Abstract
Background
Gastric cancer (GC) is confronted with limited options for precision medicine. Human epidermal growth factor receptor 2 (HER2) is the principal druggable target of GC, yet proper biomarkers for response/resistance prediction remain unveiled.
Methods
From 40 GC patients received HER2‐targeted therapy, a total of 327 peripheral blood plasma specimens was collected including baseline and treatment time points. Circulating tumor DNA (ctDNA) was extracted and sequenced with a target panel of 425 genes. Experimental validation of resistant mutations was carried out in NIH‐3T3 cell line.
Results
Genomic features, including ERBB2 copy number variation (CNV), total copy number load, and tumor mutation burdens (TMBs), dynamically changed along with the treatment process and correlated with disease progression. Plasma ctDNA‐based diagnosis was more sensitive than conventional computed tomography scanning in 40% of investigated patients, gaining additional time for clinical management. Compared to baseline, new gene alterations were emerged in 12 patients who developed drug resistance during treatment. ERBB2 mutations potentially related to Pyrotinib resistance were identified in plasma ctDNA of one patient and functional analysis of their downstream signaling pathways was carried out in NIH‐3T3 cell line. TMB exhibited more power than ERBB2 CNV in predicting treatment responses and prognosis for HER2‐targeted therapy in GC patients. Interestingly, survival analysis indicated that patients harboring both HER2 (ERBB2) positivity and high TMB might gain more therapeutic benefits from immune checkpoint inhibitors instead of HER2‐targeted regimens that required further studies and validations
Conclusions
Our work showed that the dynamic surveillance of plasma ctDNA genomic features provided instructive information for the precision medication of GC patients.
Keywords: ERBB2 copy number, gastric cancer, HER2‐targeted therapy, immunotherapy, liquid biopsy, TMB
Plasma ctDNA‐based diagnosis was more sensitive than conventional computed tomography scanning in 40% of patients, gaining time for clinical management.
Plasma tumor mutation burden was a potential marker to predict the therapeutic outcomes of human epidermal growth factor receptor 2 (HER2)‐targeted therapy in GC.
HER2‐positive patients with high TMB might gain more therapeutic benefits from immunotherapy instead of HER2‐targeted regimens that required further studies and validations.
1. INTRODUCTION
Gastric cancer (GC) is one of the leading causes of cancer death around the world. 1 As a digestive tract‐originated malignancy, GC is featured by its complicated tissue composition and high heterogeneity, which greatly hinder its early diagnosis and treatment. 2 The recent development of precision medicine has put forth the hope to expanded survival expectance or even cure for multiple types of cancer, yet the individualized treatment for GC remains to be improved.
Human epidermal growth factor receptor 2 (HER2; encoded by ERBB2) overexpression and amplification have been recognized as frequent molecular abnormalities in GC. 3 , 4 Due to its tight regulation to crucial downstream signaling nodes such as RAS/RAF/MYC and PI3K‐Akt, HER2 remains to be the prior choice of targeted drug development for patients with advanced GC. 5 To date, a variety of HER2‐targeted drugs has been developed and evaluated by clinical trials separately and combined, including HER2 antibody and its derivatives (Trastuzumab and Pertuzumab), tyrosine kinase inhibitor (Lapatinib, Neratinib, and Afatinib), and antibody‐drug conjugates (ADCs) (DS‐8201a, TDM1, and RC48). 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 Monoclonal antibody Trastuzumab is the only HER2‐targeted agent currently approved for first‐line practice of GC. 14 DS‐8201a, also known as Trastuzumab deruxtecan, is an ADC conjugated with humanized anti‐HER2 antibody, cleavable peptide‐based linker, and topoisomerase I inhibitor payload. DS‐8201 has been evaluated in a phase II trial consisted of HER2‐positive advanced GC. Both response rate and overall survival (OS) was significantly improved in DS‐8201‐treated patients compared to standard therapy. 8 TDM1(Trastuzumab emtansine) has shown to be active in HER2‐positive breast cancer patients with brain metastasis from the phase III KAMILLA trial. 13 Another HER2‐ADC, RC48‐ADC, has been carried out in phase II trials and demonstrated a clinically meaningful overall response rate (ORR) in pretreated HER2‐positivie urothelial carcinoma patients. 15 Although these agents/combinations provide supporting evidence for further investigation in HER2‐positivie GC, the response rate of HER2‐positive patients received HER2‐targeted therapy remain limited, and the frequently upcoming resistance is another reason that impaired therapeutic efficacy.
In order to overcome the unsatisfactory situation of HER2‐targeted therapy, timely evaluation of gene alterations along with the treatment progresses is of great importance so as to aid in better therapeutic management and clinical decision‐making for GC patients. However, conventional diagnostic methods, such as computed tomography (CT)‐scanned imaging and invasive endoscopic sampling, are insufficient to fulfill the need of real‐time disease surveillance. Furthermore, the genomic profiles and molecular traits of metastatic tumor tissue are sometimes inconsistent with the primary tumors, making it difficult to monitor the cancer progression on tissue level. As a type of tumor‐derived, fragmented cell free DNA, ctDNA (circulating tumor DNA) is enriched in blood (specifically, plasma) of cancer patients and reflecting the information of the entire tumor genome. By elucidating the genomic features in patient's plasma with next generation sequencing (NGS), ctDNA‐based liquid biopsy complements the shortages of tissue evaluation and has been proved to be a powerful choice to facilitate the monitoring of cancer progression. 16 , 17 ctDNA levels correlate with the tumor size, stage, and depth of invasion in patients with GC. 18 Previous study showed that GC patients with higher ctDNA levels had higher risk of peritoneal recurrence and exhibited a 5‐year global survival rate poorer than patients without detected ctDNA. 19 Meanwhile, the possibility of ctDNA monitoring in clinical response to immunotherapy and tracking HER2 resistance have been also been demonstrated in GC. 20 , 21 In this study, by evaluating the longitudinal changes of ctDNA in plasma derived from advanced GC patients during HER2‐targeted therapy, we seek to identify the genetic information that directed responses and prognosis, and to explore additional therapeutic options for advanced GC.
2. MATERIALS AND METHODS
2.1. Study design and specimen information
All patients in this study were enrolled and followed‐up by the Department of Gastrointestinal Oncology, Peking University Cancer Hospital & Institute for conventional therapy or clinical trials. Peripheral blood specimens were collected with cell‐free DNA BCT tubes (Streck Laboratories, USA). All clinical data of patients were acquired by referring to their individual medical records. For both cohorts, the inclusion criteria were as follows: patients must be diagnosed with unresectable or metastatic GC, had at least one measurable, or unmeasurable but evaluable, lesion (described according to RECIST 1.1), without history of severe heart or liver disease, psychiatric disorders, without hemorrhage or perforation of digestive tract, remained treatment naive 3 weeks before the baseline of treatment, scored with ECOG (Eastern Cooperative Oncology Group) performance status of 0/1 three days before the baseline of treatment. Exclusion criteria were as follows: patients harbored GC combined with other types of cancer, or histopathologically diagnosed as neuroendocrine tumor or squamous type of tumor.
Based on these criteria, 45 patients treated with anti‐HER2 regimens (including conventional treatment and clinical trials) during October 1, 2013 to October 1, 2018 were enrolled and 40 were included in the HER2‐targeted therapy cohort (Figure S1A). Totally, 327 peripheral blood samples from pretreatment baseline (BL) to disease progression were collected every two treatment cycles for the 40 GC patients. By referring to CT, patients’ responses to HER2‐targeted therapy were evaluated as partial response (PR), stable disease (SD), or progressed disease (PD) according to the RECIST 1.1 criteria, whereas no complete response (CR) was observed in this cohort. For each series of specimen, the time of the first response evaluation after treatment was defined as first point (FP). Additionally, 39 patients treated with immune checkpoint inhibitors (from clinical trials) during Jun 1, 2016 to Jun 1, 2020 were enrolled and 37 were included in the immunotherapy cohort (Figure S1B). Baseline blood specimens of these 37 GC patients received immunotherapy were collected for validation.
