Graphical abstract
Keywords: Lycorine, coronavirus, COVID-19, RNA-dependent RNA polymerase, cell-based reporter assay, remdesivir
Abbreviations: CDC, Centers for Disease Control and Prevention; CoV, coronavirus; COVID-19, coronavirus infectious disease 2019; DMSO, dimethyl sulfoxide; DMEM, Dulbecco's Modified Eagle's Medium; FBS, fetal bovine serum; FDA, Food and Drug Administration; HCV, hepatitis C virus; HIV, human immunodeficiency virus; KNIH, Korea National Institute of Health; MERS, Middle East respiratory syndrome coronavirus; MHV, mouse hepatitis virus; NAA, nucleoside analog antiviral; NNA, non-nucleoside antiviral; RdRp, RNA-dependent RNA polymerase; SARS, severe acute respiratory syndrome
Abstract
Background: Highly effective novel treatments need to be developed to suppress emerging coronavirus (CoV) infections such as COVID-19. The RNA dependent RNA polymerase (RdRp) among the viral proteins is known as an effective antiviral target. Lycorine is a phenanthridine Amaryllidaceae alkaloid isolated from the bulbs of Lycoris radiata (L'Hér.) Herb. and has various pharmacological bioactivities including antiviral function.
Purpose: We investigated the direct-inhibiting action of lycorine on CoV's RdRp, as potential treatment for emerging CoV infections.
Methods: We examined the inhibitory effect of lycorine on MERS-CoV, SARS-CoV, and SARS-CoV-2 infections, and then quantitatively measured the inhibitory effect of lycorine on MERS-CoV RdRp activity using a cell-based reporter assay. Finally, we performed the docking simulation with lycorine and SARS-CoV-2 RdRp.
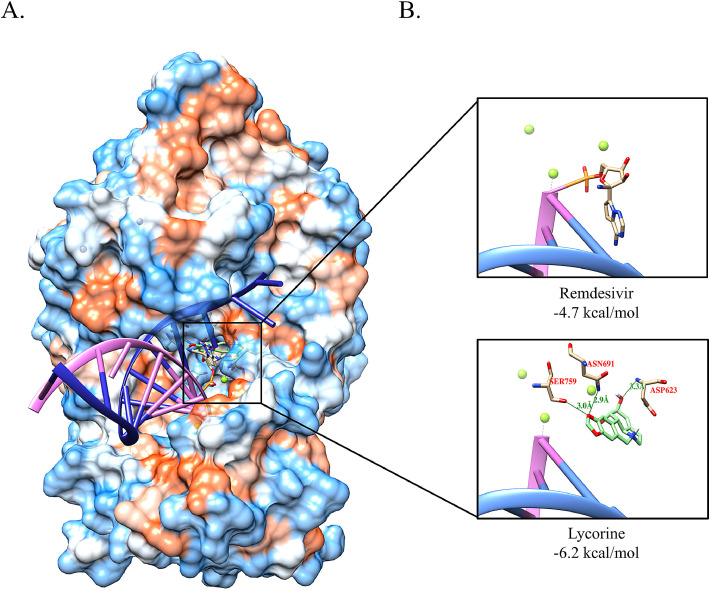
Results: Lycorine efficiently inhibited these CoVs with IC50 values of 2.123 ± 0.053, 1.021 ± 0.025, and 0.878 ± 0.022 μM, respectively, comparable with anti-CoV effects of remdesivir. Lycorine directly inhibited MERS-CoV RdRp activity with an IC50 of 1.406 ± 0.260 μM, compared with remdesivir's IC50 value of 6.335 ± 0.731 μM. In addition, docking simulation showed that lycorine interacts with SARS-CoV-2 RdRp at the Asp623, Asn691, and Ser759 residues through hydrogen bonding, at which the binding affinities of lycorine (−6.2 kcal/mol) were higher than those of remdesivir (−4.7 kcal/mol).
Conclusions: Lycorine is a potent non-nucleoside direct-acting antiviral against emerging coronavirus infections and acts by inhibiting viral RdRp activity; therefore, lycorine may be a candidate against the current COVID-19 pandemic.
Introduction
Emerging coronavirus (CoV) infections such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome coronavirus (MERS), and coronavirus infectious disease 2019 (COVID-19) have been threatening human survival. Notably, the COVID-19 pandemic has destroyed healthcare systems in various countries. As of August 2020, confirmed COVID-19 cases have increased to almost 25 million, and global deaths are at around 616 thousand (Johns Hopkins University, 2020). To solve this pandemic, many efforts have been made, and recently, the FDA authorized remdesivir as a potential COVID-19 treatment during emergency situations (US Food and Drug Administration, 2020). Although remdesivir is the first-class drug used for treating COVID-19, to date, clinical trials have shown ambiguous results regarding its clinical benefits; moreover, it has unexpected adverse effects (Grein et al., 2020; Wang et al., 2020). The Centers for Disease Control and Prevention (CDC) of South Korea recognized that remdesivir cannot lower viral propagation in the early phase of disease due to it is only used for hospitalized patients with severe disease. Therefore, the development of novel drugs with high clinical efficacy and applicability to a broad range of patient statuses is highly required.
