Abstract
Advanced studies demonstrated that hypoxic stress induced KIAA1199 expression leading to enhanced cell migration. KIAA1199 is a protein related with cancer metastasis. Hypoxia inducible factor 1α (HIF-1α) is a transcriptional factor that maintains oxygen homeostasis. Both KIAA1199 and HIF-1α were upregulated in many human cancers. In the present study, co-expression of KIAA1199 and HIF-1α was evaluated for the clinicopathological characteristics and survival in hepatocellular carcinoma (HCC). Clinical-pathological information and follow-up data were collected from 152 HCC patients. KIAA1199 and HIF-1α expression were scored based on the percentage and intensity of immunohistochemical staining in pathological slide. Correlations between clinical features and the expression of KIAA1199 and HIF-1α were evaluated by Chi-square test, Kaplan-Meier curves and multivariate Cox regression analysis. The frequency of KIAA1199 high expression was higher in HCC than adjacent tissue. KIAA1199(H)/HIF-1α(H) tumors were more frequently of TNM (P = .011), tumor size (P = .021), vascular invasion (P = .002) and HBV (P = .001). In survival analysis, KIAA1199(H)/HIF-1α(H) patients had the worst prognosis. Using the combination of the two parameters increased the prognostic value (P < .01 vs P = .03). KIAA1199 in combination with HIF-1α expression tends to indicate a more accurate prognosis.
Keywords: hepatocellular carcinoma, HIF-1α, KIAA1199, prognosis
1. Introduction
KIAA1199 was first described as an inner-ear-specific gene. KIAA1199 mutations are associated with nonsyndromic hearing loss.[1] KIAA1199, also defined as cell migration inducing protein (CEMIP), plays a role in the development and maintenance of cancer metastasis. Yoshida et al. discovered that KIAA1199 mediated depolymerization of hyaluronic acid via the cell membrane-associated clathrin-coated pit endocytic pathway, which would endow the tumor with an aggressive phenotype.[2,3]
As we know, hypoxia is one of the most important factors that related to the tumor microenvironment.[4] Because of aberrant vascularization and a poor blood supply, it is common that most solid tumors indeed have subjected to hypoxic stress permanently or transiently. The hypoxic response is mainly ascribed to hypoxia-inducible factors (HIFs).[5] Oxygen sensitive HIFs family was constituted by O2-labile alpha subunit (HIF1α, -2α and -3α) and a stable beta subunit (HIF1β, also known as ARNT).[6] Of interest, it has been reported that hypoxia promotes KIAA1199 expression in invasive cancer cells.[7] HIF-2α binds directly to the KIAA1199 promoter region resulting in increased KIAA1199 expression.[7] While HIF-2α displays a more tissue specific expression profile, HIF-1α is expressed ubiquitously under hypoxic activation.[8] The target genes of HIF-1α encode proteins that play important roles in multiple aspects of tumorigenesis.[9,10] Genome-wide chromatin immunoprecipitation reported that many loci bound both isoforms with similar affinity.[11] HIF-1α and -2α are closely related and share a consensus hypoxia responsive elements.[12]
Recently, emerging researches have demonstrated that KIAA1199 overexpression was associated with tumor progression and poor prognosis in numerous cancers, such as colorectal,[13,14] gastric,[15] breast,[16] pancreatic,[17] cervical[18] and liver,[19] cancers. It also has reported that KIAA1199 knockdown inhibited the growth and metastasis of HCC.[20] KIAA1199 plays an indispensable role in maintaining sorafenib-resistant HCC cell metastasis.[21] Although numbers of studies have revealed that increased HIF-1α level in the primary tumor was associated with increased mortality in most human cancers, the interaction of KIAA1199 and HIF-1α, which affects tumor growth and the clinical outcome of HCC patients, is still obscure to some extent.[22] The aim of our study was to investigate the effects of KIAA1199 and HIF-1α expression on the prognosis in HCC patients.