Written informed consents were obtained from all donors. These specimens and associated clinical information were approved for experimental applications by the Institutional Ethics Committee, Peking University Cancer Hospital & Institute. This study was conducted in accordance with the Declaration of Helsinki.
2.2. Plasma isolation and cfDNA extraction
Genomic DNA from white blood cells (WBCs) was extracted using DNeasy Blood & Tissue Kit (Qiagen, Germany) and used as normal control. After centrifuging whole blood samples at 4℃ for 10 min (1600 g), plasma supernatant was separated with blood cell sediment and then re‐centrifuged at 4℃ for 10 min (16 000 g). Plasma cfDNA was extracted using Qiagen QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany) following the manufacturer's protocols, qualified with Nanodrop 2000 (Thermo Fisher Scientific), and quantified by Qubit 2.0 using the dsDNA HS Assay Kit (Life Technologies) according to the manufacturer's recommendations. The median yield for the plasma cfDNA was 25.9 ng, ranging from 10.1 to 739.2 ng. Isolated blood components (plasma, serum, and PBMC) and extracted cfDNAs were maintained at –80℃ for long‐term storage.
2.3. Next generation sequencing
NGS and data processing were performed as previously described. 21 In brief, isolated ctDNA samples were processed with the KAPA Hyper Prep kit (KAPA Biosystems) for libraries construction. A customized NGS panel targeting 425 cancer‐relevant genes was used for hybridization enrichment. The list of panel genes was displayed in Table S2. Indexed DNA libraries were pooled together to a total amount of 2 μg and subjected to probe‐based hybridization using IDT xGen Lockdown reagents (IDT, Coralville, IA) and Dynabeads M‐270 (Thermo Fisher). The library was quantified by KAPA Library Quantification kit (KAPA Biosystems) according to manufacturer's instructions. Bioanalyzer 2100 (Agilent, USA) was used to determine the fragment size distribution of the final library and then sequenced on Illumina HiSeq4000 NGS (Illumina) platform following the manufacturer's instructions. An expected sequencing depth of 3000× was set for ctDNA samples.
2.4. Data processing
Trimmomatic was used for FASTQ file quality control. Leading/trailing low quality (quality reading below 30) or N bases were removed. Remaining reads were mapped to the reference sequence data (Human Genome version 19) using Burrows‐Wheeler Aligner (BWA‐mem, v0.7.12). Indels realignment and base quality score recalibration were performed with Genome Analysis Toolkit (GATK 3.4.0). Somatic mutations were detected with VarScan2. Copy number variations (CNVs) were detected using ADTEx (http://adtex.sourceforge.net) with default parameters as reported by previous studies. 21 , 22 , 23 To eliminate sequencing artifact, a local bioinformatics pipeline was performed. First, we used a local WBCs database, containing recurrent somatic alterations from WBCs of 400 patients, to eliminate the sequencing artifacts. Specifically, if a variant was detected (i.e., more than mutant reads) in >10% of the samples, it was considered a likely artifact and was removed. Second, a background denoising strategy was performed. Briefly, we performed panel sequencing with similar sequencing depth used in this study on plasma samples of 50 healthy individuals to assemble a database of alterations at each site of the panel and build a background error model. A specific alteration at a specific site was considered sequencing noise if the allele frequency (AF) and distinct supporting reads were not significantly beyond the background error probability. Only alteration with an AF over three standard deviations from the mean AF of healthy plasma ctDNAs pool remained and subjected for further analyze.
The cutoffs of gene CNVs were set to 1.4‐fold copy number for amplification (gain) and 0.65‐fold copy number for deletion (loss). Total copy number load (TCL) was summarized by counting amplifications and deletions. Tumor mutation burden (TMB) was calculated by summing all base substitutions and indels in the coding region of targeted genes, including synonymous alterations to reduce sampling noise and excluding known driver mutations as previously described. 24 Samples within the highest mutation load or CNV load tertile (top 33.3%) were classified as having high TMB or TCL, which equal to ≥8.5 mutations/Mb in TMB or ≥2 in TCL. Samples with <8.5 mutation/Mb in TMB or <2 in TCL were considered as low TMB or TCL. These cutoffs were consistently used for both the HER2‐targeted therapy and immunotherapy cohort. Neutrophil‐to‐lymphocyte (NLR) or monocyte‐to‐lymphocyte ratio (MLR) were defined as the absolute neutrophil/monocyte count divided by the absolute lymphocyte count. 25 , 26 Maximum variant allele frequency was defined as the highest allele frequency of the detected somatic variants in each sample.
2.5. Cell culture, transfection, and drug treatment
Mouse embryonic fibroblast cell line NIH‐3T3 was purchased from Cell Bank (Chinese Academy of Sciences, China). Cells were maintained in RPMI‐1640 medium (Invitrogen, Carlsbad, CA) supplemented with 1% penicillin plus streptomycin (HyClone, Logan, UT) and 10% fetal calf serum (Gibco BRL), and then incubated at 37℃ in a humidified incubator (5% CO2). Wild‐type and point mutation of ERBB2 were constructed into plasmids and transfected into cells with Lipofectamine 3000 reagent (Thermo Fisher, USA). After 36 h of transfection, cells were treated with Pyrotinib (200 nm) for additional 48 h. Pyrotinib (SHR‐1258) was purchased from Selleck.
2.6. Western blot assay
For western blotting, cells were collected and lysed in 1× SDS‐PAGE loading buffer (1% SDS, 11% glycerol, 10% β‐mercaptoethanol, 0.1 M Tris, pH 6.8) for assay. Protein samples were then probed with corresponding primary/secondary antibodies, illuminated with the Clarity Western ECL substrate (Bio‐Rad, Hercules, CA), and visualized for protein bands with Amersham Imager 600 (GE Healthcare, Chicago, IL). Antibody for HER2 (#4290), p‐S6 (S240/S244, #4858), S6 (#2217), p‐Akt (S473, #9271), Akt (#4691), p‐Erk (T202/Y204, #4370), Erk (#4695), and β‐actin (#4970) were purchased from Cell Signaling technology.
2.7. Statistical analysis
Numerical diversity between subgroups was assessed by Wilcoxon‐Mann‐Whitney test. The consistency between ctDNA genomic variations and carcinoembryonic antigen (CEA) level was assessed by Spearman rank‐order correlation coefficient. Concordance between ctDNA ERBB2 copy number and tissue HER2 expression was assessed by Kappa test. Pathway involvement and gene set enrichment were performed by referring to GO (http://geneontology.org/), DAVID (https://david.ncifcrf.gov/), and KOBAS (http://kobas.cbi.pku.edu.cn/) bioinformatics resources. Survival proportions were assessed by Kaplan‐Meier survival analysis paired with Log‐rank test. For all these tests, P < .05 was considered as statistically significant. All analyses were performed with SPSS 21.0 or R program, and then formatted with GraphPad Prism 6.
3. RESULTS
3.1. Overall characteristics of enrolled patients
Forty GC patients received HER2‐targeted therapy or HER2‐targeted therapy plus chemotherapy were enrolled in this study, with a median age of 59 (29‐75 years). Thirty‐one patients (77.5%) were male and nine (22.5%) were female. Most patients (85%; 34/40) were non‐esophagogastric junction origination, diagnosed with middle (50%; 20/40) or low (45%; 18/40) differentiation, or classified as intestinal subtype (70%; 28/40). Confirmed by immunohistochemistry (IHC) staining or fluorescence in situ hybridization (FISH), 77.5% (31/40) patients were HER2 positive, whereas the rest were recruited by HER2‐targeted clinical trials. According to regimens, 14 patients received Trastuzumab‐combined chemotherapy (Paclitaxel, Oxaliplatin, Fluorouracil, etc) treatment, 11 received the JACOB treatment (Pertuzumab plus Trastuzumab‐combined chemotherapy), eight received HER2‐targeted antibody‐drug conjugate (ADC) treatment (TDM1 or RC48), and seven received HER2‐targeted inhibitor treatment (Pyrotinib or Pirotinib). Additionally, seven patients received two lines of HER2‐targeted treatments.
