Abstract
Hypertension (HT) has recently been defined as a systolic blood pressure (BP) of ≥130 mm Hg and/or a diastolic BP of ≥80 mm Hg. It is important to further understand the pathophysiology of essential HT as its proportion is larger among most of the diagnosed HT cases. The apelin and apelin receptor (APLNR) are known to play roles in regulating BP, but the putative associations of single nucleotide polymorphisms in the APLNR gene with the risk of development of essential HT have not yet been fully investigated. Herein, we conducted a meta-analysis to investigate the relationship between single nucleotide polymorphisms in the APLNR gene and the risk of essential HT.
We conducted a search in the PubMed and Web of Science databases for eligible studies. The pooled odds ratios (ORs) with their 95% confidence intervals (CI) were calculated using random-effects models when heterogeneity was expected across the studies. Otherwise, fixed-effect models were used.
Regarding the SNP rs7119375, 5 studies were analyzed, which included a total of 3567 essential HT patients and 3256 healthy controls. Four of the 5 studies were from China and 1 was from Mexico. The meta-analysis showed the existence of a significant association between the AA genotype of rs7119375 and the risk of developing essential HT in the Chinese population, as determined using additive and recessive models (OR, 2.11; 95% CI, 1.12–3.96; I2 = 86% for AA vs GG. OR, 1.53; 95% CI, 1.21–1.94; I2 = 28% for AA vs AG. OR, 1.88; 95% CI, 1.13–3.12; I2 = 79% for AA vs AG + GG).
Our study showed, for the first time, the existence of an association between rs7119375 and the risk of development of essential HT in the Chinese population, although the sample size was small and there was considerable population heterogeneity. The apelin/APLNR system could be a novel therapeutic target for the treatment of essential HT, and more studies are warranted to further investigate the association.
Keywords: apelin, hypertension, single nucleotide polymorphism
1. Introduction
Previously, hypertension (HT) was defined as a systolic blood pressure (BP) of 140 mm Hg or higher and/or a diastolic BP (dBP) of 90 mm Hg or higher. However, the definition of HT was revised in the 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline as a systolic BP of ≥130 mm Hg and/or a dBP of ≥80 mm Hg.[1] Based on this, it is estimated that almost half of the adult population in the United States of America may have HT.[1] Essential HT (also called primary HT or idiopathic HT) is diagnosed when there are no other diseases or disorders that can cause secondary HT, and it is the most common type of HT in clinical practice (approximately 90% of all cases of HT).[1] HT complications include cardiovascular diseases, cerebrovascular diseases, and chronic kidney disease, which can be potentially fatal.[1] The World Health Organization has reported that ischemic heart disease and stroke are the leading causes of death,[2] and that high BP accounts for about 47% of ischemic heart disease cases and 54% of stroke cases worldwide.[3] Therefore, it is important to better understand the pathophysiology further and develop more effective treatments for essential HT.
Essential HT is considered a complex disorder that may be caused by multiple factors, including not only environmental factors but also genetic factors. For example, genome-wide association studies and meta-analyses have suggested that single nucleotide polymorphisms (SNPs) in the uromodulin gene[4] and the endothelial nitric oxide synthase (eNOS) gene[5,6] are associated with the risk of development of essential HT. The apelin and apelin receptor (APLNR) are known to play roles in regulating BP, and several case-control studies have been performed to investigate the associations between SNPs in the apelin and/or APLNR (also known as APJ, APJR, HG11, and AGTRL1) gene and essential HT susceptibility. Recently, a meta-analysis has found no significant correlations between SNPs in the apelin gene and the risk of development of essential HT in the Chinese population.[7] To the best of our knowledge, however, the associations between SNPs in the APLNR gene and the risk of development of essential HT have not been investigated. Therefore, we conducted this meta-analysis to determine the association between SNPs in the APLNR gene and the risk of development of essential HT.
2. Materials and methods
2.1. Search strategy and eligibility criteria
We searched for eligible studies among all papers published before January 8, 2020, without any language restrictions, in PubMed and Web of Science databases, in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.[8] Two authors (M.Y. and K.A.) performed the database searches independently, and each discrepancy was discussed until a consensus was reached. We used the following terms for the database searches: ((apelin receptor) OR APLNR OR APJ OR APJR OR HG11 OR AGTRL1) AND (polymorphism OR polymorphisms OR variant OR variants) AND (hypertension OR (blood pressure)). We also screened the references of the included studies manually. Our eligibility criteria were as follows:
-
(1)
studies that focused on the associations between SNPs in the APLNR gene and the risk of development of essential HT;
-
(2)
studies in which the participants had been classified into an essential HT patients group (systolic BP ≥140 mm Hg and/or dBP ≥90 mm Hg until the year 2017, excluding secondary HT) and a healthy control group;
-
(3)
studies that provided sufficient data to calculate the odds ratios (ORs) and 95% confidence intervals (CIs).
Family-based studies were excluded from our study.
2.2. Data extraction
From the shortlisted studies, we extracted the following necessary information: the first author's name; publication year; region and country; genotyping method used; SNPs in the APLNR gene; number of cases and controls; allele and genotype frequencies.
2.3. Quality assessment of the included studies
The Newcastle-Ottawa scale (NOS)[9] was used to assess the quality of the studies selected for inclusion in our meta-analysis. 2 authors (M.Y. and K.A.) evaluated the scores independently, and any discrepancy was discussed until a consensus was reached.
2.4. Data analysis and statistics
The Hardy-Weinberg equilibrium (HWE)[10] for each study was tested for the control group using the chi-squared (χ2) test (in Table 1). A P value of less than .05 was considered to indicate a statistically significant result in the HWE test. The heterogeneity was estimated using the Cochran Q test and the I2 statistic.[11] The meta-analyses were conducted using random-effects models when the Cochran Q test results were significant (P < .10). Otherwise, fixed-effects models were used. The heterogeneity was categorized as low if the I2 was 0% to 25%, as moderate if the I2 was 25% to 75%, or as high if the I2 was 75% to 100%.[12] The data on the OR and their 95% CIs were pooled, and forest plots were drawn using Review Manager, version 5.3. Begg[13] and Egger[14] tests were also conducted, and a funnel plot was drawn using the R software, version 3.4.0, to assess the presence of publication bias, as described previously.[15] A P value of less than 0.1 was considered as indicative of a statistically significant result in both Begg and Egger tests. We compared the genotype distributions between the case and control groups in each study using the χ2 test based on 3 × 2 tables (in Table 2). A P value of less than .05 was considered as indicative of a statistically significant result in the χ2 test.