2. Patients and methods
2.1. Study populations
This study included 152 unselected Chinese HCC patients, all of whom underwent surgery at the Affiliated Hospital of Nantong University from 2004 to 2010. No patient treated preoperatively with radiotherapy, chemotherapy, or immunotherapy. Clinical-pathological variables and follow-up data were: gender, age at diagnosis, grade, vessel invasion, TNM, hepatitis B virus (HBV) infection, tumor size, AFP value and cirrhosis. Included in this study were 117 males and 35 females. The average of age was 53.18 years. The detail Characteristics were present in Table 1. The study was approved by the Ethics Committee of the Human Research Ethics Committee of the Affiliated Hospital of Nantong University (2017-K036). Written informed consent was obtained from the patients for publication of this study.
Table 1.
Characteristics of the populations studied.
| Characteristic | Detail |
| N | 152 |
| Age (mean) | 53.18 ± 9.94 years (range 31–79 years)∗ |
| Sex | 117 male, 35 female |
| Follow-up (mean) | 44.81 ± 30.37 months∗ |
∗; range in parentheses.
2.2. KIAA1199 and HIF-1α staining
Tissue samples were collected from HCC patients who visited Affiliated Hospital of Nantong University from 2004 to 2010. A thorough histologic examination was made by tissue microarray slides from patients. Both KIAA1199 and HIF-1α staining were performed by Tissue Microarray System (Quick-Ray, UT06, UNITMA, Korea). Core tissue biopsies (2 mm in diameter) were taken from individual paraffin-embedded sections and arranged in the new recipient paraffin blocks. Immunohistochemistry (IHC) analysis was performed as previously described.[23] The slides were incubated with the primary antibodies against KIAA1199 (Cat No: 21129-1-AP, ProteinTech Group, Chicago, IL, USA) or HIF-1α (Cat No: ab1, Abcam, Cambridge, MA, USA) at 4°C overnight. The images were acquired and analyzed by the Vectra 3 System (PerkinElmer, USA). Two variables were estimated: intensity (0 to 3 as negative, weak, moderate or strong) and percentage (0% to 100%). The final staining score of each tissue sample was generated by multiplying the intensity and percentage scores. The scores ranged from 0 (no staining) to 300 (100% of cells with strong staining intensity).
The cutoff points for KIAA1199 and HIF-1α expression were set by X-tile software (http://medicine.yale.edu/lab/rimm/research/software.aspx; Rimm lab at Yale University). The degree of KIAA1199 and HIF-1α expression were quantified using a two-level grading system defined as follows: score less than or equal to cutoff point (score = 80) as low (L), otherwise defined as high (H).
2.3. Cell culture and hypoxia treatment
HepG2 cells were maintained in 1640 medium (HyClone, UT, USA) containing 10% fetal bovine serum (HyClone, UT, USA) and were cultured at 37°C in an incubator with 5% CO2. Cells were subjected to hypoxic treatment of 1% O2 – 5% CO2 in hypoxia chamber (Invivo2 400, Ruskinn Technologies, Leeds, UK) or cultured under normoxic conditions of 21% O2 to 5% CO2.
2.4. Western blot
Cells or tissues were lysed with the cell lysis buffer (Beyotime, Shanghai, China). Whole cell or tissue extracts were resolved by 10% SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membranes (Roche Diagnostics, Mannheim, Germany). The membranes were blocked, and then incubated with anti-KIAA1199 (Cat No: 21129-1-AP, ProteinTech Group, Chicago, IL), anti-β-actin (Cat No: ab8226, Abcam, Cambridge, MA) or anti-HIF-1α antibodies (Cat No: ab1, Abcam, Cambridge, MA) at 4°C overnight, followed by an incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, PA). The chemiluminescence reaction was performed using ECL reagent (Thermo Scientific, IL). The specificity of KIAA1199 and HIF1α antibodies by RNA interference were confirmed by RNA interference (Supplementary Fig. 1).
2.5. Statistical analyses
The Chi-square and Fisher exact test were used to assess correlations between clinicopathologic features and expression of KIAA1199 and HIF-1α. Survival time was calculated from date of diagnosis to date of death/censoring. Overall survival curves were constructed according to Kaplan–Meier method. And the log-rank test was applied to examine the survival difference. The univariate and multivariate survival analysis were performed with Cox regression. All P values reported are from two-sided tests and the threshold for significance was set at .05.
