41% (24/58) | + | + | + | + | + | 46% (6/13) | + | + | ||||||||||||||||||||||||
| Thyroid anomalies | 28% (5/18) | 38% (22/58) | + | + | 62% (8/13) | + | + | + | + | + | |||||||||||||||||||||||||
| Congenital heart defects | 67% (12/18) | + | + | + | + | + | 43% (25/58) | + | + | + | 77% (10/13) | + | + | + | + | + | + | + | |||||||||||||||||
| Anal anomalies (atresia/stenosis or malposition) | 33% (6/18) | + | 0% (0/58) | + | 0% (0/13) | + | + | ||||||||||||||||||||||||||||
| Genital anomalies b | 89% (16/18) | + | + | + | + | + | + | + | 40% (23/58) | + | + | + | + | + | + | 77% (10/13) | + | + | + | + | + | ||||||||||||||
| Renal anomalies c | 83% (15/18) | + | + | + | + | + | + | 5% (3/58) | 23% (3/13) | + | + | + | + | + | + | ||||||||||||||||||||
| Patellar agenesis or hypoplasia | 83% (15/18) | + | + | + | 19% (11/58) | 46% (6/13) | + | + | + | + | + | ||||||||||||||||||||||||
| Contractures (hips/knees, club feet) | 94% (17/18) | + | + | + | + | + | + | 31% (18/58) | + | 46% (6/13) | + | + | + | + | + | + | |||||||||||||||||||
| Spine anomalies (e.g. kyphosis, scoliosis) | 11% (2/18) | 0% (0/58) | + | 0% (0/13) | + | + | + | ||||||||||||||||||||||||||||
| Thoracic anomalies | 22% (4/18) | + | 0% (0/58) | + | + | 23% (3/13) | + | ||||||||||||||||||||||||||||
| Other skeletal anomalies d | 17% (3/18) | + | + | + | + | + | 7% (4/58) | + | + | + | + | 23% (3/13) | + | + | |||||||||||||||||||||
| Long thumbs/long great toes | 22% (4/18) | + | 57% (33/58) | + | + | + | + | + | + | + | + | + | + | + | + | 54% (7/13) | + | + | + | + | |||||||||||||||
| Other digital anomalies | 6% (1/18) | + | + | + | 12% (7/58) | + | + | + | + | + | 46% (6/13) | + | + | + | + | ||||||||||||||||||||
| Feeding difficulties | 44% (8/18) | + | + | + | + | 74% (43/58) | + | + | + | + | + | + | + | + | + | + | 54% (7/13) | + | + | + | + | + | + | + | |||||||||||
| Laryngomalacia/respiratory distress | 22% (4/18) | + | + | + | + | 5% (3/58) | + | + | + | 38% (5/13) | + | + | + | + | + | ||||||||||||||||||||
GPS: Genitopatellar Syndrome/SBBYSS: Say-Barber-Biesecker-Young-Simpson syndrome/NOS: not otherwise specified/NT: nuchal translucency.
Blepharophimosis and/or ptosis and mask-like facies share joint proportions for previously published individuals, as these features were often categorized together in the literature.
Genital anomalies include cryptorchidism, scrotal hypoplasia and hypospadias in males; clitoromegaly and hypoplasia of the labia minora or majora in females.
Renal anomalies include hydronephrosis, multicystic kidneys or dysplastic kidneys.
Other skeletal anomalies include pelvic anomalies, fractures or dislocations.
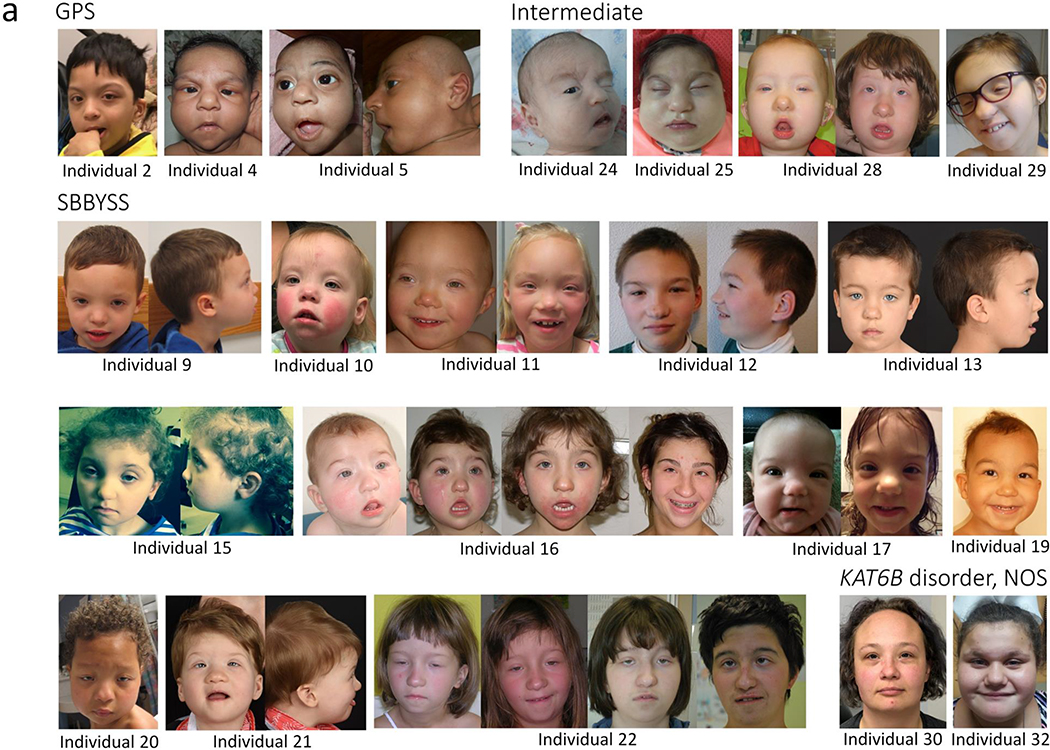
Figure 1:
Clinical photographs of 20 newly reported individuals with KAT6B disorders (GPS: Genitopatellar Syndrome/SBBYSS: Say-Barber-Biesecker-Young-Simpson Syndrome/ NOS: not otherwise specified) showing:
a) Facial features of affected individuals. Note the blepharophimosis, ptosis and mask-like facies in individuals with SBBYSS and the intermediate phenotype.
