Sołtysik et al. show that the inner nuclear membrane of nonhepatocytes harbors triglyceride synthesis enzymes and generates nuclear lipid droplets (LDs). Seipin knockdown increases nuclear LDs through upregulating lipin-1β transcription and increasing phosphatidic acid levels in the nucleus.
Abstract
Nuclear lipid droplets (LDs) in hepatocytes are derived from precursors of very-low-density lipoprotein in the ER lumen, but it is not known how cells lacking the lipoprotein secretory function form nuclear LDs. Here, we show that the inner nuclear membrane (INM) of U2OS cells harbors triglyceride synthesis enzymes, including ACSL3, AGPAT2, GPAT3/GPAT4, and DGAT1/DGAT2, and generates nuclear LDs in situ. mTOR inhibition increases nuclear LDs by inducing the nuclear translocation of lipin-1 phosphatidic acid (PA) phosphatase. Seipin, a protein essential for normal cytoplasmic LD formation in the ER, is absent in the INM. Knockdown of seipin increases nuclear LDs and PA in the nucleus, whereas seipin overexpression decreases these. Seipin knockdown also up-regulates lipin-1β expression, and lipin-1 knockdown decreases the effect of seipin knockdown on nuclear LDs without affecting PA redistribution. These results indicate that seipin is not directly involved in nuclear LD formation but instead restrains it by affecting lipin-1 expression and intracellular PA distribution.
Introduction
Lipid droplets (LDs) are cytoplasmic organelles that are made of lipid esters covered by a phospholipid monolayer. The process of LD formation is thought to begin with the deposition of lipid esters between the two leaflets of the ER membrane, which eventually bulges toward the cytoplasm as a globule (Gao et al., 2019; Henne et al., 2019; Olzmann and Carvalho, 2019; Walther et al., 2017). The deposition and globule formation of lipid esters can be reconstituted in liposomes without proteins (Ben M’barek et al., 2017), but the process of LD biogenesis in vivo requires orchestration by regulatory proteins, including seipin (Renne et al., 2020).
Seipin is an evolutionally conserved integral membrane protein that distributes in the ER domain where nascent LDs form (Salo et al., 2016; Szymanski et al., 2007). Seipin deficiency causes aberrant cytoplasmic LDs in both mammalian cells and budding yeast (Fei et al., 2008; Salo et al., 2016; Szymanski et al., 2007; Wang et al., 2016), illustrating its importance in normal LD formation. Interestingly, yeast lacking the seipin orthologue Fld1 harbor nuclear LDs (Cartwright et al., 2015; Grippa et al., 2015), whereas nuclear LDs are scarce in normal yeast even in the post-diauxic phase when triglyceride (TG) is synthesized in the inner nuclear membrane (INM; Barbosa et al., 2019). These results suggest that nuclear LDs in seipin-deficient yeast may occur as a result of disrupted seipin function in the ER. This interpretation, however, contradicts that of another study on yeast, which reported that seipin exists in the INM and is involved in nuclear LD formation (Romanauska and Köhler, 2018). It thus remains unclear how seipin deficiency increases nuclear LDs in yeast. Whether and how seipin is involved in nuclear LD formation in mammalian cells also remains unknown.
Recently, we found that nuclear LDs in hepatocytes are derived from apolipoprotein B100 (ApoB)–free lipid particles in the ER lumen, a precursor of very-low-density lipoprotein (VLDL; Sołtysik et al., 2019b). Briefly, the ApoB-free lipid particles accumulate at the end of the type I nucleoplasmic reticulum (NR), an inward extension of the nuclear cistern (Malhas et al., 2011), intrude into the nucleoplasm through breaches in the INM, and become nuclear LDs. This unique mechanism explains why hepatocytes harbor abundant nuclear LDs, but other mechanisms are likely to exist, because cells that do not have lipoprotein secretory function also have nuclear LDs (Soltysik et al., 2019a).
In the present study, we addressed how nuclear LDs form in nonhepatocytes and whether and how seipin is related to this process. We found that the INM harbors TG synthesis enzymes and generates nuclear LDs in situ. Importantly, seipin is not present in the INM, and thus, seipin is not directly involved in nuclear LD formation. Rather, seipin depletion increases nuclear LDs by the transcriptional up-regulation of lipin-1β and an increase of phosphatidic acid (PA) in the nucleus. Moreover, seipin overexpression decreases nuclear LDs and nuclear PA. These results indicate that nuclear LDs in nonhepatocytes form in the INM through a seipin-independent mechanism and that seipin restricts nuclear LD formation through its effect on lipin-1 expression and intracellular PA distribution.
Results and discussion
Nuclear LDs form in the INM
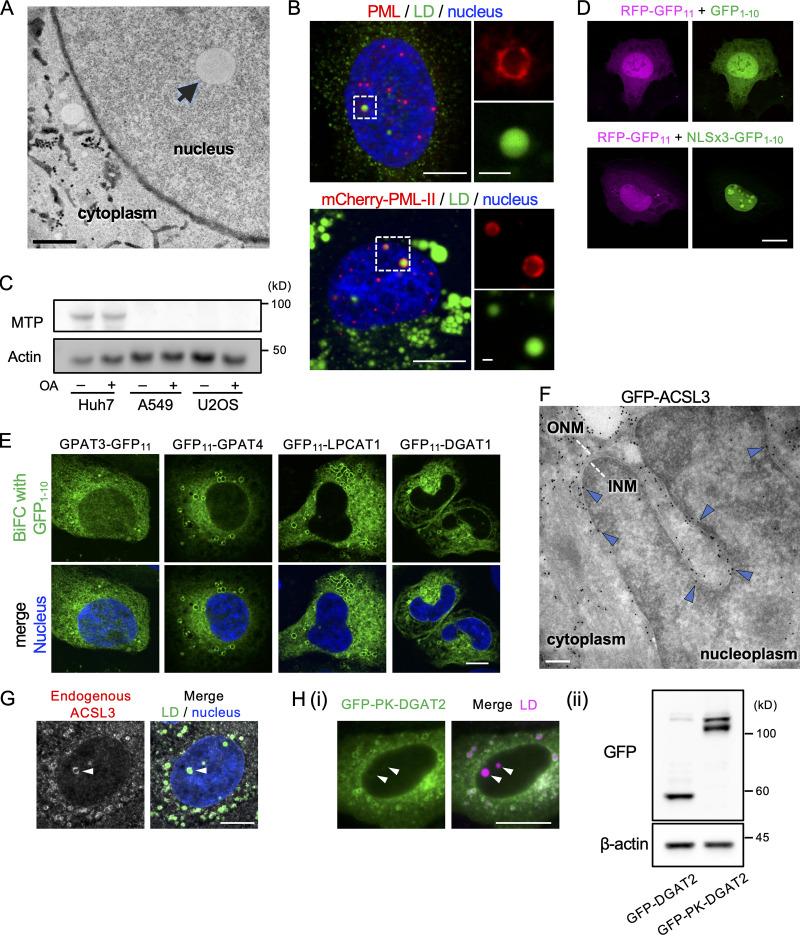
U2OS, an osteosarcoma cell line, frequently harbors nuclear LDs (Fig. S1 A; Ohsaki et al., 2016). As in Huh7, a hepatocarcinoma cell line, the number of nuclear LDs in U2OS increased when cells were cultured with oleic acid (OA), and the increase was suppressed by triacsin C, a long-chain-fatty-acid–CoA ligase (ACSL) inhibitor (Fig. 1 A). Nuclear LDs in U2OS were also similar to those in Huh7 in that they are associated with the promyelocytic leukemia–nuclear body (PML-NB; Fig. S1 B; Ohsaki et al., 2016).
Figure S1.
Characterization of nuclear LDs in U2OS. (A) Electron micrograph of U2OS cultured with OA for 1 d. Arrow indicates an LD in the nucleoplasm. The nuclear envelope and the ER lumen are delineated by DAB precipitated by HRP-KDEL. Scale bar, 0.2 µm. (B) Colocalization of PML (red) and nuclear LDs (green). (Upper panel) Endogenous PML was immunolabeled. (Lower panel) mCherry PML-II was expressed. Nucleus, blue. Scale bars, 10 µm; 2 µm (magnified photos). (C) Western blotting of MTP and actin. Total lysates of Huh7, A549 (human alveolar adenocarcinoma), and U2OS were cultured with or without OA supplementation. (D) BiFC. Green fluorescence due to complementation between GFP1–10 and RFP-GFP11 is present in both the nucleus and the cytoplasm, whereas that of NLSx3-GFP1–10 and RFP-GFP11 is confined to the nucleus. Scale bar, 10 µm. (E) BiFC with GFP1-10. GPAT3, GPAT4, LPCAT1, and DGAT1 tagged with GFP11 produce fluorescence signal in the cytoplasmic network. Nucleus, blue. Scale bar,10 µm. (F) Immuno-EM of GFP-ACSL3 using ultrathin cryosections. Immunogold labels in the INM (arrowheads). Scale bar, 0.2 µm. (G) Immunolabeling of endogenous ACSL3 in nuclear LDs (arrowhead). LD, green; nucleus, blue. Scale bar, 10 µm. (H) Comparison of GFP-DGAT2 and GFP-PK-DGAT2. (i) Distribution of GFP-PK-DGAT2 (green). LDs, magenta. Arrowheads indicate nuclear LDs. Scale bar, 10 µm. See Fig. 2 B for distribution of GFP-DGAT2. (ii) The expression level of GFP-DGAT2 and GFP-PK-DGAT2 (Western blotting).