Remdesivir acts as a potent inhibitor of RNA-dependent RNA polymerase (RdRp) in various coronaviruses including mouse hepatitis virus (MHV), MERS-CoV, and SARS-CoV-2 (Agostini et al., 2018; Gordon et al., 2020; Yin et al., 2020). Inhibition of RdRp activity to prevent viral genome replication using nucleoside analog antivirals (NAAs) has been regarded as a highly effective strategy for developing novel antivirals (De Clercq and Li, 2016). However, NAAs function by terminating chain elongation and mutation of viral progeny, which may induce viral mutation that can cause drug resistance and genetic toxicity to the host (Agostini et al., 2018; Jordheim et al., 2013). Therefore, discovery of non-nucleoside antivirals (NNAs) targeting viral polymerase against hepatitis C virus (HCV), ZIKA, and human immunodeficiency virus (HIV) infections has also been extensively studied (Ahmad et al., 2020; Costa et al., 2019; Watkins, 2019).
Lycorine is a bioactive phenanthridine alkaloid isolated from bulbs of Lycoris radiata (L'Hér.) Herb. (family Amaryllidaceae). Lycorine is known for its anti-cancer, anti-inflammatory, antibacterial, antimalarial, enzyme inhibitory, and analgesic effects (Cao et al., 2013). Lycorine has also shown suppressive activity on in vitro viral replication of flaviviruses such as Japanese encephalitis, yellow fever, dengue-4, bunya, Punta Toro, Rift Valley fever, herpes simplex 1, and poliomyelitis viruses (Gabrielsen et al., 1992; Hwang et al., 2008; Renard-Nozaki et al., 1989). Moreover, lycorine was reported to inhibit diverse coronavirus infections such as SARS-CoV, MERS-CoV, HCoV-NL63, and HCoV-OC43 both in vitro and in vivo (Li et al., 2005; Shen et al., 2019), and recent studies in cell culture show lycorine's potent inhibition of SARS-CoV-2, the pathogen that causes the current COVID-19 pandemic (Zhang et al., 2020); however, the mechanism of action of lycorine's antiviral activity still needs to be elucidated.
In this study, we confirmed and compared the antiviral effect of lycorine on emerging CoVs such as SARS-CoV, MERS-CoV, and SARS-CoV-2, and we elucidated lycorine's inhibitory effect as a NNA on RdRp activity of the CoVs by using a recently established cell-based reporter assay for MERS-CoV activity. Taken together, we report lycorine as a potent NNA that acts against emerging CoV infections by targeting RdRp.
Materials and methods
Test compounds
Lycorine hydrochloride (PubChem CID: 164943, purity ≥ 98%) was purchased from Wuhan ChemFaces Biochemical Co., Ltd. (Wuhan, China). Remdesivir (PubChem CID: 121304016, purity 97%) was purchased from LALPharm Co., Ltd. (Beijing, China). Chloroquine diphosphate was purchased from Sigma-Aldrich (St. Louis, MO, USA). Compounds were prepared as 20 mM stock solutions in 100% dimethyl sulfoxide (DMSO) (Sigma-Aldrich).
Cell lines and virus infection
Vero cells were obtained from the American Type Culture Collection (ATCCⓇ CCL-81™, Manassas, VA, USA). MERS-CoV (MERS-CoV/KOR/KNIH/002_05_2015, Genbank accession no. KT029139.1) and SARS-CoV-2 (βCoV/KOR/KCDC03/2020) were kindly provided by Korea Disease Control and Prevention Agency (KDCA). SARS-CoV (strain HK39849) was kindly provided by Prof. JSM Peiris from the University of Hong Kong. These viruses were propagated in Vero cells, and viral titers were determined by plaque assays in the Vero cells (Jeon et al., 2020; Kim et al., 2015; Peiris et al., 2003). All experiments using the above coronaviruses were performed at laboratories of the Institut Pasteur Korea in compliance with the guidelines of the Korea National Institute of Health (KNIH) using enhanced Biosafety Level 3 (BSL-3) procedures, as approved by the KDCA.
Dose-response curve (DRC) analysis by immunofluorescence staining
Vero cells were seeded in DMEM and supplemented with 2% FBS and 1x Antibiotic-Antimycotic solution (Gibco, Carlsbad, CA, USA) at a concentration of 1.2 × 104 cells/well on 384-well black μCLEAR® plates (Greiner Bio-One, Kremsmünster, Austria) for 24 h. Ten-point DRCs were generated as two-fold serially diluted compound concentrations. Subsequently, 10 µl of the diluted compounds were added to the cell plates [final DMSO concentration of 0.5% (v/v)]. Afterwards, 10 µl of MERS-CoV, SARS-CoV, and SARS-CoV-2 at a multiplicity of infection (MOI) of 0.0625, 0.05, and 0.0125, respectively were used to infect the cells.