Table 1.
Characteristics and rs7119375 genotype distributions (expressed in numbers) between the case and control groups in the studies included in our meta-analysis.
| Genotypes in HT cases | Genotypes in controls | ||||||||||||
| Author year | Region country | Geno-typing | N of cases/controls | GG | AG | AA | MAF | GG | AG | AA | MAF | HWE for controls | NOS |
| Huang 2016 | Fujian China | TaqMan | 556/475 | 271 | 220 | 65 | 0.315 | 243 | 183 | 49 | 0.296 | P = .10 | 7 |
| Li 2016 | Heilongjiang China | TaqMan | 650/645 | 361 | 256 | 33 | 0.248 | 415 | 211 | 19 | 0.193 | P = .20 | 8 |
| Liu 2014 | Heilongjiang China | PCR-LDR | 1009/756 | 517 | 403 | 89 | 0.283 | 565 | 173 | 18 | 0.138 | P = .28 | 6 |
| Niu 2010 | Shanghai China | PCR-RFLP | 969/980 | 593 | 312 | 64 | 0.227 | 601 | 339 | 40 | 0.214 | P = .36 | 7 |
| Esteban 2016 | Mexico City Mexico | TaqMan | 383/400 | 224 | 140 | 19 | 0.232 | 205 | 168 | 27 | 0.278 | P = .34 | 6 |
HT = hypertension, HWE = Hardy–Weinberg equilibrium, MAF = minor allele frequency, N = number, NOS = Newcastle-Ottawa scale, PCR-LDR = polymerase chain reaction-ligase detection reaction, PCR–RFLP = polymerase chain reaction-restriction fragment length polymorphism.
Table 2.
Genotype distributions (expressed in numbers) of each SNP between the case and control groups.
| Wild homozygote | Heterozygote | Mutant homozygote | |||||||
| SNPs in APLNR gene | Author year (sub-group) | Case | Control | Case | Control | Case | Control | χ2 | P |
| rs10501367 | Wu 2018 (male) Wu 2018 (female) | 232 143 | 115 93 | 135 113 | 72 59 | 11 11 | 16 7 | 7.5209 1.1278 | .0233 .569 |
| Esteban 2016 | 223 | 202 | 139 | 167 | 21 | 31 | 5.1562 | .0759 | |
| Niu 2010 (male) Niu 2010 (female) | 286 278 | 287 297 | 175 150 | 171 167 | 35 45 | 31 27 | 0.2407 5.7054 | .8866 .0577 | |
| rs11544374 | Wu 2018 (male) Wu 2018 (female) | 281 169 | 132 117 | 91 94 | 62 36 | 6 4 | 9 6 | 7.5663 8.9248 | .0228 .0115 |
| Liu 2014 | 548 | 637 | 381 | 110 | 80 | 9 | 180.33 | <.001 | |
| Nowzari 2018 | 13 | 12 | 32 | 37 | 15 | 21 | 0.6369 | .7273 | |
| rs9943582 | Huang 2016 (male) Huang 2016 (female) | 83 166 | 92 120 | 65 176 | 111 99 | 25 41 | 22 31 | 5.9853 2.5138 | .0502 .2845 |
| Li 2016 (male) Li 2016 (female) | 179 175 | 222 161 | 121 122 | 119 105 | 22 31 | 23 15 | 2.0863 3.8172 | .3524 .1483 | |
| Liu 2014 | 647 | 466 | 319 | 245 | 43 | 45 | 2.9851 | .2248 | |
| rs948847 | Nowzari 2018 | 18 | 26 | 26 | 29 | 16 | 15 | 0.8865 | .642 |
| Liu 2014 | 316 | 268 | 493 | 360 | 200 | 128 | 4.3103 | .1159 | |
| rs2282623 | Liu 2014 | 339 | 257 | 490 | 368 | 180 | 131 | 0.0855 | .9582 |
P values shown in bold are considered as indicative of statistical significance.
SNP = single nucleotide polymorphism.
2.5. Ethical review
Because the present study is a meta-analysis of previously published studies, additional ethical approval and patient consent were not necessary.
2.6. Data availability statement
All data analyzed during this study are available on reasonable request to the corresponding author.
3. Results
We searched in PubMed and Web of Science databases, and identified a total of 63 articles. After removing 19 duplicates, we reviewed the titles and/or abstracts, and excluded 33 articles. We then assessed the full texts of the remaining 11 articles and excluded 4 more articles. Finally, we selected 7 studies in which the associations between 6 SNPs in the APLNR gene, rs7119375, rs10501367, rs11544374, rs9943582, rs948847, and rs2282623 and the risk of developing essential HT had been investigated.[16–22] Rs7119375, rs10501367, and rs9943582 are located upstream of exon 1, rs11544374 and rs948847 are located within exon 1, and rs2282623 is located in intron between exon 1 and exon 2 of the APLNR gene.[23] A flow diagram showing our search strategy and process is presented in Figure 1.[24] However, as there were just a few studies pertaining to SNPs other than rs7119375, we decided to conduct a meta-analysis for rs7119375 alone and to discuss the other SNPs later.
Figure 1.
