Rift Valley fever virus (RVFV) is a dangerous human and animal pathogen that was ranked by the World Health Organization in 2018 as among the eight pathogens of most concern for being likely to cause wide epidemics in the near future and for which there are no, or insufficient, countermeasures. The focus of this work is to address the question of the mechanisms underlying RVFV-induced cytopathic effects that participate in RVFV pathogenicity. We demonstrate here that RVFV targets cell adhesion and the actin cytoskeleton at the transcriptional and cellular level, affecting cell mobility and inducing cell shape collapse, along with distortion of cell-cell adhesion. All these effects may participate in RVFV-induced pathogenicity, facilitate virulent RVFV dissemination, and thus constitute interesting potential targets for future development of antiviral therapeutic strategies that, in the case of RVFV, as with several other emerging arboviruses, are presently lacking.
KEYWORDS: Abl2, actin cytoskeleton, Rift Valley fever virus, innate antiviral response
ABSTRACT
Rift Valley fever virus (RVFV) is a highly pathogenic zoonotic arbovirus endemic in many African countries and the Arabian Peninsula. Animal infections cause high rates of mortality and abortion among sheep, goats, and cattle. In humans, an estimated 1 to 2% of RVFV infections result in severe disease (encephalitis, hepatitis, or retinitis) with a high rate of lethality when associated with hemorrhagic fever. The RVFV NSs protein, which is the main virulence factor, counteracts the host innate antiviral response to favor viral replication and spread. However, the mechanisms underlying RVFV-induced cytopathic effects and the role of NSs in these alterations remain for the most part unknown. In this work, we have analyzed the effects of NSs expression on the actin cytoskeleton while conducting infections with the NSs-expressing virulent (ZH548) and attenuated (MP12) strains of RVFV and the non-NSs-expressing avirulent (ZH548ΔNSs) strain, as well as after the ectopic expression of NSs. In macrophages, fibroblasts, and hepatocytes, NSs expression prevented the upregulation of Abl2 (a major regulator of the actin cytoskeleton) expression otherwise induced by avirulent infections and identified here as part of the antiviral response. The presence of NSs was also linked to an increased mobility of ZH548-infected cells compared to ZH548ΔNSs-infected fibroblasts and to strong changes in cell morphology in nonmigrating hepatocytes, with reduction of lamellipodia, cell spreading, and dissolution of adherens junctions reminiscent of the ZH548-induced cytopathic effects observed in vivo. Finally, we show evidence of the presence of NSs within long actin-rich structures associated with NSs dissemination from NSs-expressing toward non-NSs-expressing cells.
IMPORTANCE Rift Valley fever virus (RVFV) is a dangerous human and animal pathogen that was ranked by the World Health Organization in 2018 as among the eight pathogens of most concern for being likely to cause wide epidemics in the near future and for which there are no, or insufficient, countermeasures. The focus of this work is to address the question of the mechanisms underlying RVFV-induced cytopathic effects that participate in RVFV pathogenicity. We demonstrate here that RVFV targets cell adhesion and the actin cytoskeleton at the transcriptional and cellular level, affecting cell mobility and inducing cell shape collapse, along with distortion of cell-cell adhesion. All these effects may participate in RVFV-induced pathogenicity, facilitate virulent RVFV dissemination, and thus constitute interesting potential targets for future development of antiviral therapeutic strategies that, in the case of RVFV, as with several other emerging arboviruses, are presently lacking.
INTRODUCTION
Rift Valley fever virus (RVFV) is a dangerous human and animal pathogen ranked by the World Health Organization in 2018 as among the eight pathogens of most concern for being likely to cause wide epidemics in the near future and for which there are no, or insufficient, countermeasures (https://www.who.int/blueprint/priority-diseases/en/). RVFV is a mosquito-borne, zoonotic phlebovirus endemic throughout many African countries and the Arabian Peninsula (1). Animal infections cause high rates of neonatal mortality and abortion, mainly among sheep, goats, and cattle. Human infections lead to a wide range of clinical manifestations from mild flu-like symptoms to severe illness, such as encephalitis, retinitis, and hepatitis, the last of which can be fatal when associated with hemorrhagic fever. An estimated 1 to 2% of human infections result in severe disease often associated with high levels of mortality (1–3).
RVFV is an enveloped, segmented RNA virus of negative or ambisense polarity. It has a tripartite single-stranded RNA genome consisting of the large (L) and medium (M) segments of negative polarity and the small (S) segment of ambisense polarity. The L segment encodes the RNA-dependent RNA polymerase, the M segment encodes two glycoproteins as well as two nonstructural NSm proteins, and the S segment encodes the nucleoprotein N with a negative polarity and the nonstructural NSs protein with a positive polarity (2, 4, 5). The NSs protein that is produced shortly after infection (6) is not essential for the viral cycle but is considered the main virulence factor of RVFV. Indeed, the virulent ZH548 strain that expresses NSs kills wild-type mice, whereas mutant viruses carrying an NSs-deletion mutation, either natural (Clone 13) or recombinant (ZH548ΔNSs), are avirulent and nonpathogenic for mice (7, 8). NSs is characterized by its capacity to accumulate in the cell nucleus, where it forms filament-like structures (9, 10) that include NSs itself as well as several cellular transcription factors and cofactors (11, 12). NSs has been found associated with specific cellular DNA sequences, such as the pericentromeric major satellite DNA sequences associated with chromosome cohesion defects (13) and the promoter region of the gene coding for interferon beta (IFN-β), a cytokine that plays a major role in the innate antiviral response of the host, where NSs maintains the IFN-β gene (Ifnb1) in a repressed state (12). Along with inhibiting the expression of Ifnb1 (7, 12, 14), NSs also counteracts the cellular innate immune response by targeting kinase PKR for degradation (15). The inhibition of the host’s innate antiviral response directly facilitates viral replication and propagation. However, the mechanisms underlying the development of significant ultrastructural changes such as cell shrinkage, rounding, and loss of cell junctions that have been observed in vivo in the liver after RVFV infection (16), potentially participating in the pathogenicity of RVFV, remain mostly to be identified.
In previous work, we have performed a genome-wide analysis of the interactions between NSs and the host genome. For this, genome-wide chromatin immunoprecipitation (Chip) carried out using an anti-NSs antibody was combined with promoter sequence microarray hybridization (ChIP-on-chip). The DNA immunoprecipitated in these experiments was recovered from L929 murine fibroblastic cells either before or at different times postinfection (p.i.) with the pathogenic, NSs-expressing ZH548 strain of RVFV (17). Several cellular promoter regions were identified as significantly interacting with NSs and the establishment of NSs interactions with these regions was often linked to the deregulation of the expression of the corresponding genes. Among annotated NSs-interacting genes were present not only genes regulating innate immunity and inflammation, but also genes regulating cellular pathways potentially participating in RVFV pathogenicity. The functional analysis of the genes associated with these NSs-interacting regions identified cell adhesion as the biological process most significantly enriched among cellular NSs-interacting genes (17).
In order to test the effect of RVFV infection and the NSs protein in cell adhesion, we have in this work compared the effects of infections with the NSs-expressing strains that are either virulent ZH548 (ZH) or attenuated (MP12) and the non-NSs-expressing avirulent ZH548ΔNSs (ΔNSs) strain of RVFV on the actin cytoskeleton and on cell-cell adhesion at the transcriptional and cellular level. We show here that the expression of the host Abl2 protein, actin cytoskeleton organization, and cell-cell adhesion were differently affected after infection by NSs-expressing and non-NSs-expressing strains of RVFV. Abl2 is a cytosolic protein encoded by the Abl2 gene (also known as Arg for Abelson related gene) that is considered a major regulator of the actin cytoskeleton, regulating cell morphology and mobility as well as cell-cell and cell-matrix adhesion (18–23). Within its sequence, Abl2 contains a tyrosine kinase domain and two filamentous actin (F-actin) binding domains (20). Through its kinase domain, Abl2 regulates Rho GTPases resulting in a negative regulation of focal adhesion and stress fiber formation, attenuating cell contractility, and altering adhesion dynamics, leading to the negative regulation of cell migration (18) and the positive regulation of adherens junction formation (21). Through its F-actin binding domains, Abl2 affects cell shape and positively regulates lamellipodia and membrane ruffles (20, 22). In this work we have identified the upregulation of Abl2 expression as part of the cellular response meant to restrict virulence, which appears to be counteracted by the NSs protein following either infection by the ZH and MP12 strains of RVFV or the ectopic expression of NSs.
ΔNSs infection induced the activation of Abl2 expression that was correlated with a diminution of cell migration. On the contrary, ZH infection prevented Abl2 upregulation in fibroblasts, macrophages, and hepatocytes, hampering the subsequent ΔNSs-induced reduction of cell migration. Thus, fibroblasts infected with virulent ZH strain migrated faster than fibroblasts infected with the avirulent ΔNSs strain. In the case of nonmigrating hepatocytes, infection with ZH but not ΔNSs induced a strong collapse of cell shape and the distortion of cell-cell adhesion. Similar effects on Abl2 expression and cell structure were observed after infection with the NSs-expressing attenuated MP12 strain of RVFV, as well as following the ectopic expression of the RVFV NSs protein on hepatocytes, where NSs was identified as the viral factor responsible for the inhibition of the upregulation of Abl2 expression, capable also of autonomously affecting the host’s actin cytoskeleton. Interestingly, the NSs protein was found abundantly present within the cytoplasm of ZH- and MP12-infected hepatocytes, as well as within long actin-rich structures connecting neighboring cells. While working with the attenuated MP12 strain at low multiplicity of infection, these NSs-containing actin-rich structures were found associated with the dissemination of NSs from infected to noninfected cells.
We demonstrate here that infection with the virulent strain of RVFV targets cell adhesion and the actin cytoskeleton at the transcriptional and cellular level, affecting cell mobility and inducing cell shape collapse along with distortion of adherens junctions. We propose that the upregulation of Abl2 expression is part of the host strategy to restrict virulence and we discuss how its counteraction by the RVFV NSs protein, together with the NSs effects on the actin-cytoskeleton, cell-cell adhesion, and the presence of NSs within intercellular actin-rich structures, could facilitate RVFV dissemination and pathogenicity.
