Abstract
Cannabis smoking is the dominant route of delivery, with the airway epithelium functioning as the site of first contact. The endocannabinoid system is responsible for mediating the physiological effects of inhaled phytocannabinoids. The expression of the endocannabinoid system in the airway epithelium and contribution to normal physiological responses remains to be defined.
To begin to address this knowledge gap, a curated dataset of 1090 unique human bronchial brushing gene expression profiles was created. The dataset included 616 healthy subjects, 136 subjects with asthma, and 338 subjects with COPD. A 32-gene endocannabinoid signature was analysed across all samples with sex and disease-specific analyses performed. Immunohistochemistry and immunoblots were performed to probe in situ and in vitro protein expression.
CB1, CB2, and TRPV1 protein signal is detectable in human airway epithelial cells in situ and in vitro, justifying examining the downstream endocannabinoid pathway. Sex status was associated with differential expression of 7 of 32 genes. In contrast, disease status was associated with differential expression of 21 of 32 genes in people with asthma and 26 of 32 genes in people with COPD. We confirm at the protein level that TRPV1, the most differentially expressed candidate in our analyses, was upregulated in airway epithelial cells from people with asthma relative to healthy subjects.
Our data demonstrate that the endocannabinoid system is expressed in human airway epithelial cells with expression impacted by disease status and minimally by sex. The data suggest that cannabis consumers may have differential physiological responses in the respiratory mucosa.
Short abstract
The endocannabinoid system is differentially expressed in human airway epithelial cells from healthy subjects compared to those with asthma or COPD, which may be relevant for population-specific responses to inhaled cannabis smoke https://bit.ly/3cEWc2h
Introduction
Cannabis is the most commonly consumed illicit drug worldwide, with prevalence expected to increase due to trends in legalisation and innovations in medicinal applications [1]. Canada legalised cannabis in 2017 and concomitantly initiated the annual Canadian Cannabis Survey to monitor the perception and use patterns, demonstrating that 20% of the general population consumes cannabis [2–4]. Over 90% of cannabis consumers identified combusted smoke inhalation as a route of delivery, highlighting the lung as the dominant target for cannabis exposures. Within the cannabis consumer population, there is a skewed distribution of use patterns with males more frequently consuming in the past 12 months relative to females (26% versus 18%) with a subset (12%) of the population using cannabis for medicinal purposes. The prospective design of the Canadian Cannabis Surveys has demonstrated that cannabis use over time is widely accepted at a population level for recreational and medicinal purposes with no signal for decline in use post-legalisation [2–4]. Additionally, the data demonstrate a strong preference for inhalation routes of delivery and a skewing between male and female use. Therefore, a generalised understanding of how a cannabis consumer responds to smoke exposure is likely to be insufficient and should include both females and males in analyses and consider both healthy individuals and those that may have underlying medical needs.
The Canadian Cannabis Surveys report of inhalation as a dominant route of delivery confirms the importance of focusing on how the lungs respond to cannabis exposures. Early clinical exposure studies demonstrated that acute cannabis smoke exposure in healthy subjects is able to provide a sustained increase in lung function, which contrasted tobacco smoke inhalation [5, 6]. In people with asthma, cannabis smoking is able to produce a rapid reversal of exercise-induced bronchoconstriction and minimises bronchoconstriction induced by methacholine inhalation [7, 8]. Despite these objective benefits in a controlled laboratory setting, critical consideration needs to be given to the observed negative impacts on lung function in the context of increased frequency and intensity of cannabis smoking. Contrasting with acute cannabis exposure, cannabis smoking over a 2-month period was associated with a decrease in airway compliance that was correlated with quantity of cannabis consumed [9]. Further consolidating the negative impacts of chronic cannabis use on lung health, population-level analyses reveal that greater intensity of cannabis smoking is correlated with reduced lung function and increased risk of developing COPD [10, 11]. Furthermore, in multiple independent cohorts, cannabis smoking has been associated with a proinflammatory phenotype in the lung, associated with bronchitis and impaired immune cell function [12–16]. The mechanism(s) responsible for the clinical observations resulting from cannabis inhalation, whether beneficial acute or detrimental chronic exposures, remain elusive, but are likely to be influenced by the endocannabinoid system [17–19].
The endocannabinoid system is responsible for mediating the pharmacological effects of the endogenous cannabinoids anandamide and 2-arachidonylglycerol (2-AG) and some phytocannabinoids present in cannabis [20–22]. The first identified cannabinoid receptors were CB1 and CB2, both G-protein-coupled receptors that modulate downstream cyclic AMP signalling by inhibiting adenylyl cyclase activity [23, 24]. The mechanism of acute phytocannabinoid-induced bronchodilation is suggested to be mediated via CB1 receptors regulating neural control of airway tone [17, 18], which may have benefits for management of bronchoconstriction in asthma or COPD. In addition to CB1 and CB2, the transient receptor potential vanilloid-1 (TRPV1) and GPR55 have been identified as receptors for cannabinoids [25, 26]. Although the endocannabinoid signalling pathway is present throughout multiple tissues and organ systems, the lungs are of particular interest as they are the organ system targeted by cannabis inhalation. Airway epithelial cells play a critical role in lung health by acting as the first line of defence against pathogens and inhaled insults [27–29]. Airway epithelial cells carry out a number of functions such as providing a physical barrier against microbial infiltration, maintaining the inflammatory microenvironment, and releasing immune mediators to recruit leukocytes to the site of insult. It has been demonstrated that the inhalation of air pollution, tobacco smoke, and cannabis smoke can compromise airway epithelial function [30–32]. Notably, recent findings show that cannabis smoke exposure can lead to impaired airway epithelium barrier integrity, attenuated antiviral capacity, and exacerbated inflammatory response to immune challenges [31, 32]. However, the contribution of the endocannabinoid system to these observations has not been defined. The primary cannabinoid receptors, CB1 and CB2, have been shown to be expressed in the respiratory mucosa [33, 34] and human airway epithelial cells are responsive to tetrahydrocannabinol and anandamide in vitro [35, 36]. Additional components of the endocannabinoid system including MAPK, PI3K, and protein kinase-A signalling pathways downstream of receptors and enzymes responsible for cannabinoid metabolism have not been explored in an integrated fashion in human airway epithelial cells. An examination of the entire endocannabinoid system in human airway epithelial cells is required to better understand which components are dominant and likely to be functionally relevant in response to inhaled cannabis smoke. Furthermore, it remains possible that sex and disease status impact the endocannabinoid system expression, which may have functional consequences in distinct populations of cannabis consumers.
To begin our interrogation of the endocannabinoid system in human airway epithelial cells, we first generated a 32-gene endocannabinoid signature encompassing ligand recognition, signalling, and metabolism. We set out to examine if the expression of this 32-gene endocannabinoid signature was present in human airway epithelial cells and whether this was impacted by sex or disease status. The importance on examining sex and disease status on the endocannabinoid system is due to the possibility that specific populations may experience differential effects of cannabis, either positive or negative. To complete this study, we used a bioinformatic approach to analyse gene expression in 1090 unique human subject samples of airway epithelial cells isolated via bronchial brushing that included samples from healthy males and females and individuals with asthma or COPD. We complement our bioinformatic approach with validation and confirmation of CB1, CB2, and TRPV1 in human airway epithelial cells at the protein and gene level in situ and in vitro. Lastly, we validate a bioinformatic observation that TRPV1 gene expression is elevated in airway epithelial cells isolated from people with asthma by performing confirmatory immunoblot analysis on primary human airway epithelial cells. Collectively, our results demonstrate that an intact endocannabinoid system is expressed in human airway epithelial cells and that disease status impacts expression to a greater extent than sex, which may have functional consequences that lead to differential responses in distinct populations of cannabis consumers.
Methods
Human ethics
All studies using primary human lung material and blood were approved by the Hamilton Integrated Research Ethics Board or the University of British Columbia Human Research Ethics Board. No information on cannabis smoking history was available for any of the analyses described in detail below (in situ protein expression, in vitro immunoblots, in situ hybridisation, and microarray gene expression).
Primary human airway epithelial cells
Primary human airway epithelial cells isolated via bronchial brushings from consented healthy or asthmatic subjects were grown in PneumaCult ExPlus (Stemcell Technologies, Vancouver Canada) under submerged monolayer culture conditions and used in between passage 1 and 4. Where relevant, asthma diagnosis was confirmed with methacholine challenge and PC20 (provocative concentration causing a 20% fall in forced expiratory volume in 1 s) analysis as per ATS guidelines. No information on underlying medication use was collected.
Human whole-lung tissue
Noninvolved tissues from lung cancer patients were used. Lungs were homogenised using a mechanical homogeniser (Omni International, Waterbuy, CT, USA), lysed in 1× lysis buffer supplemented with complete protease inhibitors (Roche), and the supernatant was collected for immunoblots. No information on underlying medication use was collected.
Human peripheral blood mononuclear cells
Human peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood of healthy volunteers using Ficoll (Sigma-Aldrich) density centrifugation in Greiner LeucoSep-tubes (Sigma) according to the manufacturer's recommendations.
Immunohistochemistry and in situ hybridisation
Formalin-fixed paraffin-embedded human lung tissue from noninvolved regions of lung taken during resection were used for localisation of CB1, CB2, and TRPV1/VR1. Three-micron-thick sections were cut and stained for CB1 (Abcam, Ab23703, lot GR3239384-12 at 1:1000 dilution), CB2 (Abcam, Ab3561, lot GR3259180-6 at 1:50 dilution), and TRPV1/VR1 (Abcam, Ab3487, lot GR3296259-1 at 5 µg·mL−1). All staining was performed on a Leica Bond RX system with Leica Bond reagents and heat-induced antigen retrieval in citrate buffer at pH 6. To confirm CB1 and CB2 protein expression, in situ hybridisation using RNAscope® technology was used to detect CNR1 (ACD, 591528) and CNR2 (ACD, 596028) transcripts on a Leica Bond RX system. Digital slide scanning was performed using an Olympus VS120-L100 Virtual Slide System at ×40 magnification with VS-ASW-L100V2.9 software and a VC50 colour camera.
Analysis of promoter activity from the FANTOM5 dataset
The FANTOM5 promoterome dataset for the hg38 assembly was used to examine promoter activity of CNR1 and CNR2. Using the ZENBU genome browser, the nearest cap analysis of gene expression (CAGE) peak upstream and on the same strand as each of the aforementioned genes was extracted and analysed. The dataset consists of CAGE promoter activity data for 1886 primary cells, cell lines, and tissues from humans, and is quantified as normalised transcripts per million (TPM). A subset of FANTOM5 CAGE data (28 samples) is presented considering only samples related to lung tissues. Normalised TPM values for each CAGE peak, an approximation for promoter activity, were log10 transformed and separated according to tissue and cell type, and the heat map colour is proportional to these transformed normalised TPM values.
