Abstract
Liquid-liquid phase separation (LLPS) has recently emerged as a possible mechanism that enables ubiquitin-binding shuttle proteins to facilitate the degradation of ubiquitinated substrates via distinct protein quality control (PQC) pathways. Shuttle protein LLPS is modulated by multivalent interactions among their various domains as well as heterotypic interactions with polyubiquitin chains. Here, the properties of three different shuttle proteins (hHR23B, p62, and UBQLN2) are closely examined, unifying principles for the molecular determinants of their LLPS are identified, and how LLPS is connected to their functions is discussed. Evidence supporting LLPS of other shuttle proteins is also found. In this review, it is proposed that shuttle protein LLPS leads to spatiotemporal regulation of PQC activities by mediating the recruitment of PQC machinery (including proteasomes or autophagic components) to biomolecular condensates, assembly/disassembly of condensates, selective enrichment of client proteins, and extraction of ubiquitinated proteins from condensates in cells.
Keywords: autophagy, biomolecular condensates, liquid-liquid phase separation, polyubiquitin, proteasomal degradation, protein quality control, ubiquitin shuttle proteins
1. Introduction
Cells contain multiple compartments to modulate distinct physiological processes. Aside from well-known membrane-bound structures, there also exist membraneless organelles or biomolecular condensates, including cytoplasmic stress granules, processing bodies (P-bodies), the nucleolus, and PML (promyelocytic leukaemia) bodies.[1–4] Condensates form via liquid-liquid phase separation (LLPS) (Box 1).[5] Condensates can perform biological functions such as concentrating, sequestering, and buffering components. Moreover, with their microenvironments exhibiting characteristics distinct from the rest of the cell, some condensates serve as active centers of increased biochemical activity.[6] The absence of a membrane surrounding biomolecular condensates enables cells to rapidly respond to changes in their environment.
Box 1. Molecular Origins of Liquid-Liquid Phase Separation.
LLPS is a thermodynamic process that drives a solution of macromolecules (e.g., RNA, DNA, proteins, or a mixture of these) to demix into at least two distinct liquid phases. Conditions for LLPS can be modulated by physiochemical parameters, such as pH, salt, and temperature. Dysregulation of LLPS can lead to malfunctions in cells, such as disease-linked mutations that “mature” condensates into irreversible aggregated states found in disease or that mislocalize condensates from the nucleus to the cytoplasm.[21–23]
It is best to study LLPS by observing endogenously-expressed proteins in cells. More commonly, for the convenience of studying effects of disease-linked mutations on LLPS in live cells, overexpressed proteins in transfected cells are used. However, as LLPS is highly sensitive to protein concentration (Box 2), overexpression can lead to formation of non-native condensates. LLPS is also investigated using purified protein, enabling better control of experimental parameters and access to a wider range of experimental techniques, both of which lead to detailed understanding of the molecular driving forces of LLPS. However, properties elucidated through in vitro studies of LLPS might not correlate with in cell observations. Hence, there exists a need for careful examination of both in cell and in vitro LLPS findings.[24]
One useful approach to study molecular origins of LLPS employs associative polymer theory to describe macromolecules as biopolymers containing “sticker” and “spacer” segments.[25–28] “Stickers” are sites of multivalent interactions that drive LLPS, whereas the sequence and length of connecting “spacers” can modulate condensate material properties.[29–31] For proteins, “stickers” can be either short segments in intrinsically disordered regions (IDRs), interaction patches on globular proteins, or folded domains in multidomain proteins.[30,32–34] With at least 30% of the human proteome comprising IDRs,[35] and a significant population composed of multidomain proteins, it is expected that many systems phase separate under certain cellular conditions.
The extent to which a system undergoes LLPS increases with multivalency. One common mechanism for proteins to achieve multivalency is through oligomerization, which can give rise to emergent “stickers” that further drive LLPS.[26] Examples include the folded NTF2L domain of G3BP1 and BTD and BACK domains of SPOP.[36–38] Oligomerization domains can alsobe encoded in IDRs, such as the transient helical region in the C-terminal segment of TDP-43.[39] Oligomerization provides a scaffold that enhances the multivalency needed to mediate LLPS under physiological conditions. Indeed, light-induced oligomerization of RNA-binding proteins drives technologies to form membraneless organelles in cells, such as optoSGs.[9]
Dysregulated condensates can be precursors of protein inclusions commonly found in diseased neurons of patients with neurodegenerative disorders.[7–9] Many of these inclusions contain components of protein quality control (PQC) pathways, such as ubiquitin (Ub), polyubiquitin (polyUb) chains, Ub-binding shuttle proteins, autophagic components, and proteasome subunits, suggesting that dysregulation of Ub-mediated PQC pathways contributes to disease pathology.[10–13] Here, we focus on Ub-binding shuttle proteins which bind both polyUb chains and proteasome/autophagy-related components, to facilitate transport of ubiquitinated substrates to degradation centers. Notably, Ub-binding shuttle proteins UBQLN2, p62, and hHR23B undergo LLPS in vitro and in cells.[14–17] Exogenous UBQLN2 forms stress-induced puncta and endogenous UBQLN2 colocalizes with stress granules (SGs).[15,18] UBQLN2 phase separates alone, and polyUb chain interactions disassemble UBQLN2 droplets in vitro.[15] By contrast, p62 and hHR23B phase separate with polyUb chains.[17,19,20] Importantly, in each of these cases, polyUb chains directly affect the ability of shuttle proteins to form condensates.
In this essay, we propose that shuttle protein LLPS leads to spatiotemporal control of degradation of ubiquitinated substrates. First, we discuss unifying molecular properties of shuttle proteins that have been shown to phase separate. Importantly, we discriminate between those shuttle proteins that require polyUb chains to phase separate, and those that do not. These two types of phase-separating systems exhibit different cellular functions and confer richness to how protein interactions modulate LLPS. Moreover, we hypothesize that other shuttle proteins may also undergo LLPS and present evidence accordingly. We then link shuttle protein LLPS to their biological functions and propose several mechanisms for how LLPS is beneficial for PQC activities. We aim to bridge recent discoveries of shuttle protein LLPS for both the ubiquitin-proteasome system (UPS) and autophagy fields. We propose that LLPS contributes mechanistically to maintaining protein homeostasis and proper PQC in cells.
2. Shuttle Proteins Differ in Binding Affinities to Ub, Domain Architectures, Cellular Functions, and Localization
To date, three Ub-binding shuttle proteins, UBQLN2, hHR23B, and p62, are known to undergo LLPS. All three proteins contain folded N-terminal Ub-like (UBL) and C-terminal Ub-associating (UBA) domains connected by intrinsically disordered regions (IDRs), such as STI1 domains (Figure 1A). The UBL domains all interact with the proteasome, consistent with the roles of these shuttle proteins in proteasomal degradation.[40–42] p62 UBL (named PB1) also has a competing ability to form an oligomeric scaffold essential for the role of p62 in autophagy.[43] The UBA domains interact with Ub, polyUb chains, and ubiquitinated substrates. Different polyUb chain types, distinguished by covalent linkage of lysine side chain or N-terminal amine of one Ub to the C-terminus of another Ub, signal for different cellular processes.[44,45] Different chain types can exhibit significantly varied binding affinities to the three proteins and, therefore, differ in their abilities to drive shuttle protein LLPS, as discussed below. The IDRs interact with PQC components, such as HSP70, LC3, as well as client proteins marked for degradation.
Figure 1.
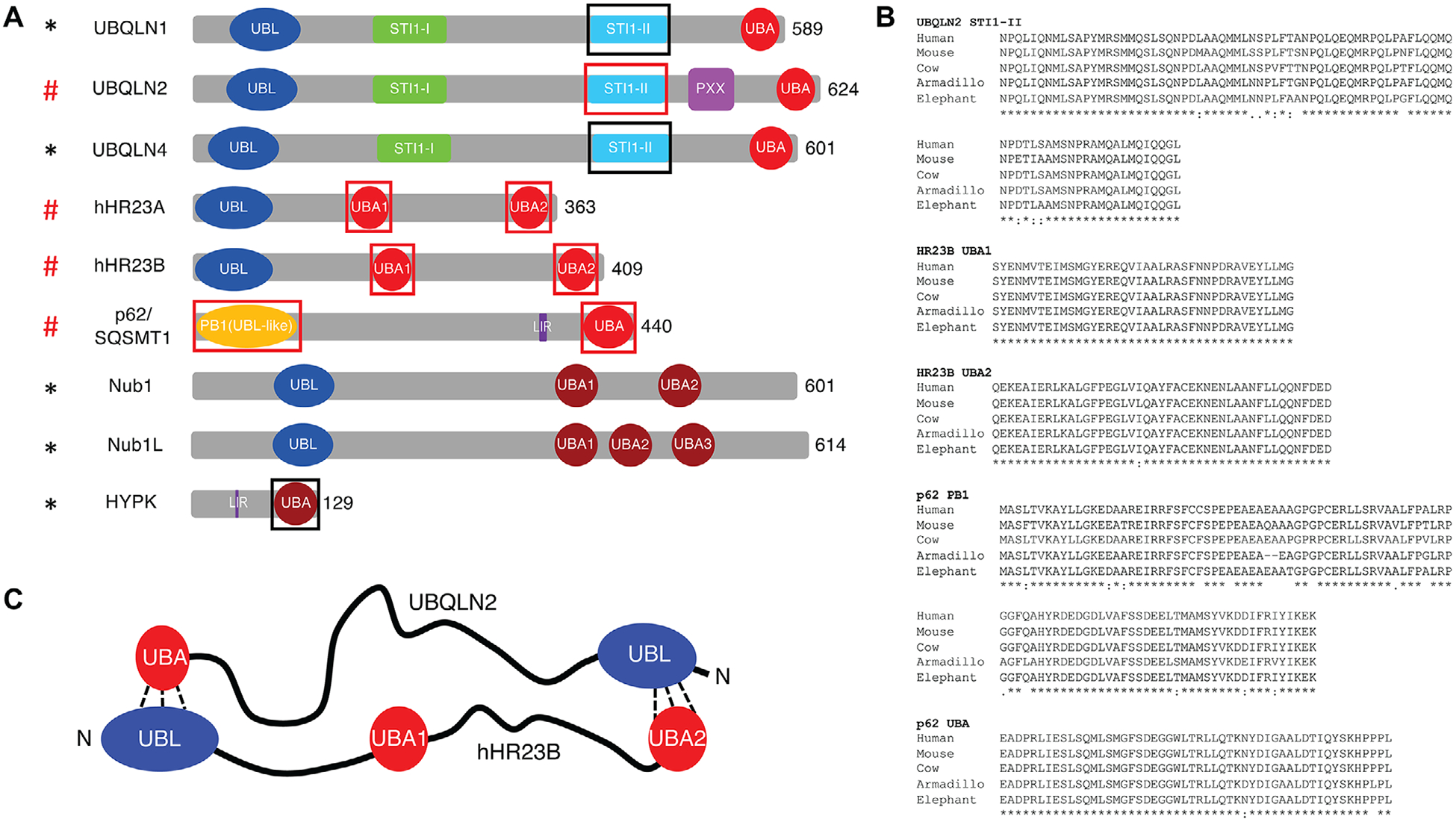
Structures and conservation of ubiquitin-binding shuttle proteins. A) Domain architecture map of shuttle proteins. UBL, ubiquitin-like domain; STI1 (STI1-like region); UBA, ubiquitin-associating domain (red) with Nedd8-associating domain (dark red); PXX, proline-rich domain; PB1 (UBL domain); LIR, LC3-interacting region. Red pound (#) signs and black asterisks (*) denote proteins that are known and hypothesized, respectively, to undergo LLPS. Regions boxed in red and black are known and hypothesized, respectively, to be necessary for promoting multivalency and phase separation of the system. B) Amino acid sequence conservation is shown for the UBQLN2, HR23B, and p62 domains that promote multivalency and LLPS for representative species in each of the five orders in the mammalia class. C) A model for how UBQLN2 and hHR23B bind to one another, adapted with permission.[64] Copyright 2007, Elsevier.