RESULTS
Avirulent and virulent strains of RVFV differently affect the expression of the gene coding for Abl2, a main regulator of actin cytoskeleton.
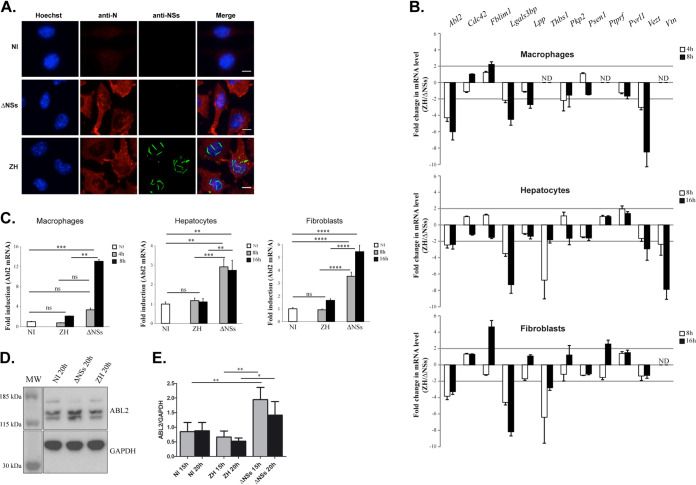
Infection with the virulent ZH548 (ZH) strain of RVFV is characterized by the presence of the nonstructural NSs protein within the nucleus of infected cells, which is absent from cells infected with the avirulent ZH548ΔNSs (ΔNSs) strain lacking NSs (Fig. 1A). As expected, though, the structural N protein encoded by RVFV was expressed in cells infected by ZH as well ΔNSs.
FIG 1.
Infection with RVFV affects the expression of genes related to cell adhesion, including Abl2 expression. (A) Murine L929 fibroblasts either noninfected (NI) or infected with the virulent ZH548 (ZH) or the avirulent ZH548ΔNSs (ΔNSs) strains of RVFV were labeled at 15 h p.i. using a DNA intercalating agent to visualize the nucleus (Hoechst, in blue), an antibody directed against the structural N protein of RVFV (N, in red), or an antibody directed against the nonstructural NSs protein of RVFV (NSs, in green). (B and C) The expression levels of 12 genes associated with cell adhesion, previously identified to interact with the NSs protein of RVFV (17), were measured in macrophages (RAW cell line), hepatocytes (AML12 cell line), and fibroblasts (L929 cell line) either noninfected (NI) or at the indicated times after infection with ZH or ΔNSs strains. Purified RNAs were analyzed by RT-qPCR using Biomark with primers specific for each gene of interest. Relative quantification of mRNA expression was calculated with respect to three reference genes corresponding to Hmbs, Ppib, and Hprt1 in the case of RAW cells; Hprt1, Ppib, and Rplp0 in the case of AML12 cells; and Ppib, Rplp0, and Utp6c in the case of L929 cells. The fold change in ZH-infected cells was calculated with respect to ΔNSs-infected cells (B) and with respect to mock-infected (NI) cells (C). Results correspond to the average of n = 3 independent experiments in the case of AML12 and L929 cells for all conditions tested (except L929 ZH 16 h, for which n = 2) and n = 2 in the case of RAW cells for all conditions tested. Data are means ± standard deviation (SD) with significance assessed by one-way ANOVA and Tukey’s multiple-comparison test. In (B), the expression level of certain genes could not be tested since the corresponding mRNA remained not detectable (ND). (D and E) Abl2 protein levels were analyzed by Western blotting in extracts from L929 cells noninfected (NI) or infected by the ΔNSs or ZH strains of RVFV. (E) Quantification using ImageJ of n = 3 independent experiments in the case of ZH and ΔNSs at 15 and 20 h p.i. and n = 6 for NI at 15 and 20 h. Data are means ± SD with significance assessed by a one-way ANOVA test with Bonferroni correction. P values: ****, <0.0001; ***, <0.001; **, <0.01; *, <0.05; ns, not significant. Images correspond to a single confocal section. Bar = 10 μm.
With the aim to investigate the effect of RVFV infection on the expression of genes regulating cell adhesion, the main biological host process represented among NSs-interacting promoter regions (17), the expression profile of 12 out of the 96 NSs-interacting genes associated with cell adhesion was analyzed in this work by quantitative PCR (qPCR) (Fig. 1B). The expression rates of these 12 genes, chosen because all are described as expressed in many different cell types, were analyzed with respect to three reference genes, in three different cell lines (fibroblasts, hepatocytes, and macrophages) under conditions of either noninfection (NI) or at two different times after infection with either the ZH or the ΔNSs strain of RVFV.
In Fig. 1B are shown the ZH/ΔNSs expression ratios for the 12 genes tested. The expression levels of all genes except Cdc42, Psen1, and Pvrl1 displayed a minimum of 2-fold differences in at least one cell line, with the expression of the Abl2 gene negatively affected in all the cell lines and at all the time points tested (Fig. 1B). Compared to NI conditions, Abl2 gene expression was enhanced after ΔNSs- but not ZH-infection in the three cell lines tested (Fig. 1C), with consistent results also observed at the protein level (Fig. 1D and E). These results suggested a potential role for Abl2 in the innate antiviral response, as its expression was induced after infection with the ΔNSs strain, which induces the innate antiviral response, whereas its expression was blocked after the ZH infection that counteracts the innate antiviral response.
Abl2 behaves as a gene associated with the cellular innate antiviral response independent of interferon beta signaling.
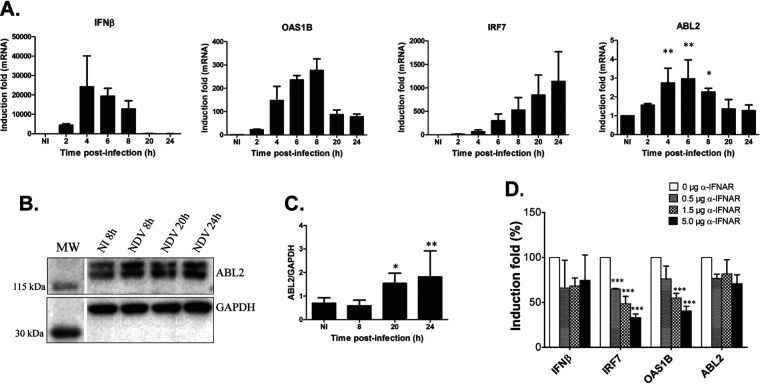
In order to consolidate the potential link between Abl2 expression and the innate antiviral response, Abl2 expression was further analyzed after infection of L929 cells (corresponding to the murine fibroblasts used in Fig. 1) with the Newcastle disease virus (NDV). NDV is a Paramyxoviridae member that, like the ΔNSs strain of RVFV, is a good inducer of the innate antiviral response in murine cells (24, 25). The expression level of the Abl2 gene was measured throughout the time course of infection by reverse transcriptase quantitative PCR (RT-qPCR) together with the expression of genes related to the IFN-β response, a main actor in the innate antiviral response (26, 27), including the Ifnb1 gene coding for IFN-β and the Oas1b and Irf7 genes corresponding to two well-characterized interferon β stimulated genes (ISGs) (Fig. 2A).
FIG 2.
Abl2 behaves as a gene of the cellular innate antiviral response independent of interferon beta signaling. (A) The expression levels of genes associated with the IFN-β response, coding for IFN-β and for two ISGs (IRF7 and OAS1B), along with the gene coding for Abl2, were measured by RT-qPCR in L929 cells either noninfected (NI) or at different times after NDV infection. Relative expression levels were determined with respect to three reference genes (Utp6c, Ppib, and Hprt1) and fold inductions were calculated with respect to noninfected (NI) cells. Results correspond to the average of n = 3 independent experiments for all conditions tested. Data are means ± SD. Statistical analysis was carried out only for Abl2 gene expression using a one-way ANOVA test with Dunnett correction with each NDV-infected condition compared to noninfected conditions. (B and C) Abl2 protein levels were analyzed by Western blotting in extracts from L929 cells noninfected (NI) or infected by NDV. (C) Quantification using ImageJ with n = 3 independent experiments for NDV infection at 8, 20, and 24 h p.i. and n = 10 for NI. Data are means ± SD. Significance was assessed by one-way ANOVA test with Dunnett correction and each NDV-infected condition compared to the noninfected condition. (D) The expression levels of genes associated with the IFN-β response, coding for IFN-β and for two ISGs (IRF7 and OAS1B), along with the gene coding for Abl2, were measured by RT-qPCR in L929 cells collected at 6 h p.i. with NDV, either untreated or treated with increasing amounts of anti-IFNAR antibody. Relative expression levels were determined with respect to three reference genes (Utp6c, Ppib, and Hprt1) and fold inductions were calculated with respect to noninfected (NI) cells considered 100%. Results correspond to the average of n = 4 independent experiments for 0 μg; n = 2 for 0.5 μg; and n = 3 for 1.5 and 5 μg of anti-IFNAR antibody. Data are means ± SD. Significance was assessed by one-way ANOVA test with Bonferroni correction for each gene separately. P values: ***, <0.001; **, <0.01; and *, <0.05.
NDV infection of L929 cells induced the expression of not only the Ifnb1, Oas1b, and Irf7 genes, as expected, but also that of the Abl2 gene (Fig. 2A) as previously observed after ΔNSs infection (Fig. 1C). Again, the effects observed at the transcriptional level were also observed at the protein level (Fig. 2B and C). While the kinetics of expression of the ISGs Oas1b and Irf7 were, as expected, delayed compared to that of the Ifnb1 gene, the kinetics of the expression of the Abl2 gene resembled more closely that of the Ifnb1 gene, suggesting that the virus-induced activation of the expression of Abl2 would be independent of IFN-β. The role of IFN-β signaling on NDV-induced Abl2 expression was tested using an anti-IFNAR antibody that, by blocking the IFN-β receptor IFNAR, antagonizes the IFN-β response, neutralizing the expression of ISGs (28). As expected, a dose-dependent inhibition of the expression of Oas1b and Irf7 was observed in the presence of the anti-IFNAR antibody (Fig. 2D). However, no effect was detected on the expression of the Abl2 gene under the same conditions, indicating that although Abl2 behaved as a gene associated with the innate antiviral response, its expression in the context of NDV infection appeared independent of IFN-β signaling.