Immunoblots
Immunoblots confirming antibody staining and protein expression in human airway epithelial cells were performed using Bio-Rad stain-free 4–20% pre-cast gradient gels and imaged on a Bio-Rad ChemiDoc XRS+ Imaging system. For each immunoblot, 40 µg of protein was added per lane. CB1 (Abcam, Ab23703 at 1:500 dilution), CB2 (Abcam, Ab3561 at 1:100 dilution), and TRPV1 (Abcam, Ab3487 at 1:500 dilution) were diluted in 5% skimmed milk/TBS with 0.1% Tween-20. Primary antibody detection was performed using an anti-rabbit–horseradish peroxidase-conjugated secondary (Cell Signalling Technology, 7074) at 1:1000 dilution for 50 min at room temperature. Visualisation was performed using Clarity™ Western ECL Substrate (Bio-Rad) (CB1 and TRPV1) and SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) (CB2). Total protein loading images were collected as a confirmation of equal protein loading between sample types [37].
Gene expression dataset curation, normalisation, and statistical analyses
Candidate genes to be analysed were selected based on known relevance in the endocannabinoid signalling pathway. Additional genes and proteins implicated to be involved in cannabis-associated disorders and novel cannabinoid-based therapeutic approaches were included and described in annotated table format (table 1) [23–26, 38–60].
TABLE 1.
Relevant endocannabinoid signalling pathway candidates
| Molecule | Gene | Associated protein | Function | Ref. |
| Receptor | CNR1 | Cannabinoid receptor 1 (CB1) | Primary receptor involved in endocannabinoid signalling | [23] |
| CNR2 | Cannabinoid receptor 2 (CB2) | Primary receptor involved in endocannabinoid signalling | [24] | |
| GABRA2 | γ-aminobutyric acid receptor subunit α-2 (GABRA2) | Implicated in cannabis dependency | [38] | |
| GPR55 | G protein-coupled receptor 55 (GPR55) | Novel cannabinoid receptor | [26] | |
| OPRM1 | Opioid receptor μ1 (MOR1) | Implicated in cannabis dependency | [38] | |
| TRPV1 | Transient receptor potential vanilloid 1 (TRPV1) | Novel cannabinoid receptor | [25] | |
| Enzyme | ABHD12 | 2-AG hydrolase ABHD12 (ABHD12) | Degradation of 2-AG | [39] |
| ABHD6 | 2-AG hydrolase ABHD6 (ABHD6) | Degradation of 2-AG | [40] | |
| ADCY3 | Adenylyl cyclase 3 (AC) | Catalyses formation of cAMP | [41] | |
| AKT1 | AKT serine/threonine kinase 1 (AKT) | Regulates cell survival | [42] | |
| COMT | Catechol-O-methyltransferase (COMT) | Degradation of dopamine | [43] | |
| CYP2C9 | Cytochrome P450 2C9 (CYP2C9) | Metabolism of THC | [44] | |
| CYP3A4 | Cytochrome P450 3A4 (CYP3A4) | Metabolism of THC | [44] | |
| DAGLA | Diacylglycerol lipase α (DAGLA) | Biosynthesis of 2-AG | [57] | |
| DAGLB | Diacylglycerol lipase β (DAGLB) | Biosynthesis of 2-AG | [57] | |
| DUSP6 | Dual-specificity phosphatase 6 (MKP3) | Regulates MAPK signalling | [45] | |
| FAAH | Fatty acid amide hydrolase (FAAH) | Degradation of AEA | [46] | |
| FAAH2 | Fatty acid amide hydrolase 2 (FAAH2) | Degradation of AEA | [58] | |
| MAPK14 | Mitogen-activated protein kinase 14 (MAPK14) | Regulates cell survival | [47] | |
| MAP2K2 | Mitogen-activated protein kinase kinase 2 (MAP2K2) | Regulates cell survival | [45] | |
| MAPK3 | Extracellular signal-regulated kinase (MAKP3) | Regulates cell survival | [48] | |
| MGLL | Monoglyceride lipase (MAGL) | Degradation of 2-AG | [49] | |
| NAAA | N-acylethanolamine acid amidase (NAAA) | Degradation of AEA | [59] | |
| NAPEPLD | N-acylphosphatidylethanolamine phospholipase D (NAPEPLD) | Biosynthesis of AEA | [60] | |
| NOS2 | Inducible nitric oxide synthase (iNOS) | Inflammatory mediator | [50] | |
| PIK3CA | Phosphatidylinositol-3-kinase (PI3 K) | Regulates cell survival | [42] | |
| PRKACA | Protein kinase-A (PKA) | Regulates cell survival | [51] | |
| PTGS2 | Cyclooxygenase-2 (COX2) | Inflammatory mediator | [52] | |
| Other proteins | ABCB1 | P-glycoprotein 1 (p-GP) | Cannabinoid transportation | [53] |
| GNAI1 | Gi/o α subunit (Gi/o) | Coupled to cannabinoid receptors | [54] | |
| NRG1 | Neuregulin 1 (NRG1) | Mediates cell–cell signalling | [55] | |
| TP53 | Tumour protein p53 (p53) | Regulates cell survival | [56] |
2-AG: 2-arachidonoylglycerol; THC: tetrahydrocannabinol; AEA: anandamide.
Public microarray experiments using Affymetrix chips (HG-U133 Plus 2, HuEx-1.0-st-v1 and HuGene-1.0-st-v1) on airway epithelial cell samples from healthy individuals or those with asthma or COPD were selected from the National Center for Biotechnology Information Gene Expression Omnibus database. Healthy samples were further filtered by removing former or current smokers. This resulted in a total of 1090 individual samples from 27 experiments (table 2) that included samples from 616 healthy subjects, 136 subjects with asthma, and 338 subjects with COPD [61–88]. Within each sample population, sex was reported for a subset of samples (healthy: 103 females/227 males; asthma: 34 females/28 males; COPD: 48 females/93 males). Within all datasets analysed, only 19 samples (from GSE4302) had information on medication use. For this reason, no stratification on medication use was possible for the bioinformatic analyses.
TABLE 2.
Gene expression omnibus datasets analysed
| GSE accession | Affymetrix chip | Asthma | COPD | Healthy | Ref. |
| GSE4302 | HG-U133 Plus 2 | 74 | 0 | 13 | [61, 62] |
| GSE4498 | HG-U133 Plus 2 | 0 | 0 | 12 (2F/10M) | [63, 64] |
| GSE5058 | HG-U133 Plus 2 | 0 | 14 (4F/10M) | 0 | [64, 65] |
| GSE7832 | HG-U133 Plus 2 | 0 | 0 | 8 (2F/6M) | [64] |
| GSE8545 | HG-U133 Plus 2 | 0 | 15 (3F/12M) | 5 (1F/4M) | [66] |
| GSE10006 | HG-U133 Plus 2 | 0 | 20 (4F/16M) | 21 (2F/19M) | [67] |
| GSE11784 | HG-U133 Plus 2 | 0 | 17 (1F/3M) | 40 (18F/22M) | [68] |
| GSE11906 | HG-U133 Plus 2 | 0 | 0 | 30 (10F/20M) | [69] |
| GSE13931 | HG-U133 Plus 2 | 0 | 0 | 19 (4F/15M) | [70] |
| GSE13933 | HG-U133 Plus 2 | 0 | 0 | 11 (6F/5M) | [71] |
| GSE14224 | HuEx-1.0-st-v2 | 0 | 0 | 11 (7F/4M) | [72] |
| GSE17905 | HG-U133 Plus 2 | 0 | 0 | 1 (1M) | [73] |
| GSE19667 | HG-U133 Plus 2 | 0 | 0 | 3 (3F) | [74] |
| GSE20257 | HG-U133 Plus 2 | 0 | 1 (1F) | 0 | [75] |
| GSE22047 | HG-U133 Plus 2 | 0 | 23 | 81 | [76] |
| GSE34450 | HG-U133 Plus 2 | 0 | 0 | 11 | [77] |
| GSE37147 | HuGene-1.0-st-v1 | 0 | 110 (35F/52M) | 8 | [100] |
| GSE40364 | HG-U133 Plus 2 | 0 | 7 | 0 | [79] |
| GSE43079 | HG-U133 Plus 2 | 0 | 0 | 16 | [80] |
| GSE43939 | HG-U133 Plus 2 | 0 | 0 | 13 | [81] |
| GSE52237 | HG-U133 Plus 2 | 0 | 0 | 2 | [82] |
| GSE64614 | HG-U133 Plus 2 | 0 | 0 | 31 | [83] |
| GSE67472 | HG-U133 Plus 2 | 62 (34F/28M) | 0 | 43 (20F/23M) | [84] |
| GSE77658 | HG-U133 Plus 2 | 0 | 0 | 6 | [85] |
| GSE84101 | HG-U133 Plus 2 | 0 | 0 | 7 | [86] |
| GSE97010 | HuGene-1.0-st-v1 | 0 | 0 | 126 (28F/98M) | [87] |
| GSE108134 | HG-U133 Plus 2 | 0 | 131 | 98 | [88] |
F: female; M: male
For all dataset samples, raw intensity values and annotation data were downloaded with the R statistical language (version 3.6.1; R Core Team, 2019) using the GEOquery R package (version 2.52.0) [89]. Probe definition files were downloaded from the Brainarray database (version 24) [90]. To obtain processed microarray gene expression values unaffected by probe CG compositional biases, the single channel array normalisation (SCAN) method was used via the SCAN.UPC R package (version 2.26.0) [91] using annotation data from the Bioconductor project (version 3.9) [92]. All log2-transformed gene expression data were unified into a single dataset, and only genes detected in all three platforms (16 543) were kept for subsequent analyses. Correction of experiment-specific batch effects was performed using the ComBat method [93] implemented in the sva R package (version 3.32.1) [94], with disease status and sex supplied as covariates. Following batch correction, all data underwent Z-score transformation to set the mean of all samples to zero and replace expression values with a measure of variance from the mean [95]. Principal-component analysis (PCA) was performed using the probabilistic PCA method in the pcaMethods R package (version 1.76.0) [96]. Gene expression levels were tested for significant differences via a t-test with a Benjamini–Hochberg multiple testing correction using the stats R package (version 3.6.1; R Core Team, 2019). Gene expression box plots, heat maps, and PCA plots were generated with the ggplot2 R package (version 3.2.1).
