Abstract
Background:
Prescreening of biopsies has the potential to improve pathologists' workflow. Tools that identify features and display results in a visually thoughtful manner can enhance efficiency, accuracy, and reproducibility. Machine learning for detection of glomeruli ensures comprehensive assessment and registration of four different stains allows for simultaneous navigation and viewing.
Methods:
Medical renal core biopsies (4 stains each) were digitized using a Leica SCN400 at ×40 and loaded into the Corista Quantum research platform. Glomeruli were manually annotated by pathologists. The tissue on the 4 stains was registered using a combination of keypoint- and intensity-based algorithms, and a 4-panel simultaneous viewing display was created. Using a training cohort, machine learning convolutional neural net (CNN) models were created to identify glomeruli in all stains, and merged into composite fields of views (FOVs). The sensitivity and specificity of glomerulus detection, and FOV area for each detection were calculated.
Results:
Forty-one biopsies were used for training (28) and same-batch evaluation (6). Seven additional biopsies from a temporally different batch were also evaluated. A variant of AlexNet CNN, used for object recognition, showed the best result for the detection of glomeruli with same-batch and different-batch evaluation: Same-batch sensitivity 92%, “modified” specificity 89%, average FOV size represented 0.8% of the total slide area; different-batch sensitivity 90%, “modified” specificity 98% and average FOV size 1.6% of the total slide area.
Conclusions:
Glomerulus detection in the best CNN model shows that machine learning algorithms may be accurate for this task. The added benefit of biopsy registration with simultaneous display and navigation allows reviewers to move from one machine-generated FOV to the next in all 4 stains. Together these features could increase both efficiency and accuracy in the review process.
Keywords: Convolutional neural network, glomeruli, image registration, machine learning, renal
INTRODUCTION
Digital pathology has many potential utilities. These include advancements in education, case archiving and retrieval, teleconsultation, and also improvements in efficiency and accuracy in the daily practice of the anatomic pathologist. Digitization of histopathology slides allows for various modes of case presentation which would be advantageous for rapid and accurate review. One such example is the simultaneous navigation of serial sections of different stains from the same tissue block. Localization and direct comparison of findings on individual cells and structures can enhance productivity over the normal microscopy procedure of switching slides and locating the same fields of views (FOVs) as well as locating the same cells or structures within those FOVs. In addition to presentation, machine learning applications on digitized specimens have been shown to be effective for the automated identification of particular organs, structures, disease processes or other features which under standard microscopy would need to be identified manually.[1,2,3,4] The combination of automated feature identification and simultaneous and synchronized multiple section display of whole slide images (WSIs) has the potential to increase both efficiency and accuracy. A similar scenario was well shown with “location-guided screening” in cervical cytology with digital results display; a process which was first approved for clinical use more than 2 decades ago.[5,6,7,8]
In the present study, the authors developed a prototype system of synchronized section display and machine learning-based feature detection in medical renal biopsies. The renal pathologist must evaluate a series of special stains on renal biopsies, with particular attention to glomerular features. Being able to automate the detection of glomeruli, display each glomerulus in 4 stains (hematoxylin and eosin, silver, trichrome, periodic acid-Schiff) simultaneously, and move all 4 screens to the next identified glomerulus in a synchronized fashion, might create a significant efficiency advantage over standard single slide microscopic review. In addition, the ability to directly compare staining features simultaneously on each glomerulus, as well as the ability to autolocate a high majority (>90%) of all glomeruli present in each specimen, could lead to a more accurate final interpretation through improved correlation of findings and fewer missed glomeruli, respectively.
This report details the processes of developing the simultaneous multiplex display for reviewing the images and the machine learning methods that were utilized to obtain a model for glomerulus localization. Preliminary data on a single institution's slide set show the effectiveness of the models in the detection of glomeruli. The best results in this study will be used in future projects across multi-institutional biopsy material in order to generalize this model to a broader application.
