Abstract
Background:
Determining the site of origin for metastatic well-differentiated neuroendocrine tumors (WDNETs) is challenging, and immunohistochemical (IHC) profiles do not always lead to a definitive diagnosis. We sought to determine if a deep-learning convolutional neural network (CNN) could improve upon established IHC profiles in predicting the site of origin in a cohort of WDNETs from the common primary sites.
Materials and Methods:
Hematoxylin and eosin (H&E)-stained tissue microarrays (TMAs) were created using 215 WDNETs arising from the known primary sites. A CNN trained and tested on 60% (n = 130) and 40% (n = 85) of these cases, respectively. One hundred and seventy-nine cases had TMA tissue remaining for the IHC analysis. These cases were stained with IHC markers pPAX8, CDX2, SATB2, and thyroid transcription factor-1 (markers of pancreas/duodenum, ileum/jejunum/duodenum, colorectum/appendix, and lung WDNET sites of origin, respectively). The CNN diagnosis was deemed correct if it designated a majority or plurality of the tumor area as the known site of origin. The IHC diagnosis was deemed correct if the most specific marker for a particular site of origin met an H-score threshold determined by two pathologists.
Results:
When all cases were considered, the CNN correctly identified the site of origin at a lower rate compared to IHC (72% vs. 82%, respectively). Of the 85 cases in the CNN test set, 66 had sufficient TMA material for IHC stains, thus 66 cases were available for a direct case-by-case comparison of IHC versus CNN. The CNN correctly identified 70% of these cases, while IHC correctly identified 76%, a finding that was not statistically significant (P = 0.56).
Conclusion:
A CNN can identify WDNET site of origin at an accuracy rate close to the current gold standard IHC methods.
Keywords: Artificial Intelligence, convolutional neural network, deep learning, neuroendocrine tumor
INTRODUCTION
Recent advances in the digital pathology spurred by the Food and Drug Administration (FDA) approval of whole-slide imaging for the primary diagnosis have led to the development of numerous computer image analysis systems and deep-learning algorithms, which have opened the door to computer-assisted diagnostics in the pathology. Prior studies have established the safe use of digital pathology in primary diagnostic reporting generally[1,2,3] and in the gastrointestinal pathology specifically.[4]
Many computer image analysis systems are based on machine learning, a term coined to express a program that can “learn” without explicit programing to achieve a task. While this may be complex, a machine learning system is often simply creating a function for a line of best fit, but in the multiple dimensions. These systems can be trained in supervised learning or unsupervised learning. Supervised learning consists of providing the system with both the data (inputs) and the result (output). As input parameters change (for example, the color or granularity of a cell's cytoplasm), the function alters the coefficients to create a more accurate function. In anatomic pathology, the system is provided with the visual data or images of a tumor and given the diagnosis (a training set of images). As the system is shown more examples of a tumor, the model adjusts its function to match the provided data. When the system is then given input data without the result (a test set of images), the model can execute the same function and come up with an output that is, ideally, accurate.
Deep learning is a subset of machine learning and is based on interconnected simple processing nodes, similar to neurons in a brain. These densely connected nodes typically operate on a feed forward (mono-directional data flow) system with the data passing through the several layers of nodes. Each node receives data from multiple nodes from the layer above it. The system then multiplies the data by the weight coefficient (that was determined in training) and sums these values. If the sum of values exceeds a threshold (also determined in training), the data propagate to the next layer (just as action potentials do in neurons); if not, it passes no data to its outgoing connections. In the training of a deep-learning system, each of the coefficients and thresholds are randomized, and data are fed into the initial layer. It then passes through the system and the output is compared to the “correct” output. The coefficients and thresholds are then adjusted over many iterations as the function evolves. As such, deep-learning algorithms have been applied to anatomic pathology due to the visual analysis that is intrinsic to anatomic pathology.
A common type of deep learning used in the image analysis is a convolutional neural network (CNN). CNNs are regularized feedforward artificial neural networks. They serve as solutions to overfitting, where the analysis fits too closely to the exact data set and is unable to fit additional data. For example, if a nonregularized system was trained to recognize a Coke can as soda, it could exclude a Pepsi can due to overfitting. CNNs have shown promise in both neoplastic and nonneoplastic anatomic pathology and have been particularly well studied in breast pathology; focused on identification, grade, hormone immunohistochemical (IHC) status, and lymph node metastases with impressive results.[1,2,5,6,7,8,9,10,11,12]
Well-differentiated neuroendocrine tumors (WDNETs) are a diverse group of tumors that share a neural crest origin and a similar histologic appearance. WDNETs can demonstrate a wide variety of architectural patterns, including solid, nested, trabecular, insular, glandular, or cribriform, often in a fibrous or hyalinized stroma. WDNETs usually have uniform nuclei, finely stippled chromatin (salt-and-pepper), granular cytoplasm, and a monotonous appearance.[13]
A review of the Surveillance, Epidemiology, and End Results Program of the National Cancer Institute found: “The greatest incidence of carcinoids were the gastrointestinal tract (67.5%) and the bronchopulmonary system (25.3%). Within the gastrointestinal tract, most carcinoid tumors occurred in the small intestine (41.8%), rectum (27.4%), and stomach (8.7%).”[14] While WDNETs from the disparate locations can have very similar morphologic appearances, they vary significantly in both response to site-specific therapy and prognosis.[15,16] The latter can especially be site dependent, as the majority of WDNET metastases of occult origin come from the ileum, jejunum, or pancreas. This contrasts with most gastric, appendiceal, and small (<1 cm) rectal WDNETs, which are usually clinically indolent.
