Persons with HIV infection are frequently coinfected with chronic herpesviruses, which periodically replicate and produce viable herpes virions, particularly in anogenital and cervical tissues. Persistent protein expression results in proliferation of CD8+ and CD4+ T cells, and the latter could potentially expand and sustain HIV tissue reservoirs. We found HSV genital shedding rates were positively correlated with HIV DNA concentrations and HIV divergence from ancestral sequences in tissues. Our work suggests that immune responses to common coinfections, such as herpesviruses, may sustain HIV tissue reservoirs during suppressive ART, suggesting future cure strategies should study interventions to suppress replication or reactivation of chronic herpes infections.
KEYWORDS: human immunodeficiency virus, HIV, herpes simplex virus 2, HSV-2, coinfection, female genital tract, HIV tissue reservoirs, ART
ABSTRACT
Antigen (Ag)-specific immune responses to chronic infections, such as herpes simplex virus type 2 (HSV-2) in HIV/HSV-coinfected persons, may sustain HIV tissue reservoirs by promoting T-cell proliferation but are poorly studied in women on antiretroviral therapy (ART). Mixed anogenital swabs and cervical secretions were self-collected by nine HIV/HSV-2-coinfected women during ART for 28 days to establish subclinical HSV DNA shedding rates and detection of HIV RNA by real-time PCR. Typical herpes lesion site biopsy (TLSB) and cervical biopsy specimens were collected at the end of the daily sampling period. Nucleic acids (NA) isolated from biopsy specimens had HIV quantified and HIV envC2-V5 single-genome amplification (SGA) and T-cell receptor (TCR) repertoires assessed. Women had a median CD4 count of 537 cells/μl (IQR: 483 to 741) at enrollment and HIV plasma viral loads of <40 copies/ml. HSV DNA was detected on 12% of days (IQR: 2 to 25%) from anogenital specimens. Frequent subclinical HSV DNA shedding was associated with increased HIV DNA tissue concentrations and increased divergence from the most recent common ancestor (MRCA), an indicator of HIV replication. Distinct predominant TCR clones were detected in cervical and TLSB specimens in a woman with frequent HSV DNA shedding, with mixing of minor variants between her tissues. In contrast, more limited TCR repertoire mixing was observed in two women with less frequent subclinical HSV DNA shedding. Subclinical HSV shedding in HIV/HSV-coinfected women during ART may sustain HIV tissue reservoirs via Ag exposure or HIV replication. This study provides evidence supporting further study of interventions targeting suppression of Ag-specific immune responses as a component of HIV cure strategies.
IMPORTANCE Persons with HIV infection are frequently coinfected with chronic herpesviruses, which periodically replicate and produce viable herpes virions, particularly in anogenital and cervical tissues. Persistent protein expression results in proliferation of CD8+ and CD4+ T cells, and the latter could potentially expand and sustain HIV tissue reservoirs. We found HSV genital shedding rates were positively correlated with HIV DNA concentrations and HIV divergence from ancestral sequences in tissues. Our work suggests that immune responses to common coinfections, such as herpesviruses, may sustain HIV tissue reservoirs during suppressive ART, suggesting future cure strategies should study interventions to suppress replication or reactivation of chronic herpes infections.
INTRODUCTION
HIV-1 (HIV) has been “cured” in two patients who received intensive chemotherapeutic regimens to treat concomitant cancer diagnoses and allogeneic hematopoietic stem cell transplant from CCR5delta32/delta32 donors, rendering newly engrafted CD4+ T cells resistant to HIV infection (1, 2). However, other patients with HIV infection and cancer treated with both high-intensity (3) and reduced-intensity conditioning followed by allogeneic transplant without CCR5delta32/delta32 donors have experienced HIV rebound (4), confirming that poorly defined HIV reservoirs are likely a barrier to a cure. Mechanisms that drive HIV persistence are poorly understood, but have largely focused on homeostatic proliferation (5) and integration into cell proliferation-related genes as mechanisms that promote HIV persistence (6, 7), with no in vivo studies examining whether antigen (Ag)-specific immune responses to chronic infections impact HIV tissue reservoirs during antiretroviral therapy (ART) for HIV infection.
Chronic herpes simplex virus (HSV) infections are prevalent in persons with HIV infection (65 to 85% of HIV-infected persons) (8–11), and episodically reactivate in tissues throughout a participant’s lifetime, even during suppressive ART for HIV (12–14). While genital HSV-2 clinical recurrences are less frequent after the first year of infection (15), clinical recurrences still occur in many individuals, and HSV DNA subclinical shedding is relatively frequent (20 to 29% of days) when participants are sampled daily (16, 17). Both clinical recurrences and subclinical shedding are accompanied by robust CD4+ and CD8+ Ag-specific cellular expansion (18–23), which persist for at least 2 weeks (20). In response to HSV-2, Ag-specific CD4+ T cells increase by approximately 10-fold (20) in tissues, compared to the slower and steadier T cell accumulation associated with homeostatic proliferation (24). We hypothesize that immune responses to HSV replication in HIV/HSV-coinfected persons might increase or sustain CD4+ T cells with integrated HIV. We undertook a pilot study to evaluate whether immune responses to HSV subclinical shedding affect HIV tissue reservoirs among women receiving suppressive ART for HIV.
RESULTS
Participant characteristics and study design.
Ten women with HIV/HSV-2 coinfection were enrolled in this pilot study (Fig. 1). One woman declined to collect any swabs after enrollment and was excluded from further analysis; 9/10 completed the entire study protocol. Nine participants had HIV RNA monitored throughout the study with HIV plasma viral load suppressed (<40 copies/ml) by three active antiretroviral medications (Table 1). Women were free from genital coinfections (trichomonas, gonorrhea, chlamydia, or yeast) and cervical dysplasia at enrollment. All women had chronic HIV infection with a median 13 years postdiagnosis (interquartile range [IQR]: 7 to 20 years), a median age of 48 years (IQR: 46 to 54), and a median CD4 count of 554 (IQR: 492 to 723) cells/μl (Table 1).
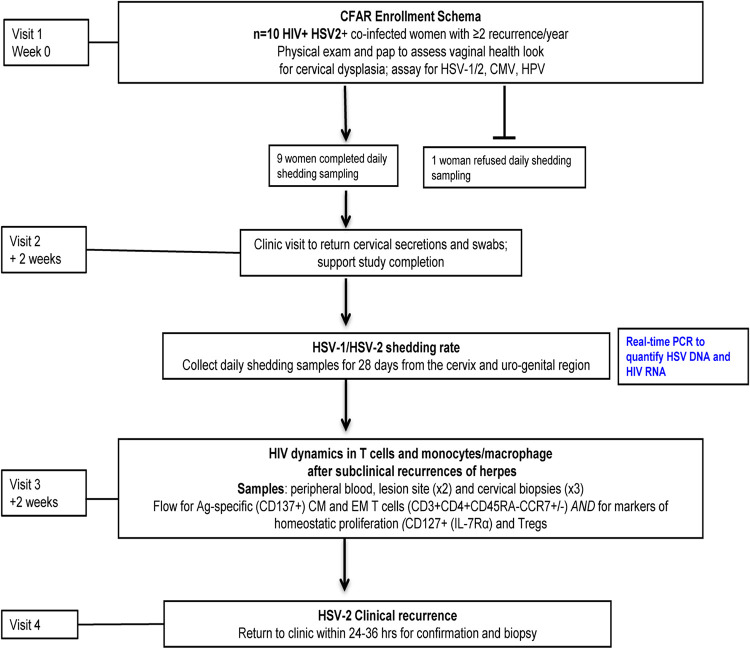
FIG 1.
Study design for the Center for AIDS Research (CFAR) pilot project. HIV/HSV-coinfected women receiving antiretroviral therapy (ART) for suppression of HIV replication with a history of ≥2 HSV-2 recurrences/year were enrolled in this pilot study. Ten women were enrolled in the current study and one woman declined to collect the 28-day at-home sampling and was not included in the later analyses.
TABLE 1.
Clinical characteristics of women with HIV and HSV-2 coinfection during antiretroviral therapy
| Participant ID | CD4 cell count | Age at enrollment (yrs) | Yrs since HIV diagnosis (Dx) | HLA typea | Antiretroviral therapyb | HSV shedding rate (no. days HSV detected/total days sampled [%])c |
|---|---|---|---|---|---|---|
| 1 | 688 | 48 | 24 | ND | DTG, 3TC, ABC | 0/27 (0) |
| 2 | 520 | 54 | 16 | ND | EFV, FTC, TDF | 1/28 (4) |
| 3a | 758 | 37 | 5 | A*02:01 A*03:01; B*35:01 B*44:01; C*05:01 C*04:01 | DTG, FTC, TDF | 3/29 (10) |
| 4a | 510 | 54 | 13 | A*03:01 A*29:01; B*07:05 B*40:01; C*03:04 C*15:05 | DTG, FTC, TDF | 4/28 (14) |
| 5 | 554 | 45 | 1 | ND | DTG, FTC, TDF | 4/26 (15) |
| 6a | 647 | 47 | 9 | A*02:01 A*03:01; B*07:02 B*53:01; C*04:01 C*07:02 | EVG, COBI, FTC, TDF | 6/33 (18) |
| 7 | 241 | 48 | 24 | ND | EFV, ABC, MVC | 8/28 (29) |
| 8 | 474 | 54 | 10 | ND | DTG, FTC, TDF | 11/36 (31) |
| 9 | 1538 | 58 | 13 | ND | EFV, FTC, TDF | 10/28 (35) |
Three women with sufficient DNA had DNA sent to Adaptive technologies for survey-level deep sequencing of their TCR repertoire in paired cervical and anogenital biopsy specimens after at-home 28-day shedding visit (V3) and, in one woman (PID number 6), from the cervix at the time of her clinical recurrence (V4). HLA, human leukocyte antigen; ND, not done.