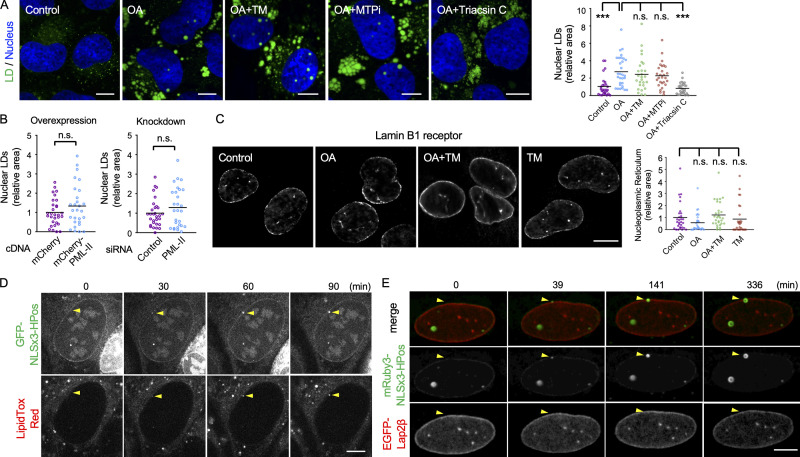
Figure 1.
Nuclear LDs in U2OS. (A) Nuclear LDs in U2OS treated for 1 d with none (control), 0.4 mM OA (OA), OA and 5 µg/ml tunicamycin (OA+TM), OA and 100 nM BAY13-9952 (OA+MTPi), or OA and 5 µM triacsin C (OA+Triacsin C). LDs, green; nucleus, blue. The area of total nuclear LDs per nucleus was quantified. The number of nuclei counted: 28 (control), 30 (OA), 27 (OA+TM), 28 (OA+MTPi), 29 (OA+Triacsin C). Pooled data from three independent experiments. Kruskal–Wallis ANOVA test followed by Dunn’s test; ***, P < 0.001. (B) The effect of PML-II overexpression and knockdown. U2OS transfected with PML-II cDNA or PML-II siRNA cultured with OA for 1 d. The number of nuclei counted: 30 (control cDNA), 28 (PML-II cDNA), 30 (control siRNA), 28 (PML-II siRNA). Pooled data from three independent experiments. Mann–Whitney test. (C) The NR immunolabeled for lamin B1 receptor. U2OS were cultured for 1 d with none (control), 0.4 mM OA (OA), OA and 5 µg/ml tunicamycin (OA+TM), or 5 µg/ml tunicamycin (TM). The area of intranuclear labels per nucleus was quantified. The number of nuclei counted: 30 (control, OA, and OA+TM), 27 (TM). Pooled data from three independent experiments. Kruskal–Wallis ANOVA test followed by Dunn’s test. (D and E) Nuclear LD formation in U2OS treated with OA for 1 d. (D) GFP-NLSx3-HPoS, nuclear LDs; LipidTox Red, general LDs. Selected frames from Video 1 are shown. Arrowheads indicate a nuclear LD forming in the nuclear periphery. (E) mRuby3-NLSx3-HPos, nuclear LD; EGFP-Lap2β, INM. Selected frames from Video 2 are shown. Arrowheads indicate a nuclear LD forming in the nuclear envelope. Scale bars, 10 µm. n.s., not significant.
Nuclear LDs in U2OS, however, were different from those in Huh7 in several respects. First, ER stressor (tunicamycin) and microsomal TG transfer protein (MTP) inhibitor (BAY13-9952), which increases and decreases nuclear LDs in Huh7, respectively (Sołtysik et al., 2019b), did not affect nuclear LDs in U2OS (Fig. 1 A). Second, overexpression and knockdown of PML-II, which increases and decreases nuclear LDs in Huh7, respectively (Ohsaki et al., 2016), did not have such effects in U2OS (Fig. 1 B). Third, the type I NR develops in accordance with nuclear LDs in Huh7 (Sołtysik et al., 2019b), but this did not occur in U2OS (Fig. 1 C).
These differences suggest that nuclear LDs in Huh7 and those in U2OS form through different mechanisms. In Huh7, nuclear LDs derive from ApoB-free lipid particles that are synthesized in the ER lumen by an MTP-dependent process (Sołtysik et al., 2019b), but in U2OS, which lacks MTP (Fig. S1C), the origin of nuclear LDs should be different. Live imaging in which nuclear LDs were visualized by the specific marker NLSx3-HPos (Sołtysik et al., 2019b) showed that nuclear LDs in U2OS originate in the nuclear envelope and grow toward the nucleoplasm (Fig. 1, D and E; and Videos 1 and 2). This result indicates that nuclear LDs in U2OS are generated in the INM in situ. Additionally, the time course observed here suggests that the nuclear LD formation in the INM is a slower process than cytoplasmic LD formation in the ER (Kassan et al., 2013; Wang et al., 2016).
Video 1.
Nuclear LD formation. U2OS treated with OA for 1 d. GFP-NLSx3-HPoS (nuclear LD marker), green; LipidTox Red (general LD stain), red. Arrowheads indicate a nuclear LD forming in the nuclear periphery. One frame captured every 2 min; displayed at 12 frames/s. Selected frames are shown in Fig. 1 D. Scale bar, 10 µm.
Video 2.
Nuclear LD formation at the INM. U2OS treated with OA for 1 d. mRuby3-NLSx3-HPos (nuclear LD marker), green; GFP-Lap2β (INM marker), red. One frame captured every 90 s; displayed at 20 frames/s. Selected frames are shown in Fig. 1 E. Scale bar, 5 µm.
TG synthetic enzymes in the INM
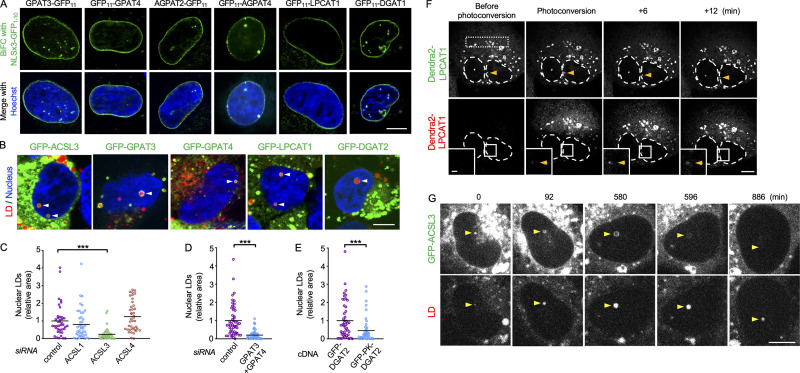
The aforementioned findings suggest that TG synthesis enzymes distribute in the INM. To test this possibility, we performed the bimolecular fluorescence complementation (BiFC) using the GFP fragment GFP1–10 appended with a nuclear localization signal (NLS; Fig. S1 D). The complementary GFP fragment GFP11, fused to glycerol-3-phosphate acyltransferase 3 (GPAT3), GPAT4, 1-acyl-sn-glycerol-3-phosphate acyltransferase 2 (AGPAT2), AGPAT4, lysophosphatidylcholine acyltransferase 1 (LPCAT1, which has lysophosphatidic acid acyltransferase activity; Shindou et al., 2009), and diacylglycerol O–acyltransferase 1 (DGAT1), showed linear fluorescence in the nuclear periphery (Fig. 2 A). In contrast, when GFP1–10 without NLS was used as the BiFC partner, GFP11-tagged proteins generated fluorescence in a cytoplasmic network pattern, validating our methodology (Fig. S1 E).
Figure 2.
TG synthesis enzymes in U2OS cultured with OA for 1 d. (A) BiFC (green) of NLSx3-GFP1–10 and GFP11-tagged enzymes. Nucleus, blue. (B) GFP-tagged enzymes (green) in nuclear LDs (arrowheads). LDs, red; nucleus, blue. (C) The effect of ACSL RNAi on nuclear LDs. The number of nuclei counted: 40 (control), 44 (ACSL1), 39 (ACSL3), or 40 (ACSL4). Pooled data from three independent experiments. Kruskal–Wallis ANOVA test followed by Dunn’s test, ***, P < 0.001. (D) The effect of GPAT3 and GPAT4 RNAi on nuclear LDs. The number of nuclei counted: 51 (control), 52 (GPAT3+GPAT4). Pooled data from three independent experiments. Mann–Whitney test; ***, P < 0.001. (E) The effect of GFP-DGAT2 and GFP-PK-DGAT2 expression on nuclear LDs in U2OS after knockdown of endogenous DGAT2. The number of nuclei counted: 48 (GFP-DGAT2), 45 (GFP-PK-DGAT2). Pooled data from three independent experiments. Mann–Whitney test; ***, P < 0.001. (F) Appearance of red Dendra2-LPCAT1 signal in nuclear LDs (arrowheads) after photoconversion in the cytoplasm (rectangular area). Selected frames from Video 3 are shown. (G) Disappearance of GFP-ACSL3 from nuclear LDs (arrowheads). Selected frames from Video 5 are shown. Scale bars, 10 µm; 2 µm (inset).
For ACSL3, BiFC did not work, but GFP-ACSL3 distributed in nuclear LDs (Fig. 2 B) and was labeled in the INM by immuno-EM (Fig. S1 F). Endogenous ACSL3 was also immunolabeled in nuclear LDs (Fig. S1 G). GFP-tagged GPAT3, GPAT4, LPCAT1, and DGAT2, all of which are distributed in cytoplasmic LDs via a hairpin domain (Wilfling et al., 2013), were also found in nuclear LDs, indicating their presence in the INM (Fig. 2 B).
The inhibitory effect of triacsin C (Fig. 1 A) indicated that ACSL is necessary for nuclear LD formation. Knockdown of individual triacsin C–sensitive ACSL isoforms (i.e., ACSL1, ACSL3, and ACSL4; Grevengoed et al., 2014) indicated that ACSL3 is essential (Fig. 2 C). Double knockdown of GPAT3 and GPAT4 also decreased the number of nuclear LDs (Fig. 2 D). To examine whether TG synthesis in the INM is critical for nuclear LD formation, we compared GFP-DGAT2 and GFP-PK-DGAT2, the latter of which contains a bulky pyruvate kinase (PK) segment that precludes its entry into the INM (Soullam and Worman, 1995). In cells depleted of endogenous DGAT2, the expression of GFP-DGAT2 induced significantly more nuclear LDs than the expression of GFP-PK-DGAT2 did (Figs. 2 E and S1 H). These results indicated that nuclear LD formation involves TG synthesized in the INM.