Metastatic WDNETs behave differently depending on the site of origin. For example, small intestinal WDNETs are associated with the greater incidence of obstruction and perforation.[17,18,19,20] In addition, the site-specific response to different chemotherapeutic regiments is well documented, with cytotoxic chemotherapies showing little response in midgut WDNETs, and targeted biologic agents (e.g., Sunitinib) demonstrating promising the results in pancreatic WDNETs but not in midgut WDNETs.[19,21] Molecular studies have demonstrated variant expression of key signaling pathway components between WDNETs of different sites, which explains this biologically divergent behavior.[15,21,22,23,24]
When metastatic WDNETs are encountered and no primary site can be identified, this can pose a significant diagnostic challenge. Despite multiple modern imaging modalities, 21% of WDNETs are occult.[25] IHC has been employed to confirm neuroendocrine differentiation and identify the site of origin.
General IHC markers of the neuroendocrine phenotype include synaptophysin, chromogranin, HISL-19, neuron specific enolase, the proprotein convertases PC2 and PC3, the lymphoreticular epitope Leu-7, and the neural cell adhesion molecule (or CD56).[26,27,28] Once a neuroendocrine phenotype is identified, additional IHC can be used to further characterize a likely site of origin. The most common WDNET IHC panel pathologists employ include caudal-type homeobox 2 (CDX2) for midgut, thyroid transcription factor-1 (TTF1) for lung, polyclonal Paired Box Gene 8 (pPAX8) or PAX6 for pancreas, and Special AT-Rich Sequence Binding Protein 2 (SATB2) for colorectal tumors.[29,30,31,32] However, some markers perform better than others. CDX2 is arguably the most sensitive and specific of the lot, with strong/diffuse staining seen in up to 90% of jejunoileal WDNETs.[31,33] Weak-to-moderate CDX2 expression can be found in duodenal and pancreatic primaries, but this usually occurs in a minority of cases (approximately 30% of duodenal primaries and 15% of pancreatic primaries).[31,33] TTF1, on the other hand, suffers in diagnostic sensitivity (30%–80% depending on the study) but is highly specific for pulmonary origin.[29,31] Polyclonal PAX8 cross-reacts with PAX6, a transcription factor critical to pancreatic islet development, and this feature has been exploited to utilize pPAX8 as a marker of pancreatic WDNETs, though it has only been shown to be 55% sensitive and duodenal primaries may express pPAX8 in up to 79% of cases.[31] Finally, SATB2 has recently demonstrated an 83%/93% sensitivity/specificity pairing for rectal WDNETs, making this a promising marker of lower gastrointestinal tract origin.[33] Given the wide-ranging accuracy metrics for current IHC markers, there is a need for improved identification of WDNET site of origin when pathologists are faced with a metastasis from an unknown primary. CNNs have proven capable in a wide range of challenging morphologic assessments and may therefore have a role in this difficult task.
MATERIALS AND METHODS
This study was performed in accordance with the Institutional Review Board requirements at the University of New Mexico (Albuquerque, New Mexico).
We obtained 215 primary WDNET formalin fixed paraffin-embedded tissue blocks from 215 individual subjects with known sites of origin from our archives. The tumors' origins were as follows: appendix (n = 13), colorectum (n = 45), duodenum (n = 21), lung (n = 25), pancreas (n = 38), small bowel (n = 43, jejunum and ileum), and stomach (n = 30). The corresponding slides from all blocks were reviewed by two gastrointestinal pathologists, who confirmed the diagnosis and selected areas for the inclusion in tissue microarrays (TMAs). A total of seven WDNET TMAs were created and two 1.5 mm cores were placed for each case. The 1.5 mm cores were removed from the donor blocks and placed into the TMA blocks using the Chemicon Tissue Micro Arrayer ATA-100. Slides were then created for each TMA block and these were stained with both hematoxylin and eosin (H&E) and the following IHC antibodies: PPAX8 (Proteintech 10336–1-AP), CDX2 (Abcam ab76541), SATB2 (Cell Marque 384R), and TTF1 (Novocastral/Leica NCL-L-TTF1).