In total, ORR, disease control rate (DCR), and median progression‐free survival (mPFS) for the cohort were 50%, 75%, and 4.4 months, respectively. The PFS of the first, second to third line HER2‐targeted treatment was shown in Figure S2A. Patients in JACOB treatment displayed better efficacy than Trastuzumab‐combined chemotherapy, ADC, or inhibitors (Figure S2B), which is complied with current clinical consensus. The clinicopathological features of all patients and specimens were concluded as Table S1.
3.2. Genomic features in ctDNA dynamically reflected treatment response during HER2‐targeted therapy
For the 40 enrolled patients, ctDNAs were extracted from 327 plasma samples and sequenced by NGS. Genomic features of 425 genes, including CNVs and mutations, were evaluated along with the proceeding of treatment. Genomic landscape with evaluated responses (BL, PR, SD, and PD) is shown in Figure 1A. The most frequently observed copy number changes were in ERBB2/CDK12/CCNE1/TERC, whereas the top 10 somatic mutations were observed in TP53/ERBB2/ALK/APC/FLT1/NOTCH2/XPC/ERBB4/STAG2/BCR (Figure 1A). The trends of ERBB2 copy number, TCL, and TMB changes in each individual case during HER2‐targeted treatment were displayed in Figure S3.
FIGURE 1.
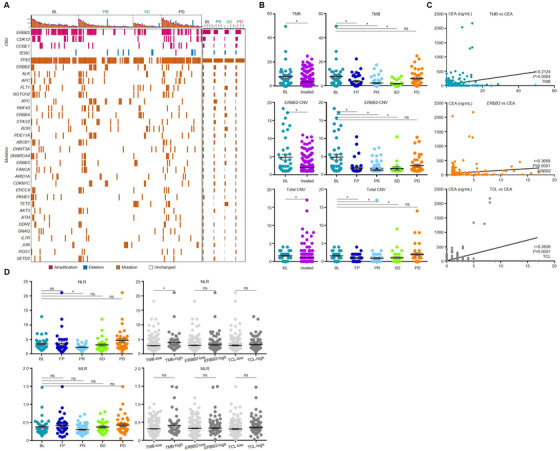
ctDNA genomic features dynamically changed in gastric cancer patients received HER2‐targeted therapy. (A) The landscape for all specimens collected at BL, PR, SD, and PD. (B) Comparison of ERBB2 copy number, total copy number load (TCL), and tumor mutation burden (TMB) across all evaluation points. (C) The Spearman correlation of ERBB2 copy number, TCL, and TMB with CEA. (D) The NLR/MLR values across all evaluation points and groups with low‐ or high‐ctDNA genomic features. *P < .05. ns, not significant
We stratified all plasma samples according to time point and compared ERBB2 CNV, TCL, and TMB status. As shown in Figure 1B, the level of ERBB2 CNV, TCL, and TMB all reduced after treatment. The reduction began at the first point after treatment (FP), reached the bottom at PR, remained low at SD, and recovered to the similar levels as BL at PD, suggesting that tumor fraction in plasma ctDNA was efficiently eliminated by HER2‐targeted treatment, yet recovered along with the progression of disease.
The correlation between these genomic features and other blood indexes were also investigated. Levels of ERBB2 copy number, TCL, and TMB were positively correlated with the tumor biomarker CEA (Figure 1C). For immune‐cell‐related indexes, the NLR was reduced at PR, yet was recovered or even elevated at PD (Figure 1D). Combining samples from different time point, NLR was only significantly higher in TMBhigh compared to TMBlow (Figure 1D). On the other hand, MLR displayed similar trend of dynamic changes to NLR, yet was insignificantly differed across all time points or genomic feature‐based classifications.
3.3. ctDNA shared a high concordance with tissue assay in identifying ERBB2/HER2 positivity
The ERBB2/HER2 status evaluated by ctDNA (NGS) and tissue assay (including IHC and FISH) displayed a high consistency. Eighteen patients had biopsied tumor tissue and matched plasma at BL (Table S3), among which a significant concordance of 94.4% (17/18) was achieved (Table S4), verifying that plasma ERBB2 could be used as an alternative method for the screening of HER2‐targeted population. Notably, for the patient (case 39) displayed inconsistency between pathological diagnosis and ctDNA assay, the record of IHC staining was negative while ERBB2 copy number was 1.7937. This inconsistency could be due to that plasma ERBB2 copy number was slightly higher than the cutoff of amplification (1.4), or that the section of tissue containing amplified ERBB2 was not captured by biopsy.
3.4. Genomic features in ctDNA forecasted disease progression earlier than CT scanning
Current evaluation standard of therapeutic response (the RECIST guidance) relies on CT scanning, which was economically inefficient for dynamic detection and usually missed occult lesions, especially in the abdominal region. To compare the plasma ctDNA testing and CT scanning in monitoring disease progression, patients who reached disease progression by CT and possessed at least three plasma specimens were used for analysis. Dynamics of the three genomic features (ERBB2 copy number, TCL, or TMB levels) along with the treatment were normalized to relative fold changes by comparing to their respective cutoff values. If either of genomic features elevated beyond the respective cutoff for at least two time points, the first point was considered as ctDNA‐based PD (Figure 2A). According to this standard, ctDNA‐based PD was detected ahead of CT‐based PD in 12 of 30 (40%) cases with an average advancement of 38.4 days (Figure 2B). Representative images for ctDNA‐based and CT‐based PD were demonstrated (Figure 2C).
FIGURE 2.

Genomic features in ctDNA facilitated the early detection of disease progression during HER2‐targeted therapy. (A) The real‐time changes of ctDNA features (ERBB2 copy number, total copy number load [TCL], and tumor mutation burden [TMB]) and CEA for 30 patients who ever reached progressed disease (PD). Computed tomography (CT)‐based PD, ctDNA‐based PD, and CEA‐based PD were marked by separate vertical lines. (B) The advancement of ctDNA‐based PD and CEA‐based PD compared with CT‐based PD in detecting disease progressions for the 30 patients received HER2‐targeted therapy. (C) CT‐scanned images for three representative cases were displayed to compare ctDNA‐based PD with CT‐based PD
Additionally, we also assessed whether changes of CEA exerted similar function to forecast disease progression. Concentrations of CEA were normalized to standard cutoff value (5 ng/mL) and were displayed in diagram as log2‐transformed indexes (Figure 2A). CEA‐based PD was detected ahead of CT‐based PD in four of 30 cases (13.3%) case with an average advancement of 10.0 days (Figure 2B), evidently less efficient than ctDNA‐based PD. Our data indicated that plasma ctDNA‐based liquid biopsy may serve as a complementary test to aid in the disease monitoring for GC patients, so as to gain further time for clinical practices.
3.5. Emerging genomic aberrations identified in ctDNA potentially contributed to therapeutic resistance
In our cohort, new genomic aberrations (compared with BL) were detected in 12 patients after developing therapeutic resistance (Figure 3A). The change of maximum variant allele frequency (MaxiVAF) in each sample during anti‐HER2 therapy from 12 patients with newly identified mutations was shown in Figures S2C and S2D. Here, we employed MaxiVAF as an indicator for the tumor fraction in the plasma ctDNA based on previous publication. 27 As shown in Figure S2, there was no significant difference in MaxiVAF between baseline and PD samples, suggesting the newly identified mutations were likely acquired during anti‐HER2 therapy. These aberrations were vigorously annotated to multiple ERBB2 downstream pathways (such as PI3K‐Akt and MAPK signaling) for patients received JACOB or Trastuzumab‐combined chemotherapy, or annotated to scattered cancerous pathways for patients received TDM1 or RC48‐ADC (Figures 3A and 3B). Patients without newly identified aberrations during treatment displayed slightly higher therapeutic responses (ORR = 52.2% vs 41.7%, DCR = 74% vs 66.7%) and slightly longer PFS (Figure S4) than patients with.
FIGURE 3.