Results
Human airway epithelial cells express CB1, CB2, and TRPV1, in situ and in vitro
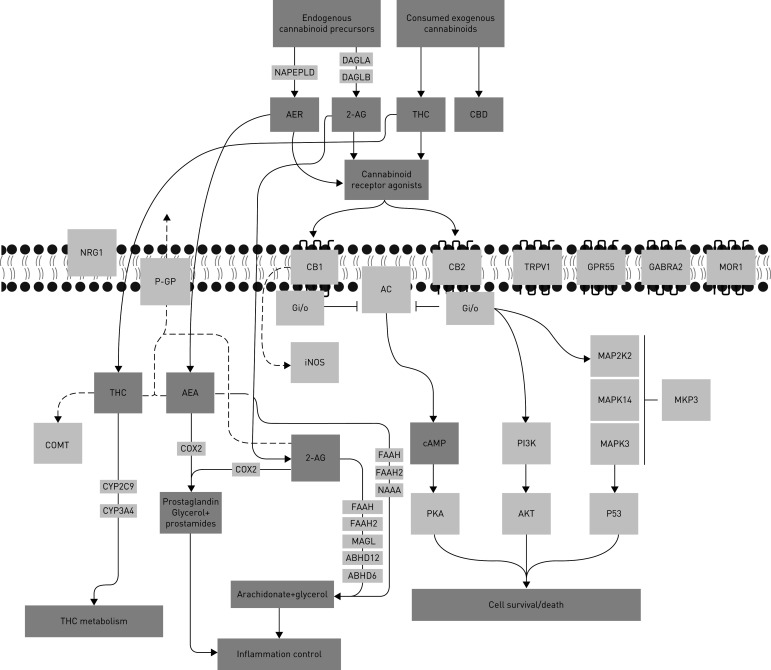
A curated list of genes involved in cannabinoid signalling was generated (herein called the 32-gene endocannabinoid signature) to provide a focused overview of this pathway in human airway epithelial cells (figure 1 and table 1).
FIGURE 1.
Visual representation a 32-gene endocannabinoid signature. Solid arrows indicate known relationship between candidates and ligands. Dotted arrows indicate proposed relationships. Blunted lines indicate inhibition. Candidate functions are annotated in table 1. THC: tetrahydrocannabinol.
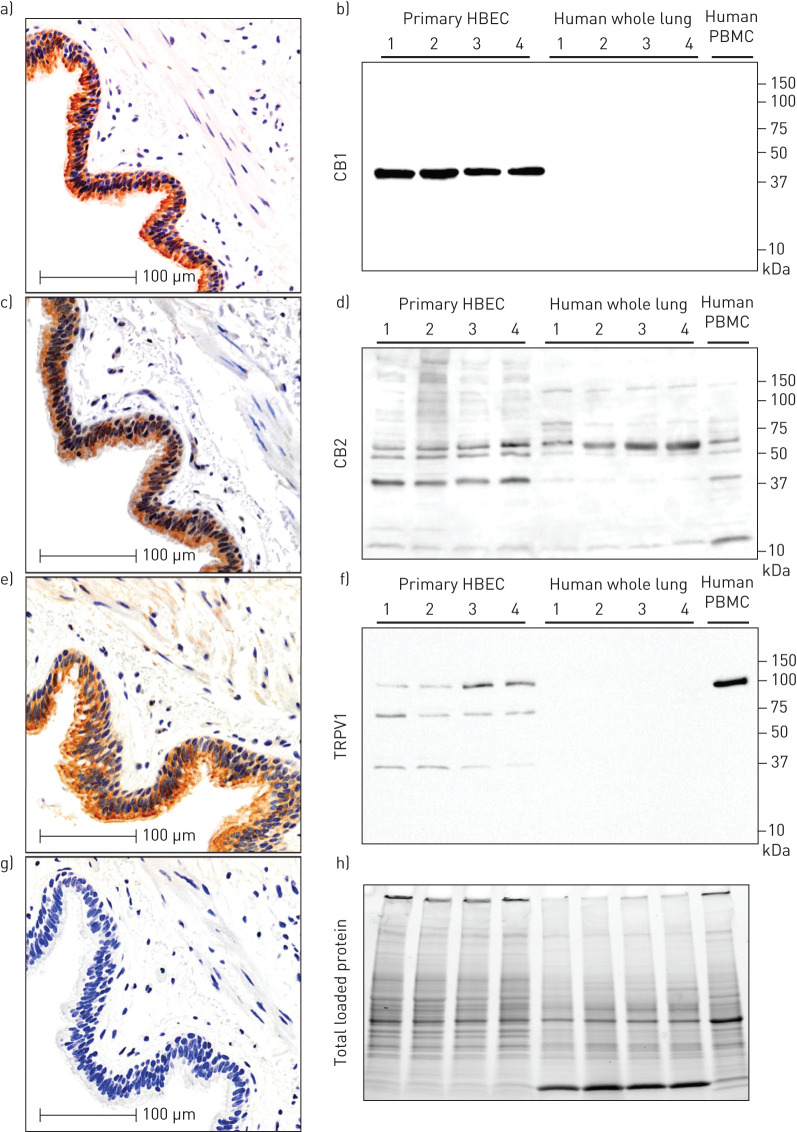
To begin our characterisation of the endocannabinoid system in human airway epithelial cells, we performed in situ localisation of CB1, CB2, and TRPV1 protein in human lung tissue (figure 2). We demonstrate that all three receptors are present at protein level in human airway epithelial cells in situ relative to negative control (figure 2a, c, e and g). To validate the staining patterns observed and antibody specificity, we performed immunoblots with primary human airway epithelial cells, whole-lung tissue, and PBMCs. A single band for CB1 was observed at approximately 45 kDa in human airway epithelial cells, but not whole human lung, or PBMCs (figure 2b). A dominant band for CB2 was observed at approximately 40 kDa and accompanied by a reported 52–55 kDa doublet [97] in human airway epithelial cells, with a similar pattern observed for PBMCs (figure 2d). In contrast, in whole human lung the dominant band was observed at 55 kDa with only a faint 40 kDa band. The band patterns observed for CB2 are consistent with glycosylation of the N terminus and processing of the full-length peptide [97]. A dominant band for TRPV1 was observed at approximately 100 kDa in human airway epithelial cells and accompanied by two lower molecular weight bands at approximately 70 kDa and 37 kDa (figure 2f). No TRPV1 bands were observed in whole human lung, while a dominant single band at 100 kDa was observed in PBMCs. Total protein loading staining from a representative blot demonstrates equal loading within replicates of the same sample type and distinct protein compositions between sample types (figure 2h).
FIGURE 2.
In situ and in vitro validation of CB1, CB2 and TRPV1 protein expression in human airway epithelial cells. Serial sections from a single patient donor that is representative of n=10, for immunohistochemistry of a) CB1, c) CB2 and e) TRPV1 with g) negative control. Immunoblots on primary human airway epithelial cells cultured in vitro: b) CB1, d) CB2, and f) TRPV1 with h) total protein loading control (n=4 airway epithelial cells (HBEC), n=4 whole-lung samples, n=1 peripheral blood mononuclear cells (PBMCs)). Molecular weights (in kilodaltons) are denoted on y-axis of immunoblots.
To extend our in situ CB1 and CB2 protein staining and to interrogate the potential of nonspecific staining generated with the CB2 antibody, we next performed in situ hybridisation for CNR1 and CNR2 gene transcripts using RNAscope® technology (figure 3). In situ hybridisation demonstrates transcripts for both CNR1 (figure 3a) and CNR2 (figure 3b) in the airway epithelium of all human lung samples examined (n=10). Serial sections from each lung sample reveal that CNR1 and CNR2 are likely to coexpress in airway epithelium.
FIGURE 3.
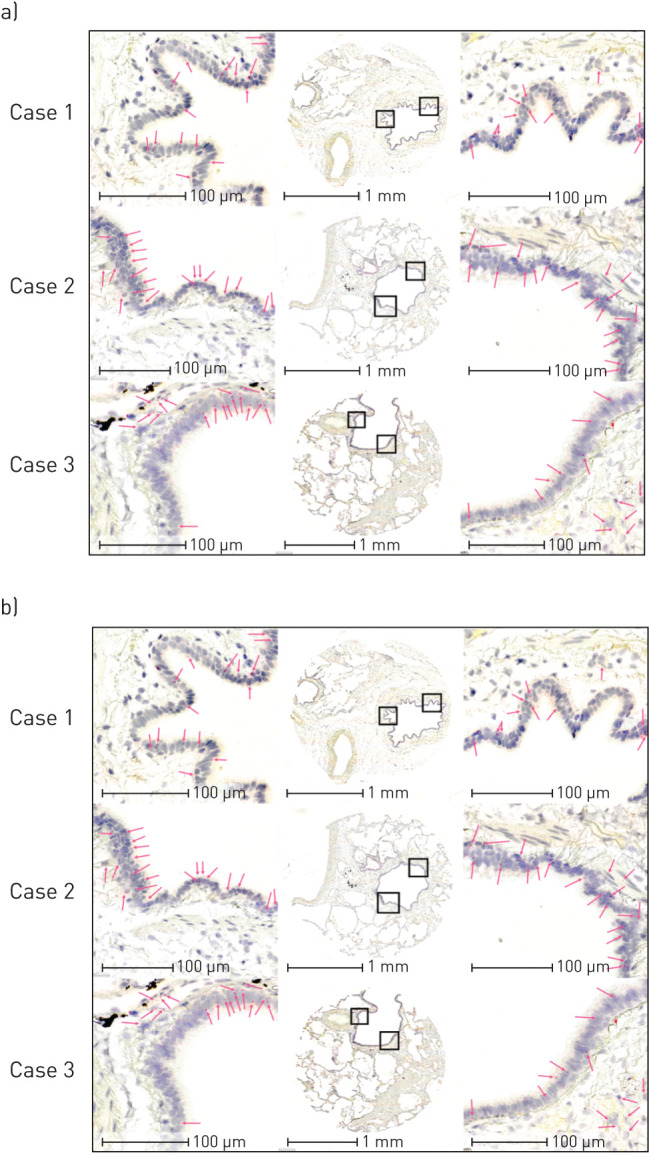
In situ hybridisation detection of CNR1 and CNR2 gene transcripts in human airway epithelial cells. In situ hybridisation of a) CNR1 and b) CNR2 in three patient donors representative of n=10. Serial sections of each of the three cases were stained for CNR1 and CNR2. Low magnification images are in the centre of a and b, with high-powered magnification regions of interest on either side highlighted by black boxes. Pink arrows correspond to positive puncta representative of mRNA transcript.