We next examined whether nuclear LDs are connected to the INM. First, using photoconvertible Dendra2-LPCAT1, red fluorescence provoked by laser illumination in the cytoplasm appeared in nuclear LDs several minutes later (Fig. 2 F and Video 3). Second, GFP-DGAT2 photobleached in nuclear LDs recovered its fluorescence (Video 4). These results indicated that nuclear LDs and the INM are connected at some point. However, the appearance of photoconverted Dendra2-LPCAT1 and the recovery of GFP-DGAT2 occurred only rarely, suggesting that the nuclear LD–INM connection is transient and that most nuclear LDs are detached from the INM. Consistently, GFP-ACSL3 in nuclear LDs faded away and thereafter those LDs shrank, suggesting that GFP-ACSL3 is not continuously imported from the INM to nuclear LDs (Fig. 2 G and Video 5). The nuclear LD–INM relationship will be examined further in a later section.
Video 3.
Appearance of red Dendra2-LPCAT1 signal in nuclear LDs (arrowheads) after photoconversion in the cytoplasm. Left: green, original color of Dendra2; middle: red, color of photoconverted Dendra2. One frame captured every 1 min; displayed at 3 frames/s. Selected frames are shown in Fig. 2 E. Scale bar, 10 µm.
Video 4.
FRAP of GFP-DGAT2 (green) in nuclear LDs (arrow). LDs are stained with LipidTox Red (red). One frame captured every 1 min; displayed at 3 frames/s. Scale bar, 10 µm.
Video 5.
Disappearance of GFP-ACSL3 (green) from nuclear LDs (arrowheads). LDs are stained with LipidTox Red (red). One frame captured every 2 min; displayed at 25 frames/s. Selected frames are shown in Fig. 2 F. Scale bar, 10 µm.
Lipins and nuclear LDs
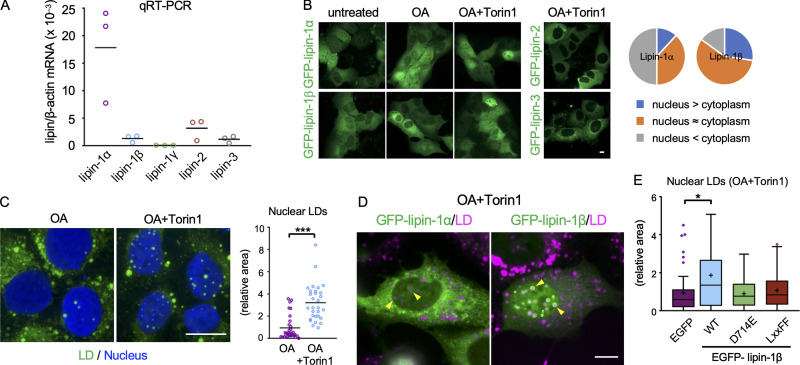
The penultimate step of TG synthesis, the conversion of PA to diacylglycerol, is catalyzed by lipins (Wang et al., 2017), which are soluble proteins that bind membranes through hydrogen bonds with PA (Zhang and Reue, 2017). Quantitative real-time PCR (qRT-PCR) showed that U2OS expresses lipin-1α, lipin-1β, lipin-2, and lipin-3 (Fig. 3 A). In adipocytes and hepatocytes, lipin-1β is confined to the cytoplasm (Bou Khalil et al., 2009; Péterfy et al., 2005), but lipin-1β in U2OS also distributed in the nucleus as in HEK293 cells (Ren et al., 2010; Fig. 3 B). The nuclear distribution of lipin-1 was increased by mTOR inhibition with Torin1 as reported previously (Peterson et al., 2011); this increase was particularly prominent for lipin-1β (Fig. 3 B). Lipin-2 and lipin-3 distributed only in the cytoplasm, even after Torin1 treatment (Fig. 3 B).
Figure 3.
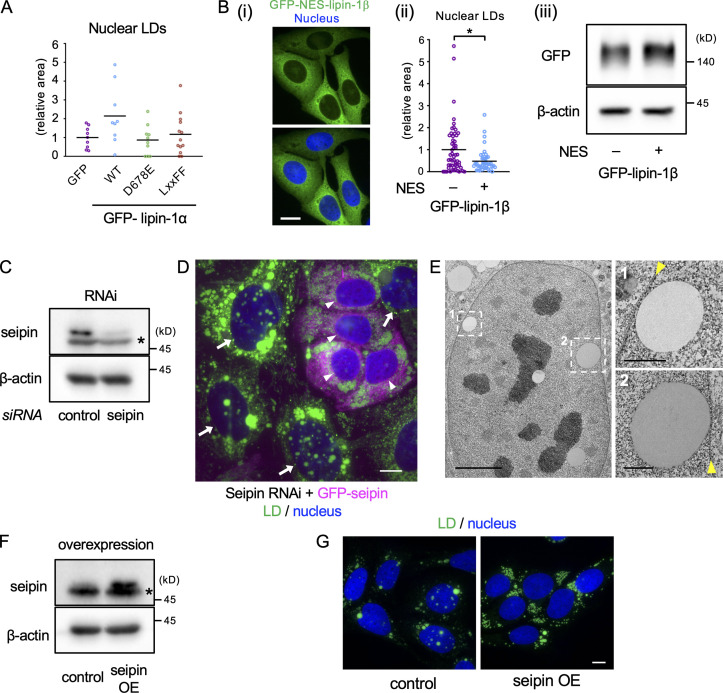
Lipins and nuclear LDs. (A) qRT-PCR. The lipin mRNA levels are expressed relative to β-actin mRNA. A representative result of two independent experiments. (B) GFP-lipin distribution in U2OS cultured in normal medium, with OA for 1 d or with OA for 1 d and OA and 0.25 µM Torin1 (OA/Torin1) for another 8 h. The pie charts show the relative distribution of GFP–lipin-1α and GFP–lipin-1β in cells treated with OA and Torin1. (C) The effect of Torin1. U2OS cultured with OA for 2 d or treated with OA for 1 d and OA/Torin1 for another day. The number of nuclei counted: 30. Pooled data from three independent experiments. Mann–Whitney test; ***, P < 0.001. (D) Distribution of GFP–lipin-1α and -1β (green; arrowheads) in cells cultured with OA for 1 d and with OA/Torin1 for another 8 h. LDs, magenta. (E) The effect of wild-type and mutant lipin-1β expression. U2OS depleted of lipin-1 was transfected with siRNA-resistant GFP–lipin-1 cDNAs and cultured with OA for 1 d and with OA/Torin1 for another 8 h. The number of nuclei counted: 49 (GFP), 58 (WT), 56 (D714E), or 44 (LxxFF). Pooled data from three independent experiments. Box boundaries indicate the 25th and 75th percentiles, and whiskers indicate Tukey type. Kruskal–Wallis ANOVA test followed by Dunn’s test; *, P < 0.05. Scale bars, 10 µm.
Nuclear LDs increased in cells treated with OA and Torin1 (Fig. 3 C), suggesting that the nuclear translocation of lipin-1 may facilitate nuclear LD formation. This idea was supported by the observation that GFP–lipin-1, particularly GFP–lipin-1β, showed transient concentration around nuclear LDs (Video 6). Accumulation of GFP–lipin-1β in nuclear LDs was observed more clearly after lipin-1 knockdown (Fig. 3 D), probably because hetero-oligomer formation between GFP–lipin-1β and endogenous lipin-1α was decreased (Liu et al., 2010).
Video 6.
Live imaging of GFP-Lipin1β (green; arrowheads) in U2OS treated with OA and Torin1. LDs, red. One frame captured every 2 min; displayed at 10 frames/s. Scale bar, 10 µm.
To examine whether lipin-1 functionality is necessary for nuclear LD formation, lipin-1β mutants that were deficient in the PA phosphatase activity alone (D714E; Harris et al., 2007) or in both PA phosphatase and transcriptional coactivator activities (LxxFF; Finck et al., 2006) were expressed after knockdown of endogenous lipin-1. The expression of wild-type lipin-1β increased nuclear LDs as expected, whereas the expression of lipin-1β (D714E) and lipin-1β (LxxFF) did not (Fig. 3 E). Similar results were obtained for wild-type lipin-1α and lipin-1α mutants (Fig. S2 A). Additionally, lipin-1β tagged with a nuclear export signal (NES) showed a smaller effect on nuclear LDs than wild-type lipin-1β (Fig. S2 B). These results indicated that the PA phosphatase activity of lipin-1 in the nucleus is involved in nuclear LD formation.
Figure S2.
Effects of lipin-1 expression and seipin knockdown and overexpression on nuclear LDs. (A) The effect of wild-type and mutant lipin-1α expression on nuclear LDs. U2OS depleted of lipin-1 was transfected with siRNA-resistant GFP–lipin-1α cDNAs and cultured with OA for 1 d and with OA and 0.25 µM Torin1 (OA/Torin1) for another 8 h. The number of nuclei counted: 9 (GFP), 9 (WT), 10 (D678E), 13 (LxxFF). A representative result of two independent experiments. (B) Comparison of lipin-1β and NES-tagged lipin-1β. (Bi) Distribution of GFP–NES–lipin-1β (green). Nucleus, blue. Scale bar, 10 µm. (Bii) The effect of GFP-NES-lipin-1β expression on nuclear LDs. U2OS depleted of lipin-1 was transfected with either GFP–lipin-1β or GFP–NES–lipin-1β cDNA and cultured with OA for 1 d and with OA/Torin1 for another 8 h. The number of nuclei counted: 54 (lipin-1) or 44 (NES–lipin-1β). Pooled data from three independent experiments. Mann–Whitney test; *, P < 0.05. (Biii) The expression level of GFP–lipin-1β and GFP–NES–lipin-1β (Western blotting). (C) Western blotting of seipin showing U2OS transfected with either control or seipin siRNA. Asterisk indicates a nonspecific band. (D) The effect of seipin re-expression in U2OS treated with seipin siRNA. U2OS after seipin knockdown was transfected with siRNA-resistant GFP-seipin cDNA and cultured with OA for 1 d. Cells expressing GFP-seipin (magenta; arrowheads) show fewer nuclear LDs than those not expressing it (arrows). LD, green; nucleus, blue. Scale bar, 10 µm. (E) EM of U2OS transfected with seipin siRNA and cultured with OA for 1 d. Arrowheads indicate the nuclear envelope. Scale bars, 5 µm; 1 µm (magnified photos). (F) Western blotting of seipin showing control U2OS without cDNA transfection and U2OS transfected with nontagged seipin cDNA. Asterisk indicates a nonspecific band. OE, overexpression. (G) Stable overexpression of seipin decreases nuclear LDs. Cells were cultured with OA for 1 d and OA and 0.25 µM Torin1 for another 1 d. LD, green; nucleus, blue. Scale bars, 10 µm.
