Abstract
For a complete understanding of a system's processes and each protein's role in health and disease, it is essential to study protein expression with a spatial resolution, as the exact location of proteins at tissue, cellular, or subcellular levels is tightly linked to protein function. The Human Protein Atlas (HPA) project is a large‐scale initiative aiming at mapping the entire human proteome using antibody‐based proteomics and integration of various other omics technologies. The publicly available knowledge resource www.proteinatlas.org is one of the world's most visited biological databases and has been extensively updated during the last few years. The current version is divided into six main sections, each focusing on particular aspects of the human proteome: (a) the Tissue Atlas showing the distribution of proteins across all major tissues and organs in the human body; (b) the Cell Atlas showing the subcellular localization of proteins in single cells; (c) the Pathology Atlas showing the impact of protein levels on survival of patients with cancer; (d) the Blood Atlas showing the expression profiles of blood cells and actively secreted proteins; (e) the Brain Atlas showing the distribution of proteins in human, mouse, and pig brain; and (f) the Metabolic Atlas showing the involvement of proteins in human metabolism. The HPA constitutes an important resource for further understanding of human biology, and the publicly available datasets hold much promise for integration with other emerging efforts focusing on single cell analyses, both at transcriptomic and proteomic level.
Keywords: antibodies, cells, Human Protein Atlas, proteomics, transcriptomics, tissues, single‐cell
1. INTRODUCTION
Human physiology is tightly linked to the complex interplay between cell type‐specific functions and molecular interactions between cells. For a full understanding of underlying disease mechanisms, it is necessary to study the tissue architecture with a single‐cell resolution. Ever since completion of the human genome sequence, several efforts have focused on mapping the entire human proteome, the functional representation of the genome. The standard approach for spatial localization of proteins in tissue is antibody‐based proteomics, for example, immunohistochemistry (IHC) that allows for in situ detection of human proteins in intact tissue samples.
Built upon this strategy, the Human Protein Atlas (HPA) project has been established, with the overall aim to reveal the spatial distribution of the entire human proteome in different cells, tissues, and organs. 1 Antibody‐based imaging is combined with transcriptomics, mass spectrometry‐based proteomics, kinetics, and systems biology, forming the basis for the comprehensive open‐source database divided into six different interconnected sub‐atlases: (a) the Tissue Atlas showing the distribution of proteins across all major tissues and organs in the human body; 2 (b) the Cell Atlas showing the subcellular localization of proteins in single cells; 3 (c) the Pathology Atlas showing the impact of protein levels for survival of patients with cancer; 4 (d) the Blood Atlas showing the expression profiles of blood cells and actively secreted proteins; 5 (e) the Brain Atlas showing the distribution of proteins in human, mouse and pig brain; 6 and (e) the Metabolic Atlas showing the involvement of proteins in human metabolism 7 (Figure 1).
FIGURE 1.
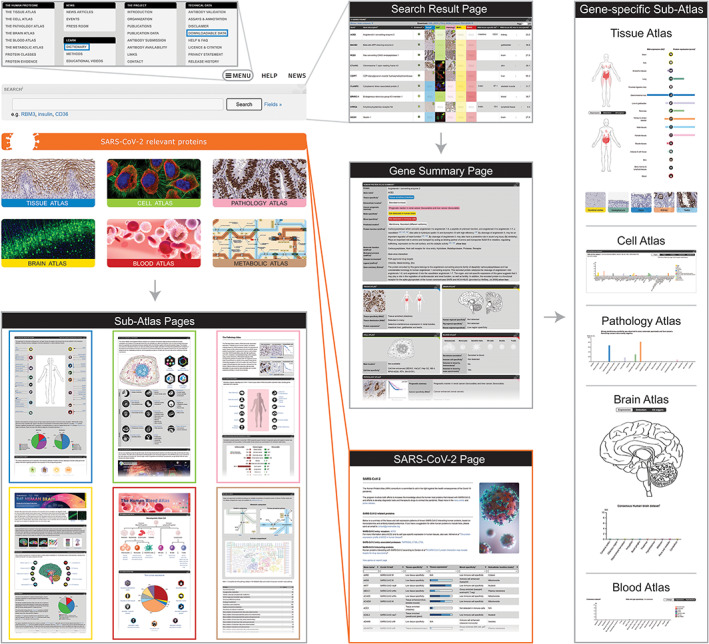
The global structure of the Human Protein Atlas web portal. Exploration of the Human Protein Atlas data is guided through a systemic and a gene‐centric approach, both subdivided into six interconnected sub‐atlases. The systemic approach entails exploration of various sub‐atlas proteomes, each specific to a group of genes belonging to a certain location or activity within the body, such as an organ, blood, or genes involved in metabolism. The sub‐atlas pages are accessed by clicking on either of the sub‐atlas images on the startpage. The other option is to search for specific genes using the startpage search field that can be combined with various different filtering options. The search leads the visitor to a search result page, wherein the gene of interest can be selected to access gene‐specific data, both summarized in the gene summary page and presented in depth in the different gene‐specific sub‐atlases. In addition, the database contains a page with data concerning SARS‐CoV‐2 relevant proteins, as well as downloadable datasets and educational material such as dictionaries, found in the menu (blue frames)
The database is updated on a yearly basis, including both new antibody characterization data in tissues and cells, as well as new functionalities. Version 19 contains >26,000 antibodies, covering >17,000 unique proteins, thereby covering ~87% of the human protein‐coding genome. The Blood Atlas, Brain Atlas, and Metabolic Atlas constitute the most recent additions to the database, providing novel insights on several different aspects of human biology. In addition to these new sections, the previous Tissue Atlas, Cell Atlas, and Pathology Atlas have been expanded with new datasets and functionalities.
HPA provides an important resource for both basic and clinical research. Here, a summary of recent updates are provided, together with examples highlighting how the data can be useful for answering different research questions, and how the six different sections are inter‐connected and complementary.
2. THE TISSUE ATLAS
The integral part of the HPA database that has been the main focus since the first release in 2005 is the Tissue Atlas, that presents the cell type‐specific spatial localization of 15,313 proteins in >40 different human tissues and organs. The analysis is combined with mRNA expression data from three different body‐wide datasets: Internally generated HPA data, 2 , 8 the genome‐based tissue expression (GTEx) consortium 9 based on RNA‐seq, and the FANTOM5 consortium 10 based on cap analysis gene expression.
In order to provide a comprehensive overview of expression levels across the entire human body and highlight genes and proteins selectively expressed in certain organs, a consensus classification was performed taking into consideration all three RNA expression datasets. Based on normalized expression levels (NX) across 61 tissues, organs, and blood cell types 5 , 11 summarized into 37 different main organ systems (Figure 2a), each gene was classified based on tissue specificity and tissue distribution. The tissue specificity is based on the fold‐change of mRNA expression levels across all analyzed 37 tissues and organs, divided into five different specificity categories: (a) Tissue enriched: One tissue has at least fourfold higher mRNA level compared to all other tissues, (b) Group enriched: A group of 2–5 tissues have at least fourfold higher mRNA levels compared to all other tissues, (c) Tissue enhanced: One tissue has at least fourfold higher mRNA level compared to the average level in all other tissues, (d) Low tissue specificity: At least one tissue has mRNA levels above cutoff, but gene does not belong to any of the above categories, or (e) Not detected: All tissues have mRNA levels below cutoff. Genes defined as elevated (tissue enriched, group enriched, or tissue enhanced) constitute particularly interesting targets for organ‐specific research, as the corresponding proteins are responsible for many of the organ‐specific functions that may be disrupted in disease (Figure 2b).
FIGURE 2.