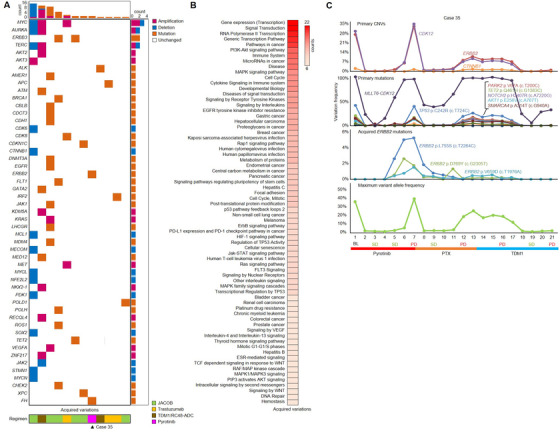
Emerging genomic aberrations potentially contributed to gastric cancer's (GC) resistance to HER2‐targeted therapy. (A) The distribution of novel gene alterations emerged in 12 patients and their correlation with HER2‐targeted regimens. (B) The biological processes and pathways involved with the emerging genomic aberrations were enriched. (C) The real‐time frequency of primary copy number variations (CNVs), primary mutations, three novel ERBB2 point mutations, and maximum variant allele frequency changed along with the treatments in case 35. PTX, paclitaxel
Notably, three ERBB2 mutations (p.V659D [c.T1976A], p.L755S [c.T2264C], and p.D769Y [c.G2305T]) were identified in the plasma of case 35 after developing resistance to Pyrotinib treatment (Figure 3C). To find out whether these mutations were associated with Pyrotinib resistance, we transfected plasmids expressing these ERBB2 mutations into NIH‐3T3 cell to study their impact on downstream molecules. Before Pyrotinib treatment, the phosphorylation of Erk (p‐Erk) was higher in three ERBB2‐mutation groups than in ERBB2 wild‐type group, whereas phosphorylation of Akt and S6 was comparable across these groups. After administration of Pyrotinib (200 nm, 48 h), p‐Akt was unaffected, S6 was evidently repressed in both ERBB2 mutation and wild‐type groups, and p‐Erk remained high for ERBB2 p.V659D and p.L755S mutation groups (Figure S5), suggesting that Erk phosphorylation and activation induced by specific ERBB2 mutations might be associated with resistance to Pyrotinib.
3.6. Plasma‐borne genomic features prior to treatment predicted responses to HER2‐targeted therapy
We then evaluated the potential of using ctDNA genomic information in guiding HER2‐targeted GC therapy. The genomic landscape at BL is shown in Figure 4A. We analyzed the genomic diversity between patients harboring different ERBB2 or TMB status and performed pathway enrichment for each fraction (Figures S6A and S6B). Genomic aberrations in ERBB2‐high or TMB‐high patients involved in much more cancerous processes and pathways than ERBB2‐low or TMB‐low patients, whereas the ERBB2‐related comparison displayed larger disparity than TMB‐related comparison, emphasizing the validity of utilizing HER2‐targeted regimens in our cohort.
FIGURE 4.
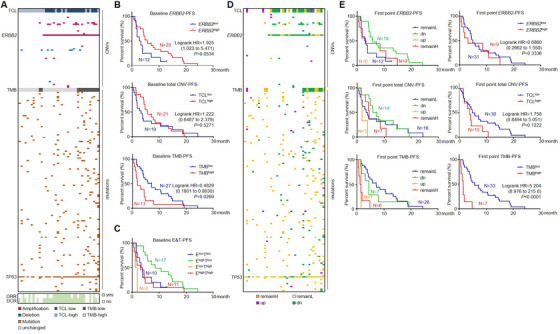
Genomic features served as predictive biomarkers for output of HER2‐targeted therapy. (A) An overview of ERBB2 copy number, total copy number load (TCL), and tumor mutation burden (TMB) for 40 patients at baseline and individually paired responses. (B) ERBB2 copy number, TCL, and TMB information at baseline were used to predict progression‐free survival (PFS) proportions of subsequent HER2‐targeted therapy. (C) PFS was calculated for patients stratified by baseline ERBB2 copy number and TMB. (D) The relative changes of ERBB2 copy number, TCL, and TMB for 40 patients at the first point after treatment started were compared with baseline status. (E) Stratified (left panel) or combined (right panel) information of ERBB2 copy number, TCL, and TMB at the first point were used to predict PFS proportions of HER2‐targeted therapy. ElowTlow, ERBB2 lowTMBlow; EhighTlow, ERBB2 highTMBlow; ElowThigh, ERBB2 lowTMBhigh; EhighThigh, ERBB2 highTMBhigh
At baseline point, 28 cases carried ERBB2 amplification (defined as ERBB2 high), with ORR = 60.7% (17/28) and DCR = 85.7% (24/28); 12 cases carried no ERBB2 amplification (ERBB2 low), with ORR = 25% (3/12) and DCR = 50% (6/12), suggesting that although responded less vigorously than HER2 positive patients, a part of HER2 negative patients was also sensitive to HER2‐targeted therapy. Considering TCL, 21 cases were defined as TCLhigh, with ORR = 61.9% (13/21) and DCR = 90.5% (19/21); 19 cases were defined as TCLlow, with ORR = 36.8% (7/19) and DCR = 57.9% (11/19). The high similarity between ERBB2 copy number‐ and TCL‐predicted ORRs/DCRs indicated that ERBB2 copy number might be the major contributor to therapeutic responses for all CNVs. From the view of TMB, 13 cases were defined as TMBhigh, with ORR = 38.5% (5/13) and DCR = 53.8% (7/13); 27 were defined as TMBlow, with ORR = 55.6% (15/27) and DCR = 85.2% (23/27), suggesting that TMBlow patients were more likely to benefit from HER2‐targeted therapy (Figure 4A). We then further classified the 40 patients into four categories by combining baseline ERBB2 and TMB status. For ERBB2 highTMBhigh patients (11 cases), ORR and DCR were 45.5% (5/11) and 63.6% (7/11) , respectively; for ERBB2 highTMBlow patients (17 cases), ORR and DCR were 70.6% (12/17) and 100% (17/17) , respectively; for ERBB2 lowTMBhigh patients (two cases), ORR and DCR were 0% (0/2) and 0% (0/2) , respectively; and for ERBB2 lowTMBlow patients (10 cases), ORR and DCR were 30% (3/10) and 60% (6/10), respectively. All above response rates are concluded in Table S5.
Kaplan‐Meier survival analysis was then performed to describe the longevity of response stratified by genomic traits. At baseline, high ERBB2 CNV and TCL status were not associated with better survival rate. Interestingly, TMBhigh at baseline was associated with poorer PFS than TMBlow (Figure 4B). This prognostic inconformity between ERBB2 and TMB indicated that HER2‐positive patients harboring high TMB may not benefit from HER2‐targeted therapy as much as expected. When combining these two parameters for stratification, ERBB2 highTMBlow patients displayed the highest response rate and the most favorable PFS; ERBB2 lowTMBhigh patients had the worst; and PFS of ERBB2 highTMBhigh and ERBB2 lowTMBlow patients lie in between (Table S5 and Figure 4C), suggesting that the combination of plasma‐borne ERBB2 copy number and TMB was a powerful option to predict patients’ response to HER2‐targeted anti‐GC therapy. It is worth mentioning that although the prognostic role of TMB was observed only in patients receiving second or higher lines of HER2‐targeted therapy, PFS of ERBB2 highTMBlow group was slightly better than ERBB2 lowTMBhigh group for patients receiving first line HER2‐targeted therapy. However, due to the limited number of patients in each group, further study is warranted to validate this observation (Figure S7).
3.7. Plasma‐borne TMB dynamically surveillant therapeutic outputs of HER2‐targeted therapy
Considering that ctDNA genomic traits dynamically changed along with HER2 targeted therapy, we investigated whether changes in plasma at the first point (FP) after treatment started provided useful information in therapeutic surveillance. Genomic features at FP were compared with those at BL. According to changes of ERBB2 copy number/TCL/TMB, patients were stratified into four categories: dn (from AMP/high at BL to non‐AMP/low at FP), up (from non‐AMP/low at BL to AMP/high at FP), remainL (non‐AMP/low at both BL and FP), and remainH (AMP/high at both BL and FP). Relative changes of these genomic features are shown as Figure 4D.