Seipin and nuclear LDs
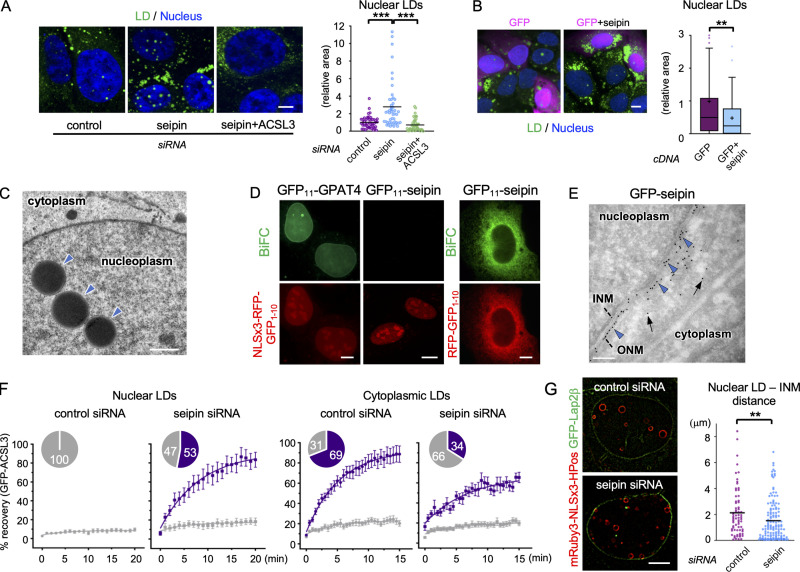
We next asked how seipin is related to nuclear LD formation. First, we found that seipin knockdown increased nuclear LDs (Figs. 4 A and S2 C), and the reintroduction of either nontagged seipin (Fig. 4 B) or GFP-seipin (Fig. S2 D) negated this effect. Knockdown of ACSL3 suppressed the increase of nuclear LDs by seipin deficiency (Fig. 4 A), as observed for OA-induced nuclear LDs (Fig. 2 C). EM confirmed that nuclear LDs in seipin-deficient cells are located in the nucleoplasm (Fig. 4 C).
Figure 4.
Seipin and nuclear LDs. (A) The effect of seipin RNAi. U2OS were transfected with control siRNA, seipin siRNA, or seipin siRNA and ACSL3 siRNA and cultured with OA for 1 d. The number of nuclei counted: 48 (control), 48 (seipin), 43 (seipin+ACSL3). Pooled data from three independent experiments. Kruskal–Wallis ANOVA test followed by Dunn’s test; ***, P < 0.001. (B) Re-expression of seipin in seipin-deficient cells. U2OS depleted of seipin was transfected with nontagged seipin cDNA and GFP cDNA (10:1 ratio) and cultured with OA for 1 d. GFP, magenta; LD, green; nucleus, blue. The number of nuclei counted: 134 (GFP), 110 (GFP+seipin). Pooled data from three independent experiments. Box boundaries indicate the 25th and 75th percentiles, and whiskers indicate Tukey type. Mann–Whitney test; **, P < 0.01. (C) EM of U2OS transfected with seipin siRNA and cultured with OA for 1 d. The nuclear envelope and the ER lumen are delineated by DAB precipitated by HRP-KDEL. Nuclear LDs, arrowheads. (D) BiFC. Both NLSx3-RFP-GFP1-10 and RFP-GFP1-10 are used for GFP11-seipin. (E) Immuno-EM of GFP-seipin in ultrathin cryosections. Immunogold labels in the outer nuclear membrane (ONM; arrowheads) and the ER (arrows). (F) FRAP of GFP-ACSL3 in nuclear and cytoplasmic LDs. U2OS transfected with either control or seipin siRNA cultured with OA for 1 d. The number of analyzed LDs: 14 (control) or 15 (seipin) for nuclear LDs; 39 (control) or 29 (seipin) for cytoplasmic LDs. Recovery plots show the mean ± SEM and the curve fitting with nonlinear regression and the exponential one-phase decay model. The pie charts show the proportion of LDs showing >40% (purple) or <40% (gray) fluorescence recovery 15 min after photobleaching. See Videos 7 and 8 for representative results. (G) Super-resolution microscopy of nuclear LDs (mRuby3-NLSx3-HPos) and the INM (GFP-Lap2β) in U2OS transfected with either control or seipin siRNA and cultured with OA for 1 d. The distance between nuclear LDs and the INM was measured; n = 70 (control), 138 (seipin). Pooled data from three independent experiments. Mann–Whitney test; **, P < 0.01. The proportion of nuclear LDs within 0.1 µm of the INM: 2.9% for control, 15.2% seipin. Scale bars, 10 µm (fluorescence micrographs); 0.2 µm (EM).
Yeast seipin was inferred to exist in the INM based on a BiFC experiment using a basket nucleoporin, Nup60, as the complementation partner (Romanauska and Köhler, 2018). To study whether seipin exists in the INM in U2OS, we also used BiFC, but with NLSx3-RFP–tagged GFP1–10, a soluble nucleoplasmic protein, as the complementation partner. NLSx3-RFP-GFP1–10 complemented GFP11-GPAT4 and generated linear green fluorescence in the nuclear periphery as expected, whereas it never did with GFP11-seipin (Fig. 4 D). The functionality of GFP11-seipin was verified by complementation with RFP-GFP1–10, which generated green fluorescence in a cytoplasmic network pattern (Fig. 4 D). Furthermore, immuno-EM of GFP-seipin showed labeling in the ER and the outer nuclear membrane, but not in the INM (Fig. 4 E; see Fig. S1 F for comparison). These results led us to conclude that seipin does not exist in the INM in U2OS. What causes the difference between yeast and mammalian results is not clear and needs further investigation.
Next, we examined the influence of seipin knockdown on nuclear and cytoplasmic LDs using FRAP of GFP-ACSL3. In agreement with the previous observation using HPos (Salo et al., 2016), the majority of control cells exhibited fluorescence recovery of GFP-ACSL3 in cytoplasmic LDs, while in seipin-deficient cells, recovery occurred more slowly and in a smaller proportion of cytoplasmic LDs (Fig. 4 F). In contrast, in nuclear LDs, the fluorescence recovery of GFP-ACSL3 was not observed in control cells (Fig. 4 F and Video 7), consistent with the low frequency of GFP-DGAT2 FRAP (Video 4). Remarkably, after seipin knockdown, recovery occurred in more than half of nuclear LDs (Fig. 4 F and Video 8), indicating that the nuclear LDs connected to the INM were increased. Consistent with this, super-resolution microscopy of mRuby3-NLSx3-HPos showed that a nuclear LD–INM association was far more common in seipin-deficient cells than in control cells (Fig. 4 G). The association of nuclear LDs with the INM was also observed by EM (Fig. S2 E).
Video 7.
FRAP of GFP-ACSL3 (left, green; arrow) in nuclear LDs (right, red) in U2OS transfected with control siRNA. One frame captured every 1 min; displayed at 3 frames/s. Quantitative results are shown in Fig. 4 F. Scale bar, 10 µm.
Video 8.
FRAP of GFP-ACSL3 (left, green; arrow) in nuclear LDs (right, red) in U2OS transfected with seipin siRNA. Quantitative results are shown in Fig. 4 F. One frame captured every 1 min; displayed at 3 frames/s. Scale bar, 10 µm.
Overexpression of seipin also caused a reciprocal change in nuclear and cytoplasmic LDs, decreasing nuclear LDs while increasing small and constant-sized cytoplasmic LDs (Fig. S2, F and G). These results indicate that seipin affects nuclear and cytoplasmic LDs differently, often in opposite ways. Considering the absence of seipin in the INM, we inferred that seipin knockdown and overexpression affect nuclear LDs indirectly through changes of seipin functionalities in the ER.