3. Results
3.1. KIAA1199 expression in HCC and adjacent normal tissue
First, we examined the KIAA1199 protein expression in 48 pairs of HCC and adjacent non-cancerous tissues. The KIAA1199 expression levels were significantly higher in tumor tissues than in non-tumor tissues (Fig. 1A and B B). We further evaluated the KIAA1199 expression in HCC and non-tumor tissues by IHC analysis. Representative images of KIAA1199 staining are shown in Figure 1C. Positive KIAA1199 staining was predominantly localized to the cellular membrane and cytoplasm. The expression level of KIAA1199 protein in adjacent tissue was clearly lower than that of cancerous tissues.
Figure 1.
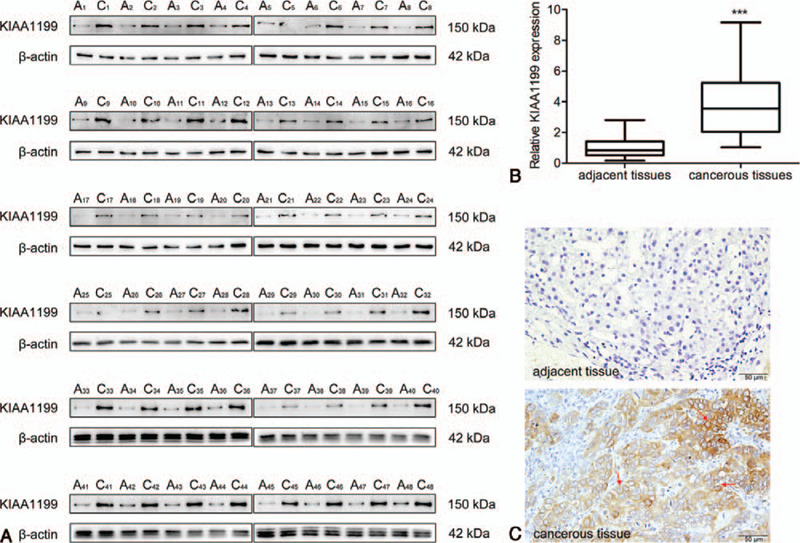
The expression of KIAA1199 in HCC and adjacent tissues. The KIAA1199 expression in individual 48 HCC patients were analyzed by Western blot (A) and quantified (B) using β-actin as control. (C) Immunohistochemistry for KIAA1199 expression in adjacent and cancerous tissue (bar = 50 μm). The KIAA1199 positive expression were indicated by red arrows. A: adjacent tissue; C: cancerous tissue; ∗∗∗P < .001 compared with the control.
3.2. Association between the expression of KIAA1199/HIF-1α and clinicopathological parameters in HCC
To determine the expression of KIAA1199 and HIF-1α in response to hypoxia, HepG2 cells were exposed to hypoxia (1% oxygen) for periods from 1 to 12 hours. As shown in Figure 2, hypoxia upregulated both the KIAA1199 and HIF-1α in a time-dependent manner. Then we analyze the relationship with the clinicopathologic features and KIAA1199 expression (Table 2). A total of 103 (67.76%) patients showed high expression of KIAA1199, while 49 (32.24%) showed low or no expression of KIAA1199. High KIAA1199 expression was significantly associated with vascular invasion (P = .024), TNM (P = .034), HBV (P = .001), tumor size (P = .034) and cirrhosis (P = .021). By contrast, no correlation (P > .05) was observed between KIAA1199 expression and other clinical parameters, such as sex, age at diagnosis, histopathology grading and AFP value.
Figure 2.