After viral infection, cells were fixed with 4% paraformaldehyde at 24 h post infection (pi) and were analyzed by immunofluorescence staining using the anti-MERS-CoV spike protein, anti-SARS-CoV spike protein, or anti-SARS-CoV-2 nucleocapsid protein primary antibodies (Sino Biological Inc., Beijing, China), Alexa Fluor 488 goat anti-rabbit IgG secondary antibody, and Hoechst 33342 (Molecular Probes/Thermo Fisher Scientific, Waltham, MA, USA). Images were acquired by Perkin Elmer Operetta imaging system (20 × ; Waltham, MA, USA). The acquired images were analyzed using an in-house-developed Image-Mining 3.0 (IM 3.0) plug-in software, as previously described (Cruz et al., 2013). The infection ratio of each well was normalized by using the average infection ratio of mock control as 0% and the average infection ratio of negative control (0.5% DMSO) as 100% in each assay plate. Cell ratio was determined by dividing the number of cells in each well by the average number of cells in the mock control, in each assay plate.
DRCs were fitted using sigmoidal dose-response models and the following equation: Y = Bottom + (Top − Bottom) / [1 + (IC50/X)Hillslope]; calculations were done using XLfit 4 Software (ID Business Solutions, Guildford, UK) or Prism 6.05 (GraphPad Software Inc., San Diego, CA, USA). The IC50 (50% inhibition concentration) was calculated using the software's non-linear regression function. All IC50 were measured in duplicate, and the quality of each assay was controlled by their Z'-factor and their coefficient of variation (%CV). Selectivity Index (SI) was calculated using the formula: SI = CC50 / IC50.
Cytotoxicity assay
Cells were seeded into 96-well plates (Thermo Fisher Scientific) and were treated with the indicated concentration for 18 h. Cytotoxicity was analyzed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation, Madison, WI, USA). Absorbance was determined at 490 nm using a GloMax® Discover Microplate Reader (Promega Corporation).
Cell-based MERS-CoV RdRp activity assay
To determine MERS-CoV RdRp activity in cells, we used a previously described method (Min et al., 2020). Briefly, HEK293T were transfecteäüd with MERS-CoV RdRp-expressing plasmid (pN-termFlag-nsp12) and reporter plasmids [p(+)FLuc-(−)UTR-NLuc]. Cells were treated with the indicated concentrations of test compounds or 0.25% DMSO (control) starting at 6 h after transfection. Firefly luciferase (FLuc) and Nano-GloⓇ (NLuc) reporter gene expression in the cells was measured using a Nano-GloⓇ Dual-LuciferaseⓇ Reporter Assay System (Promega Corporation). Relative activity of MERS-CoV RdRp was determined by normalizing the level of NLuc activity to that of FLuc (Index of NLuc / FLuc ratio).
Molecular docking simulation
AutoDock Vina (Trott and Olson, 2010) was used to simulate dockings of lycorine and remdesivir with the SARS-CoV-2 RdRp protein. The docking pocket was from previously identified active sites in complexes of the monophosphate form of remdesivir (PDB ID: 7BV2). To prepare the docking simulation, the 3D structure of lycorine (Pubchem CID: 164943) was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Images of the complexes and hydrogen-bond predictions were obtained using Chimera version 1.14 (https://www.cgl.ucsf.edu/chimera/)
Statistical analysis
Data are presented as the mean ± SEM. Non-linear regression analysis of IC50 was conducted using GraphPad PrismⓇ Software V.6.05 for Windows (GraphPad Software Inc., San Diego, CA, USA).
Results
Lycorine inhibits MERS-CoV infection in Vero cells
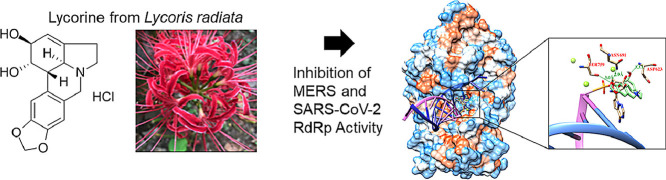
To examine the antiviral effect of lycorine (Fig. 1 A) against MERS-CoV infection, Vero cells were treated with the indicated concentrations of lycorine (0.2–100 μM) during viral infection. The infected cells were subjected to immunofluorescence analysis using an anti-MERS-CoV spike protein antibody at 24 h post infection (24 hpi) (Fig. 1B). Our data showed that IC50 values were 2.123 ± 0.053 μM (Fig. 1C), and the CC50 values were > 50 μM. These data resulted in selectivity indices (SI, CC50/IC50) of > 23.55 (Supplementary Table 1). We also confirmed the inhibitory effect of lycorine on MERS-CoV infection by plaque assay (Fig. 1D). These data suggested that lycorine treatment has potent anti-MERS-CoV infection activities.