ABC, abacavir; COB, cobicistat; DTG, dolutegravir; EFV, efavirenz; EVG, elvitegravir; FTC, emtricitabine; MVC, maraviroc; TDF, tenofovir disoproxil fumarate; 3TC, lamivudine.
HSV shedding rate was determined by daily sampling from the anogenital region using a swab that was quantified using an HSV real-time PCR method (64).
HSV DNA subclinical shedding rates and HIV detection.
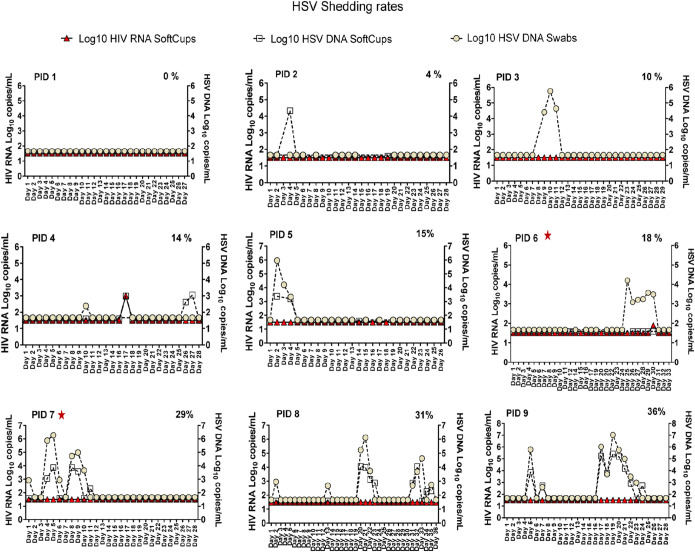
Women collected a median of 28 days of mixed anogenital swabs (IQR: 28 to 31 days). Eight of nine participants (89%) had HSV DNA shedding detected on at least one day, with a median HSV anogenital shedding rate of 12% of days (IQR: 2% to 25%) (Fig. 2). Overall, there were 27/264 days (10%) when HSV DNA was detected from cervicovagional secretions (SoftCup) and 38/264 days (14%) when HSV DNA was detected in the anogenital swabs. Concurrent HSV DNA shedding at both sites was detected in 23/264 (9%) of days. Two participants with relatively high subclinical HSV DNA shedding rates (% shedding frequency: participant identification [PID] 4, 14%; PID 6, 18%) had HIV RNA detected at relatively low concentrations, 1.9 and 3.0 log10 copies/ml, respectively, both with concurrent HSV DNA shedding (Fig. 2); also, each had a clinical recurrence detected 38 and 50 days, respectively, after their visit three (V3).
FIG 2.
Establishing herpes simplex virus DNA shedding rates from 28 days of self-collected anogenital specimens during ART for HIV. Anogenital secretions were self-collected using Dacron swabs (Fisher Scientific, Waltham, MA) (22) for a median 28 days. Specimens were reconstituted with a QIAamp 96 DNA blood kit and eluted into 100 μl of AE buffer (Qiagen, Germantown, MD). Cervical secretions were collected using a SoftCup that was designed to collect menstrual specimens worn by each participant for a median of 8 h. Left y axis shows HIV RNA log10 concentrations/ml and the right axis shows the HSV DNA log10 concentrations/ml corrected for the dilution of each specimen; the x axis shows the day of sampling indicated as day 1 up to day 36. A range of HSV DNA shedding was observed across participants and used as a predictor in subsequent analyses to evaluate the association between HSV DNA shedding. The participant identification (PID) number is indicated in the upper left of each graph. The frequency of HSV DNA shedding detected/total days sampled is presented as a percentage in the upper right side of each graph. Red stars indicate the two women with clinical recurrences identified within 38 to 50 days of the V3 biopsy specimens.
Quantification of participant tissue HIV DNA stratified by HSV DNA shedding rates.
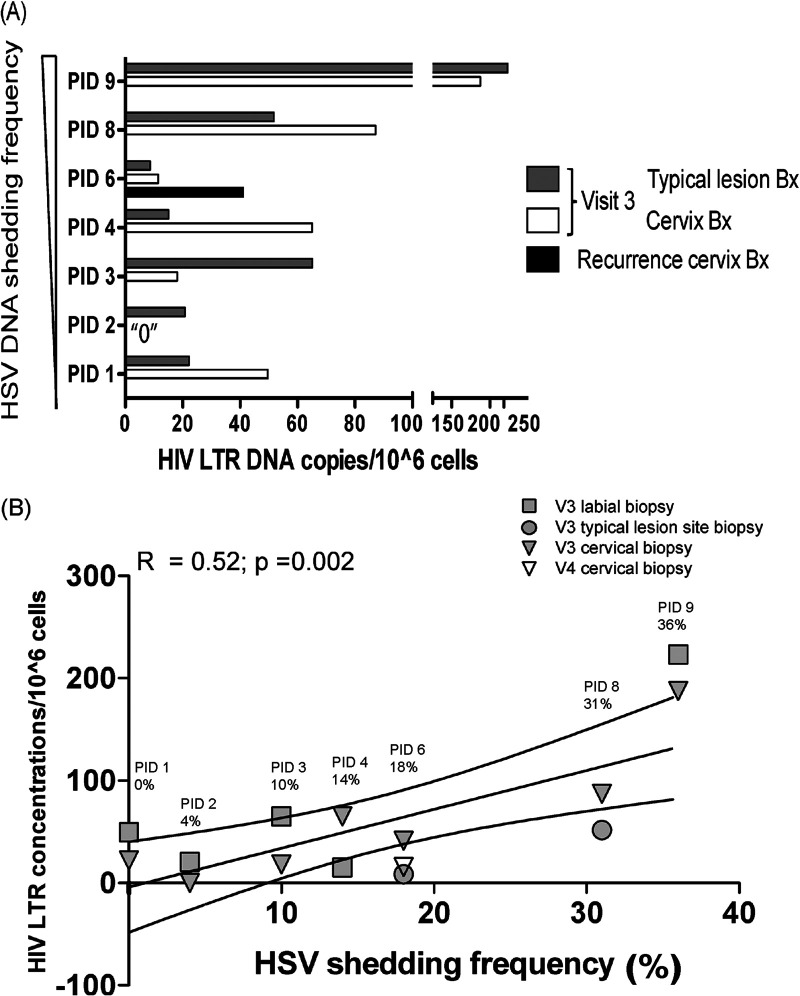
HIV DNA was quantified at V3 from 6/9 cervical specimens, 7/9 typical lesion site biopsy (TLSB) specimens, and from the actual lesion site biopsy (ALB) (V4 clinical recurrence) specimen when sample was sufficiently abundant. Women with higher HSV DNA shedding frequency had increased HIV DNA concentrations (median [IQR]: TLSB 1.92 [0.94 to 2.35] log10 copies/106 cells; cervix 1.94 [1.06 to 2.27] log10 copies/106 cells) compared to those with low frequency HSV shedding (median [IQR]: TLSB 1.32 [1.22 to 1.74] log10 copies/106 cells; cervix 1.53 [0.65 to 1.79] log10 copies/106 cells) (Fig. 3A). Across tissues, HSV shedding frequency was positively correlated with HIV DNA concentrations (R = 0.52, P = 0.002) in a simple linear regression (Fig. 3B). However, this was largely driven by participant 9; after her data were excluded, the correlation was diminished (R = 0.18, P = 0.14). In a regression analysis that accounted for repeat measurements, we observed a 4.8% increase in HIV DNA concentrations for every percentage increase in HSV DNA shedding (geometric mean = 1.048; 95% confidence interval [CI]: 1.010 to 1.088; P = 0.01). One of two participants (PID 6) with a clinical recurrence had an 8-fold increase in HIV DNA concentrations at her cervix compared with her V3 cervical biopsy specimen; however, insufficient nucleic acids (NA) were available to evaluate HIV DNA concentrations from her ALB (Fig. 3A).
FIG 3.