METHODS
This study was approved by the institutional review board of the facility from which the biopsy material was derived. Medical renal biopsies consisting of hematoxylin and eosin, silver, trichrome, and periodic acid-Schiff stained serial sections were collected from a single institution. Cases were obtained consecutively with the only limitation being adequate numbers of glomeruli being present. No selection process based on diagnosis or other glomerular feature criteria was used. The biopsies were digitized into WSIs using a Leica SCN400 scanner (Leica Biosystems Inc., Buffalo Grove, IL, USA) at ×40 magnification (0.25 μm/pixel resolution. The WSIs were entered into the Corista Quantum research platform (Corista LLC, Concord, MA, USA) where they were automatically deidentified. The Quantum platform is a single interface which allows users to annotate, train, evaluate and utilize machine learning models for use with WSI slide sets. The core functionality of Quantum used in this study is broken into 5 high-level user and system tasks: Registration of WSI serial sections, manual annotation of features, training of deep learning models, classification and evaluation of the trained models (comparison to the manual annotations), and observer review of the model's feature-identifying FOVs.
Image registration
The purpose of registering a pair of WSIs is to be able to translate any point on one registered WSI to a corresponding point on the other registered WSI. “Corresponding” in this context means belonging to the same (or nearly the same) physical location in the original 3-dimensional tissue sample. This is accomplished with the use of a spatially distributed set of affine transforms covering the entire WSI. The affine transform matrices (ATMs) provide a simple mathematical way to correlate any point on the first image with a point on the second image. A distributed set of ATMs allows for better accuracy when finding corresponding points on different parts of the images, each distorted in its own unique way.
A multi-resolution registration algorithm was developed and used to provide fine ATMs at different stages.[9] Initially, coarse ATMs are calculated for the entire WSI. In actual practice one WSI might include several tissue samples (e.g., several sections taken from the same block). Such samples may require initial alignment which differs from that of the whole WSI, as some samples may be in different positions or orientations on the slide. In such cases in addition to the coarse ATMs for the entire WSI, coarse ATMs for the discrete portions of each slide are calculated (e.g., if the WSI consists of the four discrete tissue samples (two at the top and two at the bottom of the slide), then additional coarse ATMs are calculated for the four quadrants of the slide.
In further steps, the image is recursively divided into additional subsections of smaller size, generating progressively finer ATMs for each subsection based on the previously calculated coarser ATMs, until no additional precision can be calculated or the greatest amount of subdivision is achieved (e.g., with a single coarse whole-WSI ATM with a 2 × 2 matrix of the coarse ATMs for the four tissue samples from the example above, the next step is to generate a matrix of 4 × 4 ATMs, then 8 × 8 ATMs, and so on). ATMs at each step will not always be calculated if data is insufficient, such as in the background of the slide. Once determined, these local ATMs comprise a progressively more precise pyramid of ATM levels, where higher precision fine ATMs represent smaller subsections of the images.
Initial coarse ATMs describe the alignment of whole images generated by the identification of keypoint pairs. Keypoints are distinct positions in an image, usually relying on visible features such as tissue corners or appendages. In each keypoint pair, one keypoint belongs to the first WSI and the second keypoint refers to the matching location on the second WSI. For example, the keypoint on a tissue fragment in the first stained image (such as the edge of the tissue, or a cluster of red blood cells) should correspond to the same feature in the second image [Figure 1]. For real world WSIs, there are usually more than three matching keypoint pairs identified, hence special mathematical methods are used to find the ATM that best satisfies all keypoint matches. These keypoints are identified using a feature detection algorithm. In our study, the oriented FAST and rotated BRIEFn algorithm (ORB) was used to detect keypoints, which using these algorithms are tissue “corners” such as the joined edges of an angular tissue fragment.[10]
Figure 1.