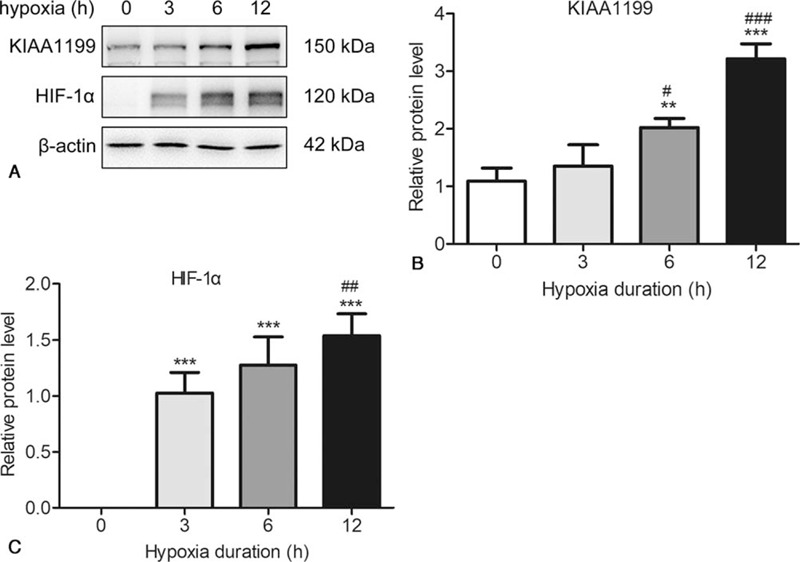
The expression of KIAA1199 and HIF-1α in respond to hypoxia. HepG2 cells were exposed to hypoxia for 0, 3, 6, 9 and 12 h. The expressions of KIAA1199 and HIF-1α were measured by Western blotting (A) and quantified (B, C) using β-actin as control. The data are presented as the means ± SD, n = 3. ∗∗P < .01 & ∗∗∗P < .001 compared to 0 hour.
Table 2.
KIAA1199 expression and clinical variables in hepatocellular carcinoma.
| KIAA1199 | |||
| Low | High | ||
| Total | 49 (32.24%) | 103 (67.76%) | P |
| Gender | .91 | ||
| female | 11 (31.43%) | 24 (68.57%) | |
| male | 38 (32.48%) | 79 (67.52%) | |
| Age | .40 | ||
| ≤ 50 | 16 (28.07%) | 41 (71.93%) | |
| > 50 | 33 (34.74%) | 62 (65.26%) | |
| Grade | .49 | ||
| well | 4 (23.53%) | 13 (76.47%) | |
| moderate | 33 (31.43%) | 72 (68.57%) | |
| poor | 12 (40.00%) | 18 (60.00%) | |
| Vascular invasion | .02∗ | ||
| no | 45 (59.21%) | 45 (59.21%) | |
| yes | 58 (76.32%) | 58 (76.32%) | |
| TNM | .03∗ | ||
| I | 28 (41.18%) | 40 (58.82%) | |
| II & III | 21 (25.00%) | 63 (75.00%) | |
| HBV | .001∗∗ | ||
| no | 15 (62.50%) | 9 (37.50%) | |
| yes | 34 (26.56%) | 94 (73.44%) | |
| Tumor size | .03∗ | ||
| ≤ 5 cm | 29 (40.85%) | 42 (59.15%) | |
| > 5 cm | 20 (24.69%) | 61 (75.31%) | |
| AFP | .48 | ||
| ≤400 μg/L | 28 (30.11%) | 65 (69.89%) | |
| >400 μg/L | 21 (25.59%) | 38 (64.41%) | |
| Cirrhosis | .02∗ | ||
| no | 22 (44.90%) | 27 (55.10%) | |
| yes | 27 (26.21%) | 76 (73.79%) | |
P < .05.
P < .01.
It has been reported that HIF-1α expression strongly was associated with clinical features in HCC patients.[24,25] Thus, we also detected the correlation of KIAA1199 and HIF-1α expression. We found that there is a strong correlation of KIAA1199 expression with levels of HIF-1α (P = .0005) (Table 3).
Table 3.
The correlation of KIAA1199 and HIF-1α expression in hepatocellular carcinoma.
| KIAA1199 | ||||
| Low (n) | High (n) | P | ||
| HIF-1α | Low (n) | 28 | 29 | .000∗∗∗ |
| High (n) | 21 | 74 | ||
P < .001.
Furthermore, we analyze the combination expression of KIAA1199 and HIF-1α in HCC (Table 4). A total of 74 (74/152, 48.68%) patients showed high expression of both KIAA1199 and HIF-1α, while 28 (28/152, 18.42%) showed low expression of both KIAA1199 and HIF-1α. KIAA1199(H)/HIF-1α(H) expression was associated with vascular invasion (P = .002), TNM (P = .011), HBV (P = .001), tumor size (P = .021) and cirrhosis (P = .032).
Table 4.