Figure 1.
Dose-dependent inhibition of MERS-CoV infection by lycorine. (A) Chemical structure of lycorine. (B) The confocal microscope images showed cell nuclei (red) and MERS-CoV spike (S) protein (green) at the indicated lycorine concentration or 8.3 μM remdesivir (RDV) after MERS-CoV infection. Scale bar = 100 μM. (C) Dose-response curve analysis by immunofluorescence staining was used to measure the antiviral effect of lycorine. Blue circles represent inhibition of SARS-CoV-2 infection (%) by lycorine, the red squares represent cell viability (%) by lycorine and black circles represent inhibition of SARS-CoV-2 infection (%) by remdesivir. Data are representative of duplicate experiments and are presented as the mean ± SEM. (D) Lycorine (6 μM) or chloroquine (40 μM, positive control) inhibition of plaque formation was determined by plaque assay at 3 days post MERS-CoV (20 PFU) infection.
Lycorine directly inhibits MERS-CoV RdRp activity
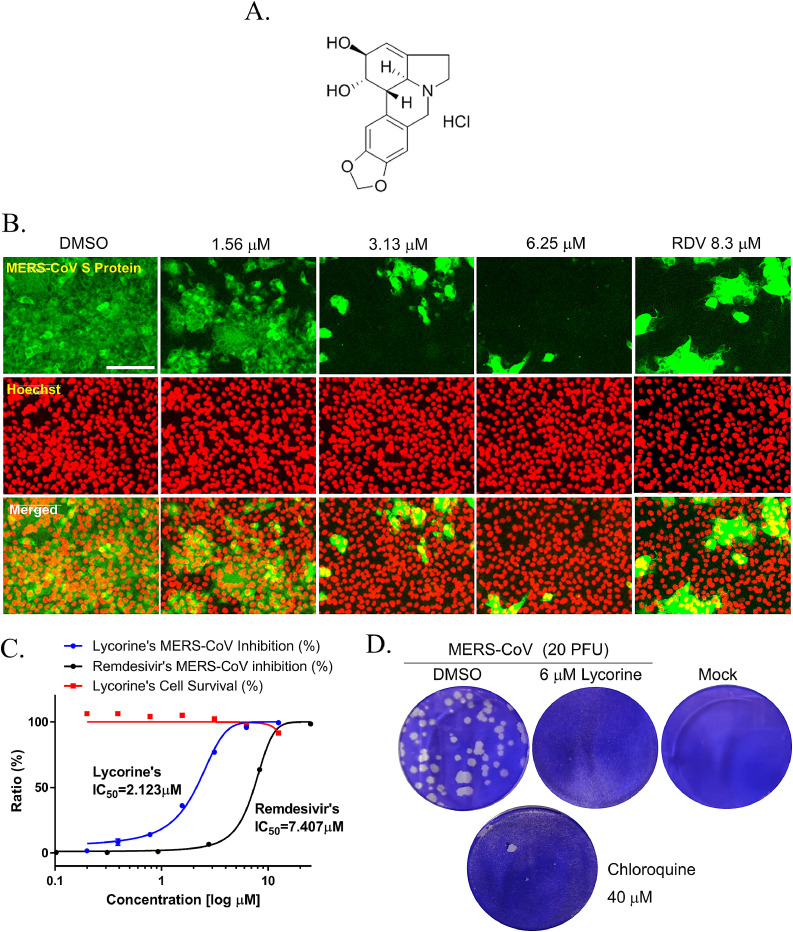
Previously, we established a cell-based MERS-CoV RdRp activity reporter assay (Min et al., 2020) by modifying the cell-based HCV RdRp activity assay (Lee et al., 2010). The cell-based reporter system consisted of the MERS-CoV nsp12 plasmid and the bicistronic reporter plasmid, p(+)FLuc-(−)UTR-NLuc which contains the sense orientation firefly luciferase gene (+)FLuc and the antisense orientation (−)NLuc, which is flanked by the antisense orientation 3′- and 5′-UTR of MERS-CoV and the hepatitis delta virus (HDV) ribozyme self-cleavage sequence. The full length of the bicistronic reporter plasmid is transcribed by the host DNA-dependent RNA polymerase, and the transcripts were then processed by HDV ribozyme self-cleavage. The exposed (-) strand of NLuc, flanked by the antisense 3′- and 5′-UTR RNA, can be replicated by MERS-CoV RdRp. Then, the replicated (+) strand of NLuc RNA is translated, and the expressed NLuc signal represents the activity of MERS-CoV RdRp. Meanwhile, the expression level of FLuc is used as the internal control to normalize NLuc activity. Since it was previously reported that lycorine inhibited the ZIKA virus by blocking RdRp activity in the in vitro RNA polymerase assay (Chen et al., 2020), we examined the effect of lycorine on MERS-CoV RdRp activity using the cell-based MERS-CoV RdRp activity reporter assay. First, we evaluated lycorine cytotoxicity in HEK293 cells. Data showed that lycorine did not induce the cytotoxic effect up to 10 μM (Fig. 2 A). Then, we measured NLuc and FLuc activities following lycorine treatment using the MERS-RdRp activity assay. Data showed that lycorine treatment reduced the relative NLuc activity in a dose-dependent manner, and that FLuc activity was maintained up to 3.6 μM. These data suggested that lycorine directly inhibits MERS-CoV RdRp activity without interfering with the host's transcriptional and translational machinery (Fig. 2B). IC50 of lycorine was 1.406 ± 0.260 μM, which was calculated by a non-linear regression analysis (Fig. 2C). Under the same conditions, we also evaluated the IC50 of remdesivir as positive control, which is known to inhibit coronavirus infection by the blocking RdRp activity (Gordon et al., 2020). The IC50 of remdesivir was 6.335 ± 0.731 μM (Fig. 2D), consistent with our previous results which showed inhibition of MERS-CoV RdRp activity in this assay (Min et al., 2020). To exclude the possibility that decreased expression of MERS-CoV RdRp by lycorine caused the decreased activity of MERS-CoV RdRp, we confirmed the consistent expression level of MERS-CoV RdRp by western blot analysis (Supplement Fig. 1). Therefore, these results show that the inhibitory effect of lycorine on MERS-CoV RdRp activity may be comparable with that of remdesivir.