b) Long thumbs and/or long great toes (upper left: individual 13; upper right: individual 15; lower left and lower right: individual 29).
c) Flexion contractures and club feet (left: individual 24; right: individual 5).
d) Absent patella in individual 5.
e) Pre-axial polydactyly of the right hand in individual 25.
f) Overlapping toes in individual 21.
g) A newborn with GPS who died after birth at 28 weeks of gestation (Individual 8). Note the high forehead, blepharophimosis, small palpebral fissures with hypertelorism, sparse eyebrows and eyelashes, small simplified ears with bilateral pits, proximal/distal arthrogryposis and contractures of the hip/knees.
Neurological findings
All eight individuals with GPS in this cohort had agenesis or hypoplasia of the corpus callosum; this is in keeping with the literature as it has been noted as a major neurologic feature of the GPS phenotype. 23 Other common neurologic findings in all KAT6B disorders include microcephaly and axial and appendicular hypotonia. Six previously reported individuals and two from this cohort presented with seizures. 13,24–28
Affected individuals, including the ones in this report, often present with other cerebral/neurological anomalies such as colpocephaly, hydrocephaly, dilated ventricles or ventriculomegaly (eleven individuals), pachygyria or simplification of cortical sulci (four individuals), delayed white matter myelination or hypomyelination (four individuals) and grey matter heterotopia (three individuals). 6,17,26,27,29–32 Each of the following anomalies were described in two individuals: periventricular gliosis, agenesis of the septum pellucidum, telencephalic or periventricular leukoencephalopathy, and hypertonia/dystonia; and other anomalies were reported in only one individual each: cortical atrophy, agenesis of the anterior commissure, cerebral palsy, lenticulostriate vasculopathy, brainstem hypoplasia, unspecified cerebellar abnormalities, spina bifida, Horner’s syndrome, nonspecific gray matter differences, partial agenesis of the cingulate gyrus, and wide Virchow-Robin spaces. 24,26,30,31,33 A newborn from this cohort presented with lissencephaly, cerebellar/cerebral hemorrhage, and hypoplasia/immaturity of the cerebellar cortex. Two individuals from the literature presented with lower limb muscle atrophy with no neuropathy identified at electromyography studies. 25
Neurodevelopmental disorder
Developmental delay and/or intellectual disability are expected features in KAT6B disorders. Language disorders have been previously reported in 18% of individuals and was also noted in 19/32 (59%) of the individuals in this cohort. 17,19,20,24–26,31,32,34,35 Of note, 5/19 (26%) of children with speech difficulties in this cohort also had hearing impairment. Behavioral and/or psychiatric issues, such as features suggestive of autism spectrum disorder, anxiety, aggressive behavior and attention problems have been noted previously in four individuals, but also in eight individuals in this report. 13,19,25,34 Additionally, we report an individual where the pathogenic variant was inherited from an affected 34-year-old-mother with reported learning disability, but no diagnosis of intellectual disability (individual 30). She lives independently, cares for her family and works as a cashier. Mosaicism was not detected in the blood, and she was heterozygous for the deleterious variant.
Hearing impairment and ocular anomalies
Conductive or sensorineural hearing loss was present in 17% of individuals from the literature and in 7/32 (22%) of individuals from this cohort. External auditory canal malformations and/or stenosis, with or without hearing loss, was present in nine individuals from the literature. 24,29–31,34,36
Visual deficits and various ocular anomalies are also frequent. Blepharophimosis, a major feature of SBBYSS that can impact visual function,23 was found in 23/32 (72%) of individuals in this cohort, and specifically in 14/15 (93%) of individuals with SBBYSS. Lacrimal duct anomalies (mainly dacryostenosis) were found in 15% and 6/32 (19%) of individuals from the literature and this cohort, respectively. Optic atrophy or hypoplasia have previously been reported in at least five individuals and were also observed in six individuals in this report. 6,13,28,30 Some children also had early-onset visual problems such as myopia, hypermetropia, astigmatism, nystagmus, strabismus, amblyopia, and visual pursuit deficits. 19,25,26,30–32,34 Telecanthus, epicanthus inversus, and hypertelorism are also sometimes noted. 17,24,25,27–29,32,36,37
Craniofacial features
A distinctive facial appearance of blepharophimosis, ptosis and/or mask-like facies characterizes most individuals with SBBYSS. Other features that can be common in both GPS and SBBYSS include low-set and posteriorly rotated ears, down-slanting palpebral fissures, flat broad nasal bridge, tubular/bulbous nose, long philtrum, thin upper vermilion, and micro/retrognathia.