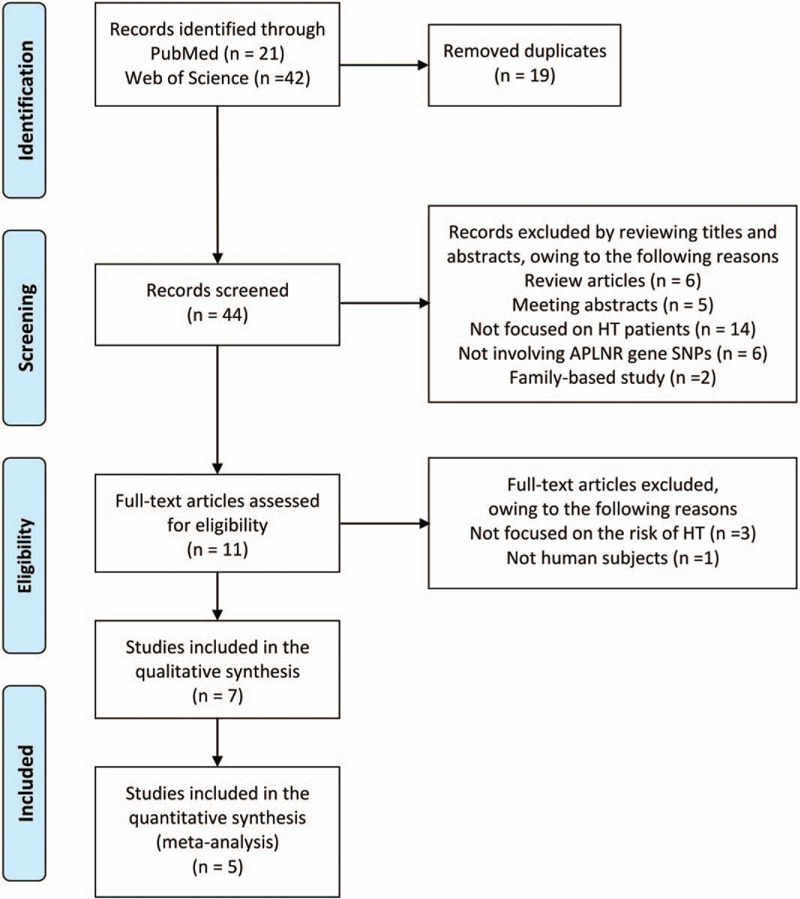
Flow diagram[24] of our search strategy and process.
A total of 5 studies, with a total of 3567 essential HT patients and 3256 controls, were included in our meta-analysis for determining the clinical significance of rs7119375 for essential HT.[16–20] The profiles of the 5 studies are shown in Table 1. Four of the 5 studies were reported from China[17–20] and 1 was conducted in Mexico.[16] Although we noticed that 3 of the 4 studies from China had been conducted by the same group or related groups,[18–20] we considered that the study populations were different from each other for the following reasons: First, the participants of Niu's study[20] were from Shanghai. Second, both, the participants in Li's[18] and Liu's[19] studies were from Heilongjiang province. But another study, [25] in which the participants were the same as those in Liu's study,[19] was accepted for publication in July 2013, and the participants of Li's study[18] were enrolled between September 2013 and October 2015. Therefore, we considered that there was no overlap in the participants of the 3 studies.[18–20] The control groups in all the 5 included studies were in accordance with the HWE (P > .05). The NOS scores in the 5 included studies are also shown in Table 1. The quality of 3 of the 4 studies conducted in the Chinese populations was considered to be high (≥7). Liu's study[19] was thought to introduce a high level of heterogeneity, as described later.
Following extraction of the data on the genotype distributions in each study, we conducted the meta-analysis by combining the OR for the risk of development of essential HT in each study using additive (Fig. 2 A, B), dominant (Fig. 2 C), recessive (Fig. 2 D), and allelic models (Fig. 2 E). We found a significant risk of essential HT using the additive model (AA vs AG) (OR, 1.42; 95% CI, 1.14–1.77; I2 = 43% as shown in Fig. 2 B) without publication bias (P values as evaluated by Begg and Egger test were 0.82 and 0.78, respectively), although we could not find any significant risk under the other models. We then performed sub-group analyses by ethnicity, and found that the risk of development of essential HT was significantly higher in the groups with the AA genotype as per the additive and recessive models, in the Chinese population (OR, 2.11; 95% CI, 1.12–3.96; I2 = 86% for AA vs GG [as shown in Fig. 2 A]; OR, 1.53; 95% CI, 1.21–1.94; I2 = 28% for AA vs AG [as shown in Fig. 2 B]; OR, 1.88; 95% CI, 1.13–3.12; I2 = 79% for AA vs AG+GG [as shown in Fig. 2 D]). Both Begg and Egger tests again showed the absence of any publication biases in the sub-group analyses as shown in Figure 2 F to J.
Figure 2.
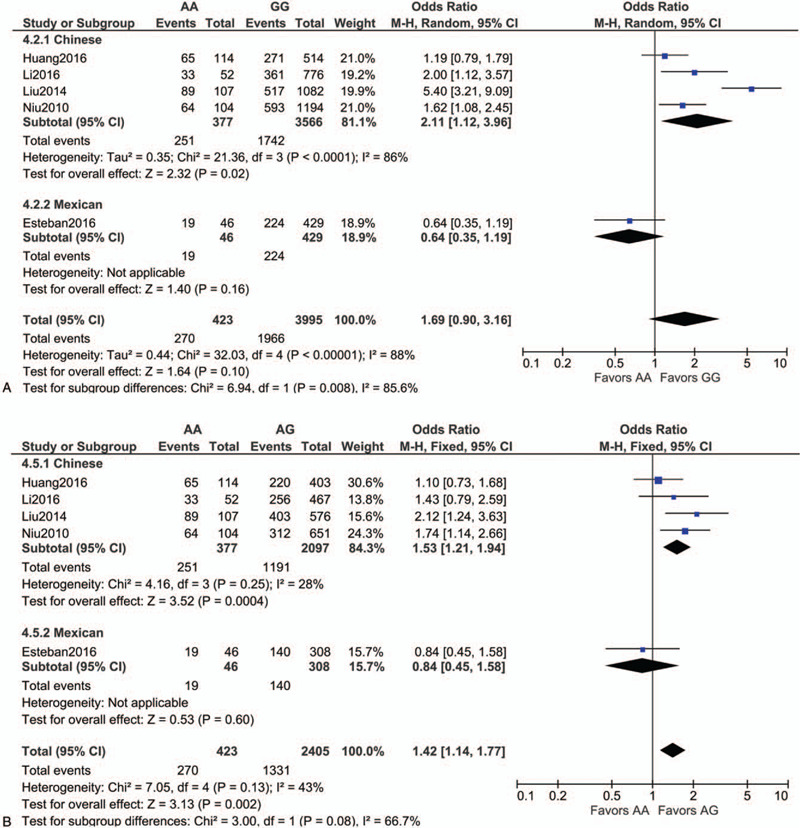
Forest plot of the risk of development of essential HT associated with the rs7119375 SNP in Chinese and Mexican populations using the (A) additive model (AA vs GG), (B) additive model (AA vs AG), (C) dominant model, (D) recessive model and (E) allelic model. (F–J) Funnel plot, Begg test, and Egger test for (A–E) in the sub-group analysis of the Chinese population. Note that the log (OR) is plotted on the horizontal axis.