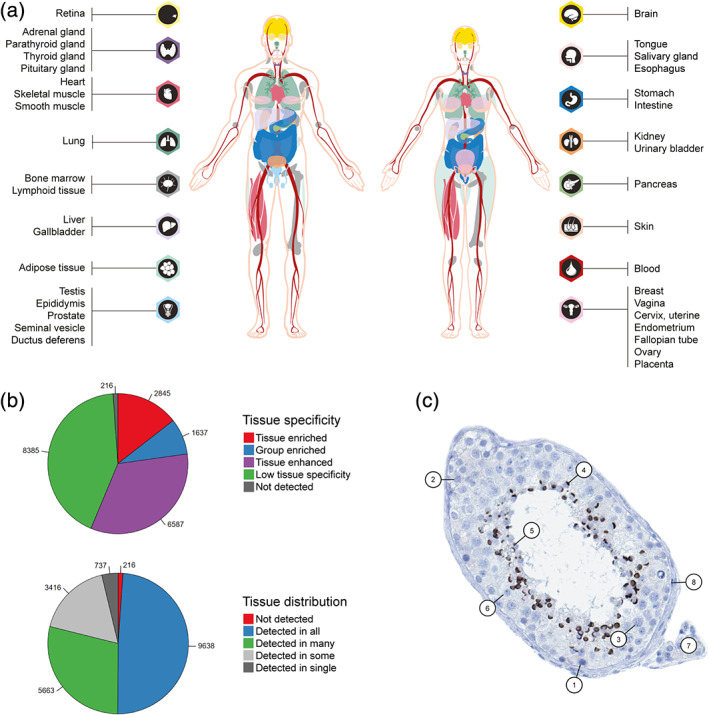
The Tissue Atlas—integration of mRNA expression with IHC. (a) Overview of the 37 main organ systems with RNA expression levels based on three different datasets (HPA, GTEx, and FANTOM5), used for classification of tissue specificity and tissue distribution. (b) Pie charts showing the number of genes belonging to each of the categories for tissue specificity and tissue distribution. (c) IHC staining of zona pellucida binding protein (ZPBP) in testis tissue, outlining the eight different cell types present in the sample: 1: spermatogonia; 2: preleptoteine spermatocytes; 3: pachytene spermatocytes, 4: round/early spermatids; 5: elongated/late spermatids; 6: Sertoli cells; 7: Leydig cells; 8: peritubular cells. ZPBP is highly expressed in round/early spermatids and elongated/late spermatids
In addition to tissue specificity, another level of categorization based on the consensus RNA expression dataset has been introduced in the most recent version of the Tissue Atlas—tissue distribution. The tissue distribution takes into consideration how many tissues that have detectable mRNA levels above cutoff, and is divided into five different categories: (a) detected in single, where only a single tissue has detectable levels, (b) detected in some, where more than one but less than one‐third of the tissues have detectable levels, (c) detected in many, where at least a third but not all tissues have detectable levels, (d) detected in all, where all 37 tissues have detectable levels, and (e) not detected, where none of the 37 tissues have detectable levels. The analysis showed that only 737 proteins are detected in a single tissue, constituting especially interesting targets for organ‐specific research (Figure 2b).
Combining the mRNA‐based tissue specificity with IHC analysis allows for further investigation of the exact location of the corresponding proteins, thereby revealing important functional context. Based on this strategy, HPA has published many separate papers focusing on tissue‐specific proteomes. 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 One organ that has proven particularly interesting is the testis, which constitutes the organ with the highest number of tissue elevated genes. 2 This is considered to be due to the complex nature of this organ, whereby thousands of genes and proteins are activated and repressed during spermatogenesis, the process where stem cells undergo several steps of mitosis and meiosis before being developed into mature sperm. Many of the corresponding proteins involved in this process have an unknown function, 27 , 28 , 29 , 30 and studying their detailed cell type‐specific locations gives important insights into their potential function in spermatogenesis, revealing targets important for further research on infertility and reproductive disorders. Previously, manual annotation of each testis image on the HPA was limited to two main cell types—cells in seminiferous ducts, and Leydig cells. This broad characterization however lacked important details on the various cell types and cell states present in this organ, and in order to perform an in‐depth analysis and add important information about exact cell type‐specific locations, the annotation was expanded to eight different cell types by involving an expert in reproductive biology (Figure 2c). This extended effort was built upon existing publicly available images on the HPA, and in a first study focusing on ~500 genes with an elevated expression in testis, 31 detailed spatial localization of a large number of previously uncharacterized proteins was provided. The proposed strategy of adding more in‐depth characterization to already existing images has large potential to be used also for the other normal tissues and organs, not only focusing on the main organ‐specific cell types, but also looking at cell types present in all organs such as endothelial cells, fibroblasts and different immune cells. During the last few years, the emerging technology of single cell RNA‐seq (scRNA‐seq) has received increased attention, 32 allowing for quantitative measurements of single cell transcriptomes across different human tissues and cell types. Further advances in this field will lead to the possibility to identify genes and proteins elevated in certain cell types that can be combined with detailed cell type‐specific characterization of protein expression levels based on IHC.
The exact cell type‐specific localization of proteins is not only important for understanding normal organ‐specific functions, but also for revealing mechanisms crucial for disease progression and therapy decisions. One example of such a study, taking advantage of the stringent antibody validation workflow implemented in the HPA, is the recently published body‐wide protein expression profile of the Angiotensin I converting enzyme 2 (ACE2), suggested to be the key protein involved in SARS‐CoV‐2 host cell entry. 33 For a full understanding of the susceptibility for SARS‐CoV‐2 infection and the progression to severe and fatal disease, it is necessary to study the cell type‐specific expression of suggested entry factors. It has previously been suggested that entry of SARS‐CoV‐2 via ACE2 would explain the severe clinical manifestations observed in various tissues and organs, including the respiratory system. In the new study, it could confidently be shown that none or only very low levels of ACE2 were present in the normal respiratory system, which was in contrast to previous studies. 34 Further studies are urgently needed to study the dynamic regulation of ACE2, and to confirm whether the low ACE2 expression in the human respiratory system is sufficient for SARS‐CoV‐2 infection, or if other factors are needed for host cell entry. 35 , 36
The protein expression profile of ACE2 is the result of the extensive knowledge gathered from the three different RNA expression datasets published on the HPA portal, combined with stringent IHC analysis and several other external datasets both on the RNA and protein level. The IHC data was generated using strategies proposed by the International Working Group for Antibody Validation (IWGAV). 37 IWGAV was formed with representatives from several major academic institutions and proposed five different pillars to be used for antibody validation, to ensure reproducibility of antibody‐based studies. Validation must be performed in an application‐specific manner, using at least one of the strategies suggested by IWGAV. HPA has implemented the criteria for enhanced validation as suggested by IWGAV, and in the present version 19 of the HPA, >10,000 antibodies have been validated using at least one of these criteria. 38 The overall aim is to increase this number in upcoming versions. For IHC, two different strategies for antibody validation can be used: (a) orthogonal strategy, based on comparison of protein expression levels using an antibody‐independent method, analyzing the expression levels of the same target across tissues expressing the target protein at different levels; or (b) independent antibody strategy, defined as a similar expression pattern observed by an independent antibody targeting a non‐overlapping region of the same protein. In the Tissue Atlas, 3,775 genes meet the criteria for enhanced validation, where the corresponding proteins have been targeted by at least one antibody validated by orthogonal or independent antibody strategy.
The Tissue Atlas workflow, that combines transcriptomics‐based tissue specificity with stringent IHC analysis, has also proven important in further characterization of “missing proteins,” defined as proteins that lack evidence of existence on the protein level according to the international initiative Human Proteome Project (HPP) and its reference knowledgebase neXtProt. 39 The current count of “missing proteins” stands at 1900, out of which a high proportion appear to be localized in special tissues not part of the standard tissue repertoire in the HPA. 40 By adding novel tissue types based on transcriptomics information from the GTEx and FANTOM5 datasets, >300 proteins specifically expressed in, for example, eye, thymus, hair follicles, and specialized regions of the brain could be characterized, out of which a large proportion corresponded to “missing proteins.” 41 Future efforts in adding more specialized tissues, based on information from external datasets both on the mRNA and protein level, hold promise for increasing the coverage of the tissue‐based map of the human proteome by including such rarely expressed proteins. In the quest for characterizing “missing proteins,” consideration should also be taken regarding the fact that some proteins may only be expressed during development, for example, expressed in fetal tissues, or under certain functional circumstances, for example, in lactating breasts. Another set of proteins highlighted by the HPP is “uncharacterized proteins” that have been confidently identified but lack functional annotation. One such group of uncharacterized proteins belong to the protein evidence category PE1, that is, have evidence on the protein level, and are thus defined as uPE1 proteins. 42 For this set of proteins, corresponding to 1,136 Ensembl genes in the HPA of which 899 have available data in the Tissue Atlas, the spatial information may guide in selecting appropriate tissues and cell types for further functional characterization.