Mechanism of nuclear LD increase in seipin deficiency
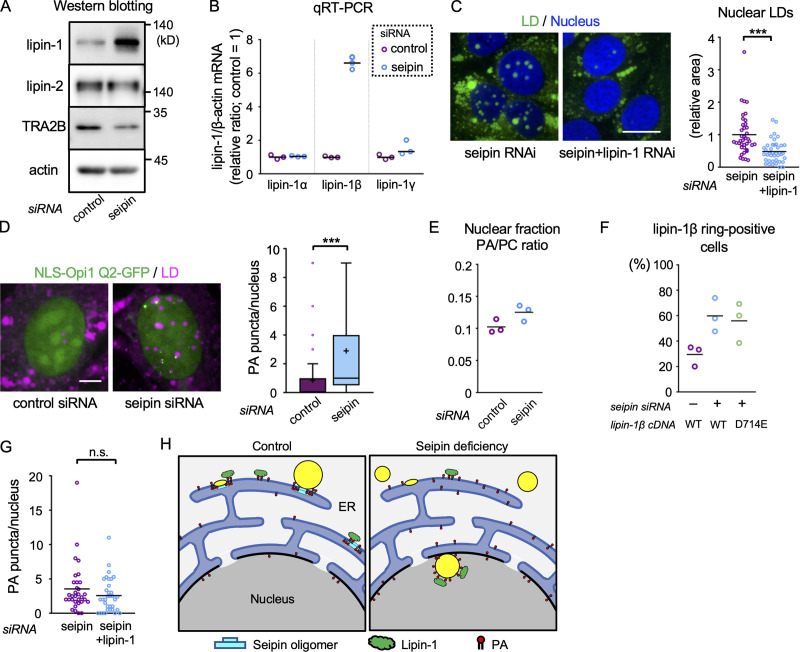
Western blotting revealed that lipin-1, but not lipin-2, increased as a result of seipin knockdown (Fig. 5 A). Lipin-1α and -1β could not be separated in Western blotting, but qRT-PCR showed that lipin-1β mRNA increased while lipin-1α mRNA did not change (Fig. 5 B), suggesting that lipin-1β protein was increased in seipin-deficient cells. It is notable that seipin knockdown also decreased splicing factor TRA2Β (SFRS10; Fig. 5 A). Reduction of TRA2Β was shown to favor the generation of lipin-1β mRNA in human obesity (Pihlajamäki et al., 2011; but see Brosch et al., 2012 for an alternative result) and thus may be the cause of lipin-1β up-regulation in seipin deficiency. The mechanism linking seipin and TRA2Β warrants further investigation in relation to abnormalities caused by seipin mutation.
Figure 5.
The effect of seipin knockdown. (A) Western blotting. U2OS transfected with control or seipin siRNA. (B) qRT-PCR. U2OS transfected with control or seipin siRNA. The lipin-1/β-actin mRNA ratio in the control sample is set to one. A representative result of three independent experiments. (C) The effect of seipin/lipin-1 double knockdown. U2OS transfected with seipin siRNA alone or with seipin siRNA and lipin-1 siRNA was cultured with OA for 1 d. The number of nuclei counted: 39 (seipin) or 40 (seipin+lipin-1). Pooled data from three independent experiments. Mann–Whitney test; ***, P < 0.001. (D) The effect of seipin knockdown on NLS-Opi1 Q2-GFP (green). U2OS were cultured with OA for 1 d. LD, magenta. The number of PA puncta/rings per nucleus was counted. The number of nuclei counted: 81 (control) or 57 (seipin). Pooled data from three independent experiments. Box boundaries indicate the 25th and 75th percentiles, and whiskers indicate Tukey type. Mann–Whitney test; ***, P < 0.001. (E) The effect of seipin knockdown on nuclear PA. PA and PC were quantified in nuclei isolated from U2OS cultured with OA for 2 d. A representative result of two independent experiments. (F) The effect of seipin knockdown on GFP–lipin-1β concentration in nuclear LDs. U2OS were cultured with OA for 1 d. Wild-type and PA phosphatase-deficient mutant (D714E) were compared. The number of nuclei counted: 60 (control siRNA+WT cDNA), 63 (seipin siRNA+WT cDNA), 41 (seipin siRNA+D714E cDNA). A summary of three independent experiments. (G) The effect of seipin/lipin-1 double knockdown on NLS-Opi1 Q2-GFP. The number of nuclei counted: 32 (seipin) or 34 (seipin+lipin-1). Pooled data from three independent experiments. Mann–Whitney test. (H) Diagram of nuclear LD formation. Nuclear LDs in normal cells will also form using nuclear PA and lipin-1. Scale bars, 10 µm. n.s., not significant.
To examine whether lipin-1 up-regulation is essential for the increase of nuclear LDs in seipin deficiency, knockdown of seipin alone and double knockdown of seipin and lipin-1 were compared. The additional knockdown of lipin-1 suppressed the increase of nuclear LDs by seipin knockdown (Figs. 5 C and S3 A), indicating that up-regulation of lipin-1 is at least partially responsible for the increase of nuclear LDs in seipin-deficient cells.
Figure S3.
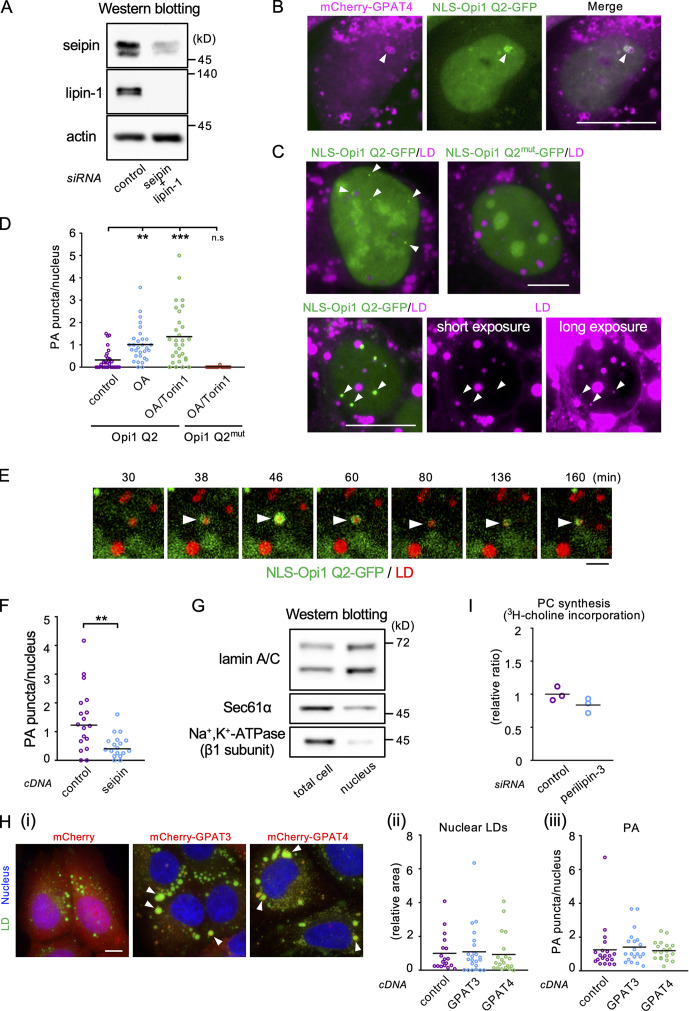
Effects of seipin knockdown on PA in the nucleus. (A) Western blotting showing the efficacy of seipin/lipin-1 double knockdown. (B) Colocalization of mCherry-GPAT4 (magenta) and NLS-Opi1 Q2-GFP (green). Scale bar, 10 µm. (C) The effect of OA/Torin1 on NLS-Opi1 Q2-GFP (green). U2OS expressing NLS-Opi1 Q2-GFP or NLS-Opi1 Q2mut-GFP were cultured with OA for 1 d and OA/Torin1 for another 8 h. The long-exposure image shows the invariable association of NLS-Opi1 Q2-GFP signals (arrowheads) with nuclear LDs (magenta). Scale bars, 10 µm. (D) The number of PA puncta in the nucleus. U2OS expressing NLS-Opi1 Q2-GFP (Opi1 Q2) was cultured either in normal medium, in medium with OA for 1 d, or in medium with OA for 1 d and with OA/Torin1 for another 8 h. Cells expressing NLS-Opi1 Q2mut-GFP (Opi1 Q2mut) were treated with OA and OA/Torin1. The number of nuclei counted: 30 (Opi1 Q2), 15 (Opi1 Q2mut). Pooled data from three (Opi1 Q2) or two (Opi1 Q2mut) independent experiments. Kruskal–Wallis ANOVA test followed by Dunn’s test; **, P < 0.01; ***, P < 0.001; n.s., not significant. (E) Live imaging of NLS-Opi1 Q2-GFP (green) in U2OS treated with OA for 1 d. LDs, red. Selected frames from Video 9 are shown. Arrows indicate a nuclear LD with fluctuating PA signals. Scale bar, 2 µm. (F) The effect of seipin overexpression on the number of PA puncta in the nucleus. The number of nuclei counted: 18. A representative result of two independent experiments. Welch’s unpaired t test; **, P < 0.01. (G) Western blotting for verification of nuclear isolation. Equal amounts of proteins were loaded for the total cell and isolated nucleus samples. (H) The effect of GPAT3/4 overexpression. (Hi) Large cytoplasmic LDs surrounded with mCherry-tagged GPAT3 and GPAT4 (arrowheads). Scale bar,10 µm. (Hii) Nuclear LDs. The number of nuclei counted: 18 (mCherry), 23 (mCherry-GPAT3), 20 (mCherry-GPAT4). A representative result of two independent experiments. Mann–Whitney test. (Hiii) Nuclear PA puncta. The number of nuclei counted: 20. A representative result of two independent experiments. Mann–Whitney test. (I) The effect of perilipin-3 knockdown on PC synthesis in U2OS. Incorporation of 3H-choline to PC was measured 3 d after perilipin-3 siRNA transfection. A representative result of two independent experiments.
An important change in seipin deficiency occurs in the intracellular distribution of PA (Fei et al., 2011; Han et al., 2015; Wolinski et al., 2015). We hypothesized that seipin deficiency increases nuclear LD formation by increasing nuclear PA, thereby facilitating lipin-1 recruitment (Eaton et al., 2013; Ren et al., 2010). To test this idea, we used the fluorescent biosensor NLS-Opi1 Q2-GFP, which binds to nuclear PA (Romanauska and Köhler, 2018). Although the Opi1 Q2 domain may have a preference for some PA species (Kassas et al., 2017), the accumulation of NLS-Opi1 Q2-GFP in mCherry-GPAT4–positive nuclear LDs supports its functionality as a PA sensor (Fig. S3 B). NLS-Opi1 Q2-GFP distributed diffusely in untreated U2OS, but in U2OS cultured with OA alone or with OA and Torin1, puncta besides nuclear LDs and rings around them increased, probably reflecting an increase of PA synthesis (Menon et al., 2017; Fig. S3, C and D). In live imaging, NLS-Opi1 Q2-GFP showed blinking and punctum-to-ring changes, indicating fluctuation of nuclear PA (Fig. S3 E and Video 9). NLS-Opi1 Q2mut-GFP, mutated in the PA-binding domain, distributed only diffusely in the nucleus, verifying the specificity of the PA biosensor (Fig. S3, C and D).