For IHC staining, all TMA blocks were sectioned at 4–5 μ, and the sections were mounted on charged (+) slides. Slides were baked at 60°C for 60 min. The Ventana Discovery platform was used for deparaffinization and staining. Prior to the application of the antibodies, slides were treated with Discovery Cell Conditioner #1 (Ventana 950–500) for 64 min at 100°C (pPAX8), for 32 min at 100°C (CDX2 and TTF1), and for 36 min at 95°C (SATB2). Each antibody dilution/incubation time and temperature was as follows: PPAX8 = 1:500/32 min at 36°C, CDX2 = 1:1000/32 min at 36°C, SATB2 = 1:25/16 min at 37°C, TTF1 = 1:200/24 min at 37°C. All antibody incubation was followed by DAB CM detection. Slides were counterstained with hematoxylin (Ventana 760–2021) and bluing solution (Ventana 760–2037), removed from the autostainer and hand-coverslipped.
The stained slides (IHC and H&E) were then digitized by scanning with the Aperio VERSA 200 slide scanner (Leica, Wetzlar, Germany) at ×20 and imported into a computer containing a 12 Core, 2.2 GHz Intel Xeon Processor E5-2650 chip and a Nvidia Titan XP graphics card. HALO-AI image analysis software (Indica Labs, Albuquerque, New Mexico) was used to perform the training and testing on H and E slides only. HALO-AI is a CNN that uses a fully convolutional version of the VGG architecture.
All 215 H&E-stained cases were then manually annotated on the software. This was performed by the consensus of two gastrointestinal pathologists and one pathology resident viewing the digitized TMAs. The tissue was annotated to include only the tumor cells and nonneuroendocrine intratumoral cells inherent to the tumor (e.g., tumor-associated endothelial cells and indistinct stroma) and to exclude any areas that might be informative as to the site of origin [Figure 1] (e.g., native background epithelium and smooth muscle). In the cases which proved insufficient on the TMAs, either due to the lack of tumor or other artifact, the most representative H and E slides from the original resection or biopsy were scanned and annotated separately, with the total annotation area not exceeding that of the TMA cores. Only six cases (2 from stomach, 1 from pancreas, 1 from duodenum, 1 from colorectal, and 1 from appendix) required TMA core replacement with biopsy/resection tissue.
Figure 1.
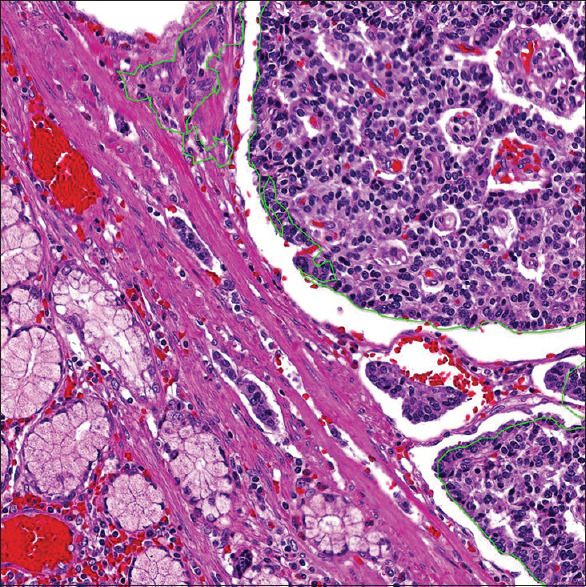
A duodenal example of how the WDNETs was annotated for CNN analysis. The green outline includes only the tumor cells and indistinct stromal/vascular tissue for analysis. The normal duodenal epithelium and smooth muscle are not outlined and are thus excluded from all CNN analysis. WDNETs: Well-differentiated neuroendocrine tumors, CNN: Convolutional neural network
Of the 215 cases, 130 (60%) were randomly selected for the inclusion in the CNN training set and 85 (40%) were randomly included in the test set [Table 1]. Training was performed by HALO-AI only on the annotated areas of the TMAs, consisting of the aggregate of the two cores or biopsy/resection slides, broken down into “image patches” of 400 × 400 pixels (where 1 pixel = 1 μm) at resolutions corresponding to a × 5.5, a × 10, and a × 20 digital view magnification. The image patches were generated by the automated selection of random points and cropping a patch around the point. The patches were further augmented with random rotations and random shifts to hue, saturation, contrast, and brightness. Training was performed for a total of 325,790 analytic iterations using RMSProp at × 5.5, 451,176 at × 10, and 534,805 at × 20. During these iterations, the algorithm would continuously change the node-weighted values based off the annotation of the two gastrointestinal pathologists. The HALO-AI operator stopped the algorithm once an error rate/cross entropy rate of <0.01 was achieved for the × 5.5 and × 10 and < 0.02 at × 20.
Table 1.