Antibody‐based data is currently not considered for evidence of protein existence in neXtProt, but future integrative studies with mass spectrometry and IHC are planned in order to increase the understanding of which types of proteins that can be confidently detected by one method and not the other. This is of special interest in the quest for “missing proteins.”
3. THE PATHOLOGY ATLAS
In order to compare the protein expression levels under normal circumstances with a disease state, the Pathology Atlas was introduced, with main emphasis on cancer. The major release in 2017 presented the association of all human protein‐coding genes with clinical outcome in 17 major cancer types, based on genome‐wide expression data from the Cancer Genome Atlas. 4 This systems level approach allows for exploration of genes that correlate with favorable or unfavorable prognosis, that is, genes where high RNA expression levels are associated with either longer or shorter survival time. All primary data is summarized in an interactive survival scatter plot for each gene, which gives a unique opportunity to study the correlation between expression levels and survival time for each individual patient, or focusing separately on factors that could potentially affect the survival data, such as gender, dead/alive or a certain cancer stage. In version 18.1 released at the end of 2018, additional features were added to the existing survival scatter plots. Both axes were complemented with kernel density curves demonstrating the data density over the axes, which facilitates interpretation (Figure 3a). The top density plot shows the distribution of expression levels among dead and alive patients, while the right density plot visualizes the data density of the number of years the dead patients survived based on high or low expression levels, respectively. In addition to the density curves, a P‐score landscape is shown together with dead median separation (Figure 3b), that defines the difference in median mRNA expression between dead patients with high and low expression. This is intended to aid the user in visually exploring custom cutoffs and the associated P‐scores and dead/median separation.
FIGURE 3.
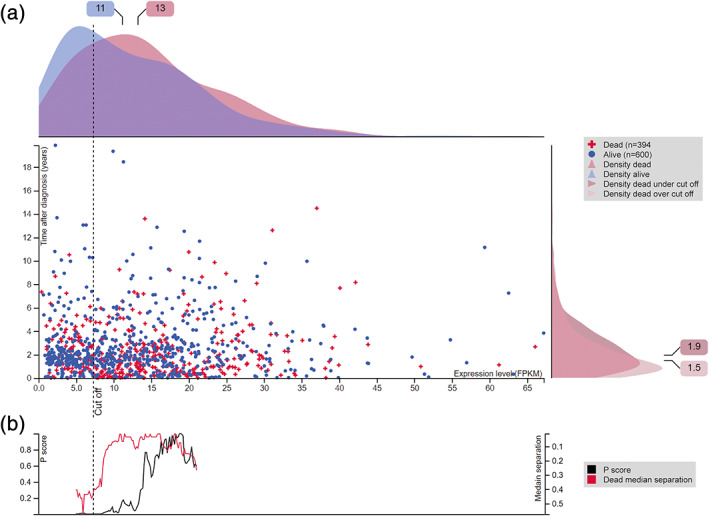
The Pathology Atlas—association of gene expression with clinical outcome. (a) Interactive survival scatter plot of cyclin dependent kinase 1 (CDK1) in lung cancer. The plot shows the clinical status (i.e., dead or alive) for all individuals in the patient cohort. The top kernel density plot demonstrates the expression level (FPKM) distribution among dead (red) and alive (blue) patients. The right density plot shows the data density of the survived years of dead patients with high and low expression levels, respectively, stratified using the cutoff indicated by the vertical dashed line. The cutoff is automatically generated, representing the lowest P score. (b) The P score landscape (black curve; left axis) is shown together with dead median separation (red curve; right axis), highlighting the difference in median mRNA expression between patients who have died with high and low expression, respectively
Exploration of the potentially prognostic genes based on mRNA data can be complemented by studying the >5,000 images of IHC stained cancer tissues of 20 different cancer types, providing the basis for further cancer research. The expression levels in cancer can also be compared with corresponding normal tissue or cell types in the Tissue Atlas, identifying proteins that could serve as diagnostic biomarkers. Due to the importance of IHC in diagnostic pathology, analysis of the cell type‐specific expression patterns of potential prognostic and diagnostic biomarkers gives an unique opportunity to search for novel candidates related to cancer types where the currently used markers in the clinic are not enough to guide treatment modalities or predict patient outcome.
The data from the Pathology Atlas has proven to be useful to validate findings in a large number of studies focusing on various aspects of cancer research, including therapeutic efforts 43 , 44 , 45 , 46 or more basic understanding of tumor biology, such as tumor heterogeneity. 47 , 48
4. THE CELL ATLAS
Taking the spatial distribution of proteins to an even more detailed level includes determining the location of all human proteins at a subcellular level, such as different organelles. The Cell Atlas presents the subcellular distribution of 12,390 proteins based on high‐resolution immunofluorescence images of cell lines. The proteins are mapped to 32 different subcellular structures corresponding to 13 main organelles. Since the major update in 2016, further efforts have focused on increasing the number of enhanced validated antibodies, and presently, 1,484 genes meet the criteria for enhanced validation. Three main strategies are used for enhanced validation of protein expression patterns in cell lines: (a) Genetic validation/siRNA, where the staining procedure is repeated on siRNA transfected U‐2 OS cells in order to knock‐down the expression of the protein, 49 (b) Recombinant expression validation, where the antibody is further analyzed in HeLa cell lines stably expressing a GFP‐tagged target protein, and (c) Independent antibody validation, where two or more independent antibodies directed towards non‐overlapping regions of the target protein are used to assess the reliability of the staining.
Further development of the Cell Atlas includes characterization of the cell cycle specific proteome, providing a comprehensive map of spatiotemporal heterogeneity of the humane proteome. 50 The analysis was based on integrating proteomics at subcellular resolution with scRNA‐seq and pseudotime measurements of individual cells within the cell cycle. It was shown that 17% of the human proteome displays cell‐to‐cell variability, of which 26% has a connection to cell cycle progression. The spatially resolved proteomic map of the cell cycle will be integrated in the upcoming version 20 of the HPA database, presenting the first evidence for 235 novel cell cycle‐associated proteins, and serving as a valuable tool for accelerating molecular studies of the human cell cycle and cell proliferation.
In addition to the vast amount of biological information provided in the HPA database, the massive image collection of >10 million annotated images serves a valuable resource as benchmark datasets for further development of deep learning models for image classification and segmentation. The HPA has been engaged in two such different projects based on the high‐resolution images provided in the Cell Atlas. One project focused on a general audience through a groundbreaking gamified citizen science effort, 51 in which the gaming community was involved through integration of subcellular classification tasks in EVE Online, a popular massively multiplayer online game. The resulting mini‐game called “Project Discovery” was played by >300,000 citizen scientists that achieved the major milestone of together generating >33 million classifications of protein subcellular location. The classifications were both compared with and used for boosting an artificial intelligence (AI) model for automated prediction of subcellular location, the first generalized tool for annotating proteins with multiple locations. The citizen science project constitutes a unique example of a workflow to rapidly leverage the output of large‐scale science efforts.
In another project, the computational community was involved through an open Kaggle challenge, 52 where >2000 teams participated over a period of three months in predicting multiple subcellular labels per image. This classification is easy to a trained eye, but challenging to automate. The presented solutions showed a wide variety of applied strategies, and the winning model outperformed the previous effort for multi‐label classification by ~20%. The presented models can be used for new image classifications or feature extractions for a wide range of biological applications.