The functionalities of hHR23B, UBQLN2, and p62 are different (Figure 2). hHR23 proteins participate in DNA damage response and proteasomal degradation pathways.[46–48] hHR23s typically exist in the nucleus, but their subcellular localization is cell-cycle dependent. UBQLN2 functions in cellular stress response via both proteasomal degradation and autophagy pathways, and may act as chaperones for misfolded proteins via HSP70-mediated interactions.[49–51] Like hHR23B, UBQLN2’s subcellular localization is cell-cycle dependent, although UBQLN2 is primarily found in the cytoplasm. Cytoplasmic p62 is best known to participate in selective autophagy, whereby cellular components are brought into autophagosomes, double membrane-bound structures that fuse with lysosomes for degradation. p62 directly interacts with polyubiquitinated cargo to localize them into autophagosomes.[43]
Figure 2.
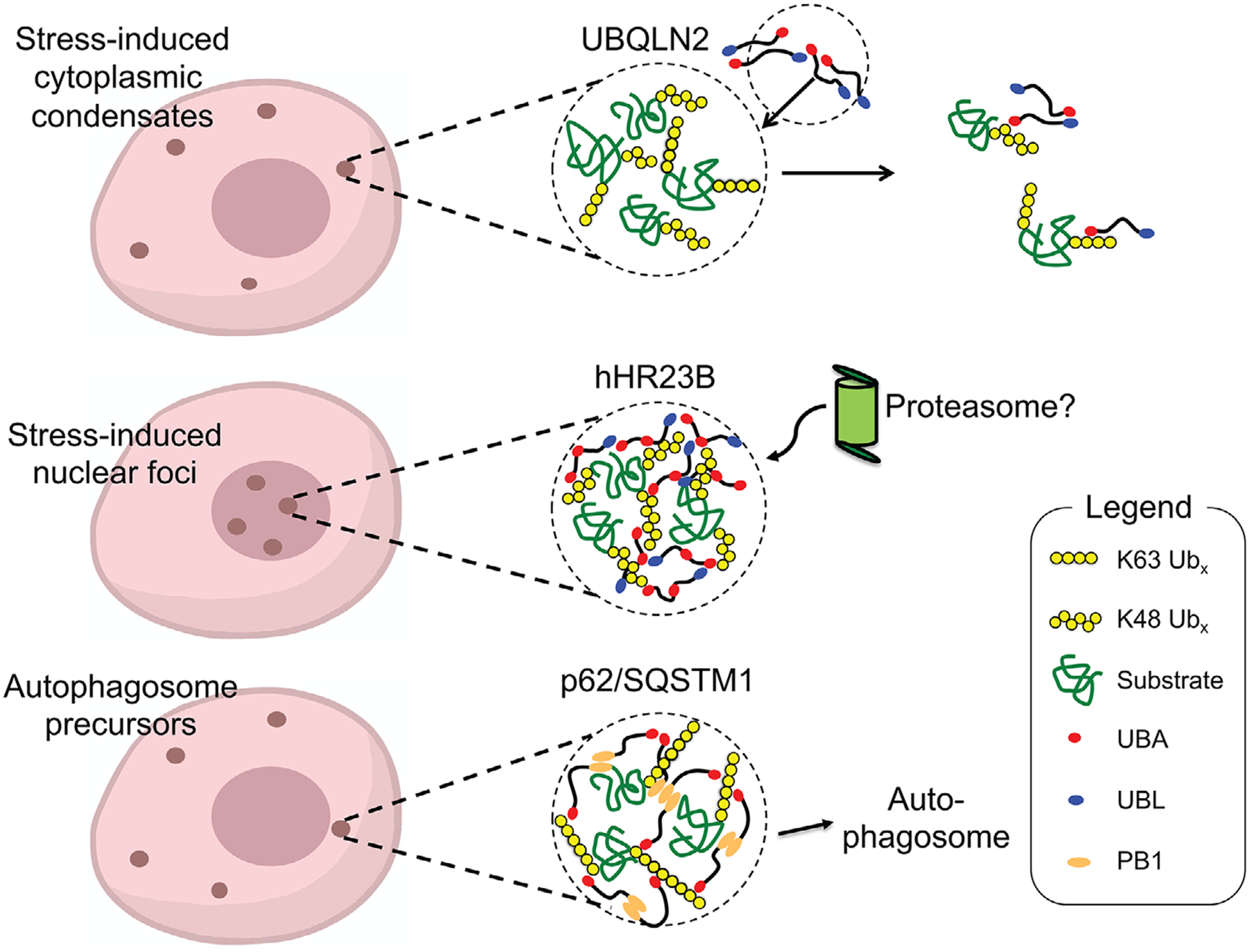
Ub-binding shuttle proteins localize to condensates for PQC functions. (Top) UBQLN2 phase separates and colocalizes with stress-induced condensates in the cytoplasm. Interactions between UBQLN2 and Ub/polyUb disassemble UBQLN2-containing condensates, perhaps providing a mechanism whereby UBQLN2 can shuttle ubiquitinated substrates out of condensates. (Middle) Proteasomal shuttle protein hHR23B condenses mainly with K48-linked polyUb. hHR23B colocalizes with polyubiquitinated substrates and proteasomes in stress-induced nuclear condensates. These proteasome-containing foci are active degradation centers. (Bottom) Autophagy adaptor p62 condenses mainly with K63-linked polyUb chains to form cytoplasmic condensates that may serve as precursors of autophagosomes. In the case of hHR23B and p62, shuttle proteins may provide multivalent interactions to promote formation of condensates with polyUb.
Interestingly, prior to the discoveries that each protein undergoes LLPS, all three proteins were shown to localize to round punctate structures in cells.[52,53] The challenge in analyzing LLPS of these proteins in vivo stems from the heterogeneity in punctate structures and dynamics, and the need for careful experimental controls to evaluate the biophysical properties of the cellular compartment (e.g., membrane-bound vs membraneless). This problem is exemplified by p62, which is found in both autophagosomes and condensates. Sun et al. cleverly used autophagy-deficient cells that lack autophagosomes and lysosomes to observe overexpressed p62 in membraneless compartments.[19] UBQLN2 is found in both autophagosomes and condensates (SGs when endogenously expressed or stress-induced puncta when overexpressed).[54–56] Therefore, techniques like that used by Sun et al. will be necessary to isolate different UBQLN2 populations for study. These observations highlight the need to revisit literature for evidence of LLPS in other systems.
3. Oligomerization Contributes to LLPS of Ub-Binding Shuttle Proteins
Oligomerization propensity has emerged as a common molecular determinant underlying shuttle proteins LLPS (Box 1). Shuttle protein oligomerization is mediated by both folded domains and IDRs, which are highly conserved in mammals, consistent with their functional importance (Figure 1B). Here, we discuss specific contributions of each domain to the abilities of UBQLN2, hHR23B, and p62 to undergo LLPS.