Well-differentiated neuroendocrine tumors: HALO-AI training and testing
| Site of origin (n) | Training set | Test set |
|---|---|---|
| Appendix (13) | 8 | 5 |
| Colorectal (45) | 27 | 18 |
| Duodenum (21) | 13 | 8 |
| Lung (25) | 15 | 10 |
| Pancreas (38) | 23 | 15 |
| Small Bowel (43) | 26 | 17 |
| Stomach (30) | 18 | 12 |
| Total (215) | 130 | 85 |
After training was completed, for the test set, the annotated areas (performed on the two TMA cores for each case) were arbitrarily designated with colored digital labels according to the HALO-AI site of origin diagnosis. Appendiceal origin was given a magenta digital label, colorectal origin a brown label, pancreatic origin a yellow label [Figure 2], duodenal origin a bright green label [Figure 3], lung origin a blue label, small bowel origin a red label, and stomach origin an olive-green label. HALO-AI analyzed these cases blindly, assigning each annotated region (“patch”) a likelihood score for that area, which corresponded to the most probable diagnostic call. The output for each test case was ultimately X% area (WDNET favored site of origin) versus Y% area (additional preferred site of origin) based on these calls. These percentages are termed area distribution (AD) in this paper, with each case receiving a mosaic of %AD calls, comprised of various percentages adding up to 100%.
Figure 2.
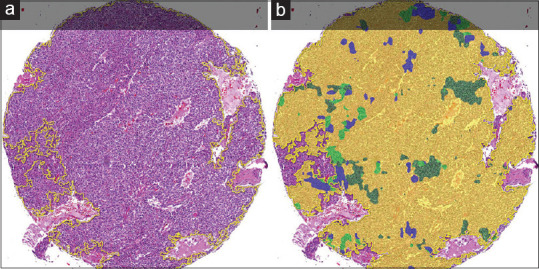
An entire pancreatic WDNET TMA core (a) and its corresponding HALO-AI colored AD (b). The majority of the tumor was assigned a yellow AD by HALO-AI. Because yellow corresponded to the pancreas site of origin, the CNN was credited with a correct diagnosis. Minority ADs in this case are the stomach (olive green), duodenum (bright green), and lung (blue). WDNET: Well-differentiated neuroendocrine tumor, CNN: Convolutional neural network, TMA: Tissue microarray, AD: Area distribution
Figure 3.
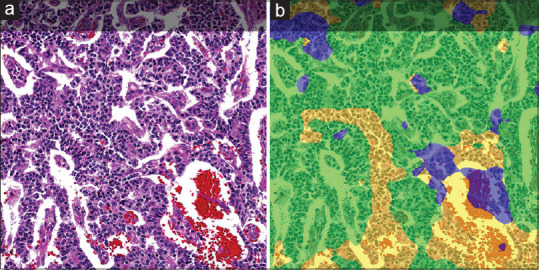
A duodenal WDNET (a) and its corresponding HALO-AI-colored AD (b). The majority of the tumor was assigned a bright green AD by HALO-AI. Because bright green corresponded to the duodenum site of origin, the CNN was credited with a correct diagnosis. Minority ADs in this case are the pancreas (yellow) and lung (blue). WDNET: Well-differentiated neuroendocrine tumor, CNN: Convolutional neural network, AD: Area distribution
The ADs were compared against the known site of origin to determine if the correct diagnosis was achieved. If the majority or plurality of a sample was assigned the correct AD by HALO-AI, this was considered a correct diagnosis in the binary correct or incorrect classification. Majority was defined as >50% of the sample called correctly and plurality was defined as the greatest AD called overall if a majority AD was not achieved.
The IHC-stained slides were annotated in the similar fashion with representative areas of tumor selected for cytonuclear analysis by the HALO Cytonuclear Analysis Module (this module is not a CNN), with the cut-off staining optical density values (negative, low, moderate, and strong) determined by a consensus of both GI pathologists. The total number of cases for IHC analysis was reduced as the IHC was only performed on cases with adequate TMAs (the biopsy/resection-only cases were excluded due to financial limitations). In addition, some samples within TMAs were exhausted during preparation prohibiting analysis for all four IHC stains. Accordingly, 179 cases remained for IHC analysis.
H-score (a measure of nuclear immunoreactivity, determined by the calculation 3 × percentage of strongly staining nuclei + 2 × percentage of moderately staining nuclei + 1 × percentage of weakly staining nuclei, giving a range of 0–300)[29] cutoffs for positivity were designated at ≥60 for pPAX8, ≥30 for TTF1, ≥20 for SATB2, and ≥10 for CDX2 by a consensus of the GI pathologists as values that represent “real-world positivity”[30,31,32] [Figure 4]. To elaborate, the pathologists found that the pPAX8 displayed the weakest nuclear staining and most nonspecific background staining, resulting in the highest H-score cutoff compared to the other stains. Conversely, the pathologists found the CDX2 stain was the most crisp, resulting in the lowest H-score cutoff. All H-score cutoffs were established without knowledge of the CNN results.