5. THE BRAIN ATLAS
While the Tissue Atlas includes four different regions of the human brain (cerebral cortex, hippocampus, caudate, and cerebellum), these parts do not fully represent the heterogeneous nature of the brain, with many nuclei and cell types organized in complex networks. In an extended effort, an integrated view of one‐to‐one orthologs of proteins located in different regions of the human, mouse, and pig brain was added as the new Brain Atlas, 24 released in 2019. The regional expression in the brain of these three mammalian species was profiled based on 1710 human brain samples, 119 pig brain samples, and 67 mouse brain samples. A gene classification was based on 10 main anatomical regions of each mammalian brain (Figure 4a). As many as 2,587 genes were elevated in the brain compared to other organs when taking into consideration the highest normalized expression value within these 10 regions in comparison to the rest of the human body (Figure 4b). This number of elevated genes is the highest of all human organs, thereby highlighting the brain together with the testis as the organs with most proteins potentially involved in organ‐specific functions. A separate classification was then performed for the brain, in order to identify genes elevated in individual brain regions. Interestingly, many of the top genes regionally elevated in the brain have not been previously described in neural cells. Some of these genes were not elevated from a whole‐body perspective, and many of the previously identified “signature genes” for brain specific cell types were shown to have higher expression in certain peripheral tissues. Two such examples are ankyrin 1 (ANK1), a transport protein elevated in skeletal muscle, and transcription factor AP‐2 beta (TFAP2B), enriched in epididymis. In brain, both of these proteins were specifically expressed in cerebellum (Figure 4c). Another important finding in the Brain Atlas is that many key genes were differentially expressed between the three species, calling for caution when results from animal models are translated into research on the human brain.
FIGURE 4.
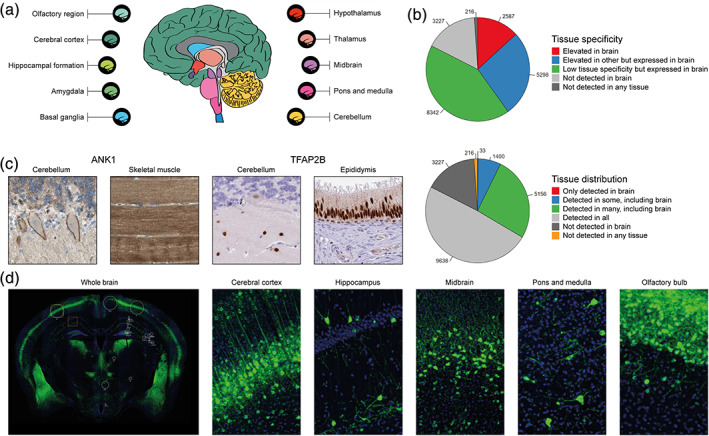
The Brain Atlas—exploration of gene expression in the mammalian brain. (a) Overview of the 10 main regions with RNA expression data from all three species (human, pig, and mouse), used for classification of genes according to expression levels. (b) Pie charts showing the number of genes belonging to each of the categories for tissue specificity and tissue distribution, comparing brain to other human tissues. (c) IHC images of ANK1 and TFAP2B that on a body‐wide level are elevated in other tissue types, but in the brain showed regional expression only in cerebellum. (d) Immunofluorescent images of whole mouse brain, as well as five specific regions detailing the protein expression of NECAB1 at a cell type‐specific level
As a complement to the IHC images of human brain in the Tissue Atlas, the Brain Atlas also includes immunofluorescently stained whole sections of mouse brain for a selection of proteins involved in normal brain physiology, brain development and neuropathological disorders. This allows localization of the proteins to specific structures that are challenging to target when assessing human samples, and as many as 129 different areas and subfields of the brain are covered for 271 different brain‐relevant proteins. One example is the N‐terminal EF‐hand calcium binding protein 1 (NECAB1), specifically expressed in soma and dendrites of neuronal cells (Figure 4d).
6. THE BLOOD ATLAS AND THE HUMAN SECRETOME
Transportation of cells and proteins throughout the human circulatory system is vital to our survival. Physiological functions such as the immune system, systems‐level control of homeostasis, transport of nutrients and hormone regulation are all dependent on the circulating cells and proteins found in the blood. Since blood is easily accessible and a rich source of systemic health‐related information, it has become the most commonly used material for clinical and medical research analysis. The effort of the HPA program to map the complete human proteome was regarding blood previously limited to gene expression in peripheral blood mononuclear cells (PBMCs). The complete group of actively secreted human proteins to blood or elsewhere, also known as the “human secretome,” was another research area in need of further exploration. A tandem of efforts was therefore undertaken to map the complete proteome of blood cells as well as the proteins secreted by human cells. 5 , 53 The resulting data have been included into the HPA website in the form of a new Blood Atlas released in 2019, as well as integrated into the general exploration of protein expression.
A genome‐wide single‐cell transcriptomics study was performed to map the expression of all protein‐coding genes in flow cytometric sorted blood immune cell populations. 5 Data from the study can be found summarized in the Blood Atlas part of the HPA website, as well as integrated into the individual gene pages. In the “human blood cell”‐part of the Blood Atlas, expression profiles have been created for the 18 blood cell types and six lineages analyzed in the study, highlighting the specificity and distribution of the expression of all protein‐coding genes among the blood cell populations (Figure 5a). The mRNA levels of individual protein‐coding genes in multiple blood immune cell populations can be explored in the Blood Atlas gene pages of HPA, where four different transcriptomic datasets have been made available. In addition to the HPA dataset of 18 blood cell types, two additional datasets have been imported from other recent transcriptomics studies, including the analysis in 15 blood cell types by Schmiedel et al. 54 and 29 blood cell types as well as total PBMCs by Monaco et al. 55 A normalized “consensus dataset” was also created from the HPA blood cell dataset to enable between‐sample‐comparison with mRNA data from tissues.
FIGURE 5.
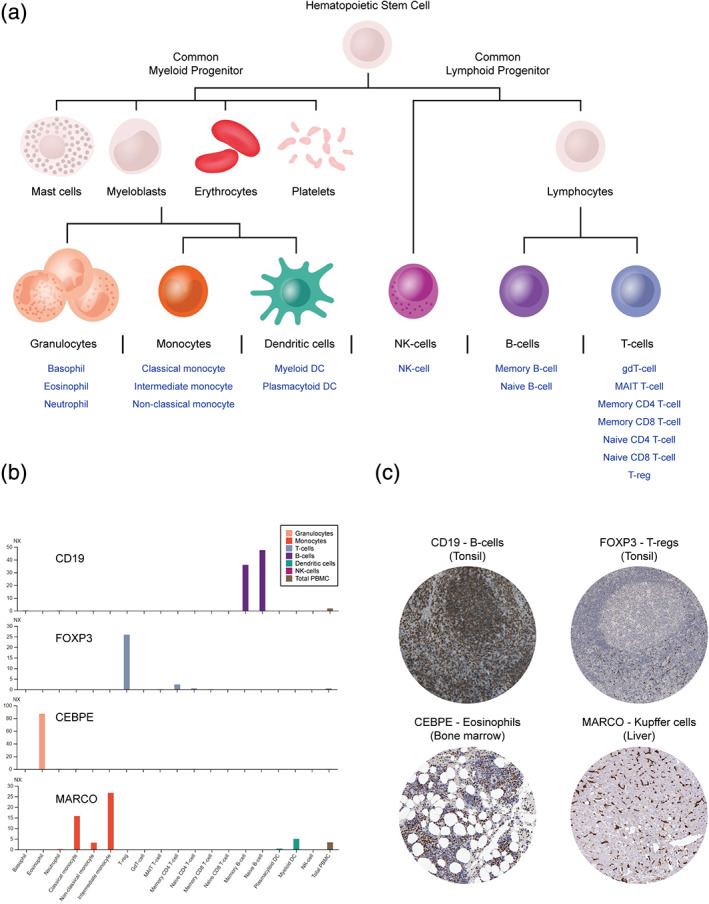
The Blood Atlas—exploration of gene expression in human blood cells. (a) A schematic visualization of the tree of hematopoiesis is found in the Blood Atlas, including six lineages and 18 blood cell types that are clickable to enable access to expression profiles with information regarding expression‐related specificity and distribution. (b) RNA expression levels of known protein markers for specific blood cell types, and (c) representative IHC images of corresponding protein expression patterns in human tissues
Global expression analysis using the data from the study found blood cells to be most highly correlated with bone marrow and lymphoid tissues, both of which are rich in immune cells, as well as revealing a relatively large portion of intracellularly expressed proteins among the blood cells compared to organs with a high degree of actively secreted proteins, such as the liver and the pancreas. Concerning expression specificity, genes with elevated expression in certain blood cell types or lineages are indicated in the Blood Atlas expression profiles along with lists of the genes that have the highest level of enriched expression in each cell population. The analysis confirmed the expression specificity of many of the markers used to distinguish different blood cell types. In Figure 5b, RNA expression levels of four such example genes are provided, complemented with IHC‐based location in human tissues (Figure 5c). These examples include the canonical cell surface receptor CD19, found to be exclusively expressed in B‐cells, the transcriptional regulator forkhead box P3 (FOXP3), that was found to be mainly expressed on regulatory T cells (Tregs), as well as two proteins with enriched expression in cells of myeloid origin: the CCAAT/enhancer binding protein epsilon (CEBPE), expressed in eosinophils, and macrophage receptor with collagenous structure (MARCO), expressed in monocytes. CEBPE is a transcriptional activator linked to the primary immunodeficiency specific granule deficiency (SGD), where granulocyte differentiation is impaired. 56 IHC images of CEPBE in HPA show clear expression of the protein in bone marrow, where granulocyte differentiation occurs (Figure 5c). MARCO is a macrophage‐associated pattern recognition receptor (PRR) involved in the innate recognition and phagocytosis of Gram‐positive and Gram‐negative bacteria. 57 Based on mRNA levels, MARCO was expressed in two types of monocytes, the intermediate and the classical type. Monocytes are progenitors to both macrophages and dendritic cells, a relationship which was supported by the mRNA expression of MARCO in myeloid type dendritic cells as well as IHC‐based protein expression in resident macrophages, including Kupffer cells of the liver (Figure 5b,c). Overall, neutrophils, basophils, and plasmacytoid dendritic cells were found to have the highest amount of genes with cell type‐enriched expression.
