Cellular signal peptidase (SPase) cleavage represents an important step in maturation of viral envelope proteins. Fine tuning of this system allows for establishment of concerted folding and processing processes in different enveloped viruses. We report here on SPase processing of the Erns-E1-E2 glycoprotein precursor of pestiviruses. Erns-E1 cleavage is delayed and only executed efficiently when the complete E1 sequence is present. C-terminal truncation of the Erns-E1 precursor impairs processing and leads to significant secretion of the protein. The latter is not detected when internal deletions preserving the E1 carboxy terminus are introduced, but also these constructs show impaired processing. Moreover, Erns-E1 is only processed after cleavage at the E1/E2 site. Thus, processing of the pestiviral glycoprotein precursor by SPase is done in an ordered way and depends on the integrity of the proteins for efficient cleavage. The functional importance of this processing scheme is discussed in the paper.
KEYWORDS: Erns protein, pestiviruses, protein processing, signal peptidase
ABSTRACT
Like other enveloped viruses, pestiviruses employ cellular proteases for processing of their structural proteins. While typical signal peptidase cleavage motifs are present at the carboxy terminus of the signal sequence preceding Erns and the E1/E2 and E2/P7 sites, the Erns-E1 precursor is cleaved by signal peptidase at a highly unusual structure, in which the transmembrane sequence upstream of the cleavage site is replaced by an amphipathic helix. As shown before, the integrity of the amphipathic helix is crucial for efficient processing. The data presented here demonstrate that the E1 sequence downstream of this cleavage site is also important for the cleavage. Carboxy-terminal truncation of the E1 moiety as well as internal deletions in E1 reduced the cleavage efficiency to less than 30% of the wild-type (wt) level. Moreover, the C-terminal truncation by more than 30 amino acids resulted in strong secretion of the uncleaved fusion proteins. The reduced processing and increased secretion were even observed when 10 to 5 amino-terminal residues of E1 were left, whereas extensions by 1 or 3 E1 residues resulted in reduced processing but no significantly increased secretion. In contrast to the E1 sequences, a 10-amino-acid c-myc tag fused to the Erns C terminus had only marginal effect on secretion but was also not processed efficiently. Mutation of the von Heijne sequence upstream of E2 not only blocked the cleavage between E1 and E2 but also prevented the processing between Erns and E2. Thus, processing at the Erns-E1 site is a highly regulated process.
IMPORTANCE Cellular signal peptidase (SPase) cleavage represents an important step in maturation of viral envelope proteins. Fine tuning of this system allows for establishment of concerted folding and processing processes in different enveloped viruses. We report here on SPase processing of the Erns-E1-E2 glycoprotein precursor of pestiviruses. Erns-E1 cleavage is delayed and only executed efficiently when the complete E1 sequence is present. C-terminal truncation of the Erns-E1 precursor impairs processing and leads to significant secretion of the protein. The latter is not detected when internal deletions preserving the E1 carboxy terminus are introduced, but also these constructs show impaired processing. Moreover, Erns-E1 is only processed after cleavage at the E1/E2 site. Thus, processing of the pestiviral glycoprotein precursor by SPase is done in an ordered way and depends on the integrity of the proteins for efficient cleavage. The functional importance of this processing scheme is discussed in the paper.
INTRODUCTION
Pestiviruses are important pathogens of livestock, and Pestivirus is classified as one genus within the family Flaviviridae together with the genera Flavivirus, Hepacivirus, and Pegivirus. Within the genus Pestivirus, 11 species are found (pestivirus A to K), with pestivirus A, B, C, and D representing the long-known type species bovine viral diarrhea virus type 1 (BVDV-1) and type 2 (BVDV-2), classical swine fever virus (CSFV), and border disease virus (BDV), respectively, while the other species comprise more exotic viruses (1, 2).
As for other family members, the pestivirus genome consists of a positive-strand RNA, which is single stranded and comprises one long open reading frame (ORF) (3). Translation of the genomic RNA results in a polyprotein of ∼4,000 amino acids (aa). It is cotranslationally and posttranslationally processed by cellular and viral proteases into at least 12 mature proteins (3, 4), present in the polyprotein in the order NH2-Npro, C, Erns, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B-COOH. Pestivirus particles contain the proteins C, Erns, E1, and E2 (5, 6), with the glycoproteins Erns, E1, and E2 exposed on the host cell-derived lipid membrane surrounding the viral capsid composed of C and the viral genomic RNA. E2 represents the receptor binding protein and forms disulfide-linked heterodimers with E1. It is the main target for neutralizing antibodies, whereas antibodies directed against Erns have only limited neutralizing activity (7–10).
E1, E2, and also Erns represent essential structural proteins of pestiviruses (11, 12). Erns forms disulfide-linked homodimers of ∼90 kDa, about half of which are due to glycosylation (5, 13, 14). Interestingly, homodimer formation is dispensable for virus viability but correlated with virus virulence (15, 16). The Erns protein is unique among viral surface proteins since it contains an active site sequence motif and a three-dimensional (3D) structure typical for RNases of the T2 superfamily (11, 17–19). It was shown to degrade preferentially single-stranded RNA but is also able to cleave double-stranded RNA, though with reduced activity (20–23). The RNase activity of Erns is dispensable for virus viability but correlates with virus virulence (24, 25), most likely because it is involved in blocking the host type 1 interferon response to infection (26–29). A further unusual feature of this protein is its membrane anchor consisting of a long carboxy-terminal amphipathic helix that aligns in plane with the membrane surface (30–33). This kind of membrane association has been reported for only one other surface protein so far, the GP3 protein of the arterivirus porcine respiratory and reproductive syndrome virus (PRRSV) and seems to be responsible for the secretion of these two proteins (34). In the case of Erns, about 5% of the protein translated in an infected cell is secreted into the cell-free supernatant (14, 30–32). Erns secretion is regarded as crucial for its inhibitory effect on the interferon system, which is especially important for establishment of persistent pestivirus infections (26, 35, 36). In the infected animals, considerable amounts of Erns are found in the blood (27).
Processing of the structural protein region of the pestivirus polyprotein is done by cellular proteases. The cleavage sites identified at the amino-terminal ends of Erns, E2, p7, and NS2 (14, 37, 38) meet the requirements of signal peptidase (SPase) cleavage sites, namely, a positively charged N-terminal (n-) region and a central hydrophobic (h-) region followed by a more hydrophilic part (c-region). The latter domain contains the actual cleavage site with small and uncharged residues at positions −3 and −1 (39–42). SPase processing at these sites was confirmed by mutagenesis analyses (3, 14, 37, 38). In contrast, the site separating Erns and E1 does not comply with the above-described features of SPase cleavage sites because the hydrophobic transmembrane region preceding the cleavage site with the von Heijne motif is missing. As we have shown before, the long amphipathic helix located upstream of the von Heijne motif at the Erns carboxy terminus serves as Erns membrane anchor and substitutes for the TM region of the standard SPase cleavage site so that the Erns/E1 site is also processed by SPase (43). However, cleavage at this site is delayed when compared to the other processing steps so that an Erns-E1 precursor is readily detected in infected or transfected cells (14, 43). Because of its unusual membrane anchor, the complete Erns protein including its carboxy terminus stays on the luminal side of the endoplasmic reticulum (ER) (30, 32).
In the work presented here, we analyzed the role of the E1 sequence for cleavage at the Erns/E1 site. We show that efficient processing at this site depends on the presence of full-length E1 and can occur only after cleavage of the E1/E2 site so that it follows a hierarchical order.
RESULTS
E1 sequences influence Erns-E1 processing.