To further corroborate our in situ CB1 and CB2 protein and gene transcript staining, promoter activity data for CNR1 and CNR2 were extracted and analysed from the FANTOM5 dataset, which includes 1886 primary cells, cell lines, and tissue sample types. We selected all samples that included “lung”, “nasal”, “trachea”, “bronchial”, “airway”, or “alveolar” to identify lung-specific sample types (n=28). CDH1 promoter activity was analysed as a positive control. Consistent with our observed gene expression analysis in airway epithelial cells, normalised TPM values for each CAGE peak demonstrate that CNR1 and CNR2 promoter activity was present but modest across airway epithelial cells and lung tissue samples (supplementary figure 1). Both microarray gene expression analysis and promoter activity were consistent with results of candidate gene and protein expression in airway epithelial cells via in situ hybridisation and immunohistochemistry.
Collectively, our in situand in vitro data are supportive for expression of CB1, CB2, and TRPV1 protein and gene in human airway epithelial cells.
Expression of endocannabinoid system genes in human airway epithelial cells from healthy male and female subjects
Sex differences in CB1 and CB2 expression levels have been reported [98], which could impact downstream responses to cannabinoid exposures. Furthermore, sex and gender differences in cannabis consumption practices have been reported [2–4]. Collectively, these two factors could interact and contribute to differential responses to cannabinoids in distinct populations.
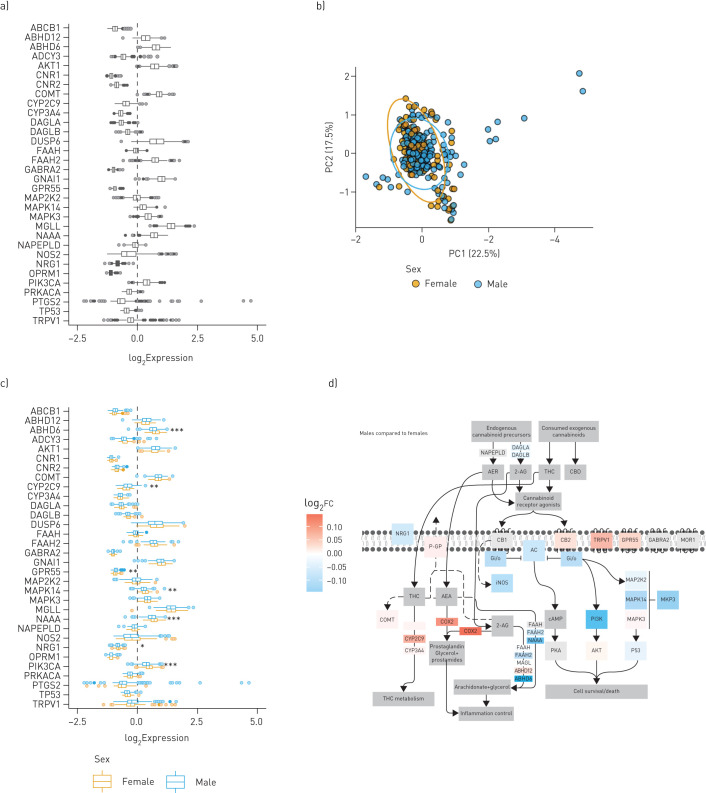
To examine sex differences in the endocannabinoid system, we analysed our 32-gene endocannabinoid signature in a curated dataset of airway epithelial cells from 616 unique healthy subjects, where the identifier of sex was available for 103 females and 227 males (table 2). The 32-gene endocannabinoid signature was first analysed in all 616 subjects to show overall trends for each gene (figure 4a). Subsequently, a PCA plot was performed for all samples with sex as an identifier (figure 4b). The PCA plot reveals clustering of samples from males within a larger space occupied by the female samples. Statistical analysis at the individual gene level revealed a difference in 7 of 32 genes (figure 4c). Five genes (ABHD6, MAPK14, NAAA, NRG1, and PIK3CA) were downregulated in males relative to females, while two genes (CYP2C9 and GPR55) were upregulated in males relative to females. The gene expression patterns were overlaid on the endocannabinoid signalling pathway for qualitative visualisation of global changes in the 32-gene endocannabinoid signature in males relative to females (figure 4d).
FIGURE 4.
Impact of sex status on endocannabinoid system gene expression in human airway epithelial cells from healthy individuals. a) Gene expression data for 616 healthy subjects with no history of smoking or chronic respiratory disease. b) Principal-component (PC) analysis plot of healthy females (n=103) and males (n=227) generated by expression patterns of the 32-gene endocannabinoid signature. The first (22.5%) and second (17.5%) PCs were used. Ellipses were added to represent 95% confidence intervals per sex. c) Healthy samples with metadata defining sex were further divided into male and female groups and plotted separately as blue and orange-outlined box plots, respectively. For both a and c, log2-transformed expression values were plotted as box plots. The dashed line at zero represents the global baseline of expression for the entire set of genes. d) Visual representation of the differences between healthy females and males in the 32-gene endocannabinoid signature. Colour coding is reflective of log2 fold change of males relative to females. THC: tetrahydrocannabinol. *: p<0.05; **: p<0.01; ***: p<0.001.
Collectively, our data confirm that the endocannabinoid system is expressed at gene level in human airway epithelial cells, suggesting that signalling downstream of receptors is intact, with mild sex differences observed in endocannabinoid system gene expression.
The endocannabinoid system is dysregulated in human airway epithelial cells from individuals with asthma and COPD
In addition to sex, disease status may also impact the expression of the endocannabinoid system in airway epithelial cells, as specific phenotypes are observed in cells isolated from people with asthma and individuals with COPD [99–101]. We therefore tested the hypothesis that the 32-gene endocannabinoid signature was dysregulated in asthma and COPD.
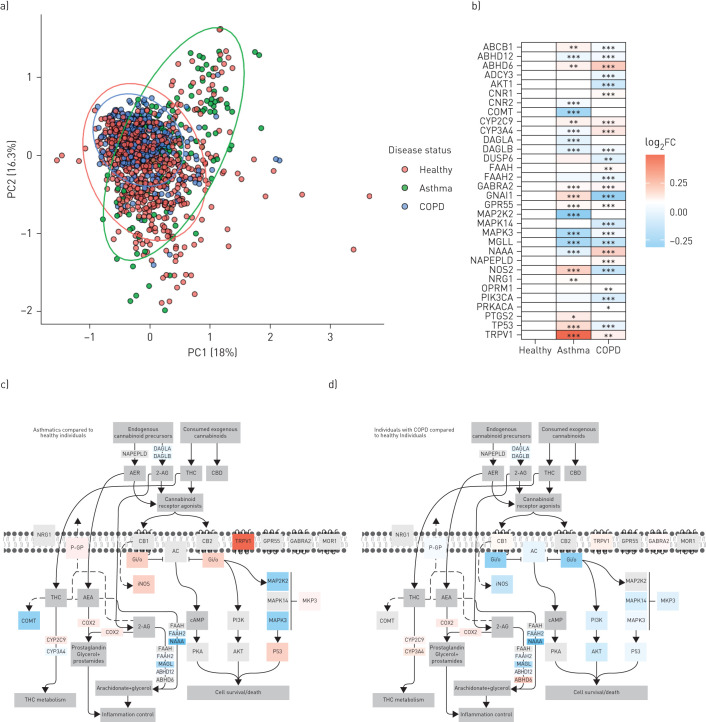
To test this hypothesis, we curated all 1090 samples that included 616 healthy subjects, 136 subjects with asthma, and 338 subjects with COPD. A PCA plot reveals clustering of samples from healthy, asthmatic, and COPD subjects, with samples from people with asthma separating from both healthy subjects and those with COPD (figure 5a). Statistical analysis at the individual gene level revealed changes in 21 of 32 genes in people with asthma and 26 of 32 genes in those with COPD (figure 5b). In people with asthma, 11 of 21 dysregulated genes were upregulated (ABCB1, ABHD6, CYP2C9, GABRA2, GNAI1, GPR55, NOS2, NRG1, PTGS2, TP53, and TRPV1), while 10 of 21 were downregulated (ABHD12, CNR2, COMT, CYP3A4, DAGLA, DAGLB, MAP2K2, MAPK3, MGLL, and NAAA). In COPD subjects, 11 of 26 dysregulated genes were upregulated (ABHD6, CNR1, CYP2C9, CYP3A4, FAAH, GABRA2, GPR55, NAAA, NAPEPLD, OPMRI, and TRPV1), while 15 of 26 were downregulated (ABCB1, ABHD12, ABCY3, AKT1, DAGLB, DUSP6, FAAH2, GNAI1, MAPK14, MAPK3, MGLL, NOS2, PIK3CA, PRKACA, and TP53). The most dysregulated gene was TRPV1, with the largest upregulation observed in the samples from people with asthma relative to healthy controls. The differential gene expression patterns were overlaid on the endocannabinoid signalling pathway for qualitative visualisation in asthma (figure 5c) and COPD (figure 5d).
FIGURE 5.
Impact of disease status on endocannabinoid system gene expression analysis in human airway epithelial cells from healthy individuals, people with asthma, and individuals with COPD. a) Principal-component (PC) analysis plot of healthy subjects (n=616), people with asthma (n=136) and individuals with COPD (n=338) generated by expression patterns of the 32-gene endocannabinoid signature. The first (18%) and second (16.3%) PCs were used. Ellipses were added to represent 95% confidence intervals per sex. b) Gene expression data of the 32 genes were compared between healthy, asthmatic and COPD samples. The log2-transformed mean expression values were compared to that of the healthy samples and shown as log2 fold change (FC). Visual representation of the differences in the 32-gene endocannabinoid signature between c) healthy subjects and people with asthma and d) healthy subjects and individuals with COPD. Colour coding is reflective of log2FC relative to healthy subjects. THC: tetrahydrocannabinol. *: p<0.05; **: p<0.01; ***: p<0.001.
Collectively, our data demonstrate that underlying chronic respiratory disease status is associated with a dysregulation of the endocannabinoid system at the gene level in human airway epithelial cells.
Impact of sex on endocannabinoid system gene expression in human airway epithelial cells from people with asthma and those with COPD
Sex differences in incidence, age of onset, and pathology are observed in both asthma and COPD [102, 103]. Cannabinoid exposures have been explored in the context of both asthma and COPD management for immunomodulatory and bronchodilation purposes [7, 8, 104]. To date, the potential interaction of sex status and endocannabinoid system expression in chronic respiratory disease has not been addressed.