KIAA1199/HIF-1α expression and clinical variables in hepatocellular carcinoma.
| KIAA1199(L)/HIF-1α(L) | KIAA1199(L)/HIF-1α(H) | KIAA1199(H)/HIF-1α(L) | KIAA1199(H)/HIF-1α(H) | ||
| n (%) | n (%) | n (%) | n (%) | P | |
| Total | 28 (18.42) | 21 (13.82) | 29 (19.08) | 74 (48.68) | |
| Gender | .16 | ||||
| female | 7 (20.00) | 4 (11.43) | 11 (31.43) | 13 (37.14) | |
| male | 21 (17.95) | 17 (14.53) | 18 (15.38) | 61 (52.14) | |
| Age | .03 | ||||
| ≤ 50 | 9 (15.79) | 7 (12.28) | 9 (15.79) | 32 (56.14) | |
| > 50 | 19 (20.00) | 14 (14.74) | 20 (21.05) | 42 (44.21) | |
| Grade | .02 | ||||
| well & moderate | 20 (16.39) | 17 (13.93) | 22 (18.03) | 63 (51.64) | |
| poor | 8 (26.67) | 4 (13.33) | 7 (23.33) | 11 (36.67) | |
| Vascular invasion | .002∗∗ | ||||
| no | 20 (26.32) | 11 (14.47) | 19 (25.00) | 26 (34.21) | |
| yes | 8 (10.53) | 10 (13.16) | 10 (13.16) | 48 (63.16) | |
| TNM | .011∗ | ||||
| I | 17 (25.00) | 11 (16.18) | 17 (25.00) | 23 (33.82) | |
| II & III | 11 (13.10) | 10 (11.90) | 12 (14.29) | 51 (60.71) | |
| HBV | .001∗∗ | ||||
| no | 10 (41.67) | 5 (20.83) | 5 (20.83) | 4 (16.67) | |
| yes | 18 (14.06) | 16 (12.50) | 24 (18.75) | 70 (54.69) | |
| Tumor size | .02∗ | ||||
| ≤ 5 cm | 17 (23.94) | 12 (16.90) | 17 (23.94) | 25 (35.21) | |
| > 5 cm | 11 (13.58) | 9 (11.11) | 12 (14.81) | 49 (60.49) | |
| AFP | .58 | ||||
| ≤ 400 μg/L | 16 (17.20) | 12 (12.90) | 21 (22.58) | 44 (47.31) | |
| > 400 μg/L | 12 (20.34) | 9 (15.25) | 8 (13.56) | 30 (50.85) | |
| Cirrhosis | .03∗ | ||||
| no | 15 (30.61) | 7 (14.29) | 10 (20.41) | 17 (34.69) | |
| yes | 13 (12.62) | 14 (13.59) | 19 (18.45) | 57 (55.34) |
H = high, L = low.
P < .05.
P < .01.
3.3. Survival analysis according to KIAA1199/HIF-1α expression for HCC patients
Finally, we carried out survival analyses in these HCC patients. Patients in the high KIAA1199 expression group had worse overall survival than the low expression group (Fig. 3A). The 5-year overall survival rate for patients with high expression of KIAA1199 and for patients with low expression of KIAA1199 was 42.22% and 68.57% (P = 0.028), respectively. While, there was no difference in 1-year, 3-year and 5-year overall survival rate (Table 5). As shown in Figure 3B, we found that patients with high expression levels for both KIAA1199 and HIF-1α had the worst overall survival rate. The 3- and 5-year overall survival rate of KIAA1199(H)/HIF-1α(H) patients are much worse than that of KIAA1199(L)/HIF-1α(L) group (P = .041 and .004, respectively) (Table 6). Thus, using the combination with the two parameters increased the prognostic value, compared with KIAA1199 alone.
Figure 3.
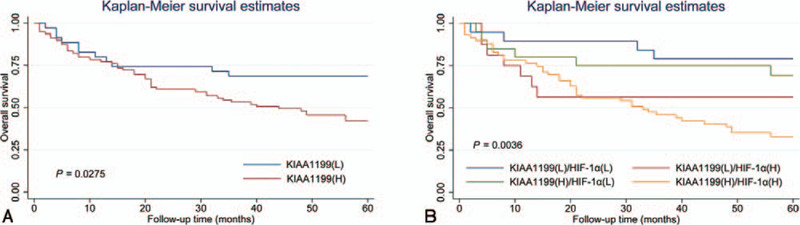
Survival curves for HCC patients, in relation to KIAA1199 and HIF-1αexpressions. Overall survival of HCC patient calculated by the Kaplan-Meier method. Overall survival in dependence on KIAA1199 (A) and in KIAA1199/HIF-1α (B). L: low; H: high.