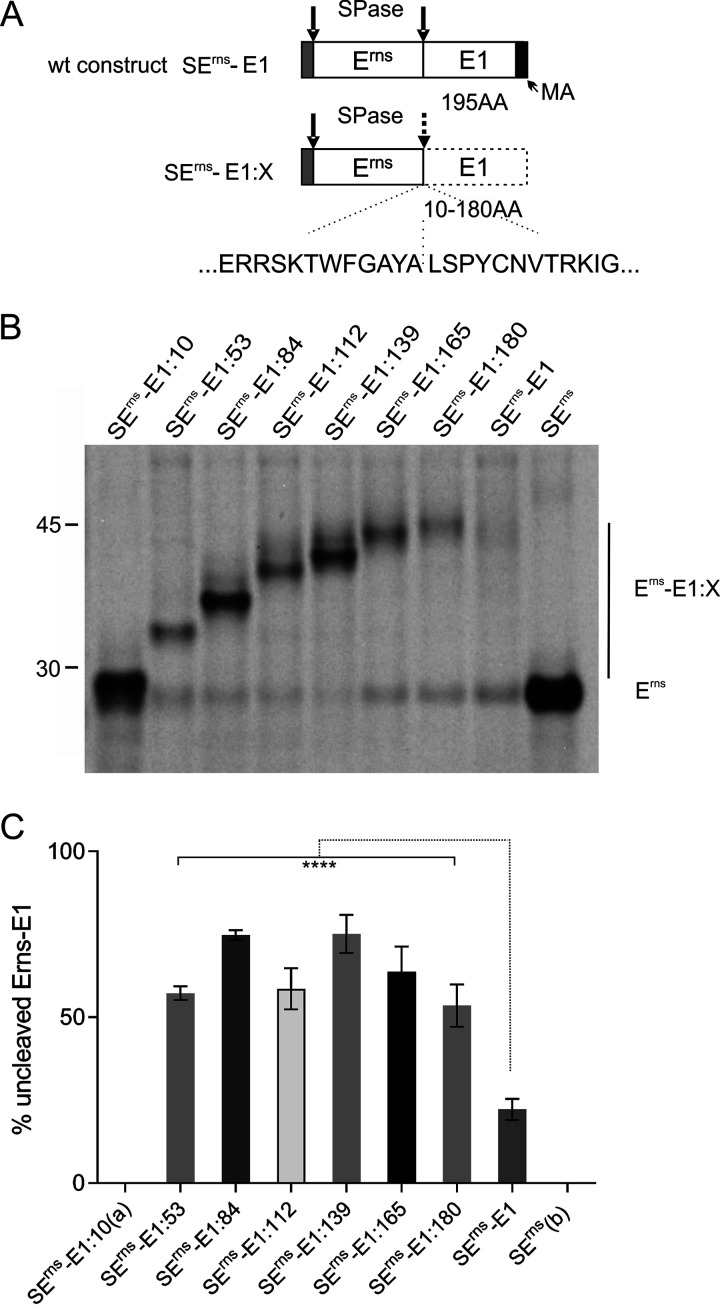
We have shown in a previous publication that the integrity of the amphipathic helix preceding the Erns/E1 cleavage site is crucial for the cleavage at the Erns carboxy terminus (43). This finding indicated structural requirements necessary to present this unusual cleavage site to SPase in a cleavable conformation at the appropriate position with respect to the ER membrane. This hypothesis raised the question of whether the sequence downstream of the cleavage site also had a major impact on Erns-E1 processing. To analyze the influence of E1 sequences on the processing step, we established a series of constructs expressing the Erns-E1 fusion protein of CSFV with increasing carboxy-terminal truncations of E1. Full-length E1 consists of 195 amino acids. Already, a truncation by 15 carboxy-terminal residues increased the amount of Erns-E1 precursor within the cell from ca. 20% uncleaved product for the wild-type (wt) construct to more than 50% for the truncated version (Fig. 1). The level of the precursor increased up to 75% when E1 was further truncated so that the residual fragment encompassed only 139 amino acids. Truncation beyond that point in steps preserving 112, 84, or 53 residual residues of E1 resulted in somewhat lower processing impairment for the 112 and 53 constructs, but the lowest level was still in the range of 60% of uncleaved precursor protein and, thus, considerably higher than Erns fused to full-length E1.
FIG 1.
Effect of C-terminal truncation on processing of the Erns-E1 precursor protein. (A) Schematic representation of the proteins expressed from constructs encoding wild-type Erns-E1 (SErns-E1) or Erns-E1 with carboxy-terminal truncations (SErns-E1:X) of CSFV. The signal sequence preceding Erns is shown as gray bar, whereas Erns and E1 sequences are represented by white bars. The carboxy-terminal hydrophobic region of E1 representing the putative membrane anchor (MA) is indicated by a dark gray bar in the wt construct. The dotted line surrounding the E1 sequence in SErns-E1:X indicates that the E1 sequence in these constructs is incomplete due to the truncations. The length of the E1 sequence of constructs SErns-E1:X is 10 to 180 amino acids (aa) as given below the E1 bar and in the name of the constructs. Arrows indicate the signal peptidase (SPase) cleavage sites in the expressed proteins with a dotted arrow indicating the impaired cleavage between Erns and E1 in the truncation constructs. Below the scheme, the amino acid sequence flanking the Erns/E1 cleavage site in CSFV Alfort/Tübingen is given. The location of the cleavage site is indicated with a dotted vertical line. (B) Result of an immunoprecipitation experiment using Erns-specific monoclonal antibody 24/16 with PNGase F treatment of precipitation products prior to separation by PAGE. The transiently expressed constructs are given on top. The constructs code for SErns-E1 (wild-type Erns-E1 fusion protein), SErns (Erns alone), and the carboxy-terminal truncation mutants (Erns plus the indicated residual amino acids of E1). The position of size marker bands is given on the left, and the location of unprocessed precursor (Erns-E1:X) and processed Erns (Erns) are indicated. (C) Diagram summarizing the results of immunoprecipitation experiments. The bars represent the percentage of uncleaved Erns-E1 precursor determined in at least 3 independent experiments. For calculation, the radioactivity measured for processed Erns was multiplied with a factor correcting the different numbers of labeled residues in Erns versus the fusion proteins Erns-E1:X. This corrected value plus the signal determined for the uncleaved Erns-E1:X was set as 100% recovered expression product from which the percent uncleaved product was calculated. Error bars are indicated, and the P value for the results determined for the mutants with respect to the full-length Erns-E1 is given (****, P < 0.0001). (a) For residual 10 amino acids of E1, a correct differentiation of precursor and processing product is not possible. (b) There is no cleavage in SErns since it encodes only Erns.
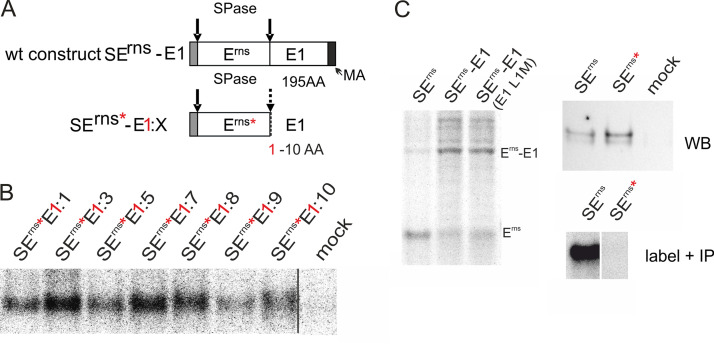
When truncation was done in a way that only the 10 amino-terminal residues of E1 were left in the precursor, the size difference between precursor and processed Erns was too low to allow for fully reliable quantification of the two possible products, even though the location of the bands on the gel indicated that most of the protein was not processed. To get further support for hampered processing of the plus 10 amino acid product and to be able to analyze products with even shorter E1 extensions, we replaced, in a synthetic construct, all methionine codons in the Erns coding sequence except for the translational start codon by leucine codons and mutated the leucine codon on position 1 of the E1 gene to a methionine triplet (Fig. 2A). Labeling of the proteins encoded by the constructs shown in Fig. 2A with only methionine allowed detection of all of the expressed proteins containing E1-derived extensions of 1 to 10 residues (Fig. 2B). This result was not due to the block of Erns-E1 processing introduced by the Leu to Met exchange at position 1 of the E1 sequence since the SErns-E1 control construct carrying this mutation was cleaved efficiently (Fig. 2C, left). According to the Sherman rules (44), the initiator methionine of the expressed Erns is cleaved off so that transient expression and labeling of the encoded protein with methionine would result in an unlabeled Erns protein even in case of incomplete processing of the signal sequence. Erns carrying a short E1 extension could only be detected when the E1 extension was not cleaved off. Indeed, upon transfection of a control plasmid coding for Erns containing the mutations but no E1 extension, Erns could not be detected when only a [35S]Met label was used, even though the mutated protein was detected via Western blotting (Fig. 2C, right).Thus, at least part of the proteins detected in steady state represent fusion proteins of Erns with a short E1 extension. The amount of radioactivity present in the [35S]Met-labeled protein was quite low so that quantitative evaluation of the results was difficult. However, comparison of the signals determined via phosphorimager analysis for the mutated proteins carrying only one labeled residue and the corresponding polypeptides encompassing a wt Erns sequence indicated that the amount of the mutated product (only unprocessed protein detectable) was in a similar range as the protein with a wt Erns sequence (total protein amount including processed and unprocessed form) (data not shown). Thus, processing of the precursor proteins with small E1 extensions is obviously strongly hampered.
FIG 2.
Influence of short E1 extensions on signal peptidase processing at the Erns C terminus. (A) Schematic representation of the constructs. The red asterisks indicate that all methionine residues were removed from the Erns moiety of the expressed constructs. The red “1” shows that the first amino acid of the E1 part was changed to methionine. The carboxy-terminal hydrophobic region of E1 representing the putative membrane anchor (MA) is indicated by a dark gray bar in the wt construct. (B) Results of immunoprecipitation of Erns-E1 precursors after labeling with 35S-methionine only. Please notice that the only labeled residue present in the precipitated proteins is the first amino acid of E1. Thus, the processed product would not be detected here, and the labeling intensity is very poor, resulting in an image with lots of pixels. As indicated by the black line, the samples and the mock control were run on the same gel but not directly next to each other. (C) The left part shows the results of an immunoprecipitation comparing processing of wt Erns-E1 and a mutant with leucine at position 1 of E1 replaced by methionine. As a control, the processed Erns is shown. The upper gel on the right shows a Western blot demonstrating expression of Erns without methionine and wt, whereas the gel below shows the immunoprecipitated products after methionine labeling. The samples shown in the bottom panel on the right were run on the same gel but not in neighboring lanes.
Truncated E1 sequences fused to Erns lead to increased secretion.