Taking the same approach as for healthy subjects, we analysed a curated dataset of airway epithelial cells from 136 unique asthmatic subjects, where the identifier of sex was available for 34 females and 28 males. For COPD, we analysed a curated dataset from 338 unique COPD subjects, where the identifier of sex was available for 48 females and 93 males (see table 2 for study group compositions).
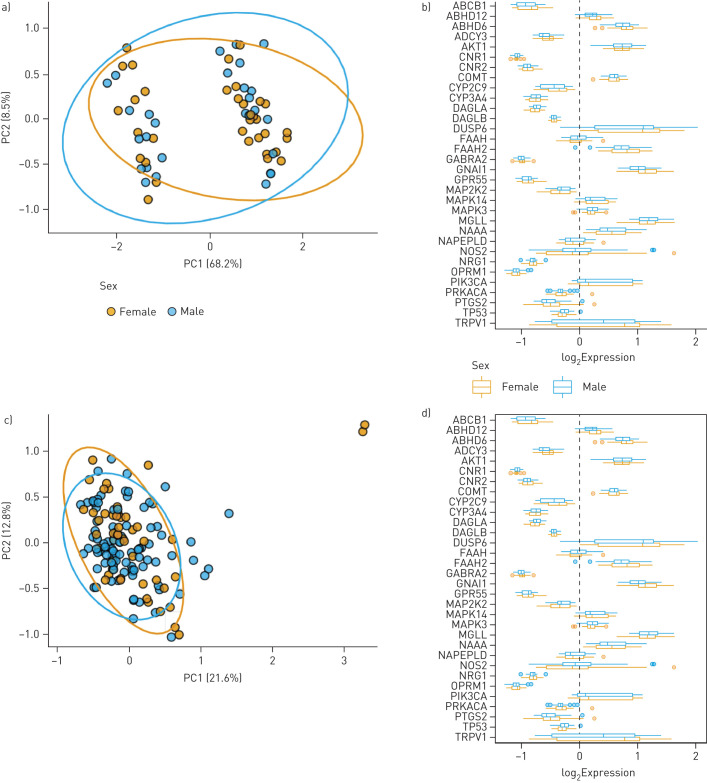
In both asthma and COPD samples, PCA plots revealed no separation between sexes with clustering of samples overlapping between disease groups (figure 6a and c). At the individual gene level, no sex-dependent differences were observed for any gene in the 32-gene endocannabinoid signature in either disease groups (figure 6b and d).
FIGURE 6.
Impact of sex status on endocannabinoid system gene expression in human airway epithelial cells from individuals with chronic respiratory disease. a) Principal-component analysis (PCA) plot of asthmatic females (n=34) and males (n=28) generated by expression patterns of the 32-gene endocannabinoid signature. The first (68.2%) and second (8.5%) principal components (PCs) were used. b) Asthmatic samples divided into female and male, and plotted separately. c) PCA plot of females (n=48) and males (n=93) with COPD generated by expression patterns of the 32-gene endocannabinoid signature. PC1 (21.6%) and PC2 (12.8%) used. d) COPD samples divided into female and male, and plotted. For both a and c, ellipses were added to represent 95% confidence intervals per sex. For both b and d, log2-transformed expression values were plotted as boxplots of log2 fold change. The dashed line at zero represents the global baseline of expression for the entire set of genes.
Collectively, our data do not support sex differences in the endocannabinoid system in human airway epithelial cells from people with asthma or subjects with COPD.
TRPV1 is upregulated in airway epithelial cells from people with asthma
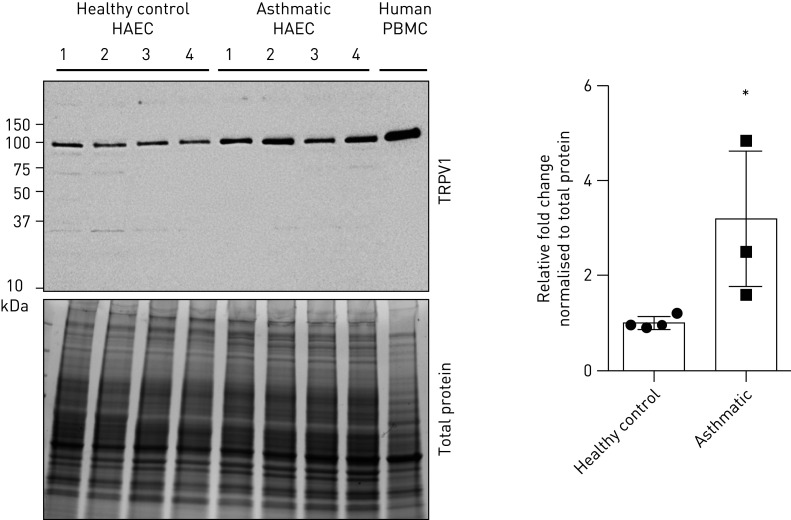
As our bioinformatic interrogation of the 32-gene endocannabinoid signature was restricted to genes, we next performed confirmatory protein expression analysis. The candidate we chose for validation was TRPV1, a confirmed receptor for cannabinoids that was the most differentially expressed candidate between our comparisons examining sex or disease status.
Using primary human airway epithelial cells from healthy donors or those with physician-diagnosed asthma, protein was isolated from cells grown under submerged monolayer culture conditions. Immunoblot analysis confirmed TRPV1 protein expression in human airway epithelial cells and revealed an increase in cells from people with asthma (figure 7). In closing, our protein analysis is consistent with the bioinformatic analysis that revealed elevations in the TRPV1 gene in human airway epithelial cells from people with asthma.
FIGURE 7.
TRPV1 protein is elevated in human airway epithelial cells (HAECs) from people with asthma. Immunoblot of HAECs from healthy subjects (n=4), people with asthma (n=4) and peripheral blood mononuclear cell (PBMCs) (control) were analysed for TRPV1 and quantified as fold change over healthy subjects, normalised to total protein loading. *: p<0.05.
Discussion
The dominant route of delivery of cannabis is via inhalation of combustion smoke, resulting in exposure to phytocannabinoids and activation of the endocannabinoid system [2–4]. The airway epithelium represents the first line of defence in the human lung against inhaled insults, including cannabis smoke. To better understand how the epithelium is able to respond to cannabis smoke exposure in the context of the endocannabinoid system, we performed a characterisation study using bioinformatic and complementary protein analysis approaches. We provide data supporting expression of CB1, CB2, and TRPV1 at the protein level in the human airway epithelium in situ and in vitro. We support our observations of protein expression by demonstrating that CNR1 and CNR2 transcripts are present in human airway epithelial cells using in situ hybridisation methods. The demonstration that these receptors were present warranted an exploration into the endocannabinoid system downstream of the receptors. Using 1090 unique patient samples of airway epithelial cells curated from publicly available datasets, we demonstrated in healthy subjects that the gene expression levels of the endocannabinoid system show minor differences between females relative to males. In contrast, we demonstrated with samples from people with asthma or individuals with COPD that disease status appears to be a strong driver of endocannabinoid system gene expression, with no interaction with sex. Lastly, we validated the bioinformatic approach by demonstrating that TRPV1, a top candidate upregulated at the gene level in our studies, is also upregulated at the protein level in people with asthma. Collectively, our results confirm the expression of the endocannabinoid system in human airway epithelial cells and that disease status impacts expression to a greater extent than sex, which may have functional consequences in distinct populations of cannabis consumers.
The legalisation of cannabis in multiple jurisdictions on a global scale reduces barriers for individuals to consume cannabis for either medicinal or recreational purposes. The dominant route of cannabis delivery is through inhalation of smoke from plant combustion [2–4]. Inhaled cannabis smoke travels through the upper and lower airways, with airway epithelial cells being a major site of first contact. We have demonstrated with an in vitro model of airway epithelial cell culture that cannabis smoke induces a concentration-dependent reduction in airway epithelial cell viability, barrier function, while promoting proinflammatory cytokine secretion [31, 32]. Complementary profiling of human epithelial cells isolated via bronchial brushings has demonstrated cannabis consumption-dependent elevations in TLR5, TLR6, and TLR9 gene expression [105]. A limitation of these studies is the lack of mechanistic interrogation into the role that the endocannabinoid system contributed to the observed functional consequences of cannabis smoke. To begin to implicate the endocannabinoid system in epithelial cell functions, an in vitro approach has used direct cannabinoid administration to airway epithelial cells independent of combustion, showing functional consequences with altered barrier function mediated by mechanisms dependent and independent of cannabinoid receptors [35, 36]. Our demonstration that multiple endocannabinoid system components are expressed at the gene level in airway epithelial cells with CB1, CB2, and TRPV1 confirmed at the protein level, is consistent with this system have the potential to play a role in mediating cannabis smoke-induced effects. Collectively, our data and existing literature suggest that the dominant form of cannabis consumption, cannabis smoke inhalation, is able to induce functional responses in airway epithelial cells that may be influenced by the endocannabinoid system.
Cannabis use patterns have been reported to differ among males and females, with females consuming less in overall quantity and frequency [2–4]. The endocannabinoid system is in turn regulated by sex hormones with diverse interactions at levels of receptors, enzymes, and signalling molecules [106, 107]. In the context of lung health and disease, sex-dependent lung physiology is observed as well as asthma and COPD disease incidence and progression [102, 103]. In light of the potential for interactions between sex, user practices, and the endocannabinoid system, we performed a bioinformatic analysis that stratified for male and female sex status. In healthy individuals, we observe that only 7 of 32 of our endocannabinoid gene signature candidates were differentially regulated between males versus females (2 upregulated, and 5 downregulated). In our analysis, the most significant differentially expressed gene between sexes was ABHD6 (increased in female samples). Increased female expression of ABHD6 has been previously reported in immune cells, with only female cells showing oestrogen- or progesterone-dependent induction of ABHD6 gene levels [108]. Our observations of minor sex-dependent effects on the expression of the 32-gene endocannabinoid signature contrast with sex differences in the expression of CB1 and CB2 protein expression in heart tissue, where CB1 receptors are more highly expressed in females and CB2 receptors more highly expressed in males [98]. Our observations of limited impact of sex on the 32-gene endocannabinoid signature expression in healthy subjects were conserved in people with asthma and individuals with COPD. In contrast to sex status, disease status drove a large effect for differential expression of the 32-gene endocannabinoid signature, with the majority of genes dysregulated (21 of 32 genes in people with asthma and 26 of 32 genes in those with COPD). Collectively, these findings suggest that asthma or COPD status impacts expression of the endocannabinoid system, whereas sex status plays a comparatively smaller role.