Figure 2 (Continued).
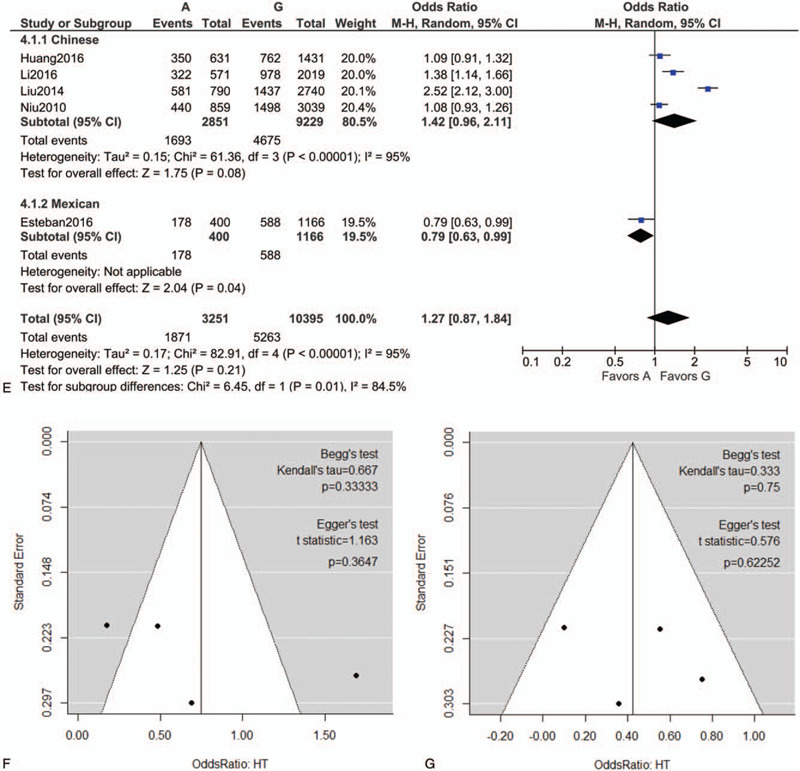
Forest plot of the risk of development of essential HT associated with the rs7119375 SNP in Chinese and Mexican populations using the (A) additive model (AA vs GG), (B) additive model (AA vs AG), (C) dominant model, (D) recessive model and (E) allelic model. (F–J) Funnel plot, Begg test, and Egger test for (A–E) in the sub-group analysis of the Chinese population. Note that the log (OR) is plotted on the horizontal axis.
Figure 2 (Continued).
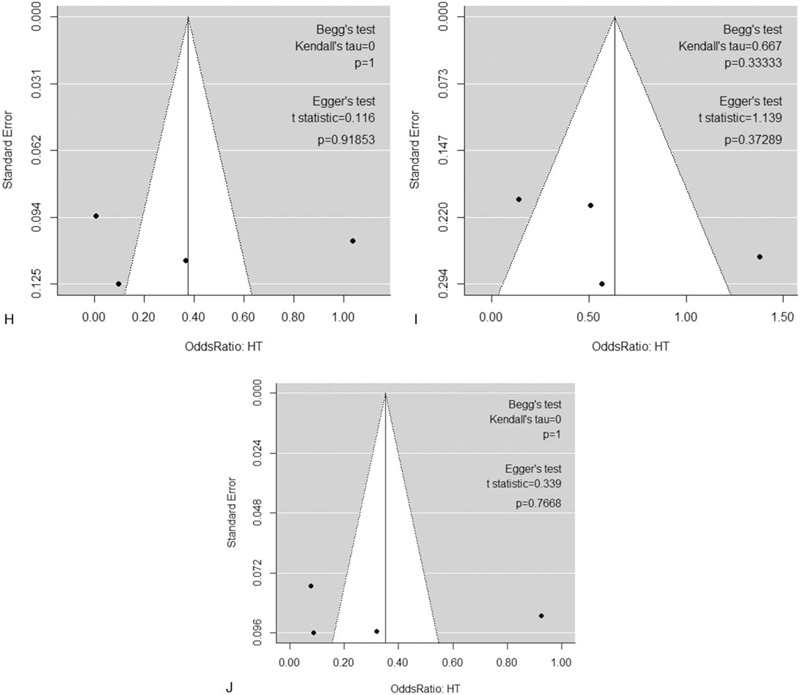
Forest plot of the risk of development of essential HT associated with the rs7119375 SNP in Chinese and Mexican populations using the (A) additive model (AA vs GG), (B) additive model (AA vs AG), (C) dominant model, (D) recessive model and (E) allelic model. (F–J) Funnel plot, Begg test, and Egger test for (A–E) in the sub-group analysis of the Chinese population. Note that the log (OR) is plotted on the horizontal axis.
The heterogeneity levels across the studies included in our meta-analysis were very high. We performed sensitivity analysis by excluding each of the 4 studies[17–20] in our sub-group analysis of the Chinese population using the additive model (I2 = 86% for AA vs GG) and recessive model (I2 = 79%), and found that when we excluded Liu's study,[19] the heterogeneity decreased and the ORs were still significant in both models (OR, 1.50; 95% CI, 1.15–1.94; I2 = 13% for AA vs GG [as shown in Fig. 3A]; OR, 1.45; 95% CI, 1.12–1.86; I2 = 9% for AA vs AG+GG [as shown in Fig. 3B], respectively). Therefore, inclusion of Liu's study[19] was thought to increase the heterogeneity in our sub-group analysis conducted for the Chinese population.