Video 9.
Live imaging of NLS-Opi1 Q2-GFP (green) in U2OS treated with OA for 1 d. LDs, red. Arrows indicate a nuclear LD with fluctuating PA signals. One frame captured every 2 min; displayed at 5 frames/s. Selected frames are shown in Fig. S3 E. Scale bar, 10 µm.
Remarkably, the number of PA puncta/rings significantly increased in seipin-deficient cells (Fig. 5 D) and decreased in cells overexpressing seipin (Fig. S3 F). The increase of nuclear PA in seipin-deficient cells was confirmed by enzyme-coupled fluometric assay of PA in isolated nuclei (Figs. 5 E and S3 G; Morita et al., 2009). Accumulation of GFP–lipin-1β in nuclear LDs increased in seipin-deficient cells, irrespective of its PA phosphatase activity (Fig. 5 F), indicating that increased PA facilitated lipin-1 recruitment, although a PA-independent mechanism might be also involved. The nuclear PA signal in seipin-deficient cells was not affected by additional knockdown of lipin-1 (Fig. 5 G). The result indicated that the increase of nuclear PA occurs independently from the increase of lipin-1 expression.
GPAT3 and GPAT4, when overexpressed, were mostly distributed in the cytoplasm and induced large cytoplasmic LDs, as previously reported (Pagac et al., 2016), but they did not increase nuclear LDs or nuclear PA puncta (Fig. S3 H). This result suggests that, in the presence of seipin, PA that is produced by GPAT3/4 is used efficiently for LD biogenesis in the ER and scarcely increases PA in the INM. Diffusion of PA to the INM may also be restricted by binding to seipin (Yan et al., 2018). Moreover, under normal conditions, seipin down-regulates GPAT3 and GPAT4 (Pagac et al., 2016). When these effects are lost by seipin depletion, freely diffusible PA should increase in the ER and reach the INM. We speculate that PA that is increased in the INM in this manner recruits up-regulated lipin-1, thereby enhancing TG synthesis and nuclear LD formation (Fig. 5 H), although a mechanism unrelated to PA may be also involved. The increased PA in seipin-deficient cells may also increase the INM-connected nuclear LDs by making LD budding inefficient (Ben M’barek et al., 2017) and stabilizing the connecting membrane bridge (Choudhary et al., 2018).
Nuclear LDs in hepatocytes recruit choline-phosphate cytidylyltransferase A (CCTα) and activate phosphatidylcholine (PC) synthesis (Sołtysik et al., 2019b). Nuclear LDs in U2OS also recruit CCTα (Lee et al., 2020), but knockdown of perilipin-3, which up-regulates PC synthesis in Huh7 by increasing CCTα recruitment, did not have the same effect in U2OS (Fig. S3 I), suggesting that the impact of nuclear LDs on CCTα regulation may be smaller in U2OS. On the other hand, nuclear LDs may modulate PML-NB functions (Lee et al., 2020; Soltysik et al., 2019a). Interestingly, the nuclear translocation of lipin-1 induced by mTOR inhibition causes down-regulation of sterol regulatory element–binding proteins (Peterson et al., 2011), which may distribute in PML-NBs (Zoumi et al., 2005). Increased nuclear LDs may be involved in this regulatory process. The present study provides a solid basis to probe this and other fascinating possibilities by showing how nuclear LDs form and how they can be manipulated experimentally.
Materials and methods
Antibodies
Rabbit anti-human perilipin-3 antibody was raised against a peptide of human perilipin-3 segment (amino acids 305–318) and affinity purified by using a peptide column (Ohsaki et al., 2006). Rabbit anti-GFP and rabbit anti-PML antibodies were donated by Dr. Masahiko Watanabe (Hokkaido University, Sapporo, Japan) and Dr. Kun-San Chang (The University of Texas MD Anderson Cancer Center, Houston, TX), respectively. Mouse anti-ACSL3 (H00002181-B01P; Abnova), mouse anti-Na+,K+-ATPase β 1 (NB300-147SS; Novus Biologicals), rabbit anti-GPAT3 (AP18013c; ABGENT), rabbit anti-GPAT4 (NB100-2390SS; Novus Biologicals), rabbit anti-DGAT1 (NB110-41487SS; Novus Biologicals), rabbit anti-seipin (NBP1-79336; Novus Biologicals; 23846; Cell Signaling), rabbit anti–lipin-1 (HPA038021; Atlas Antibodies), rabbit anti–lipin-2 (A303-703A; Bethyl Lab), rabbit anti–lamin B receptor (GWB-C7CA28; Genway Biotech), rabbit anti-PML (M041-3; Medical and Biological Laboratories), rabbit anti-actin (A2066; Sigma), rabbit anti-TRA2B (GTX114752; GeneTex), rabbit anti–lamin A/C (GTX101127; GeneTex), goat anti-MTP (sc-33116; Santa Cruz Biotechnology), and goat anti-Sec61α (sc-12322; Santa Cruz Biotechnology) antibodies were obtained from the indicated suppliers. Secondary antibodies conjugated to fluorochromes (Invitrogen and Jackson ImmunoResearch Lab) and protein A conjugated with colloidal gold (Utrecht University Medical Center) were also purchased.
Cell lines and reagents
U2OS, Huh7, and A549 cells were kindly donated by Dr. Hidemasa Goto (Aichi Cancer Center, Nagoya, Japan), Dr. Eija Jokitalo (University of Helsinki, Helsinki, Finland), and Dr. Takashi Takahashi (Nagoya University, Nagoya, Japan), respectively. Cells were cultured in DMEM (U2OS and A549) or minimum essential medium (Huh7) supplemented with 10% FBS and antibiotics at 37°C in a humidified atmosphere of 95% air and 5% CO2. In some experiments, OA (Sigma) in complex with fatty acid–free BSA (Wako) at a molar ratio of 6:1 was added. BAY13-9952 (Implitapide; Bayer Healthcare), triacsin C (Santa Cruz Biotechnology), tunicamycin, and Torin1 (Sigma) were purchased from the indicated suppliers.
Plasmids and siRNA
pEGFP-C3-PML-II was donated by Dr. P. Hemmerich (Fritz-Lipmann Institute, Jena, Germany; Weidtkamp-Peters et al., 2008). The sequences encoding human GPAT3 (NCBI RefSeq accession no. NM_032717.5), human GPAT4 (NM_178819.4), human AGPAT2 (NM_006412.4), human AGPAT4 (NM_020133.3), human LPCAT1 (NM_024830.5), human ACSL3 (NM_004457.5), human seipin (NM_001122955.4), human lipin-2 (NM_014646.2), and human lipin-3 (NM_022896.3) were amplified using standard PCR procedures with cDNA from U2OS, Huh7, or HeLa cells and subcloned into pEGFP-C1, pEGFP-N3, pmCherry-C1, or pDendra2-C vectors (Clontech). Mouse DGAT1 (NM_010046.3) and mouse DGAT2 (NM_026384.3) were subcloned from the Flag mouse DGAT1 and Venus mouse DGAT2 vectors, which were donated by Dr. Ho Yi Mak (Hong Kong University of Science and Technology, Hong Kong; Xu et al., 2012). Human lipin-1α (NM_145693.4), lipin-1β (NM_001349204.2), and lipin-1γ (NM_001349200.2) were subcloned from pET-28b(+) plasmids that were donated by Dr. George Carman (Rutgers University, New Brunswick, NJ; Han and Carman, 2010). Plasmids encoding lipin-1 mutants, lipin-1α (D678E, LxxFF) and lipin-1β (D714E, LxxFF) were generated by PCR using primers with appropriate nucleotide substitutions. Split GFPs for BiFC, GFP1–10, and GFP11 were subcloned from pcDNA3.1-GFP(1–10) (#70219; Addgene) and pEGFP-GFP11-clathrin light chain (#70217; Addgene), which were gifts of Dr. Bo Huang (University of California, San Francisco, San Francisco, CA; Kamiyama et al., 2016). A chicken muscle pyruvate kinase segment lacking the first 16 amino acids was subcloned from pEGFP-C1-PK (#24163; Addgene), which was a gift of Dr. Warner Greene (Gladstone Institutes, San Francisco, CA), to construct a plasmid for expression of GFP-PK-DGAT2. For fluorescent PA biosensor NLS-tagged Opi1 Q2-GFP, Opi1 Q2 (amino acids 103–189 of Opi1; NM_001179100.1) was amplified by standard PCR procedures from Saccharomyces cerevisiae (SEY6210) cDNA and subcloned into pEGFP-N3; NLS-Opi1 Q2mut-GFP was generated by substituting eight amino acids with alanine (L124R, Y127A, L129R, M131A, I133R, K136A, K137A, and R138A) as previously described (Romanauska and Köhler, 2018). GFP-NLSx3-Hpos, a nuclear LD marker, was constructed by inserting three NLSs in tandem before Hpos (MDVLVPLLQLLVLLLTLPLHLLALGCWQPLFEAIGKIFSNIRISTQKEI; Kassan et al., 2013) as previously described (Sołtysik et al., 2019b). NLS (KRPAATKKAGQAKKKK) and NES (NELALKLAGLDINKT) were taken from Xenopus nucleoplasmin (GenBank accession no. X04766.1) and human cAMP-dependent protein kinase inhibitor α (NCBI RefSeq accession no. NM_006823.4), respectively.
siRNAs were synthesized by Japan Bio Service, Inc. Target nucleotide sequences of siRNAs are shown in Table S1.
Plasmids and siRNAs were transfected using Lipofectamine 2000 and Lipofectamine RNAiMAX (Thermo Fisher), respectively, according to the manufacturer’s instruction. The cells were analyzed 2 d (cDNA) and 3 d (siRNA) after transfection.