Activation of Abl2 expression in the context of ΔNSs but not ZH infection is correlated with the negative regulation of cell migration.
In order to investigate the potential implication of the opposing effects of ΔNSs and ZH infection on Abl2 expression, we compared the effects of these two strains of RVFV on cell migration that is negatively regulated by Abl2 (18, 19, 23).
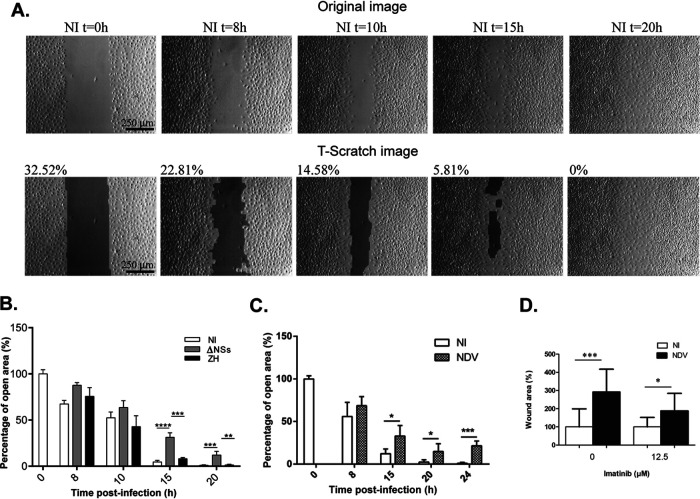
Wound-healing assays (scratch tests) were carried out on monolayers of confluent L929 fibroblasts, which are fast migrating cells, that were either NI or infected by the ΔNSs or ZH strains of RVFV. The coverage of the open (scratched) cell-free area by migrating fibroblasts was assessed over time using bright-field imaging. The percentage of open, cell-free area was determined by an automated quantitative analysis using the TScratch software (29). Wounding, which corresponded to T = 0 h, was performed 24 h after plating the cells on glass coverslips and immediately before infection. Cell counting performed at the beginning (T = 0 h) and the end (T = 20 h) of the assay indicated that confluent cells did not divide during the time lapse of the experiment (data not shown).
Under these experimental conditions, NI L929 fibroblasts reached complete coverage of the open area at 20 h after wounding (Fig. 3A). TScratch analysis of the percentage of open area showed that infection with the ΔNSs and ZH strains of RVFV differently affected cell migration (Fig. 3B). Infection with the ΔNSs strain significantly inhibited cell migration, with ΔNSs-infected cells displaying a significantly higher percentage of open area than NI cells starting at 15 h p.i. (Fig. 3B). Infection with the ZH strain hampered the inhibitory effect of ΔNSs on cell migration. ZH-infected cells migrated faster than ΔNSs-infected cells, with ZH-infected cells displaying a significantly lower percentage of open area than ΔNSs-infected cells starting at 15 h p.i. (Fig. 3B). The inhibitory effect of the ΔNSs strain of RVFV on cell migration was also observed after NDV infection, with NDV-infected cells displaying a significantly higher percentage of open area than NI cells starting at 15 h p.i. (Fig. 3C). Overall, these results were in agreement with the role of Abl2 as a negative regulator of cell migration (18), as ΔNSs- and NDV-infected cells that expressed the largest amount of Abl2 corresponded to the slowest migrating cells with the highest percentages of open area.
FIG 3.
Infection with ΔNSs or NDV reduces cell migration. (A) Representative bright-field images of a monolayer of confluent noninfected (NI) L929 cells taken at different times after introducing a scratch wound with a pipette tip. Images are shown before (original images) and after automated analysis by the TScratch program. The percentage of open areas as determined by the TScratch program are indicated, with 100% corresponding to the area of the entire image. (B to D) Scratch-wound closure of L929 cells was monitored over time in noninfected (NI) cells and after infection with ΔNSs or ZH (B) or NDV (C) or NDV at 15 h p.i. in the presence or absence of imatinib (D). Wound closure is expressed as the % of remaining open area uncovered by cells as determined by the TScratch program, with 100% corresponding in (B) and (C) to the scratch area of noninfected (NI) cells at time point 0 h and in (D) to the scratch area of noninfected cells at 15h in the absence of imatinib. (B) For each condition (NI, ΔNSs, and ZH), results correspond to the average of n = 18 images analyzed from 3 independent experiments for 0, 8, 15, and 20 h and n = 12 from 2 independent experiments for 10 h. (C) For each condition (NI and NDV), results correspond to the average of n = 12 images analyzed from 2 independent experiments for 0, 8, 15, 20, and 24 h. (D) Results correspond to the average of n = 9 images analyzed from 3 independent experiments. Data are means ± SD. Significance was assessed by one-way ANOVA test with Bonferroni correction for each time point separately in (B), for all samples in (D), and by Student’s t test in (C). P value: ****, <0.0001; ***, <0.001; **, <0.01; and *, <0.05.
In order to question the role of Abl2 on the capacity to slow down cell migration in the context of viral infection, we used imatinib, which is an inhibitor of the Abl2 kinase activity that has been previously used to counteract Abl2-dependent inhibition of cell migration (30). The percentage of wound area under NI and NDV-infected conditions, in the presence or absence of imatinib, was measured 15 h after wounding with 100% corresponding to the open wound area under NI conditions in the absence of imatinib (Fig. 3D). As previously observed, in the absence of imatinib the percentage of open wound area was significantly higher after NDV infection than in NI cells (Fig. 3D), which agrees with NDV-dependent reduction of cell migration. In the presence of imatinib, the capacity of NDV infection to negatively affect cell migration was significantly diminished (Fig. 3D), indicative of a role for Abl2 kinase activity in NDV-induced inhibition of cell migration.
Virulent RVFV infection affects cell morphology and adherens junctions.
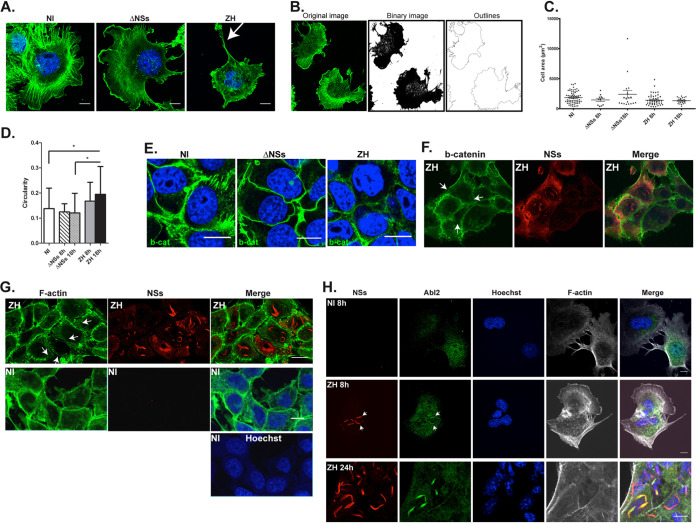
While through its kinase activity Abl2 negatively regulates cell migration in line with the attenuation of cell contractility and focal adhesion dynamics (18), it positively regulates cell spreading through its two F-actin binding domains, promoting the formation of actin-rich structures at the lamellipodia (22, 31, 32). The impact of ΔNSs and ZH infection on cell morphology was analyzed independently of cell migration by fluorescence and confocal microscopy using phalloidin as a marker of F-actin. Experiments were carried out on hepatocytes (AML12 cell line) that constitute one of the main cell types targeted during RVFV infection, with liver damage in association with hemorrhagic fever being the most frequent cause of death in humans (1–3). Under our experimental conditions, AML12 cells did not display migratory capacity neither before nor after infection with either ΔNSs or ZH strains of RVFV (data not shown), thus allowing the analysis of cell structure independently of cell migration. Despite the propensity of AML12 cells to adhere to each other, isolated cells were identified that were used here to analyze effects on cell structure (Fig. 4A to D). Important changes in cell morphology were consistently observed after infection with the ZH strain compared to NI and ΔNSs-infected cells, for which representative images are shown in Fig. 4A. While NI and ΔNSs-infected cells displayed significant cell spreading, ZH-infected cells displayed a reduction of cell spreading, lamellipodia and membrane ruffles, and a disorganization of actin fibers visible from 8 h p.i. (Fig. 4A). Also, long F-actin-rich structures (indicated by an arrow in Fig. 4A) were often seen among isolated ZH-infected cells. ImageJ software was used to quantify changes in cell shape and area based on phalloidin staining of NI, ΔNSs-, and ZH-infected cells. Original images were transformed into binary images (Fig. 4B) and the outlines of the binary images were used to measure cell area (Fig. 4C) and circularity (Fig. 4D). Even though not statistically significant, a trend toward an enhancement of cell area was observed 18 h p.i. under conditions of ΔNSs infection compared to NI and ZH infection (Fig. 4C), this being in agreement with the upregulation of Abl2 expression observed in ΔNSs-infected cells with respect to NI and ZH-infected cells (Fig. 1C). In addition, a significant increase in cell circularity was observed after ZH infection at 18 h p.i. with respect not only to ΔNSs-infected cells but also NI cells (Fig. 4D).
FIG 4.