Cleft lip and palate and high-arched palate were present in 27% of published individuals and in 14/32 (44%) of individuals in this cohort. The uvula was reported to be bifid in five individuals from the literature and this cohort and absent in one individual in this cohort. 25,30 A Pierre-Robin sequence was previously reported once and has also been observed in four individuals of this cohort. 24
Dental anomalies such as delayed eruption of teeth and absent/hypoplastic teeth were previously seen in 36% of individuals and also in 8/32 (25%) of individuals in this cohort.
Affected individuals, including the ones in this report, also presented with other craniofacial anomalies such as sagittal craniosynostosis (two individuals), overriding skull bones (one individual), dolichocephaly (four individuals), and scaphocephalic skull shape (one individual). 25,35,36
Genito-urinary and renal anomalies
Genital anomalies were frequently present in all subtypes of KAT6B disorders, and 7/8 individuals with GPS in this cohort had genital anomalies. In males, this can include cryptorchidism, retractile testis, scrotal hypoplasia, micropenis, unilateral testicular agenesis and hypospadias. In females, clitoromegaly and hypoplasia of the labia minora/majora are observed. 5–7,17,24,25,29,30,36 One child with SBBYSS from this cohort had a bicornuate uterus.
Renal anomalies are more commonly observed in GPS, with hydronephrosis and multicystic kidneys being most common. 6,7,13,26,27,30,36 Kim et al reported a girl with GPS who had multicystic dysplastic kidneys leading to renal failure and pulmonary hypoplasia/hypertension. 27 Two newborns with GPS in this cohort also presented with an oligohydramnios sequence due to bilateral renal hypoplasia or dysplasia. One individual had stage III bilateral vesicoureteral reflux with renal hypoplasia and another had unilateral ureteropelvic junction obstruction. One individual had lower urinary tract obstruction with reflux and dysplastic cystic kidneys.
Gastrointestinal anomalies
Anal anomalies have been previously noted in six individuals, specifically anteriorly positioned anus and anal atresia or stenosis, 6,26,33 and were also present in four individuals in this cohort. Intestinal malrotation, with potential fatal complications, has been reported previously in four individuals, and in two from this cohort. 6,26,30 Feeding difficulties, gastroesophageal reflux and/or recurrent emesis are frequent in affected individuals, and were present in nine individuals from this cohort. 25,33 Chronic constipation has been previously reported in one individual and was also noted in four individuals in this series. 17 Other reported findings include diaphragmatic eventration in one individual from this cohort and necrotising enterocolitis in a 30-week premature newborn from the literature. 36.
Musculoskeletal features
Patellar anomalies, including agenesis, hypoplasia, delayed ossification and displacement of the patella, were present in 8/32 (25%) of individuals in this cohort. Club feet and flexion contractures of the knees and/or hips are also frequently present (6/8 individuals with GPS in this cohort). Patellar anomalies and contractures both constitute major features of GPS. Contractures of the wrists or elbows were also found in three individuals in this cohort.
Long thumbs and/or long great toes are often observed in individuals with SBBYSS (in 12/15 (80%) in this cohort). Other digital anomalies were observed in 16% of previously reported individuals and in 12/32 (38%) of this cohort. These anomalies included preaxial or postaxial polydactyly, camptodactyly, clinodactyly, arachnodactyly, brachydactyly, overlapping fingers or toes, proximally implanted thumbs, and syndactyly of toes. 13,17,19,24–26,29,31,32,34,36,37
Other musculoskeletal findings occasionally observed in the literature and in this cohort included: kyphosis and scoliosis (six individuals), 6 thoracic anomalies such as pectus excavatum and narrow thorax (eleven individuals), 6,25,27,34,36 pelvic anomalies (six individuals), 6,7, and joint hypermobility (seven individuals). 17,25,30,31,34 Other rarely reported features included hypoplastic heels, 27 dolichostenomelia, 24 coxa valga, 25 genu valgum, 17 femoral fracture and dislocated hips and knees, 26 and exophytic lesions on the scapular spine. 34
Cardiovascular anomalies
In this cohort, 15/32 (47%) of individuals had congenital heart defects, compared to 53% in previously reported individuals. These anomalies include mainly atrial and/or ventricular septal defects, patent foramen ovale and patent ductus arteriosus. 5–7,13,17,24–32,36,38 In this cohort, one individual had an abnormal aortic arch with a single left vertebral artery exiting between the left common carotid and left subclavian arteries as well as a retro-esophageal right subclavian artery. Another individual had recurrent episodes of supraventricular tachycardia.