Figure 2.
Effects of lycorine and remdesivir on MERS-CoV RNA-dependent RNA polymerase activity in the cell-based reporter assay system. (A) Viability of lycorine-treated HEK293T cells was determined after treatment with the indicated concentrations of lycorine for 18 h. (B) HEK293T cells were transiently transfected with MERS-CoV RdRp-expressing plasmid and bicistronic reporter plasmid, p(+)FLuc-(−)UTR-NLuc. After 6 h, cells were treated with lycorine at the indicated concentrations for 18 h, and the NLuc/FLuc ratio was calculated. (C) IC50 value of lycorine was determined through non-linear regression analysis (n = 5; t4.426 = 39.54, p < 0.0001). (D) IC50 value of remdesivir was calculated through non-linear regression analysis. Data, which are representative of at least three independent experiments, are presented as mean ± SEM.
Lycorine efficiently inhibits SARS-CoV and SARS-CoV-2 infection
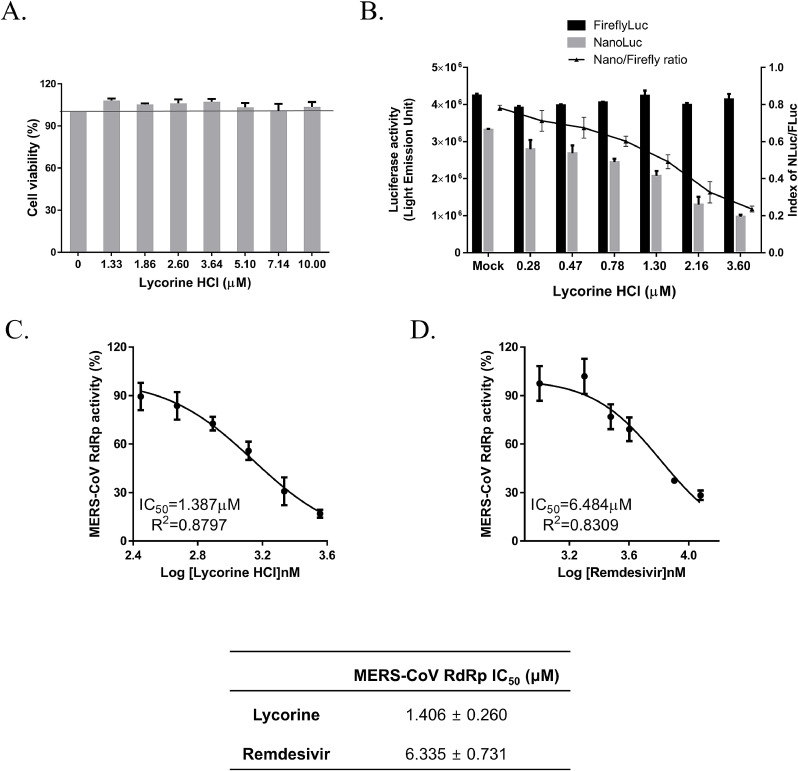
We also examined the antiviral effect of lycorine against the other emerging coronavirus infections such as SARS-CoV and SARS-CoV-2. Vero cells were treated with 2-fold serially diluted concentrations: 0.2–100 μM of lycorine for SARS-CoV and 0.1–50 μM for SARS-CoV-2 infection. Infected cells were subjected to immunofluorescence analysis with an anti-SARS-CoV spike protein antibody (Fig. 3 A) or with a SARS-CoV-2 nucleocapsid protein antibody (Fig. 3B), respectively, at 24 hpi. Data on SARS-CoV infection showed that IC50 was 1.021 ± 0.025 μM (Fig. 3C). In the case of SARS-CoV-2 infection, data showed that IC50 was 0.878 ± 0.022 μM (Fig. 3D). These data resulted in SI of > 48.97 for SARS-CoV infection and > 56.95 SARS-CoV-2 infection (Supplementary Table 1). Under our conditions, we determined that the IC50 of remdesivir is 4.084 ± 0.248 and 6.499 ± 0.256 μM for SARS-CoV and SARS-CoV-2, respectively (Supplementary Table 1). We also confirmed the inhibitory effect of lycorine on SARS-CoV infection (Fig. 3E) and SARS-CoV-2 infection (Fig. 3F) by plaque assay. Therefore, our data demonstrated that lycorine has potent anti-SARS-CoV and SARS-CoV-2 infection abilities comparable with that of remdesivir. Moreover, we showed that the dose-dependent inhibition of human coronavirus OC43 spike and nucleoprotein expression by lycorine using western blot analysis (Supplement Fig. 2).