Immunofluorescence microscopy and data analysis
Cells were fixed with 3–4% formaldehyde in 0.1 M phosphate buffer for 15 min and permeabilized with either 0.01% digitonin in PBS for 30 min or 0.5% Triton X-100 in PBS for 10 min. Blocking and antibody dilution were performed with 1–3% BSA in PBS or 4% normal goat serum in PBS. Nuclei were labeled with Hoechst 33342 (Sigma), whereas LDs were labeled with either BODIPY493/503 or LipidTox Red (Thermo Fisher). Samples were mounted in Mowiol 4–88 containing 2.5% 1,4-diazabicyclo-[2,2,2]-octane.
Images were captured using a Zeiss Axio Imager M2 with Apotome2 using Plan-Neofluar 100×/1.30 and Plan-Apochromat 63×/1.40 lenses. For super-resolution microscopy, a Nikon Eclipse Ti with N-SIM module equipped with an Andor DU-897 camera, an Apo TIRF 100×/1.49 Oil DIC N2 lens, and a Nikon NIS-Elements 5.20.02 software was used. The color, brightness, and contrast of presented images were adjusted using Adobe Photoshop CS3 or Zeiss Zen. Quantification was performed as described previously (Ohsaki et al., 2016). The number of LDs was counted manually, and the area was measured using ImageJ. The total nuclear LD area and the total intranuclear lamin B1 receptor–positive area were divided by the total nuclear area to obtain the relative nuclear LD area and the relative NR area, respectively. For NR measurement, signals in the nuclear periphery were excluded.
Live imaging
Live confocal images (except for those shown in Video 2) were obtained using a Cell Voyager CV1000 spinning-disk confocal laser system (Olympus) equipped with 488-nm and 561-nm diode lasers. An electron multiplying charge-coupled device camera (1,000 × 1,000 pixels, ImagEM C9100-14 and 512 × 512 pixels, ImagEM C9100-13; Hamamatsu Photonics) and oil immersion objective lenses (UPLSAPO 100×/1.4 and UPLSAPO 60×/1.35; Olympus) were used. For Video 2, an Olympus SpinSR10 spinning-disk confocal super-resolution microscope equipped with a Hamamatsu ORCA-Flash 4.0 camera, a UPLAPO OHR 60×/1.50 lens, and an Olympus cellSens Dimension 2.3 software was used. Cells were kept at 37°C in a culture chamber filled with 95% air and 5% CO2. Cells were placed in DMEM and 10% FBS for most experiments, except for the experiment shown in Video 2, in which they were placed in FluoroBrite DMEM (Thermo Fisher) supplemented with 10% FBS and 4 mM L-glutamine. For each point, multiple z-stack sections with intervals <1.0 µm were obtained. Images were processed using the Fiji software application.
FRAP and photoconversion
U2OS cells expressing each of the respective fluorescent proteins were cultured with OA for 1 d and observed with a confocal laser-scanning microscope (TiE-A1R; Nikon) equipped with an A1-DUG GaAsP Multi Detector Unit and a Plan Apo ×100/1.45 oil immersion lens. For FRAP, one nuclear LD or up to three cytoplasmic LDs were bleached 10 times at 63-ms intervals using the 561-nm laser at 100% intensity. Images were captured for 20 min at 1-min intervals (nuclear LDs) or for 15 min at 30-s intervals (cytoplasmic LDs). For photoconversion, a cytoplasmic area was illuminated 10 times at 63-ms intervals using the 488-nm laser at 100% intensity. All experiments were performed at 37°C in a humidified atmosphere of 95% air and 5% CO2. Florescence recovery in regions of interest was quantified using ImageJ and normalized to fluorescence in the LDs before bleaching. Results are presented as the mean ± SEM, and curves fitting with nonlinear regression and the exponential one-phase decay model were applied (GraphPad Prism 8).
PC synthesis activity
Cells cultured with 1 µCi/ml 3H-choline (PerkinElmer) for 30 min were incubated with hexane/isopropanol (9/1) for 30 min to selectively extract PC among choline-containing metabolites (Vassar et al., 2007). The 3H-choline radioactivity was measured using a liquid scintillation counter (Aloka) and normalized according to the protein concentration.
EM
Cells were fixed with a mixture of 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) and postfixed with a mixture of 1% osmium tetroxide and 0.1% potassium ferrocyanide in 0.1 M sodium cacodylate buffer. Cells expressing HRP-KDEL were incubated in an enzyme histochemical reaction solution to form DAB precipitates before osmification (Brown and Farquhar, 1989; Jokitalo et al., 2001). Samples were embedded in epoxy resin, and ultrathin sections were observed using a JEOL JEM1011 electron microscope operated at 100 kV.
Immuno-EM of ultrathin cryosections
Cells were fixed with 4% formaldehyde and 0.25% glutaraldehyde for 60 min, infiltrated with 2.3 M sucrose in 0.1 M phosphate buffer, pH 7.4, and frozen in liquid nitrogen. Ultrathin cryosections were prepared in an ultramicrotome with an attached cryochamber FC7 (Leica), immunolabeled, stained with 2% neutral uranyl acetate, and embedded in 2% methyl cellulose plus 0.4% uranyl acetate (Tokuyasu, 1980).
Western blotting
Cells were either directly dissolved in SDS sample buffer or treated with RIPA buffer (1% Triton X-100, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% SDS, and 0.5% sodium deoxycholate) supplemented with protease inhibitor (Nacalai), and the supernatant was used. The reaction obtained with Super Signal West Dura Substrate (Thermo Fisher) was captured using a Fusion Solo S instrument (Vilber Lourmat) and analyzed using Fusion-Capt Advance Software version 16.15.
qRT-PCR
Total RNA was extracted from U2OS using the ReliaPrep RNA cell miniprep system (Promega). cDNA was obtained using Superscript III reverse transcription (Thermo Fisher). qRT-PCR was performed using Brilliant II SYBR with a Low Rox Master Mix and an Mx3005P RT-PCR system (Agilent Technologies) according to the manufacturer’s instructions. The variances in the cDNA amount were normalized to the level of β-actin cDNA. The primer sequences are shown in Table S2.
PA and PC measurement in isolated nuclei
The nuclear fraction was isolated using a sucrose method (Nabbi and Riabowol, 2015), and lipids were extracted by the Folch method (Folch et al., 1957). The concentrations of PA and PC were measured as previously described (Morita et al., 2009, 2010). Briefly, lipids were incubated with lipoprotein lipase of Pseudomonas sp. (129–04501; FUJIFILM Wako Pure Chemical) and then with glycerol-3-phosphate oxidase (T-60; Asahi Kasei Pharma) for PA, and with glycerophospholipid-specific phospholipase D from Streptomyces sp. (T-39; Asahi Kasei Pharma) and then with choline oxidase from Alcaligenes sp. (037–14401; FUJIFILM Wako Pure Chemical) for PC. Hydrogen peroxide generated by glycerol-3-phosphate oxidase or choline oxidase was reacted with Amplex Red (Invitrogen) in the presence of HRP (Oriental Yeast) to form the fluorescent reporter resorufin, which was measured using a microplate reader (SpectraMax iD3; Molecular Devices).
Statistical analysis and plots
Normality was assessed by the Shapiro–Wilk test. Statistical differences between samples were examined by Kruskal–Wallis ANOVA test followed by Dunn’s test for the data in Fig. 1, A and C; Fig. 2 C; Fig. 3 E; Fig. 4 A; and Fig. S3 D; Mann–Whitney test for the data in Fig. 1 B; Fig. 2, D and E; Fig. 3 C; Fig. 4, B and G; Fig. 5, C, D, and G; Fig. S2 B; and Fig. S3 H; or Welch’s t test for the data in Fig. S3 F. Pooled data obtained in three independent experiments were analyzed to produce the results shown in Fig. 1, A–C; Fig. 2, C–E; Fig. 3, C and E; Fig. 4, A, B, and G; and Fig. 5, C, D, and G. A representative result of two or three independent experiments is shown for all other data. The bars in each scatter plot indicate the average. Box plots are used when the sample number exceeds 40. In box plots, the center line indicates the median, box boundaries indicate the 25th and 75th percentiles, whiskers are Tukey type, and the average is marked as “+.” Statistical analyses were performed with Prism 8 (GraphPad).
Online supplemental material
Fig. S1 shows characterization of nuclear LDs in U2OS. Fig. S2 shows effects of lipin-1 expression and seipin knockdown and overexpression on nuclear LDs. Fig. S3 shows characterization of NLS-Opi1 Q2-GFP and effects of seipin knockdown on PA in the nucleus. Table S1 lists the sequences of siRNAs. Table S2 lists the sequences of RT-PCR primers. Videos 1 and 2 show nuclear LD formation at the nuclear envelope in U2OS treated with OA for 1 d. Video 3 illustrates the appearance of red Dendra2-LPCAT1 signal in nuclear LDs after photoconversion in the cytoplasm. Video 4 demonstrates FRAP of GFP-DGAT2 in nuclear LDs. Video 5 shows the disappearance of GFP-ACSL3 from nuclear LDs. Video 6 displays live imaging of GFP-Lipin1β in U2OS treated with OA and Torin1. Videos 7 and 8 demonstrate FRAP of GFP-ACSL3 in nuclear LDs in U2OS transfected with control siRNA and seipin siRNA, respectively. Video 9 shows live imaging of NLS-Opi1 Q2-GFP in U2OS treated with OA for 1 d.
Supplementary Material
lists siRNA sequences.
lists RT-PCR primer sequences.
Acknowledgments
We thank the staff members of the Division for Medical Research Engineering and the Radioisotope Research Center of Nagoya University Graduate School of Medicine for their skillful support, Dr. Takaki Miyata (Nagoya University, Nagoya, Japan) and Dr. Noboru Mizushima (The University of Tokyo, Tokyo, Japan) for generously permitting the use of their microscopes and reagents, and many other colleagues for the donation of valuable materials.