Infection with the virulent ZH but not the avirulent ΔNSs strain affects cell morphology and adherens junctions. Murine hepatocytes (AML12 cell line) either noninfected (NI) or infected with the avirulent ΔNSs or the virulent ZH strains of RVFV were labeled at 8 h (A, B, F, and H) and 18 h (E, G, and H) p.i. using a DNA intercalating agent to visualize the nucleus (Hoechst, in blue), phalloidin to visualize filamentous actin (F-actin in green or gray in A, B, G, and H), an anti-NSs antibody (NSs, in red), an anti-beta-catenin antibody (b-cat in green in E and F), and an anti-Abl2 (Abl2 in green in H). White arrows indicate long F-actin-rich extensions in (A), dissolution of adherens junctions in (F) and (G), and colocalization of Abl2 with NSs in (H). (B) The analyze particle function of ImageJ software was used to transform the images into binary images and determine the cell outlines. (C and D) Main parameters such as the cell area and perimeter were determined and cell shape was analyzed by calculating the circularity = 4π × (area/perimeter2). Results, from a minimum of 2 independent experiments for each condition, correspond to the average of n = 70 cells analyzed for NI; n = 11 for ΔNSs at 8 h p.i.; n = 18 for ΔNSs at 18 h p.i.; n = 48 for ZH at 8 h p.i.; and n = 21 for ZH at 18 h p.i. Data are means ± SD. Significance was assessed by one-way ANOVA test with Dunnett correction with ΔNSs and ZH compared to NI and ΔNSs compared to ZH. P value: *, <0.05. Images correspond to single confocal sections. Bar = 10 μm.
AML12 cells have a strong propensity to establish cell-cell contacts forming adherens junctions that contain β-catenin. Abl2 has been described to positively regulate β-catenin stability (33) and maintain adherens junctions (19, 21). In agreement with distinct effects on Abl2 expression, infections with ΔNSs and ZH strains differently affected the stability of β-catenin-containing cell junctions (Fig. 4E). At late times after infection (18 h p.i.), ΔNSs-infected cells appeared to sustain β-catenin-containing cell junctions, while a systematic dissolution of β-catenin-containing cell junctions was observed after infection with ZH (Fig. 4E). At earlier (8 h) times p.i., disruption of β-catenin-containing adherens junctions was evident among ZH-infected cells with accumulated NSs (Fig. 4F). Dissolution of adherens junctions among ZH-infected AML12 cells resulted in loosening of cell-to-cell contact, visible after F-actin labeling in ZH-infected NSs-expressing cells with respect to NI cells (Fig. 4G), with the strongest effect observed among the cells displaying the largest amount of NSs (indicated by white arrows in Fig. 4G).
Interestingly, while analyzing the distribution of Abl2 in ZH-infected cells with respect to the NSs protein, we observed a colocalization of Abl2 with NSs that started to be visible at 8 h p.i. (indicated by arrows in Fig. 4H) and was clearly enhanced 24 h p.i. (Fig. 4H). Though not visible among all NSs-expressing cells, Abl2 colocalization with NSs appeared associated with dissolution of adherens junctions (24 h p.i.) (Fig. 4H).
NSs-expressing MP12 strain of RVFV affects Abl2 expression and cell morphology similarly to the ZH strain.
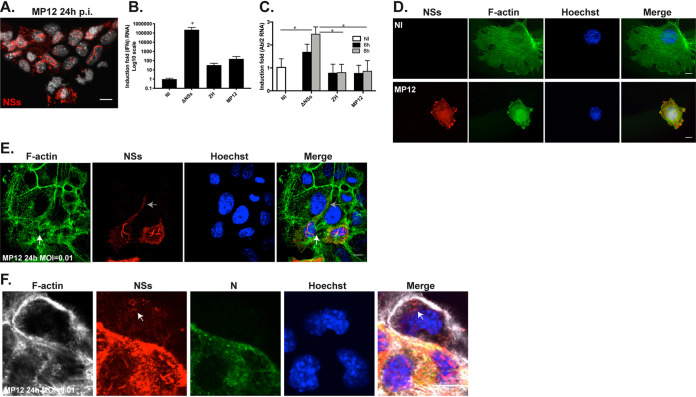
To further confirm the main role of NSs in targeting the actin cytoskeleton of the host at the transcriptional and cellular level, we carried out infections with the RVFV MP12 strain that, despite being attenuated by several mutations in its S, M, and L segments, still encodes a functional NSs protein (34, 35). The NSs protein encoded by the MP12 strain formed filamentous structures in the nuclei of AML12-infected cells (Fig. 5A) similarly to the ZH-encoded NSs protein (Fig. 4). The NSs-dependent inhibition of IFN-β gene expression is a major trait observed after ZH infection (7, 8, 12, 23). Comparison of IFN-β expression in NI, ΔNSs-, ZH-, and MP12-infected AML12 cells indicated that, as in the case of ZH infection, MP12 infection strongly inhibited the expression of the IFN-β gene otherwise induced by ΔNSs infection (Fig. 5B). Along with measuring IFN-β expression, we also measured the level of Abl2 mRNA in NI, ΔNSs-, ZH-, and MP12-infected cells (Fig. 5C). The significant ΔNSs-dependent induction of Abl2 expression previously observed in AML12 cells (Fig. 1C), and reproduced here, was significantly hampered not only after infection with the ZH strain, in agreement with our previous observations, but also after MP12 infection (Fig. 5C). Consistently, as in ZH-infected cells, the expression of NSs in MP12-infected cells was correlated with a strong alteration of cell shape and spreading (Fig. 5D) and with a loosening of adherens junctions (Fig. 5E), for which representative images are shown here. Results obtained after MP12 infection at low multiplicity of infection (MOI) confirmed the correlation between NSs expression and disruption of β-catenin-rich adherens junctions (as indicated by a white arrow in Fig. 5E) compared to non-NSs-expressing cells. Overall, the results obtained, which are in agreement with previously reported functional similarity between MP12- and ZH-encoded NSs proteins (34), confirmed the capacity of NSs protein to affect the actin cytoskeleton by hampering the upregulation of Abl2 expression and affecting cell shape and cell-to-cell adhesion.
FIG 5.
Infection with the NSs-expressing MP12 strain of RVFV affects Abl2 expression and cell morphology. Murine AML12 hepatocytes infected with the MP12 strain of RVFV were labeled at 8 h (D) and 24 h (A, E, and F) p.i. using a DNA intercalating agent to visualize the nucleus (Hoechst, in gray or blue), antibodies directed against the nonstructural NSs (in red) or the N (in green) proteins of RVFV and phalloidin to visualize filamentous actin (F-actin in green or gray). In (E) and (F), MP12 infection was carried out at an MOI = 0.01. (B and C) The expression of the genes coding for IFN-β (B) and Abl2 (C) was measured by RT-qPCR in AML12 cells either noninfected (NI) or at the indicated times after infection with ΔNSs, ZH, or MP12 strains. Relative expression levels were determined with respect to Rplp0 as a reference gene and fold inductions were calculated with respect to noninfected (NI) cells. Results correspond to the average of n = 2 independent experiments. Data are means ± SD. Significance was assessed by one-way ANOVA and Tukey’s multiple-comparison test. (E) Gray arrow indicates NSs dissemination and white arrow indicates dissolution of adherens junctions. (F) White arrow indicates NSs dissemination. P value: *, <0.05. In (E) and (F), images correspond to single confocal sections. Bar = 10 μm.
ZH- and MP12-encoded NSs proteins are found within long actin-rich structures in association with intercellular NSs dissemination.
AML12 cells infected with MP12 displayed not only nuclear but also a cytoplasmic distribution of NSs. In Fig. 5E, the cytoplasmic staining of NSs extended more than 30 μm away from the nucleus of the corresponding NSs-expressing cell, reaching toward non-NSs-expressing cells through weakened adherens junctions (as indicated by a gray arrow in Fig. 5E). Cytoplasmic NSs protein was also detected in noninfected cells (indicated by an arrow in Fig. 5F), suggesting a dispersion of NSs from neighboring infected cells expressing N and NSs toward noninfected cells favored by disrupted adherens junctions.
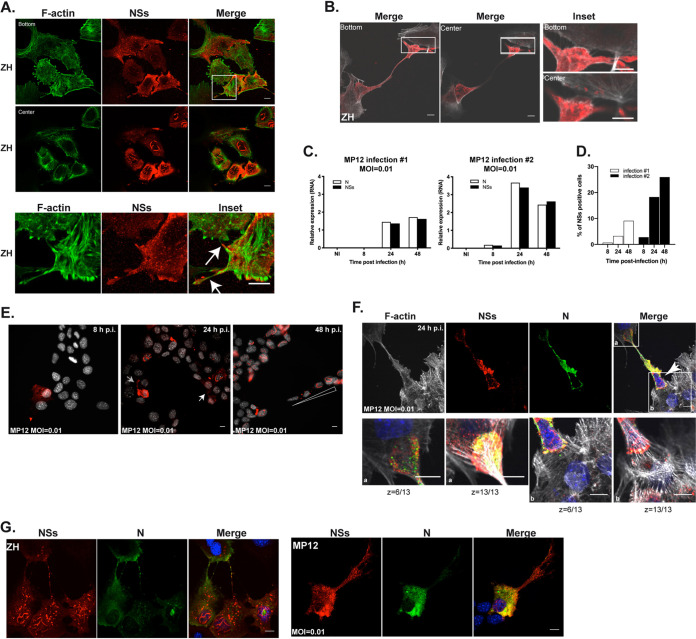
The presence of cytoplasmic NSs aggregates was a consistent trait observed not only among MP12-infected cells but also among ZH-infected cells, and that persisted throughout infection and remained visible at 18 h p.i. (Fig. 6A). Among ZH-infected cells, the NSs protein was often visible within F-actin-rich extensions, as indicated by white arrows in representative Fig. 6A. Occasionally, these actin-rich extensions extended over a long distance, interconnecting cells more than 100 μm apart (Fig. 6B), terminating in an apparent delivery process of the NSs protein into the cell body of a neighboring cell (Fig. 6B, insets), suggesting a cell-to-cell spreading of NSs.
FIG 6.