Growth and endocrine anomalies
Functional or structural thyroid abnormalities such as hypothyroidism and thyroid agenesis or hypoplasia are present in many affected individuals. Hypothyroidism was present in six individuals from this cohort. Growth retardation, short stature, failure to thrive/poor weight gain and delayed bone age have also been reported in nine individuals from the literature and six from this cohort. 13,26,29–31,35,36,39 Delayed puberty with primary amenorrhea has also been reported. 26 Li et al. reported one individual with pituitary hypoplasia and severe growth hormone deficiency. 39 One individual in this series had hyperphagia and obesity.
Respiratory findings
Respiratory distress exacerbated by tracheomalacia and/or laryngomalacia was present in 12/32 (38%) of individuals in this cohort and in 13 % of individuals in the literature. Laryngeal cleft was previously reported in one individual.30 Central or obstructive sleep apnea was previously reported once 13 but was noted in five individuals in this cohort.
Cutaneous findings
Cutaneous anomalies are infrequent. Among all individuals reported to date including this cohort, three had café-au-lait macules, one had cutaneous hemangiomas, six had abnormal palmar creases, five had widely spaced nipples, and four had hypoplastic nails.17,26,36,37 A webbed neck with redundant skin was observed in one individual but the result of the nuchal translucency was not specified. 27
Prenatal findings
One child from this cohort had increased nuchal translucency prenatally and three had cystic hygroma, one of which was persistent into the second trimester. However, increased nuchal translucency and cystic hygroma have not been reported in individuals with KAT6B disorders in the literature. Kraft et al reported an individual with a RASopathy-like phenotype with a chromosomal translocation disrupting the KAT6B gene. 2 The result of the nuchal translucency scan was not specified. Additionally, in this cohort, two pregnancies were complicated by polyhydramnios, which has been previously reported in five individuals in the literature. 25,32,35,36
Other findings
Knight et.al reported a child with GPS who developed a stage I neuroblastoma extending into the portal vein. While it was poorly differentiated, it had an overall favorable histopathology. 33
One individual from this cohort had severe unexplained anemia at birth, poor extra-hepatic hematopoiesis for gestational age on autopsy examination, and polysplenia.
Mortality
Kim et al reported on a girl with GPS with early-onset seizures, multicystic dysplastic kidneys, renal failure, and pulmonary hypoplasia. 27 She died at 8 months of age due to tracheostomy tube displacement. Gannon et al reported a boy who was stillborn at 36 weeks of gestation and a girl who died from bronchiolitis at three years of age. 30 In this cohort of 32 individuals, one pregnancy affected by GPS was medically terminated, one child with an intermediate phenotype died at 2 years of age, and three children with GPS died in infancy. The first infant died from pulmonary hypoplasia due to renal hypoplasia/dysplasia. The second, with multiple malformations including renal hypoplasia, also had severe anemia, and disseminated intravascular coagulation; she died shortly after birth at 28 weeks of gestation. The third died from Influenza H1N1 infection at four months of age.
Update on pathogenic variants
We previously generated a LOVD database of all reported KAT6B pathogenic variants and have now updated it (Figure 2). We report 23 novel variants, including 12 small intragenic deletions, two small intragenic duplications and nine single nucleotide variants. These include the following types of variants at the protein level: fourteen frameshift, six nonsense, one missense and two intronic variants predicted to affect splicing. Most variants are located in exon 18, but one is in exon 4, seven in exon 16 and two in exon 17. The two splicing variants were located in intron 7 and 17. When parental samples were available, variants were de novo, with the exception of the splicing defect in intron 7 that was maternally inherited.
Figure 2:
Distribution of all pathogenic variants in KAT6B reported in literature (upper panel) and 23 new variants reported in this article (lower panel) with the associated phenotype(s). GPS: Genitopatellar Syndrome/SSBYSS: Say-Barber-Biesecker-Young-Simpson Syndrome. NOS: not otherwise specified. The variant c.5624_5625del/p.Ala1876Leufs*3 reported by Gannon et al in 2015 has not been included in this figure, because of discordance between the reported cDNA and protein change.
Genotype-phenotype analysis
Variants leading to GPS occur within the proximal portion of exon 18, always between amino acids 1150 and 1515 in this cohort and the literature (Figure 2). Variants within this region are associated with GPS in 60% of cases, SBBYSS in 19% of cases, and the intermediate phenotype in 19% of cases. Variants outside of this region cause SBBYSS and the intermediate phenotype in 84%, and 13 % of cases, respectively, and never cause GPS.
Variants associated with SBBYSS occur throughout the gene, but more often in exons 16–17 or distally in exon 18. In general, individuals with variants occurring in exons 1–16 tend to be more mildly affected, as observed in the three families with inherited variants, whereas individuals with variants occurring more distally seem to present with a more severe phenotype including more congenital anomalies. For instance, genitourinary anomalies and digital malformations are more frequent in individuals with variants in exons 17 and 18.
No variant reported in the literature or in our cohort has been associated with both GPS and SBBYSS. However, six variants have been reported so far both in individuals with GPS or SBBYSS and in individuals with an intermediate phenotype.