The potential disease-specific response to cannabis exposure as a result of dysregulated endocannabinoid signalling is relevant as cannabinoids have and are being pursued for bronchodilatory and immunomodulatory properties [7, 8, 104, 109]. Furthermore, in populations of people with asthma and those with COPD, cannabis consumption is not avoided and is shown to interact with disease progression. The reported benefits of cannabis exposure on lung function in people with asthma may be selective for this population based on endocannabinoid signalling. Indeed, if TRPV1 is a dominant receptor for responses downstream of inhaled cannabis, the signalling mediated in airway epithelial cells from people with asthma may be augmented relative to healthy controls. Our observation of elevated TRPV1 in airway epithelial cells from people with asthma is consistent with a previous report demonstrating correlation with asthma severity [110]. A limitation of our study is that we did not explore other cell types for TRPV1, which is also expressed in airway smooth muscle cells and can modulate smooth muscle contraction [111]. In contrast, the lack of observed benefit on lung function and association with advanced pathology in COPD subjects may be a result of a distinct expression profile of the endocannabinoid system [10, 11, 104]. Our results and those in the literature suggest that a universal response to inhaled cannabis by healthy subjects and those with asthma or COPD should not be assumed and cautions translation of safety and efficacy studies performed in healthy individuals to those with underlying asthma or COPD.
Our study is heavily focused on using deposited datasets generated by microarray gene expression technology. Microarrays are low cost, cover large numbers of transcripts, and benefit from a standardised format for analysis and public deposition of data. As a result of these benefits, gene expression data for large, independent cohorts are available for curation and data-mining purposes if appropriate measures are taken to normalise and integrate datasets from diverse user groups. Despite benefits of curated microarray datasets, there exist limitations of our approach. Specifically, microarray technology is susceptible to a poor signal-to-noise ratio, making transcripts expressed at low levels difficult to detect, suggesting that we may be under-estimating the signal or introducing high variance of these transcripts. To address this limitation, the SCAN method deployed functions to reduce technical and across-sample variation and to increase the signal-to-noise ratio while maintaining the ability to detect differentially expressed genes [91]. As in all analyses, the sample size will dictate the statistical power, which can only be as large as the available datasets. In our case, the sample sizes for females and males in our asthma and COPD datasets were smaller than our healthy cohorts, which may have limited our ability to detect any sex-specific effects on endocannabinoid gene expression. Finally, in order to compare data across multiple experiments, the microarray chip technology used needs to be considered with additional normalisation methods required for data generated on different platforms. Different microarray technologies will have unique probe compositions and quality control protocols which can lead to systematic biases between experiments, though these batch-specific effects can be addressed using batch correction techniques and normalisation [112]. Despite these highlighted limitations, the curation of multiple microarray gene expression datasets from diverse cohorts of study subjects can be aligned with normalisation methods to minimise batch and cross-platform effects and maximising sample sizes to detect differential gene expression patterns.
The advances of our study are accompanied by several limitations that need to be discussed. All analyses performed used human lung samples or datasets that did not contain information on cannabis consumption practices of the study subjects or body mass index, and we did not stratify by age. Furthermore, we broadly categorised people with asthma and COPD without stratifying by disease severity. It is possible that cannabis consumption impacts the expression of endocannabinoid system components examined in our study, a possibility we are not able to interrogate. Related to this possibility, a key finding of our study is the elevation of TRPV1 in people with asthma, which may be regulated differently with and without cannabis exposure [59]. Future studies examining healthy subjects and those with asthma or COPD in the context of cannabis exposure will address this limitation. We also only focused on airway epithelial cells, a dominant aspect of the respiratory mucosal defence system in the lungs. Observations in airway epithelial cells may be cell-specific and not translate to other structural or immune cells. Lastly, our archived human lung tissue samples used for immunohistochemistry and in situ hybridisation were collected from noninvolved regions of the lung taken during resection in individuals undergoing surgery as part of lung cancer management. It is possible that the underlying status of lung cancer impacts the expression profiling of the endocannabinoid system. Our analysis of nonlung cancer microarray datasets and immunoblot confirmation using healthy and diseased (asthma) samples attempted to corroborate observations made from the samples taken during lung resections.
In summary, we demonstrate that endocannabinoid system components are expressed in human airway epithelial cells at the gene level and CB1, CB2, and TRPV1 are expressed at the protein level. We demonstrate that gene expression patterns for a 32-gene endocannabinoid signature are differentially expressed between healthy individuals and those with asthma or COPD, whereas only minor differences are observed between sexes. We confirm that our bioinformatic approach for analysis of gene expression has the potential to reflect corresponding protein expression level changes as demonstrated by elevated TRPV1 protein expression in human airway epithelial cells from people with asthma relative to healthy controls. Our study lays a foundation with primary human lung samples from well-defined patient populations to justify exploring the functional consequences of endocannabinoid system signalling in human airway epithelial cells in both health and disease. The complete functions of the endocannabinoid system in airway epithelial cells remain to be defined.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 Detection of CNR1 and CNR2 promoter activity in human lung tissues and lung epithelial cell samples. Heat map of FANTOM5 CAGE promoter activity data for CNR1, CNR2, and CDH1 (positive control) are shown for samples related to lung tissues (n=28). Heat map colour is proportional to promoter activity, depicted as log10-transformed normalized transcripts per million (TPM). Grey represents no detection. 00128-2020.figureS1 (411.2KB, pdf)
Acknowledgements
The authors acknowledge Olivia Marcello (freelance graphic artist) for her help in figure design. Additionally, the authors acknowledge James MacKillop and Allan Fein (Centre for Medicinal Cannabis Research at McMaster University) for administrative support, and graduate student stipend support (M.F. Fantauzzi and A. Chandiramohan) from the Centre for Medicinal Cannabis Research at McMaster University.
Footnotes
This article has supplementary material available from openres.ersjournals.com.
Data availability: All data are available upon request.
Support statement: This work was supported by the McMaster Centre for Medicinal Cannabis Research and the Ontario Lung Association. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: M.F. Fantauzzi has nothing to disclose.
Conflict of interest: J.A. Aguiar has nothing to disclose.
Conflict of interest: B.J-M. Tremblay has nothing to disclose.
Conflict of interest: M.J. Mansfield has nothing to disclose.
Conflict of interest: T. Yanagihara has nothing to disclose.
Conflict of interest: A. Chandiramohan has nothing to disclose.
Conflict of interest: S. Revill has nothing to disclose.
Conflict of interest: M. Hyung Ryu has nothing to disclose.
Conflict of interest: C. Carlsten has nothing to disclose.
Conflict of interest: K. Ask has nothing to disclose.
Conflict of interest: M. Stämpfli has nothing to disclose.
Conflict of interest: A.C. Doxey has nothing to disclose.
Conflict of interest: J.A. Hirota has nothing to disclose.
References
- 1.World Health Organization World Drug Report 2018. United Nations publication SNEX. [Google Scholar]
- 2.Health Canada The Cannabis Survey: Methodological Report. http://epe.lac-bac.gc.ca/100/200/301/pwgsc-tpsgc/por-ef/health/2017/102-16-e/index.html Date last accessed: November 24, 2020. Date last updated: November 24, 2020 2017.
- 3.Health Canada Canadian Cannabis Survey. http://epe.lac-bac.gc.ca/100/200/301/pwgsc-tpsgc/por-ef/health/2018/006-18-e/index.html Date last accessed: November 24, 2020. Date last updated: November 24, 2020 2018.
- 4.Health Canada Canadian Cannabis Survey. https://www.canada.ca/en/health-canada/services/publications/drugs-health-products/canadian-cannabis-survey-2019-summary.html Date last accessed: November 24, 2020. Date last updated: November 24, 2020 2019.