ZH- and MP12-encoded NSs proteins are found within long actin-rich structures in association with intercellular dissemination. Murine hepatocytes (AML12 cell line) infected with the NSs-expressing ZH and MP12 strains of RVFV at an MOI of 0.01 in the case of MP12 were labeled at 8 h (B, E, and G), 18 h (A), 24 h (E and F), and 48 h (E) p.i. using a DNA intercalating agent to visualize the nucleus (Hoechst in gray or blue in E, F, and G), phalloidin to visualize filamentous actin (F-actin in green or gray in A, B, and F), an anti-NSs antibody (NSs, in red), and an anti-N antibody (N in green in F and G). White arrows indicate NSs-containing actin-rich structures in (A), NSs dissemination in (E), and infected, NSs-expressing cells in F. White triangle indicates a gradient staining of NSs in (E). (C) The expression levels of N and NSs RNAs were measured by RT-qPCR in AML12 cells either noninfected (NI) or at the indicated times after infection with MP12 at an MOI of 0.01. Relative expression levels were determined with respect to Rplp0. (D) The percentage of cells displaying NSs labeling was randomly determined at the indicated times following MP12 infection at an MOI of 0.01. In (A), (B), (F), and (G), images correspond to single confocal sections. Bar = 10 μm.
To further investigate the capacity of NSs to disseminate from NSs-expressing to non-NSs-expressing cells, we carried out infections with the attenuated strain of MP12 at a low MOI. Viral replication and the percentage of NSs-positive cells were jointly analyzed throughout infection. Viral replication was determined by measuring the relative expression of N and NSs RNA levels by RT-qPCR and the percentage of NSs-expressing cells was analyzed after fluorescent labeling of cells with Hoechst and anti-NSs antibodies. In order to carry out a random analysis of NSs+ cells, cells were counted according to Hoechst staining before further analyis for NSs staining. Measurements were done throughout two independent infections, with each infection carried out with a different batch of MP12 RVFV (Fig. 6C and D). Though differences in the levels of N and NSs expression and in the percentage of NSs-expressing cells were observed between the two infections, the values evolved according to the same pattern (Fig. 6C and D). MP12 RVFV stopped replicating at 24 h p.i., with the relative expression of N and NSs RNAs remaining constant (infection number 1) or slightly diminishing (infection number 2) between 24 and 48 h p.i. (Fig. 6C). The percentage of NSs-positive cells was proportional to the amount of N and NSs RNA between the two infections, as well as between 8 and 24 h p.i. for each infection. However, notwithstanding a cessation in the increase of N and NSs RNA expression, the percentage of NSs+ cells continued to increase between 24 and 48 h p.i. in both infections (Fig. 6D). MP12 infection has been shown to induce cell cycle arrest (36), similar to what we have previously shown in the case of ZH infection (13), thus the increase of NSs+ cells observed between 24 and 48 h p.i. suggested the existence of a cell-cycle-independent mechanism of dissemination of NSs from NSs-expressing toward non-NSs-expressing cells.
The distribution of NSs+ cells was further analyzed among microscope field of views specifically selected for NSs staining. In Fig. 6E are shown representative images of fields of views selected for NSs staining at 8, 24, and 48 h p.i. in the case of infection number 1. At 8 h p.i., no more than one NSs+ cell was observed within a field, while at 24 h p.i. clusters of NSs+ cells appeared (indicated by a white arrow in Fig. 6E) with an average of 11.6% of NSs+ cells present per field. The clustered distribution of NSs+ cells was further enhanced at 48 h p.i. so that an average of 58.4% of NSs+ cells were present per field selected for NSs staining (Fig. 6E). Yet, when counting NSs+ cells in a random selection of field views, we observed only an average of 3.25% of NSs+ cells at 24 h, with several fields displaying none, and then rising to 7.6% per random field at 48 h p.i. (Fig. 6D).
A cytoplasmic spreading of NSs more than 30 μm away from the cell nucleus was again observed 24 h p.i. (indicated by a gray arrow in Fig. 6E). Also, a gradient-like distribution of NSs was observed 48 h p.i. (indicated by a triangle in Fig. 6E), suggesting cell-to-cell spreading of NSs. In order to further confirm the spreading of NSs from NSs+ toward non-NSs-expressing cells, we looked for NSs dissemination events under conditions for which only a small percentage of NSs+ cell were detected in a field view, such as was the case during infection number 1 of AML12 cells infected by MP12 at a low MOI 24 h p.i. Under these conditions, NSs dissemination events were indeed identified, such as the one shown in Fig. 6F, where an infected, NSs-expressing cell (indicated by an arrow) bridges two noninfected, non-NSs-expressing cells through long actin-rich NSs-containing structures that culminated in an NSs delivery process (Fig. 6F). Though the N protein of RVFV was also visible within actin-rich extensions, only NSs was delivered to noninfected cells, as observed previously (Fig. 5F).
The RVFV N protein was consistently found within long actin-rich structures in the context of ZH infection but to a lesser extent following MP12 infection at low MOI (Fig. 6G). Since NSs protein was similarly present within long actin-rich extensions in the context of ZH and low MOI MP12 infections, the differential staining observed here for the RVFV N protein in long actin-rich extensions, for the most part not colocalizing with NSs, suggested that the N protein of RVFV was not necessary for NSs to be present within these structures.
Ectopic expression of NSs protein mimics ZH infection.
The presence of the nonstructural NSs protein within intercellular actin-rich protrusions raised the question of the potential capacity of NSs to convey virulent phenotypes autonomously, independently of other viral factors.
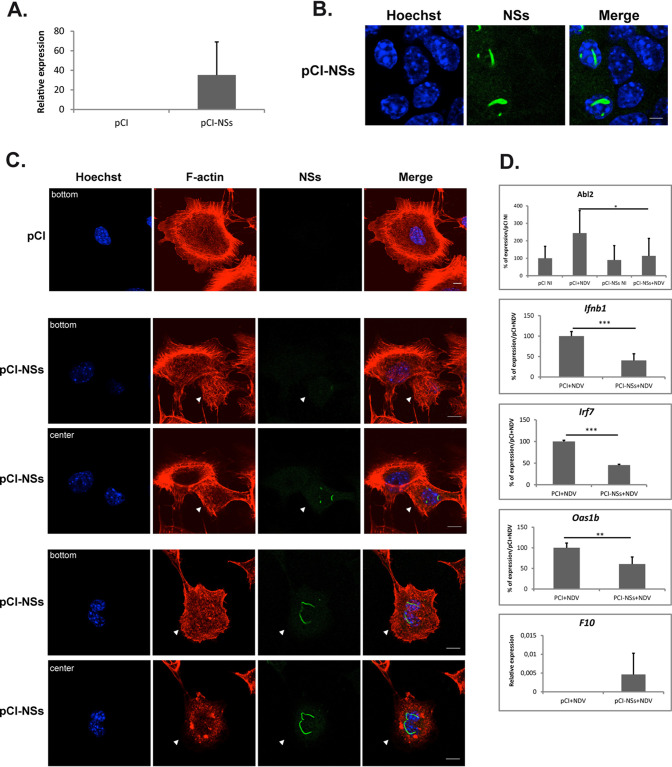
The capacity of the ectopically expressed NSs protein to induce ZH-like phenotypes was tested by transfecting noninfected AML12 cells with either a plasmid carrying the cDNA coding for NSs (pCI-NSs) or the empty vector (pCI). Twenty-four hours after transfection, the expression of NSs RNA was detected in AML12 cells transfected with pCI-NSs but not in pCI-transfected cells (Fig. 7A). The expression of NSs RNA was calculated here with respect to three reference genes (Ppib, Utp6c, and Hprt1), for which the corresponding expression rates did not vary after transfection with either pCI-NSs or pCI, indicating that at least 24 h after its expression, NSs did not significantly affected the transcriptional capacity of AML12 cells. This lack of effect on the transcriptional capacity of the host observed here 24 h after NSs expression is in agreement with our previous observations in ZH-infected L929 cells (17) and with more recently reported results in MP12-infected HEK293 cells (37). Notwithstanding variations observed from one transfection assay to another, 10 to 30% of the pCI-NSs-transfected cells were identified at the single cell level as expressing NSs protein by immunofluorescence using an anti-NSs antibody. In these cells, the ectopically expressed NSs protein formed nuclear filamentous structures (Fig. 7B) resembling those observed after infection with the virulent ZH strain of RVFV. However, compared to ZH- and MP12-infected AML12 cells, the ectopically expressed NSs protein displayed an exclusively nuclear distribution with no NSs detected outside the nucleus, suggesting that a viral component other than NSs could be required to induce a cytoplasmic NSs distribution in AML12 cells.
FIG 7.
The ectopic expression of the NSs protein affects cell morphology and gene expression mimicking ZH infection. AML12 cells were transfected with a plasmid encoding the NSs protein of RVFV (pCI-NSs) or with the empty vector (pCI). Twenty-four hours after transfection, the presence of NSs mRNA was quantified by RT-qPCR (A) and the presence of NSs protein was analyzed by fluorescence (B) using an anti-NSs antibody (NSs, in green) and a DNA intercalating agent to visualize the nucleus (Hoechst, in blue). (C) Cell shape was analyzed by fluorescence using phalloidin to visualize filamentous actin (F-actin, in red) and a DNA intercalating agent to visualize the nucleus (Hoechst, in blue). (D) Twenty four hours after transfection with plasmids pCI or pCI-NSs, transfected cells were either noninfected (NI) or infected with NDV. Four hours after infection, RNA was collected and the expression levels of RNA coding for NSs, Abl2, IFN, Irf7, Oas1b, and F10 were measured by RT-qPCR. (A and D) Relative expression levels were calculated with respect to three reference genes (Ppib, Utp6c, and Hprt1). In the case of the Abl2, Ifnb1, Irf7, and Oas1b genes, the percentage of expression was determined with respect to pCI+NDV conditions that were considered 100%. (A and D) Results correspond to an average of n = 3 independent experiments except for the Ifnb1 and Oas1b genes, for which n = 4. Data are means ± SD. (D) Bar graphs of relative expression levels. Student’s t test with P values: ***, <0.001; **, <0.01; and *, <0.05. Images correspond to single confocal sections. Bar = 10 μm.