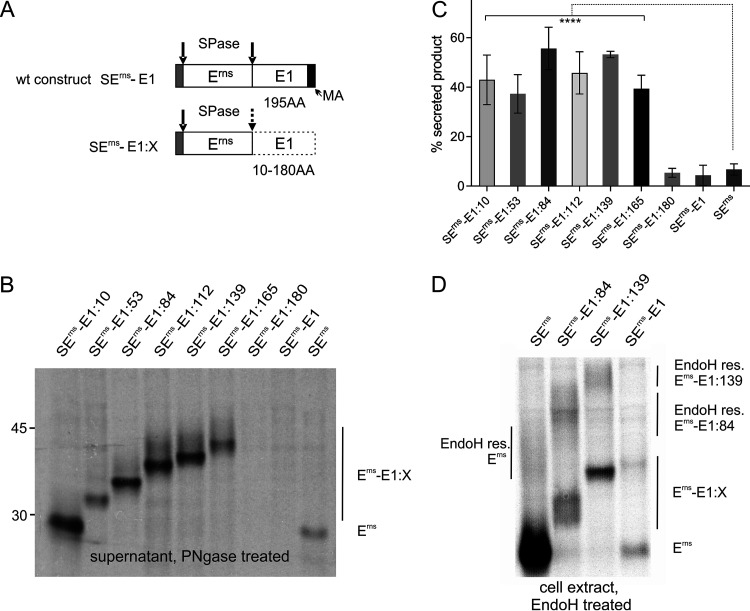
Part of the Erns protein synthesized within infected cells is secreted into the supernatant (14). We have shown before that secretion is also observed after transient expression of Erns. The unusual membrane anchor of Erns is important for secretion, and mutations affecting the nature of the amphipathic helix lead to dramatically increased release of Erns into the cell-free medium (30–33). Since truncated E1 sequences fused to the Erns carboxy terminus strongly impair processing of the Erns/E1 cleavage site, we were wondering whether these changes also influence the equilibrium of Erns retention/secretion. We, therefore, analyzed the supernatant of cells transiently expressing the Erns-E1 precursor protein with carboxy-terminal deletions of E1 for the presence of secreted proteins (Fig. 3). For all of our proteins truncated by 30 or more carboxy-terminal residues, we found significant secretion that by far exceeded the level determined for wt Erns without E1 sequences or a fusion protein composed of both full-length Erns and E1 (Fig. 3, SErns or SErns-E1, respectively). Importantly, only the Erns-E1 truncated fusion proteins were found in significant amounts in the supernatant, whereas even after prolonged exposure times the processed Erns was not clearly detected, which can be a result of the low amounts of processed product generated in the cells (compare Erns band of the truncated mutants or SErns-E1 in the cell extract with the intensity of the Erns signal derived from the SErns construct in Fig. 1 and the weak band observed in the supernatant for this construct in Fig. 3). It has to be mentioned that this fact changes, for the truncated E1 fusion constructs, the ratio of processed versus unprocessed product to even higher levels of unprocessed product because the amount of precursor found within the cells plus the signal in the supernatant (total uncleaved precursor) have to be compared with the processed Erns in the cells. Taking this into account, 26% of total uncleaved precursor can be determined for SErns-E1 versus 73% to 89% for the proteins with truncated E1.
FIG 3.
E1 extensions at the Erns carboxy terminus lead to increased secretion of the precursor protein. (A) Schematic representation of the constructs. (B) PAGE of proteins precipitated from the supernatant of cells transiently expressing the indicated viral proteins. The precipitation products were treated with PNGase F before electrophoresis. (C) Diagram summarizing the results of at least three independent immunoprecipitation experiments quantified by phosphorimager analysis. The bars represent the amount of secreted proteins as a percent of total recovered expression product. For calculation, the results for extra- and intracellular proteins (Erns counts corrected for lower number of labeled residues) were set to 100% expression product as basis for calculation of the secretion value. Error bars are indicated as well as the P value of Erns, with E1 extension compared to Erns without E1 (construct SErns). ****, P < 0.0001. Please note that secretion of processed Erns was negligible for the constructs with extension. Neither full-length Erns-E1 expressed from SErnsE1 nor the truncated version stemming from SErns-E1:180 revealed significantly different secretion rates in comparison with SErns. (D) PAGE of proteins precipitated from the extracts of cells transiently expressing the indicated viral proteins. The precipitation products were treated with EndoH before electrophoresis. The location of the highly glycosylated proteins in the upper region of the gel with EndoH-resistant carbohydrates is indicated. The bands below represent ER resident proteins deglycosylated by the EndoH treatment.
To verify that the observed increase in product amounts in the supernatant was due to increased transport instead of more efficient release of the fusion proteins from a cell surface-bound state, we analyzed the products present in extracts of transfected cells via immunoprecipitation and subsequent endoglycosidase H (EndoH) treatment. Since carbohydrates on glycoproteins acquire an EndoH-resistant phenotype when passing the Golgi apparatus, the detection of higher amounts of EndoH-resistant products demonstrates increased transport from the ER toward the plasma membrane. After SDS-PAGE, a significantly higher amount of EndoH-resistant protein was detected for the two analyzed truncated fusion proteins compared to Erns alone (6% for SErns alone versus 44% or 37% for SErns-E1:84 or SErns-E1:139, respectively) (Fig. 3D). As expected in the light of its very low secretion rate, EndoH-resistant carbohydrates could not be identified for the full-length SErns-E1. Taken together, truncated E1 extensions fused to the Erns carboxy terminus strongly impair Erns-E1 processing and interfere with intracellular retention of the fusion protein despite the presence of the known Erns retention signal (45).
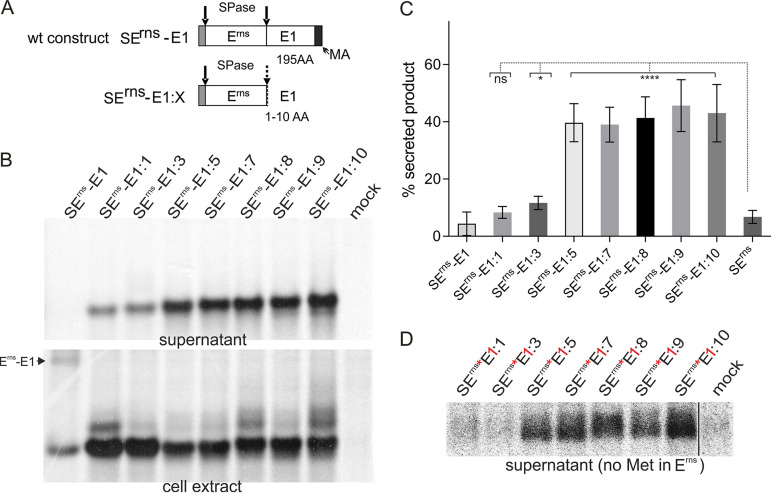
Furthermore, we wanted to test the influence of very short E1 sequences fused to Erns on intracellular retention of the protein. Therefore, we analyzed the supernatant of cells transfected with the constructs SErns-E1:1 to SErns-E1:10. As shown in Fig. 2, products expressed from these constructs preserved at least a considerable percentage of the E1 sequences bound to Erns. For all of these constructs, we observed elevated secretion rates. The difference was not significant for SErns-E1:1. In contrast, the other constructs yielded increases with P values of 0.011 for the 3-amino-acid extension and P values of <0.0001 for the other constructs compared to the Erns protein without extension. The secretion level increased with the length of the E1 sequence, and extensions of 5 and more residues resulted in very strongly elevated secretion rates of up to 50% for SErns-E1:9 compared with the rate of ca. 5% determined for SErns (Fig. 4).
FIG 4.
Very short E1 extensions at the Erns carboxy terminus lead to increased secretion of the precursor protein. (A) Schematic representation of the constructs. (B) PAGE of proteins precipitated from the supernatant or extracts of cells transiently expressing the indicated viral proteins. The precipitation products were treated with PNGase F before electrophoresis. (C) Diagram summarizing the results of at least three independent immunoprecipitation experiments quantified by phosphorimager analysis. The bars represent the amount of secreted proteins as percent of total recovered expression product. For calculations, the results for extra- and intracellular proteins were set to 100% expression product as the basis for calculation of the secretion value. Error bars are indicated as well as the P value of Erns with E1 extension compared to Erns without E1 (construct SErns; see Fig. 1 and 3 for PAGE pictures). ****, P < 0.0001; *, P = 0.011. Please note that secretion of full-length Erns-E1 and Erns expressed from SErnsE1was detected only after prolonged exposure time. The secretion rate determined for SErnsE1:1 was not significantly different compared to that of SErns. (D) PAGE with secretion products precipitated from supernatants of cells expressing the indicated proteins with Erns containing no methionine (indicated by red asterisk) and the first residue of the E1 moiety replaced by methionine (indicated by red “1”). The expressed protein was labeled with [35S]-methionine alone, resulting in low intensity of the bands and visible pixels. Only Erns proteins carrying a carboxy-terminal E1 extension are detected here. As indicated by the black line, the samples and the mock control were run on the same gel but not directly next to each other.
To verify that the secreted Erns contained the E1-derived extensions, we expressed the mutant proteins with only one methionine (first residue of the E1 moiety; see Fig. 2) and analyzed the secreted products after [35S]Met labeling. For extensions of 5 to 10 residues, clear signals were observed, whereas Erns proteins with 1 or 3 E1 residues were difficult to detect (Fig. 4D). As mentioned above, quantification of the obtained weak signals is not precise enough, but the analyses indicated that the apparent secretion rate of the methionine-depleted proteins was higher than that of the corresponding polypeptides with a wt number of methionine residues. This observation can be explained with partial cleavage at the Erns/E1 site, resulting in unlabeled Erns for the former constructs. As a result, a lower total protein amount (sum of intracellular and secreted protein) would be determined after [35S]Met labeling, leading to an apparent shift toward the secreted protein if predominantly the unprocessed precursor was secreted as seen before (see Fig. 3).
Taken together, the results of our analyses also show that very short E1-derived sequences fused to the Erns C terminus are mostly not cleaved off. The fusion proteins with 5 or more E1 residues are strongly secreted from the cells.
Presence of the E1 membrane anchor prevents secretion of Erns/E1 but is not sufficient for its efficient processing.