Higher HSV shedding rates during ART are associated with increased HIV DNA concentrations in tissues. HIV nucleic acids (NA) were isolated from typical lesion site biopsy (TLSB) specimens and uterine cervical biopsy specimens at the 28-day postshedding visit (V3) and at clinical recurrence (V4) for a single participant. HIV RNA and HSV DNA were quantified using a real-time PCR assay from anogenital swabs and cervical secretions. (A) HIV DNA concentrations were stratified by HSV DNA shedding frequency and are presented as the quantity of HIV LTR DNA/10̂6 cells. (B) Linear regression using HSV shedding rate as the predictor shows a positive correlation with HIV DNA concentrations. Symbols indicate the tissue and the time point sampled: gray squares, V3 labial biopsy specimens; gray circles, V3 TLSB specimens; gray triangles, V3 cervical biopsy specimens; open triangles, V4 clinical recurrence. The three lines represent the mean and the 95% confidence interval of the regression line.
HIV DNA sequences from genital tissues show increased HIV divergence from the most recent ancestor in women with higher subclinical HSV DNA shedding compared to those with lower HSV shedding rates.
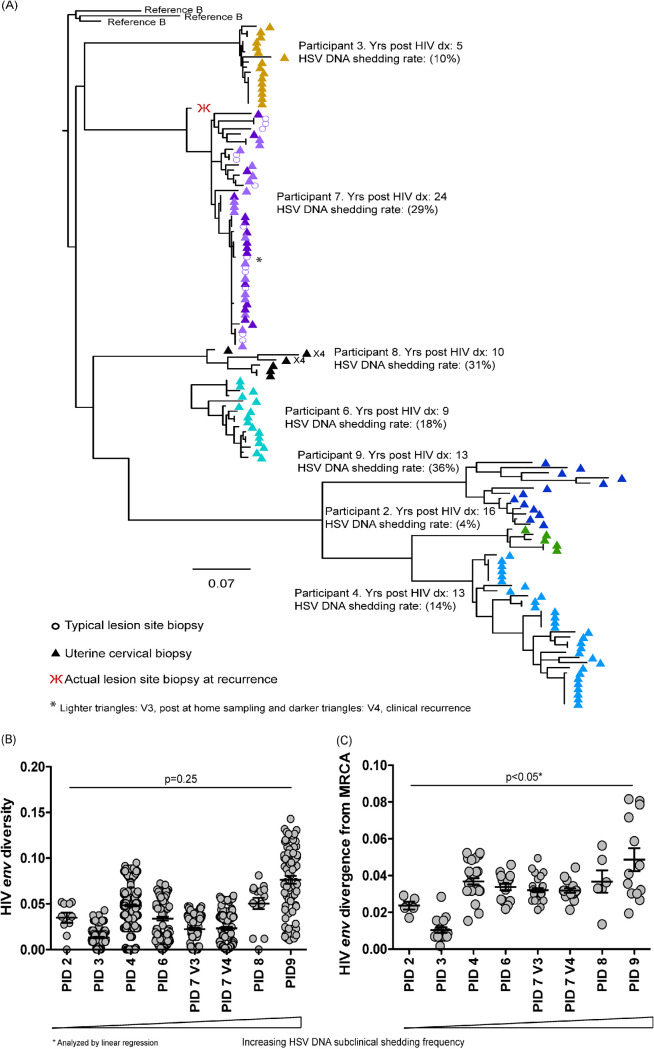
Uterine cervical biopsy specimens from 7/9 participants had sufficient NA to submit for single genome amplification (SGA) of HIV envC2-V5, and from one woman’s ALB (PID 7). All sequences were present in participant-specific clades (Fig. 4A). One woman had two HIV sequences predicted to use X4, but all other sequences were predicted to use R5 (25). When women were stratified by HSV shedding rate, we observed variable changes in HIV env diversity across HSV shedding rates that were not statistically significant (P = 0.25) (Fig. 4B). In contrast, women with higher HSV DNA shedding frequency had significantly higher (P < 0.05) HIV divergence from the most recent common ancestor (MRCA) compared to those with lower HSV shedding rates (Fig. 4C). Similar to the analysis of the HIV DNA concentrations, this was driven by the lower divergence in two participants (PID 2 and 3); when these data were removed from the analysis, there was no longer a significant difference in the remaining participants (P > 0.05).
FIG 4.
Higher HSV shedding is associated with increased divergence from the most common recent ancestor (MRCA). Nucleic acids from typical lesion site (TLSB) and uterine cervical biopsy specimens at V3 and for one clinical recurrence (at V4) were submitted for single genome amplification (SGA) of HIV envC2-V5 and aligned in Geneious. (A) A phylogenetic tree displaying sequences from all seven participants with successful HIV SGA were rooted to Reference B sequences. Most participants’ sequences were generated from their cervical tissues at V3 (participants 2, 3, 4, 6, 8, and 9) and are represented as different colored triangles by participant. Participant 7 has sequences amplified from her TLSB and cervical tissues from V3 represented by light colored purple triangles for her cervical sequences and by open light purple circles for her TLSB sequence; her clinical recurrence cervical sequences are represented as the darker purple triangles with one HIV DNA sequence from her actual lesion site biopsy specimen (ALB) (red psi symbol). Sequences were rooted using representative sequences for subtype B from the GenBank database (clade B: B.US.JRFL, B.US.90.WEAU160, B.FR.83.HXB2). The scale bar (horizontal line) indicates the number of substitutions per site. (B and C) Phylogenetic pairwise distance analyses were generated in DIVEIN (25) and diversity (B) and divergence (C) data were visualized using GraphPad Prism (GraphPad).
HIV envC2-V5 sequences were successfully amplified from one woman’s skin biopsy specimen (PID 7) and from 7/7 cervical biopsy specimens (Fig. 4A). For this patient, the TLSB sequences were often identical to her cervical sequences at V3 and V4 (large lower clade), with the two upper clades showing more diversity with similar mixing between tissues and between visits (Fig. 4A). The ALB (V4) specimen had lower quantity NA, but we amplified a single HIV envC2-V5 that clearly clustered with her other sequences (Fig. 4A); however, it was on a distant branch relative to her V3 TLSB and cervical sequences. This sequence was generated from a labial biopsy specimen, whereas her V3 TLSB specimen was taken from her right buttocks, which could account for the increased genetic distance.
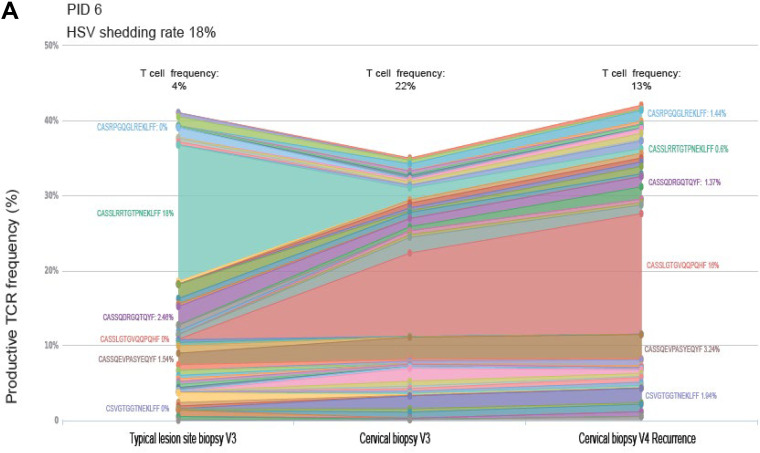
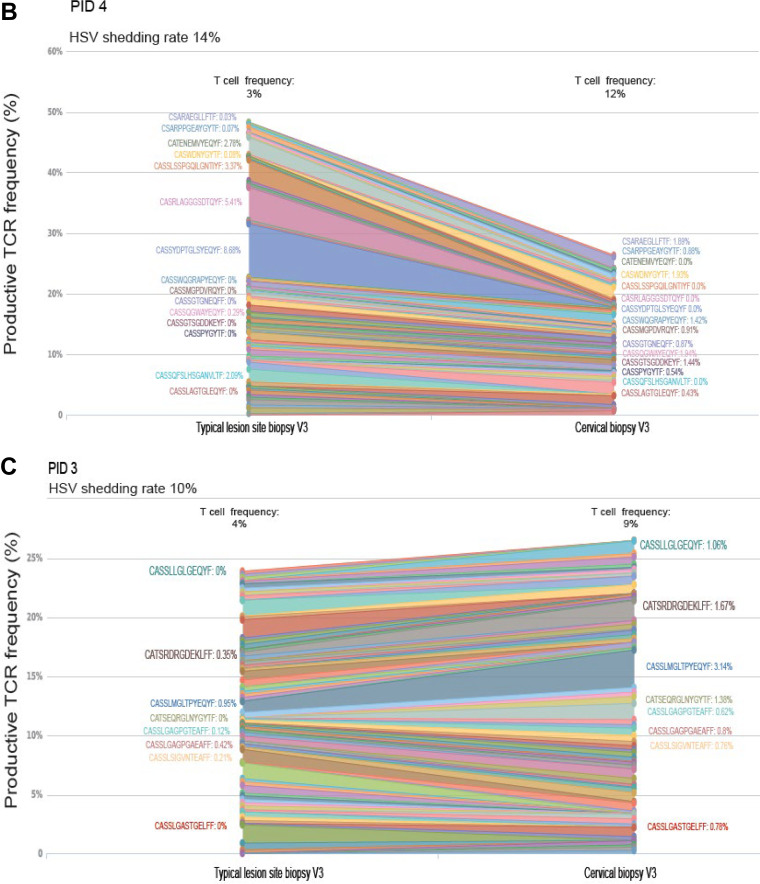
Predominant TCR clones are detected in participants with higher HSV shedding frequencies, with minor variants mixing between skin and cervix, whereas women with low HSV DNA shedding had less mixing of TCR clones between tissues.