Figure 3.
Dose-dependent inhibition of SARS-CoV and SARS-CoV-2 infection by lycorine. (A and B) The confocal microscope images showed SARS-CoV spike (S) protein (green) and cell nuclei (red) at the indicated lycorine concentration or 8.3 μM remdesivir (RDV) after SARS-CoV infection (A) and SARS-CoV-2 nucleocapsid (N) protein (green) and nuclei (red) at the indicated lycorine concentration or 5 μM remdesivir (RDV) after SARS-CoV-2 infection (B). Scale bar = 100 μM. (C and D) Dose-response curve analysis by immunofluorescence staining was used to measure anti-SARS-CoV (C) and anti-SARS-CoV-2 (D) infection effects of lycorine. Blue circles represent inhibition of viral infection (%) and red squares represent cell viability (%) by lycorine. Black circles represent inhibition of SARS-CoV-2 infection (%) by remdesivir. Data are representative of duplicate independent experiments and are presented as the mean ± SEM. (E and F) lycorine (2 μM) or remdesivir (15 μM) inhibition of plaque formation was determined by plaque assay at 3 days post SARS-CoV (70 PFU) infection (E) or at 4 days post SARS-CoV-2 (50 PFU) infection (F).
Lycorine can occupy the catalytic active site of SARS-CoV-2 RdRp protein
We next examined possible mechanisms for lycorine-induced ectopic localization of the SARS-CoV-2 RdRp protein through in silico modeling. Remdesivir is a known selective RdRp inhibitor and acts as a prodrug that is converted to the triphosphate form within cells, (Siegel et al., 2017) afterwards, the metabolized monophosphate form of remdesivir inhibits the RdRp protein by covalently linking to the primer strand of the viral RNA at the center of the catalytic active site of SARS-CoV-2 (Yin et al., 2020).
Docking simulations showed that lycorine was properly located at the same binding sites as remdesivir in the RdRp protein. The docking score of lycorine was −6.2 kcal/mol, as measured by the AutoDock Vina program, whereas docking score of the monophosphate form of remdesivir was −4.7 kcal/mol (Fig. 4 A). This suggested that the binding affinity of lycorine to the SARS-CoV-2 RdRp protein is stronger than that of remdesivir. Based on the docking results, the lycorine position overlapped with the nucleoside ring of remdesivir in the pocket site (Fig. 4B). In addition, there were three hydrogen-bond interactions between lycorine and the SARS-CoV-2 RdRp protein such as Asp623, Asn691, and Ser759 (Fig. 4B). These suggested that these hydrogen bonds could increase binding affinity with the SARS-CoV-2 RdRp protein and that lycorine could interrupt the activity of SARS-CoV-2 RdRp protein, thereby blocking viral RNA replication.
Figure 4.
Docking simulation between SARS-CoV-2 RdRp and lycorine. (A) Surface view of the RdRp active site with viral RNA (blue: viral RNA template, pink: replicated RNA. (B) The zoomed-in zone inside the rectangle indicates the binding pocket of lycorine and remdesivir. The green-colored chemical structure represents lycorine, which is located in the major pocket of RdRp protein. The gray-colored chemical structure shows remdesivir covalently bound to the RNA.
Discussion
Lycorine is a bioactive pyrrolidine alkaloid isolated from the bulbs of L. radiata and is reported to have various pharmacological bioactivities including antiviral function (Cao et al., 2013). Lycorine was identified to possess an inhibitory ability on the SARS-CoV infection (Li et al., 2005) and on four other CoV infections such as HCoV-OC43, MERS-CoV, HCoV-NL63, and MHV-A59 (Shen et al., 2019). Thus, we measured the inhibitory effect of lycorine on MERS-CoV, SARS-CoV, and SARS-CoV-2 infection and found that lycorine efficiently inhibited these CoVs with IC50 values of 2.123 ± 0.053, 1.021 ± 0.025, and 0.878 ± 0.022 μM, respectively, which is comparable with the inhibitory effect of remdesivir in our conditions. Our data suggested that lycorine may be more effective against SARS-CoV or SARS-CoV-2 infections than in MERS-CoV infections, and that lycorine is more effective than remdesivir, which is currently the most promising anti-SARS-CoV-2 drug.