This study was supported by the Japan Society for the Promotion of Science KAKENHI grants to Y. Ohsaki (18K06829) and T. Fujimoto (15H05902 and 18H04023), and Advanced Bioimaging Support (JP16H06280).
The authors declare no competing financial interests.
Author contributions: K. Sołtysik, Y. Ohsaki, and T. Fujimoto designed the experiments and analyzed the results. K. Sołtysik, Y. Ohsaki, T. Tatematsu, J. Cheng, A. Maeda, and S. Morita performed the experiments. T. Fujimoto wrote the manuscript.
References
- Barbosa A.D., Lim K., Mari M., Edgar J.R., Gal L., Sterk P., Jenkins B.J., Koulman A., Savage D.B., Schuldiner M., et al. . 2019. Compartmentalized synthesis of triacylglycerol at the inner nuclear membrane regulates nuclear organization. Dev. Cell. 50:755–766.e6. 10.1016/j.devcel.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben M’barek K., Ajjaji D., Chorlay A., Vanni S., Forêt L., and Thiam A.R.. 2017. ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell. 41:591–604.e7. 10.1016/j.devcel.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Bou Khalil M., Sundaram M., Zhang H.Y., Links P.H., Raven J.F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D.N., and Yao Z.. 2009. The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 50:47–58. 10.1194/jlr.M800204-JLR200 [DOI] [PubMed] [Google Scholar]
- Brosch M., von Schönfels W., Ahrens M., Nothnagel M., Krawczak M., Laudes M., Sipos B., Becker T., Schreiber S., Röcken C., et al. . 2012. SFRS10--a splicing factor gene reduced in human obesity? Cell Metab. 15:265–266, author reply :267–269. 10.1016/j.cmet.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Brown W.J., and Farquhar M.G.. 1989. Immunoperoxidase methods for the localization of antigens in cultured cells and tissue sections by electron microscopy. Methods Cell Biol. 31:553–569. 10.1016/S0091-679X(08)61626-X [DOI] [PubMed] [Google Scholar]
- Cartwright B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V., Golani G., Joshi A.S., Cottier S., Schneiter R., Prinz W.A., and Kozlov M.M.. 2018. Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr. Biol. 28:915–926.e9. 10.1016/j.cub.2018.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton J.M., Mullins G.R., Brindley D.N., and Harris T.E.. 2013. Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J. Biol. Chem. 288:9933–9945. 10.1074/jbc.M112.441493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. . 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck B.N., Gropler M.C., Chen Z., Leone T.C., Croce M.A., Harris T.E., Lawrence J.C. Jr., and Kelly D.P.. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4:199–210. 10.1016/j.cmet.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., and Sloane Stanley G.H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- Gao M., Huang X., Song B.L., and Yang H.. 2019. The biogenesis of lipid droplets: Lipids take center stage. Prog. Lipid Res. 75:100989 10.1016/j.plipres.2019.100989 [DOI] [PubMed] [Google Scholar]
- Grevengoed T.J., Klett E.L., and Coleman R.A.. 2014. Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 34:1–30. 10.1146/annurev-nutr-071813-105541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., and Carman G.M.. 2010. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285:14628–14638. 10.1074/jbc.M110.117747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Binns D.D., Chang Y.F., and Goodman J.M.. 2015. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16:29 10.1186/s12860-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.E., Huffman T.A., Chi A., Shabanowitz J., Hunt D.F., Kumar A., and Lawrence J.C. Jr. 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282:277–286. 10.1074/jbc.M609537200 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Reese M.L., and Goodman J.M.. 2019. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 38:e101816 10.15252/embj.2019101816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokitalo E., Cabrera-Poch N., Warren G., and Shima D.T.. 2001. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154:317–330. 10.1083/jcb.200104073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama D., Sekine S., Barsi-Rhyne B., Hu J., Chen B., Gilbert L.A., Ishikawa H., Leonetti M.D., Marshall W.F., Weissman J.S., and Huang B.. 2016. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7:11046 10.1038/ncomms11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. . 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassas N., Tanguy E., Thahouly T., Fouillen L., Heintz D., Chasserot-Golaz S., Bader M.F., Grant N.J., and Vitale N.. 2017. Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292:4266–4279. 10.1074/jbc.M116.742346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Salsman J., Foster J., Dellaire G., and Ridgway N.D.. 2020. Lipid-associated PML structures assemble nuclear lipid droplets containing CCTα and Lipin1. Life Sci. Alliance. 3:e202000751 10.26508/lsa.202000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H., Qu J., Carmack A.E., Kim H.B., Chen C., Ren H., Morris A.J., Finck B.N., and Harris T.E.. 2010. Lipin proteins form homo- and hetero-oligomers. Biochem. J. 432:65–76. 10.1042/BJ20100584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A., Goulbourne C., and Vaux D.J.. 2011. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 21:362–373. 10.1016/j.tcb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Menon D., Salloum D., Bernfeld E., Gorodetsky E., Akselrod A., Frias M.A., Sudderth J., Chen P.H., DeBerardinis R., and Foster D.A.. 2017. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem. 292:6303–6311. 10.1074/jbc.M116.772988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S.Y., Ueda K., and Kitagawa S.. 2009. Enzymatic measurement of phosphatidic acid in cultured cells. J. Lipid Res. 50:1945–1952. 10.1194/jlr.D900014-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S.Y., Takeuchi A., and Kitagawa S.. 2010. Functional analysis of two isoforms of phosphatidylethanolamine N-methyltransferase. Biochem. J. 432:387–398. 10.1042/BJ20100490 [DOI] [PubMed] [Google Scholar]
- Nabbi A., and Riabowol K.. 2015. Isolation of pure nuclei using a sucrose method. Cold Spring Harb. Protoc. 2015:773–776. 10.1101/pdb.prot083741 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Maeda T., Maeda M., Tauchi-Sato K., and Fujimoto T.. 2006. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem. Biophys. Res. Commun. 347:279–287. 10.1016/j.bbrc.2006.06.074 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Kawai T., Yoshikawa Y., Cheng J., Jokitalo E., and Fujimoto T.. 2016. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212:29–38. 10.1083/jcb.201507122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., and Carvalho P.. 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20:137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. . 2016. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péterfy M., Phan J., and Reue K.. 2005. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 280:32883–32889. 10.1074/jbc.M503885200 [DOI] [PubMed] [Google Scholar]
- Peterson T.R., Sengupta S.S., Harris T.E., Carmack A.E., Kang S.A., Balderas E., Guertin D.A., Madden K.L., Carpenter A.E., Finck B.N., and Sabatini D.M.. 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 146:408–420. 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamäki J., Lerin C., Itkonen P., Boes T., Floss T., Schroeder J., Dearie F., Crunkhorn S., Burak F., Jimenez-Chillaron J.C., et al. . 2011. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 14:208–218. 10.1016/j.cmet.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Federico L., Huang H., Sunkara M., Drennan T., Frohman M.A., Smyth S.S., and Morris A.J.. 2010. A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol. Biol. Cell. 21:3171–3181. 10.1091/mbc.e10-01-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne M.F., Klug Y.A., and Carvalho P.. 2020. Lipid droplet biogenesis: A mystery “unmixing”? Semin. Cell Dev. Biol.:S1084-9521(18)30318-5 In press. [DOI] [PubMed] [Google Scholar]
- Romanauska A., and Köhler A.. 2018. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell. 174:700–715.e18. 10.1016/j.cell.2018.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou H., Hishikawa D., Harayama T., Yuki K., and Shimizu T.. 2009. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J. Lipid Res. 50(Suppl):S46–S51. 10.1194/jlr.R800035-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysik K., Ohsaki Y., and Fujimoto T.. 2019a Duo in a mystical realm—Nuclear lipid droplets and the inner nuclear membrane. Contact (Thousand Oaks). 2:1–11. 10.1177/251525641989696531777772 [DOI] [Google Scholar]
- Sołtysik K., Ohsaki Y., Tatematsu T., Cheng J., and Fujimoto T.. 2019b Nuclear lipid droplets derive from a lipoprotein precursor and regulate phosphatidylcholine synthesis. Nat. Commun. 10:473 10.1038/s41467-019-08411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B., and Worman H.J.. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27. 10.1083/jcb.130.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K.T. 1980. Immunochemistry on ultrathin frozen sections. Histochem. J. 12:381–403. 10.1007/BF01011956 [DOI] [PubMed] [Google Scholar]
- Vassar V., Hagen C., Ludwig J., Thomas R., and Zhou J.. 2007. One-step method of phosphatidylcholine extraction and separation. Biotechniques. 42:442, 444. 10.2144/000112435 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Chung J., and Farese R.V. Jr. 2017. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. . 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Airola M.V., and Reue K.. 2017. How lipid droplets “TAG” along: Glycerolipid synthetic enzymes and lipid storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10 Pt B, 10 Pt B):1131–1145. 10.1016/j.bbalip.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T.G., Schwanitz G., Hoischen C., Maul G., Dittrich P., and Hemmerich P.. 2008. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 121:2731–2743. 10.1242/jcs.031922 [DOI] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. . 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Xu N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V. Jr., and Mak H.Y.. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911. 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN binds anionic phospholipids. Dev. Cell. 47:248–256.e4. 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Zhang P., and Reue K.. 2017. Lipin proteins and glycerolipid metabolism: Roles at the ER membrane and beyond. Biochim. Biophys. Acta Biomembr. 1859(9 Pt B, 9 Pt B):1583–1595. 10.1016/j.bbamem.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoumi A., Datta S., Liaw L.H., Wu C.J., Manthripragada G., Osborne T.F., and Lamorte V.J.. 2005. Spatial distribution and function of sterol regulatory element-binding protein 1a and 2 homo- and heterodimers by in vivo two-photon imaging and spectroscopy fluorescence resonance energy transfer. Mol. Cell. Biol. 25:2946–2956. 10.1128/MCB.25.8.2946-2956.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists siRNA sequences.
lists RT-PCR primer sequences.