Paired TLSB and cervical biopsy specimen NA were available from 3/7 participants for evaluation of T-cell receptor (TCR) repertoires as an exploratory analysis. More mixing of TCR clonotypes between TLSB and cervix was observed in one woman with more frequent HSV DNA shedding rate (PID 6; HSV shedding 18% of days), compared to those with lower HSV DNA shedding rates (PID 4 and 3) (see Data Set S1 in the supplemental material). In PID 6, unique clonotypes were detected in both TLSB and cervical biopsy specimens from V3 and V4 at a frequency of 24.9% or 26.0% during quiescence and at clinical recurrence, respectively. In PID 3 and 4, in comparison, the fraction of overlapping clonotypes was slightly lower (20.8 and 22.1%) (Fig. 5).
FIG 5.
T-cell receptor (TCR) β chain sequencing shows increased mixing between tissues in women with more frequent subclinical HSV DNA shedding compared with women with lower shedding and are positively correlated within the uterine cervix over time. Three women had nucleic acids (NA) extracted from paired typical lesion site biopsy (TLSB) specimens and cervical biopsy specimens. (A) PID 6 had higher HSV DNA shedding frequency (18%) and a clinical recurrence. A predominant TCR clone was detected at her V3 in her TLSB specimen that is distinct from the predominant clone detected in her cervical biopsy specimen from the same visit, which expanded as a predominant clone in her cervix at her clinical recurrence visit sampled 5 weeks later (V4), although we were unable to evaluate whether this clone was detected in her actual lesion site biopsy specimen (ALB) at the time of her clinical recurrence. Mixing of minor TCR clones was detected in both her TLSB and cervical tissues at V3. (B and C) Two women without recurrences with relatively lower shedding rates (14% [B] and 10% [C]) showed minimal mixing between TLSB and cervical biopsy specimens, with each having smaller predominant TCR clones. T-cell frequencies are indicated above each study visit and represent the T cell estimate in the sample normalized to the number of templates in the DNA specimen. (D) TCR repertoires are positively correlated within the uterine cervix over time in a woman with higher HSV DNA shedding frequency, with little correlation or overlap between the uterine cervix and skin biopsy specimens regardless of HSV DNA shedding rate. Three women had nucleic acids (NA) extracted from paired TLSB and cervical biopsy specimens. One woman with higher HSV DNA subclinical shedding rates had a positive correlation between TCR repertoires in her uterine cervix across study visits, with less correlation between her tissue and cervical biopsy specimens at her V3 post-28-day sampling visit. Women with lower HSV DNA shedding had little correlation between their tissue and cervical biopsy specimens. Assessment of specimen overlap, as assessed by the Morisita Index (Adaptive Immunoseq Analyzer), confirm there is a high specimen overlap within the uterine cervical biopsy specimens for a woman between her V3 and V4 recurrence visits, but with less overlap between the cervix and TLSB specimens. In contrast, women with lower subclinical HSV DNA shedding have relatively low Morisita’s scores, suggesting there is less mixing between these two tissues.
The TCR repertoires from TLSB and cervix specimens in PID 6 each contained a single unique predominant TCR clonotype representing up to 20% of her repertoire. In her TLSB specimen, CASSLRRTGTPNEKLFF represented 18% of detected clonotypes, but was a minor variant in her cervical biopsy specimen at the same visit (Fig. 5A and Data Set S1 in the supplemental material). In her cervical biopsy specimen at V3, a predominant TCR clone (CASSLGTGVQQPQHF; 11% of repertoire) was detected that expanded to represent 16% of the clones detected during clinical recurrence (V4). This clone was also detected, although at low frequency (<1%), in her TLSB specimen at V3. She had minor TCR clonotypes (<5%) detected at her V3 in both TLSB and cervical biopsy specimens (CASSQEVPASYEQYF and CASSQDRQTQYF) that remained detectable in her cervical biopsy specimen at her clinical recurrence (Fig. 5B and Data Set S1). A smaller predominant clone (CASSYDPTGLSYEQYF; 9% of repertoire) was detected from PID 4 TLSB that was not detected in her cervical biopsy specimen at the same visit. PID 3 had multiple low-frequency TCR clonotypes (≤3%) detected in her TLSB specimen that were either absent or at very low frequency in her cervical biopsy specimen (Fig. 5C and Data Set S1). When comparing the frequency of overlapping clonotypes between biopsy specimens, there was a strong correlation noted within the TCR clonotypes over time in the uterine cervix, with little correlation or overlap (Morisita’s index = ≤0.25) in the TCR frequencies between the TLSB and cervical biopsy specimens regardless of subclinical HSV DNA shedding frequency (Fig. 5D).
Although in silico technology is rapidly evolving, it is not possible to determine the antigen specificity of a CDR3 in the absence of paired TCRα/β sequencing (requiring single-cell sequencing) and either direct or synthetic TCR antigen testing was beyond the scope of the current study. Thus, published TCRB sequence databases were queried for matching CDR3 sequences with in vitro-verified specificities associated with at least one of a participant’s HLA haplotypes. PID 6 had 2 to 3 TCRB clonotypes that matched published CDR3 sequences specific for HIV Gag, HSV, cytomegalovirus (CMV), Epstein-Barr virus (EBV), and influenza (Data Set S1); however, none of these were among the expanded clones identified in our current study.
CD4+ effector memory and antigen-specific T cell frequencies are more prevalent in uterine cervical cytobrush specimens compared to PBMC.
Uterine cervical cytobrush and peripheral blood mononuclear cell (PBMC) specimens were available from enrollment and V3 in 6/9 women. T cell subsets from blood were compared to the corresponding T cell subset in the cervical cytobrush sample at enrollment and at V3 (post 28-day daily sampling) to determine whether more potential T-cell reservoir targets were detected in tissues compared to blood. Naïve CD3+ CD4+ T cells (CD45RA+ CCR7+) were higher in PBMCs compared to cervical tissues at V3 (P < 0.05), but not at the enrollment or at clinical recurrence (Fig. 6). Central memory (CM) T cells (CD4+ CD45RA− CCR7+) were similar between the two tissues. In contrast, effector memory (EM) T cells (CD4+ CD45RA− CCR7−) were significantly increased in cervical tissues compared with PBMCs at all study visits (P < 0.001). Terminally differentiated CD3+ CD4+ T cells (CD45RA+ CCR7−) were low in blood and tissues and CD3+ CD4+ Treg cells (CD25+ CD127[low]) were generally more prevalent in the cervical tissues, but not significantly (Fig. 6). Lastly, CD3+ CD4+ CD137+ (Ag-specific) T cells were significantly increased in cervical tissues at V3 compared to PBMCs (P = 0.05); and were increased at times of clinical recurrence, although this did not reach statistical significance.
FIG 6.
Effector memory T cells and antigen-specific (CD137+) CD4+ cells are increased in tissues. Cytobrush and PBMC samples were collected at enrollment and at the 28-day postshedding visit (V3). Six paired PBMC and cervical cytobrush specimens were available for flow cytometry analysis of T cell frequencies. CD4+ effector memory (EM) cells were consistently significantly increased in the uterine cervix compared to the same population from PBMCs. Of note, Ag-specific T cells (detected as CD137+ cells) were more frequent in the CD3+ CD4+ cervical tissues compared to the PBMCs at V3.
The number of observations for T cell frequencies was small, so when the T cell frequencies were compared across HSV shedding frequency, there were no significant correlations across T cell populations (data not shown).
DISCUSSION
The current study, to our knowledge, is the first to evaluate “directly ex vivo” associations between subclinical genital HSV shedding and HIV reservoirs in the female genital tract among HIV/HSV-coinfected women during suppressive ART for HIV. Specifically, we evaluated HIV DNA reservoir size, HIV populations, and TCR repertoire clonotypes in two distinct mucosal tissues stratified by HSV subclinical shedding frequencies along with T cell populations in blood and cervical cytobrush specimens. We present several novel observations as follows: Women with higher subclinical HSV DNA shedding had (i) higher HIV DNA concentrations at mucosal tissues and (ii) significantly increased HIV DNA divergence from the MRCA in uterine cervical specimens. Additionally, we (iii) identified predominant TCR clones in TLSB specimens that were distinct from predominant TCR clonotypes detected in the uterine cervix, and that expanded during a clinical recurrence which potentially represents Ag-specific cellular proliferation. (iv) The women with higher HSV DNA shedding had more mixing of TCR clones between the TLSB and uterine cervical biopsy specimens compared to women with lower shedding frequency. Lastly, (v) there were increased EM- and Ag-specific CD4+ T cells in uterine cervix cytobrush specimens compared to blood, which may be cells associated with HIV persistence.