Table 5.
Relationship between KIAA1199 expression and survival rate in hepatocellular carcinoma.
| KIAA1199 density† | |||
| Survival measurement | Low | High | P |
| 1-year overall survival (%) | 80.00 ± 6.67 | 77.22 ± 4.72 | .70 |
| 3-year overall survival (%) | 68.57 ± 7.85 | 53.16 ± 5.61 | .16 |
| 5-year overall survival (%) | 68.57 ± 7.85 | 42.22 ± 5.66 | .03∗ |
†±SE.
P < 0.05.
Table 6.
Relationship between KIAA1199/HIF-1α expression and survival rate in hepatocellular carcinoma.
| KIAA1199/HIF-1α density† | |||
| Survival measurement | KIAA1199(L)/HIF-1α(L) | KIAA1199(H)/HIF-1α(H) | P |
| 1-year overall survival (%) | 89.47 ± 7.04 | 76.27 ± 5.54 | .53 |
| 3-year overall survival (%) | 78.95 ± 9.35 | 45.76 ± 6.49 | .04∗ |
| 5-year overall survival (%) | 78.95 ± 9.35 | 33.01 ± 6.26 | .004∗∗ |
†± SE.
P < .05.
P < .01.
Next, a multivariate Cox regression analysis was performed to search for an independent prognostic factor (Table 7). After adjustment for clinical variables, cirrhosis (HRadj = 2.26; 95% CI = 1.11 – 4.62) and HIF-1α (HRadj = 2.61; 95% CI = 1.29–5.28) were all found to be independent prognostic factors.
Table 7.
Cox regression analysis of prognostic factors for 5-year survival in hepatocellular carcinoma.
| Univariate analysis | Multivariate analysis | |||||
| P | HR | 95% CI | P | HR | 95% CI | |
| Gender | .33 | 0.75 | 0.42–1.34 | |||
| male vs female | ||||||
| Age | .44 | 1.24 | 0.72–2.14 | |||
| < 55 vs ≥ 55 | ||||||
| Grade | .48 | 0.84 | 0.52 –1.37 | |||
| well & moderate vs poor | ||||||
| Vessel invasion | .61 | 1.15 | 0.68–1.95 | |||
| no vs yes | ||||||
| TNM | .74 | 1.10 | 0.64–1.87 | .33 | 1.20 | 0.83–1.72 |
| I vs II & III | ||||||
| HBV | .56 | 1.35 | 0.49–3.74 | |||
| no vs yes | ||||||
| Tumor size | .051 | 1.72 | 1.00–2.98 | |||
| ≤ 5 cm vs > 5 cm | ||||||
| AFP | .51 | 1.19 | 0.71–2.02 | |||
| ≤ 400 μg/L vs > 400 μg/L | ||||||
| Cirrhosis | .02∗ | 2.42 | 1.18–4.94 | .03∗ | 2.26 | 1.11–4.62 |
| no vs yes | ||||||
| HIF-1α | .001∗∗ | 3.09 | 1.55–6.13 | .007∗∗ | 2.61 | 1.29–5.28 |
| low vs high | ||||||
| KIAA1199 | .03∗ | 2.06 | 1.06–3.99 | .17 | 1.60 | 0.81–3.14 |
| low vs high | ||||||
P < .05.
P < .01.
4. Discussion
HCC is the most frequent primary liver cancer and represents a major medical problem.[26,27] Only surgery provides a suitable therapeutic method for HCC to prolong life span or provide better prognosis in the last few years. However, it is difficult to deal with the cases that at terminal stage of HCC. Therefore, the identification of biological markers would clearly be of great benefit. Our research revealed that increased expression of KIAA1199/HIF-1α was significantly correlated with vascular invasion, tumor TNM stage, HBV infection, tumor size and cirrhosis. KIAA1199/HIF-1α high staining was associated with poor prognosis in HCC patients. The disparity in the prognosis between KIAA1199(H)/HIF-1α(H) and KIAA1199(L)/HIF-1α(L) patient was much more apparent than the disparity observed when evaluating KIAA1199 alone.