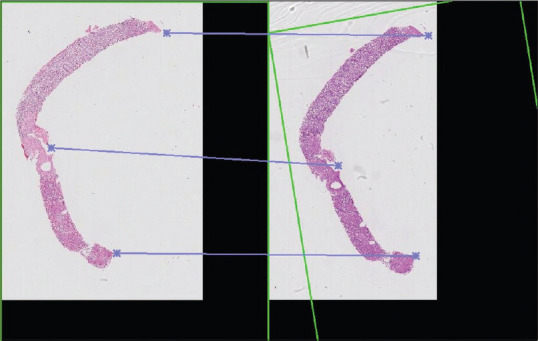
An example of the keypoints portion of image registration. Areas of distinct features (e.g., edges, appendages, points) are denoted by the asterisks. These features are aligned and allow serial sections to be matched and rotated (see green box indicating rotation of the section on the right) for simultaneous review
The keypoints are paired using a matching algorithm with an evaluation criterion. This algorithm is the Brute-Force Matcher and the evaluation distance is the Hamming distance which is a measure of the differences between two feature vectors.[11] Next, the keypoint matches are analyzed for consistency using a mean-shift clustering algorithm, with unfit or poorly correlated matches removed. This was accomplished by: (1) joining closely located keypoints using the Density-based spatial clustering of applications with noise clustering algorithm (DBSCAN) to reduce the total number of keypoints and hence lower the computational complexity of further steps;[12] (2) creating all possible triplets of matches; (3) eliminating triplets composed of keypoints located too close to each other; (4) calculating candidate ATMs for each of the keypoint triplets; (5) using an iterative mean-shift clustering algorithm to eliminate poorly correlated candidate ATMs, which produces an average ATM as a center of the largest cluster.[13] In each iteration the bandwidth parameter is reduced (thus reducing the size of the biggest cluster) until the standard deviation of the coordinate translation errors produced by the ATMs in the largest cluster reaches a pre-defined empirical value. Essentially this step selects the subset of well-correlated keypoints and uses only those to produce the averaged ATM.
It is possible that some areas of the WSI do not contain any detectable features (e.g., white background, missing portions of tissue). For these areas, the keypoints cannot be found, hence the ATMs cannot be calculated. For such feature-less subsections of the WSI, the local ATMs are computed by approximation from the previously computed nearby ATMs, including the lower precision ATMs calculated for the larger area, or from the entire coarse WSI ATM.
After the coarser ATMs were calculated via the keypoint-based method providing basic alignment of the two WSIs, a parametric, intensity-based registration algorithm was applied to bolster the coarse registration's precision. This algorithm compares the pixels of the two images instead of finding keypoints, matching up comparable areas based on a defined similarity metric and assessing the differences in the images in an iterative stochastic optimization process. This algorithm requires initial placement of the compared areas to be relatively precise, hence the requirement of the keypoint WSI registration as a first step. Before intensity-based registration, the second image is warped to fit the first image in the registration, producing a new result image. This result image can be overlayed on the first image to assess the quality of the registration, where greater nonoverlapping area suggests a lower quality registration.[14,15]
The keypoints- and intensity-based registrations both may repeat on progressively smaller areas of WSIs, with higher resolution, as needed to obtain optimal registration. The combination of the most precise ATMs for any region blended into a single structure creates a field of ATMs which provide a continuous and mathematically smooth way of finding corresponding locations on several slides.
The registered serial sections of the tissue block are displayed in a 4-panel viewer in which each stain is visualized in its own area (quarter panel). Navigation of each panel is synchronized for both movement and magnification. The resultant full panel display is illustrated in Figure 2.
Figure 2.
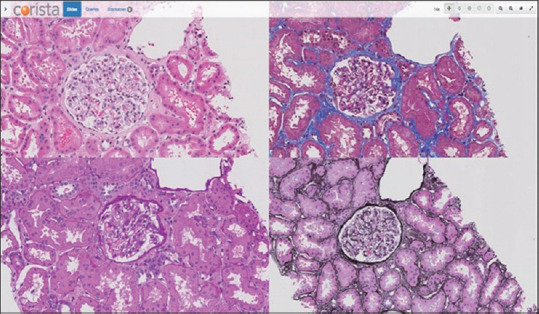
A 4 stain registered panel display. Each panel navigates and changes magnification in a linked fashion to allow for examination of each glomerulus simultaneously in all 4 stains
Glomerulus annotation
Using a freehand or circle/oval annotation tool available in the Quantum platform, pathologists outlined each glomerulus identified in all 4 stains in each renal biopsy [Figure 3]. Each section was annotated by 2 pathologists with truth for the study being any annotated structure. Double annotation was not required as truth because glomeruli are such distinct morphologic structures. The double annotation process was to ensure that all glomeruli were captured. A third pathologist reviewed all “single-annotated” structures to ensure that they were in fact glomeruli (and all were deemed to be so). Any portion of each WSI containing tissue (not background) which does not contain an annotation is assigned a default classification (“not glomerulus”), which is used as such by the system during training.
Figure 3.