For all 40 patients at FP, considering ERBB2 copy number, ERBB2‐dn cases possessed higher ORR and DCR (63.2% and 94.7%, respectively) than ERBB2‐remainH (55.6% and 66.7%, respectively) and ERBB2‐remainL (25% and 50%, respectively) cases, whereas no patients were defined as ERBB2‐up; considering TCL, TCL‐dn cases possessed higher ORR and DCR (64.2% and 92.9%, respectively) than TCL‐remainH (57.1% and 85.7%, respectively), TCL‐remainL (43.8% and 62.5%, respectively), and TCL‐up (0% and 33.3%, respectively) case; considering TMB, TMB‐remainL cases possessed higher ORR and DCR (57.7% and 88.5%, respectively) than TMB‐dn (42.9% and 71.4%, respectively), TMB‐remainH (33.3% and 33.3%, respectively), and TMB‐up (0% and 0%, respectively) case. In general, patients with reduced or remained‐low TMB at FP were better responders to HER2‐targeted therapy, whereas changes of ERBB2 and TCL at FP were less efficient in predicting therapeutic responses. All these response rates are concluded in Table S6.
We then performed Kaplan‐Meier analysis for those categories at FP. For ERBB2 stratification, PFS of ERBB2‐dn patients were slightly longer than ERBB2‐remainL or ERBB2‐remainH patients, suggesting a favorable prognostic trend to HER2‐targeted regimens. For TCL stratification, TCL‐dn/TCL‐remainL patients had the longest and TCL‐up patients had the shortest PFS. For TMB stratification, TMB‐remainL and TMB‐dn patients had longer PFS than TMB‐remainH and TMB‐up patients (Figure 4E, left panel). We further combined all groups of “up” with “remainH” and “dn” with “remainL” into a two‐dimensional stratification manner. Status of ERBB2 and TCL did not correlate with PFS, whereas high level of TMB predicted a poor PFS (Figure 4E, right panel). Combining both BL and FP conditions, we concluded that compared with ERBB2 copy number or TCL, plasma TMB was a better option to predict and dynamically surveillant the therapeutic outputs of HER2‐targeted therapy in GC.
3.8. Immunotherapy may serve as a better therapeutic option than HER2‐targeted therapy for ERBB2 highTMBhigh GC patients
As we have demonstrated, ERBB2 highTMBhigh patients were unlikely to benefit much from HER2‐targeted therapy if they carried high TMB (Table S5 and Figure 4C). To mark a mechanistic insight, we performed enrichment analysis for the pathways potentially disturbed by plasma‐borne genomic aberrances. The numbers of enriched signaling were larger in ERBB2 highTMBhigh than in ERBB2 highTMBlow patients at BL (272 vs 17) and FP (195 vs 58). Combining BL and FP, the most enriched pathways were displayed in Figure 5A. Apart from several major cancer‐related pathways, immunity‐related pathways were also found more enriched in ERBB2 highTMBhigh patients. The enrichment diversity between these two subgroups suggested that accumulation of tumor mutations dysregulated cancerous signaling and disrupted tumor immune responses.
FIGURE 5.
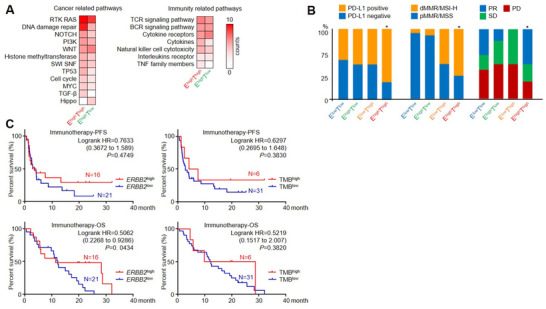
ERBB2 highTMBhigh patients displayed a more favorable outlook for immunotherapy. (A) Through investigating the ctDNA genomic changes in the HER2‐targeted cohort, the major cancer‐related and immunity‐related pathways displayed a higher enrichment in the ERBB2 highTMBhigh group. For the immunotherapy cohort, (B) the correlation of PD‐L1 expression, MSI status, and responses after received immune checkpoint inhibitors were compared across ERBB2‐TMB‐stratified patients. (C) Progression‐free survival (PFS) and overall survival (OS) for patients with different ERBB2/TMB status were demonstrated
We then explored whether these HER2‐positive/ERBB2 amplified plus TMB‐high patients were apt for immunotherapy. We referred to an immunotherapy cohort consisted of 37 GC patients recruited by PD‐1/PD‐L1/CTLA‐4‐targeted clinical trials (patient information is shown in Table S7). The ORR and DCR for the immunotherapy cohort were 29.7% (11/37) and 56.8% (21/37), respectively, inferior to the HER2‐targeted cohort (50% and 75%), whereas mPFS and mOS were 3.53 and 11.9 months, respectively. In this immunotherapy cohort, ctDNAs for baseline samples were extracted and sequenced for ERBB2 copy number and TMB. With the same cutoff standard as in the HER2‐targeted cohort, 12 TMBlow ERBB2 high and four TMBhigh ERBB2 high patients were identified.
The positive rates for PD‐L1 (75% vs 50%) and dMMR/MSI‐H (66.7% vs 9.1%) were higher in ERBB2 highTMBhigh patients than in ERBB2 highTMBlow patients (Figure 5B). However, PFS and OS showed no significant differences between TMBhigh and TMBlow group, which might due to the limited numbers of patients. ERBB2 high patients showed a better OS but not PFS than ERBB2 low patients (Figure 5C). Additionally, ERBB2 highTMBhigh patients displayed higher response rates (ORR = 50% vs 16.7%; DCR = 75% vs 50%) and a trend of longer PFS/OS than ERBB2 highTMBlow patients after receiving immune checkpoint inhibitors (Figure S8). Compared with the previous HER2‐targeted cohort, the mPFS of ERBB2 highTMBhigh patients received immune checkpoint inhibitors (19.85 month) was much longer than received HER2‐targeted regimens (3.07 month), whereas the mPFS of ERBB2 highTMBlow patients received immune checkpoint inhibitors (2.53 month) was inferior to patients received HER2‐targeted regimens (8.5 month). The overall workflow and major findings in this study are summarized in Figure 6.
FIGURE 6.
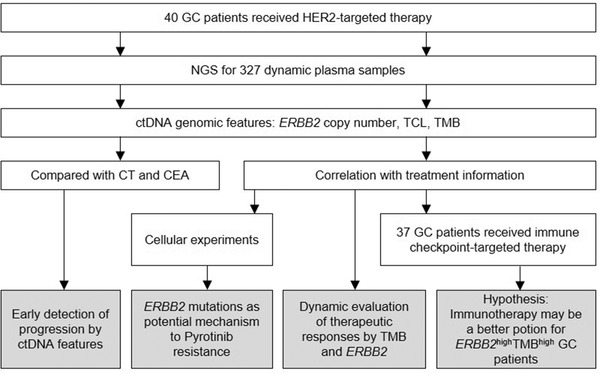
An overview of the workflow and the major conclusions in this study. Through tracking the serial changes of plasma ctDNA from GC patients underwent HER2‐targeted therapy or immunotherapy, our work provided four section of clues that directed the clinical decision‐making of GC
4. DISCUSSION
GC remains to be one of the most prevalent and deadly types of cancer around the world. Due to the high heterogeneity and cavitary characteristic of the digestive tract, the development of precision medicine in GC lag behind other cancer types. Here, we analyzed plasma‐borne genomic information throughout the whole treatment processes and discovered that ERBB2 copy number and TMB showed strong correlation with responses to HER2‐targeted therapy. As shown by our data, patients with high ctDNA ERBB2 copy number at baseline exerted better response to HER2‐targeted therapy, which complied with other reported studies based on tissue HER2 positivity. 14 , 28 However, the reduction of ERBB2 copy number at the first point after regimen administration was insufficient to mark an optimistic therapeutic output. On the other hand, patients with high TMB at baseline or first point both displayed poor response, suggesting that ctDNA‐derived TMB was a more powerful prognostic biomarker than ERBB2 copy number. It is worth to be mentioned that 62% of patients received combination of chemotherapy and HER2‐targeted therapy. Chemotherapy has been demonstrated to lead to ctDNA reduction in GC, 29 thus may have some impact on our interpretation of ctDNA analysis in this study.