DISCUSSION
Our study has allowed further delineation of the clinical spectrum of KAT6B disorders. The description of additional individuals further challenges the original syndrome delineation and highlights that GPS, SBBYSS and the intermediate phenotype are all part of a broad clinical spectrum. We are indeed moving toward naming these conditions as only KAT6B disorder in the future. However, clinicians still use the terms GPS and SBBYSS in their practice at the present time, so we thought it was premature to stop using them in the present article. Moreover, we suspected it might still be clinically relevant to subdivide affected individuals into KAT6B disorder subtypes as they might present with different clinical outcomes, developmental impairment, cognitive profile and medical complications. We have thus classified the individuals in this cohort into four different KAT6B disorder clinical subtypes and have suggested an approach to do so. More research is needed to address these questions. Nonetheless, this case series has highlighted that the major features previously defined by Campeau et al 23 remain the prominent clinical findings of KAT6B disorders, but also identified additional features that should raise clinical suspicion of this diagnosis and influence management.
In addition to abnormalities of the corpus callosum, almost half of affected individuals had other cerebral anomalies. A thorough neurological evaluation with a cerebral MRI and clinical surveillance for seizures is thus indicated in all individuals with suspicion of or a molecularly confirmed KAT6B disorder. Ocular anomalies and visual deficits were also found in many individuals with KAT6B disorders. Of note, optic nerve hypoplasia appeared to be more frequent than previously reported. This reinforces the importance of an ophthalmological evaluation. Additionally, periodic hearing examination should be performed as a significant proportion of affected individuals had sensorineural or conductive hearing loss. Intestinal malrotation has been described in several individuals and clinicians involved in the care of affected individuals should keep a high clinical suspicion for this. Currently, further evidence of utility is required to recommend screening by a barium enema study. We also highlight that some individuals presented with neurobehavioral problems suggestive of autism spectrum disorder. Screening and follow-up for these disorders with appropriate therapy should be considered. Again, we again observe that hypothyroidism is frequent, thus periodic thyroid function testing is indicated. Finally, routine evaluation of individuals with suspected or confirmed KAT6B disorders should also include an echocardiogram and a renal ultrasound. The presence of contractures, mostly of the lower limbs, and other skeletal anomalies (including the spine) should be assessed by a clinical evaluation with X-Rays and/or orthopedics consultation if needed.
We demonstrated that cystic hygroma can complicate the pregnancy of fetuses affected by KAT6B disorders. Thirteen percent of this cohort presented prenatally with an increased nuchal translucency or cystic hygroma. Interestingly, given the high frequency of Noonan syndrome in cohorts of fetuses with increased nuchal translucency and cystic hygroma, 40 KAT6B haploinsufficiency was shown to upregulate the RAS-MAP Kinase pathway (the pathway implicated in Noonan syndrome). 2 We also confirmed that polyhydramnios is an associated anomaly. KAT6B disorders should thus be included in the differential diagnosis of these prenatal findings. Additionally, we identified two fetuses with severe renal anomalies leading to renal failure and four children with Pierre-Robin sequence. One child with Pierre-Robin sequence (individual 26) had initially been clinically diagnosed with Toriello-Carey syndrome, suggesting that when this syndrome is suspected, KAT6B disorders should also be considered in addition to the other genetic anomalies previously associated with Toriello-Carey syndrome (i.e. chromosomal anomalies and variants in PGAP3, DDX3X, and UBE3B). 41 Digital anomalies, other than long thumbs and/or great toes, were also commonly observed. These included pre-axial or post-axial polydactyly, camptodactyly, and brachydactyly. To date, the only neoplasm reported in an individual with a KAT6B disorder is a neuroblastoma. Although there might not be a direct relationship between KAT6B alteration and the development of this malignancy, it is interesting to note that a tumor suppressor role of KAT6B through histone H3 Lys23 acetyltransferase activity was suggested after observing KAT6B genomic loss in small cell lung cancer cell lines.42 Finally, we identified an individual with a maternally inherited variant, the third familial case of a molecularly confirmed KAT6B disorder reported so far. The proband’s mother presented an attenuated phenotype and was functioning at a higher level than most reported individuals. These findings expand the phenotypic spectrum of KAT6B disorders.
Our work provides a comprehensive review and expansion of the genotypic and phenotypic spectrum of KAT6B disorders that will assist clinicians in the assessment, counseling and management of affected individuals. Research involving functional studies is required to investigate the hypothesized molecular mechanisms leading to KAT6B disorders.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the families for their participation and Natascia Anastasio for revision of the manuscript. We thank the Canadian Institutes of Health Research and Fonds de la recherche en santé du Québec for clinician-scientist awards to P.M.C. GL is supported by a Children’s Hospital Academic Medical Organization clinical fellowship award through the Children’s Hospital of Eastern Ontario. This work was partly supported by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, the Ontario Research Fund, Genome Alberta, Genome Quebec, Genome British Columbia and CHEO Foundation. Sequencing and analysis was partly provided by the University of Washington Center for Mendelian Genomics (funded by grants UM1 HG006493 and U24 HG008956), the Roberts Individualized Medical Genetics Centre of the Children’s Hospital of Philadelphia, and the Baylor-Hopkins Center for Mendelian Genomics (grants UM1 HG006542, R01NS058529 and R35NS105078) to J.R.L). We thank the European Reference Network for Developmental Anomalies and Intellectual Disability for its contribution.
Footnotes
CONFLICT OF INTEREST
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. J.R.L. serves on the Scientific Advisory Board of BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor Genetics Laboratories.
Contributor Information
Li Xin Zhang, Sainte-Justine Hospital Research Center, University of Montreal, Montreal, Canada.