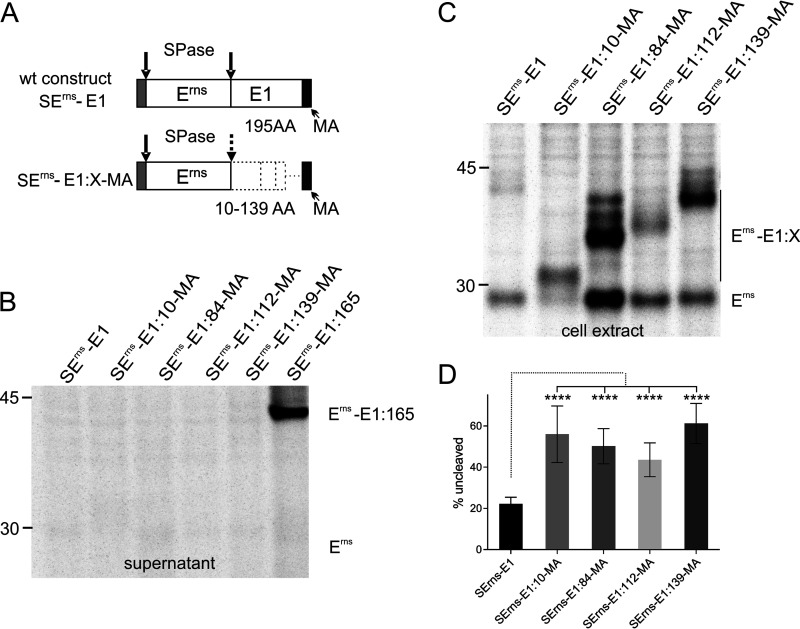
One possible explanation for the above-mentioned results concerning the secretion of the C-terminally truncated Erns-E1 fusion proteins is the loss of membrane anchoring of E1. The length of the E1 membrane anchor and its membrane topology have not yet been studied in detail, but in analogy to the situation in hepatitis C virus (HCV) (46), it is likely that the last ca. 30 residues of the protein are responsible for membrane binding and processing at the E1/E2 site. The deletion of the last 16 codons introduced into construct SErns-E1:180 preserves at least part of this hypothetical membrane binding region and might serve as a stop transfer signal, whereas the next truncated protein (SErns-E1:165) has lost all of it. As shown before, secretion of the SErns-E1:180 protein is in the same close to the background range as the full-length product. In order to test whether the absence of a carboxy-terminal membrane anchor is responsible for the increased secretion of the other truncated Erns-E1 proteins, we established constructs SErns-E1:10-MA, SErns-SE1:53-MA, SErns-E1:84-MA, SErns-E1:112-MA, and SErns-E1:139-MA coding for proteins with the indicated amino-terminal fragments of E1 followed by the carboxy-terminal 30 amino acids of this protein. Transient expression studies revealed no significantly increased secretion of either of these proteins in comparison with wt Erns-E1, whereas a positive control (truncated protein without membrane anchor; see also Fig. 3) was clearly visible (Fig. 5B). However, the efficiency of processing at the Erns/E1 site was still compromised, though not in all cases to the same extent as found before for the truncated proteins without membrane anchor (Fig. 5C and D compared to Fig. 1B and C). These results fit with the data presented for SErns-E1:180 in Fig. 1 and 3, showing absence of strong secretion but impairment of processing for this construct so that it can be concluded that a decrease of processing and increase of secretion of Erns proteins carrying truncated E1 sequences are not directly linked. We also checked whether processing efficiency could be restored upon coexpression of truncated Erns-E1 with full-length E1. We selected constructs SErns-E1:84 and SErns-E1:139-MA for this test and used an E1 expression construct coding for E1 with an amino-terminal hemagglutinin (HA) tag. However, despite the presence of full-length E1, processing efficiency was low and equivalent to what we measured before (data not shown). Thus, it can be concluded that efficient processing of the Erns-E1 precursor is dependent on a full-length E1 present in the precursor protein.
FIG 5.
The presence of the proposed membrane anchor of E1 prevents increased secretion but cannot restore efficient processing of truncated Erns-E1 proteins. (A) Schematic representation of the expressed constructs. The carboxy-terminal 30 amino acids of E1 containing the predicted E1 membrane anchor is shown as a black bar. The different lengths of the internal deletions in the E1 sequences (symbolized by the horizontal dotted line for the construct with the shortest deletion) are indicated. The constructs are equivalent with the truncation products shown in Fig. 1 and 3 except for the presence of the 30 carboxy-terminal amino acids of E1. (B) PAGE of proteins precipitated from the supernatant or extracts of cells transiently expressing the indicated viral proteins. Construct SErns-E1:165 is shown here as a positive control for a strongly secreted truncation mutant. The precipitation products were treated with PNGase F before electrophoresis. (C) PAGE with proteins precipitated from extract of cells expressing the indicated proteins. The positions of Erns-E1 precursors and processed Erns are indicated. The upper bands of lower intensity seen for SErns-E1:84-MA and SErns-E1:139-MA are due to incomplete deglycosylation. (D) Diagram summarizing the results of at least three independent immunoprecipitation experiments quantified by phosphorimager analysis. The bars represent the amount of precursor proteins as a percent of total recovered expression product. The calculation including correction for lower numbers of labeled residues in Erns was done as described in the Fig. 1 legend. Since no detectable secretion of the proteins occurred, protein from the supernatant did not have to be taken into account. Error bars are indicated as well as the P value of Erns with E1 extension compared to Erns with full-length E1 (construct SErns-E1). ****, P < 0.0001.
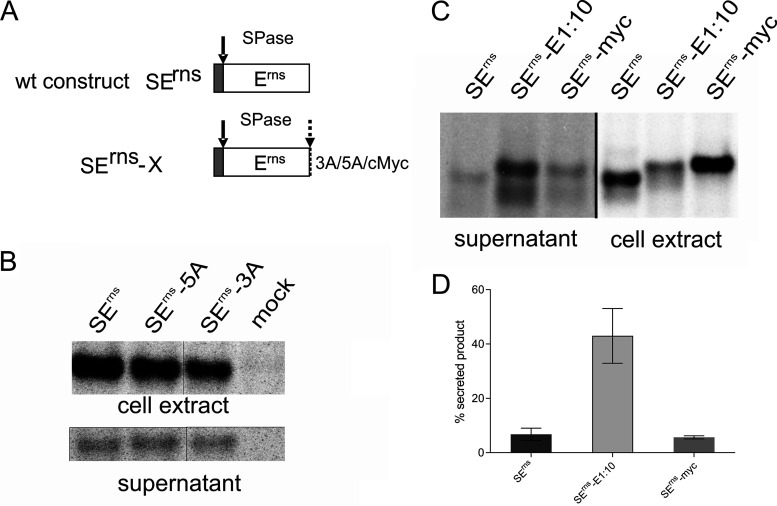
Not all sequences fused to the Erns carboxy terminus lead to strongly increased secretion in the absence of the E1 membrane anchor.
We were interested to analyze whether the presence of additional amino acids at the Erns carboxy terminus generally leads to impaired processing and increased secretion of the protein if a carboxy-terminal membrane anchor is missing. To test whether the primary sequence of the extension plays a role, we established constructs coding for Erns with 3 and 5 extra alanine residues at the carboxy terminus (SErns-3A and SErns-5A, respectively). In transient experiments, about 7% of the expressed proteins were secreted, which is in the same range as the wt level (Fig. 6). As a further step, we analyzed a construct containing the Erns gene fused to a sequence coding for the 10-amino-acid c-Myc tag. In contrast to Erns-E1:10 for which more than 40% of secretion was determined, only significantly less than 10% of the Erns-Myc protein was detected in the supernatant (Fig. 6). For the c-Myc tag construct, only one band was detected that comigrated with Erns-E1:10, at a position somewhat higher than Erns. Thus, the difference between the two constructs with extension with regard to secretion cannot be due to more efficient processing of the c-Myc fusion protein. The Erns c-Myc fusion protein is less efficiently secreted, maybe because its membrane binding and/or interaction with retention-relevant partners is less impaired. Taken together, these results show that a C-terminal extension fused to Erns does not generally lead to a high secretion level. However, processing at the Erns carboxy terminus is strongly hampered in the absence of a full-length E1, showing again the importance of the downstream sequence for cleavage at this site.
FIG 6.
Extensions of 3 or 5 alanine residues or a c-Myc tag fused to the Erns carboxy terminus do not provoke strong secretion, but at least cleavage of c-Myc is very poor. (A) Schematic representation of the constructs with SErns-X containing the given extensions fused to Erns. (B) Results of immunoprecipitation of transiently expressed wt (SErns) or alanine extended Erns from cell extract and supernatant. (C) Results of immunoprecipitation of transiently expressed wt (SErns) or Erns with either 10 residues of E1 or the 10-amino-acid c-Myc tag from cell extracts or cell-free supernatant. (D) Bar diagram representing the secretion rate quantified from at least 3 independent experiments as shown exemplarily in panel C. Error bars are given. The difference between wt SErns and SErns-myc is not significant, whereas P < 0.0001 was determined for SErns-E1:10 in relation to the other two constructs.
Mutation analysis reveals that the conserved amino-terminal sequence of E1 is not crucial for Erns-E1 processing.