- 5.Vachon L, FitzGerald MX, Solliday NH, et al. . Single-dose effects of marihuana smoke. Bronchial dynamics and respiratory-center sensitivity in normal subjects. N Engl J Med 1973; 288: 985–989. doi: 10.1056/NEJM197305102881902 [DOI] [PubMed] [Google Scholar]
- 6.Tashkin DP, Shapiro BJ, Frank IM. Acute pulmonary physiologic effects of smoked marijuana and oral (Delta)9 -tetrahydrocannabinol in healthy young men. N Engl J Med 1973; 289: 336–341. doi: 10.1056/NEJM197308162890702 [DOI] [PubMed] [Google Scholar]
- 7.Tashkin DP, Shapiro BJ, Frank IM. Acute effects of smoked marijuana and oral delta9-tetrahydrocannabinol on specific airway conductance in asthmatic subjects. Am Rev Respir Dis 1974; 109: 420–428. [DOI] [PubMed] [Google Scholar]
- 8.Tashkin DP, Shapiro BJ, Lee YE, et al. . Effects of smoked marijuana in experimentally induced asthma. Am Rev Respir Dis 1975; 112: 377–386. [DOI] [PubMed] [Google Scholar]
- 9.Tashkin DP, Shapiro BJ, Lee YE, et al. . Subacute effects of heavy marihuana smoking on pulmonary function in healthy men. N Engl J Med 1976; 294: 125–129. doi: 10.1056/NEJM197601152940302 [DOI] [PubMed] [Google Scholar]
- 10.Tan WC, Bourbeau J, Aaron SD, et al. . The effects of marijuana smoking on lung function in older people. Eur Respir J 2019; 54: 1900826. doi: 10.1183/13993003.00826-2019 [DOI] [PubMed] [Google Scholar]
- 11.Tan WC, Lo C, Jong A, et al. . Marijuana and chronic obstructive lung disease: a population-based study. CMAJ 2009; 180: 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fligiel SE, Roth MD, Kleerup EC, et al. . Tracheobronchial histopathology in habitual smokers of cocaine, marijuana, and/or tobacco. Chest 1997; 112: 319–326. [DOI] [PubMed] [Google Scholar]
- 13.Gong H Jr, Fligiel S, Tashkin DP, et al. . Tracheobronchial changes in habitual, heavy smokers of marijuana with and without tobacco. Am Rev Respir Dis 1987; 136: 142–149. [DOI] [PubMed] [Google Scholar]
- 14.Roth MD, Arora A, Barsky SH, et al. . Airway inflammation in young marijuana and tobacco smokers. Am J Respir Crit Care Med 1998; 157: 928–937. doi: 10.1164/ajrccm.157.3.9701026 [DOI] [PubMed] [Google Scholar]
- 15.Hancox RJ, Shin HH, Gray AR, et al. . Effects of quitting cannabis on respiratory symptoms. Eur Respir J 2015; 46: 80–87. doi: 10.1183/09031936.00228914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldwin GC, Tashkin DP, Buckley DM, et al. . Marijuana and cocaine impair alveolar macrophage function and cytokine production. Am J Respir Crit Care Med 1997; 156: 1606–1613. doi: 10.1164/ajrccm.156.5.9704146 [DOI] [PubMed] [Google Scholar]
- 17.Calignano A, Katona I, Desarnaud F, et al. . Bidirectional control of airway responsiveness by endogenous cannabinoids. Nature 2000; 408: 96–101. doi: 10.1038/35040576 [DOI] [PubMed] [Google Scholar]
- 18.Grassin-Delyle S, Naline E, Buenestado A, et al. . Cannabinoids inhibit cholinergic contraction in human airways through prejunctional CB1 receptors. Br J Pharmacol 2014; 171: 2767–2777. doi: 10.1111/bph.12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turcotte C, Blanchet MR, Laviolette M, et al. . Impact of cannabis, cannabinoids, and endocannabinoids in the lungs. Front Pharmacol 2016; 7: 317. doi: 10.3389/fphar.2016.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devane WA, Hanus L, Breuer A, et al. . Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992; 258: 1946–1949. doi: 10.1126/science.1470919 [DOI] [PubMed] [Google Scholar]
- 21.Stella N, Schweitzer P, Piomelli D. A second endogenous cannabinoid that modulates long-term potentiation. Nature 1997; 388: 773–778. doi: 10.1038/42015 [DOI] [PubMed] [Google Scholar]
- 22.Pertwee RG, Howlett AC, Abood ME, et al. . International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB(1) and CB(2). Pharmacol Rev 2010; 62: 588–631. doi: 10.1124/pr.110.003004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda LA, Lolait SJ, Brownstein MJ, et al. . Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 1990; 346: 561–564. doi: 10.1038/346561a0 [DOI] [PubMed] [Google Scholar]
- 24.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993; 365: 61–65. doi: 10.1038/365061a0 [DOI] [PubMed] [Google Scholar]
- 25.Zygmunt PM, Petersson J, Andersson DA, et al. . Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nature 1999; 400: 452–457. doi: 10.1038/22761 [DOI] [PubMed] [Google Scholar]
- 26.Ryberg E, Larsson N, Sjogren S, et al. . The orphan receptor GPR55 is a novel cannabinoid receptor. Br J Pharmacol 2007; 152: 1092–1101. doi: 10.1038/sj.bjp.0707460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiemstra PS, McCray PB Jr, Bals R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur Respir J 2015; 45: 1150–1162. doi: 10.1183/09031936.00141514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota JA, Knight DA. Human airway epithelial cell innate immunity: relevance to asthma. Curr Opin Immunol 2013; 24: 740–746. doi: 10.1016/j.coi.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 29.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 2011; 45: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huff RD, Carlsten C, Hirota JA. An update on immunologic mechanisms in the respiratory mucosa in response to air pollutants. J Allergy Clin Immunol 2019; 143: 1989–2001. [DOI] [PubMed] [Google Scholar]
- 31.Aguiar JA, Huff RD, Tse W, et al. . Transcriptomic and barrier responses of human airway epithelial cells exposed to cannabis smoke. Physiol Rep 2019; 7: e14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huff RD, Aguiar JA, Tse W, et al. . Effect of long-acting β-agonists/glucocorticoids on human airway epithelial cell cytokine, transcriptomic and oxidative stress responses to cannabis smoke. ERJ Open Res 2020; 6: 00265-2019. doi: 10.1183/23120541.00265-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galiegue S, Mary S, Marchand J, et al. . Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem 1995; 232: 54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x [DOI] [PubMed] [Google Scholar]
- 34.Corrado A, Battle M, Wise SK, et al. . Endocannabinoid receptor CB2R is significantly expressed in aspirin-exacerbated respiratory disease: a pilot study. Int Forum Allergy Rhinol 2018; 8: 1184–1189. doi: 10.1002/alr.22163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang VC, Kendall DA, Roberts RE. Delta(9)-tetrahydrocannabinol reverses TNF-α-induced increase in airway epithelial cell permeability through CB2 receptors. Biochem Pharmacol 2016; 120: 63–71. doi: 10.1016/j.bcp.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 36.Shang VC, O'Sullivan SE, Kendall DA, et al. . The endogenous cannabinoid anandamide increases human airway epithelial cell permeability through an arachidonic acid metabolite. Pharmacol Res 2016; 105: 152–163. doi: 10.1016/j.phrs.2016.01.023 [DOI] [PubMed] [Google Scholar]
- 37.Gurtler A, Kunz N, Gomolka M, et al. . Stain-free technology as a normalization tool in western blot analysis. Anal Biochem 2013; 433: 105–111. doi: 10.1016/j.ab.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 38.Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction 2009; 104: 518–532. doi: 10.1111/j.1360-0443.2009.02504.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol 2007; 14: 1347–1356. doi: 10.1016/j.chembiol.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrs WR, Blankman JL, Horne EA, et al. . The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat Neurosci 2010; 13: 951–957. doi: 10.1038/nn.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kano M, Ohno-Shosaku T, Hashimotodani Y, et al. . Endocannabinoid-mediated control of synaptic transmission. Physiol Rev 2009; 89: 309–380. doi: 10.1152/physrev.00019.2008 [DOI] [PubMed] [Google Scholar]
- 42.Galve-Roperh I, Rueda D, Gomez del Pulgar T, et al. . Mechanism of extracellular signal-regulated kinase activation by the CB(1) cannabinoid receptor. Mol Pharmacol 2002; 62: 1385–1392. doi: 10.1124/mol.62.6.1385 [DOI] [PubMed] [Google Scholar]
- 43.Henquet C, Rosa A, Krabbendam L, et al. . An experimental study of catechol-o-methyltransferase Val158Met moderation of delta-9-tetrahydrocannabinol-induced effects on psychosis and cognition. Neuropsychopharmacology 2006; 31: 2748–2757. doi: 10.1038/sj.npp.1301197 [DOI] [PubMed] [Google Scholar]
- 44.Watanabe K, Yamaori S, Funahashi T, et al. . Cytochrome P450 enzymes involved in the metabolism of tetrahydrocannabinols and cannabinol by human hepatic microsomes. Life Sci 2007; 80: 1415–1419. doi: 10.1016/j.lfs.2006.12.032 [DOI] [PubMed] [Google Scholar]
- 45.Powles T, te Poele R, Shamash J, et al. . Cannabis-induced cytotoxicity in leukemic cell lines: the role of the cannabinoid receptors and the MAPK pathway. Blood 2005; 105: 1214–1221. doi: 10.1182/blood-2004-03-1182 [DOI] [PubMed] [Google Scholar]
- 46.Kathuria S, Gaetani S, Fegley D, et al. . Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med 2003; 9: 76–81. doi: 10.1038/nm803 [DOI] [PubMed] [Google Scholar]
- 47.Onwuameze OE, Nam KW, Epping EA, et al. . MAPK14 and CNR1 gene variant interactions: effects on brain volume deficits in schizophrenia patients with marijuana misuse. Psychol Med 2013; 43: 619–631. doi: 10.1017/S0033291712001559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosier B, Lambert DM, Hermans E. Reciprocal influences of CB1 cannabinoid receptor agonists on ERK and JNK signalling in N1E-115 cells. FEBS Lett 2008; 582: 3861–3867. doi: 10.1016/j.febslet.2008.10.022 [DOI] [PubMed] [Google Scholar]
- 49.Dinh TP, Carpenter D, Leslie FM, et al. . Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA 2002; 99: 10819–10824. doi: 10.1073/pnas.152334899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cinar R, Iyer MR, Liu Z, et al. . Hybrid inhibitor of peripheral cannabinoid-1 receptors and inducible nitric oxide synthase mitigates liver fibrosis. JCI insight 2016; 1: e87336. doi: 10.1172/jci.insight.87336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castillo PE, Younts TJ, Chavez AE, et al. . Endocannabinoid signaling and synaptic function. Neuron 2012; 76: 70–81. doi: 10.1016/j.neuron.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruhaak LR, Felth J, Karlsson PC, et al. . Evaluation of the cyclooxygenase inhibiting effects of six major cannabinoids isolated from Cannabis sativa. Biol Pharm Bull 2011; 34: 774–778. doi: 10.1248/bpb.34.774 [DOI] [PubMed] [Google Scholar]
- 53.Kebir O, Lafaye G, Blecha L, et al. . ABCB1 C3435 T polymorphism is associated with tetrahydrocannabinol blood levels in heavy cannabis users. Psychiatry Res 2018; 262: 357–358. doi: 10.1016/j.psychres.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 54.Bondar A, Lazar J. The G protein Gi1 exhibits basal coupling but not preassembly with G protein-coupled receptors. J Biol Chem 2017; 292: 9690–9698. doi: 10.1074/jbc.M116.768127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hryhorowicz S, Walczak M, Zakerska-Banaszak O, et al. . Pharmacogenetics of cannabinoids. Eur J Drug Metab Pharmacokinet 2018; 43: 1–12. doi: 10.1007/s13318-017-0416-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim HR, Son BH, Lee SY, et al. . The role of p53 in marijuana smoke condensates-induced genotoxicity and apoptosis. Environ Health Toxicol 2012; 27: e2012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carey CE, Agrawal A, Zhang B, et al. . Monoacylglycerol lipase (MGLL) polymorphism rs604300 interacts with childhood adversity to predict cannabis dependence symptoms and amygdala habituation: evidence from an endocannabinoid system-level analysis. J Abnorm Psychol 2015; 124: 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sirrs S, van Karnebeek CD, Peng X, et al. . Defects in fatty acid amide hydrolase 2 in a male with neurologic and psychiatric symptoms. Orphanet J Rare Dis 2015; 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Petrocellis L, Ligresti A, Moriello AS, et al. . Effects of cannabinoids and cannabinoid-enriched cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br J Pharmacol 2011; 163: 1479–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Hilton DA, Hanemann CO, et al. . Cannabinoid receptor and N-acyl phosphatidylethanolamine phospholipase D: evidence for altered expression in multiple sclerosis. Brain Pathol 2011; 21: 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodruff PG, Boushey HA, Dolganov GM, et al. . Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA 2007; 104: 15858–15863. doi: 10.1073/pnas.0707413104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woodruff PG, Modrek B, Choy DF, et al. . T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med 2009; 180: 388–395. doi: 10.1164/rccm.200903-0392OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harvey BG, Heguy A, Leopold PL, et al. . Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med 2007; 85: 39–53. doi: 10.1007/s00109-006-0103-z [DOI] [PubMed] [Google Scholar]