Gabrielle Lemire, Division of Medical Genetics, Department of Pediatrics, CHU Sainte-Justine, University of Montreal, Montreal, Canada.
Claudia Gonzaga-Jauregui, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Sirinart Molidperee, Sainte-Justine Hospital Research Center, University of Montreal, Montreal, Canada.
Carolina Galaz-Montoya, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
David S. Liu, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Alain Verloes, Department of Genetics and INSERM UMR1141, APHP-Nord Université de Paris, Robert DEBRE Hospital, Paris and ERN-ITHACA, France.
Amelle G. Shillington, Department of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Kosuke Izumi, Division of Human Genetics, Department of Pediatrics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Alyssa L. Ritter, Division of Human Genetics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Beth Keena, Division of Human Genetics, Department of Pediatrics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Elaine Zackai, Division of Human Genetics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Dong Li, Center for Applied Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Elizabeth Bhoj, Division of Human Genetics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Jennifer M. Tarpinian, Roberts Individualized Medical Genetics Center, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Emma Bedoukian, Roberts Individualized Medical Genetics Center, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Mary K. Kukolich, Cook Children’s Health Care System, Fort Worth, TX, USA.
A. Micheil Innes, Department of Medical Genetics and Alberta Children’s Hospital Research Institute, University of Calgary, Calgary, Canada.
Grace U. Ediae, Children’s Hospital of Eastern Ontario Research Institute, Ottawa, Canada
Sarah L. Sawyer, Department of Genetics, Children’s Hospital of Eastern Ontario, Ottawa, Canada.
Karippoth Mohandas Nair, Department of Pediatrics, Government Medical College, Kozhikode, Kerala, India.
Para Chottil Soumya, Department of Pediatrics, Government Medical College, Kozhikode, Kerala, India.
Kinattinkara R. Subbaraman, Department of Pediatrics, Government Medical College, Kozhikode, Kerala, India.
Frank J. Probst, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA. Texas Children’s Hospital, Houston, TX, USA.
Jennifer A. Bassetti, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA. Texas Children’s Hospital, Houston, TX, USA.
Reid V. Sutton, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA. Texas Children’s Hospital, Houston, TX, USA.
Richard A. Gibbs, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA..
Chester Brown, Baylor College of Medicine, Houston, TX, USA..
Philip M. Boone, Division of Genetics and Genomics, Boston Children’s Hospital, Harvard Medical school, Boston, MA, USA.
Ingrid A. Holm, Division of Genetics and Genomics, Boston Children’s Hospital, Harvard Medical school, Boston, MA, USA.
Marco Tartaglia, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, Rome, Italy.
Giovanni Battista Ferrero, Department of Public Health and Pediatrics, University of Torino, Turin, Italy.
Marcello Niceta, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, Rome, Italy.
Maria Lisa Dentici, Medical Genetics, Academic Department of Pediatrics, Ospedale Pediatrico Bambino Gesù, IRCCS, Rome, Italy.
Francesca Clementina Radio, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, Rome, Italy.
Boris Keren, Genetic department, AP-HP, Sorbonne Université, Paris, France.
Constance F. Wells, Service de Génétique Clinique, CHU de Montpellier, Montpellier, France.
Christine Coubes, Service de Génétique Clinique, Département de Génétique Médicale, Maladies Rares et Médecine Personnalisée, CHU de Montpellier, Montpellier, France.
Annie Laquerrière, Department of Pathology, Centre for Genomic and Personalized Medicine, UNIROUEN Normandie University, Inserm U1245, Normandy, 76000 Rouen, France.
Jacqueline Aziza, Département anatomie et cytologie pathologiques, CHU Toulouse, France.
Charlotte Dubucs, Département anatomie et cytologie pathologiques, CHU Toulouse, France.
Sheela Nampoothiri, Department of Pediatric Genetics, Amrita Institute of Medical Sciences and Research Centre, Cochin, Kerala, India.
David Mowat, Centre for Clinical Genetics, Sydney Children’s Hospital Randwick, Sydney, Australia.
Millan Patel, BC Children’s Hospital Research Institute and Department of Medical Genetics, University of British Columbia, Vancouver, Canada.
Ana Bracho, Genetic Research Institute, University of Zulia, Maracaibo, Venezuela.
Francisco Cammarata-Scalisi, Medical Genetics, University of Los Andes, Mérida, Venezuela.
Alper Gezdirici, Department of Medical Genetics, Istanbul Health Science University, Kanuni Sultan Suleyman Training and Research Hospital, Istanbul, Turkey.
Alberto Fernandez-Jaen, Department of Pediatric Neurology, Hospital Quirónsalud School of Medicine, Universidad Europea, Madrid, Spain.
Natalie Hauser, Inova Health system, Falls Church, VA, USA.
Yuri A. Zarate, Department of Pediatrics, Section of Genetics and Metabolism, University of Arkansas for Medical Sciences, Little Rock, AR, USA.
Katherine A. Bosanko, Department of Pediatrics, Section of Genetics and Metabolism, University of Arkansas for Medical Sciences, Little Rock, AR, USA.
Klaus Dieterich, Medical Genetics, CHU Grenoble Alpes, Université Grenoble Alpes, Inserm, U1216, GIN, 38000 Grenoble, France.