HSV DNA subclinical shedding was used as a surrogate for Ag stimulation that potentially increases HIV tissue reservoir size during suppressive ART for HIV. This study represents a novel in vivo approach to evaluate HIV tissue reservoirs in the context of a chronic herpes infection. Subclinical HSV DNA shedding rates reported here were slightly lower across women than reported for participants solely HSV infected and not on ART (11, 17, 22, 26), but were higher than in other studies of HIV-infected persons on ART (27). HSV shedding rate has been shown to be a surrogate marker for HSV clinical disease activity in persons without HIV infection (28); in this study, the women with the higher HSV shedding rates also had clinical recurrences during the follow-up period. While there are other chronic human herpesvirus coinfections that likely contribute to maintaining HIV reservoirs (29, 30), we chose to evaluate HSV coinfection because participants can be trained to identify clinical recurrences at mucosal sites and return to the clinic for biopsy of the ALB and cervical tissues (20, 22, 31). Despite the small number of women enrolled in this pilot project, we were able to detect increases in HIV tissue concentrations that were associated with higher HSV shedding rates, which should be evaluated in larger studies to confirm our findings.
HIV DNA concentrations in mucosal tissues were significantly increased in women with more frequent HSV DNA shedding compared to those with lower HSV DNA shedding. While our HIV long terminal repeat (LTR) real-time PCR assay likely overestimates the actual size of replication-competent HIV (32), we nevertheless found a positive correlation between HIV DNA concentrations and HSV subclinical shedding, and noted an 8-fold increase in samples from one woman’s cervical tissue during clinical HSV recurrence. These data represent an important first step toward improving our understanding of the impact of herpes coinfections on HIV tissue reservoirs, which has not been studied during suppressive ART for HIV in women. HIV DNA reservoir size has been reported to have slower decay in participants with active cytomegalovirus (CMV) replication in men on suppressive ART for HIV (29, 33), and more recently confirmed in a study of chronically HIV-infected persons coinfected with CMV during suppressive ART (30). These studies focused on HIV reservoir size in peripheral blood, whereas we evaluated genital tissues in HIV/HSV-coinfected women, a potentially important site for HIV transmission but also a source of HIV reservoirs, and detected similar increases in HIV DNA (34).
An unexpected result from this study was that HIV divergence from the most common recent ancestor (MRCA) was increased in women with higher HSV DNA shedding. While divergence may be a phylogenetic indicator of HIV replication (34–36), HIV does not typically replicate during suppressive ART and modern therapies do not allow for full cycles of replication (37, 38). Thus, it is unclear whether HSV replication promotes HIV replication. A few studies allow us to speculate about possible mechanisms that would allow HIV replication. First, HSV replication may decrease availability of tenofovir (TDF) and other antiretrovirals, as suggested by in vitro studies (39–41). Tenofovir, a medication taken by a majority (75%) of women in our study (Table 1), penetrates well into the female genital tract (FGT) (42). Both viruses’ polymerases interact with TDF, which might decrease local concentrations and allow both viruses to replicate (43, 44). A second potential mechanism might be that expression of HSV proteins (ICP0, ICP4, and ICP27) promote HIV “replication” (45–47), presumably via activation of NF-κB (48). Another in vitro study demonstrated that herpes replication promoted dendritic cell (DC) anergy, resulting in the release of inflammatory cytokines (IL-6, IL-10, and TNF-α) (49), which can activate HIV LTRs (50, 51) and lead to production of HIV RNA. In the current study, we did not assay for p24, drug concentrations, or cytokines at time points where TLSB or cervical biopsy specimens were collected. However, it is of interest to note that we were able to detect HIV RNA during the daily at-home sampling period in two women, although infrequently. However, detection of HIV RNA could also simply be the result of HIV RNA expression.
Participants in the current study had variable changes in HIV diversity across HSV DNA shedding rates. The differences in HIV diversity were likely not significant due to the small number of women evaluated in this cohort. Previous studies of other human herpesviruses, such as CMV, and HIV coinfection have not included HIV phylogenetic analysis to determine whether CMV replication diversifies the HIV reservoir (30). Recent work in a larger cohort examined molecular diversity and showed increases in HIV diversity associated with CMV and Epstein-Barr virus (EBV) shedding (33). The large variation in HIV diversity by HSV shedding frequency observed in the current study may be due to the uneven depth of sequencing across women given limited sample sizes, where a larger sample will be required to confirm these findings.
The distribution of shared TCR repertoire clones between TLSB and cervical biopsy specimens and over time suggests robust trafficking between these tissues in one woman with higher HSV shedding rates and clinical recurrence. In silico analyses of Ag-specificity employed here was exploratory, but supports the hypothesis that T-cell expansion and infiltration into tissues during HSV-related inflammation can be detected via TCRB CDR3 sequencing that includes a mixture of resident and transient Ag-specific populations (41, 52). Previous work has primarily focused on the immune responses to HSV subclinical shedding and clinical recurrences in the skin associated with subclinical shedding or clinical recurrences (20, 53). Our data are consistent with Posavad et al., who demonstrated in women without HIV infection (54) that there is a robust immune response in the uterine cervix that can be shared, and that is unique from that observed at the TLSB and HSV lesion sites and noted to persist over years (21).
While we were not able to examine differences in T-cell frequencies associated with HSV shedding rates, we compared T cell subsets in PBMCs to paired cervical tissues and found significant increases in both CD3+ CD4+ EM and Ag-specific T-cell populations in the uterine cervix compared to the blood. Previous work has shown that most HIV DNA detected during ART was in the CM population (5), with a more modest detection in the EM subset; however, more recent work suggests EM may be a more relevant cellular reservoir (55, 56). Our results show EM are more prevalent in the uterine cervix compared to blood, similar to gut and lymph node tissues where EM cells frequently have identical HIV DNA that was more closely related to plasma HIV RNA (55, 57), suggesting they may contain replication-competent virions. The small number of biopsy specimens obtained for this study limited the number of assessments performed per specimen, e.g., making it difficult to perform viral outgrowth assays (32) or more detailed studies of replication competency that could be explored in future studies.
Evaluating HIV reservoirs in tissue sites in women during ART is not well studied, in part due to the limitations of obtaining sufficient tissue samples. However, it is important to note that the uterine cervix is a site readily biopsied that, as our study demonstrates, can provide valuable information about HIV reservoirs in women receiving ART. These procedures are routinely performed in both women with and without HIV infection and this pilot study suggests that intensive tissue-based studies are possible and relevant.
Limitations of the current pilot study include the small number of women enrolled. The study focused on HSV and not other common viruses such as cytomegalovirus (CMV), which also appears to enhance HIV shedding and potential reservoirs (29, 33). Variation in the size of the tissue biopsy specimens and NA yield limited the number of assays we could uniformly perform across all 9 women, and this limited our power to detect significant differences in outcomes and predictors of HIV tissue reservoir, but has been informative in planning future studies. We were limited by poor yields of NA from skin tissues, where it has been previously reported that NA isolation from skin biopsy specimens is difficult (58). We did not determine the Ag specificity of predominant TCR clones identified herein to show whether they were HSV-specific, in part because the available databases have limited epitopes for HSV. Ideally, comparisons of the parameters evaluated in this cohort of HIV/HSV-coinfected women would be compared to women with HIV infection who are not coinfected with HSV-2, or would involve suppression of HSV using antivirals to decrease HSV replication.
In summary, this study provides the first evidence that subclinical HSV DNA shedding in genital sites from women during suppressive ART affects HIV DNA tissue reservoirs and presents evidence that HSV replication might promote HIV divergence in tissue sites, potentially by inducing HIV replication. These mechanisms should be further explored in future studies to fully elucidate the impact that viral coinfections have on HIV tissue reservoirs and are of critical importance to study for HIV cure strategies. If these findings are confirmed in larger studies, strategies to suppress herpesvirus replication to prevent Ag-specific proliferation of HIV-infected cells may be considered in future HIV cure research.
MATERIALS AND METHODS
Study design.
HIV/HSV-2-coinfected women who were 18 years of age or older were enrolled at the University of Washington (UW) Virology Research Clinic (VRC) in Seattle, WA. Eligible women were receiving a three-drug regimen of ART for HIV with plasma HIV RNA levels of <40 copies/ml and had a clinical history of ≥2 HSV-2 recurrences/year and were not taking anti-HSV therapy. Women with CD4 counts of <200 cells/mm3, were pregnant or breastfeeding, or those who were immunosuppressed due to reasons other than HIV infection or using long-term oral steroids were excluded. Women were screened for gonorrhea and chlamydia by vaginal nucleic acid amplification testing (NAAT) and for BV and trichomoniasis by wet mount/Amsel criteria and were treated appropriately prior to enrollment. HSV-2 serostatus was confirmed by Western blotting (59). Screening for cervical cancer was performed by Papanicolau (Pap) test if clinically indicated; those with cervical dysplasia within the past year were not eligible. Participants were trained to identify HSV genital lesions and instructed to come into the clinic within 24 to 36 h to have the lesion biopsied (22). All were followed for 4 months after the 28-day sample collection. The women were asked to keep daily diaries recording any genital symptoms and to report intercourse (31, 60). All participants provided written informed consent in a protocol that was approved by Institutional Review Boards at the UW (IRB number 00006878) and the Seattle Children’s Research Institute (SCRI) in Seattle, WA.