We have shown before that the C terminus of Erns with five out of six residues conserved among pestivirus species is crucial for SPase cleavage (43). Since the utmost N-terminal sequence of E1 also contains highly conserved residues (Fig. 7), we tested whether, in addition to dependence on a full-length E1, these conserved residues also have a major impact on Erns-E1 processing. We, therefore, introduced several mutations into the 5′ terminal part of the E1 coding sequence of SErns-E1. In the first series of three mutants, we changed the residues at positions 3, 4, and 5 of E1 (P, Y, and C) to A, T, and S, respectively. A second set contained AAA for PYC (positions 3 to 5) or EQKL instead of LSP (positions 1 to 3 plus one extra amino acid), with EQKL representing the amino-terminal sequence of the c-Myc tag. After transient expression, these exchanges did not lead to significantly impaired Erns-E1 processing or Erns secretion rates, indicating that conservation of the sequence was not due to a function in processing or Erns membrane binding (not shown). To check for another role of the E1 amino terminus in the virus life cycle, we transferred these changes into our infectious full-length cDNA clone and tried to recover viruses upon the transfection of RNA transcribed from the mutant constructs. The mutants with only one exchange were easily recovered, leading to 100% positive results when fresh cells were infected with freeze-thaw extracts of the electroporated cells (Table 1). However, analysis of the recovered viruses revealed reversion of the Y498T and C499S mutants regenerating the Tyr and Cys residues at their original positions. In contrast, the P497A mutation was still present. The triple mutant PYC to AAA was negative when reinfection of fresh cells was tried, whereas the variant with the 4 c-myc deduced residues resulted in 100% infection. Sequence analysis revealed that the recovered virus had regenerated the Pro residue at position 497 (Table 1). Taken together, the amino-terminal residues of E1 are not important for Erns-E1 processing when full-length E1 is present, but the conserved residues play a role in production of infectious viruses.
FIG 7.
Conservation of amino acid sequences flanking the Erns/E1 cleavage site in pestiviruses. The graph generated with the WebLogo 3 software (http://weblogo.threeplusone.com/manual.html) (86, 87) shows the results of an alignment of 62 pestivirus sequences representing 9 of the 11 International Committee on Taxonomy of Viruses (ICTV)-listed species (species pestivirus A to I) in one letter code. The size of the letters corresponds to the degree of conservation among the 62 sequences. The two Trp residues and the Cys contained in the displayed part of the sequence are the most conserved amino acids.
TABLE 1.
Reversion analysis for E1 amino-terminal mutations introduced into the viral genomea
| Mutation | RNA rep | Percentage infection of fresh cells | Sequence of recovered virus |
|---|---|---|---|
| P497A | + | 100 | 497A |
| Y498T | + | 100 | 498Y |
| C499 | + | 100 | 499C |
| PYC/AAA | + | 0 | NA |
| LSP/EQKL | + | 100 | EQKP |
Results of electroporation of viral genome-like RNA with mutations affecting the first 5 amino acids (positions 495 to 499 in the polyprotein) of E1. The introduced changes are given in the left column, followed by the results of immunofluorescence analysis with antibody A18 against E2 24 h postelectroporation. Extracts of electroporated cells were used for infection of fresh cells that were again analyzed by immunofluorescence 48 h later; the percentage of positive cells is given in column 3. The results of sequence analyses of cDNA obtained after RT-PCR with RNA from infected cells are given as encoded amino acids in the right column. Reverted amino acids are given in bold letters. NA, not applicable since no virus present.
Processing at the Erns/E1 site is prevented when E1/E2 cleavage is blocked.
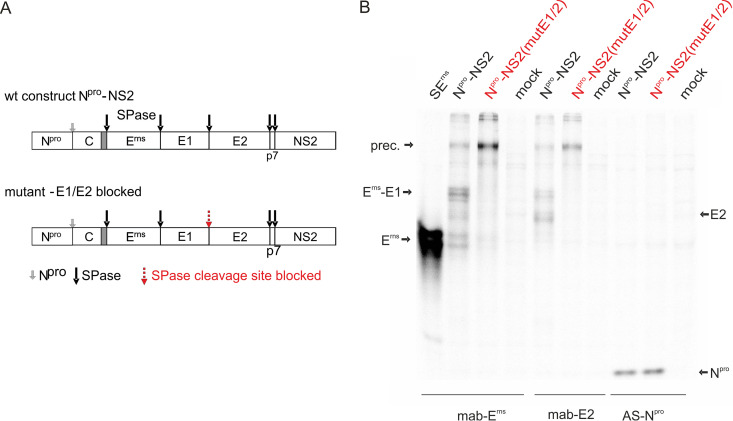
In addition to the mature glycoproteins Erns, E1, and E2, two precursor proteins were detected in cells infected with a recombinant vaccinia virus expressing the CSFV proteins Npro-C-Erns-E1-E2 (vaccinia virus recombinant 3.8 [14]). These precursor proteins encompass all three glycoproteins (130 kDa) or the already mentioned Erns-E1 (73 kDa), respectively. Equivalent precursors were also identified in BVDV-infected cells (gp116 and gp62, respectively), with gp116 representing a weak band that is rather rapidly cleaved into the Erns-E1 precursor and E2 (47, 48). Expression of a cDNA construct coding for Npro-C-Erns-E1-E2-p7-NS2 via a T7 RNA polymerase-driven transcription through vaccinia virus MVA T7 (49) and immunoprecipitation with the Erns-specific monoclonal antibody (MAb) 24/16 resulted in detection of the glycoprotein precursor Erns-E1-E2, the Erns-E1 precursor, Erns, and E2 (Fig. 8). In contrast, neither Erns nor other expected processing products were detected in significant amounts when an equivalent construct was expressed, in which SPase cleavage at the E1/E2 site was blocked by replacement of the small amino acid at position −3 of the von Heijne motif by an arginine (42) (Fig. 8). Thus, it can be concluded that processing of the glycoprotein region of the pestivirus polyprotein follows a hierarchical order, in which cleavage at the E1/E2 site represents the first step and prerequisite for the delayed Erns-E1 cleavage.
FIG 8.
Blocking cleavage at the E1/E2 site impairs processing at the Erns/E1 site. (A) Schematic drawing of constructs encompassing the CSFV coding sequence from the start codon of the long ORF to the end of the NS2 gene. The proteolytic cleavage sites are indicated by arrows (small gray arrow for autoproteolytic cleavage of Npro, black large arrow for SPase). In contrast to the wt, the mutant construct contains a blocked E1/E2 cleavage site. (B) Results of immunoprecipitation of proteins from cell extracts transiently expressing the constructs shown in panel A with MAb 24/16 (Erns, left), MAb A18 (E2, middle), and a rabbit antiserum against Npro (right). Names and positions of the important products are given. Due to heterodimer formation between E1 or the E1 part in Erns-E1 and E2 coprecipitation of Erns-E1 with the E2 MAb and of E2 with the Erns-specific MAb, precursor in analogy to the E1-E2 heterodimer can be detected. Please note that the glycosylated products are shown here so that multiple and broad bands are detected due to different carbohydrate side chains. Prec., Erns-E1-E2 (14, 47, 48).
DISCUSSION
Processing of the polyproteins expressed from positive-strand RNA virus genomes is a critical step during replication of these viruses. Incomplete or delayed cleavage at certain sites leads to the generation of precursors composed of two or more viral proteins, some of which are functionally important, such as, e.g., the 3CD protein of picornaviruses or the NS2-3 protein of pestiviruses (50–58). In pestiviruses, the envelope proteins can be detected in the form of a Erns-E1-E2 precursor that is rapidly cleaved into Erns-E1 and E2 followed by a delayed processing of the former to give rise to Erns and E1 (14). According to the published data, part of the E2 or Erns-E1-E2 precursor proteins present in the cell should contain the p7 polypeptide at the carboxy terminus (37). Cleavage of the precursor proteins at the C/Erns, Erns/E1, E1/E2, and E2/p7 sites is done by SPase (14, 43). After SPase cleavage, the signal sequence responsible for translocation of Erns into the ER represents the C-terminal part of the C protein precursor. It is subsequently cleaved by signal peptide peptidase, which generates the carboxy terminus of the mature C protein (59). Except for the Erns/E1 site, all of the SPase processing sites follow the standard scheme with n, h, and c domain, in which the C domain with a short unstructured region followed by the von Heijne cleavage site motif is preceded by an α-helical transmembrane region (h domain) (39–42). Erns does not contain a transmembrane sequence but is anchored in the membrane by a long amphipathic helix (30–33). Therefore, the cleavage site Erns/E1 is unusual with a von Heijne motif preceded by the amphipathic helix, which binds in plane to the membrane surface. This site is nevertheless cleaved by SPase (43), and the integrity of the amphipathic helix as well as the conserved utmost C-terminal residues are crucial for Erns-E1 processing. Since changes affecting the amphipathic helix of separately expressed Erns also led to highly increased secretion of Erns (32), we speculated that insertion of the helix into the membrane and signal peptidase processing of Erns-E1 are directly linked processes. Accordingly, the time needed for initiation of the membrane interaction and establishment of the correct protein fold at the site of cleavage could be responsible for the delayed Erns-E1 processing.