The distribution of gene expression conveys which and how many of the genes that are expressed or not expressed in the various cell populations. Blood cells were together found to express about half (~10,000) of the protein‐coding genes and almost two‐thirds of the amount of genes expressed in tissues (~14,000–16,600), demonstrating the diversity of the multicellular composite tissues compared to single cells. A large amount (889) of interesting genes were also identified based on the fact that they had enriched gene expression and were only found to be expressed in a single cell type, indicating the possibility of those genes having important biological functions linked to the phenotypes of respective cell type.
A large‐scale effort of annotating the complete set of actively secreted proteins, the “human secretome,” was performed in parallel to the transcriptomic study of blood cells. 53 A group of 2,641 genes were predicted to have at least one secreted protein isoform and consequently selected for deep analysis to determine a final location, a site of origin and the relative abundance of each candidate. The analysis was in large based on existing data from both internal and external sources, including antibody and mass spectrometry‐based methods. The resulting human secretome data can be found in the Blood Atlas, as a complement to the human blood cell exploration, as many of the secreted proteins have blood related functions. All of the predicted actively secreted proteins have been classified according to their final location in the human body, based on manual annotation of antibody‐based spatial proteomics combined with published literature, bioinformatics analysis, and experimental evidence. The classes include three main categories: intracellular, secreted to blood, and secreted locally, where the latter was divided into seven location‐based sub‐categories (Figure 6a). The resulting nine classes of secretome proteins can be explored in the human secretome‐part of the Blood Atlas, wherein the group of proteins belonging to each class are described in terms of functions, specificity and distribution of mRNA expression across 37 analyzed tissues, and tissue origin of the predicted secreted proteins (Figure 6a–d).
FIGURE 6.
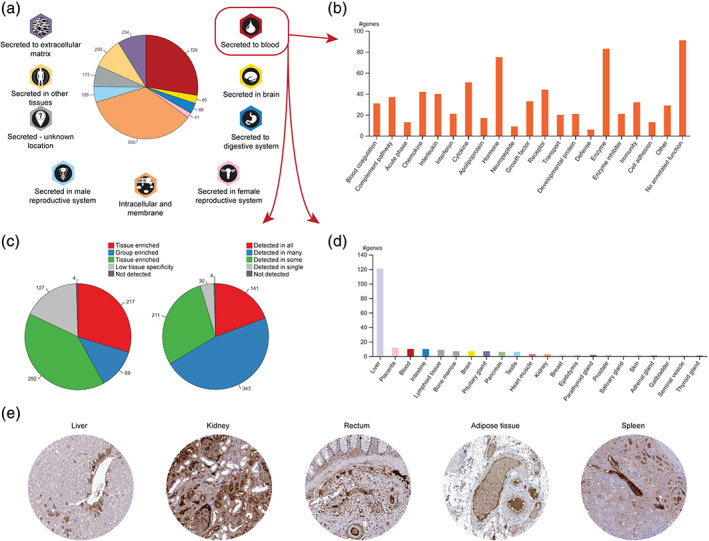
The Human Secretome—annotation of actively secreted proteins. (a) The complete set of predicted actively secreted proteins were annotated and divided into nine different classes based on their final location in the body. Each class was analyzed regarding (b) protein function, (c) expression specificity and distribution, and (d) tissue of origin among tissue enriched proteins (exemplified by the class “secreted to blood”). (e) Spatial IHC analysis of F2 in five tissues, a protein that is annotated as secreted to blood by the liver, with exclusive mRNA expression in the liver
The analysis found that a somewhat surprisingly small portion (729) of the proteins are secreted to blood, while many of the predicted secreted proteins (933) were found to be retained intracellularly or fused to cell membranes, bringing the number of actively secreted proteins down to 1,709. Among the proteins secreted to blood, 72 are products or targets of FDA‐approved drugs. Apart from well‐known groups of proteins such as cytokines, interleukins, interferons, and chemokines (154), complement and coagulation factors (68), hormones (75), growth factors (33) and enzymes (83), in‐depth analysis of the proteins secreted to blood revealed about 100 proteins with unknown functions, and hence potentially attractive candidates for future studies (Figure 6b). A large portion (217) of the proteins secreted to blood are tissue enriched (Figure 6c), where more than half originate from the liver (Figure 6d). One such protein is coagulation factor 2, or thrombin (F2), exclusively expressed by the liver and subsequently secreted to the blood where it exerts its function during coagulation, in part by cleaving fibrinogen into the coagulation agent fibrin. Spatial IHC analysis reveals presence of F2 in a subset of liver hepatocytes as well as high expression in plasma, visible in tissues from all parts of the body (Figure 6e).
The presence and quantity of the proteins predicted to be actively secreted was investigated by compiling experimental data from mass spectrometry, antibody‐based assays and proximity extension assay (PEA). A large majority (99%) of the combined protein mass was found to be contributed by a small number of proteins, while the majority of the proteins were detected at low concentrations. Areas in need of further exploration were identified, as it was not clear whether many of the proteins detected at low concentration were actively secreted or leaked from dying cells. As many as 142 of the proteins predicted to be secreted to blood could not be detected with the current methods. The complete set of plasma concentrations of blood proteins, compiled from various sources, based on antibody and mass spectrometry‐based methods, can be found in the “human plasma proteome”‐part of the Blood Atlas.
7. THE METABOLIC ATLAS
The Metabolic Atlas is a collaborative effort with Chalmers University of Technology, to map and facilitate a holistic understanding of human metabolism by combining a newly created genome‐scale metabolic model (GEM): Human1, with visual metabolic maps and HPA multi‐omics data, parts of which have been incorporated into the HPA. 7 Human1 was created based on the components and information from three human GEMs (HMR2, iHsa, and Recon3D), 58 , 59 , 60 and constitutes the most extensive model of human metabolism, taking into account 13,417 reactions, 4,164 metabolites, and 3,625 genes. As a means to overcome issues of reproducibility and transparency, details of the extensive curation of Human1 has been stored into a public Git‐repository on GitHub to allow for research community‐driven development of the model. The Metabolic Atlas (metabolicatlas.org) is an open‐access website that provides the infrastructure for the exploration of Human1 and other GEMs through 2D and 3D visualization of metabolic maps together with integrated data concerning the thousands of reactions, compounds, and genes that form the metabolic systems. HPA transcriptomic data corresponding to 37 different human tissues has been integrated into the metabolic maps of Human1, showing relative mRNA expression of each enzyme in the displayed metabolic pathway.
Parts of the Metabolic Atlas features, including Human1‐based metabolic maps and metabolic pathway data, have recently been incorporated into HPA as a Metabolic Atlas sub‐atlas, providing information regarding the tool together with lists of all of the human metabolic pathways and associated enzymes (Figure 7a). The metabolic data is also integrated into the general exploration of the proteome via the Tissue Atlas gene pages, displaying a metabolic summary of associated reaction pathways and cellular compartments for proteins involved in human metabolism (Figure 7b). Metabolic maps of each reaction pathway are also imported from metabolicatlas.org and are accompanied with heatmaps displaying mRNA expression of all pathway‐associated genes across 37 human tissues.