3.1. UBQLN2 LLPS Is Driven by Its STI1-II Region and Modulated by UBL and UBA Domains
Secondary structure and disorder algorithms predict middle segment of UBQLN2 (residues 110–580) to be intrinsically disordered, whereas the UBL and UBA domains are well-folded.[15] Via biophysical experiments including small-angle scattering and size exclusion chromatography, we showed that UBQLN2 STI1-II (Figure 1), containing residues 379–462, was necessary for both oligomerization and LLPS.[15] The STI1-II segment exhibits a prion-rich and low-complexity amino acid composition characteristic of other phase separating IDRs. Exclusion of STI1-II rendered UBQLN2 monomeric and unable to phase separate. We created a minimal construct (450–624) that oligomerized and underwent LLPS in vitro. Nuclear magnetic resonance (NMR) studies revealed that resonances for STI1-II residues 450–470 exhibited extensive exchange broadening with increasing protein concentration, consistent with oligomerization behavior. Importantly, NMR revealed that residues 450–580 are intrinsically disordered, confirming secondary structure predictions.[15]
Important to UBQLN2 self-association are also the proline-rich (PXX) and UBA domains that transiently contact the STI1-II region.[15,21] The PXX region harbors most of the disease-linked mutations.[57] Disease-linked UBQLN2 450–624 mutants exhibiting increased oligomerization propensity phase separate at lower protein concentrations than WT, and undergo liquid-to-solid phase transitions in vitro.[21] Deletion of the UBA domain significantly affects the ability of UBQLN2 to phase separate in vitro and when overexpressed in cells.[15] The UBL domain also modulates UBQLN2 LLPS since UBQLN2 379–624, which lacks the UBL domain, phase separates to a lesser extent when UBL is added in trans. We hypothesize that UBL effects on LLPS are mediated via weak UBA-UBL interactions with a binding affinity around 175 μm.[58] Therefore, we suggest that the STI1-II is the main driver of UBQLN2 oligomerization and LLPS with further modulation by the PXX region and UBL and UBA domains.[59–61]
3.2. hHR23B LLPS Is Driven by Interactions between Its UBA Domains and Polyub Chains and Modulated by Its UBL Domain
In contrast to UBQLN2, in vitro experiments show that hHR23B does not phase separate alone, but via interactions between hHR23B UBA domains and polyUb chains (Figure 2).[17] For hHR23B, the UBA domains are essential for LLPS with polyUb. hHR23B exists in an inactive dimeric state such that two copies of the UBL-UBA protein interact in a head-to-tail fashion.[62,63] NMR studies of homologous hHR23A show that the UBL interacts with each of the two UBA domains.[63] A NMR dynamics study revealed that hHR23A did not behave as a monomer unless the UBL domain was deleted.[63] Notably, removal of the UBL domain also altered hHR23B-polyUb LLPS in vitro.[17] Instead of being round and liquid-like, hHR23B-polyUb droplets appeared amorphous and aggregated. This suggests that the UBL domain modulates hHR23B/polyUb LLPS without directly interacting with polyUb. Furthermore, hHR23A UBA2 interacts with UBQLN2 UBL,[64] thus providing a possible multivalent platform for LLPS involving multiple UBL and UBA domains across Ub-binding shuttle proteins (Figure 1C).
3.3. p62 LLPS Can Be Driven by Oligomerization of Its UBL Domain and Interactions between Its UBA Domain and PolyUb Chains
The molecular basis for p62/polyUb LLPS lies with the ability of its structured p62 PB1 domain to homo-oligomerize to form a filamentous scaffold.[65,66] Without this domain, overexpressed p62 does not form filaments or condense with polyUb.[19] It is proposed that the PB1 oligomeric scaffold in p62 filaments allows the C-terminal UBA domains to extend out of the scaffold, enabling dynamic interactions with polyUb to mediate p62/polyUb LLPS.
p62 can also phase separate without its PB1 domain and polyUb.[67] Histone chaperone DAXX interacts with p62 at intrinsically-disordered residues 246–300 to promotes p62 oligomerization and LLPS in vitro and in cells. Therefore, p62 can readily phase separate with interacting partners that significantly enhance the multivalency needed for LLPS (Box 1). Since the LLPS-promoting interactions between p62 and its binding partners (i.e., polyUb and DAXX) occur at separate p62 regions, it is likely that simultaneous interactions lower the p62 concentration needed for LLPS (Box 2). Having multiple LLPS-driving binding partners also enables rapid formation of p62 condensates in response to different cellular stimuli.
Box 2. Principles of Phase Separation by Homotypic and Heterotypic Interactions.
Phase separation by homotypic interactions occurs when a single macromolecular component spontaneously undergoes LLPS in response to physicochemical changes. These interactions can be driven by self-associating “sticker” IDRs and folded domains, as observed for the UBQLN2 system described here. Phase separation by homotypic interactions is characterized by a saturation concentration (csat) threshold above which the system phase separates, such that the dense phase concentration (cdense) is significantly higher than that of the dilute phase (cdilute = csat) (Figure 3A).[68] Below csat, the protein solution exists in a single, miscible phase. At a single temperature, as the total protein concentration is increased, cdilute and cdense remain fixed, while the relative volumes of the dilute and dense phases change. This is reflected in a horizontal tie line (Figure 3A). This behavior is further confirmed in cells.[69]
In contrast, phase separation by heterotypic interactions requires binding between two or more distinct macromolecular components, such as DNA-protein, RNA-protein coacervates, or the hHR23B-polyUb and p62-polyUb systems described here (Figure 3B). Unlike what is observed for homotypic LLPS, there is no fixed csat for each component to phase separate.[25,70] Instead, multicomponent LLPS depends on binding stoichiometry and the strengths of the self- and cross-interactions among the components.[71] These interaction strengths are reflected both in the shapes of the concentration-concentration phase diagrams and the slopes of the tie lines. Important to LLPS driven by heterotypic interactions is reentrant phase behavior, whereby increasing concentration of one component can drive both assembly and dissolution of the condensate (Figure 3B). LLPS can be hindered if the number of binding sites on one component matches or is an integer multiple of the number of binding sites on the other, inhibiting the formation of networks of interconnected molecules.[72,73] These principles have been illustrated in the Rubisco-EPYC1 system,[72] but likely extend to the shuttle proteins discussed here.
3.4. Identification of “Sticker” Regions Is Key to Elucidating Determinants of Shuttle Protein LLPS
We suggest that folded domains and IDRs that mediate shuttle protein oligomerization are LLPS-promoting “stickers” (Box 1). Identifying “stickers” in shuttle proteins via experimental and computational methods will be important to further test this hypothesis.[31] NMR has proven to be a powerful approach to examine sites of self-association on a residue-by-residue basis for UBQLN2.[15,21,31] Additionally, the domains contributing most to mediating LLPS differ among shuttle proteins, given the variations in shuttle protein multidomain architecture and whether polyUb is necessary for LLPS. However, polyUb chains do not have to be the only promoter of p62 or hHR23B LLPS, as p62 interactions with DAXX can also promote p62 LLPS. This raises the important point that both p62 and hHR23B could condense with other proteins or even small molecules that increase oligomerization propensity and promote LLPS. While our focus here has been on “stickers,” “spacers” may also modulate shuttle protein LLPS. In general, we found substitutions to “spacer” residues to have little effects on the overall UBQLN2 phase diagram. However, certain single amino acid substitutions, such as Asp and Glu, at both “sticker” and “spacer” sites substantially increase the UBQLN2 concentrations needed to promote LLPS.[31]
4. PolyUb Induces Phase Transitions
The effects of polyUb on UBQLN2, hHR23B, and p62 phase transitions are examples of polyphasic linkage, defined as ligand binding that changes the equilibrium between phases, for example, phase-separated and miscible states.[74] For UBQLN2, in vitro Ub or K48-linked polyUb binding favors the diffuse, miscible state.[15] In other words, the UBQLN2 saturation concentration (csat) required for LLPS is shifted to higher values (Figure 3C). UBQLN2 is unique as it is the only known system whose condensates are disassembled via noncovalent interactions with Ub. For hHR23B and p62, polyUb binding favors the phase-separated state.[17,19,20] Other examples of polyphasic linkage include profilin interacting with huntingtin fragments to shift csat for huntingtin LLPS and assembly formation[75] and Karyopherin-β1 interacting with nuclear localization sequences on RNA-binding proteins to disassemble RNA-binding protein aggregates.[76]
Figure 3.
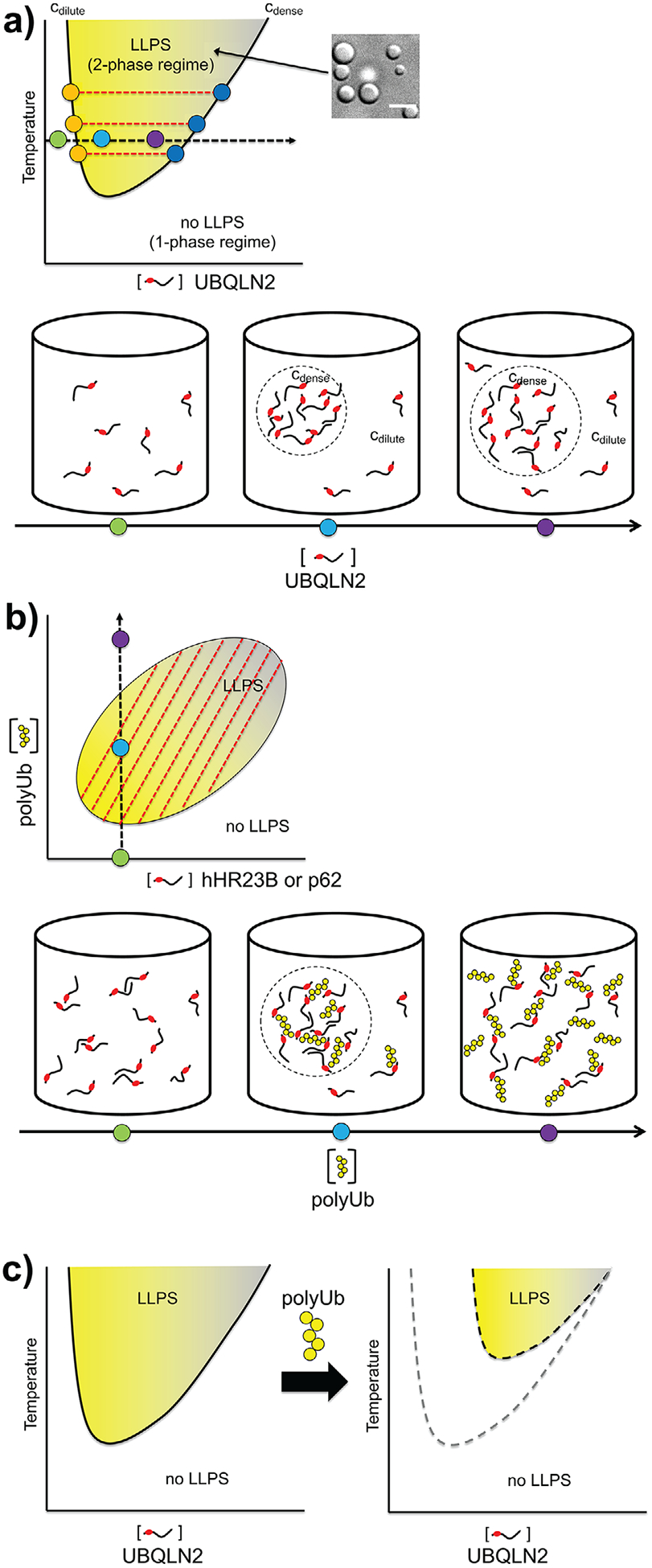
Mapping LLPS behavior of Ub-binding shuttle proteins. A) Temperature-concentration phase diagram of UBQLN2. UBQLN2 phase separates as temperature increases,[15] characteristic of systems with LCST (lower critical solution temperature) phase behavior. Colored dots on cdilute and cdense arms of phase diagram indicate protein concentrations in dilute and dense phases, respectively, as a function of temperature. Red dashed lines are tie lines connecting cdilute and cdense at a specific temperature. Example depictions of protein solution behavior are indicated by colored dots on black dashed line as overall UBQLN2 concentration is increased. B) Concentration-concentration phase diagram for shuttle protein/polyUb condensates driven by heterotypic interactions (e.g., hHR23B or p62) at a particular temperature. Red dashed tie lines do not need to be horizontal to determine dilute and dense phase concentrations of components. Positive slopes indicate mutual interactions between components that drive both components into the phase-separated state. For details, refer to ref. [25]. Based on current knowledge, neither component (polyUb or shuttle protein) is able to phase separate on its own. Example depictions of protein solution behavior are indicated by colored dots on black dashed line as overall polyUb concentration is increased at specified shuttle protein concentration and temperature. C) Illustration of polyphasic linkage, that is, the effect of Ub (or K48-linked polyUb) binding on shifting the LLPS phase boundaries of UBQLN2. As polyUb is titrated into UBQLN2 solution, the UBQLN2 concentration needed for LLPS is increased to higher values.