Open questions concerned the contribution of the E1 sequence downstream of the cleavage site to generation of a cleavable conformation of the SPase substrate and the possible role of the E1 transmembrane region in hooking the Erns-E1 precursor to the membrane, which would give time for establishing the membrane contact of the Erns amphipathic helix. To address the latter point, we analyzed the effect of increasing carboxy-terminal truncations of the E1 on Erns-E1 processing. Processing of the truncated proteins was severely impaired, and the uncleaved precursors were strongly secreted. This finding would be in agreement with the hypothesis that the Erns membrane anchor needs time to fold and get inserted into the lipid bilayer before SPase cleavage can occur. The presence of the E1 membrane anchor would support this process by preventing release of the protein from the membrane. Indeed, fusion of the last 30 amino acids of E1 to the carboxy terminus of the truncated Erns-E1 proteins prevented increased secretion. However, the above described hypothesis was in part challenged by the finding that the presence of the E1 membrane anchor was not able to restore efficient processing of the fusion protein. Thus, processing at the Erns/E1 site and maybe also the establishment of a stable membrane contact of the Erns amphipathic helix is not only a matter of time, since the presence of a full-length E1 is obviously essential. Even the loss of only 15 C-terminal residues of E1 reduced processing efficiency significantly, although this truncation leaves a stretch of hydrophobic residues at the C terminus that is sufficient for preventing Erns-E1 secretion. Since constructs with internal deletions preserving the original C-terminal sequence or the expression of full-length E1 in trans also do not lead to efficient processing, the most logical explanation of our findings is that E1 has to fold properly in order to generate a cleavable structure at the Erns/E1 site and that this fold is very sensitive to changes affecting E1. E1 seems to have an active role in promoting processing instead of just hindering cleavage when truncated since very short E1-derived extensions or the c-Myc tag that can hardly be imagined to impose a structural block on the cleavage site are also not cleaved off efficiently. A somewhat similar finding was described for gp160 of human immunodeficiency virus (HIV) (60). The signal peptide inducing translocation of the gp160 precursor into the ER is cleaved off only after termination of translation and folding of the soluble subunit gp120 to an almost native conformation. This effect is due to a signal sequence, in which the α-helix of the h region is extended so that h and c regions overlap. As a result, the SPase cleavage site is initially positioned within the membrane and not accessible for SPase so that gp120 is tethered to the membrane via its amino-terminal transmembrane signal sequence as a type II membrane protein, which gives time for correct folding (60). A late folding event, which probably implies establishment of a hairpin conformation via interaction of the proteins N and C termini, is believed to induce a conformational change in the N terminus leading to a cleavable structure. This idea is supported by the fact that the N terminus of gp120, which is suggested to be α-helical in the beginning, is a β-strand in the native protein. Moreover, carboxy-terminally truncated gp120 showed only poor signal sequence cleavage (60). Several aspects of the HIV story are reminiscent of our present findings, namely, delayed cleavage, dependency of efficient cleavage on the presence of the C-terminal residues of the protein to be processed on its N terminus, and the apparent importance of (nearly) complete folding of the protein prior to cleavage.
Another case of delayed SPase cleavage was reported for human cytomegalovirus (HCMV) US11, a protein involved in viral immune suppression. Part of the newly synthesized US11 molecules retain the signal sequence. The delayed cleavage is dependent on the four N-terminal residues of the signal sequence and the transmembrane sequence close to the C terminus of the protein. Thus, also in this case, sequence elements located close to the cleavage site seem to interact with the carboxy-terminal membrane anchor and influence SPase cleavage (61).
A further example for delayed and coordinated signal sequence cleavage is known for members of the genus Flavivirus where SPase cleavage generating the amino terminus of prM can only occur when the viral serine protease has removed the signal sequence from the carboxy terminus of the capsid protein C (62–64). This ordered process is dependent on specific features of the signal peptide and the downstream prM sequence (65). The delay of SPase cleavage is functionally important, as optimization of the processing in yellow fever virus was lethal (66).
The expression of Erns-E1 with increasing internal deletions in E1 demonstrated that processing at the Erns/E1 site and intracellular retention of the protein are not directly linked. Interestingly, the correct amino-terminal sequence of E1 is obviously not essential for efficient cleavage since even exchange of the conserved residues in the N-terminal part of E1 had no significant impact on processing but impaired recovery of viable viruses. Thus, the (delayed) cleavage of Erns-E1 seems not to be dependent on the utmost primary sequence downstream of the cleavage site.
It, therefore, can be concluded that a certain fold at the cleavage site, maybe together with interaction with the E1 carboxy terminus, is necessary for efficient Erns-E1 cleavage. Truncated E1 sequences prevent both membrane binding and adaptation of a cleavable structure, whereas the c-Myc tag allows membrane binding but still prevents the cleavage, which again supports the conclusion that full-length E1 actively promotes processing. Based on the available data, the following processing scheme for the amino-terminal third of the pestivirus polyprotein can be proposed: precursors of pestiviral structural proteins containing the Npro protein have never been reported so that self-cleavage of Npro represents most likely the first step. Similarly, the presence of a precursor protein encompassing capsid protein C and envelope protein(s) was not proven in cells expressing pestivirus proteins so that the cleavage at the C/Erns site is also apparently done already during translocation of the glycoprotein precursor. In contrast, a rather stable Erns-E1-E2 fusion protein is generated that is clearly detectable in the cells. As a next step, the E1/E2 site in this precursor is processed leading to the Erns-E1 fusion protein bound to the membrane via the E1 membrane anchor. According to our results, release of E2 has to occur in order to allow further (efficient) processing, for which correct folding of E1 is necessary. The importance of the cleavage at the E1/E2 site as the first step of the process could be due to hinderance of proper E1 folding by the fused E2. Alternatively, generation of a free E1 carboxy terminus is necessary to allow its interaction with the amino-terminal cleavage site region in analogy to HIV gp160 or HCMV US11 (60, 61). After release of E2, E1 is able to establish a correct structure, which initiates membrane binding of the Erns amphipathic helix and cleavage at the Erns/E1 site. Our earlier data showed that the conformation of the amphipathic helix is very important for processing and intracellular retention of Erns (32, 43). Thus, the correct conformation of the proteins on both sides of the cleavage site and maybe also its interaction with the E1 carboxy terminus is a crucial prerequisite for processing, which proves that SPase can accept this unusual substrate only under very restricted conditions.
An important question is whether the delayed processing at the Erns/E1 site is functionally important. This was shown for HIV gp160. Forced cotranslational cleavage in consequence of introduction of a helix-breaking proline upstream of the von Heijne motif resulted in localized misfolding of gp120 and reduced viral fitness (60). For pestiviruses, one possible scenario could be that Erns-E1 processing is inhibited as long as the two proteins have not established their final 3D structure. The cleavage could represent kind of a maturation step activating, e.g., a putative membrane fusion domain that has to be shielded by proper folding to prevent its premature activation. There are numerous examples of such mechanisms in viral fusion proteins (see references 67–70 for recent reviews), but it is important to notice that Erns is obviously dispensable for pestivirus membrane fusion (71, 72) and fusion activity of E1 has been hypothesized but so far not proven (73–76). In HCV, folding of E1 and E2 occurs slowly and in a concerted action, which implies not only cellular chaperons but also a reciprocal chaperoning between E1 and E2 (77, 78). Such a process could also be necessary for Erns-E1 folding with the integrity of the Erns-E1 precursor representing a prerequisite for this mechanism. However, Erns expressed in the absence of any other viral protein is able to bind to membranes and achieve efficient retention in the ER (32, 45). Moreover, the formation of correctly folded E1-E2 heterodimers in HCV is not dependent on expression of a common precursor (79). Similarly, mutants with large deletions affecting either the Erns or the E1 coding region of the pestiviral genome can be rescued on complementing cell lines (12, 80, 81). Thus, the generation of an Erns-E1 fusion protein anchored in the membrane via the E1 carboxy terminus cannot be a prerequisite for generation of a membrane bound Erns or the production of infectious pestivirus particles. However, a bicistronic construct expressing Erns from a second ORF resulted in retarded virus growth compared with a similar construct, in which the second ORF coded for Erns-E1, arguing in favor of an advantage for a virus being able to express an Erns-E1 fusion protein (81). Thus, a functional role of the delayed and apparently coordinated Erns-E1 processing is not definitely proven but seems highly probable.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells (kindly provided by T. Rümenapf, Veterinärmedizinische Universität, Wien, Austria) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and nonessential amino acids. The modified vaccinia virus strain Ankara containing the phage T7 RNA polymerase (MVA-T7) vaccinia virus MVA-T7 was kindly provided by B. Moss (National Institutes of Health, Bethesda, MD) and G. Sutter (Ludwig-Maximilians-Universität, München, Germany) (49, 82).
Construction of recombinant plasmids.
Restriction and subcloning were done according to standard procedures (83). Unless stated otherwise, all restriction and modifying enzymes were purchased from New England BioLabs (Frankfurt, Germany) and Thermo Fisher (Karlsruhe, Germany). Synthetic DNA oligonucleotides were purchased from Metabion (München, Germany).
Plasmids SSeqErns and SSeqErns-E1 containing the Erns and Erns-E1 coding sequences of CSFV Alfort/Tübingen (43) served as templates for PCR leading to most of the mutant constructs (renamed here to SErns and SErns-E1, respectively). The 3′ terminally truncated versions of SSeqErns-E1 were generated by PCR with primer Ol-pCITE and an antisense primer generating the desired 3′ end plus translational stop codon and an XbaI site for cloning. PCR fragments were restricted with NcoI and XbaI and inserted into plasmid pCITE 2a+ (AGS, Heidelberg, Germany), cleaved with the same enzymes. Point mutations were introduced with standard PCR-based methods, with thermostable Pfu polymerase (Promega, Heidelberg, Germany) and synthetic primers (QuikChange mutagenesis protocol, Promega). Constructs with large deletions within the E1 coding region were established from two PCR fragments with first fragment extending from 5′ NcoI site to the 5′ border of the desired deletion and the second fragment covering the part from the 3′ end of the deletion to the 3′ end of the E1 gene. Fusion of these two fragments was done via EcoRI sites introduced into both fragments leading to two extra amino acids (Glu and Thr) at the fusion site.