FIGURE 7.
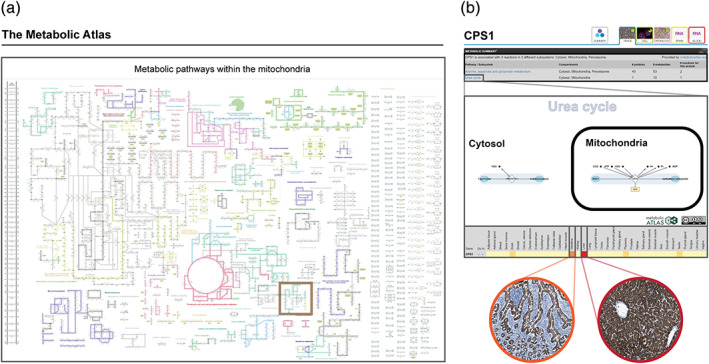
The Metabolic Atlas—integration of metabolic pathways with tissue‐specific proteomics. Features of the Metabolic Atlas (metabolicatlas.org) have been integrated into HPA as (a) a sub‐atlas with summarized information and maps regarding all human metabolic pathways (example showing the mitochondrial subcellular compartment with the Urea cycle indicated by a brown box) and (b) integrated into the Tissue Atlas part of the exploration of individual genes, including a summary of gene‐associated metabolic information, maps of associated metabolic pathways and heatmaps showing the mRNA expression of all pathway‐associated genes in 37 tissues
In addition to being tools for visualization of metabolic pathways, GEMs are also used to compare metabolic network structures, predict gene essentiality and simulate flow of metabolites, flux, through a reaction network. Flux simulations can be used to predict the complex behavior of metabolic systems in response to various internal and external changes, such as pathological imbalances, and thus get predictions of outcomes, such as changes in growth rates. GEMs thus have the potential to discern novel metabolic therapeutic targets that other less complex methods are unable to recognize. Various ways of utilizing Human1 were demonstrated by the developers, 7 including a comparative study of transcriptome‐based metabolic reaction structures among healthy and cancer tissues. It revealed a closer metabolic relationship between a cancer type and the healthy tissue type from where the cancer originates than between various cancer types originating from different tissues. This suggests that cancer development is somewhat constrained by the metabolic reaction structures of the tissue from which it originates, which means that acquired alterations to the metabolism during cancer development are significantly shaped by the varying nature of metabolism in different tissues of the body.
8. DATA AVAILABILITY AND OUTREACH
The main findings from each of the six different sub‐atlases are comprehensively summarized on landing pages with clickable figures, tables, lists, and examples, for quick access to results related to each aspect of the human proteome. Users of the HPA can also combine personal advanced search queries based on, for example, general gene and protein data, protein classes, tissue or cell type‐specific expression and validation criteria, using information from each of the different sub‐atlases. The results from these queries are presented as gene‐centric lists that are both clickable and downloadable in different formats. Another approach to access the extensive datasets found in the HPA is through 26 different downloadable files (https://www.proteinatlas.org/about/download) containing genome‐wide data across various assays, allowing for large‐scale bioinformatic studies.
The HPA spends considerable efforts on outreach, and the database is sustained through community contribution to European infrastructure ELIXIR, 61 where the HPA is listed as one of the core data resources with importance for the wider life‐science community. Another important aspect of outreach is educational material. One such example is production of a series of “Movie of the month” during 2020 and 2021. These educational movies allow for taking a journey into the body through 3D videos that transport you deep inside various organs. The imaging is based on antibody‐based profiling of tissues and light sheet microscopy. The videos are available at the HPA website as well as on a YouTube channel. The HPA educational material also includes a histology dictionary, 62 that serves as a helpful tool for both students and scientists interested in gaining further knowledge on the different tissue and cell structures that form the basis for the tissue and cell type‐specific protein expression patterns. By knowing how to interpret the >10 million high‐resolution images publicly available on www.proteinatlas.org, the visitors on the HPA can answer novel research questions not yet elucidated by the HPA team, for example, protein expression in rare structures or cell types, or patterns of regional expression. In HPA20, released in the autumn of 2020, a major update of the normal and pathology tissue dictionary is provided, allowing for free exploration of entire large‐scale tissue sections stained with hematoxylin and eosin, where all major cell types and structures have been highlighted and described.
To aid research related to the SARS‐CoV‐2 pandemic and allow researchers to quickly access all gene and protein expression data on SARS‐CoV‐2 related proteins, a dedicated page on the HPA database has been created (Figure 1). The clickable list allows for filtering of all SARS‐CoV‐2 related proteins based on tissue specificity or subcellular location, and studying their expression pattern in situ.
Several future national and international efforts are planned to expand the educational impact of the HPA resource even further, including more educational videos, Wikipedia summaries, and integration in student textbooks as well as tutorials related to both lab protocols and how to navigate the HPA database. Feedback from users is an integral part of the daily workflow, and many HPA researchers are active in responding to contact emails and participating in discussions on social media, thereby contributing to the scientific community and informing on recent progress in the field of antibody‐based proteomics.
9. DISCUSSION
For further understanding of cell and tissue heterogeneity, differentiation, diseases, and various biological processes involved in health and disease, it is necessary to study proteins at a single cell level. While various emerging technologies based on mass spectrometry are being developed in order to detect and quantify proteomes in single cells, 63 , 64 , 65 , 66 , 67 none of these currently have the resolution provided by IHC. By linking the identification of proteins with in situ location at tissue, cellular or subcellular level, important aspects of spatio‐temporal expression are provided, since the location is tightly linked to a proteins' function. This is particularly important as a large proportion of human proteins still have an unknown function. There are four main approaches for spatial proteomics: in situ mass spectrometry, fractionated cell lysates, proximity labeling, or imaging‐based proteomics. 68 , 69 The clear advantage of imaging‐based proteomics is that the proteins are analyzed in the native location with a single‐cell resolution.
The major initiative that aims to systematically study protein expression and localizations based on spatial proteomics is the HPA project. Initiated in 2003, the HPA approach allows the dissection of the human proteome from different perspectives, such as identifying all proteins localized to a certain cell type or organelle, housekeeping proteins, proteins related to certain pathways, or proteins enriched in a particular organ. In 2010, the HPA achieved a major milestone with protein expression data covering >50% of the protein‐coding genome. 70 During the last decade, the HPA open‐access knowledge resource (www.proteinatlas.org) has grown into one of the world's most visited biological databases with more than 300,000 visitors per month. The available data and images allows scientists from both academia and industry to freely explore the human proteome, and the massive image collection of >10 million high‐resolution images serves as an important resource for the computer vision community as benchmark datasets to develop deep learning models for image classification and segmentation. Several peer‐reviewed publications are published by external groups every day using data generated as part of the HPA project. The HPA has thus contributed to several thousands of external publications in the field of human biology and disease, including both basic and clinical research. The database is updated on a yearly basis, and in 2019, three new major sections were added: the Brain Atlas, the Blood Atlas and the Metabolic Atlas, providing novel insights into various aspects of the human proteome. The HPA will continue to evolve in future releases, adding both novel data and functionalities, refining the details of the human proteome.
The HPA spends a considerable effort on antibody validation and has implemented application‐specific criteria for enhanced validation as suggested by IWGAV. There are, however, still challenges in this field. Antibody‐based proteomics has a narrow dynamic range, the results are only semi‐quantitative, and cross‐reactivity is a well‐known issue. To reduce the risk of cross‐reactivity, all internally generated HPA antibodies have been affinity purified and analyzed with protein arrays. Antibodies that do not bind specifically to the intended antigen among other randomly selected protein fragments are not approved for further use. In addition, a manual evaluation of the staining pattern at tissue, cellular, and subcellular level is performed, resulting in the assignment of a reliability score. While this still does not rule out that cross‐reactivity may occur, transparent display of the results from these quality steps divide the available data into various categories, highlighting which set of proteins that have been most confidently validated. Further developments in the field of antibody validation are clearly warranted, and integration with other methods on the single cell level will likely lead to increased understanding of antibody specificity.