Apart from tumor mutations, the accumulation of CNVs could also lead to cancer development and therapeutic resistance. TCL was reported to be inversely correlated with prognosis in isocitrate dehydrogenase (IDH)‐mutant astrocytoma, 30 whereas the total CNV burden in metastatic prostate cancer was significantly higher than that in nonmetastatic prostate cancer. 31 CNV burden was also considered as a pan‐cancer prognostic factor associated with recurrence and death. 32 , 33 However, the prognostic efficiency of TCL was inferior to TMB and ERBB2 copy number in our cohort. We inferred that the intrinsic genomic heterogeneity of GC as well as the limited information provided by panel‐sequencing compared to whole exome/genome sequencing might have some impact on the precise evaluation of overall CNVs in the plasma ctDNA. The detectability of mutations that is highly affected by the plasma‐ctDNA frequency abundance is another factor that may impact TMB interpretation. The mutations detected in plasma ctDNA sometimes underrepresent the mutations harbored in tumor tissue, which leads to inaccurate TMB calculation. Thus, plasma TMB need to be interpreted with caution in clinical setting when identifying patients for better therapeutic outcomes. Nevertheless, because several studies have already explored the possibility to accurately estimate TMB from target sequencing using various gene panel across different cancer type, 34 , 35 , 36 the relatively small panel size (425 candidate genes) of targeted sequencing does not discolored the findings and conclusions concerning TMB in our study.
HER2 is the most important therapeutic target in GC. Around 10‐20% GC patients are identified as HER2 positive, for whom Trastuzumab plus chemotherapy is the only regimen approved as the first‐line treatment. 37 In order to expand the armory of therapy against GC, the development of additional HER2‐targeted compounds and regimens so as to dealing with emerging Trastuzumab tolerance/acquired resistance is an urgent need. Besides patients accepting conventional Trastuzumab‐based therapy, those who were administrated with other HER2‐targeted regimens (JACOB, TDM1, RC48‐ADC, and Pyrotinib) initiated by our center were also included in this study. 28 Considering the limited case number (40 in total) and complicated therapeutic background (several patients were treated twice with HER2‐targeted regimens), we did not perform further stratification based on these regimens, but still, a consistent trend of mutation and copy number changes has been observed across all cases. On the other hand, given that this cohort is mainly consisted of HER2‐positive cases, the conclusion and observation we found in this study may not be applied to all GC cases. Meanwhile, this cohort bias may also affect the concordance with tumor tissue analysis of GC in other study, due to which limited HER2‐negative cases were included.
IHC staining combined with FISH of tissue has been recognized as the golden standard in identifying GC HER2 positivity. 38 However, the requirement for formalin‐Fixed Paraffin‐Embedded (FFPE)‐embedded tissue slides overwhelms tissue‐based assay for quick diagnosis and dynamic surveillance of disease progression. In the era of precision medicine, omics technique‐powered liquid biopsy is being embraced for guiding therapeutic decisions and monitoring relapse. For example, nonsmall cell lung cancer patients tested positive for plasma EGFR mutation have been approved eligible for EGFR tyrosine kinase inhibitors. 39 On the other hand, a study (TARGET) that aimed to allocate patients with advanced cancer to suitable clinical trials based on ctDNA assay also showed high concordance with tissue on aspects of both testing and therapeutic outcomes. 40 Nevertheless, for GC, although several studies have reported liquid biopsy's efficacy in detecting disease and tracking therapeutic responses, 41 , 42 , 43 the longitudinal plasma‐borne genomic details along with HER2‐targeted therapy remain largely uninvestigated. Our data provided clues that ctDNA copy number‐defined ERBB2 positivity shared a high concordance with HER2 expression‐ or ERBB2 copy number‐defined positivity in paired tissues, which emphasized the potential to utilize ctDNA assay as an auxiliary diagnosis and an alternative method in noninvasive, dynamic evaluation. On the other hand, although it has not been evaluated in this study, NGS‐quantified plasma‐borne TMB (pTMB or bTMB) status has been proved to be correlated with tissue TMB status, suggesting that plasma‐based NGS sequencing is feasible for TMB estimation and subsequent clinical evaluation. 44 , 45
CT scanning serves as the most common noninvasive method to diagnose and evaluate cancer progression. 46 Originated from cavitary organs, gastrointestinal cancers involve in abdominal region and are generally highly metastatic. Therefore, the detection of lesions by CT has a low sensitivity in gastrointestinal cancer than in other solid tumors and accompanied with time delays. 47 Although NGS‐based liquid biopsy for ctDNA has been reported as a powerful tool in screening, detecting, and monitoring cancer development, 48 the advantage of liquid biopsy in predicting therapeutic responses is not well characterized yet. Our study demonstrated that plasma ctDNA‐based PD was able to be detected more than a month earlier than the conventional CT scan in 40% of GC patients, saving enough time for clinical decisions. However, there were some PD patients who were not covered by plasma ctDNA testing in this cohort, suggesting ctDNA may serve as a complementary rather than substitute test.
It has been pointed out by several studies that mutations of ctDNA ERBB2 correlated with HER2‐targeted resistance in cancer. ERBB2 L869R mutation was reported to contributed to Trastuzumab resistance in breast cancer. 49 ERBB2 S310F, S310Y, and E321G mutants exhibited poor response to Trastuzumab in lung cancer. 50 Previous reports indicated that D769Y was an activation mutation form of HER2, 51 whereas L755S was known to confer Lapatinib resistance. 52 Our findings indicated that specific ERBB2 mutations (V659D and L755S) might contribute to acquired resistance to Pyrotinib therapy, yet further studies are warranted to confirm these results and to reveal the phenotypical changes induced by these mutations.
In addition to prompting early diagnosis and pre‐imaging evaluation, optimizing other regimens for patients is also a key point in improving therapeutic efficiency. By targeting immune‐suppressive checkpoint molecules (e.g., PD1, PD‐L1, and CTLA‐4), immunotherapy rescued patients’ immune system and prevented cancer cells from evading the immune surveillance, and thus exerted a consistent antitumor efficacy for heterogenic patients. 53 Apart from single‐agent therapy, the combination of ICI agents with chemotherapy, targeted therapy, or additional ICI agents was being testified by various of clinical trials. 54 , 55 Recently, CheckMate 649 trial has revealed that patients with HER2‐negative GCs displayed significantly improved PFS and OS from combinational nivolumb(nivo, PD‐1 inhibitor) and chemotherapy(chemo) compared to chemotherapy alone in Western populations. This observed result was also supported by ATTRACTION 4 trial that evaluated efficacy of nivo + chemo in Asian HER2‐negative GC patients. 56 , 57 Both studies showed that ICI combined with chemo could be a potential first‐line treatment option for patients with GC.
It has been shown in multiple tumors, indexes such as PD‐L1 positivity, dMMR/MSI, and EBV infection are benefitable biomarkers from ICI therapy, yet TMB has not been considered valid in instructing immunotherapy in GC. 59 , 60 , 61 In this study, considering patients in ICI and HER2‐targeted cohorts that shared identical HER2/ERBB2 and TMB status and similar clinical features (comparable age and gender distribution, both in advanced stage), we speculated that immunotherapy might be a better option than HER2‐targeted therapy for advanced GC patients with positive HER2 and high TMB. Moreover, it has been shown by a series of studies that HER2 inhibition strengthened the antibody‐dependent cellular cytotoxicity while enhancing the activation of T and NK cells, potentially synergized with immune checkpoint blockade approaches. 62 Currently, combining ICI inhibitors with HER2 inhibitors (such as Trastuzumab or Margetuximab) has been expected to display promising therapeutic efficacy against HER2‐positive breast and GC, 63 , 64 , 65 and thus we inferred that the feasibility of inhibiting both HER2 and immune checkpoints in ERBB2 highTMBhigh GC patients also deserved further investigation, whereas for ERBB2 highTMBlow GC patients, our data suggested that HER2‐targeted therapy remained to be a more suitable option than immunotherapy. On the other hand, the ORR, DCR, and mPFS of the two ERBB2 low (non‐AMP/negative) groups were slightly better after received immunotherapy (33.33% [7/21], 57.14% [12/21], and 3.53 months, respectively) than received HER2‐targeted therapy (25% [3/12], 50% [6/12], and 3.05 months, respectively), suggesting that immunotherapy might also be a potential therapeutic strategy for HER2‐negative patients. Yet still, these therapeutic indications remained to be validated in larger population of GC patients.