KIAA1199 has been reported to not be expressed only in tumor tissues [1]. While, tumor hypoxia also represents a remarkably exploitable target for cancer therapy, because hypoxia encountered in solid tumors are not observed elsewhere in healthy tissues.[28,29] Therefore, it is possible to develop reagents inhibiting KIAA1199 expression by a hypoxia-triggered liposome platform to avoid side effects.
Some research reported that KIAA1199 could promote cancer cell migration and invasion through different signaling pathways.[18,30,31] In contrast, Tiwari et. al, argued that KIAA1199 overexpression reduced SW480 cell invasive ability and cell proliferation, but did not alter migratory ability.[32] Similarly, HIF-1α is overexpressed in many human cancers, such as brain (oligodendroglioma), breast, cervix, oropharynx, ovary and uterus (endometrial) cancer. Whereas it was reported that HIF-1α overexpression patients had increased survival time in oropharyngeal, head and neck, non-small-cell lung and ovarian cancer. This discrepancy of KIAA1199 and HIF-1α overexpression could be due to cancer type, experimental models, cell lines, and ectopic/endogenous expression, and the presence or absence of genetic alterations. Furthermore, the effects of gain or loss of protein function can vary according to the stage of cancer progression. Although biopsy immunohistochemistry is a common approach to analyze the association between altered expression of proteins and clinical features, it does not reveal whether the protein carries any mutations or has been post-translationally modified, which could affect its function. Combination with the two or more parameters would significantly improve the prognostic value and diagnostic accuracy.
Several factors could limit the outcomes of this study. Firstly, the sample sizes were small. In such cases, large sample sizes will be necessary for reliable interpretation. Secondly, this study doesn’t contain the variables like race.
5. Conclusion
Here, we concluded that KIAA1199 expression correlation with clinical variables in HCC, and in combination with HIF-1α expression tends to indicate a more accurate prognosis.
Author contributions
Conceptualization: Dan Wang, Hui Zhao.
Data curation: Dan Wang, Shu Lu, Xiaojing Zhang, Linlin Huang.
Formal analysis: Dan Wang, Shu Lu.
Funding acquisition: Hui Zhao, Dan Wang.
Investigation: Dan Wang, Shu Lu.
Methodology: Xiaojing Zhang, Linlin Huang.
Project administration: Dan Wang, Shu Lu, Xiaojing Zhang.
Resources: Hui Zhao.
Software: Dan Wang.
Supervision: Hui Zhao.
Validation: Dan Wang, Shu Lu, Xiaojing Zhang, Linlin Huang, Hui Zhao.
Writing – original draft: Dan Wang.
Writing – review & editing: Hui Zhao.
Supplementary Material
Footnotes
Abbreviations: HCC = hepatocellular carcinoma, HIF-1α = hypoxia inducible factor 1α.
How to cite this article: Wang D, Lu S, Zhang X, Huang L, Zhao H. Co-expression of KIAA1199 and hypoxia-inducible factor 1α is a biomarker for an unfavorable prognosis in hepatocellular carcinoma. Medicine. 2020;99:50(e23369).
DW and SL contributed equally in this study.
The study was partially funded by National Natural Science Foundation of China (Grant Nos. 81702874).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental digital content is available for this article.