Figure 2 (Continued).
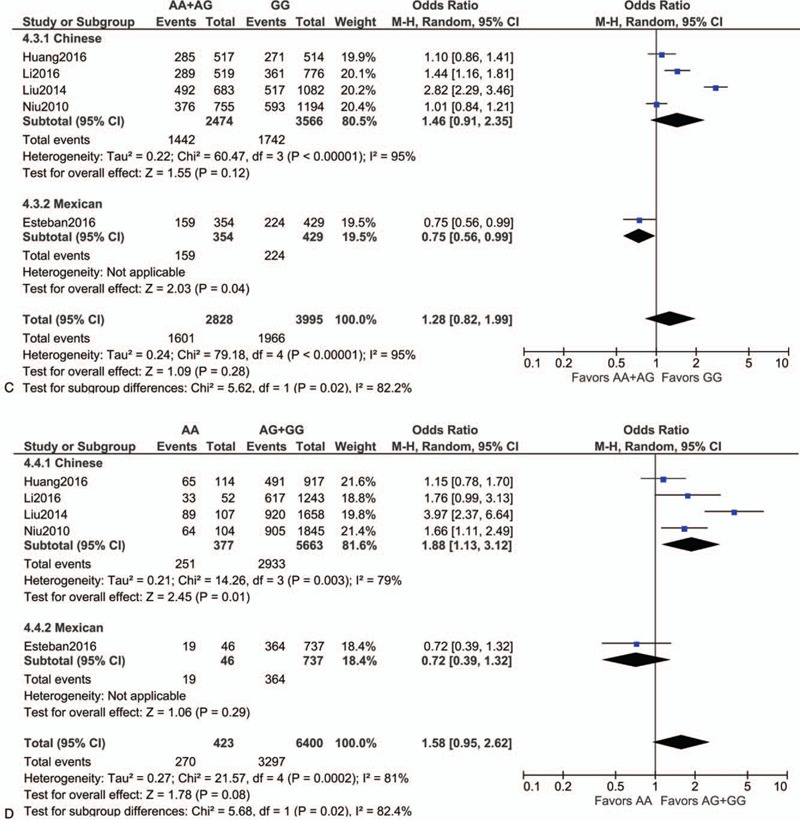
Forest plot of the risk of development of essential HT associated with the rs7119375 SNP in Chinese and Mexican populations using the (A) additive model (AA vs GG), (B) additive model (AA vs AG), (C) dominant model, (D) recessive model and (E) allelic model. (F–J) Funnel plot, Begg test, and Egger test for (A–E) in the sub-group analysis of the Chinese population. Note that the log (OR) is plotted on the horizontal axis.
Figure 3.
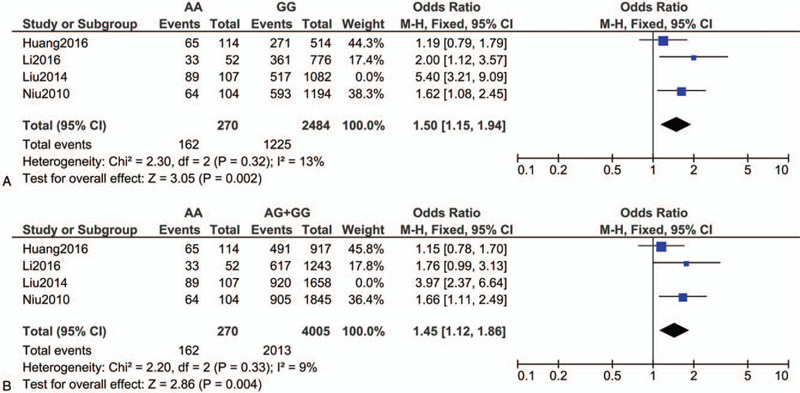
Forest plot of the risk of development of essential HT associated with the rs7119375 SNP in the sub-group analysis of the Chinese population using the (A) additive model (AA vs GG) and (B) recessive model after excluding Liu's study.
As there were only a few studies in which the association of the rs10501367, rs11544374, rs9943582, rs948847, and rs2282623 SNPs with the risk of development of essential HT had been investigated, we did not include these 5 SNPs in our meta-analysis. Instead, we compared the genotype distributions between the case and control groups in each study using the χ2 tests (Table 2). Among the studies, in Nowzari's study,[21] which was based on the Iranian population, the participants were classified into four groups: group 1, consisting of patients with both coronary artery disease (CAD) and HT (systolic BP ≥140 mm Hg and/or dBP≥90 mm Hg); group 2, consisting of patients with CAD but without HT; group 3, consisting of patients without CAD but with HT; group 4, consisting of patients with neither CAD nor HT. We decided to include the data of group 3 as HT cases and those of group 4 as healthy controls (Table 2). Overall, as shown in Table 2, analysis using the χ2 test showed that none of these SNP genotypes were significantly associated with the prevalence of essential HT (P > .05), except for rs11544374 in 2 Chinese studies (Liu's[19] and Wu's[22] study).
4. Discussion
To the best of our knowledge, this is the first meta-analysis investigating the association between SNPs in the APLNR gene and the risk of development of essential HT. Although the sample size was small, our meta-analysis showed that the AA genotype of rs7119375, as compared to the AG or the GG genotypes, increased the risk of development of essential HT in the Chinese population. None of the other SNP genotypes seemed to be significantly associated with the prevalence of essential HT, except for rs11544374 as shown in Table 2. However, whether these SNPs are actually associated with the risk of development of essential HT or not is still unclear, because of the small number of related studies.