4.1. How Does PolyUb Drive Both Assembly and Disassembly of Shuttle Protein Condensates?
This phenomenon occurs due to the differences in the mechanisms that drive shuttle protein LLPS. UBQLN2 phase separates independently of polyUb, but noncovalent interactions with polyUb chains disassemble UBQLN2 droplets in vitro.[15] By contrast, p62 and hHR23B require polyUb chains to undergo LLPS. We classify these different phase behaviors as driven by homotypic (UBQLN2) and heterotypic (p62 and hHR23B) interactions (Figures 2 and 3, Box 2). For UBQLN2, Ub interactions with two “sticker” regions (residues 592–594, 616–620) in the UBQLN2 UBA effectively reduces the number of “stickers” needed for LLPS by homotypic interactions.[15] Consequently, this drives the csat required for LLPS to higher values (Figure 3C).[30] By contrast, polyUb interactions with hHR23B or p62 are necessary to provide emergent “stickers” for LLPS. Furthermore, the conditions for LLPS no longer necessitate that a component’s concentration is above a fixed csat but depend on the concentrations of both hHR23B (or p62) and polyUb, as expected of LLPS by heterotypic interactions (Figure 3B, Box 2). High valency of the components (provided by the two UBA domains in hHR23B and/or long polyUb chains of six or more Ub) promotes LLPS driven by heterotypic interactions.[17,19,20] Importantly, the driving forces for LLPS can be switched from heterotypic to homotypic and vice versa, and this has major implications for how condensates function in vivo.[70,77] For example, polyUb chains of different linkages (K48 vs K63) could bind differently to shuttle proteins or induce conformational changes that alter the driving forces for LLPS (see below).
Systems phase separating via heterotypic interactions are subject to reentrant phase behavior (Figure 3B).[78,79] At a given [B] (e.g., hHR23B or p62), the system transitions into two-phase behavior (i.e., phase separates) as [A] (e.g., polyUb) increases. However, as [A] increases significantly beyond [B], there are not enough [B] molecules to promote two-phase behavior, and thus the system becomes single-phase again.[21] While reentrant phase behavior has not yet been shown for hHR23B-polyUb condensates, this principle is somewhat illustrated in vitro by polyUb effects on the morphology of p62 clusters. Without polyUb, p62 forms filaments. At substoichiometric ratios of octaUb to p62, p62 filaments shorten and form roundish 20 nm-sized clusters.[65] However, excess amounts of polyUb dissolve p62 filaments.[65] Therefore, increasing polyUb concentration promotes a p62 phase transition from filamentous structures into spherical assemblies, but as polyUb concentration is further increased to excess, the system becomes single phase again, consistent with reentrant behavior.
4.2. What Are the Functional Consequences of Shuttle Protein LLPS Driven by Homotypic or Heterotypic Interactions?
We hypothesize that the ability of UBQLN2 to phase separate via homotypic interactions enables it to be a scaffold in assembling stress-induced condensates. We find the UBQLN2 constructs that phase separate in vitro are also ones that form stress-induced condensates when overexpressed in cells, and vice versa.[15] Additionally, UBQLN2 condensates could potentially fuse with other biomolecular condensates, as we and others find endogenously expressed UBQLN2 in SGs.[15,18] Upon engagement with a polyubiquitinated substrate (e.g., covalently modified with a K48-Ub4 tag), UBQLN2 may remove substrate from the condensate as polyUb dissolves UBQLN2 condensates in vitro.
Unlike UBQLN2, hHR23B and p62 require ubiquitinated substrates to form condensates, suggesting an important signaling mechanism for condensate assembly only when enough polyubiquitinated proteins are present, and for condensate dissolution when polyubiquitinated proteins are degraded.[17] This property enables interactions between endogenously expressed hHR23B and polyUb to drive rapid assembly of osmotic stress-induced nuclear foci, which fail to form in hHR23B knockout cells or when polyubiquitination is impaired. Aside from hHR23B and polyubiquitinated substrates, these foci also contain protein degradation machinery, including the p97 segregase, 26S proteasome, and UBE3A ligase, consistent with their role as active proteolytic compartments. Indeed, proteasomal inhibition significantly delays clearance of these nuclear foci. Similarly, overexpressed p62 forms condensates that sequester polyubiquitinated cargo and signal cargo’s degradation by selective autophagy. p62/polyUb condensates may act as scaffolding to recruit LC3 and other autophagy-related proteins to form autophagosomes, which later fuse with lysosomes to degrade the material inside.[19,20] Removal of p62 UBA eliminated LLPS with polyUb and reduced the number of p62 puncta seen in cells.[20] Therefore, the heterotypic interactions that form hHR23B and p62 condensates are critical to the beginning stages of PQC, specifically proteasome-mediated degradation in the nucleus, as well as selective autophagy mechanisms in the cytoplasm. While we presented evidence that suggests shuttle proteins can be scaffolds of condensates, we must emphasize that shuttle proteins can be both scaffolds and clients of condensates, depending on the cellular conditions and compositions (e.g., UBQLN2 as a client in being recruited to SGs).
4.3. How Is Shuttle Protein LLPS Regulated?
We postulate that there are many layers of controls, including 1) binding affinity, 2) polyUb chain linkage and length, 3) conformational changes, and 4) post-translational modifications (PTMs). Dissociation constants (Kd) between Ub and UBA domains vary substantially (1–500 μm), adding complexity to how Ub-binding drives shuttle protein oligomerization and LLPS. For example, p62 UBA exists as monomers or homodimers, which bind monoUb with significantly different Kd’s of ≈40 and 500 μm, respectively,[80] suggesting that polyUb-mediated p62 LLPS is more efficient when p62 UBA domain is monomeric.
Longer polyUb chains increase valency of polyUb, but linkage type modulates binding affinity with UBA domains. Binding preferences for polyUb chain types vary significantly among the three proteins. hHR23A UBA2 domain (91% identity with hHR23B UBA2) binds monoUb, K48- and K63-linked Ub4 with Kd’s of 540, 8, and 28 μm, respectively.[81,82] Structurally, a single UBA2 engages two Ub units in K48-linked chains in a sandwich-like mode, whereas Ub units in K63-linked chains and UBA2 interact with 1:1 stoichiometry.[81] Consistent with binding affinities, lesser amounts of K48- than K63-linked polyUb were required to induce in vitro hHR23B LLPS.[17] Binding preference of p62 UBA for polyUb chain linkages is M1 > K63 > K48.[42,83,84] Correspondingly, purified p62 phase separates more readily with K63-linked polyUb compared to K48-linked polyUb.[19] The binding preferences of hHR23B and p62 for K48-and K63-linked chains, respectively, are consistent with the roles of hHR23B in proteasomal degradation and p62 in autophagy. However, we note that K48- and K63-linked polyUb chains do not exclusively determine pathway choice between UPS and autophagy, which is far more complex and highly dependent on the oligomerization states of shuttle proteins.[84,85] Unlike hHR23B and p62, UBQLN2 does not exhibit preferences to either chain in studies that used isolated UBA domains rather than full-length proteins.[82,86] Recently, full-length UBQLN1, a UBQLN2 paralog, was found to exhibit a preference for K63-linked polyUb, although the authors cautioned against overinterpretation of these results as GST-UBQLN constructs were employed and GST is known to dimerize.[87] In light of observations made for hHR23B and p62 and involvement of UBQLN2 in both proteasomal degradation and autophagy, it is important to compare the effects of K48- and K63-linked chain on UBQLN2 LLPS. We showed that K48-linked Ub4 eliminates UBQLN2 LLPS in vitro at equimolar ratios.[15] Depending on the binding affinity and mode of binding (see below) between UBQLN2 and K63-linked chains, K63-linked chains could eliminate UBQLN2 LLPS in a similar manner as K48-linked polyUb, or engage in heterotypic interactions with UBQLN2 to enhance UBQLN2 LLPS at low molar ratios before eliminating LLPS at high molar ratio due to reentrant phase behavior (Box 2, Figure 3B).