Specimen collection and processing.
Each woman was instructed at enrollment how to perform at-home sampling of the mixed anogenital region (using Dacron swabs; Fisher Scientific, Waltham MA) and cervix for 28 consecutive days (22). Cervical secretions were collected using a SoftCup (Flow Company, Venice, CA) worn for an average of 8 h daily and stored in a 50-ml conical tube in home freezers after collection. These specimens were used to quantify HSV DNA to establish a participant’s genital HSV shedding rate (61) (Fig. 1). Women returned 2 weeks after enrollment to return specimens and to encourage study retention. After the 28-day at home sampling period, women returned to clinic for visit 3 (V3) to have three cervical biopsy specimens collected corresponding to the hour hand of a clock (3, 6, and 9) from the uterine cervix with the os at the center and the sacrum at 6 o’clock, after application of lidocaine spray, using Tischler cervical biopsy forceps (Howard Medical Company, Chicago, IL), along with typical lesion site biopsy specimens (TLSB) (22, 62) (Fig. 1). Two cervical and one TLSB were snap-frozen in optimal cutting temperature (OCT) freezing medium. At enrollment and after 28-day sampling visits, a cervical cytobrush (Copan, Murietta, CA) was inserted into the uterine cervical os and rotated 360 degrees to collect cervical cells. Peripheral blood mononuclear cells (PBMCs) were collected into EDTA tubes and isolated by Ficoll separation (62).
Nucleic acid extraction from anogenital swabs and cervical secretions for HSV DNA quantification to establish genital HSV shedding rates.
Nucleic acids (NA) were isolated from the anogenital swabs (63) and cervical secretions from the SoftCup. Briefly, anogenital swabs were reconstituted with PCR buffer and HSV DNA was extracted from 200 μl of swab specimen and from 50 μl of cervical secretions (diluted 1:3 with 1× phosphate-buffered saline [PBS]) using a QIAamp 96 DNA blood kit and eluted into 100 μl of AE buffer (Qiagen, Germantown, MD). NA extract (10 μl) was submitted in duplicate for HSV DNA quantification by real-time PCR to detect the HSV gB gene with a limit of detection of 3 copies/reaction (64). Each 30-μl PCR contained 10 μl of nucleic acid extract, 833 nM primers, 100 nM probe, 15 μl of 2× QuantiTect Multiplex PCR master mix, with 0.03 units of uracil DNA glycosylase enzyme (UNG) to eliminate PCR carryover (New England BioLabs, Ipswich, MA). To monitor PCR inhibition, an exogenous (EXO) internal control was spiked into all reactions. Thermocycling conditions were 50°C for 2 min, 95°C for 15 min, followed by 45 cycles of 94°C for 1 min and 60°C for 1 min.
Isolation and quantification of HIV RNA from cervical secretions.
HIV RNA was extracted from 50 μl of cervical secretions from a SoftCup that was diluted 1:16 for a final volume of 800 μl and eluted into 50 μl of extraction buffer. Samples were quantified using a CLIA-compliant RealTime HIV-1 PCR (Abbott Park, Illinois) that detects HIV integrase with a lower limit of quantification (LLQ) of 40 copies/ml.
Extraction of cell-associated HIV DNA from uterine cervical and typical lesion site biopsy specimens.
DNA was extracted from four to eight 40-micron sections from nonsorted (bulk OCT frozen tissues) cervical biopsy, typical legion site biopsy (TLSB), and actual lesion biopsy (ALB) specimens using the Wizard Genomic DNA purification kit (Promega, Madison WI) following the manufacturer’s instructions. These NA were used for quantification of HIV DNA concentrations, single genome amplification (SGA) of the HIV envC2-V5 region (62), and for TCR repertoire analysis (described below).
Quantification of HIV DNA from uterine cervix, typical lesion site and actual lesion site biopsy specimens.
HIV DNA was quantified from nonsorted (bulk) uterine cervix and TLSB (V3) and ALB specimens by a real-time PCR that amplifies a region of HIV LTR with reproducible detection of 3 copies per PCR (65).
Single genome amplification of the HIV envC2-V5 region.
Single genome HIV envC2-V5 sequences were generated from cervical, TLSB, and ALB NA that were diluted to single copy (66) for direct sequencing (36, 67). HIV envC2-V5 sequences were aligned using the MUSCLE algorithm in Geneious (BioMatters, Newark, NJ), with maximum-likelihood trees generated using PhyML in DIVEIN (https://indra.mullins.microbiol.washington.edu/DIVEIN/) (25). Identical sequences were identified in several women and were included in all analyses, as these likely represent cellular proliferation. Coreceptor usage of viral variants was predicted using a position-specific scoring matrix (PSSM) of the HIV env V3 loop (https://indra.mullins.microbiol.washington.edu/webpssm/) for clade B viruses, the dominant strain in the US, using the X4R5 matrix (25).
Amplification of the TCR CDR3 gene for T cell repertoire analysis.
Between 2.7 to 3.4 μg/reaction of NA from nonsorted (bulk) uterine cervix, TLSB, and ALB specimens were submitted to Adaptive Technologies (Adaptive Technologies, Seattle, WA) for survey sequencing of the TCRBeta CDR3 region (68). There were >2,000 templates per reaction for each specimen, which allows for detection of clonotypes present at low (0.1%) frequency, and also sufficiently samples the TCR population. TCR sequences were analyzed using the ImmunoSEQ Analyzer program (Adaptive Biotechnologies, Seattle WA) and graphed using Excel software (Microsoft, Redmond WA) and R Studio (R 3.4.1, R Core Team, Vienna, Austria). Participant TCRB CDR3 sequences were matched to published sequences to identify candidate sequences from three participants with paired CDR3 in anogenital tissues and cervix after the 28-day shedding period. Previously associated reactivity to either HSV or HIV in the context of a shared HLA, and as comparison sequences with published reactivity to CMV, EBV, and influenza, were included. Three sources of published CDR3 associations were queried: LymphoSeq, McPAS-TCR, and Adaptive ImmuneAccess (https://clients.adaptivebiotech.com/login).
Flow-cytometry evaluation of CD4+ and CD3+ CD4− populations from cervical cytobrush and PBMCs at enrollment, after 28 days of at-home sampling, and at clinical recurrence.
An eight-color panel was designed to evaluate the frequency of naive (CD45RA+ CCR7+), central memory (CM) (CD45RA− CCR7+) and effector memory (EM) (CD45RA− CCR7−) and terminally differentiated (TEMRA) (CD45RA+ CCR7−) T-cell populations in cervical cytobrush and PBMCs using the following antibodies (Ab) as Ag-fluorochrome and clone: CD3 APC-H7 (clone SK7); CD4 PercP-Cy5 (clone SK3); CD45RA FITC (clone H1100); CCR7 Alexa Fluor 647 (clone G043H7); CD127 BV605 (clone HIL-7R M21); CD25 (clone MA-251); PE and CD137 BV421 (clone 4B4-1) and Aqua Live/Dead Cell Stain kit (Thermo Scientific, Waltham, MA). All Ab were purchased from Becton, Dickinson (BD) (Franklin Lakes, New Jersey). Nonfixed PBMC and cervical cytobrush specimens were evaluated for T-cell frequencies using a BSL-3 BD FACS ARIA–II sorting cytometer (BD). The same number of events was collected for each woman; for PBMC there were 200,000 CD4+ events and for cervical cytobrush there were 100,000 CD3+ events.
Statistical Analysis.
HSV shedding rate was calculated per person by dividing the number of days with HSV detected by the total number of days with swabs/secretions collected. HSV shedding rate was used as the predictor for multiple outcomes: (i) HIV DNA concentrations were analyzed by linear regression using STATA and GraphPad Prism5 (Stata SE V12.1; StataCorp., College Station, Texas and GraphPad, San Diego, CA, respectively); and (ii) population diversity (estimated as the average pairwise genetic distances) and divergence from the most recent common ancestor (MRCA) were generated using DIVEIN (25) (https://indra.mullins.microbiol.washington.edu/DIVEIN/). Differences in HIV diversity were evaluated using an online t test implemented in DIVEIN and HIV divergence was evaluated by a paired t test in Graph Pad. TCR clone correlations and sample overlap were calculated using the Adaptive ImmunoSeq program (https://www.adaptivebiotech.com/products-services/immunoseq/immunoseq-analyzer/), using a Spearman’s correlation and by calculating a Morisita’s index for specimen overlap, respectively. Cervical cytobrush and PBMC T-cell frequencies were compared using a paired t test (GraphPad). P values of <0.05 were considered significant.