Our results suggest an important clinical implication of rs7119375 because it is located in the promoter region of the APLNR gene[23] and may regulate gene expression.[7] Although the included studies in our meta-analysis did not measure circulating apelin levels or expressing APLNR levels, a meta-analysis showed that circulating apelin levels were significantly lower in patients with CAD.[26] The APLNR gene localizes in chromosome 11q12.1[27] and encodes for the G protein-coupled receptor for which apelin is an endogenous ligand.[28] APLNR is expressed in various organs, such as the blood vessels, heart, and kidneys, and in multiple cell types, such as endothelial cells (ECs), smooth muscle cells, and cardiomyocytes.[29] The apelin/APLNR system induces vasodilation via activation of eNOS pathway in intact ECs under physiological conditions whereas the presence of APLNR in smooth muscle cells can cause vasoconstriction under conditions where ECs are damaged.[30] Therefore, although apelin has complex vasomotor effects, the apelin/APLNR system may be clinically recognized as a novel therapeutic target for HT,[29–31] especially among patients with SNPs in the promoter region of the APLNR gene after the relationships between the SNPs and apelin/APLNR expressions are investigated in future researches. Actually, when administered intravenously in rats, apelin peptides lowered the mean arterial pressure by approximately 10 to 20 mmHg.[32] When infused intra-arterially in healthy volunteers, apelin peptides caused arterial vasodilation in human forearm vessels.[33] These effects of apelin peptides were suppressed by nitric oxide inhibitors. Although plasma half-lives of apelin peptides are short (less than 10 minutes),[31,33] novel APLNR agonists with longer biological half-lives and higher activities have been developed.[30] In addition, angiotensin-converting enzyme inhibitors and angiotensin II type 1 receptor blockers may be effective especially among hypertensive patients with SNPs in the promoter region of the APLNR, because these drugs can also activate eNOS pathway.[34] But this hypothesis needs to be verified.
The heterogeneity levels across the studies included in our meta-analysis were very high. The small number of studies could be the main reason for the high heterogeneity. In addition, inclusion of Liu's study[19] was thought to be one of the reasons for the high levels of heterogeneity. In Liu's study,[19] the fasting blood glucose levels and body mass index in the essential HT group were relatively higher than those in the other studies (Table 3). Moreover, subjects with diabetes were excluded in Huang's study,[17] and both the fasting blood glucose levels and body mass index were comparable between the case and control groups in Li's study.[18] Matching of the characteristics between the case and control groups is important in non-randomized studies, as described in the NOS scoring system.[9] We hypothesize that the case group in Liu's study[19] might not be representative of essential HT patients because of the relatively higher number of essential HT patients with underlying diabetes and/or obesity. For example, the apelin/APLNR system could be involved in the glucose uptake via the eNOS and/or PI3K/Akt pathway.[35–37] Therefore, we infer that the presence of diabetic patients in the case group might have influenced the allele frequencies of the APLNR gene in Liu's study.[19]
Table 3.
Fasting blood glucose and body mass index in the group with essential HT in each study (data shown as means ± standard deviations).
| Author year | Fasting Glucose Levels (mmol/L) | |||
| sub-group | Male CT group | Female CT group | Male HT group | Female HT group |
| Huang 2016 | 4.9 ± 0.5 | 5.0 ± 0.5 | 5.1 ± 0.6 | 5.1 ± 0.6 |
| Li 2016 | 5.77 ± 1.76 | 5.77 ± 1.80 | 5.73 ± 1.84 | 5.92 ± 1.92 |
| Liu 2014 | 5.33 ± 1.12 | 6.14 ± 2.15 | ||
| Niu 2010 | 4.94 ± 0.67 | 4.91 ± 0.60 | 5.47 ± 1.52 | 5.54 ± 2.01 |
| Author year | Body Mass Index (kg/m2) | |||
| sub-group | Male CT group | Female CT group | Male HT group | Female HT group |
| Huang 2016 | 23.0 ± 3.1 | 22.5 ± 2.9 | 25.0 ± 4.0 | 24.9 ± 3.2 |
| Li 2016 | 25.29 ± 3.02 | 25.14 ± 3.48 | 25.41 ± 3.31 | 24.72 ± 3.40 |
| Liu 2014 | 23.18 ± 3.77 | 27.89 ± 6.29 | ||
| Niu 2010 | 23.85 ± 3.36 | 23.21 ± 3.03 | 25.75 ± 2.83 | 25.10 ± 3.68 |
CT = Control, HT = hypertension.
The results of Ebstein's study[16] conducted in Mexico were conflicting with the results of the Chinese studies[17–20] as shown in Figure 2 . The authors inferred that this difference might be because of the heterogeneous genetic background of cases in Mexico City where a lot of immigration has occurred in the last century.[16] Otherwise, we infer that this discrepancy may be related to the ethnic differences of daily salt intake between East Asia and Latin America.[38] East Asian people have higher salt intake, which can lead to higher salt sensitivity of BP.[39] Consistently, the GenSalt study showed that 2 SNPs in the APLNR gene (rs2282623 and rs746886) were significantly associated with BP response to low salt diet among the Chinese population with daily high dietary salt intake.[40] Anyway, it is of great interest to investigate whether our findings in the Chinese population can be generalized to other ethnic groups, especially among East Asian populations because rs671 in ALDH2 gene, for example, is considered to be associated with essential HT among Chinese and Japanese populations[41,42] as well as a Korean population.[43] Moreover, a meta-analysis has shown that rs9943582 in APLNR gene is associated with a marginally increased risk of CAD among Chinese, Japanese, and Korean populations.[26] However, our systematic literature search showed that there were only 7 studies (5 in China, 1 in Mexico, and 1 in Iran) in which the association between SNPs in the APLNR gene and the risk of development of essential HT had been investigated,[16–22] and we agree with Liu et al that more studies in other ethnic groups and populations should investigate whether the associations can be generalized to non-Chinese groups.[19]
The apelin/APLNR system plays various roles in many physiological processes including cardiovascular functions, fluid homeostasis, and energy metabolism,[44] and could also be involved in inflammatory responses. The lipopolysaccharide-induced pro-inflammatory cytokines, interleukin-6 (IL-6), and interferon-gamma, activated enteric apelin expression via the Jak/Stat signaling pathway in the rodent gastrointestinal tract.[45] Jak/Stat is also a downstream signaling pathway activated by granulocyte colony stimulating factor.[46] In experimental rodents with colitis, administration of exogenous apelin stimulated colonic epithelial proliferation,[47] and decreased the expression of pro-inflammatory cytokines including IL-6 by improving intestinal lymphatic drainage function.[48] This indicated that the apelin/APLNR system may play regenerative and supportive roles in inflammatory bowel disease.[49] In Dahl salt-sensitive rats loaded with high salt, administration of exogenous ELABELA, a novel endogenous ligand of APLNR, suppressed high salt-induced HT and decreased the expression of pro-inflammatory cytokines including IL-6 and INF-γ in the kidney.[50] Therefore, although the precise mechanisms are yet to be elucidated, the apelin/APLNR system could be a therapeutic target for inflammatory-related diseases as well.