PolyUb chains of different linkages also adopt dynamic but distinct conformations.[45] Ub units on K63-linked polyUb arrange in a beads-on-string scaffold that could promote LLPS, whereas K48-linked polyUb can be compact and sequester Ub “sticker” regions.[44,45] These different chain conformations can impact mode of binding between Ub units and shuttle proteins to either promote or inhibit LLPS. Moreover, shuttle proteins may undergo polyUb-induced conformational changes that modify the multivalent scaffold necessary for LLPS. hHR23A exhibits binds conformational changes upon binding to proteasomal subunit s5a.[63] UBQLN2 is postulated to switch conformations between its substrate-unbound (“inactive”) and substrate-bound (“active”) states.[60] The effects of different polyUb chains on shuttle protein LLPS are likely sensitive to affinities between shuttle proteins and chains, chain conformation, and conformational changes of shuttle proteins upon binding polyUb. A systematic, quantitative mapping of polyUb/shuttle protein phase diagrams with cdilute and cdense of all components as well as detailed structural and dynamic studies using methods like NMR could shed light on the contributions of these different factors.
Last, PTMs can also regulate LLPS.[88,89]Domains important for shuttle protein LLPS can be phosphorylated (p62 UBA, hHR23A/B UBL) or ubiquitinated (p62 UBA).[90,91] Phosphomimetic substitution (S403E) on p62 UBA promotes stronger binding to polyUb and LLPS.[19] We are only just beginning to determine how these four (or more) parameters regulate shuttle protein LLPS. Deeper understanding of these regulatory mechanisms requires work on both experimental and theoretical levels.
5. Do Other Ub-Binding or Ub-Like Binding Shuttle Proteins Function via LLPS?
Prior to the appreciation of LLPS in driving condensate formation, certain Ub-binding shuttle proteins were found in liquid-like assemblies. We presented evidence above that UBQLN2, hHR23B, and p62 all form puncta in cells prior to their recent classification as phase-separating proteins. Many Ub-like proteins exist in cell, including NEDD8, FAT10, SUMO, among others. Some of these proteins are also covalently-linked together via enzymatic cascades similar to how polyUb is made. Therefore, we explore the possibility that these systems also undergo LLPS for cellular functions. Upon close examination, we propose that four Ub/Ub-like binding shuttle proteins, UBQLN1, UBQLN4, NUB1, and HYPK, may undergo LLPS with other components in cells. UBQLN1 and UBQLN4 are UBQLN2 paralogs and bind to Ub. HYPK and NUB1 both comprise UBA domains that are distinct from Ub-binding UBA domains but bind specifically to NED D8 and neddylated substrates to mediate PQC. Interestingly, neddylation parallels ubiquitination in that they both signal for proteasomal degradation and autophagy, among other pathways. Based on their functions, HYPK and NUB1 may be the equivalents of p62 and UBQLN2, respectively, in targeting neddylated substrates for degradation pathways.
5.1. Do UBQLN1 and UBQLN4 Phase Separate?
UBQLN1 and UBQLN4 are 74% and 56% identical to UBQLN2, respectively. The largest difference between UBQLN2 and UBQLNs is the presence of the PXX region between the STI1-II and UBA domains (Figure 1). Importantly, UBQLN2 still phase separates without the PXX region.[15] Moreover, like UBQLN2, UBQLN1 and UBQLN4 localize into puncta in cells.[92,93] Given these observations, we propose that UBQLN1 and UBQLN4 undergo LLPS and form membraneless condensates in cells. However, UBQLNs also associate with membrane-bound autophagosomes; UBQLN4 and UBQLN1 colocalize with autophagosomal marker LC3.[94] UBQLN4 colocalizes with polyUb in aggresome structures[95] and is recruited to sites of DNA damage,[96] where UBQLN4 form puncta that colocalize with 53BP1. Therefore, UBQLNs are found in diverse types of cellular structures. While we hypothesize that all UBQLNs can condense into membraneless structures, careful work is necessary to delineate which puncta are liquid-like condensates, aggresomes, autophagosomes or others.
Notably, UBQLN paralogs have different subcellular localizations depending on cellular conditions. UBQLN2 is generally cytoplasmic; UBQLN1 is in the nucleus and cytosol; UBQLN4 is in the nucleus, ER, and cytoplasm.[49,92] Therefore, different spatial arrangements and expression levels under certain cellular signals could be ways to control when and where UBQLN condensates form. UBQLN STI1-I and STI1-II domains interact with many clients including RNA-binding proteins, autophagosome marker LC3, and membrane proteins.[18,61,95,97] Therefore, UBQLN condensate behavior could be regulated by different client protein interactions, depending on the UBQLN paralog. Moreover, UBQLN2 forms heterodimers with UBQLN1 and UBQLN4.[59,94] It remains to be seen how interactions among UBQLN paralogs modulate UBQLN LLPS in cells. Interactions among client proteins and UBQLNs may lead to client sequestration into UBQLN condensates where clients are protected from degradation before possible recruitment of PQC machinery to enable client degradation (see below).
5.2. HYPK Could Phase Separate to Form Autophagosomes around Polyneddylated Aggregates
HYPK is a multifunctional protein that binds and sequesters Htt97Qexon1, SOD1-G93A, and α-Syn-A53T aggregates, acts as a chaperone to reduce aggregates and apoptosis, and drives the formation of autophagosomes to clear huntingtin aggregates via autophagy, among other aggregation-reducing roles.[98–101] HYPK is a small, mostly disordered protein comprising LIR and UBA domains that bind to LC3 and NEDD8, respectively (Figure 1).[101] HYPK self-associates via its UBA domain.[100,102] UBA domain exists as metastable seeds that drive formation of nanometer-sized HYPK annular structures, which resemble in vitro protein droplets.[100,103] Interestingly, overexpressed HYPK UBA forms highly dynamic cytoplasmic bodies.[97] Although these results suggest that HYPK could undergo LLPS, more stringent studies are needed to test this hypothesis.
HYPK is involved in many pathways, but mechanisms by which HYPK carries out its functions are still largely unknown. Ghosh and colleagues proposed that HYPK binds to polyneddylated protein aggregates and recruits LC3 to form autophagosomes around aggregates to initiate aggrephagy.[101] In yeast, several autophagy proteins undergo LLPS to form preautophagosomal structures.[104] HYPK might form condensates with other autophagy proteins and then recruit polyneddylated aggregates or vice versa, akin to how p62 condenses with polyubiquitinated proteins (see above). It will be interesting to determine the effects of polyNEDD8 chains on HYPK and its oligomeric structures.
5.3. NUB1 Could Phase Separate to Facilitate Both Proteasomal Degradation and Autophagy
NUB1 binds to and reduces cellular levels of NEDD8 and neddylated subtrates.[105] Its longer splicing variant, NUB1L, also binds to and facilitate FAT10 degradation.[106] NUB1 and NUB1L contain two and three UBA domains, respectively, and an N-terminal proteasome-binding UBL domain (Figure 1).[107,108] These properties enable NUB1/L to facilitate proteasomal degradation of NEDD8, FAT10, and their substrates.[109,110] NUB1 specifically interacts with p97/VCP to join p97UFD1/NPL4 complex and mediate the delivery of NEDD8 to the proteasome.[110] Interestingly, upon proteasomal inhibition, increased NUB1 expression leads to increased LC3B and p62 levels, and autophagosome size and number, suggesting a role for NUB1 in facilitating a switch from proteasomal degradation to autophagy.[111] Moreover, NUB1 and p62 interact and colocalize into cytoplasmic puncta. As discussed above, p62 phase separates and both p97 and p62 are recruited to SGs, suggesting that NUB1 could also phase separate, and potentially use LLPS as a mechanism to partition into other condensates to carry out its functions.
NUB1 also directly interact with substrate proteins, independent of NEDD8 or FAT10, reminiscent of how UBQLNs interact with clients (see above). NUB1 colocalizes with synphilin-1 and α-synuclein in Lewy bodies in brains of patients with dementia or Parkinson’s disease.[112,113] In HEK293 cells, NUB1 is found in synphilin-1-positive foci and NUB1 overexpression lowers synphilin-1 level. NUB1 interacts directly with and might shuttle synphilin-1 to the proteasome for degradation.[112] NUB1 interacts with tau and is recruited to tau-positive microtubule bundles and inclusions, reducing tau aggregation.[114] Moreover, NUB1 specifically interacts with telomeric protein TRF1 to facilitate its proteasomal degradation.[115] Interestingly, the NUB1 binding proteins just described are all found in phase-separated compartments: synphilin-1 and α-synuclein undergo LLPS before maturing into insoluble Lewy bodies-like aggregates[116,117]; tau LLPS may be an intermediate step toward aggregate formation[118]; telomeres and TRF1 cluster in a class of PML bodies specialized in telomere length maintenance.[119] NUB1 comprises multiple UBA domains and 13 cysteines, providing the mulitivalency and oligomerization (from potential disulfide bonds) needed for LLPS. Therefore, NUB1 might undergo LLPS to be recruited to condensates where it interacts with and shuttle aggregated proteins to the proteasome for degradation.
While evidence supporting NUB1 LLPS is indirect, NUB1 shares properties of other phase-separating Ub/Ub-like binding shuttle proteins, such as the potential to oligomerize/self-associate and colocalize with proteins found in condensates. Indeed, there exists crosstalk among ubiquitination, neddylation, and FATylation signaling pathways and components. The ease and speed at which biomolecular condensates can form, disassemble, and change would be efficient mechanisms for cells to communicate different signaling events.