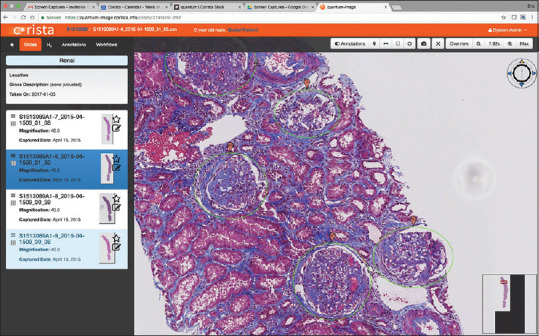
An example of glomerulus annotation – pathologists circled individual structures for the determination of “ground truth” against which the convolutional neural net model developed was tested for accuracy and precision of detection
Training models for feature identification
Using the annotated WSIs, several models were trained to detect glomeruli in the tissue sections of each of the 4 stain types. For each WSI, the system produces a series of image patches (smaller, overlapping segments of each image that can more readily be processed by a machine learning algorithm) and then assigns each patch a classification based on the annotation (i.e., “glomerulus” or “not glomerulus”). Performance is influenced by the marking precision of the glomerulus annotations, and the specific annotated regions of each particular class can be used as a mask to produce accurate training patches. During training, the system can augment the amount of available training data and patches by modifying and reanalyzing the patches via rotation, flipping, scaling, adding noise, or blurring. Such maneuvers increase the overall size and variety of the dataset used for machine learning training. The set of annotated patches is then provided as training data to each machine learning algorithm, which then produces a model for each algorithm.
During the initial phases of this study, a variety of automatic feature generation methods (where the machine learning algorithm includes the processing to generate its own features, such as with a convolutional neural net [CNN]) operated on the raw image data, and other ML algorithms which included a preprocessing step were used before employing an ML algorithm. These included feature detection such as ORB, color normalization and other computer vision algorithms, but they did not prove beneficial as pre-processing steps comparing to use of the raw image data fed to CNN. Other methods such as background removal (color saturation-based) and resampling of the training patches were found useful and included in the overall system data flow.
The best performing algorithm was a CNN derived from AlexNet,[16] which is a well-established, verified, and universally applicable CNN model. Other CNNs were considered but compared to AlexNet, presented their own set of operational “quirks” and usage specifics. In this study AlexNet consisted of two or more convolutional layers, pooling layers to reduce the dimensionality of the data and prevent overfitting, nonlinearity layers (e.g., rectified linear unit [ReLU] layers) to increase the nonlinearity properties of the network, dropout layers to prevent overfitting, one or more fully connected layers, and an output classification layer. The classification layer is a softmax classifier with cross-entropy loss function, leveraging stochastic gradient descent with adaptive moment estimation as an optimization algorithm.[17,18] Trained CNN models were snapshotted and stored for further evaluation and determining their performance.
Evaluating new whole slide image slide sets (testing of the model)
Testing of the model created by the above process evaluates the classification of a new specimen (one not used for training). This was performed on 2 batches of slides not used for training. One batch (labeled “same”) was from the same group temporally as the training set. The other batch (labeled “different”) was from the same institution but from a different time. Evaluation results in the assignment of a class for each identified ROI with a confidence score based on classes used in training of the model. The classes are referred to as “default,” indicating the absence of glomeruli, and “nondefault” indicating the presence of glomeruli. Once the classification model had been created on the training data for the CNN, it was applied to “test” data. Similar to the training process, normalized and otherwise preconditioned image patches were supplied to the model. The pipeline selected for the classifier accepts patches of a predetermined size as input and results in a classification and scoring of each patch based on the classes identified during training.
The test WSIs were also annotated for the presence of glomeruli and the results of the classification process were compared to the annotations to determine accuracy and ROI size (calculated as a percentage of the total area of the slide). In binary classification (i.e., detection vs. nondetection), in which a phenomenon (feature) may be found in an ROI (positive or nondefault classification) or absent from an ROI (negative or default classification), the results may be evaluated via standard metrics such as sensitivity (correct positive percentage) or specificity (correct negative percentage).
For the specific task (utilizing multiple stain-specific WSIs in a slide set), the outputs from the various stain-specific classifiers were combined and evaluated using correlation techniques. The outputs from multiple input sets and multiple models were combined to boost the detection of the features and reduce false detections. The output of the classification step is a set of regions of interest (ROIs), each of which will have a classification and an associated confidence metric (or score).