One emerging technology that likely will lead to important implications for proteomics and holds promise for integration with IHC is scRNA‐seq. This approach allows for studying mRNA transcripts expressed in smaller subsets of cells, and is excellent for studying cell heterogeneity. The Human Cell Atlas consortium 32 is a large‐scale international initiative aiming to create a comprehensive map of all human organs and cells based on scRNA‐seq, by a coordinated effort comprising >1,000 different institutes across >70 countries. scRNA‐seq is especially interesting to compare with IHC, as the methods allow for direct comparisons of cell type‐specific expression patterns. In addition to the Human Cell Atlas, several other large‐scale initiatives aim at characterizing human organs and cells based on scRNA‐seq, including the Human BioMolecular Atlas program and the Chan Zuckerberg Initiative, among others. Some tissue‐specific projects are international collaborations spanning over several of these initiatives, involving multiple research groups and combining expertise on established proteomic and transcriptomic techniques with novel technologies for spatial localization of mRNA transcripts, or various methods for analysis of multiple proteins in the same sample (multiplex).
Also in the mass spectrometry field, powerful technologies are being developed with the aim to allow detection and quantification of proteins in single cells. It is an exciting era of “big data” that will transform medicine and increase our understanding of human biology at entirely new levels. The large‐scale spatial proteomics datasets provided by the HPA hold much promise for integration with other ongoing and future efforts using both transcriptomics and quantitative proteomics methods, for a complete understanding of the human proteome in health and disease.
AUTHOR CONTRIBUTIONS
Andreas Digre: Investigation; visualization; writing‐original draft. Cecilia Lindskog: Conceptualization; investigation; supervision; visualization; writing‐original draft; writing‐review and editing.
ACKNOWLEDGMENTS
The project was funded by the Knut and Alice Wallenberg Foundation. Pathologists and staff at the Department of Clinical Pathology, Uppsala University Hospital, are acknowledged for providing the tissues used for RNA‐seq and immunohistochemistry. The authors would also like to thank all staff of the Human Protein Atlas for their work. The authors declare no conflicts of interest.
Digre A, Lindskog C. The Human Protein Atlas—Spatial localization of the human proteome in health and disease. Protein Science. 2021;30:218–233. 10.1002/pro.3987
Funding information Knut och Alice Wallenbergs Stiftelse
REFERENCES
- 1. Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018;27:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue‐based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 3. Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 4. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 5. Uhlen M, Karlsson MJ, Zhong W, et al. A genome‐wide transcriptomic analysis of protein‐coding genes in human blood cells. Science. 2019;366:eaax9198. [DOI] [PubMed] [Google Scholar]
- 6. Sjostedt E, Zhong W, Fagerberg L, et al. An atlas of the protein‐coding genes in the human, pig, and mouse brain. Science. 2020;367:eaay5947. [DOI] [PubMed] [Google Scholar]
- 7. Robinson JL, Kocabas P, Wang H, et al. An atlas of human metabolism. Sci Signal. 2020;13:eaaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue‐specific expression by genome‐wide integration of transcriptomics and antibody‐based proteomics. Mol Cell Proteomics. 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keen JC, Moore HM. The Genotype‐Tissue Expression (GTEx) project: Linking clinical data with molecular analysis to advance personalized medicine. J Pers Med. 2015;5:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu NY, Hallstrom BM, Fagerberg L, et al. Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium. Nucleic Acids Res. 2015;43:6787–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uhlen M, Hallstrom BM, Lindskog C, Mardinoglu A, Ponten F, Nielsen J. Transcriptomics resources of human tissues and organs. Mol Syst Biol. 2016;12:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson S, Nilsson K, Fagerberg L, et al. The transcriptomic and proteomic landscapes of bone marrow and secondary lymphoid tissues. PLoS One. 2014;9:e115911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Danielsson A, Ponten F, Fagerberg L, et al. The human pancreas proteome defined by transcriptomics and antibody‐based profiling. PLoS One. 2014;9:e115421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Djureinovic D, Fagerberg L, Hallstrom B, et al. The human testis‐specific proteome defined by transcriptomics and antibody‐based profiling. Mol Hum Reprod. 2014;20:476–488. [DOI] [PubMed] [Google Scholar]
- 15. Habuka M, Fagerberg L, Hallstrom BM, et al. The kidney transcriptome and proteome defined by transcriptomics and antibody‐based profiling. PLoS One. 2014;9:e116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kampf C, Mardinoglu A, Fagerberg L, et al. Defining the human gallbladder proteome by transcriptomics and affinity proteomics. Proteomics. 2014;14:2498–2507. [DOI] [PubMed] [Google Scholar]
- 17. Kampf C, Mardinoglu A, Fagerberg L, et al. The human liver‐specific proteome defined by transcriptomics and antibody‐based profiling. FASEB J. 2014;28:2901–2914. [DOI] [PubMed] [Google Scholar]
- 18. Lindskog C, Fagerberg L, Hallstrom B, et al. The lung‐specific proteome defined by integration of transcriptomics and antibody‐based profiling. FASEB J. 2014;28:5184–5196. [DOI] [PubMed] [Google Scholar]
- 19. Mardinoglu A, Kampf C, Asplund A, et al. Defining the human adipose tissue proteome to reveal metabolic alterations in obesity. J Proteome Res. 2014;13:5106–5119. [DOI] [PubMed] [Google Scholar]
- 20. Edqvist PH, Fagerberg L, Hallstrom BM, et al. Expression of human skin‐specific genes defined by transcriptomics and antibody‐based profiling. J Histochem Cytochem. 2015;63:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gremel G, Wanders A, Cedernaes J, et al. The human gastrointestinal tract‐specific transcriptome and proteome as defined by RNA sequencing and antibody‐based profiling. J Gastroenterol. 2015;50:46–57. [DOI] [PubMed] [Google Scholar]
- 22. Lindskog C, Linne J, Fagerberg L, et al. The human cardiac and skeletal muscle proteomes defined by transcriptomics and antibody‐based profiling. BMC Genomics. 2015;16:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Hurley G, Busch C, Fagerberg L, et al. Analysis of the human prostate‐specific proteome defined by transcriptomics and antibody‐based profiling identifies TMEM79 and ACOXL as two putative, diagnostic markers in prostate cancer. PLoS One. 2015;10:e0133449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sjostedt E, Fagerberg L, Hallstrom BM, et al. Defining the human brain proteome using transcriptomics and antibody‐based profiling with a focus on the cerebral cortex. PLoS One. 2015;10:e0130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zieba A, Sjostedt E, Olovsson M, et al. The human endometrium‐specific proteome defined by transcriptomics and antibody‐based profiling. OMICS. 2015;19:659–668. [DOI] [PubMed] [Google Scholar]
- 26. Bergman J, Botling J, Fagerberg L, et al. The human adrenal gland proteome defined by transcriptomics and antibody‐based profiling. Endocrinology. 2017;158:239–251. [DOI] [PubMed] [Google Scholar]
- 27. Jumeau F, Com E, Lane L, et al. Human spermatozoa as a model for detecting missing proteins in the context of the chromosome‐centric human proteome project. J Proteome Res. 2015;14:3606–3620. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y, Li Q, Wu F, et al. Tissue‐based proteogenomics reveals that human testis endows plentiful missing proteins. J Proteome Res. 2015;14:3583–3594. [DOI] [PubMed] [Google Scholar]
- 29. Vandenbrouck Y, Lane L, Carapito C, et al. Looking for missing proteins in the proteome of human spermatozoa: An update. J Proteome Res. 2016;15:3998–4019. [DOI] [PubMed] [Google Scholar]
- 30. Carapito C, Duek P, Macron C, et al. Validating missing proteins in human sperm cells by targeted mass‐spectrometry‐ and antibody‐based methods. J Proteome Res. 2017;16:4340–4351. [DOI] [PubMed] [Google Scholar]
- 31. Pineau C, Hikmet F, Zhang C, et al. Cell type‐specific expression of testis elevated genes based on transcriptomics and antibody‐based proteomics. J Proteome Res. 2019;18:4215–4230. [DOI] [PubMed] [Google Scholar]