6. What Are the Functional Consequences of Ub-Binding Shuttle Protein LLPS?
Above, we established that many Ub-binding shuttle proteins undergo LLPS, either independently or facilitated by interactions with polyUb (and ubiquitinated substrates). Insights from other systems show that LLPS enables finelytuned responses to cellular stresses under which levels and subcellular organization of macromolecular components can quickly change. What are the unique functions that LLPS imparts on Ub-binding shuttle proteins? Below, we speculate on four potential biological functions of shuttle protein-containing condensates.
6.1. Shuttle Protein LLPS May Lead to Recruitment of PQC Components to Condensates
Instead of ubiquitinated cargo being “shuttled” to the proteasome or autophagosomes, shuttle protein LLPS can selectively enrich cargo and PQC components inside condensates, as shown for p62 and hHR23B. Overexpressed p62 condenses with ubiquitinated proteins to initiate a liquid-like structure that subsequently recruits LC3 to form autophagosomes.[19,20] Under osmotic stress, endogenous hHR23B phase separates with ubiquitinated substrates and recruits proteasomes to condensates which effectively act as active degradation centers.[17] These proteasome condensates fail to form in the absence of hHR23B, leading to apoptosis under mild hyperosmotic stress. In this case, hHR23B is essential for cell survival under stress.
Green alga Chlamydomonas reinhardtii also contains proteasome clusters that reside at the ER membrane, have liquid-like properties, are surrounded by Cdc48 and hypothesized to facilitate direct degradation of protein components translocated from the ER lumen.[120] Other examples of proteasome clusters include cytoplasmic proteasome storage granules (PSGs) that form in yeast and Arabidopsis in response to carbon starvation and are proposed to protect proteasomes from proteaphagy.[121,122] The driving forces for the formation of ER-associated proteasome clusters and PSGs are currently unknown, but we speculate that Ub-binding shuttle proteins may also be enriched at these locations.
Last, overexpressed adaptor protein SPOP phase separates with substrates, such as DAXX, to form nuclear condensates and recruit Ub E3 ligases into condensates to ubiquitinate substrates fated for degradation.[36] E3 ligase E6AP interacts with UBQLN1, UBQLN2, and hHR23A.[49,123] We speculate that interaction with these shuttle proteins enables recruitment of E6AP into condensates, where E6AP or another E3 ligase ubiquitinates substrate proteins. Depending on spatiotemporal cellular signals and condensate composition, these condensates could serve as sequestration compartments and/or active protein degradation centers.
6.2. Shuttle Protein LLPS Could Regulate Condensate Assembly or Disassembly
Dysregulation of assembly/disassembly processes results in persistent and/or aberrant membraneless organelles linked to neurodegenerative disease states.[124,125] p62 and UBQLN2 are associated with stress granules (SGs), along with many other PQC components including chaperone proteins (e.g., HSP70).[125,126] The ability of p62 and UBQLN2 to phase separate may enable them to partition into and mix with SGs. p62 interacts with C9orf72 to promote autophagic degradation of SGs.[127] Persistent SGs, which can be modeled by long light-induced oligomerization of a modified version of the SG scaffold protein G3BP, recruit p62 and VCP/p97 at late times.[9] UBQLN2 also negatively affects SGs as depletion of UBQLN2 increased SG number and size.[18] Moreover, UBL-containing multidomain protein ZFAND1 regulates SGs by interacting with the 26S proteasome and p97, and recruiting them to facilitate clearance of arsenite-induced SGs.[128] Therefore, shuttle proteins may partition into condensates to efficiently recruit PQC components to enable condensate disassembly by autophagy or to selectively remove condensate components by the UPS. Lack of condensate regulation by shuttle proteins can lead to persistent or less liquid-like SGs that can mature into toxic aggregates.
In contrast, other proteins, such as Ub-binding UBAP2L, can positively regulate SG formation.[37,129] UBAP2L knockdown results in smaller SGs, possibly inhibiting the sequestration of some cellular components that contribute to SG functionality and/or that need to be protected against degradation. UBAP2L contains a N-terminal UBA domain followed by long IDRs. RGG motifs and the C-terminal IDR of UBAP2L mediate SG recruitment and self-association.[37,130] The UBA domain appears dispensable for SG assembly from deletion studies,[130] but UBAP2L is enriched in Ub-containing aggregates after proteasome inhibition.[131] Therefore, we speculate that polyUb chain association with UBAP2L could further regulate maturation processes of condensates, such as liquid-to-solid changes in condensate material properties.
6.3. Shuttle Protein LLPS Could Enable Non-Ubiquitinated Client Proteins to Undergo Phase Separation and Be Sequestered inside Condensates
UBQLN1 engages with mitochondrial protein BCLb via its STI domains.[61] The middle region of UBQLN4 interacts with hydrophobic domains of mitochondrial proteins.[95] The middle region of UBQLN2 interacts with RNA-binding proteins,[18] including hnRNPA1, which phase separates and localizes to SGs.[23] Moreover, hHR23s can engage client proteins to protect them from degradation.[132] These findings illustrate the multifaceted features of shuttle proteins, particularly those functions not necessarily affiliated with protein degradation, but rather protein stabilization and/or chaperone-like abilities.[97] With the new knowledge that these shuttle proteins phase separate, we hypothesize that shuttle proteins condense with specific client proteins to shield them from degradation. Based on different binding and/or conformational states of shuttle proteins in response to cellular signals, these condensates could later recruit PQC machinery to mediate client protein degradation.[60,97] The concept of client protein sequestration is not without precedent. Nuclear phase-separating protein NPM1 interacts with misfolded proteins to convert the nucleolus into a PQC compartment.[133]
6.4. Shuttle Protein LLPS May Mediate Extraction of Ubiquitinated Proteins from Condensates
Ub and K48-linked polyUb disassemble UBQLN2 condensates in vitro.[54] We speculate that substrate proteins could be ubiquitinated with K48-linked polyUb inside UBQLN2-containing condensates and extracted from condensates upon noncovalent interactions between the polyUb chain and UBQLN2. This process resembles a versatile mechanism for UBQLN2 to transport cargo out of condensates for subsequent processing or other functions. While not entirely analogous, RNA-binding protein hnRNPA2 may use a similar PTM-driven mechanism to release mRNA cargo from hnRNPA2-containing transport granules.[134] hnRNPA2 undergoes LLPS, and tyrosine phosphorylation promotes disassembly of hnRNPA2 condensates.
In summary, we are just beginning to realize the existence of shuttle protein condensates in cells and determine their functions in PQC. We presented evidence that highlights how engagement of shuttle proteins with polyUb can both assemble and disassemble condensates, depending on whether LLPS is mediated by heterotypic or homotypic interactions. Similar principles likely apply to other PQC-related proteins that interact with Ub-like proteins such as NEDD8 and SUMO. Importantly, the myriad of interactions among shuttle proteins, PQC components, and client proteins offer a rich, energetic landscape that likely tunes condensate formation and dissolution in cells. Shuttle protein LLPS could therefore be an elegant spatiotemporal mechanism for executing a dynamic PQC response, particularly in times of cell stress or in cell types with unique protein homeostasis needs such as neurons.
7. Conclusions and Future Perspectives
Important, but seemingly unconnected, recent observations in the autophagy and UPS subfields of PQC highlight the emerging principle that LLPS drives formation of condensates specialized in PQC activities, such as proteasome-enriched active protein degradation centers and autophagosomes. Here, we highlight the common theme that Ub/Ub-like binding shuttle proteins undergo LLPS mediated by both folded UBA/UBL domains and IDRs. This ability to phase separate enables recruitment of shuttle proteins or PQC components to condensates. Shuttle proteins exhibit different expression levels and subcellular localizations, enabling spatiotemporal behavior of condensates and dynamic PQC response to different cellular stresses.
What is the connection between LLPS and mechanisms by which shuttle proteins perform their functions? Most shuttle proteins exhibit preferences for client proteins, suggesting that clients can modulate shuttle protein LLPS. We speculate that client-binding domains of shuttle proteins can affect LLPS, condensate material properties, and assembly/disassembly processes. The combination of clients and different polyUb linkages offers a rich landscape for protein–protein interactions to dynamically regulate shuttle protein LLPS. Resolving protein–protein interactions at a single-molecule level will contribute to quantification of multicomponent phase diagrams. The concepts of polyphasic linkage and theory concerning multi-component LLPS are important to elucidate how these systems respond to different interactions.
It is also critical to determine the structures that shuttle proteins can form in cells. Improvements in super-resolution microscopy and cryo-EM/cryo-ET technologies will help reveal the organization of these condensates. These experiments are increasingly powerful as more interacting partners are identified and labeled for imaging. These experiments will determine if condensates form via LLPS or other mechanisms and if shuttle protein LLPS is a requirement for their functions or just a general phenomenon. Regardless, to fully understand the role and mode of action of shuttle proteins in PQC, a multifaceted approach on cellular, molecular, and computational levels is needed.
Acknowledgements
This work was supported by ALS Association grant 18-IIP-400, the National Science Foundation (CAREER award 1750462), and NIH R01 GM136946 to C.A.C. The authors also acknowledge stimulating discussions with Tanja Mittag, J. Paul Taylor, and Rohit Pappu over the years, in addition to recent thoughtful meetings with Conrad Weihl. The authors would like to apologize to those whose works could not be included here due to citation limits.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Thuy P. Dao, Departments of Biology and Chemistry, Syracuse University, Syracuse, NY 13244, USA
Carlos A. Castañeda, Departments of Biology and Chemistry, Syracuse University, Syracuse, NY 13244, USA Bioinspired Institute, Syracuse University, Syracuse, NY 13244, USA; Interdisciplinary Neuroscience Program, Syracuse University, Syracuse NY 13244, USA.