Results were expected to differ between the four different special stains in each case set. The distance between serial sections could affect localization of an individual glomerulus, as could unique tinctorial characteristics of each stain. Hence, in our implementation of correlation, classification and scoring results were obtained separately from the WSIs of the four stains and then combined to give a composite localization result. The per-staining detected feature results were thresholded by a confidence score and combined together using inter-WSI registration in order to align the feature locations. The results were further filtered by removing spatial locations with positive detections on fewer than 3 of individual stain WSIs. The remaining positive classification data and scores were merged into a score-heatmap, from which local combined score maxima are found, which in turn become the centers of newly found ROIs. Classification data may be further used after additional filtering to aggregate around the found local maxima and create clusters that would define the sizes of the newly identified ROIs. With this approach the distance between serial sections is less important.
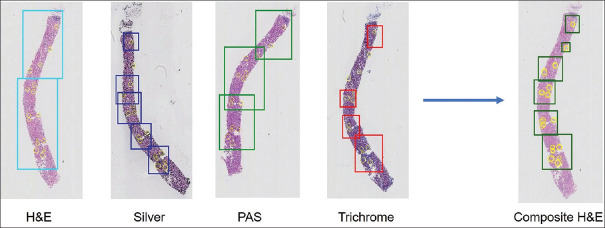
Using the combined classification results from multistain WSIs, with enhanced registration of WSI pairs to correlate the localities of the positively classified areas results in significant improvement of the detection characteristics compared to ROIs generated using a single stain WSI [Figure 4].
Figure 4.
In each of the 4 stains on the left, the rectangles are the region of interest identified by the convolutional neural net model. Yellow circles represent the manually annotated glomeruli. By strengthening strong and reducing weak signals from the individual stains, a final composite region of interest is created as displayed on the H and E stained slide on the right. Note that this process reduces the size of the region of interests that encompass annotated glomeruli, indicating a more precise localization of glomeruli
Results reporting parameters
The sensitivity and a “modified” specificity for the detection of glomeruli compared to manual annotation was reported for the best model developed (variant of AlexNet). The sensitivity indicates how many annotated glomeruli were identified by each model (true positive detections); and the “modified” specificity indicates how many ROI from each model showed no glomeruli (inverse of false positive detections). A classic true specificity calculation could not be performed in this setting because no ROI were generated for areas not containing glomeruli (which would be all nonannotated areas of the slide-which would represent true negative results). Hence a “modified” specificity-like measurement was chosen that indicates how many ROI did not contain annotated glomeruli. Overall this metric gives an indication of the specificity of the model in the detection of annotated glomeruli, although it is understood that it is not a true specificity measurement in the classic sense. Both sensitivity and “modified” specificity metrics were expressed as percent values; the higher the value the better is the performance. The FOV average area as a percentage of the overall slide size is noted for the best model. The area measurement is proportional to the robustness of the detection, with smaller FOVs indicating a more robust localization.
RESULTS
The model was trained on trained on 28 stain sets of 4 slides each, making 112 WSIs. The WSIs were preprocessed as 512,000 training samples with reuse by inverting, rotating, and focusing. Evaluation was performed on 6 stain sets of the same batch as the original training slide sets, and again on 7 slide sets from a different batch from the same institution.
For the best CNN model (variant of AlexNet), the sensitivity and “modified” specificity for the detection of glomeruli in the combined ROIs were 92% and 89%, respectively for the same-batch WSIs, and 90% and 98% for the different-batch WSIs [Table 1]. For the best model the average FOV area was 0.8% and 1.6% of the total slide area for the same batch and different batch cohorts, respectively. These area numbers are meaningless when viewed by themselves but form the basis for comparisons with other models on other cohorts of slides to be tested in the future.
Table 1.