Figure 4.
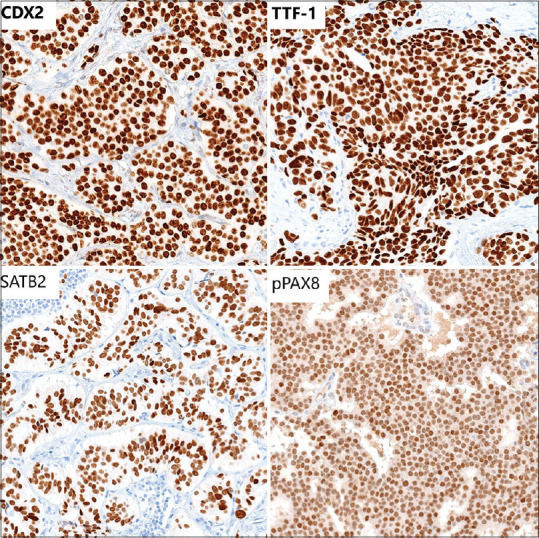
A composite image of representative IHC stains. CDX2, TTF1, and SATB2 demonstrated relatively crisp expression. Hence, they required lower H-score cutoffs to determine a true positive result. The pPAX8 stain displayed the weakest nuclear expression and showed some nonspecific cytoplasmic positivity, necessitating a higher H-score cutoff to determine a true positive result. IHC: Immunohistochemistry
The H-score values were tabulated and compared to the IHC characteristics of specific tumor origin as previously described in the literature, except in the case of gastric WDNETs which are not well described.[33] The IHC diagnosis was deemed correct if the most specific marker for a particular site of origin met the appropriate H-score cutoff and was deemed incorrect if it did not meet this cutoff. This was done to ensure that nonspecific staining for an additional incorrect IHC marker could not be used to artificially reduce the IHC accuracy calculations. For example, because CDX2 can show variable expression in pancreatic and appendiceal/colorectal WDNETs but is not as specific for these sites as pPAX8 and SATB2, respectively; CDX2 expression was considered irrelevant in these sites, and the more specific markers (pPAX8 and SATB2) were given precedence. In such an example, a pancreatic tumor that was positive for pPAX8 and CDX2 would be deemed correct by IHC due to the specific pPAX8 expression. Alternatively, a colorectal WDNET that was negative for SATB2 but positive for CDX2 would be deemed incorrect by IHC because this combination is more common in jejunoileal tumors. Accordingly, IHC characteristics used to define a correct diagnosis for each primary site were as follows: Appendix/Colorectum = SATB2+, Lung = TTF1+, Pancreas = pPAX8+, Small Bowel (Ileum/Jejunum) = CDX2+, Duodenum = CDX2 + or pPAX8+.
RESULTS
HALO-AI
The CNN performed similarly at all magnifications with insignificant performance improvement as magnification increased. The 10x classifier performance was thus chosen as the optimal magnification because it offered a good balance of speed and accuracy when compared to the × 5 and × 20 classifiers. HALO-AI correctly identified 72% of all cases in the test set [Table 2]. As Table 2 demonstrates, the CNN performed best with colorectal cases, correctly identifying 94%. The small bowel was likewise accurate at 88% correct identification. The stomach and duodenum performed similarly at 81% and 80%, respectively. Lung and appendix were both correctly classified 60% of the time. The worst performer was the pancreas at 31% correct, which prompted a return to the original materials to confirm that no annotation errors had occurred. No errors were identified.
Table 2.
HALO-AI test set accuracy, all available cases
| Site of origin (n) | Correct HALO-AI diagnosis, n (%) |
|---|---|
| Appendix (5) | 3 (60) |
| Colorectal (18) | 17 (94) |
| Duodenum (8) | 6 (80) |
| Lung (10) | 6 (60) |
| Pancreas (16) | 5 (31) |
| Small Bowel (17) | 15 (88) |
| Stomach (11) | 9 (81) |
| Total (85) | 61 (72) |
Immunohistochemistry
The IHC accuracy of the 179 WDNETs available for staining is summarized in Table 3. The gastric cases were excluded from the IHC analysis as no gastric WDNET IHC profile is well-established. The IHC performed well, identifying 146 of the 179 WDNET sites of origin correctly (82%). With our established H-score cutoffs, the IHC performed best on the colorectal cases (98% correct) and the small bowel cases (100% correct), similar to HALO-AI which also performed best at these sites of origin. The duodenal and appendiceal WDNETs were correctly identified in 94% and 92% of the cases, respectively. The lung site of the origin was identified in 82% of the WDNETs. Once again, the pancreas proved difficult to identify with only 14 of the 40 (35%) cases demonstrating sufficient pPAX8 expression to be considered correctly classified.