References
- [1].Banani SF, Lee HO, Hyman AA, Rosen MK, Nat. Rev. Mol. Cell Biol 2017, 18, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA, Science 2009, 324, 1729. [DOI] [PubMed] [Google Scholar]
- [3].Brangwynne CP, Mitchison TJ, Hyman AA, Proc. Natl. Acad. Sci. USA 2011, 108, 4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Protter DSW, Parker R, Trends Cell Biol 2016, 26, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alberti S, Dormann D, Annu. Rev. Genet 2019, 53, 171. [DOI] [PubMed] [Google Scholar]
- [6].Holehouse AS, Pappu RV, Biochemistry 2018, 57, 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nedelsky NB, Taylor JP, Nat. Rev. Neurol 2019, 15, 272. [DOI] [PubMed] [Google Scholar]
- [8].Takahashi J, Fujigasaki H, Iwabuchi K, Bruni AC, Uchihara T, El Hachimi KH, Stevanin G, Dürr A, Lebre AS, Trottier Y, de Thé H, Tanaka J, Hauw JJ, Duyckaerts C, Brice A, Neurobiol. Dis 2003, 13, 230. [DOI] [PubMed] [Google Scholar]
- [9].Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, Taylor JP, eLife 2019, 8, e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lowe J, Blanchard A, Morrell K, Lennox G, Reynolds L, Billett M,Landon M, Mayer RJ, J. Pathol 1988, 155, 9. [DOI] [PubMed] [Google Scholar]
- [11].Manetto V, Perry G, Tabaton M, Mulvihill P, Fried VA, Smith HT,Gambetti P, Autilio-Gambetti L, Proc. Natl. Acad. Sci. USA 1988, 85, 4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morimoto D, Walinda E, Fukada H, Sou Y-S, Kageyama S, Hoshino M, Fujii T, Tsuchiya H, Saeki Y, Arita K, Ariyoshi M, Tochio H, Iwai K, Namba K, Komatsu M, Tanaka K, Shirakawa M, Nat. Commun 2015, 6, 6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Riley BE, Kaiser SE, Shaler TA, Ng ACY, Hara T, Hipp MS,Lage K, Xavier RJ, Ryu K-Y, Taguchi K, Yamamoto M, Tanaka K,Mizushima N, Komatsu M, Kopito RR, J. Cell Biol 2010, 191, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bergink S, Severijnen L-A, Wijgers N, Sugasawa K, Yousaf H, Kros JM, van Swieten J, Oostra BA, Hoeijmakers JH, Vermeulen W, Willemsen R, Neurobiol. Dis 2006, 23, 708. [DOI] [PubMed] [Google Scholar]
- [15].Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, Castañeda CA, Mol. Cell 2018, 69, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang H, Yue H-W, He W-T, Hong J-Y, Jiang L-L, Hu H-Y, FASEBJ. 2018, 32, 2923. [DOI] [PubMed] [Google Scholar]
- [17].Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernández-Busnadiego R, Tanaka K, Saeki Y, Nature 2020, 578, 296. [DOI] [PubMed] [Google Scholar]
- [18].Alexander EJ, Niaki AG, Zhang T, Sarkar J, Liu Y, Nirujogi RS,Pandey A, Myong S, Wang J, Proc. Natl. Acad. Sci. USA 2018, 115, E11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun D, Wu R, Zheng J, Li P, Yu L, Cell Res. 2018, 28, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R, Tarafder AK, Sachse C, Martens S, EMBO J. 2018, 37, 98308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dao TP, Martyniak B, Canning AJ, Lei Y, Colicino EG, Cosgrove MS, Hehnly H, Castañeda CA, Structure 2019, 27, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL, Cell 2012, 149, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ,Mittag T, Taylor JP, Cell 2015, 163, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alberti S, Gladfelter A, Mittag T, Cell 2019, 176, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Choi J-M, Dar F, Pappu RV, PLoS Comput. Biol 2019, 15, e1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choi J-M, Holehouse AS, Pappu RV, Annu. Rev. Biophys 2020, 49, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rubinstein M, Colby RH, Polymer Physics, OUP, Oxford, UK: 2003. [Google Scholar]
- [28].Rubinstein M, Dobrynin AV, Trends Polym. Sci 1997, 6, 181. [Google Scholar]
- [29].Harmon TS, Holehouse AS, Rosen MK, Pappu RV, eLife 2017, 6, e30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang J, Choi J-M, Holehouse AS, Lee HO, Zhang X, Jahnel M,Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA, Cell 2018, 174, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang Y, Jones HB, Dao TP, Castañeda CA, J. Phys. Chem. B 2019, 123, 3618. [DOI] [PubMed] [Google Scholar]
- [32].Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK, Cell 2016, 166, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M,Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang Q-X,Nixon BT, Rosen MK, Nature 2012, 483, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T, Science 2020, 367, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daugh-drill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM, Chem. Rev 2014, 114, 6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X,Schulman BA, Mittag T, Mol. Cell 2018, 72, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei M-T,Whitney G, Lyons SM, Anderson P, Jacobs WM, Ivanov P, Brangwynne CP, Cell 2020, 181, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang P, Mathieu C, Kolaitis R-M, Zhang P, Messing J, Yurt-sever U, Yang Z, Wu J, Li Y, Pan Q, Yu J, Martin EW, Mittag T, Kim HJ, Taylor JP, Cell 2020, 181, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Conicella AE, Zerze GH, Mittal J, Fawzi NL, Structure 2016, 24, 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ, Structure 2016, 24, 1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E, Walters KJ, J. Mol. Biol 2019, 431, 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR,Wooten MW, Mol. Cell. Biol 2004, 24, 8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Itakura E, Mizushima N, J. Cell Biol 2011, 192, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pickart CM, Fushman D, Curr. Opin. Chem. Biol 2004, 8, 610. [DOI] [PubMed] [Google Scholar]
- [45].Castañeda CA, Chaturvedi A, Camara CM, Curtis JE, Krueger S, Fushman D, Phys. Chem. Chem. Phys 2016, 18, 5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen L, Madura K, FEBS Lett. 2006, 580, 3401. [DOI] [PubMed] [Google Scholar]
- [47].Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K, Nature 1998, 391, 715. [DOI] [PubMed] [Google Scholar]
- [48].Yokoi M, Hanaoka F, Gene 2017, 597, 1. [DOI] [PubMed] [Google Scholar]
- [49].Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM, Mol. Cell 2000, 6, 409. [DOI] [PubMed] [Google Scholar]
- [50].Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D,Rothenberg C, Wang H, Ye Y, Fang S, Monteiro MJ, J. Cell Biol 2009, 187, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].N’Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ, EMBO Rep. 2009, 10, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Laço MN, Cortes L, Travis SM, Paulson HL, Rego AC, PLoS One 2012, 7, e43563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Picher-Martel V, Dutta K, Phaneuf D, Sobue G, Julien J-P, Mol. Brain 2015, 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dao TP, Kolaitis R-M, Kim HJ, O’Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, Castañeda CA, Mol. Cell 2018, 69, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wu JJ, Cai A, Greenslade JE, Higgins NR, Fan C, Le NTT,Tatman M, Whiteley AM, Prado MA, Dieriks BV, Curtis MA,Shaw CE, Siddique T, Faull RLM, Scotter EL, Finley D, Monteiro MJ, Proc. Natl. Acad. Sci. USA 2020, 117, 15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Picher-Martel V, Dutta K, Phaneuf D, Sobue G, Julien J-P, Mol. Brain 2015, 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T, Nature 2011, 477, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nguyen K, Puthenveetil R, Vinogradova O, Biochem. Biophys. Rep 2017, 9, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ford DL, Monteiro MJ, Biochem. J 2006, 399, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, Osborne G,Bates GP, Glickman MH, Trost M, Knebel A, Marchesi F, Kurz T, Cell 2016, 166, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kurlawala Z, Shah PP, Shah C, Beverly LJ, J. Cell. Biochem 2017, 118, 2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI, Nat. Struct. Biol 2001, 8, 417. [DOI] [PubMed] [Google Scholar]
- [63].Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM, Proc. Natl. Acad. Sci. USA 2003, 100, 12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kang Y, Zhang N, Koepp DM, Walters KJ, J. Mol. Biol 2007, 365, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJH, Johansen T, Sachse C, Cell Rep. 2015, 11, 748. [DOI] [PubMed] [Google Scholar]
- [66].Jakobi AJ, Huber ST, Mortensen SA, Schultz SW, Palara A,Kuhm T, Shrestha BK, Lamark T, Hagen WJH, Wilmanns M,Johansen T, Brech A, Sachse C, Nat. Commun 2020, 11, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yang Y, Willis TL, Button RW, Strang CJ, Fu Y, Wen X, Grayson PRC, Evans T, Sipthorpe RJ, Roberts SL, Hu B, Zhang J, Lu B, Luo S, Nat. Commun 2019, 10, 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Posey AE, Holehouse AS, Pappu RV, in Methods in Enzymology (Ed: Rhoades E), Academic Press, New York: 2018, pp. 1–30. [DOI] [PubMed] [Google Scholar]
- [69].Bracha D, Walls MT, Wei M-T, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP, Cell 2018, 175, 1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, Brangwynne CP, Nature 2020, 581, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dignon GL, Best RB, Mittal J, Annu. Rev. Phys. Chem 2020, 71, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P,Plitzko JM, Förster F, Wingreen NS, Engel BD, Mackinder LCM, Jonikas MC, Cell 2017, 171, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu B, He G, Weiner BG, Ronceray P, Meir Y, Jonikas MC, Wingreen NS, arXiv:1901.09352 2019. [Google Scholar]