A major limitation of the present meta-analysis was the small number of studies. Moreover, 4 of the 5 studies were from China, and 3 of the 4 were conducted by the same or related groups, although we considered, as described above, that the participants in the 3 studies were not duplicated. The high heterogeneity of the study populations was also one of the limitations of our study. Therefore, more attention to the risk of bias is needed when interpreting the results of the present meta-analysis. Nevertheless, at least in the Chinese population, our meta-analysis showed, for the first time, a significant association between the AA genotype of the APLNR gene polymorphism rs7119375 and the risk of development of essential HT. More studies with larger sample sizes and in other populations are necessary to further investigate the effects of SNPs in the apelin/APLNR system on the risk of development of essential HT.
Acknowledgments
Our manuscript has been proofread by a native English speaker using Wolters Kluwer Author Services.
Author contributions
Conceptualization: Masahiro Yoshikawa.
Data curation: Masahiro Yoshikawa, Kensuke Asaba.
Formal analysis: Masahiro Yoshikawa.
Investigation: Masahiro Yoshikawa.
Methodology: Masahiro Yoshikawa.
Project administration: Tomohiro Nakayama.
Software: Masahiro Yoshikawa.
Supervision: Tomohiro Nakayama.
Validation: Tomohiro Nakayama.
Visualization: Masahiro Yoshikawa.
Writing – original draft: Masahiro Yoshikawa.
Writing – review & editing: Masahiro Yoshikawa, Kensuke Asaba, Tomohiro Nakayama.
Footnotes
Abbreviations: APLNR = apelin receptor, BP = blood pressure, CAD = coronary artery disease, CI = confidence interval, dBP = diastolic BP, ECs = endothelial cells, eNOS = endothelial nitric oxide synthase, HT = hypertension, HWE = Hardy-Weinberg equilibrium, IHD = ischemic heart disease, IL-6 = interleukin-6, NOS = Newcastle-Ottawa scale, OR = odds ratio, SNP = single nucleotide polymorphism.
How to cite this article: Yoshikawa M, Asaba K, Nakayama T. The APLNR gene polymorphism rs7119375 is associated with an increased risk of development of essential hypertension in the Chinese population: a meta-analysis. Medicine. 2020;99:50(e22418).
The authors have no funding and conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Whelton PK, Carey RM, Aronow WS, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018;71:e13–15. [DOI] [PubMed] [Google Scholar]
- [2].Lippi G, Plebani M. Biomarker research and leading causes of death worldwide: a rather feeble relationship. Clin Chem Lab Med 2013;51:1691–3. [DOI] [PubMed] [Google Scholar]
- [3].Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet 2008;371:1513–8. [DOI] [PubMed] [Google Scholar]
- [4].Padmanabhan S, Melander O, Johnson T, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet 2010;6:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Salvi E, Kutalik Z, Glorioso N, et al. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension 2012;59:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xie X, Shi X, Xun X, et al. Endothelial nitric oxide synthase gene single nucleotide polymorphisms and the risk of hypertension: a meta-analysis involving 63,258 subjects. Clin Exp Hypertens 2017;39:175–82. [DOI] [PubMed] [Google Scholar]
- [7].Wang T, Liu C, Jia L, et al. The association between apelin polymorphisms and hypertension in China: a meta-analysis. J Renin Angiotensin Aldosterone Syst 2019;20:1470320319827204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [10].Zhao J, Chen F, Lu L, et al. Effect of 106PEAR1 and 168PTGS1 genetic polymorphisms on recurrent ischemic stroke in Chinese patient. Medicine (Baltimore) 2019;98:e16457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 2006;11:193–206. [DOI] [PubMed] [Google Scholar]
- [12].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- [14].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yoshikawa M, Takase O, Tsujimura T, et al. Long-term effects of low calcium dialysates on the serum calcium levels during maintenance hemodialysis treatments: a systematic review and meta-analysis. Sci Rep 2018;8:5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Esteban-Martínez RL, Pérez-Razo JC, Vargas-Alarcón G, et al. Polymorphisms of APLN-APLNR system are associated with essential hypertension in Mexican-Mestizo individuals. Exp Mol Pathol 2016;101:105–9. [DOI] [PubMed] [Google Scholar]
- [17].Huang F, Zhu P, Huang Q, et al. Associations between gene polymorphisms of the apelin-APJ system and the risk of hypertension. Blood Press 2016;25:257–62. [DOI] [PubMed] [Google Scholar]
- [18].Li G, Sun X, Zhao D, et al. A promoter polymorphism in APJ gene is significantly associated with blood pressure changes and hypertension risk in Chinese women. Oncotarget 2016;7:86257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu R, Zhao H, Wang Y, et al. The contributory role of angiotensin receptor-like 1 gene multiple polymorphisms in hypertension among northeastern Han Chinese. PLoS One 2014;9:e86095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Niu W, Wu S, Zhang Y, et al. Validation of genetic association in apelin-AGTRL1 system with hypertension in a larger Han Chinese population. J Hypertens 2010;28:1854–61. [DOI] [PubMed] [Google Scholar]
- [21].Nowzari Z, Masoumi M, Nazari-Robati M, et al. Association of polymorphisms of leptin, leptin receptor and apelin receptor genes with susceptibility to coronary artery disease and hypertension. Life Sci 2018;207:166–71. [DOI] [PubMed] [Google Scholar]
- [22].Wu XD, Zhang N, Liang M, et al. Gender-specific association between Apelin/APJ gene polymorphisms and hypertension risk in Southeast China. J Cell Physiol 2018;233:5180–8. [DOI] [PubMed] [Google Scholar]