Table 3.
Accuracy of immunohistochemistry, all available cases
| Site of origin with paired specific IHC marker (n) | IHC expression ≥H-score cutoff (correct IHC diagnosis), n (%) |
|---|---|
| Appendix/SATB2 (13) | 12 (92) |
| Colorectal/SATB2 (42) | 41 (98) |
| Duodenum/CDX2 or pPAX8 (18) | 17 (94) |
| Lung/TTF1 (22) | 18 (82) |
| Pancreas/pPAX8 (40) | 14 (35) |
| Small bowel/CDX2 (44) | 44 (100) |
| Total (179) | 146 (82) |
IHC: Immunohistochemistry
HALO-AI versus immunohistochemistry
Of the 85 cases in the HALO-AI test set, 66 had sufficient TMA material for IHC stains. Thus, 66 cases were available for a direct case-by-case comparison of IHC versus CNN to determine which method was most accurate in identifying site of origin. In this direct comparison, IHC and HALO-AI performances were similar. The IHC panel correctly identified 50 of the 66 cases (76%) while HALO-AI correctly identified 46 of the same 66 cases (70%) [Table 4]. Fisher's exact test found no significant difference between these accuracy rates (P = 0.56). Regarding subgroup analysis, IHC performed better than HALO-AI at all primary sites except pancreas [Table 4]; though IHC's diagnostic advantage only amounted to 1–2 cases per site of origin. Interestingly, HALO-AI correctly identified 31% of pancreatic tumors compared to 13% for IHC in this head-to-head comparison.
Table 4.
Accuracy of HALO-AI versus immunohistochemistry, head-to-head cases
| Site of origin (n) | HALO-AI accuracy, n (%) | IHC accuracy, n (%) |
|---|---|---|
| Appendix (5) | 3 (60) | 5 (100) |
| Colorectal (15) | 14 (93) | 15 (100) |
| Duodenum (6) | 5 (83) | 6 (100) |
| Lung (8) | 5 (63) | 6 (75) |
| Pancreas (16) | 5 (31) | 2 (13) |
| Small bowel (16) | 14 (88) | 16 (100) |
| Total (66) | 46 (70) | 50 (76) |
IHC: Immunohistochemistry
DISCUSSION
WDNETs are enigmatic neoplasms in that their morphologic appearances are similar across primary sites but their biologic behavior is quite different and often depends on their site of origin. Not surprisingly, therapeutic treatments for WDNETs are increasingly becoming site dependent. It is the pathologist's job to identify a primary site when faced with a metastatic WDNET of unknown origin. To accomplish this, pathologists rely on IHC stains. These primarily include pPAX8 for the pancreas, CDX2 for jejunoileum, TTF1 for lung, and SATB2 for colorectum/appendix. While some of these stains exhibit adequate sensitivity as discussed previously, others, such as pPAX8 and TTF1, often fall short. Accordingly, we hypothesized that a CNN could potentially identify subtle morphologic differences in WDNETs from the various sites and possibly improve upon the current clinical gold standard (IHC) in identifying the site of origin.
Our CNN (HALO-AI) trained on 130 WDNETs from seven different primary sites. We then tested the algorithm on 85 subsequent cases, finding a site-of-origin accuracy rate of 72% [Table 2]. This was somewhat comparable to the IHC accuracy rate of 82% [Table 3], but was not an improvement.
The vast majority of WDNETs to present as metastases of unknown origin would be expected to originate from either the small bowel or the pancreas.[34] In looking at these site-specific accuracy rates, the CNN again demonstrated slightly inferior performance compared to IHC as HALO-AI correctly identified 31% of pancreatic tumors and 88% of small-bowel tumors; while IHC correctly identified 35% and 100%, respectively [Tables 2 and 3]. However, the aforementioned CNN/IHC site-of-origin accuracy rates were derived from all cases available to each testing method. Nineteen of the cases in the initial HALO-AI test set ultimately did not have sufficient TMA material for IHC stains, and therefore, our initial comparisons do not reflect a true case-by-case test of IHC vs. CNN. When we included only those cases that were interrogated by both IHC and the CNN [n = 66, Table 4], we discovered that their respective accuracy rates compared so well (70% HALO-AI vs. 76% IHC) that the small difference was considered insignificant (P = 0.56). Regarding the most clinically significant WDNET sites of origin, pancreas and small bowel, HALO-AI actually outperformed IHC in the pancreatic tumors (31% HALO-AI vs. 13% IHC) but remained inferior to IHC in the small bowel tumors (88% HALO-AI vs. 100% IHC).