John C. Carey, Division of Medical Genetics, Department of Pediatrics, University of Utah Health, Salt Lake City, UT, USA.
Jessica X. Chong, Department of Pediatrics, University of Washington, Seattle, WA 98195, USA; Brotman-Baty Institute for Precision Medicine, Seattle, WA 98195, USA.
Deborah A. Nickerson, Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA; Brotman-Baty Institute for Precision Medicine, Seattle, WA 98195, USA.
Michael J. Bamshad, Department of Pediatrics, University of Washington, Seattle, WA 98195, USA; Brotman-Baty Institute for Precision Medicine, Seattle, WA 98195, USA.
Brendan H. Lee, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA..
Xiang-Jiao Yang, Goodman Cancer Center, Department of Medicine, McGill University, Montreal, Canada.
James R. Lupski, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA; Texas Children’s Hospital, Houston, TX, USA.
Philippe M. Campeau, Division of Medical Genetics, Department of Pediatrics, CHU Sainte-Justine, University of Montreal, Montreal, Canada.
REFERENCES
- 1.Voss AK, Thomas T. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays. 2009;31(10):1050–1061. [DOI] [PubMed] [Google Scholar]
- 2.Kraft M, Cirstea IC, Voss AK, et al. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Invest. 2011;121(9):3479–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyon Y, Cayrou C, Ullah M, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Molecular cell. 2006;21(1):51–64. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ. MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease. Biochimica et biophysica acta. 2015;1853(8):1818–1826. [DOI] [PubMed] [Google Scholar]
- 5.Clayton-Smith J, O’Sullivan J, Daly S, et al. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet. 2011;89(5):675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campeau PM, Kim JC, Lu JT, et al. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am J Hum Genet. 2012;90(2):282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson MA, Deshpande C, Dafou D, et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause Genitopatellar syndrome. Am J Hum Genet. 2012;90(2):290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier-Daire V, Chauvet ML, Lyonnet S, Briard ML, Munnich A, Le Merrer M. Genitopatellar syndrome: a new condition comprising absent patellae, scrotal hypoplasia, renal anomalies, facial dysmorphism, and mental retardation. J Med Genet. 2000;37(7):520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohdo S, Madokoro H, Sonoda T, Hayakawa K. Mental retardation associated with congenital heart disease, blepharophimosis, blepharoptosis, and hypoplastic teeth. Journal of medical genetics. 1986;23(3):242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Say B, Barber N. Mental retardation with blepharophimosis. J Med Genet. 1987;24(8):511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biesecker LG. The Ohdo blepharophimosis syndrome: a third case. J Med Genet. 1991;28(2):131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young ID, Simpson K. Unknown syndrome: abnormal facies, congenital heart defects, hypothyroidism, and severe retardation. J Med Genet. 1987;24(11):715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlckova M, Simandlova M, Zimmermann P, et al. A patient showing features of both SBBYSS and GPS supports the concept of a KAT6B-related disease spectrum, with mutations in mid-exon 18 possibly leading to combined phenotypes. Eur J Med Genet. 2015;58(10):550–555. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K, Khajavi M, Ohyama T, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36(4):361–369. [DOI] [PubMed] [Google Scholar]
- 15.Coban-Akdemir Z, White JJ, Song X, et al. Identifying Genes Whose Mutant Transcripts Cause Dominant Disease Traits by Potential Gain-of-Function Alleles. Am J Hum Genet. 2018;103(2):171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herriges JC, Dugan SL, Lamb AN. Clinical and molecular cytogenetic characterization of a novel 10q interstitial deletion: a case report and review of the literature. Molecular cytogenetics. 2019;12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preiksaitiene E, Tumiene B, Maldziene Z, et al. Features of KAT6B-related disorders in a patient with 10q22.1q22.3 deletion. Ophthalmic genetics. 2017;38(4):383–386. [DOI] [PubMed] [Google Scholar]
- 18.Campeau PM, Lu JT, Dawson BC, et al. The KAT6B-related disorders genitopatellar syndrome and Ohdo/SBBYS syndrome have distinct clinical features reflecting distinct molecular mechanisms. Human mutation. 2012;33(11):1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YR, Park JB, Lee YJ, Hong MJ, Kim HT, Kim HJ. Identifying the KAT6B Mutation via Diagnostic Exome Sequencing to Diagnose Say-Barber-Biesecker-Young-Simpson Syndrome in Three Generations of a Family. Annals of rehabilitation medicine. 2017;41(3):505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates TM, Langley CLM, Grozeva D, Raymond FL, Johnson DS. Novel KAT6B proximal familial variant expands genotypic and phenotypic spectrum. Clin Genet. 2019;95(2):334–335. [DOI] [PubMed] [Google Scholar]