- 32. Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. Elife. 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16:e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aguiar JA, Tremblay BJ, Mansfield MJ, et al. Gene expression and in situ protein profiling of candidate SARS‐CoV‐2 receptors in human airway epithelial cells and lung tissue. Eur Respir J. 2020;56:2001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nawijn MC, Timens W. Can ACE2 expression explain SARS‐CoV‐2 infection of the respiratory epithelia in COVID‐19? Mol Syst Biol. 2020;16:e9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edfors F, Hober A, Linderback K, et al. Enhanced validation of antibodies for research applications. Nat Commun. 2018;9:4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Omenn GS, Lane L, Overall CM, et al. Progress on identifying and characterizing the human proteome: 2019 metrics from the HUPO Human Proteome Project. J Proteome Res. 2019;18:4098–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adhikari S, Nice EC, Deutsch EW, et al. A high‐stringency blueprint of the human proteome. Nat Commun. 2020;11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sjostedt E, Sivertsson A, Hikmet Noraddin F, et al. Integration of transcriptomics and antibody‐based proteomics for exploration of proteins expressed in specialized tissues. J Proteome Res. 2018;17:4127–4137. [DOI] [PubMed] [Google Scholar]
- 42. Paik YK, Overall CM, Corrales F, Deutsch EW, Lane L, Omenn GS. Toward completion of the human proteome parts list: Progress uncovering proteins that are missing or have unknown function and developing analytical methods. J Proteome Res. 2018;17:4023–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jiang Y, Sun A, Zhao Y, et al. Proteomics identifies new therapeutic targets of early‐stage hepatocellular carcinoma. Nature. 2019;567:257–261. [DOI] [PubMed] [Google Scholar]
- 46. Vasaikar S, Huang C, Wang X, et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berglund E, Maaskola J, Schultz N, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stanta G, Bonin S. Overview on clinical relevance of intra‐tumor heterogeneity. Front Med. 2018;5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stadler C, Hjelmare M, Neumann B, et al. Systematic validation of antibody binding and protein subcellular localization using siRNA and confocal microscopy. J Proteomics. 2012;75:2236–2251. [DOI] [PubMed] [Google Scholar]
- 50. Mahdessian D, Cesnik AJ, Gnann C, et al. Spatiotemporal dissection of the cell cycle with single‐cell proteogenomics. bioRxiv. 2020;5432311. [DOI] [PubMed] [Google Scholar]
- 51. Sullivan DP, Winsnes CF, Akesson L, et al. Deep learning is combined with massive‐scale citizen science to improve large‐scale image classification. Nat Biotechnol. 2018;36:820–828. [DOI] [PubMed] [Google Scholar]
- 52. Ouyang W, Winsnes CF, Hjelmare M, et al. Analysis of the Human Protein Atlas Image Classification competition. Nat Methods. 2019;16:1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uhlen M, Karlsson MJ, Hober A, et al. The human secretome. Sci Signal. 2019;12:eaaz0274. [DOI] [PubMed] [Google Scholar]
- 54. Schmiedel BJ, Singh D, Madrigal A, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Monaco G, Lee B, Xu W, et al. RNA‐seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26:1627–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lekstrom‐Himes JA, Dorman SE, Kopar P, Holland SM, Gallin JI. Neutrophil‐specific granule deficiency results from a novel mutation with loss of function of the transcription factor CCAAT/enhancer binding protein epsilon. J Exp Med. 1999;189:1847–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Elomaa O, Sankala M, Pikkarainen T, et al. Structure of the human macrophage MARCO receptor and characterization of its bacteria‐binding region. J Biol Chem. 1998;273:4530–4538. [DOI] [PubMed] [Google Scholar]
- 58. Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J. Genome‐scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non‐alcoholic fatty liver disease. Nat Commun. 2014;5:3083. [DOI] [PubMed] [Google Scholar]
- 59. Blais EM, Rawls KD, Dougherty BV, et al. Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat Commun. 2017;8:14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brunk E, Sahoo S, Zielinski DC, et al. Recon3D enables a three‐dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drysdale R, Cook CE, Petryszak R, et al. The ELIXIR Core Data Resources: Fundamental infrastructure for the life sciences. Bioinformatics. 2020;36:2636–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kampf C, Bergman J, Oksvold P, et al. A tool to facilitate clinical biomarker studies—A tissue dictionary based on the Human Protein Atlas. BMC Med. 2012;10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heath JR, Ribas A, Mischel PS. Single‐cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Magness AJ, Squires JA, Griffiths B, et al. Multiplexed single cell protein expression analysis in solid tumours using a miniaturised microfluidic assay. Convergent Sci Phys Oncol. 2017;3:024003. [Google Scholar]
- 65. Palii CG, Cheng Q, Gillespie MA, et al. Single‐cell proteomics reveal that quantitative changes in co‐expressed lineage‐specific transcription factors determine cell fate. Cell Stem Cell. 2019;24:812–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Specht H, Emmott E, Koller T, Slavov N. High‐throughput single‐cell proteomics quantifies the emergence of macrophage heterogeneity. bioRxiv. 2019;6653071. [Google Scholar]
- 67. Kagan J, Moritz RL, Mazurchuk R, et al. National Cancer Institute Think‐Tank Meeting Report on Proteomic cartography and biomarkers at the single‐cell level: Interrogation of premalignant lesions. J Proteome Res. 2020;19:1900–1912. [DOI] [PubMed] [Google Scholar]
- 68. Gingras A‐C, Abe KT, Raught B. Getting to know the neighborhood: using proximity‐dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol. 2019;48:44–54. [DOI] [PubMed] [Google Scholar]
- 69. Lundberg E, Borner GH. Spatial proteomics: a powerful discovery tool for cell biology. Nat Rev Mol Cell Biol. 2019;20:285–302. [DOI] [PubMed] [Google Scholar]
- 70. Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge‐based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The main findings from each of the six different sub‐atlases are comprehensively summarized on landing pages with clickable figures, tables, lists, and examples, for quick access to results related to each aspect of the human proteome. Users of the HPA can also combine personal advanced search queries based on, for example, general gene and protein data, protein classes, tissue or cell type‐specific expression and validation criteria, using information from each of the different sub‐atlases. The results from these queries are presented as gene‐centric lists that are both clickable and downloadable in different formats. Another approach to access the extensive datasets found in the HPA is through 26 different downloadable files (https://www.proteinatlas.org/about/download) containing genome‐wide data across various assays, allowing for large‐scale bioinformatic studies.
The HPA spends considerable efforts on outreach, and the database is sustained through community contribution to European infrastructure ELIXIR, 61 where the HPA is listed as one of the core data resources with importance for the wider life‐science community. Another important aspect of outreach is educational material. One such example is production of a series of “Movie of the month” during 2020 and 2021. These educational movies allow for taking a journey into the body through 3D videos that transport you deep inside various organs. The imaging is based on antibody‐based profiling of tissues and light sheet microscopy. The videos are available at the HPA website as well as on a YouTube channel. The HPA educational material also includes a histology dictionary, 62 that serves as a helpful tool for both students and scientists interested in gaining further knowledge on the different tissue and cell structures that form the basis for the tissue and cell type‐specific protein expression patterns. By knowing how to interpret the >10 million high‐resolution images publicly available on www.proteinatlas.org, the visitors on the HPA can answer novel research questions not yet elucidated by the HPA team, for example, protein expression in rare structures or cell types, or patterns of regional expression. In HPA20, released in the autumn of 2020, a major update of the normal and pathology tissue dictionary is provided, allowing for free exploration of entire large‐scale tissue sections stained with hematoxylin and eosin, where all major cell types and structures have been highlighted and described.
To aid research related to the SARS‐CoV‐2 pandemic and allow researchers to quickly access all gene and protein expression data on SARS‐CoV‐2 related proteins, a dedicated page on the HPA database has been created (Figure 1). The clickable list allows for filtering of all SARS‐CoV‐2 related proteins based on tissue specificity or subcellular location, and studying their expression pattern in situ.
Several future national and international efforts are planned to expand the educational impact of the HPA resource even further, including more educational videos, Wikipedia summaries, and integration in student textbooks as well as tutorials related to both lab protocols and how to navigate the HPA database. Feedback from users is an integral part of the daily workflow, and many HPA researchers are active in responding to contact emails and participating in discussions on social media, thereby contributing to the scientific community and informing on recent progress in the field of antibody‐based proteomics.