Data availability.
Sequences are available in GenBank under accession numbers MN886969 to MN887101.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the volunteers who participated in this pilot study. We thank Adaptive Technologies, specifically Manda Williams and Martin Suchorolski, for their assistance with the TCR repertoire analysis and for excellent technical support.
We have no conflicts of interest to disclose.
This work was supported by the University of Washington Center for AIDS Research New Investigator Award P30 AI027757 (PI: Bull), http://depts.washington.edu/cfar/; NIH/NIAID R01 AI124770-01 (PI: Bull); Program Grant NIH/NIAID AI030371 (PI: Wald), https://www.niaid.nih.gov/about/daids; and the AIDS Clinical Trials Group (ACTG) Laboratory Center AI106701 (PI: Coombs), https://actgnetwork.org/.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RK, Abdul-Jawad S, McCoy LE, Mok HP, Peppa D, Salgado M, Martinez-Picado J, Nijhuis M, Wensing AMJ, Lee H, Grant P, Nastouli E, Lambert J, Pace M, Salasc F, Monit C, Innes AJ, Muir L, Waters L, Frater J, Lever AML, Edwards SG, Gabriel IH, Olavarria E. 2019. HIV-1 remission following CCR5Delta32/Delta32 haematopoietic stem-cell transplantation. Nature 568:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummins NW, Rizza S, Litzow MR, Hua S, Lee GQ, Einkauf K, Chun TW, Rhame F, Baker JV, Busch MP, Chomont N, Dean PG, Fromentin R, Haase AT, Hampton D, Keating SM, Lada SM, Lee TH, Natesampillai S, Richman DD, Schacker TW, Wietgrefe S, Yu XG, Yao JD, Zeuli J, Lichterfeld M, Badley AD. 2017. Extensive virologic and immunologic characterization in an HIV-infected individual following allogeneic stem cell transplant and analytic cessation of antiretroviral therapy: a case study. PLoS Med 14:e1002461. doi: 10.1371/journal.pmed.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. 2014. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strick LB, Wald A, Celum C. 2006. Management of herpes simplex virus type 2 infection in HIV type 1-infected persons. Clin Infect Dis 43:347–356. doi: 10.1086/505496. [DOI] [PubMed] [Google Scholar]
- 9.Gupta R, Warren T, Wald A. 2007. Genital herpes. Lancet 370:2127–2137. doi: 10.1016/S0140-6736(07)61908-4. [DOI] [PubMed] [Google Scholar]
- 10.Baeten JM, Strick LB, Lucchetti A, Whittington WL, Sanchez J, Coombs RW, Magaret A, Wald A, Corey L, Celum C. 2008. Herpes simplex virus (HSV)-suppressive therapy decreases plasma and genital HIV-1 levels in HSV-2/HIV-1 coinfected women: a randomized, placebo-controlled, cross-over trial. J Infect Dis 198:1804–1808. doi: 10.1086/593214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnabas RV, Celum C. 2012. Infectious co-factors in HIV-1 transmission herpes simplex virus type-2 and HIV-1: new insights and interventions. Curr HIV Res 10:228–237. doi: 10.2174/157016212800618156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posavad CM, Wald A, Kuntz S, Huang ML, Selke S, Krantz E, Corey L. 2004. Frequent reactivation of herpes simplex virus among HIV-1-infected patients treated with highly active antiretroviral therapy. J Infect Dis 190:693–696. doi: 10.1086/422755. [DOI] [PubMed] [Google Scholar]
- 13.Tobian AA, Grabowski MK, Serwadda D, Newell K, Ssebbowa P, Franco V, Nalugoda F, Wawer MJ, Gray RH, Quinn TC, Reynolds SJ, Rakai Health Sciences P . 2013. Reactivation of herpes simplex virus type 2 after initiation of antiretroviral therapy. J Infect Dis 208:839–846. doi: 10.1093/infdis/jit252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford ES, Magaret AS, Spak CW, Selke S, Kuntz S, Corey L, Wald A. 2018. Increase in HSV shedding at initiation of antiretroviral therapy and decrease in shedding over time on antiretroviral therapy in HIV and HSV-2 infected persons. AIDS 32:2525–2531. doi: 10.1097/QAD.0000000000002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benedetti J, Corey L, Ashley R. 1994. Recurrence rates in genital herpes after symptomatic first-episode infection. Ann Intern Med 121:847–854. doi: 10.7326/0003-4819-121-11-199412010-00004. [DOI] [PubMed] [Google Scholar]
- 16.Wald A, Zeh J, Selke S, Ashley RL, Corey L. 1995. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med 333:770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- 17.Tronstein E, Johnston C, Huang ML, Selke S, Magaret A, Warren T, Corey L, Wald A. 2011. Genital shedding of herpes simplex virus among symptomatic and asymptomatic persons with HSV-2 infection. JAMA 305:1441–1449. doi: 10.1001/jama.2011.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, Triezenberg SJ. 1994. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J Virol 68:2803–2810. doi: 10.1128/JVI.68.5.2803-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Koelle DM, Cao J, Vazquez J, Huang ML, Hladik F, Wald A, Corey L. 2007. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J Exp Med 204:595–603. doi: 10.1084/jem.20061792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Hladik F, Woodward A, Klock A, Peng T, Johnston C, Remington M, Magaret A, Koelle DM, Wald A, Corey L. 2009. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med 15:886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Peng T, Johnston C, Phasouk K, Kask AS, Klock A, Jin L, Diem K, Koelle DM, Wald A, Robins H, Corey L. 2013. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature 497:494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston C, Zhu J, Jing L, Laing KJ, McClurkan CM, Klock A, Diem K, Jin L, Stanaway J, Tronstein E, Kwok WW, Huang ML, Selke S, Fong Y, Magaret A, Koelle DM, Wald A, Corey L. 2014. Virologic and immunologic evidence of multifocal genital herpes simplex virus 2 infection. J Virol 88:4921–4931. doi: 10.1128/JVI.03285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roychoudhury P, Swan DA, Duke E, Corey L, Zhu J, Dave V, Spuhler LR, Lund JM, Prlic M, Schiffer JT. 2020. Tissue-resident T cell-derived cytokines eliminate herpes simplex virus-2-infected cells. J Clin Invest 130:2903–2919. doi: 10.1172/JCI132583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min B. 2018. Spontaneous T cell proliferation: a physiologic process to create and maintain homeostatic balance and diversity of the immune system. Front Immunol 9:547. doi: 10.3389/fimmu.2018.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond S, Mullins JI. 2010. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 48:405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mark KE, Wald A, Magaret AS, Selke S, Olin L, Huang ML, Corey L. 2008. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J Infect Dis 198:1141–1149. doi: 10.1086/591913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan DH, Raboud JM, Kaul R, Walmsley SL. 2014. Antiretroviral therapy is not associated with reduced herpes simplex virus shedding in HIV coinfected adults: an observational cohort study. BMJ Open 4:e004210. doi: 10.1136/bmjopen-2013-004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agyemang E, Magaret AS, Selke S, Johnston C, Corey L, Wald A. 2018. Herpes simplex virus shedding rate: surrogate outcome for genital herpes recurrence frequency and lesion rates, and phase 2 clinical trials end point for evaluating efficacy of antivirals. J Infect Dis 218:1691–1699. doi: 10.1093/infdis/jiy372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gianella S, Anderson CM, Var SR, Oliveira MF, Lada SM, Vargas MV, Massanella M, Little SJ, Richman DD, Strain MC, Perez-Santiago J, Smith DM. 2016. Replication of Human Herpesviruses Is Associated with Higher HIV DNA Levels during Antiretroviral Therapy Started at Early Phases of HIV Infection. J Virol 90:3944–3952. doi: 10.1128/JVI.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen-Quick A, Massanella M, Frick A, Rawlings SA, Spina C, Vargas-Meneses M, Schrier R, Nakazawa M, Anderson C, Gianella S. 2019. Subclinical cytomegalovirus DNA is associated with CD4 T cell activation and impaired CD8 T cell CD107a expression in people living with HIV despite early antiretroviral therapy. J Virol 93:e00179-19. doi: 10.1128/JVI.00179-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wald A, Zeh J, Selke S, Warren T, Ashley R, Corey L. 2002. Genital shedding of herpes simplex virus among men. J Infect Dis 186 Suppl 1:S34–9. doi: 10.1086/342969. [DOI] [PubMed] [Google Scholar]