- 21.Philippakis AA, Azzariti DR, Beltran S, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Human mutation. 2015;36(10):915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemire G, Campeau PM, Lee BH. KAT6B Disorders In: Adam MP, Ardinger HH, Pagon RA, et al. , eds. GeneReviews((R)). Seattle (WA): University of Washington, Seattle University of Washington, Seattle; GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.; 1993. [PubMed] [Google Scholar]
- 24.Lonardo F, Lonardo MS, Acquaviva F, Della Monica M, Scarano F, Scarano G. Say-Barber-Biesecker-Young-Simpson syndrome and Genitopatellar syndrome: Lumping or splitting? Clin Genet. 2019;95(2):253–261. [DOI] [PubMed] [Google Scholar]
- 25.Brea-Fernandez A, Dacruz D, Eiris J, Barros F, Carracedo A. Novel truncating variants expand the phenotypic spectrum of KAT6B-related disorders. American journal of medical genetics Part A. 2019;179(2):290–294. [DOI] [PubMed] [Google Scholar]
- 26.Okano S, Miyamoto A, Fukuda I, et al. Genitopatellar syndrome: the first reported case in Japan. Human genome variation. 2018;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim BR, Han JH, Shin JE, et al. Genitopatellar Syndrome Secondary to De Novo KAT6B Mutation: The First Genetically Confirmed Case in South Korea. Yonsei medical journal. 2019;60(4):395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez R, Delea M, Dain L, Rittler M. A novel pathogenic frameshift variant of KAT6B identified by clinical exome sequencing in a newborn with the Say-Barber-Biesecker-Young-Simpson syndrome. Clinical dysmorphology. 2019. [DOI] [PubMed] [Google Scholar]
- 29.Szakszon K, Salpietro C, Kakar N, et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B in two patients with Say-Barber/Biesecker/Young-Simpson syndrome. American journal of medical genetics Part A. 2013;161a(4):884–888. [DOI] [PubMed] [Google Scholar]
- 30.Gannon T, Perveen R, Schlecht H, et al. Further delineation of the KAT6B molecular and phenotypic spectrum. Eur J Hum Genet. 2015;23(9):1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yilmaz R, Beleza-Meireles A, Price S, et al. A recurrent synonymous KAT6B mutation causes Say-Barber-Biesecker/Young-Simpson syndrome by inducing aberrant splicing. American journal of medical genetics Part A. 2015;167a(12):3006–3010. [DOI] [PubMed] [Google Scholar]
- 32.Lundsgaard M, Le VQ, Ernst A, et al. De novo KAT6B Mutation Identified with Whole-Exome Sequencing in a Girl with Say-Barber/Biesecker/Young-Simpson Syndrome. Mol Syndromol. 2017;8(1):24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight S, VanHouwelingen L, Cervi D, et al. Genitopatellar syndrome and neuroblastoma: The multidisciplinary management of a previously unreported association. Pediatr Blood Cancer. 2018;65(12):e27373. [DOI] [PubMed] [Google Scholar]
- 34.Marangi G, Di Giacomo MC, Lattante S, et al. A novel truncating variant within exon 7 of KAT6B associated with features of both Say-Barber-Bieseker-Young-Simpson syndrome and genitopatellar syndrome: Further evidence of a continuum in the clinical spectrum of KAT6B-related disorders. American journal of medical genetics Part A. 2018;176(2):455–459. [DOI] [PubMed] [Google Scholar]
- 35.Niida Y, Mitani Y, Kuroda M, Yokoi A, Nakagawa H, Kato A. A Say-Barber-Biesecker-Young-Simpson variant of Ohdo syndrome with a KAT6B 10-base pair palindromic duplication: A recurrent mutation causing a severe phenotype mixed with genitopatellar syndrome. Congenital anomalies. 2017;57(3):86–88. [DOI] [PubMed] [Google Scholar]
- 36.Bashir RA, Dixit A, Goedhart C, et al. Lin-Gettig syndrome: Craniosynostosis expands the spectrum of the KAT6B related disorders. American journal of medical genetics Part A. 2017;173(10):2596–2604. [DOI] [PubMed] [Google Scholar]
- 37.Yu HC, Geiger EA, Medne L, Zackai EH, Shaikh TH. An individual with blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) and additional features expands the phenotype associated with mutations in KAT6B. Am J Med Genet A. 2014;164a(4):950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radvanszky J, Hyblova M, Durovcikova D, et al. Complex phenotypes blur conventional borders between Say-Barber-Biesecker-Young-Simpson syndrome and genitopatellar syndrome. Clin Genet. 2017;91(2):339–343. [DOI] [PubMed] [Google Scholar]
- 39.Li G, Li N, Li J, et al. De Novo Mutation of KAT6B Gene Causing Atypical Say-Barber-Biesecker-Young-Simpson Syndrome or Genitopatellar Syndrome. Fetal and pediatric pathology. 2017;36(2):130–138. [DOI] [PubMed] [Google Scholar]
- 40.Ali MM, Chasen ST, Norton ME. Testing for Noonan syndrome after increased nuchal translucency. Prenatal diagnosis. 2017;37(8):750–753. [DOI] [PubMed] [Google Scholar]
- 41.Toriello HV, Colley C, Bamshad M. Update on the Toriello-Carey syndrome. Am J Med Genet A. 2016;170(10):2551–2558. [DOI] [PubMed] [Google Scholar]
- 42.Simo-Riudalbas L, Perez-Salvia M, Setien F, et al. KAT6B Is a Tumor Suppressor Histone H3 Lysine 23 Acetyltransferase Undergoing Genomic Loss in Small Cell Lung Cancer. Cancer Res. 2015;75(18):3936–3945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.