In conclusion, our data showed that plasma‐borne ERBB2 copy number and TMB were of great potential as progression predictors or prognostic biomarkers for HER2‐targeted in ERBB2‐postive GC. Further studies are warranted to validate our observation for better therapeutic management of GC patients.
AUTHOR CONTRIBUTIONS
Cheng Zhang, Jing Gao, and Lin Shen contributed to the study conception and design. Cheng Zhang, Zuhua Chen, Xiaoyi Chong, Zhenghang Wang, and Xiaotian Zhang contributed to the data acquisition. Zuhua Chen, Xiaoxi Chen, and Yang Shao contributed to the data interpretation. Cheng Zhang, Xiaoyi Chong, Yang Chen, Ruoying Yu, and Tingting Sun contributed to the statistical analysis. All authors read and approved the final manuscript.
CONFLICT OF INTERESTS
Ruoying Yu and Xiaoxi Chen are the employees of Geneseeq Technology Inc Canada. Tingting Sun and Yang Shao are the employee of Nanjing Geneseeq Technology Inc. The remaining authors have no conflicts of interest to declare.
AVAILABILITY OF DATA AND MATERIALS
The datasets used in the current study are available from the corresponding authors on reasonable request.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGMENTS
We deeply appreciate Drs Yanyan Li, Congcong Ji, Ying Wu, and Qian Zhao (Peking University Cancer Hospital & Institute), Dr Haixing Wang (The First Affiliated Hospital of Xiamen University), and Dr Zhentao Liu (Peking University Third Hospital) for kind suggestions and generous help in providing experimental materials during this study. This work was supported by the National Key Research and Development Program of China (2018YFC1313304), the National Natural Science Foundation of China (No. 81802327 and No. 81872341), the Major Program of National Natural Science Foundation of China (No. 91959205), and the Capital's Funds for Health Improvement and Research (No. 2020‐1‐1022).
Zhang C, Chen Z, Chong X, et al. Clinical implications of plasma ctDNA features and dynamics in gastric cancer treated with HER2‐targeted therapies. Clin Transl Med. 2020;10:e254 10.1002/ctm2.254
Cheng Zhang, Zuhua Chen, and Xiaoyi Chong contributed equally to this work.
Contributor Information
Xiaotian Zhang, Email: zhangxiaotianmed@163.com.
Jing Gao, Email: gaojing_pumc@163.com.
Lin Shen, Email: shenlin@bjmu.edu.cn.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654‐2664. [DOI] [PubMed] [Google Scholar]
- 3. Dagogo‐Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81‐94. [DOI] [PubMed] [Google Scholar]
- 4. Kurokawa Y, Matsuura N, Kimura Y, et al. Multicenter large‐scale study of prognostic impact of HER2 expression in patients with resectable gastric cancer. Gastric Cancer. 2015;18(4):691‐697. [DOI] [PubMed] [Google Scholar]
- 5. Abrahao‐Machado LF, Scapulatempo‐Neto C. HER2 testing in gastric cancer: an update. World J Gastroenterol. 2016;22(19):4619‐4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao JJ, Osgood CL, Gong Y, et al. FDA approval summary: pertuzumab, trastuzumab, and hyaluronidase‐zzxf injection for subcutaneous use in patients with HER2‐positive breast cancer. Clin Cancer Res. 2020. 10.1158/1078-0432.Ccr-20-3474. [DOI] [PubMed] [Google Scholar]
- 7. Huober J, Weder P, Veyret C, et al. PERNETTA: a non‐comparative randomized open label phase II trial of pertuzumab (P) + trastuzumab (T) with or without chemotherapy both followed by T‐DM1 in case of progression, in patients with HER2‐positive metastatic breast cancer (MBC): (SAKK 22/10 / UNICANCER UC‐0140/1207). Ann Oncol. 2018;29:viii93. [Google Scholar]
- 8. Shitara K, Bang YJ, Iwasa S, et al. Trastuzumab deruxtecan in previously treated HER2‐positive gastric cancer. N Engl J Med. 2020;382(25):2419‐2430. [DOI] [PubMed] [Google Scholar]
- 9. Xu H, Sheng X, Yan X, et al. A phase II study of RC48‐ADC in HER2‐negative patients with locally advanced or metastatic urothelial carcinoma. J Clin Oncol. 2020;38(15_suppl):e17113‐e. [Google Scholar]
- 10. Makatsoris T, Samantas E, Manousou K, et al. Afatinib in combination with cisplatin and 5‐fluorouracil (5‐FU) as first line treatment in inoperable gastric and gastro‐esophageal junction (GEJ) cancer: a phase II study by the Hellenic Cooperative Oncology Group. Ann Oncol. 2018;29:viii227‐viii8. [Google Scholar]
- 11. Tabernero J, Hoff PM, Shen L, et al. Pertuzumab plus trastuzumab and chemotherapy for HER2‐positive metastatic gastric or gastro‐oesophageal junction cancer (JACOB): final analysis of a double‐blind, randomised, placebo‐controlled phase 3 study. Lancet Oncol. 2018;19(10):1372‐1384. [DOI] [PubMed] [Google Scholar]
- 12. Saura C, Oliveira M, Feng YH, et al. Neratinib plus capecitabine versus lapatinib plus capecitabine in HER2‐positive metastatic breast cancer previously treated with >/ = 2 HER2‐directed regimens: phase III NALA trial. J Clin Oncol. 2020;38(27):3138‐3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montemurro F, Delaloge S, Barrios CH, et al. Trastuzumab emtansine (T‐DM1) in patients with HER2‐positive metastatic breast cancer and brain metastases: exploratory final analysis of cohort 1 from KAMILLA, a single‐arm phase IIIb clinical trial(). Ann Oncol. 2020;31(10):1350‐1358. [DOI] [PubMed] [Google Scholar]
- 14. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet. 2010;376(9742):687‐697. [DOI] [PubMed] [Google Scholar]
- 15. Sheng X, Zhou A‐P, Yao X, et al. A phase II study of RC48‐ADC in HER2‐positive patients with locally advanced or metastatic urothelial carcinoma. J Clin Oncol. 2019;37(15_suppl):4509. [Google Scholar]
- 16. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics‐driven oncology. Nat Rev Genet. 2019;20(2):71‐88. [DOI] [PubMed] [Google Scholar]
- 17. Pantel K, Alix‐Panabières C. Liquid biopsy and minimal residual disease ‐ latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409‐424. [DOI] [PubMed] [Google Scholar]
- 18. Kim K, Shin DG, Park MK, et al. Circulating cell‐free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014;86(3):136‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang WL, Lan YT, Huang KH, et al. Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer. 2016;138(12):2974‐2983. [DOI] [PubMed] [Google Scholar]
- 20. Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD‐1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449‐1458. [DOI] [PubMed] [Google Scholar]
- 21. Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2‐positive gastric cancer. Gut. 2019;68(7):1152‐1161. [DOI] [PubMed] [Google Scholar]
- 22. Yang W, You N, Jia M, et al. Undetectable circulating tumor DNA levels correlate with low risk of recurrence/metastasis in postoperative pathologic stage I lung adenocarcinoma patients. Lung Cancer. 2020;146:327‐334. [DOI] [PubMed] [Google Scholar]
- 23. Tong L, Ding N, Tong X, et al. Tumor‐derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9(19):5532‐5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang W, Ma Y, Yin JC, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti‐PD‐(L)1 therapies in non‐small cell lung cancer. Clin Cancer Res. 2019;25(16):5015‐5026. [DOI] [PubMed] [Google Scholar]
- 25. Feng F, Sun L, Zheng G, et al. Low lymphocyte‐to‐white blood cell ratio and high monocyte‐to‐white blood cell ratio predict poor prognosis in gastric cancer. Oncotarget. 2017;8(3):5281‐5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kao SC, Pavlakis N, Harvie R, et al. High blood neutrophil‐to‐lymphocyte ratio is an indicator of poor prognosis in malignant mesothelioma patients undergoing systemic therapy. Clin Cancer Res. 2010;16(23):5805‐5813. [DOI] [PubMed] [Google Scholar]