References
- [1].Abe S, Usami S, Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters’ cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet 2003;48:564–70. [DOI] [PubMed] [Google Scholar]
- [2].Yoshida H, Nagaoka A, Nakamura S, et al. N-Terminal signal sequence is required for cellular trafficking and hyaluronan-depolymerization of KIAA1199. FEBS Lett 2014;588:111–6. [DOI] [PubMed] [Google Scholar]
- [3].Yoshida H, Nagaoka A, Kusaka-Kikushima A, et al. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci U S A 2013;110:5612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Semenza GL. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim Biophys Acta 2016;1863:382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Petrova V, Annicchiarico-Petruzzelli M, Melino G, et al. The hypoxic tumour microenvironment. Oncogenesis 2018;7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol 2016;18:356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Evensen NA, Li Y, Kuscu C, et al. Hypoxia promotes colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP). Oncotarget 2015;6:20723–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nordgren IK, Tavassoli A. Targeting tumour angiogenesis with small molecule inhibitors of hypoxia inducible factor. Chem Soc Rev 2011;40:4307–17. [DOI] [PubMed] [Google Scholar]
- [9].Semenza GL. Regulation of Oxygen Homeostasis by Hypoxia-Inducible Factor 1. Physiology 2009;24:97–106. [DOI] [PubMed] [Google Scholar]
- [10].Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003;3:721–32. [DOI] [PubMed] [Google Scholar]
- [11].Mole DR, Blancher C, Copley RR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 2009;284:16767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 2011;12:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang D, Zhao L, Shen Q, et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer 2017;140:2298–309. [DOI] [PubMed] [Google Scholar]
- [14].Fink SP, Myeroff LL, Kariv R, et al. Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget 2015;6:30500–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Matsuzaki S, Tanaka F, Mimori K, et al. Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann Surg Oncol 2009;16:2042–51. [DOI] [PubMed] [Google Scholar]
- [16].Evensen NA, Kuscu C, Nguyen HL, et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst 2013;105:1402–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Suh HN, Jun S, Oh AY, et al. Identification of KIAA1199 as a Biomarker for Pancreatic Intraepithelial Neoplasia. Sci Rep 2016;6:38273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shostak K, Zhang X, Hubert P, et al. NF-kappaB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun 2014;5:5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gu CJ, Ni QC, Ni K, et al. [Expression and clinical significance of KIAA1199 in primary hepatocellular carcinoma]. Zhonghua Yi Xue Za Zhi 2018;98:1609–13. [DOI] [PubMed] [Google Scholar]
- [20].Liu J, Han P, Gong J, et al. Knockdown of KIAA1199 attenuates growth and metastasis of hepatocellular carcinoma. Cell Death Discov 2018;4:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu Y, Xu H, Li M, et al. KIAA1199 promotes sorafenib tolerance and the metastasis of hepatocellular carcinoma by activating the EGF/EGFR-dependent epithelial-mesenchymal transition program. Cancer Lett 2019;454:78–89. [DOI] [PubMed] [Google Scholar]
- [22].Semenza GL. Oxygen Sensing, Hypoxia-Inducible Factors, and Disease Pathophysiology. Ann Rev Pathol 2014;9:47–71. [DOI] [PubMed] [Google Scholar]
- [23].Sun R, Wang X, Zhu H, et al. Prognostic value of LAMP3 and TP53 overexpression in benign and malignant gastrointestinal tissues. Oncotarget 2014;5:12398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Luo D, Wang Z, Wu J, et al. The role of hypoxia inducible factor-1 in hepatocellular carcinoma. Biomed Res Int 2014;2014:409272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zheng SS, Chen XH, Yin X, et al. Prognostic significance of HIF-1alpha expression in hepatocellular carcinoma: a meta-analysis. PLoS One 2013;8:e65753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:436. [DOI] [PubMed] [Google Scholar]
- [27].Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301–14. [DOI] [PubMed] [Google Scholar]
- [28].Verwilst P, Han J, Lee J, et al. Reconsidering azobenzene as a component of small-molecule hypoxia-mediated cancer drugs: A theranostic case study. Biomaterials 2017;115:104–14. [DOI] [PubMed] [Google Scholar]
- [29].Li Y, Lu A, Long M, et al. Nitroimidazole derivative incorporated liposomes for hypoxia-triggered drug delivery and enhanced therapeutic efficacy in patient-derived tumor xenografts. Acta Biomater 2018. [DOI] [PubMed] [Google Scholar]
- [30].Zhao L, Zhang D, Shen Q, et al. KIAA1199 promotes metastasis of colorectal cancer cells via microtubule destabilization regulated by a PP2A/stathmin pathway. Oncogene 2018. [DOI] [PubMed] [Google Scholar]
- [31].Jami MS, Hou J, Liu M, et al. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer 2014;14:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tiwari A, Schneider M, Fiorino A, et al. Early insights into the function of KIAA1199, a markedly overexpressed protein in human colorectal tumors. PLoS One 2013;8:e69473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