Lycorine was reported to suppress viral RNA replication and viral protein synthesis of poliovirus (Hwang et al., 2008), EV71 virus (Liu et al., 2011) and avian influenza virus H5N1 (Liu et al., 2011). Recently, it was suggested that lycorine could inhibit Zika virus viral RNA synthesis and bind to the Zika RdRp protein (Chen et al., 2020). RdRp of RNA viruses is known to be one of the most important viral proteins for viral RNA synthesis. Viral RdRp has been suggested as a potential target for the development of anti-CoV therapeutics, as it only exists in the virus and does not exist in the host cells; moreover, it is highly conserved in the RNA virus, with 98% amino acid similarity between SARS-CoV and SARS-CoV-2 RdRp proteins (Shannon et al., 2020).
Previously, we established a cell-based MERS-CoV RdRp activity assay system. It is composed of the nsp12-expressing plasmid and the bicistronic MERS-CoV RdRp reporter plasmid. This assay determined MERS-CoV RdRp activity based on the expression level of NLuc (with FLuc expression as an internal control) in cells using this system. We measured the effect of lycorine and remdesivir on the MERS-CoV RdRp activity and compared the inhibitory activity of lycorine with that of remdesivir. Our data suggested that lycorine inhibited the activity of MERS-CoV RdRp in a dose-dependent manner and that the IC50 of lycorine (1.406 ± 0.260 μM) is lower compared with the IC50 of remdesivir is (6.335 ± 0.731 μM), indicating that lycorine has an efficient inhibitory ability on the activity of MERS-CoV RdRp. Moreover, docking simulations showed that the binding affinity of lycorine to the SARS-CoV-2 RdRp protein is stronger than that of remdesivir (binding free energy change of −6.2 for lycorine vs. −4.7 kcal/mol for remdesivir). The binding scores obtained from lycorine and remdesivir were correlated with the IC50 values of SARS-CoV-2 infection in our system (0.878 ± 0.022 μM for lycorine vs. 6.499 ± 0.256 μM for remdesivir). Remdesivir is known to inhibit SARS-CoV-2 RdRp activity through non-obligate RNA chain termination by targeting the center of the catalytic active site on the RdRp protein (Yin et al., 2020). Lycorine has a similar binding position which overlaps with the nucleoside rings of remdesivir in the same pocket region of the catalytic active site on the SARS-CoV-2 RdRp protein. Lycorine was found to form hydrogen bonds with RdRp at Asp623, Asn691, and Ser759 which is similar to remdesivir. We therefore suggest that those hydrogen bonds could increase the binding affinity with the SARS-CoV-2 RdRp protein. Finally, it results in an inhibition of viral RdRp activity and interrupts viral replication during viral infection. Taken together, we elucidated the inhibitory effect of lycorine on MERS-CoV, SARS-CoV, and SARS-CoV-2 infection. We identified lycorine as a direct inhibitor of RdRp through a cell-based reporter assay for MERS-CoV RdRp activity and a docking simulation with lycorine and SARS-CoV-2 RdRp protein, suggesting that lycorine is more efficient as an inhibitor compared with remdesivir. Therefore, our results suggest that lycorine could be a potential broad spectrum anti-CoV drug as it can act as a MERS-CoV, SARS-CoV, and SARS-CoV-2 therapeutic by directly inhibiting viral RdRp.
Author contributions
Young-Hee Jin: conceptualization, methodology, investigation, formal analysis, visualization, supervision, project administration, writing-original draft preparation, writing-review and editing Jung Sun Min: methodology, investigation, data curation, formal analysis Sangeun Jeon: data curation, validation, visualization Jihye Lee: data curation, validation, visualization Seungtaek Kim: methodology, validation Tamina Park: formal analysis, visualization Daeui Park: formal analysis, visualization Min Seong Jang: validation Chul Min Park: validation Jong Hwan Song: validation Hyoung Rae Kim: validation, resources Sunoh Kwon: conceptualization, methodology, investigation, data curation, formal analysis, visualization, supervision, project administration, writing-review and editing, resources, funding acquisition. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
This study was supported by the National Research Council of Science & Technology (NST) grant [grant numbers CRC-16-01-KRICT, NSN1622460, CAP-20-01-KRIBB and NSN2011460] funded by the Korea government (MSIT). J.S.M. was supported by the ‘National Research Council of Science & Technology (NST)–Korea Institute of Oriental Medicine (KIOM)’ Postdoctoral Research Fellowship for Young Scientists at the Korea Institute of Oriental Medicine in South Korea. The pathogen resource (NCCP43326) for this study was provided by the National Culture Collection for Pathogens.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.phymed.2020.153440.