For establishment of the SErns*-E1:X constructs, in which all methionine codons in the Erns gene were replaced by leucine codons, a synthetic Erns coding sequence with the desired changes was ordered from GeneArt/Invitrogen (Karlsruhe, Germany). This construct served as a basis for establishment of the plasmids coding for Erns with the different E1-derived extensions via PCR with appropriate primers that also introduced the L to M change at position 1 of the E1 sequence.
The cloned PCR products or synthetic cDNA were all verified by nucleotide sequencing with the BigDye Terminator cycle sequencing kit (PE Applied Biosystems, Weiterstadt, Germany). Sequence analysis and alignments were done with Geneious Prime software (Geneious Prime 2019.2.3).
Further details of the cloning procedures and the sequences of the primers used for cloning and mutagenesis are available upon request.
Transient expression, immunoprecipitation, and quantification of proteins.
BHK-21 cells were infected with vaccinia virus MVA-T7, subsequently transfected with the desired cDNA construct using SuperFect (Qiagen, Hilden, Germany), and labeled with Tran35S-Label, Met-Label, or [35S]methionine (ICN-MP Biochemicals, Eschwege, Germany; Hartmann Analytic, Göttingen, Germany) as described earlier (15). Supernatant of the cell cultures was harvested for determination of secreted proteins, and the cells were washed twice with phosphate-buffered saline (PBS) before cell extracts were prepared under denaturing conditions. Protein expression levels were determined in equivalent amounts of cell-free supernatant and cell extract, analyzed via immunoprecipitation as described before (32) using monoclonal antibody 24/16 (9) for detection of Erns, monoclonal antibody A18 for detection of E2, and rabbit antiserum G1 (84) for precipitation of Npro. For removing N-linked carbohydrates, the precipitates were treated before electrophoresis with 1 μl peptide-N-glycosidase F (PNGase F) or EndoH (New England BioLabs) for 1 h at 37°C as suggested by the supplier and subsequently separated by 10% PAGE (gel system as published [39]) and Erns quantified with a CR-35 Bio image plate scanner, and intensities of the signals were determined with AIDA Image Analyzer 5 software (equipment and software from Elysia-Raytest, Straubenhardt, Germany). The signals determined for Erns in supernatant and cell extract were combined and set as 100% expression product for determination of the percentage of secreted protein. Similarly, cleavage efficiency was calculated by summing up the signals of precursor and Erns (correlated for the lower number of labeled residues) to obtain 100% of expression products as the basis for determination of the percentage of the uncleaved precursor. The data presented here represent the averages of at least 3 independent experiments. Statistical analysis in the form of a two-tailed t test was done using the GraphPad Prism software (Statcon GmbH, Witzenhausen, Germany).
Recovery and analysis of viruses from cloned sequences.
The desired mutations were introduced into the full-length CSFV cDNA clone (85) with standard procedures. In vitro transcription of RNA from the engineered plasmids and electroporation of cells were done as described before (15). Replication of RNA and protein expression thereof was detected via immunofluorescence with MAb A18 (10) against E2 and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse serum (Dianova, Hamburg, Germany). Freeze-thaw extracts were prepared from positive cultures and used for infection of fresh cells. Infection of these cells was detected via immunofluorescence as above. RNA was isolated from the infected cells and subjected to reverse transcriptase PCR (RT-PCR) with primers Ol-E05S and Ol-HPS28R and analyzed by nucleotide sequencing as described before (15).
ACKNOWLEDGMENTS
We thank Gaby Stooß, Maren Ziegler, and Petra Wulle for excellent technical assistance.
The project described in the manuscript was supported by grants Me1367/4 and Me1367/7 of the Deutsche Forschungsgemeinschaft (DFG). Yu Mu was supported by a grant from the China Scholarship Council (CSC).
REFERENCES
- 1.Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Scott Muerhoff A, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P, Becher P. 2017. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol 98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tautz N, Tews BA, Meyers G. 2015. The molecular biology of pestiviruses. Adv Virus Res 93:47–160. doi: 10.1016/bs.aivir.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Murray CL, Thiel HJ, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM (ed), Fields virology, vol 6 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Thiel HJ, Stark R, Weiland E, Rümenapf T, Meyers G. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J Virol 65:4705–4712. doi: 10.1128/JVI.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiland F, Weiland E, Unger G, Saalmüller A, Thiel HJ. 1999. Localization of pestiviral envelope proteins E(rns) and E2 at the cell surface and on isolated particles. J Gen Virol 80:1157–1165. doi: 10.1099/0022-1317-80-5-1157. [DOI] [PubMed] [Google Scholar]
- 7.Bolin SR, Moennig V, Kelso Gourley NE, Ridpath J. 1988. Monoclonal antibodies with neutralizing activity segregate isolates of bovine viral diarrhea virus into groups. Arch Virol 99:117–123. doi: 10.1007/BF01311029. [DOI] [PubMed] [Google Scholar]
- 8.Donis RO, Corapi W, Dubovi EJ. 1988. Neutralizing monoclonal antibodies to bovine viral diarrhoea virus bind to the 56K to 58K glycoprotein. J Gen Virol 69:77–86. doi: 10.1099/0022-1317-69-1-77. [DOI] [PubMed] [Google Scholar]
- 9.Weiland E, Ahl R, Stark R, Weiland F, Thiel HJ. 1992. A second envelope glycoprotein mediates neutralization of a pestivirus, hog cholera virus. J Virol 66:3677–3682. doi: 10.1128/JVI.66.6.3677-3682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiland E, Stark R, Haas B, Rümenapf T, Meyers G, Thiel HJ. 1990. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol 64:3563–3569. doi: 10.1128/JVI.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulst MM, Moormann RJ. 2001. Erns protein of pestiviruses. Methods Enzymol 342:431–440. doi: 10.1016/s0076-6879(01)42564-x. [DOI] [PubMed] [Google Scholar]
- 12.Reimann I, Meyers G, Beer M. 2003. Trans-complementation of autonomously replicating bovine viral diarrhea virus replicons with deletions in the E2 coding region. Virology 307:213–227. doi: 10.1016/s0042-6822(02)00129-0. [DOI] [PubMed] [Google Scholar]
- 13.König M, Lengsfeld T, Pauly T, Stark R, Thiel HJ. 1995. Classical swine fever virus: independent induction of protective immunity by two structural glycoproteins. J Virol 69:6479–6486. doi: 10.1128/JVI.69.10.6479-6486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rümenapf T, Unger G, Strauss JH, Thiel HJ. 1993. Processing of the envelope glycoproteins of pestiviruses. J Virol 67:3288–3294. doi: 10.1128/JVI.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tews BA, Schürmann EM, Meyers G. 2009. Mutation of cysteine 171 of pestivirus Erns RNase prevents homodimer formation and leads to attenuation of classical swine fever virus. J Virol 83:4823–4834. doi: 10.1128/JVI.01710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucakov AK, Yavuz S, Schurmann EM, Mischler M, Klingebeil A, Meyers G. 2018. Restoration of glycoprotein Erns dimerization via pseudoreversion partially restores virulence of classical swine fever virus. J Gen Virol 99:86–96. doi: 10.1099/jgv.0.000990. [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi H, Yanai K, Takagi M, Yano K, Wakabayashi E, Sanda A, Mine S, Ohgi K, Irie M. 1988. Primary structure of a base non-specific ribonuclease from Rhizopus niveus. J Biochem 103:408–418. doi: 10.1093/oxfordjournals.jbchem.a122284. [DOI] [PubMed] [Google Scholar]
- 18.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel HJ. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 19.Krey T, Bontems F, Vonrhein C, Vaney MC, Bricogne G, Rümenapf T, Rey FA. 2012. Crystal structure of the pestivirus envelope glycoprotein Erns and mechanistic analysis of its ribonuclease activity. Structure 20:862–873. doi: 10.1016/j.str.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Hausmann Y, Roman-Sosa G, Thiel HJ, Rümenapf T. 2004. Classical swine fever virus glycoprotein Erns is an endoribonuclease with an unusual base specificity. J Virol 78:5507–5512. doi: 10.1128/jvi.78.10.5507-5512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulst MM, Himes G, Newbigin E, Moormann RJM. 1994. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology 200:558–565. doi: 10.1006/viro.1994.1218. [DOI] [PubMed] [Google Scholar]
- 22.Windisch JM, Schneider R, Stark R, Weiland E, Meyers G, Thiel HJ. 1996. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J Virol 70:352–358. doi: 10.1128/JVI.70.1.352-358.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lussi C, Sauter KS, Schweizer M. 2018. Homodimerisation-independent cleavage of dsRNA by a pestiviral nicking endoribonuclease. Sci Rep 8:8226. doi: 10.1038/s41598-018-26557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer C, Von Freyburg M, Elbers K, Meyers G. 2002. Recovery of virulent and RNase-negative attenuated type 2 bovine viral diarrhea viruses from infectious cDNA clones. J Virol 76:8494–8503. doi: 10.1128/jvi.76.16.8494-8503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers G, Saalmüller A, Büttner M. 1999. Mutations abrogating the RNase activity in glycoprotein Erns of the pestivirus classical swine fever virus lead to virus attenuation. J Virol 73:10224–10235. doi: 10.1128/JVI.73.12.10224-10235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers G, Ege A, Fetzer C, von Freyburg M, Elbers K, Carr V, Prentice H, Charleston B, SchüRmann E-M. 2007. Bovine viral diarrhoea virus: prevention of persistent foetal infection by a combination of two mutations affecting the Erns RNase and the Npro protease. J Virol 81:3327–3336. doi: 10.1128/JVI.02372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magkouras I, Mätzener P, Rümenapf T, Peterhans E, Schweizer M. 2008. RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J Gen Virol 89:2501–2506. doi: 10.1099/vir.0.2008/003749-0. [DOI] [PubMed] [Google Scholar]