In our experience working with CNN algorithms, the HALO-AI performance in this task is impressive as the algorithm's site-of-origin accuracy rate on a direct case-by-case comparison was not significantly different than the existing clinical gold standard IHC panel, despite an admittedly limited number of site-specific WDNETs available for training. In addition, one could argue that our method of determining correct IHC identification overestimates the accuracy of IHC, given that an incorrect marker was not counted against the IHC method if the more specific marker was also positive. CNN algorithms depend on the large image sets to hone their diagnostic accuracy. Accordingly, our data suggest strong potential for such algorithms to accurately identify WDNET site of origin that could rival IHC if trained on many more cases. We suggest this as a further avenue of research for those institutions that archive more of these tumors. In addition, we acquiesce that a more robust study would include only metastatic WDNETs as opposed to the primary tumors on which we relied. However, this would require large repositories of metastatic WDNETs with clearly defined sites of origin and could likely only be accomplished at the largest academic medical centers with vast WDNET repositories.
There is a myriad of benefits to CNN diagnosis over IHC including: Reduced turnaround time (as no staining would be required) and decreased recurring costs (the upfront cost of the CNN and scanning equipment could potentially be lower than IHC panels over time). In addition, the CNN could be configured to continually improve over time to further increase its accuracy while the accuracy of IHC panels generally remains static unless a new stain comes to the market.
CONCLUSION
We have demonstrated that a CNN can identify WDNET site of origin at an accuracy rate close to the current clinical gold standard IHC methods. We draw this conclusion based on the comparable CNN/IHC accuracy rates despite a limited number of cases available for CNN training.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2020/11/1/32/297571
REFERENCES
- 1.Gavrielides MA, Conway C, O'Flaherty N, Gallas BD, Hewitt SM. Observer performance in the use of digital and optical microscopy for the interpretation of tissue-based biomarkers. Anal Cell Pathol (Amst) 2014;2014:157308. doi: 10.1155/2014/157308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavrielides MA, Gallas BD, Lenz P, Badano A, Hewitt SM. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch Pathol Lab Med. 2011;135:233–42. doi: 10.1043/1543-2165-135.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology – New tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loughrey MB, Kelly PJ, Houghton OP, Coleman HG, Houghton JP, Carson A, et al. Digital slide viewing for primary reporting in gastrointestinal pathology: A validation study. Virchows Arch. 2015;467:137–44. doi: 10.1007/s00428-015-1780-1. [DOI] [PubMed] [Google Scholar]
- 5.Bejnordi BE, Zuidhof G, Balkenhol M, Hermsen M, Bult P, Ginneken BV, et al. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J Med Imaging (Bellingham) 2017;4:044504. doi: 10.1117/1.JMI.4.4.044504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masmoudi H, Hewitt SM, Petrick N, Myers KJ, Gavrielides MA. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans Med Imaging. 2009;28:916–25. doi: 10.1109/TMI.2009.2012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NN, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS One. 2017;12:e0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejnordi BE, Veta M, Van Diest PJ, Ginneken BV, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol. 2018;42:1636–46. doi: 10.1097/PAS.0000000000001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller B, Chen W, Gavrielides MA. Quantitative assessment and classification of tissue-based biomarker expression with color content analysis. Arch Pathol Lab Med. 2012;136:539–50. doi: 10.5858/arpa.2011-0195-OA. [DOI] [PubMed] [Google Scholar]
- 12.Keay T, Conway CM, O'Flaherty N, Hewitt SM, Shea K, Gavrielides MA. Reproducibility in the automated quantitative assessment of HER2/neu for breast cancer. J Pathol Inform. 2013;4:19. doi: 10.4103/2153-3539.115879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearse AG. The cytochemistry and ultrastructure of polypeptide hormone-producing cells of the APUD series and the embryologic, physiologic and pathologic implications of the concept. J Histochem Cytochem. 1969;17:303–13. doi: 10.1177/17.5.303. [DOI] [PubMed] [Google Scholar]
- 14.Modlin IM, Lye KD, Kidd M. A 5-decade analysis of 13,715 carcinoid tumors. Cancer. 2003;97:934–59. doi: 10.1002/cncr.11105. [DOI] [PubMed] [Google Scholar]