- 32.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaillon A, Nakazawa M, Rawlings SA, Curtin G, Caballero G, Scott B, Anderson C, Gianella S. 2020. Subclinical CMV and EBV shedding is associated with increasing HIV DNA molecular diversity in peripheral blood during suppressive antiretroviral therapy. J Virol 94:e00927-20. doi: 10.1128/JVI.00927-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boritz EA, Douek DC. 2017. Perspectives on human immunodeficiency virus (HIV) cure: HIV persistence in tissue. J Infect Dis 215:S128–S133. doi: 10.1093/infdis/jix005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73:10489–10502. doi: 10.1128/JVI.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. JVI 79:9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 69:5087–5094. doi: 10.1128/JVI.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskey SB, Siliciano RF. 2014. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat Rev Microbiol 12:772–780. doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- 39.Palu G, Benetti L, Calistri A. 2001. Molecular basis of the interactions between herpes simplex viruses and HIV-1. Herpes 8:50–55. [PubMed] [Google Scholar]
- 40.Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noel-Romas L, Grobler A, Westmacott G, Xie IY, Butler J, Mansoor L, McKinnon LR, Passmore JS, Abdool Karim Q, Abdool Karim SS, Burgener AD. 2017. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356:938–945. doi: 10.1126/science.aai9383. [DOI] [PubMed] [Google Scholar]
- 41.Rollenhagen C, Lathrop MJ, Macura SL, Doncel GF, Asin SN. 2014. Herpes simplex virus type-2 stimulates HIV-1 replication in cervical tissues: implications for HIV-1 transmission and efficacy of anti-HIV-1 microbicides. Mucosal Immunol 7:1165–1174. doi: 10.1038/mi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trezza CR, Kashuba AD. 2014. Pharmacokinetics of antiretrovirals in genital secretions and anatomic sites of HIV transmission: implications for HIV prevention. Clin Pharmacokinet 53:611–624. doi: 10.1007/s40262-014-0148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parisi SG, Andreis S, Mengoli C, Scaggiante R, Ferretto R, Manfrin V, Cruciani M, Giobbia M, Boldrin C, Basso M, Andreoni M, Palu G, Sarmati L. 2012. Baseline cellular HIV DNA load predicts HIV DNA decline and residual HIV plasma levels during effective antiretroviral therapy. J Clin Microbiol 50:258–263. doi: 10.1128/JCM.06022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrazzo JM, Rabe L, Kelly C, Richardson B, Deal C, Schwartz JL, Chirenje ZM, Piper J, Morrow RA, Hendrix CW, Marzinke MA, Hillier SL, VOICE Study Team . 2019. Tenofovir gel for prevention of herpes simplex virus type 2 acquisition: findings from the VOICE trial. J Infect Dis 219:1940–1947. doi: 10.1093/infdis/jiz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrove JM, Leonard J, Weck KE, Rabson AB, Gendelman HE. 1987. Activation of the human immunodeficiency virus by herpes simplex virus type 1. J Virol 61:3726–3732. doi: 10.1128/JVI.61.12.3726-3732.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Megazzini KM, Sinkala M, Vermund SH, Redden DT, Krebs DW, Acosta EP, Mwanza J, Goldenberg RL, Chintu N, Bulterys M, Stringer JS. 2010. A cluster-randomized trial of enhanced labor ward-based PMTCT services to increase nevirapine coverage in Lusaka, Zambia. AIDS 24:447–455. doi: 10.1097/QAD.0b013e328334b285. [DOI] [PubMed] [Google Scholar]
- 47.Vlach J, Pitha PM. 1993. Differential contribution of herpes simplex virus type 1 gene products and cellular factors to the activation of human immunodeficiency virus type 1 provirus. J Virol 67:4427–4431. doi: 10.1128/JVI.67.7.4427-4431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gimble JM, Duh E, Ostrove JM, Gendelman HE, Max EE, Rabson AB. 1988. Activation of the human immunodeficiency virus long terminal repeat by herpes simplex virus type 1 is associated with induction of a nuclear factor that binds to the NF-kappa B/core enhancer sequence. J Virol 62:4104–4112. doi: 10.1128/JVI.62.11.4104-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanidou M, Ramos I, Mas Casullo V, Trepanier JB, Rosenbaum S, Fernandez-Sesma A, Herold BC. 2013. Herpes simplex virus 2 (HSV-2) prevents dendritic cell maturation, induces apoptosis, and triggers release of proinflammatory cytokines: potential links to HSV-HIV synergy. J Virol 87:1443–1453. doi: 10.1128/JVI.01302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingsman SM, Kingsman AJ. 1996. The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem 240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 51.Goletti D, Kinter AL, Coccia EM, Battistini A, Petrosillo N, Ippolito G, Poli G. 2002. Interleukin (IL)-4 inhibits phorbol-ester induced HIV-1 expression in chronically infected U1 cells independently from the autocrine effect of endogenous tumour necrosis factor-alpha, IL-1beta, and IL-1 receptor antagonist. Cytokine 17:28–35. doi: 10.1006/cyto.2001.0989. [DOI] [PubMed] [Google Scholar]
- 52.Carr DJ, Tomanek L. 2006. Herpes simplex virus and the chemokines that mediate the inflammation. Curr Top Microbiol Immunol 303:47–65. doi: 10.1007/978-3-540-33397-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruocco V, Ruocco E, Brunetti G, Russo T, Gambardella A, Wolf R. 2014. Wolf's post-herpetic isotopic response: infections, tumors, and immune disorders arising on the site of healed herpetic infection. Clin Dermatol 32:561–568. doi: 10.1016/j.clindermatol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Posavad CM, Zhao L, Mueller DE, Stevens CE, Huang ML, Wald A, Corey L. 2015. Persistence of mucosal T-cell responses to herpes simplex virus type 2 in the female genital tract. Mucosal Immunol 8:115–126. doi: 10.1038/mi.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, Faria NR, Lemey P, Albert J, Hunt P, Loeb L, Pilcher C, Poole L, Hatano H, Somsouk M, Douek D, Boritz E, Deeks SG, Hecht FM, Palmer S. 2015. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis 212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, Massanella M, Brassard N, Alsahafi N, Delgado GG, Routy JP, Walker BD, Finzi A, Chomont N, Kaufmann DE. 2016. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 20:368–380. doi: 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee E, von Stockenstrom S, Morcilla V, Odevall L, Hiener B, Shao W, Hartogensis W, Bacchetti P, Milush J, Liegler T, Sinclair E, Hatano H, Hoh R, Somsouk M, Hunt P, Boritz E, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2019. Impact of antiretroviral therapy duration on HIV-1 infection of T cells within anatomic sites. J Virol 94:e01270-19. doi: 10.1128/JVI.01270-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidorova JV, Biderman BV, Nikulina EE, Sudarikov AB. 2012. A simple and efficient method for DNA extraction from skin and paraffin-embedded tissues applicable to T-cell clonality assays. Exp Dermatol 21:57–60. doi: 10.1111/j.1600-0625.2011.01375.x. [DOI] [PubMed] [Google Scholar]
- 59.Ashley RL, Militoni J, Lee F, Nahmias A, Corey L. 1988. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol 26:662–667. doi: 10.1128/JCM.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langenberg A, Benedetti J, Jenkins J, Ashley R, Winter C, Corey L. 1989. Development of clinically recognizable genital lesions among women previously identified as having “asymptomatic” herpes simplex virus type 2 infection. Ann Intern Med 110:882–887. doi: 10.7326/0003-4819-110-11-882. [DOI] [PubMed] [Google Scholar]
- 61.Magaret AS, Johnston C, Wald A. 2009. Use of the designation “shedder” in mucosal detection of herpes simplex virus DNA involving repeated sampling. Sex Transm Infect 85:270–275. doi: 10.1136/sti.2008.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bull M, Learn G, Genowati I, McKernan J, Hitti J, Lockhart D, Tapia K, Holte S, Dragavon J, Coombs R, Mullins J, Frenkel L. 2009. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One 4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wald A, Huang ML, Carrell D, Selke S, Corey L. 2003. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J Infect Dis 188:1345–1351. doi: 10.1086/379043. [DOI] [PubMed] [Google Scholar]
- 64.Jerome KR, Huang ML, Wald A, Selke S, Corey L. 2002. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J Clin Microbiol 40:2609–2611. doi: 10.1128/jcm.40.7.2609-2611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arvold ND, Ngo-Giang-Huong N, McIntosh K, Suraseranivong V, Warachit B, Piyaworawong S, Changchit T, Lallemant M, Jourdain G, Perinatal HIV Prevention Trial (PHPT-1), Thailand . 2007. Maternal HIV-1 DNA load and mother-to-child transmission. AIDS Patient Care STDS 21:638–643. doi: 10.1089/apc.2006.0169. [DOI] [PubMed] [Google Scholar]
- 66.Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retroviruses 13:737–742. doi: 10.1089/aid.1997.13.737. [DOI] [PubMed] [Google Scholar]
- 67.Bull ME, Learn GH, McElhone S, Hitti J, Lockhart D, Holte S, Dragavon J, Coombs RW, Mullins JI, Frenkel LM. 2009. Monotypic human immunodeficiency virus type 1 genotypes across the uterine cervix and in blood suggest proliferation of cells with provirus. J Virol 83:6020–6028. doi: 10.1128/JVI.02664-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. 2009. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences are available in GenBank under accession numbers MN886969 to MN887101.