- 28.Mätzener P, Magkouras I, Rümenapf T, Peterhans E, Schweizer M. 2009. The viral RNase Erns prevents IFN type-I triggering by pestiviral single- and double-stranded RNAs. Virus Res 140:15–23. doi: 10.1016/j.virusres.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Iqbal M, Poole E, Goodbourn S, McCauley JW. 2004. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J Virol 78:136–145. doi: 10.1128/jvi.78.1.136-145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aberle D, Muhle-Goll C, Bürck J, Wolf M, Reißer S, Luy B, Wenzel W, Ulrich AS, Meyers G. 2014. Structure of the membrane anchor of pestivirus glycoprotein Erns, a long tilted amphipathic helix. PLoS Pathog 10:e1003973. doi: 10.1371/journal.ppat.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fetzer C, Tews BA, Meyers G. 2005. The carboxy-terminal sequence of the pestivirus glycoprotein Erns represents an unusual type of membrane anchor. J Virol 79:11901–11913. doi: 10.1128/JVI.79.18.11901-11913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tews BA, Meyers G. 2007. The pestivirus glycoprotein Erns is anchored in plane in the membrane via an amphipathic helix. J Biol Chem 282:32730–32741. doi: 10.1074/jbc.M706803200. [DOI] [PubMed] [Google Scholar]
- 33.Aberle D, Oetter KM, Meyers G. 2015. Lipid binding of the amphipathic helix serving as membrane anchor of pestivirus glycoprotein Erns. PLoS One 10:e0135680. doi: 10.1371/journal.pone.0135680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M, Veit M. 2018. Differences in signal peptide processing between GP3 glycoproteins of Arteriviridae. Virology 517:69–76. doi: 10.1016/j.virol.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Zürcher C, Sauter KS, Mathys V, Wyss F, Schweizer M. 2014. Prolonged activity of the pestiviral RNase Erns as an interferon antagonist after uptake by clathrin-mediated endocytosis. J Virol 88:7235–7243. doi: 10.1128/JVI.00672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zurcher C, Sauter KS, Schweizer M. 2014. Pestiviral Erns blocks TLR-3-dependent IFN synthesis by LL37 complexed RNA. Vet Microbiol 174:399–408. doi: 10.1016/j.vetmic.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 37.Elbers K, Tautz N, Becher P, Stoll D, Rümenapf T, Thiel HJ. 1996. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J Virol 70:4131–4135. doi: 10.1128/JVI.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada T, Tautz N, Thiel HJ. 2000. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J Virol 74:9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson I, Johnson AE, von Heijne G. 2002. Cleavage of a tail-anchored protein by signal peptidase. FEBS Lett 516:106–108. doi: 10.1016/S0014-5793(02)02511-5. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson I, Whitley P, von Heijne G. 1994. The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J Cell Biol 126:1127–1132. doi: 10.1083/jcb.126.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Heijne G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Heijne G. 1990. The signal peptide. J Membr Biol 115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 43.Bintintan I, Meyers G. 2010. A new type of signal peptidase cleavage site identified in an RNA virus polyprotein. J Biol Chem 285:8572–8584. doi: 10.1074/jbc.M109.083394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman F, Stewart JW, Tsunasawa S. 1985. Methionine or not methionine at the beginning of a protein. Bioessays 3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- 45.Burrack S, Aberle D, Bürck J, Ulrich AS, Meyers G. 2012. A new type of intracellular retention signal identified in a pestivirus structural glycoprotein. FASEB J 26:3292–3305. doi: 10.1096/fj.12-207191. [DOI] [PubMed] [Google Scholar]
- 46.Cocquerel L, Op De BA, Lambot M, Roussel J, Delgrange D, Pillez A, Wychowski C, Penin F, Dubuisson J. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J 21:2893–2902. doi: 10.1093/emboj/cdf295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collett MS, Larson R, Belzer S, Retzel E. 1988. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology 165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 48.Collett MS, Wiskerchen MA, Welniak E, Belzer SK. 1991. Bovine viral diarrhea virus genomic organization. Arch Virol Suppl 3:19–27. doi: 10.1007/978-3-7091-9153-8_3. [DOI] [PubMed] [Google Scholar]
- 49.Sutter G, Ohlmann M, Erfle V. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett 371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 50.Lattwein E, Klemens O, Schwindt S, Becher P, Tautz N. 2012. Pestivirus virion morphogenesis in the absence of uncleaved nonstructural protein 2-3. J Virol 86:427–437. doi: 10.1128/JVI.06133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isken O, Langerwisch U, Jirasko V, Rehders D, Redecke L, Ramanathan H, Lindenbach BD, Bartenschlager R, Tautz N. 2015. A conserved NS3 surface patch orchestrates NS2 protease stimulation, NS5A hyperphosphorylation and HCV genome replication. PLoS Pathog 11:e1004736. doi: 10.1371/journal.ppat.1004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klemens O, Dubrau D, Tautz N. 2015. Characterization of the determinants of NS2-3-independent virion morphogenesis of pestiviruses. J Virol 89:11668–11680. doi: 10.1128/JVI.01646-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubrau D, Tortorici MA, Rey FA, Tautz N. 2017. A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis. PLoS Pathog 13:e1006134. doi: 10.1371/journal.ppat.1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubrau D, Schwindt S, Klemens O, Bischoff H, Tautz N. 2019. Determination of critical requirements for classical swine fever virus NS2-3-independent virion formation. J Virol 93:e00679-19. doi: 10.1128/JVI.00679-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. 2012. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A 109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yost SA, Marcotrigiano J. 2013. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr Opin Virol 3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmenberg AC. 1990. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol 44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 58.Sun D, Chen S, Cheng A, Wang M. 2016. Roles of the picornaviral 3C proteinase in the viral life cycle and host cells. Viruses 8:82. doi: 10.3390/v8030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heimann M, Roman-Sosa G, Martoglio B, Thiel HJ, Rümenapf T. 2006. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J Virol 80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snapp EL, McCaul N, Quandte M, Cabartova Z, Bontjer I, Kallgren C, Nilsson I, Land A, von Heijne G, Sanders RW, Braakman I. 2017. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. Elife 6:e26067. doi: 10.7554/eLife.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rehm A, Stern P, Ploegh HL, Tortorella D. 2001. Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J 20:1573–1582. doi: 10.1093/emboj/20.7.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amberg SM, Nestorowicz A, McCourt DW, Rice CM. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol 68:3794–3802. doi: 10.1128/JVI.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lobigs M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci U S A 90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamshchikov VF, Compans RW. 1994. Processing of the intracellular form of the west Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J Virol 68:5765–5771. doi: 10.1128/JVI.68.9.5765-5771.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stocks CE, Lobigs M. 1998. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J Virol 72:2141–2149. doi: 10.1128/JVI.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee E, Stocks CE, Amberg SM, Rice CM, Lobigs M. 2000. Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage in vitro is lethal for virus production. J Virol 74:24–32. doi: 10.1128/jvi.74.1.24-32.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Earp LJ, Delos SE, Park HE, White JM. 2005. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol 285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kielian M. 2014. Mechanisms of virus membrane fusion proteins. Annu Rev Virol 1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- 70.Rey FA, Lok SM. 2018. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172:1319–1334. doi: 10.1016/j.cell.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronecker S, Zimmer G, Herrler G, Greiser-Wilke I, Grummer B. 2008. Formation of bovine viral diarrhea virus E1-E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane domains. J Gen Virol 89:2114–2121. doi: 10.1099/vir.0.2008/001792-0. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Nie Y, Wang P, Ding M, Deng H. 2004. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology 330:332–341. doi: 10.1016/j.virol.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 73.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. 2013. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep 3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holinka LG, Largo E, Gladue DP, O'Donnell V, Risatti GR, Nieva JL, Borca MV. 2016. Alteration of a second putative fusion peptide of structural glycoprotein E2 of classical swine fever virus alters virus replication and virulence in swine. J Virol 90:10299–10308. doi: 10.1128/JVI.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Modis Y. 2014. A novel membrane fusion protein family in Flaviviridae? Trends Microbiol 22:176–182. doi: 10.1016/j.tim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Wang J, Kanai R, Modis Y. 2013. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc Natl Acad Sci U S A 110:6805–6810. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lavie M, Goffard A, Dubuisson J. 2007. Assembly of a functional HCV glycoprotein heterodimer. Curr Issues Mol Biol 9:71–86. [PubMed] [Google Scholar]
- 78.Vieyres G, Dubuisson J, Pietschmann T. 2014. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses 6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi: 10.1128/jvi.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reimann I, Semmler I, Beer M. 2007. Packaged replicons of bovine viral diarrhea virus are capable of inducing a protective immune response. Virology 366:377–386. doi: 10.1016/j.virol.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 81.Wegelt A, Reimann I, Zemke J, Beer M. 2009. New insights into processing of bovine viral diarrhea virus glycoproteins Erns and E1. J Gen Virol 90:2462–2467. doi: 10.1099/vir.0.012559-0. [DOI] [PubMed] [Google Scholar]
- 82.Wyatt LS, Moss B, Rozenblatt S. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 83.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 84.Stark R, Meyers G, Rümenapf T, Thiel HJ. 1993. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol 67:7088–7095. doi: 10.1128/JVI.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meyers G, Thiel HJ, Rümenapf T. 1996. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol 70:1588–1595. doi: 10.1128/JVI.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]