- [23].Li WW, Niu WQ, Zhang Y, et al. Family-based analysis of apelin and AGTRL1 gene polymorphisms with hypertension in Han Chinese. J Hypertens 2009;27:1194–201. [DOI] [PubMed] [Google Scholar]
- [24].Moher D, Liberati A, Tetzlaff J, et al. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Qi Y, Zhao H, Wang Y, et al. Replication of the top 10 most significant polymorphisms from a large blood pressure genome-wide association study of northeastern Han Chinese East Asians. Hypertens Res 2014;37:134–8. [DOI] [PubMed] [Google Scholar]
- [26].Chen T, Wu B, Lin R. Association of apelin and apelin receptor with the risk of coronary artery disease: a meta-analysis of observational studies. Oncotarget 2017;8:57345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].O’Dowd BF, Heiber M, Chan A, et al. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene 1993;136:355–60. [DOI] [PubMed] [Google Scholar]
- [28].Tatemoto K, Hosoya M, Habata Y, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun 1998;251:471–6. [DOI] [PubMed] [Google Scholar]
- [29].Luo X, Liu J, Zhou H, et al. Apelin/APJ system: a critical regulator of vascular smooth muscle cell. J Cell Physiol 2018;233:5180–8. [DOI] [PubMed] [Google Scholar]
- [30].Mughal A, O’Rourke ST. Vascular effects of apelin: mechanisms and therapeutic potential. Pharmacol Ther 2018;190:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huang Z, He L, Chen Z, et al. Targeting drugs to APJ receptor: from signaling to pathophysiological effects. J Cell Physiol 2018;234:61–74. [DOI] [PubMed] [Google Scholar]
- [32].Tatemoto K, Takayama K, Zou MX, et al. The novel peptide apelin lowers blood pressure via a nitric oxide-dependent mechanism. Regul Pept 2001;99:87–92. [DOI] [PubMed] [Google Scholar]
- [33].Japp AG, Cruden NL, Amer DA, et al. Vascular effects of apelin in vivo in man. J Am Coll Cardiol 2008;52:908–13. [DOI] [PubMed] [Google Scholar]
- [34].Li H, Förstermann U. Uncoupling of endothelial NO synthase in atherosclerosis and vascular disease. Curr Opin Pharmacol 2013;13:161–7. [DOI] [PubMed] [Google Scholar]
- [35].Bertrand C, Valet P, Castan-Laurell I. Apelin and energy metabolism. Front Physiol 2015;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dray C, Knauf C, Daviaud D, et al. Apelin stimulates glucose utilization in normal and obese insulin-resistant mice. Cell Metab 2008;8:437–45. [DOI] [PubMed] [Google Scholar]
- [37].Zhu S, Sun F, Li W, et al. Apelin stimulates glucose uptake through the PI3K/Akt pathway and improves insulin resistance in 3T3-L1 adipocytes. Mol Cell Biochem 2011;353:305–13. [DOI] [PubMed] [Google Scholar]
- [38].Powles J, Fahimi S, Micha R, et al. Global, regional and national sodium intakes in 1990 and 2010: a systematic analysis of 24 h urinary sodium excretion and dietary surveys worldwide. BMJ Open 2013;3:e003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sogunuru GP, Kario K, Shin J, et al. Morning surge in blood pressure and blood pressure variability in Asia: evidence and statement from the HOPE Asia Network. J Clin Hypertens (Greenwich) 2019;21:324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao Q, Hixson JE, Rao DC, et al. Genetic variants in the apelin system and blood pressure responses to dietary sodium interventions: a family-based association study. J Hypertens 2010;28:756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang SY, Chan SW, Zhou X, et al. Meta-analysis of association between ALDH2 rs671 polymorphism and essential hypertension in Asian populations. Herz 2015;40: Suppl 2: 203–8. [DOI] [PubMed] [Google Scholar]
- [42].Wu Y, Ni J, Cai X, et al. Positive association between ALDH2 rs671 polymorphism and essential hypertension: a case-control study and meta-analysis. PLoS One 2017;12:e0177023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cho Y, Kwak S, Lewis SJ, et al. Exploring the utility of alcohol flushing as an instrumental variable for alcohol intake in Koreans. Sci Rep 2018;8:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wysocka MB, Pietraszek-Gremplewicz K, Nowak D. The role of apelin in cardiovascular diseases, obesity and cancer. Front Physiol 2018;9:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Han S, Wang G, Qi X, et al. Involvement of a Stat3 binding site in inflammation-induced enteric apelin expression. Am J Physiol Gastrointest Liver Physiol 2008;295:G1068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dwivedi P, Greis KD. Granulocyte colony-stimulating factor receptor signaling in severe congenital neutropenia, chronic neutrophilic leukemia, and related malignancies. Exp Hematol 2017;46:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Han S, Wang G, Qiu S, et al. Increased colonic apelin production in rodents with experimental colitis and in humans with IBD. Regul Pept 2007;142:131–7. [DOI] [PubMed] [Google Scholar]
- [48].Ge Y, Li Y, Chen Q, et al. Adipokine apelin ameliorates chronic colitis in Il-10-/- mice by promoting intestinal lymphatic functions. Biochem Pharmacol 2018;148:202–12. [DOI] [PubMed] [Google Scholar]
- [49].Weidinger C, Ziegler JF, Letizia M, et al. Adipokines and their role in intestinal inflammation. Front Immunol 2018;9:1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xu C, Wang F, Chen Y, et al. ELABELA antagonizes intrarenal renin-angiotensin system to lower blood pressure and protects against renal injury. Am J Physiol Renal Physiol 2020;318:F1122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed during this study are available on reasonable request to the corresponding author.