The effect of the ectopically expressed NSs protein on the actin cytoskeleton and cell morphology was analyzed by fluorescence and confocal microscopy using phalloidin to label F-actin along with anti-NSs antibodies. Cells transfected with the empty vector (Fig. 7C, pCI) displayed a morphology and an actin cytoskeleton organization similar to that of nontransfected, noninfected AML12 cells (Fig. 4A). Among AML12 cells transfected with pCI-NSs (Fig. 7C, pCI-NSs), those cells that ectopically expressed NSs (indicated by arrowheads in Fig. 7C) displayed a strong reduction of cell spreading and a disorganization of F-actin fibers similarly to ZH- and MP12-infected AML12 cells (Fig. 4A and Fig. 5D). Therefore, the nonstructural NSs protein alone, in the absence of any other RVFV component, was able to induce changes in cell morphology similar to those induced by the virulent ZH strain. The effect of the ectopically expressed NSs protein on the expression of the Abl2 gene was also analyzed. No significant changes in Abl2 gene expression were observed among cells transfected with pCI-NSs compared to pCI (Fig. 7D). This was similar to the lack of effect of ZH and MP12 infection on the level of Abl2 expression displayed by NI AML12 cells (Fig.1C and Fig. 5C, respectively) and further confirmed the absence of effect of NSs expression on the host transcriptional capacity. However, the ectopic expression of NSs was able to significantly inhibit the expression of the Abl2 gene induced after infection with NDV (Fig. 7D), similarly to the previously observed capacity of ZH and MP12 infection to prevent the increase of Abl2 gene expression otherwise induced by ΔNSs (Fig.1C and Fig. 5C, respectively), further confirming the capacity of NSs protein to mimic ZH-induced effects independently of any other protein encoded by RVFV.
The capacity of the ectopically expressed NSs protein to mimic the effects of ZH infection on ΔNSs-induced cellular gene expression was further analyzed for genes participating in the IFN-β response and the coagulation cascade whose expressions have been previously reported to be affected after ZH infection as compared to ΔNSs infection (17, 24). As shown in Fig. 7D, the ectopic expression of NSs in AML12 cells significantly inhibited the NDV-induced activation of the expression of the genes related to the IFN-β response (such as the Ifnb1 gene coding for IFN-β and the two ISGs Irf7 and Oas1b), while it activated the expression of the F10 gene coding for coagulation factor F10, as was previously observed after infection with the ZH strain compared to ΔNSs-infected cells (17).
Overall, these results demonstrated that the NSs protein encoded by RVFV had the capacity to autonomously induce phenotypes similar to those observed after infection with the pathogenic virulent ZH strain of RVFV, raising the question of the potential deleterious effects that could be conveyed by the intercellular spreading of the NSs protein.
DISCUSSION
We show here that the viral NSs protein, which is the major virulence factor of RVFV, affects the actin cytoskeleton of the host at the transcriptional and cellular levels. At the transcriptional level, NSs expression counteracted the upregulation of Abl2 expression, a major regulator of the actin cytoskeleton that, in this work, was found associated with the cellular response to a viral infection. At the cellular level, the presence of NSs was associated with strong changes in cell morphology that were reminiscent of the effects on cell and tissue structures observed after ZH infection in vivo (16).
Besides a major role for NSs in cell structure and actin cytoskeleton organization, we describe here for the first time an effect of the RVFV NSs-expressing ZH strain on cell mobility, with ZH-infected cells migrating faster than ΔNSs-infected cells. In the case of influenza virus infection, restriction of cell mobility was found to be highly protective (38). Similarly, we hypothesize that the reduction of cell mobility induced here after avirulent ΔNSs infection, in correlation with an upregulation of Abl2 expression, would be necessary to limit viral dissemination by migrating innate immune cells such as macrophages, dendritic cells, neutrophils, and microglia that are infected by RVFV in vitro and/or in vivo (39, 40). Hampering reduction of cell mobility, as observed here following virulent ZH infection, could favor virulence and pathogenicity. In agreement with this hypothesis, a recent work has shown a major role for the migration into the central nervous of innate immune cells carrying a viral antigen in lethal RVFV encephalitis in Lewis rats (40).
Abl2 has been shown to negatively regulate cell mobility in line with the negative regulation of cell contractility and focal adhesion dynamics resulting from Abl2 regulation of the activity of Rho GTPases (18–20). Also, Abl2 positively regulates cell-cell adhesion (in line with its negative effect on cell contractility and the stabilization of β-catenin) (19, 21, 33) and cell spreading, in line with its capacity to directly interact with F-actin (22, 31, 32). The effects on cell mobility observed after ΔNSs infection with respect to NI and ZH-infected cells were indeed those expected for an upregulation of Abl2 expression. Also in agreement with ΔNSs-induced upregulation of Abl2 expression, a trend toward cell spreading and consolidation of adherens junctions was observed in ΔNSs-infected compared to noninfected cells. If the effects of ΔNSs infection on cell morphology remained weak, it is probably because NI AML12 cells expressing Abl2 displayed substantial cell spreading and consistent formation of β-catenin-rich adherens junctions before ΔNSs infection, independently of an upregulation of Abl2 expression. Therefore, no more than limited effects on AML12 cell morphology, with no further significant enhancement of cell spreading and cell-cell adhesion, are expected following ΔNS-induced upregulation of Abl2.
In the case of either ZH infection, MP12 infection, or ectopic expression of NSs, the morphological changes induced in line with NSs expression resembled those expected for Abl2-deficient cells. However, these changes were not always correlated with differences in Abl2 expression. In fact, strong morphological differences were observed not only between ΔNSs- and ZH- or MP12-infected cells, but also between ZH-infected and noninfected cells, while both displayed similar levels of Abl2 expression. However, it is plausible that the association of Abl2 with NSs filaments highlighted in this work could impact the capacity of Abl2 to interact with the actin cytoskeleton and thus participate in the establishment of the NSs-dependent morphological changes resembling Abl2-deficient phenotypes.
Nonetheless, though NSs-dependent effects on cell mobility and cell morphology resembled those expected for Abl2-deficient phenotypes, the direct link between the targeting of Abl2 by NSs and NSs-dependent effects on actin cytoskeleton shown in this work remains to be demonstrated.
The presence of long intercellular actin-rich NSs-containing structures, associated with the dissemination of the NSs protein, was another feature of ZH- and MP12-infected AML12 cells. These structures are reminiscent of the actin-rich intercellular connections that have been described to facilitate virus spreading to neighboring cells, as in the case of influenza A virus (41), HIV-1 (42), or lymphocytic choriomeningitis virus (LCMV) (43). Cell-to-cell transmission of the LCMV through actin-rich intercellular structures was facilitated by virus-induced enhanced cell migration (43), an interesting feature with respect to the capacity of ZH infection to hamper negative regulation of cell mobility. Actin-rich intercellular structures have also been described to facilitate the dissemination of prion and prion-like proteins (44, 45). In vivo, only a very small fraction of the host cells are primary infected cells and the efficiency of the host’s antiviral response largely depends on the paracrine IFN-β signaling triggered by the small number of cells initially infected (46). NSs has been proven here to be able to affect the host’s IFN-β response alone, independently of any other viral compound. Therefore, the dissemination of NSs from NSs-expressing to non-NSs-expressing cells that we have shown here to occur at a low MOI between neighboring cells, through disrupted adherens junctions or long actin-rich structures, could have strong negative effects on the capacity of the host to stop viral dissemination in vivo.
The presence of NSs within actin-rich extensions and NSs dissemination was observed here in ZH- and MP12-infected AML12 cells that displayed a dual nuclear/cytoplasmic distribution of NSs, but was not observed in cells ectopically expressing NSs that displayed only a nuclear NSs distribution. A recent work by Li et al. (10) has demonstrated an important role in vivo for the dual nuclear and cytoplasmic distribution of NSs in RVFV virulence and pathogenicity. In order to question the possible role of the subcellular distribution of NSs protein in regulating intercellular NSs spreading, it would be necessary to analyze the effect of the different rRVFVs established by Li et al. (10) on NSs dissemination. Li et al. (10) have described cytoplasmic versus nuclear distribution of NSs in relation to specific regions of NSs. This suggests a role for NSs structure and/or NSs interaction with another protein in the regulation of the nuclear/cytoplasmic distribution of NSs. The absence of cytoplasmic NSs among cells ectopically expressing NSs suggests a role for another viral component in this process.
At the transcriptional level, NSs altered not only the expression of Abl2 but also that of other genes involved in cell adhesion. For instance, we showed here (Fig. 1) that, compared to ΔNSs, ZH infection negatively impacted Lgals3bp gene expression that codes for an extracellular matrix protein promoting integrin-mediated cell-matrix and cell-cell adhesion (47, 48). Also, while analyzing alterations in the host transcriptome following infection with the ZH strain of RVFV with respect to noninfected cells, Pinkham et al. (49) identified the interleukin-linked kinase (ILK) pathway as one of the main host pathways altered after infection. ILK connects the extracellular matrix to the actin cytoskeleton by interacting with the cytoplasmic domain of β integrins, regulating several functions related to actin cytoskeleton organization. Interestingly, the cytoplasmic tail of β3 integrin has been shown to directly interact with Abl2 (22), thus connecting Abl2 with ILK signaling and further establishing cell adhesion as a main biological processes targeted by RVFV.
In summary, we have shown here that infection with the virulent ZH strain of RVFV prevented the upregulation of the expression of Abl2, counteracted ΔNSs-induced inhibition of cell migration, affected cell shape, and induced the dissolution of adherens junctions. These effects were shown here to be dependent on the expression of NSs, which itself was shown to be present within long actin-rich structures involved in the dissemination of NSs to neighboring, non-NSs-expressing cells. These NSs-dependent effects on the host actin cytoskeleton are likely to participate in RVFV-induced pathogenicity by facilitating virulent RVFV dissemination, and thus could constitute interesting potential targets in future development of antiviral therapeutic strategies that, in the case of RVFV as with several other emerging arboviruses, are presently lacking.