- 27. Zhou C, Yuan Z, Ma W, et al. Clinical utility of tumor genomic profiling in patients with high plasma circulating tumor DNA burden or metabolically active tumors. J Hematol Oncol. 2018;11(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tabernero J, Hoff PM, Shen L, et al. Pertuzumab plus trastuzumab and chemotherapy for HER2‐positive metastatic gastric or gastro‐oesophageal junction cancer (JACOB): final analysis of a double‐blind, randomised, placebo‐controlled phase 3 study. Lancet Oncol. 2018;19(10):1372‐1384. [DOI] [PubMed] [Google Scholar]
- 29. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715‐1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mirchia K, Sathe AA, Walker JM, et al. Total copy number variation as a prognostic factor in adult astrocytoma subtypes. Acta Neuropathol Commun. 2019;7(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu B, Lu X, Shen H, et al. Intratumoral heterogeneity and genetic characteristics of prostate cancer. Int J Cancer. 2020;146(12):3369‐3378. [DOI] [PubMed] [Google Scholar]
- 32. Hieronymus H, Murali R, Tin A, et al. Tumor copy number alteration burden is a pan‐cancer prognostic factor associated with recurrence and death. eLife. 2018;7:e37294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hieronymus H, Schultz N, Gopalan A, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci USA. 2014;111(30):11139‐11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fang W, Ma Y, Yin JC, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti‐PD‐(L)1 therapies in non‐small cell lung cancer. Clin Cancer Res. 2019;25(16):5015‐5026. [DOI] [PubMed] [Google Scholar]
- 35. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim J, Kim B, Kang SY, et al. Tumor mutational burden determined by panel sequencing predicts survival after immunotherapy in patients with advanced gastric cancer. Front Oncol. 2020;10:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong J, Liu T, Fan Q, et al. Optimal regimen of trastuzumab in combination with oxaliplatin/ capecitabine in first‐line treatment of HER2‐positive advanced gastric cancer (CGOG1001): a multicenter, phase II trial. BMC Cancer. 2016;16:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanayama K, Imai H, Yoneda M, Hirokawa YS, Shiraishi T. Significant intratumoral heterogeneity of human epidermal growth factor receptor 2 status in gastric cancer: a comparative study of immunohistochemistry, FISH, and dual‐color in situ hybridization. Cancer Sci. 2016;107(4):536‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non‐small cell lung cancer? Ann Transl Med. 2017;5(3):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738‐743. [DOI] [PubMed] [Google Scholar]
- 41. Wang Y, Zhao C, Chang L, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab‐resistant mechanism in advanced gastric cancer. EBioMedicine. 2019;43:261‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2‐positive gastric cancer. Gut. 2019;68(7):1152‐1161. [DOI] [PubMed] [Google Scholar]
- 43. Chen Z, Zhang C, Zhang M, et al. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. 2019;10(10):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non‐small cell lung cancer with use of a next‐generation sequencing cancer gene panel. JAMA Oncol. 2019;5(5):696‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fenizia F, Pasquale R, Roma C, Bergantino F, Iannaccone A, Normanno N. Measuring tumor mutation burden in non‐small cell lung cancer: tissue versus liquid biopsy. Transl Lung Cancer Res. 2018;7(5):668‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smyth EC, Verheij M, Allum W, Cunningham D, Cervantes A, Arnold D. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2016;27:v38‐v49. [DOI] [PubMed] [Google Scholar]
- 47. Burbidge S, Mahady K, Naik K. The role of CT and staging laparoscopy in the staging of gastric cancer. Clin Radiol. 2013;68(3):251‐255. [DOI] [PubMed] [Google Scholar]
- 48. Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell‐free DNA methylomes. Nature. 2018;563(7732):579‐583. [DOI] [PubMed] [Google Scholar]
- 49. Chen Z, Sun T, Yang Z, et al. Monitoring treatment efficacy and resistance in breast cancer patients via circulating tumor DNA genomic profiling. Mol Genet Genomic Med. 2019;8(2):e1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Greulich H, Kaplan B, Mertins P, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci USA. 2012;109(36):14476‐14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kancha RK, von Bubnoff N, Bartosch N, Peschel C, Engh RA, Duyster J. Differential sensitivity of ERBB2 kinase domain mutations towards lapatinib. PLoS ONE. 2011;6(10):e26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zayac A, Almhanna K. Esophageal, gastric cancer and immunotherapy: small steps in the right direction? Transl Gastroenterol Hepatol. 2020;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taieb J, Moehler M, Boku N, et al. Evolution of checkpoint inhibitors for the treatment of metastatic gastric cancers: current status and future perspectives. Cancer Treat Rev. 2018;66:104‐113. [DOI] [PubMed] [Google Scholar]
- 55. Dolcetti R, De Re V, Canzonieri V. Immunotherapy for gastric cancer: time for a personalized approach. Int J Mol Sci. 2018;19(6):1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moehler M, Shitara K, Garrido M, et al. LBA6_PR Nivolumab (nivo) plus chemotherapy (chemo) versus chemo as first‐line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer (GC/GEJC)/esophageal adenocarcinoma (EAC): first results of the CheckMate 649 study. Ann Oncol. 2020;31:S1191. [Google Scholar]
- 57. Boku N, Ryu MH, Oh DY, et al. LBA7_PR Nivolumab plus chemotherapy versus chemotherapy alone in patients with previously untreated advanced or recurrent gastric/gastroesophageal junction (G/GEJ) cancer: aTTRACTION‐4 (ONO‐4538‐37) study. Ann Oncol. 2020;31:S1192. [Google Scholar]
- 58. Long J, Lin J, Wang A, et al. PD‐1/PD‐L blockade in gastrointestinal cancers: lessons learned and the road toward precision immunotherapy. J Hematol Oncol. 2017;10(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee B, Hutchinson R, Wong HL, et al. Emerging biomarkers for immunomodulatory cancer treatment of upper gastrointestinal, pancreatic and hepatic cancers. Semin Cancer Biol. 2018;52:241‐252. [DOI] [PubMed] [Google Scholar]
- 60. Myint ZW, Goel G. Role of modern immunotherapy in gastrointestinal malignancies: a review of current clinical progress. J Hematol Oncol. 2017;10(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Le DT, Uram JN, Wang H, et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med. 2015;372(26):2509‐2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12(4):237‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Myint ZW, Goel G. Role of modern immunotherapy in gastrointestinal malignancies: a review of current clinical progress. J Hematol Oncol. 2017;10(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Loi S, Giobbie‐Hurder A, Gombos A, et al. Pembrolizumab plus trastuzumab in trastuzumab‐resistant, advanced, HER2‐positive breast cancer (PANACEA): a single‐arm, multicentre, phase 1b‐2 trial. Lancet Oncol. 2019;20(3):371‐382. [DOI] [PubMed] [Google Scholar]
- 65. Quagliariello V, Passariello M, Coppola C, et al. Cardiotoxicity and pro‐inflammatory effects of the immune checkpoint inhibitor Pembrolizumab associated to Trastuzumab. Int J Cardiol. 2019;292:171‐179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION
Data Availability Statement
The datasets used in the current study are available from the corresponding authors on reasonable request.