- 15.Capdevila J, Meeker A, García-Carbonero R, Pietras K, Astudillo A, Casanovas O, et al. Molecular biology of neuroendocrine tumors: From pathways to biomarkers and targets. Cancer Metastasis Rev. 2014;33:345–51. doi: 10.1007/s10555-013-9468-y. [DOI] [PubMed] [Google Scholar]
- 16.Oberg K, Casanovas O, Castaño JP, Chung D, Fave GD, Denèfle P, et al. Molecular pathogenesis of neuroendocrine tumors: Implications for current and future therapeutic approaches. Clin Cancer Res. 2013;19:2842–9. doi: 10.1158/1078-0432.CCR-12-3458. [DOI] [PubMed] [Google Scholar]
- 17.Boudreaux JP, Putty B, Frey DJ, Woltering E, Anthony L, Daly I, et al. Surgical treatment of advanced-stage carcinoid tumors: Lessons learned. Ann Surg. 2005;241:839–45. doi: 10.1097/01.sla.0000164073.08093.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Givi B, Pommier SJ, Thompson AK, Diggs BS, Pommier RF. Operative resection of primary carcinoid neoplasms in patients with liver metastases yields significantly better survival. Surgery. 2006;140:891–7. doi: 10.1016/j.surg.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 19.Kerström G, Hellman P, Hessman O. Midgut carcinoid tumours: Surgical treatment and prognosis. Best Pract Res Clin Gastroenterol. 2005;19:717–28. doi: 10.1016/j.bpg.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Sutton R, Doran HE, Williams EM, Vora J, Vinjamuri S, Evans J, et al. Surgery for midgut carcinoid. Endocr Relat Cancer. 2003;10:469–81. doi: 10.1677/erc.0.0100469. [DOI] [PubMed] [Google Scholar]
- 21.Raymond E, Dahan L, Raoul JL, Bang YJ, Borbath I, Lombard-Bohas C, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:501–13. doi: 10.1056/NEJMoa1003825. [DOI] [PubMed] [Google Scholar]
- 22.Boudreaux JP, Klimstra DS, Hassan MM, Woltering EA, Jensen RT, Goldsmith SJ, et al. The NANETS consensus guideline for the diagnosis and management of neuroendocrine tumors: Well-differentiated neuroendocrine tumors of the Jejunum, Ileum, Appendix, and Cecum. Pancreas. 2010;39:753–66. doi: 10.1097/MPA.0b013e3181ebb2a5. [DOI] [PubMed] [Google Scholar]
- 23.Kulke MH, Anthony LB, Bushnell DL, De Herder WW, Goldsmith SJ, Klimstra DS, et al. NANETS treatment guidelines: Well-differentiated neuroendocrine tumors of the stomach and pancreas. Pancreas. 2010;39:735–52. doi: 10.1097/MPA.0b013e3181ebb168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faivre S, Niccoli P, Castellano D, Valle JW, Hammel P, Raoul JL, et al. Sunitinib in pancreatic neuroendocrine tumors: Updated progression-free survival and final overall survival from a phase III randomized study. Ann Oncol. 2017;28:339–43. doi: 10.1093/annonc/mdw561. [DOI] [PubMed] [Google Scholar]
- 25.Wang SC, Parekh JR, Zuraek MB, Venook AP, Bergsland EK, Warren RS, et al. Identification of unknown primary tumors in patients with neuroendocrine liver metastases. Arch Surg. 2010;145:276–80. doi: 10.1001/archsurg.2010.10. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd RV, Mervak T, Schmidt K, Warner TF, Wilson BS. Immunohistochemical detection of chromogranin and neuron-specific enolase in pancreatic endocrine neoplasms. Am J Surg Pathol. 1984;8:607–14. doi: 10.1097/00000478-198408000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd RV. Practical markers used in the diagnosis of neuroendocrine tumors. Endocr Pathol. 2003;14:293–301. doi: 10.1385/ep:14:4:293. [DOI] [PubMed] [Google Scholar]
- 28.Chejfec G, Falkmer S, Grimelius L, Jacobsson B, Rodensjö M, Wiedenmann B, et al. Synaptophysin.A new marker for pancreatic neuroendocrine tumors. Am J Surg Pathol. 1987;11:241–7. [PubMed] [Google Scholar]
- 29.Ishibashi H, Suzuki T, Suzuki S, Moriya T, Kaneko C, Takizawa T, et al. Sex steroid hormone receptors in human thymoma. J Clin Endocrinol Metab. 2003;88:2309–17. doi: 10.1210/jc.2002-021353. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Xu B, Zheng C, Gong Y, Garibaldi J, Soria D, et al. An end-to-end deep learning histochemical scoring system for breast cancer TMA. IEEE Trans Med Imaging. 2019;38:617–28. doi: 10.1109/TMI.2018.2868333. [DOI] [PubMed] [Google Scholar]
- 31.Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CL, Bolon B, et al. The gold standard paradox in digital image analysis: Manual versus automated scoring as ground truth. Arch Pathol Lab Med. 2017;141:1267–75. doi: 10.5858/arpa.2016-0386-RA. [DOI] [PubMed] [Google Scholar]
- 32.Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. doi: 10.1186/1746-1596-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellizzi AM. Assigning site of origin in metastatic neuroendocrine neoplasms: A clinically significant application of diagnostic immunohistochemistry. Adv Anat Pathol. 2013;20:285–314. doi: 10.1097/PAP.0b013e3182a2dc67. [DOI] [PubMed] [Google Scholar]
- 34.Bellizzi AM. Immunohistochemistry in the diagnosis and classification of neuroendocrine neoplasms: What can brown do for you? Hum Pathol. 2020;96:8–33. doi: 10.1016/j.humpath.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