MATERIALS AND METHODS
Virus and cells.
Stocks of RVFV ZH548, ZH548ΔNSs, and MP12 were produced under BSL3 conditions by infecting Vero cells at an MOI of 10−3 and harvesting the medium at 72 h p.i. Cell lines were cultured at 37°C under 5% CO2 in medium supplemented with 10 μg/ml penicillin/streptomycin (Gibco, Thermo Scientific) and 10% (vol/vol) fetal calf serum (FCS, Gibco). Murine fibroblastic L929 (ATCC) cells were grown in minimal essential medium (MEM) (Gibco). Murine hepatocyte, alpha mouse liver 12 (AML12) cells (ATCC ref. CRL-2254) were cultured in DMEM-F12 medium (Gibco) supplemented with 0.005 mg/ml insulin, 0.005 mg/ml transferrin, 5 ng/ml selenium (Sigma), and 40 ng/ml dexamethasone (Gibco, Thermo Scientific). Noninfected cells (NI) correspond to mock-infected cells that were treated as infected cells except that no virus was added. When indicated, antibody directed against the mouse IFN a/b receptor 1 (clone MAR1-5A3, BD Biosciences, no. 561183) was added directly to the medium 15 h before infection and kept in the medium during the entire time of infection. All infections were carried out at an MOI = 5 unless otherwise indicated.
Antibodies.
Primary antibodies used for immunofluorescence and/or Western blotting were mouse anti-NSs and rabbit anti-N polyclonal antibodies raised against the entire NSs or N protein of RVFV, respectively (9); anti-β-catenin monoclonal antibody (BD Transduction laboratories, catalog number 610154); anti-Abl2 (anti-Arg ref 07–262) from Merck Millipore and anti-GAPDH (ref 3781) from ProSci. Secondary antibodies used for immunofluorescence were Alexa fluor 488-conjugated chicken anti-mouse from Invitrogen (A21200) and Alexa fluor 555-conjugated donkey anti-rabbit from Invitrogen (A31572). Secondary antibodies used for Western blotting were ECL mouse IgG, horseradish peroxidase (HRP)-linked whole Ab (NA931) and ECL rabbit IgG, HRP-linked whole Ab (NA934) from GE Healthcare Life Sciences.
Immunofluorescence.
Cells grown in 6-well plates on coverslips were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized with 1% Triton X-100 in PBS for 20 min. The cells were incubated for 1 h at room temperature with the corresponding primary antibodies diluted in PBS-5% bovine serum albumin. After washing with PBS, the cells were next incubated for 45 min at room temperature with the corresponding secondary antibodies or phalloidin-FITC (Sigma P5282).
Image analysis and quantification.
Samples were analyzed at room temperature by confocal laser scanning microscopy using an AxioImager Z2 (Zeiss LSM710 confocal system). This system is equipped with a 63× lens and 1.4-numerical-aperture oil immersion lens (Plan Neofluor). For oil immersion microscopy, we used oil with a refractive index of 1.518 (Zeiss). Images were captured in the z-axis corresponding to the optical axis of the microscope at 0.37 μm intervals, with the z-axis going through the image planes. LSM Zen imaging software was used for image capture. The images were analyzed by the LSM5 Image browser or Image J software. All images are representative of a minimum of two independent infections for all conditions described in this work. The Analyze Particle plugin of Image J was use to analyze and quantify cell shape and area.
Cell transfection.
Plasmid pCI-NSs used for heterologous NSs protein expression contains the cDNA sequence of NSs from ZH548 RVFV strain (50). Twenty-four hours before transfection, AML12 cells were plated in 6-well plates at a density of 2 × 105 cells per well. Mix A, containing 2 μg of plasmid diluted in 50 μl of low fetal bovine serum Opti-MEM medium, was added to mix B containing 5 μl of Lipofectamine 2000 (ref 11668-027, Invitrogen) diluted in 195 μl of Opti-MEM and incubated at room temperature for 20 min before being dispensed into the well. Transfected cells were further incubated as previously described for 24 h before treatment.
Scratch wound healing assay.
Murine fibroblast L929 cells were plated on coverslips in 6-well plates at a density of 600,000 cells/well and cultured as previously described for 24 h to reach full confluence before scratch. Cell monolayers were scratched with a P10 pipette tip, incubated in fresh MEM medium supplemented with 10% fetal bovine serum (FBS) and treated as indicated in Fig. 3. After treatment, cells were fixed in 3.7% formaldehyde for 20 min. Images were collected at the indicated times under a wide-field microscope and quantitatively analyzed using the T-Scratch software (30).
RT-qPCR.
Total RNA was extracted using Tri Reagent (Sigma) according to the manufacturer’s protocol. One microgram of total RNA was reverse transcribed using a High Capacity cDNA Reverse transcription kit (Applied Biosystems) according to the manufacturers' recommendations using random primers. qPCR was performed using SYBR green (Thermo Fisher Scientific) reagents with the following sequence: 95°C for 15 min, then 40 cycles of 95°C 15 s, 60°C 30 s, 72°C 15 s, followed by a dissociation step. Relative quantification of mRNA expression was calculated using the threshold cycle (ΔΔCT) method using three reference genes among Ppib, Hprt1, Utp6c, and Rplp0. For fold changes lower than 1 (translating to a repression of the gene expression), values were set such that a (½) fold change corresponds to −2, a (¼) corresponds to −4 and so on. Sequences (5′–3′) of primers used for RT-qPCR analysis were as follows. Abl2 F: GAGCCACCGTTTTACATTGTGA and R: CTCGCCCACTAGGCAGTTC; Cdc42 F: CTTCTTCGGTTCTGGAGGCT and R: GGGATCTGAAGGCTGTCAAG; Fblim1 F: TAGCCGTGAGTGAGGAAGTG and R: CAGAGAGTGAGGCATTGGTCT; Hprt1 F: TCCTCCTCAGACCGCTTTT and R: CCTGGTTCATCATCGCTAATC; Ifnb1 F: ATGAACAACAGGTGGATCCTCC and R: AGGAGCTCCTGACATTTCCGAA; Irf7 F: CAGCGAGTGCTGTTTGGAGAC and R: AAGTTCGTACACCTTATGCGG; Lgals3bp F: TGCTGGTTCCAGGGACTCAA and R: CCACCGGCCTCTGTAGAAGA; Lpp F: GCAACCAAGAAGTCCGCAAC and R: GCCGTAGTATAGGAGGCTGGA; N protein of RVFV F: AAGGCAAAGCAACTGTGGAG and R: CAGTGACAGGAAGCCACTCA; NSs protein of RVFV F: GCACCTCCACCAGCGAAGCC and R: CCAGTGAGGGTTCTCCAAGAGGC; Oas1b F: GAGGTGCCGACGGAGGT and R: TCCAGATGAAGTCTTCCCAAAG; Pkp2 F: GCCGAGTGTGGCTACATCC and R: CTGCTGGTTCGGTGAAGGTT; Ppib F: GGAGATGGCACAGGAGGAAA and R: CCGTAGTGCTTCAGTTTGAAGTTCT; Psen1 F: ATACCTGCACCTTTGTCCTACT and R: GCTCAGGGTTGTCAAGTCTCT; Ptprf F: CTGCTCTCGTGATGCTTGGTT and R: ATCCACGTAATTCGAGGCTTG; Pvrl1 F: GACTCCATGTATGGCTTCATCG and R: CACTCGTTTCTCGTAGGGAGG; Rplp0 F: CACTGGTCTAGGACCCGAGAAG and R: GGTGCCTCTGGAGATTTTCG; Thbs1 F: GGGGAGATAACGGTGTGTTTG and R: CGGGGATCAGGTTGGCATT; Utp6c F: TTTCGGTTGAGTTTTTCAGGA and R: CCCTCAGGTTTACCATCTTGC; Vezt F: AACCCCGAACACTTGGGATG and R: GAAGGCTCGGCTATTGGCTG; Vtn F: CCCCTGAGGCCCTTTTTCATA and R: CAAAGCTCGTCACACTGACA.
Western blotting.
Total protein extracts were prepared from cells grown in 6-well plates in 250 μl/well of radioimmunioprecipitation assay (RIPA) lysis buffer containing phosphatase inhibitor PhoSTOP (Roche, 04 906 856 001) and protease inhibitor (Roche, 04 693 132 001). After centrifugation at 14,000 rpm for 10 min at 4°C, lysates were heated in Laemmli buffer and loaded in NuPAGE 4 to 12% sodium dodecyl sulfate-polyacrylamide precast gels (Life Technologie). Loaded proteins were transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were incubated overnight at 4°C with primary antibodies, washed with TBS-T (Tris base sodium Tween 0.1%) and incubated with the corresponding secondary antibodies for 1 h at room temperature. The imager ImageQuant LAS4000 was used for chemiluminescent protein detection. Relative quantification of proteins was carried out using Image J software.
ACKNOWLEDGMENTS
Agence Nationale de la Recherche (ANR) provided funding to Eliette Bonnefoy, Marie Flamand, and Jean-Jacques Panthier under grant number ANR-11-BSV3-007. The DIM-Malinf project from Région Ile-de-France provided funding to Vasco Marcato.