- [74].Wyman J, Gill SJ, Proc. Natl. Acad. Sci. USA 1980, 77, 5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Posey AE, Ruff KM, Harmon TS, Crick SL, Li A, Diamond MI,Pappu RV, J. Biol. Chem 2018, 293, 3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, Zhang ZC, Coughlin M, Sweeny EA, DeSantis ME, Jackrel ME, Rodell CB, Burdick JA, King OD, Gitler AD, Lagier-Tourenne C, Pandey UB,Chook YM, Taylor JP, Shorter J, Cell 2018, 173, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Onuchic PL, Milin AN, Alshareedah I, Deniz AA, Banerjee PR, Sci. Rep 2019, 9, 12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Alshareedah I, Kaur T, Ngo J, Seppala H, Kounatse L-AD, Wang W, Moosa MM, Banerjee PR, J. Am. Chem. Soc 2019, 141, 14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA, Angew. Chem., Int. Ed 2017, 56, 11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B,Williamson MP, Layfield R, Searle MS, J. Mol. Biol 2010, 396, 178. [DOI] [PubMed] [Google Scholar]
- [81].Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D, Mol. Cell 2005, 18, 687. [DOI] [PubMed] [Google Scholar]
- [82].Raasi S, Varadan R, Fushman D, Pickart CM, Nat. Struct. Mol. Biol 2005, 12, 708. [DOI] [PubMed] [Google Scholar]
- [83].Cabe M, Rademacher DJ, Karlsson AB, Cherukuri S, Bakowska JC, Biochem. Biophys. Res. Commun 2018, 503, 2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wurzer B, Zaffagnini G, Fracchiolla D, Turco E, Abert C, Romanov J, Martens S, eLife 2015, 4, e08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lu K, den Brave F, Jentsch S, Nat. Cell Biol 2017, 19, 732. [DOI] [PubMed] [Google Scholar]
- [86].Zhang D, Raasi S, Fushman D, J. Mol. Biol 2008, 377, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Harman CA, Monteiro MJ, Biochim. Biophys. Acta – Gen. Subj 2019, 1863, 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O’Meally R, Dignon GL, Conicella AE, Zheng W, Best RB, Cole RN, Mittal J, Shewmaker F, Fawzi NL, EMBO J. 2017, 36, 2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A,Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ, Mol. Cell 2015, 57, 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Peng H, Yang J, Li G, You Q, Han W, Li T, Gao D, Xie X, Lee B-H, Du J, Hou J, Zhang T, Rao H, Huang Y, Li Q, Zeng R, Hui L,Wang H, Xia Q, Zhang X, He Y, Komatsu M, Dikic I, Finley D, Hu R, Cell Res. 2017, 27, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liang R-Y, Chen L, Ko B-T, Shen Y-H, Li Y-T, Chen B-R, Lin K-T, Madura K, Chuang S-M, J. Mol. Biol 2014, 426, 4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mah AL, Perry G, Smith MA, Monteiro MJ, J. Cell Biol 2000, 151, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Regan-Klapisz E, Sorokina I, Voortman J, de Keizer P, Roovers RC, Verheesen P, Urbé S, Fallon L, Fon EA, Verkleij A, Benmerah A, van Bergen en Henegouwen PMP, J. Cell Sci 2005, 118, 4437. [DOI] [PubMed] [Google Scholar]
- [94].Yun Lee D, Arnott D, Brown EJ, EMBO Rep. 2013, 14, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Suzuki R, Kawahara H, EMBO Rep. 2016, 17, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Jachimowicz RD, Beleggia F, Isensee J, Velpula BB, Goergens J, Bustos MA, Doll MA, Shenoy A, Checa-Rodriguez C, Wiederstein JL, Baranes-Bachar K, Bartenhagen C, Hertwig F, Teper N, Nishi T, Schmitt A, Distelmaier F, Lüdecke H-J, Albrecht B,Krüger M, Schumacher B, Geiger T, Hoon DSB, Huertas P, Fischer M, Hucho T, Peifer M, Ziv Y, Reinhardt HC, Wieczorek D, et al. , Cell 2019, 176, 505. [DOI] [PubMed] [Google Scholar]
- [97].Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS, Mol. Cell 2016, 63, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Arnesen T, Starheim KK, Damme PV, Evjenth R, Dinh H, Betts MJ, Ryningen A, Vandekerckhove J, Gevaert K, Anderson D, Mol. Cell. Biol 2010, 30, 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Choudhury KR, Raychaudhuri S, Bhattacharyya NP, PLoS One 2012, 7, e51415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ghosh DK, Roy A, Ranjan A, J. Mol. Biol 2018, 430, 963. [DOI] [PubMed] [Google Scholar]
- [101].Ghosh DK, Roy A, Ranjan A, bioRxiv 2019, 780379. [Google Scholar]
- [102].Ghosh DK, Kumar A, Ranjan A, Biochim. Biophys. Acta – Gen. Subj 2018, 1862, 2846. [DOI] [PubMed] [Google Scholar]
- [103].Ghosh DK, Roy A, Ranjan A, Biochemistry 2018, 57, 2009. [DOI] [PubMed] [Google Scholar]
- [104].Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, May AI, Knorr RL, Suzuki K, Ohsumi Y, Noda NN, Nature 2020, 578, 301. [DOI] [PubMed] [Google Scholar]
- [105].Kito K, Yeh ETH, Kamitani T, J. Biol. Chem 2001, 276, 20603. [DOI] [PubMed] [Google Scholar]
- [106].Hipp MS, Raasi S, Groettrup M, Schmidtke G, J. Biol. Chem 2004, 279, 16503. [DOI] [PubMed] [Google Scholar]
- [107].Rani N, Aichem A, Schmidtke G, Kreft SG, Groettrup M, Nat. Commun 2012, 3, 749. [DOI] [PubMed] [Google Scholar]
- [108].Tanji K, Tanaka T, Kamitani T, Biochem. Biophys. Res. Commun 2005, 337, 116. [DOI] [PubMed] [Google Scholar]
- [109].Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ETH, J. Biol. Chem 2001, 276, 46655. [DOI] [PubMed] [Google Scholar]
- [110].Liu S, Yang H, Zhao J, Zhang Y-H, Song A-X, Hu H-Y, J. Biol. Chem 2013, 288, 31339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Guarascio R, Salih D, Yasvoina M, Edwards FA, Cheetham ME,van der Spuy J, Hum. Mol. Genet 2020, 29, 80. [DOI] [PubMed] [Google Scholar]
- [112].Tanji K, Tanaka T, Mori F, Kito K, Takahashi H, Wakabayashi K, Kamitani T, Am. J. Pathol 2006, 169, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Tanji K, Mori F, Kito K, Kakita A, Mimura J, Itoh K, Takahashi H,Kamitani T, Wakabayashi K, J. Neuropathol. Exp. Neurol 2011, 70, 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Richet E, Pooler AM, Rodriguez T, Novoselov SS, Schmidtke G,Groettrup M, Hanger DP, Cheetham ME, van der Spuy J, Hum. Mol. Genet 2012, 21, 5254. [DOI] [PubMed] [Google Scholar]
- [115].Jeong YY, Her J, Chung IK, FEBS Lett. 2016, 590, 1776. [DOI] [PubMed] [Google Scholar]
- [116].Narayanan A, Meriin A, Andrews JO, Spille J-H, Sherman MY,Cisse II, eLife 2019, 8, e39695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ray S, Singh N, Pandey S, Kumar R, Gadhe L, Datta D, Patel K, Mahato J, Navalkar A, Panigrahi R, Chatterjee D, Maiti S, Bhatia S, Mehra S, Singh A, Gerez J, Chowdhury A, Kumar A, Padinhateeri R, Riek R, Krishnamoorthy G, Maji SK, bioRxiv 2019, 619858. [Google Scholar]
- [118].Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T,Commins C, Vanderburg C, Roe AD, Fan Z, Molliex AM, Hernandez-Vega A, Muller D, Hyman AA, Mandelkow E, Taylor JP, Hyman BT, EMBO J. 2018, 37, 98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Cho NW, Dilley RL, Lampson MA, Greenberg RA, Cell 2014, 159, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Albert S, Wietrzynski W, Lee C-W, Schaffer M, Beck F, Schuller JM, Salomé PA, Plitzko JM, Baumeister W, Engel BD, Proc. Natl. Acad. Sci. USA 2020, 117, 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Gu ZC, Wu E, Sailer C, Jando J, Styles E, Eisenkolb I, Kuschel M,Bitschar K, Wang X, Huang L, Vissa A, Yip CM, Yedidi RS, Friesen H, Enenkel C, Mol. Biol. Cell 2017, 28, 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Marshall RS, Vierstra RD, eLife 2018, 7, e34532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [123].Kumar S, Talis AL, Howley PM, J. Biol. Chem 1999, 274, 18785. [DOI] [PubMed] [Google Scholar]
- [124].Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, Pansarasa O, Cereda C, Poletti A, Alberti S, Carra S, Mol. Cell 2016, 63, 796. [DOI] [PubMed] [Google Scholar]
- [125].Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, Alberti S, EMBO J. 2017, 36, 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Buchan JR, Kolaitis R-M, Taylor JP, Parker R, Cell 2013, 153, 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G,Savard A, Palidwor G, Barry FA, Zinman L, Keith J, Rogaeva E,Robertson J, Lavallée-Adam M, Woulfe J, Couture J-F, Côté J, Gibbings D, Nat. Commun 2018, 9, 2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Turakhiya A, Meyer SR, Marincola G, Böhm S, Vanselow JT,Schlosser A, Hofmann K, Buchberger A, Mol. Cell 2018, 70, 906. [DOI] [PubMed] [Google Scholar]
- [129].Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY,Luo E-C, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao F-B, Bennett EJ, Lécuyer E, Yeo GW, Cell 2018, 172, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Huang C, Chen Y, Dai H, Zhang H, Xie M, Zhang H, Chen F, Kang X, Bai X, Chen Z, Cell Death Differ. 2020, 27, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wilde IB, Brack M, Winget JM, Mayor T, J. Proteome Res 2011, 10, 1062. [DOI] [PubMed] [Google Scholar]
- [132].Ng JMY, Vermeulen W, van der Horst GTJ, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JHJ, Genes Dev. 2003, 17, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS, Science 2019, 365, 342. [DOI] [PubMed] [Google Scholar]
- [134].Ryan VH, Perdikari TM, Naik MT, Saueressig CF, Lins J, Dignon GL, Mittal J, Hart AC, Fawzi NL, bioRxiv 2020, 10.1101/2020.03.15.992768. [DOI] [PMC free article] [PubMed] [Google Scholar]