The sensitivity and “modified” specificity for detection of glomeruli and the average size of the regions of interest identified for the best convolutional neural net model
| Sensitivity for detection of annotated glomeruli (TP/TP + FN) | “Modified” specificity for detection of annotated glomeruli (1 − FP/[TP + FP]) | Average ROI area (as a measure of the total slide area), % | |
|---|---|---|---|
| Same-batch 6 stain sets, 24 WSIs | 92% (192/192+16) | 89% (1−24/[192+24]) | 0.8 |
| Different-batch 7 stain sets, 28 WSIs | 90% (236/236+25) | 98% (1−4/[236+4]) | 1.6 |
ROI: Region of interest, TP: Generated ROI containing an annotated glomerulus, FN: Annotated glomerulus without a generated ROI, FP: Generated ROI not containing an annotated glomerulus, TN: Cannot be calculated for this study because no ROI were generated for “not-annotated as glomerulus” areas, WSIs: Whole slide images
CONCLUSIONS
The use of machine learning for glomerulus detection has the potential to substantially improve the pathologist experience for the examination of medical renal tissue biopsies. In addition, when automated localization is combined with multiplex display of different stains, with navigation and magnification synchronization, both efficiency and accuracy might be improved. Automated identification can save time and with high detection sensitivity the possibility of missing an important glomerulus would be minimized. While high specificity is important to save review time by minimizing the examination of FOVs not containing the sought after features, in actual practice this time loss would be minimal as trained pathologists would easily dismiss these FOVs as not important to their review. The ability to review multiple synchronized digital serial sections with different stains might further enhances both efficiency and accuracy. The equivalent of 4 slides can be reviewed in the time it might take to examine a single slide using a standard microscope. In addition, the features of each glomerulus could be directly compared across all 4 digital stains simultaneously which obviates the need to continually switch between analog slides and relocate glomeruli for a direct comparison. Accurate and rapid review might therefore accrue from such a platform. The pathologist experience for accuracy and efficiency was not tested in this presentation of system development. The results herein show that feature identification and multiplex presentation of multiple special stains is possible. Obtaining actual performance metrics will be the focus of a future clinical study.
In this proof of concept study, preliminary results using deep learning to identify glomeruli in all 4 stains of a standard medical renal biopsy are reported. Prior studies have also sought to use deep learning for the detection of glomeruli. Olivier et al. found an approximately 90% precision for detection of glomeruli in PAS stains from 8 human biopsies from a single institution. Bukowy et al., using renal biopsies from rats found a 97% accuracy in detecting glomeruli, again using a PAS stain only.[19] Kannan et al. found a 93% accuracy for detection of glomeruli in cropped digital images of trichrome stains in human renal biopsies.[20] Hermson et al. using 40 PAS-stained transplant and nephrectomy specimens was able to identify 92% of manually annotated glomeruli with a false positive rate of 10%.[21] In the present study, the extension of this prior work to a practical application of all 4 stains in a human renal biopsy makes this a prototypical example of results which might be expected in a real-world setting. Using a deep learning approach applied to all 4 stains simultaneously with combination of the detection signals into a final composite FOV ROI allows CNN “errors” to be reduced by eliminating weak probability areas that might occur on one (but not other) stains, and strengthening areas of high probability that occur in more than one of the stains. Ninety percent accuracy might be considered a lower limit for a clinically usable device, but further training of the CNN model with additional 4 stain sets should improve the accuracy going forward. The addition of the synchronized display adds significant practical utility to this system. Although this is a limited study on a small number of cases, the concept appears sound and the results were encouraging.
Models that use training slide sets from a single institution risk not having the ability to generalize the findings when slides from other institutions are run through the system. Significant differences in tissue processing, preparation, and staining are expected to be present between institutions and as such results would be expected to degrade without training on slides having those different features. A hint of this difference can be gleaned from the difference in the “same” and “different” batch results from the present study. Sensitivity degrades slightly (92%–90%) and average ROI size increases by 100% (0.8%–1.6%), meaning that the precision of the detection degrades. This may be the result of changes in slide preparation and staining that might have occurred at that institution between the time of collection of the “same” and “different” batches of slides.
The extension of the present study will test potential inter-institutional differences further by adding slides from several centers to the training and validation sets with retraining and testing of the resultant cross-institutional model. When accurate glomerulus detection is well-established, further investigation into pathologic classification of glomerular abnormalities by deep learning will become the ultimate goal.
The utility of digital pathology is growing as more institutions acquire the technology and infrastructure to digitize their slide base and experience is gained in pathologist interpretation from digital viewing stations. However, adoption remains slow because the cost of equipment and the practical value to users is still limited. Applications in training, continuing education, and teleconsultation are growing in value, but day to day primary interpretation still lags behind standard microscopy for efficiency, despite being shown to be as accurate.[22,23] The addition of machine learning for autolocation of features and ultimately interpretation will greatly increase the value of digital pathology for primary users. Along with this development of innovative workflow changes, such as the synchronized multiplex display reported herein, digital pathology has the potential to greatly increase the value proposition for users. When such advancements play out, digital pathology should move into the mainstream of the pathologist's daily work.