REFERENCES
- 1.Wright D, Kortekaas J, Bowden TA, Warimwe GM. 2019. Rift Valley fever: biology and epidemiology. J Gen Virol 100:1187–1199. doi: 10.1099/jgv.0.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikegami T. 2012. Molecular biology and genetic diversity of Rift Valley fever virus. Antiviral Res 95:293–310. doi: 10.1016/j.antiviral.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae:Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott RM, Brennan B. 2014. Emerging phleboviruses. Curr Opin Virol 5:50–57. doi: 10.1016/j.coviro.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreher F, Tamietti C, Gommet C, Guillemot L, Ermonval M, Failloux AB, Panthier JJ, Bouloy M, Flamand M. 2014. The Rift Valley fever accessory proteins NSm and P78/NSm-GN are distinct determinants of virus propagation in vertebrate and invertebrate hosts. Emerg Microbes Infect 3:e71. doi: 10.1038/emi.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikegami T, Won S, Peters CJ, Makino S. 2005. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J Virol 79:12106–12111. doi: 10.1128/JVI.79.18.12106-12111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J Virol 75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadani FZ, Kohl A, Préhaud C, Billecocq A, Bouloy M. 1999. The carboxy-terminal acidic domain of Rift Valley Fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J Virol 73:5018–5025. doi: 10.1128/JVI.73.6.5018-5025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Zhu X, Guan Z, Huang W, Zhang Y, Kortekaas J, Lozach PY, Peng K. 2019. NSs Filament Formation Is Important but Not Sufficient for RVFV Virulence In Vivo. Viruses 11:834. doi: 10.3390/v11090834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- 12.Le May N, Mansuroglu Z, Léger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog 4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. 2010. Nonstructural NSs protein of rift valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J Virol 84:928–939. doi: 10.1128/JVI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuerth JD, Weber F. 2016. Phleboviruses and the type I interferon response. Viruses 8:174. doi: 10.3390/v8060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol 83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed C, Steele KE, Honko A, Shamblin J, Hensley LE, Smith DR. 2012. Ultrastructural study of Rift Valley fever virus in the mouse model. Virology 431:58–70. doi: 10.1016/j.virol.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Benferhat R, Josse T, Albaud B, Gentien D, Mansuroglu Z, Marcato V, Souès S, Le Bonniec B, Bouloy M, Bonnefoy E. 2012. Large-scale chromatin immunoprecipitation with promoter sequence microarray analysis of the interaction of the NSs protein of Rift Valley fever virus with regulatory DNA regions of the host genome. J Virol 86:11333–11344. doi: 10.1128/JVI.01549-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peacock JG, Miller AL, Bradley WD, Rodriguez OC, Webb DJ, Koleske AJ. 2007. The Abl-related gene tyrosine kinase acts through p190RhoGAP to inhibit actomyosin contractility and regulate focal adhesion dynamics upon adhesion to fibronectin. Mol Biol Cell 18:3860–3872. doi: 10.1091/mbc.e07-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley WD, Koleske AJ. 2009. Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J Cell Sci 122:3441–3454. doi: 10.1242/jcs.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khatri A, Wang J, Pendergast AM. 2016. Multifunctional Abl kinases in health and disease. J Cell Sci 129:9–16. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zandy NL, Playford M, Pendergast AM. 2007. Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Proc Natl Acad Sci U S A 104:17686–17691. doi: 10.1073/pnas.0703077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Lyu W, Yu J, Koleske AJ. 2018. Abl2 is recruited to ventral actin waves through cytoskeletal interactions to promote lamellipodium extension. Mol Biol Cell 29:2863–2873. doi: 10.1091/mbc.E18-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torsello B, De Marco S, Bombelli S, Chisci E, Cassina V, Corti R, Bernasconi D, Giovannoni R, Bianchi C, Perego RA. 2019. The 1ALCTL and 1BLCTL isoforms of Arg/Abl2 induce fibroblast activation and extra cellular matrix remodelling differently. Biol Open 8:bio038554. doi: 10.1242/bio.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcato V, Luron L, Laqueuvre LM, Simon D, Mansuroglu Z, Flamand M, Panthier JJ, Souès S, Massaad C, Bonnefoy E. 2016. β-Catenin upregulates the constitutive and virus-induced transcriptional capacity of the interferon beta promoter through T-cell factor binding sites. Mol Cell Biol 36:13–29. doi: 10.1128/MCB.00641-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higashi Y. 1985. Changes of chromatin conformation around mouse interferon-beta gene associated with induction of interféron synthesis. Nucleic Acids Res 13:5157–5172. doi: 10.1093/nar/13.14.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubel P, Urban C, Bergant V, Schneider WM, Knauer B, Stukalov A, Scaturro P, Mann A, Brunotte L, Hoffmann HH, Schoggins JW, Schwemmle M, Mann M, Rice CM, Pichlmair A. 2019. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat Immunol 20:493–502. doi: 10.1038/s41590-019-0323-3. [DOI] [PubMed] [Google Scholar]
- 28.Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, Schreiber RD. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res 26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 29.Gebäck T, Schulz MM, Koumoutsakos P, Detmar M. 2009. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 30.Procaccia V, Nakayama H, Shimizu A, Klagsbrun M. 2014. Gleevec/imatinib, an ABL2 kinase inhibitor, protects tumor and endothelial cells from semaphorin-induced cytoskeleton collapse and loss of cell motility. Biochem Biophys Res Commun 448:134–138. doi: 10.1016/j.bbrc.2014.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Miller AL, Mooseker MS, Koleske AJ. 2001. The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc Natl Acad Sci U S A 98:14865–14870. doi: 10.1073/pnas.251249298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller AL, Wang Y, Mooseker MS, Koleske AJ. 2004. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J Cell Biol 165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu JJ, Rouse C, Xu X, Wang J, Onaitis MW, Pendergast AM. 2016. Inactivation of ABL kinases suppresses non-small cell lung cancer metastasis. JCI Insight 1:e89647. doi: 10.1172/jci.insight.89647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalveram B, Lihoradova O, Indran SV, Ikegami T. 2011. Using reverse genetics to manipulate the NSs gene of the Rift Valley fever virus MP-12 strain to improve vaccine safety and efficacy. J Vis Exp 57:e3400. doi: 10.3791/3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikegami T, Hill TE, Smith JK, Zhang L, Juelich TL, Gong B, Slack OA, Ly HJ, Lokugamage N, Freiberg AN. 2015. Rift Valley fever virus MP-12 vaccine is fully attenuated by a combination of partial attenuations in the S, M, and L segments. J Virol 89:7262–7276. doi: 10.1128/JVI.00135-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baer A, Austin D, Narayanan A, Popova T, Kainulainen M, Bailey C, Kashanchi F, Weber F, Kehn-Hall K. 2012. Induction of DNA damage signaling upon Rift Valley fever virus infection results in cell cycle arrest and increased viral replication. J Biol Chem 287:7399–7410. doi: 10.1074/jbc.M111.296608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Havranek KE, White LA, Lanchy JM, Lodmell JS. 2019. Transcriptome profiling in Rift Valley fever virus infected cells reveals modified transcriptional and alternative splicing programs. PLoS One 14:e0217497. doi: 10.1371/journal.pone.0217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jennings RT, Strengert M, Hayes P, El-Benna J, Brakebusch C, Kubica M, Knaus UG. 2014. RhoA determines disease progression by controlling neutrophil motility and restricting hyperresponsiveness. Blood 123:3635–3645. doi: 10.1182/blood-2014-02-557843. [DOI] [PubMed] [Google Scholar]
- 39.Gommet C, Billecocq A, Jouvion G, Hasan M, Zaverucha do Valle T, Guillemot L, Blanchet C, van Rooijen N, Montagutelli X, Bouloy M, Panthier JJ. 2011. Tissue tropism and target cells of NSs-deleted rift valley fever virus in live immunodeficient mice. PLoS Negl Trop Dis 5:e1421. doi: 10.1371/journal.pntd.0001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albe JR, Boyles DA, Walters AW, Kujawa MR, McMillen CM, Reed DS, Hartman AL. 2019. Neutrophil and macrophage influx into the central nervous system are inflammatory components of lethal Rift Valley fever encephalitis in rats. PLoS Pathog 15:e1007833. doi: 10.1371/journal.ppat.1007833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts KL, Manicassamy B, Lamb RA. 2015. Influenza A virus uses intercellular connections to spread to neighboring cells. J Virol 89:1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bracq L, Xie M, Benichou S, Bouchet J. 2018. Mechanisms for cell-to-cell transmission of HIV-1. Front Immunol 9:260. doi: 10.3389/fimmu.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labudová M, Čiampor F, Pastoreková S, Pastorek J. 2018. Cell-to-cell transmission of lymphocytic choriomeningitis virus MX strain during persistent infection and its influence on cell migration. Acta Virol 62:424–434. doi: 10.4149/av_2018_411. [DOI] [PubMed] [Google Scholar]
- 44.Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, Olivo-Marin JC, Männel D, Zurzolo C. 2009. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 45.Victoria GS, Zurzolo C. 2017. The spread of prion-like proteins by lysosomes and tunneling nanotubes: implications for neurodegenerative diseases. J Cell Biol 216:2633–2644. doi: 10.1083/jcb.201701047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patil S, Fribourg M, Ge Y, Batish M, Tyagi S, Hayot F, Sealfon SC. 2015. Single-cell analysis shows that paracrine signaling by first responder cells shapes the interferon-β response to viral infection. Sci Signal 8:ra16. doi: 10.1126/scisignal.2005728. [DOI] [PubMed] [Google Scholar]
- 47.Loimaranta V, Hepojoki J, Laaksoaho O, Pulliainen AT. 2018. Galectin-3-binding protein: a multitask glycoprotein with innate immunity functions in viral and bacterial infections. J Leukoc Biol 104:777–786. doi: 10.1002/JLB.3VMR0118-036R. [DOI] [PubMed] [Google Scholar]
- 48.Hepojoki J, Strandin T, Hetzel U, Sironen T, Klingström J, Sane J, Mäkelä S, Mustonen J, Meri S, Lundkvist A, Vapalahti O, Lankinen H, Vaheri A. 2014. Acute hantavirus infection induces galectin-3-binding protein. J Gen Virol 95:2356–2364. doi: 10.1099/vir.0.066837-0. [DOI] [PubMed] [Google Scholar]
- 49.Pinkham C, Dahal B, de la Fuente CL, Bracci N, Beitzel B, Lindquist M, Garrison A, Schmaljohn C, Palacios G, Narayanan A, Campbell CE, Kehn-Hall K. 2017. Alterations in the host transcriptome in vitro following Rift Valley fever virus infection. Sci Rep 7:14385. doi: 10.1038/s41598-017-14800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]