- 64.Tilley AE, Harvey BG, Heguy A, et al. . Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2009; 179: 457–466. doi: 10.1164/rccm.200705-795OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carolan BJ, Heguy A, Harvey BG, et al. . Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res 2006; 66: 10729–10740. doi: 10.1158/0008-5472.CAN-06-2224 [DOI] [PubMed] [Google Scholar]
- 66.Ammous Z, Hackett NR, Butler MW, et al. . Variability in small airway epithelial gene expression among normal smokers. Chest 2008; 133: 1344–1353. doi: 10.1378/chest.07-2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carolan BJ, Harvey BG, De BP, et al. . Decreased expression of intelectin 1 in the human airway epithelium of smokers compared to nonsmokers. J Immunol 2008; 181: 5760–5767. doi: 10.4049/jimmunol.181.8.5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tilley AE, O'Connor TP, Hackett NR, et al. . Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS ONE 2011; 6: e22798. doi: 10.1371/journal.pone.0022798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raman T, O'Connor TP, Hackett NR, et al. . Quality control in microarray assessment of gene expression in human airway epithelium. BMC Genomics 2009; 10: 493. doi: 10.1186/1471-2164-10-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carolan BJ, Harvey BG, Hackett NR, et al. . Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers. Respir Res 2009; 10: 111. doi: 10.1186/1465-9921-10-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turetz ML, O'Connor TP, Tilley AE, et al. . Trachea epithelium as a “canary” for cigarette smoking-induced biologic phenotype of the small airway epithelium. Clin Transl Sci 2009; 2: 260–272. doi: 10.1111/j.1752-8062.2009.00129.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schembri F, Sridhar S, Perdomo C, et al. . MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA 2009; 106: 2319–2324. doi: 10.1073/pnas.0806383106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang G, Wang R, Ferris B, et al. . Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium. Respir Res 2010; 11: 150. doi: 10.1186/1465-9921-11-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strulovici-Barel Y, Omberg L, O'Mahony M, et al. . Threshold of biologic responses of the small airway epithelium to low levels of tobacco smoke. Am J Respir Crit Care Med 2010; 182: 1524–1532. doi: 10.1164/rccm.201002-0294OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaykhiev R, Otaki F, Bonsu P, et al. . Cigarette smoking reprograms apical junctional complex molecular architecture in the human airway epithelium in vivo. Cell Mol Life Sci 2011; 68: 877–892. doi: 10.1007/s00018-010-0500-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butler MW, Fukui T, Salit J, et al. . Modulation of cystatin A expression in human airway epithelium related to genotype, smoking, COPD, and lung cancer. Cancer Res 2011; 71: 2572–2581. doi: 10.1158/0008-5472.CAN-10-2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, Xu Z, Wang R, et al. . Genes associated with MUC5AC expression in small airway epithelium of human smokers and non-smokers. BMC Med Genomics 2012; 5: 21. doi: 10.1186/1755-8794-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boudewijn IM, Faiz A, Steiling K, et al. . Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression. Respir Res 2017; 18: 213. doi: 10.1186/s12931-017-0696-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao C, Tignor NL, Salit J, et al. . HEFT: eQTL analysis of many thousands of expressed genes while simultaneously controlling for hidden factors. Bioinformatics 2014; 30: 369–376. doi: 10.1093/bioinformatics/btt690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buro-Auriemma LJ, Salit J, Hackett NR, et al. . Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum Mol Genet 2013; 22: 4726–4738. doi: 10.1093/hmg/ddt326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hessel J, Heldrich J, Fuller J, et al. . Intraflagellar transport gene expression associated with short cilia in smoking and COPD. PLoS ONE 2014; 9: e85453. doi: 10.1371/journal.pone.0085453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walters MS, De BP, Salit J, et al. . Smoking accelerates aging of the small airway epithelium. Respir Res 2014; 15: 94. doi: 10.1186/s12931-014-0094-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J, Zuo WL, Fukui T, et al. . Smoking-dependent distal-to-proximal repatterning of the adult human small airway epithelium. Am J Respir Crit Care Med 2017; 196: 340–352. doi: 10.1164/rccm.201608-1672OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Christenson SA, Steiling K, van den Berge M, et al. . Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2015; 191: 758–766. doi: 10.1164/rccm.201408-1458OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou H, Brekman A, Zuo WL, et al. . POU2AF1 functions in the human airway epithelium to regulate expression of host defense genes. J Immunol 2016; 196: 3159–3167. doi: 10.4049/jimmunol.1502400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walters MS, Salit J, Ju JH, et al. . Waterpipe smoking induces epigenetic changes in the small airway epithelium. PLoS ONE 2017; 12: e0171112. doi: 10.1371/journal.pone.0171112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Billatos E, Faiz A, Gesthalter Y, et al. . Impact of acute exposure to cigarette smoke on airway gene expression. Physiol Genomics 2018; 50: 705–713. doi: 10.1152/physiolgenomics.00092.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O'Beirne SL, Shenoy SA, Salit J, et al. . Ambient pollution-related reprogramming of the human small airway epithelial transcriptome. Am J Respir Crit Care Med 2018; 198: 1413–1422. doi: 10.1164/rccm.201712-2526OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007; 23: 1846–1847. doi: 10.1093/bioinformatics/btm254 [DOI] [PubMed] [Google Scholar]
- 90.Dai M, Wang P, Boyd AD, et al. . Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005; 33: e175. doi: 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piccolo SR, Sun Y, Campbell JD, et al. . A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012; 100: 337–344. doi: 10.1016/j.ygeno.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huber W, Carey VJ, Gentleman R, et al. . Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015; 12: 115–121. doi: 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118–127. doi: 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 94.Leek JT, Johnson WE, Parker HS, et al. . The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28: 882–883. doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheadle C, Vawter MP, Freed WJ, et al. . Analysis of microarray data using Z score transformation. J Mol Diagn 2003; 5: 73–81. doi: 10.1016/S1525-1578(10)60455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stacklies W, Redestig H, Scholz M, et al. . pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007; 23: 1164–1167. doi: 10.1093/bioinformatics/btm069 [DOI] [PubMed] [Google Scholar]
- 97.Zhang R, Kim TK, Qiao ZH, et al. . Biochemical and mass spectrometric characterization of the human CB2 cannabinoid receptor expressed in Pichia pastoris--importance of correct processing of the N-terminus. Protein Exp Purif 2007; 55: 225–235. doi: 10.1016/j.pep.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 98.Piotrowska Z, Niezgoda M, Lebkowski W, et al. . Sex differences in distribution of cannabinoid receptors (CB1 and CB2), S100A6 and CacyBP/SIP in human ageing hearts. Biol Sex Differ 2018; 9: 50. doi: 10.1186/s13293-018-0209-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kicic A, Sutanto EN, Stevens PT, et al. . Intrinsic biochemical and functional differences in bronchial epithelial cells of children with asthma. Am J Respir Crit Care Med 2006; 174: 1110–1118. doi: 10.1164/rccm.200603-392OC [DOI] [PubMed] [Google Scholar]
- 100.Steiling K, van den Berge M, Hijazi K, et al. . A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med 2013; 187: 933–942. doi: 10.1164/rccm.201208-1449OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuo CS, Pavlidis S, Loza M, et al. . A Transcriptome-driven analysis of epithelial brushings and bronchial biopsies to define asthma phenotypes in U-BIOPRED. Am J Respir Crit Care Med 2017; 195: 443–455. doi: 10.1164/rccm.201512-2452OC [DOI] [PubMed] [Google Scholar]
- 102.Postma DS. Gender differences in asthma development and progression. Gend Med 2007; 4: Suppl. B, S133–S146. doi: 10.1016/S1550-8579(07)80054-4 [DOI] [PubMed] [Google Scholar]
- 103.Sorheim IC, Johannessen A, Gulsvik A, et al. . Gender differences in COPD: are women more susceptible to smoking effects than men? Thorax 2010; 65: 480–485. doi: 10.1136/thx.2009.122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abdallah SJ, Smith BM, Ware MA, et al. . Effect of vaporized cannabis on exertional breathlessness and exercise endurance in advanced chronic obstructive pulmonary disease. A randomized controlled trial. Ann Am Thorac Soc 2018; 15: 1146–1158. doi: 10.1513/AnnalsATS.201803-198OC [DOI] [PubMed] [Google Scholar]
- 105.Bailey KL, Wyatt TA, Katafiasz DM, et al. . Alcohol and cannabis use alter pulmonary innate immunity. Alcohol 2019; 80: 131–138. doi: 10.1016/j.alcohol.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ayakannu T, Taylor AH, Marczylo TH, et al. . The endocannabinoid system and sex steroid hormone-dependent cancers. Int J Endocrinol 2013; 2013: 259676. doi: 10.1155/2013/259676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pagotto U, Marsicano G, Cota D, et al. . The emerging role of the endocannabinoid system in endocrine regulation and energy balance. Endocr Rev 2006; 27: 73–100. doi: 10.1210/er.2005-0009 [DOI] [PubMed] [Google Scholar]
- 108.Drehmer MN, Muniz YCN, Marrero AR, et al. . Gene expression of ABHD6, a key factor in the endocannabinoid system, can be modulated by female hormones in human immune cells. Biochem Genet 2019; 57: 35–45. doi: 10.1007/s10528-018-9871-8 [DOI] [PubMed] [Google Scholar]
- 109.Klein TW. Cannabinoid-based drugs as anti-inflammatory therapeutics. Nat Rev Immunol 2005; 5: 400–411. doi: 10.1038/nri1602 [DOI] [PubMed] [Google Scholar]
- 110.McGarvey LP, Butler CA, Stokesberry S, et al. . Increased expression of bronchial epithelial transient receptor potential vanilloid 1 channels in patients with severe asthma. J Allergy Clin Immunol 2014; 133: 704–712 e704. doi: 10.1016/j.jaci.2013.09.016 [DOI] [PubMed] [Google Scholar]
- 111.Yocum GT, Chen J, Choi CH, et al. . Role of transient receptor potential vanilloid 1 in the modulation of airway smooth muscle tone and calcium handling. Am J Physiol Lung Cell Mol Physiol 2017; 312: L812–L821. doi: 10.1152/ajplung.00064.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo J, Schumacher M, Scherer A, et al. . A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J 2010; 10: 278–291. doi: 10.1038/tpj.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
FIGURE S1 Detection of CNR1 and CNR2 promoter activity in human lung tissues and lung epithelial cell samples. Heat map of FANTOM5 CAGE promoter activity data for CNR1, CNR2, and CDH1 (positive control) are shown for samples related to lung tissues (n=28). Heat map colour is proportional to promoter activity, depicted as log10-transformed normalized transcripts per million (TPM). Grey represents no detection. 00128-2020.figureS1 (411.2KB, pdf)