Financial support and sponsorship
Project was supported by Corista LLC.
Conflicts of interest
Wilbur, Andryushkin, Wingard, and Wirch are employees of Corista LLC.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2020/11/1/37/300283
REFERENCES
- 1.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 2.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong F, Irshad H, Oh EY, Lerwill MF, Brachtel EF, Jones NC, et al. Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS One. 2014;9:e114885. doi: 10.1371/journal.pone.0114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abels E, Pantanowitz L, Aeffner F, Zarella MD, van der Laak J, Bui MM, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the Digital Pathology Association. J Pathol. 2019;249:286–94. doi: 10.1002/path.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koss LG, Lin E, Schreiber K, Elgert P, Mango L. Evaluation of the PAPNET cytologic screening system for quality control of cervical smears. Am J Clin Pathol. 1994;101:220–9. doi: 10.1093/ajcp/101.2.220. [DOI] [PubMed] [Google Scholar]
- 6.Wilbur DC, Black-Schaffer WS, Luff RD, Abraham KP, Kemper C, Molina JT, et al. The Becton Dickinson FocalPoint GS Imaging System: Clinical trials demonstrate significantly improved sensitivity for the detection of important cervical lesions. Am J Clin Pathol. 2009;132:767–75. doi: 10.1309/AJCP8VE7AWBZCVQT. [DOI] [PubMed] [Google Scholar]
- 7.Eichhorn JH, Buckner L, Buckner SB, Beech DP, Harris KA, McClure DJ, et al. Internet-based gynecologic telecytology with remote automated image selection: Results of a first-phase developmental trial. Am J Clin Pathol. 2008;129:686–96. doi: 10.1309/GRAV16QP8JR5XTPF. [DOI] [PubMed] [Google Scholar]
- 8.Biscotti CV, Dawson AE, Dziura B, Galup L, Darragh T, Rahemtulla A, et al. Assisted primary screening using the automated ThinPrep Imaging System. Am J Clin Pathol. 2005;123:281–7. [PubMed] [Google Scholar]
- 9.Zhang Z, Han D, Dezert J, Yang Y. A new image registration algorithm based on evidential reasoning. Sensors (Basel) 2019;19:1091. doi: 10.3390/s19051091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rublee E, Rabaud V, Konolige K, Bradski G. ORB: An efficient alternative to SIFT or SURF.Proceedings of the IEEE International Conference on Computer Vision. 2011 2564-2571 101109/ICCV20116126544. [Google Scholar]
- 11.Huang W, Shi Y, Zhang S, Zhu Y. The communication complexity of the Hamming distance problem. Inf Process Lett. 2006;99:149–53. [Google Scholar]
- 12.Ester M, Kriegel HP, Sander J, Xu X. Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining. Portland, OR: AAAI Press; 1996. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise; pp. 226–31. [Google Scholar]
- 13.Comaniciu D, Meer P. Mean shift: A robust approach toward feature space analysis. IEEE Trans Pattern Anal Mach Intell. 2002;24:603–19. [Google Scholar]
- 14.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 15.Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M, et al. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Front Neuroinform. 2013;7:50. doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90. [Google Scholar]
- 17.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–6. [Google Scholar]
- 18.Diederik K, Adam BJ. A method for stochastic optimization. San Diego: 2015. arxivorg/abs/14126980. [Google Scholar]
- 19.Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol. 2018;29:2081–8. doi: 10.1681/ASN.2017111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannan S, Morgan LA, Liang B, Cheung MG, Lin CQ, Mun D, et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep. 2019;4:955–62. doi: 10.1016/j.ekir.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, et al. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30:1968–79. doi: 10.1681/ASN.2019020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: A multicenter blinded randomized noninferiority study of 1992 cases (pivotal study) Am J Surg Pathol. 2018;42:39–52. doi: 10.1097/PAS.0000000000000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borowsky AD, Glassy EF, Wallace WD, Kallichanda NS, Behling CA, Miller DV, et al. Digital Whole Slide Imaging Compared With Light Microscopy for Primary Diagnosis in Surgical Pathology. Arch Pathol Lab Med. 2020;144:1245–1253. doi: 10.5858/arpa.2019-0569-OA. doi: 10.5858/arpa.2019-0569-OA. PMID: 32057275. [DOI] [PubMed] [Google Scholar]