Appendix. Supplementary materials
Reference
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N., Rehman A.U., Badshah S.L., Ullah A., Mohammad A., Khan K. Molecular dynamics simulation of zika virus NS5 RNA dependent RNA polymerase with selected novel non-nucleoside inhibitors. J. Mol. Struct. 2020;1203 [Google Scholar]
- Cao Z., Yang P., Zhou Q. Multiple biological functions and pharmacological effects of lycorine. Sci. China Chem. 2013;56:1382–1391. doi: 10.1007/s11426-013-4967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lao Z., Xu J., Li Z., Long H., Li D., Lin L., Liu X., Yu L., Liu W. Antiviral activity of lycorine against Zika virus in vivo and in vitro. Virology. 2020;564:88–97. doi: 10.1016/j.virol.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa G., Rocca R., Corona A., Grandi N., Moraca F., Romeo I., Talarico C., Gagliardi M.G., Ambrosio F.A., Ortuso F. Novel natural non-nucleoside inhibitors of HIV-1 reverse transcriptase identified by shape-and structure-based virtual screening techniques. Eur. J. Med. Chem. 2019;161:1–10. doi: 10.1016/j.ejmech.2018.10.029. [DOI] [PubMed] [Google Scholar]
- Cruz D.J.M., Bonotto R.M., Gomes R.G., Da Silva C.T., Taniguchi J.B., No J.H., Lombardot B., Schwartz O., Hansen M.A., Freitas-Junior L.H. Identification of novel compounds inhibiting chikungunya virus-induced cell death by high throughput screening of a kinase inhibitor library. PLOS Negl. Trop. Dis. 2013;7:e2471. doi: 10.1371/journal.pntd.0002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen B., Monath T.P., Huggins J.W., Kefauver D.F., Pettit G.R., Groszek G., Hollingshead M., Kirsi J.J., Shannon W.M., Schubert E.M. Antiviral (RNA) activity of selected Amaryllidaceae isoquinoline constituents and synthesis of related substances. J. Nat. Prod. 1992;55:1569–1581. doi: 10.1021/np50089a003. [DOI] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.-C., Chu J.J.-H., Yang P.L., Chen W., Yates M.V. Rapid identification of inhibitors that interfere with poliovirus replication using a cell-based assay. Antivir. Res. 2008;77:232–236. doi: 10.1016/j.antiviral.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. e00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University . Johns Hopkins University; 2020. Coronavirus COVID-19 Global Cases by Johns Hopkins CSSE. [Google Scholar]
- Jordheim L.P., Durantel D., Zoulim F., Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nature Rev. Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Cho Y.-J., Kim D.-W., Yang J.-S., Kim H., Park S., Han Y.W., Yun M.-r, Lee H.S., Kim A.-R. Complete genome sequence of Middle East respiratory syndrome coronavirus KOR/KNIH/002_05_2015, isolated in South Korea. Genome Announc. 2015;3 doi: 10.1128/genomeA.00787-15. e00787-00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-C., Tseng C.-k., Chen K.-J., Huang K.-J., Lin C.-K., Lin Y.-T. A cell-based reporter assay for inhibitor screening of hepatitis C virus RNA-dependent RNA polymerase. Anal. Biochem. 2010;403:52–62. doi: 10.1016/j.ab.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S., Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang Y., Xu Y., Ma C., Qin C., Zhang L. Lycorine reduces mortality of human enterovirus 71-infected mice by inhibiting virus replication. Virol. J. 2011;8:483. doi: 10.1186/1743-422X-8-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.S., Kim G.W., Kwon S., Jin Y.H. A cell-based reporter assay for screening inhibitors of MERS coronavirus RNA-dependent RNA polymerase activity. J. Clin. Med. 2020;9:2399. doi: 10.3390/jcm9082399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard-Nozaki J., Kim T., Imakura Y., Kihara M., Kobayashi S. Effect of alkaloids isolated from Amaryllidaceae on herpes simplex virus. Res. Virol. 1989;140:115–128. doi: 10.1016/s0923-2516(89)80089-5. [DOI] [PubMed] [Google Scholar]
- Shannon A., Le N.T.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93 doi: 10.1128/JVI.00023-19. e00023-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Compu. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration . 2020. Coronavirus (COVID-19) Update: FDA issues emergency use authorization for potential COVID-19 treatment. [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins W.J. Evolution of HCV NS5B non-nucleoside inhibitors. HCV: The Journey from Discovery to a Cure (Springer): Volume I. 2019:171–191. [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-N., Zhang Q.-Y., Li X.-D., Xiong J., Xiao S.-Q., Wang Z., Zhang Z.-R., Deng C.-L., Yang X.-L., Wei H.-P. Gemcitabine, lycorine and oxysophoridine inhibit novel coronavirus (SARS-CoV-2) in cell culture. Emerg. Microbes Infect. 2020;9:1170–1173. doi: 10.1080/22221751.2020.1772676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.