Abstract
Nucleation-promoting proteins tightly regulate actin polymerization in cells. Whereas many of these proteins bind actin monomers directly, formins use the actin-binding protein profilin to dynamically load actin monomers onto their flexible Formin Homology 1 (FH1) domains. Following binding, FH1 domains deliver profilin-actin complexes to filament ends. To investigate profilin’s role as an adaptor protein in formin-mediated elongation, we engineered a chimeric formin that binds actin monomers directly via covalent attachment of profilin to its binding site in the formin. This formin mediates slow filament elongation owing to a high probability of profilin binding at filament ends. Varying the position at which profilin is tethered to the formin alters the elongation rate by modulating profilin occupancy at the filament end. By regulating the availability of the barbed end, we propose that profilin binding establishes a secondary point of control over the rate of filament elongation mediated by formins. Profilin’s differential affinities for actin monomers, barbed ends and polyproline are thus tuned to adaptively bridge actin and formins and optimize the rate of actin polymerization.
Keywords: cytoskeleton, polymerization, kinetics, fluorescence microscopy
Introduction
A diverse cohort of nucleation-promoting proteins initiates and regulates the assembly of actin monomers into filaments by lowering the energy barriers for filament nucleation [1–5]. Among these proteins, formins are unique in their ability to remain bound to the growing ends of actin filaments, enabling them to regulate the rate of subunit addition [6–8]. Actin filaments assembled by formins are incorporated into a number of essential cellular structures, including cytokinetic rings, filopodia, actin cables and stress fibers [9, 10]. Most eukaryotes express multiple formin isoforms [11, 12], which typically possess unique polymerization activities [13–15]. The cellular roles of formin isoforms expressed in the same organism are often non-overlapping [16–18], suggesting that each formin’s polymerization activity is optimized for its function.
Formins interact with actin filaments primarily via their formin homology 2 (FH2) domain [6]. FH2 domains dimerize into toroidal complexes that bind filament barbed ends [19–21] and processively step onto incoming actin monomers to incorporate them into the filament [6, 7]. FH2-bound barbed ends fluctuate between polymerization-competent “open” and polymerization-incompetent “closed” conformations, thereby regulating the rate at which actin monomers can bind the filament end [13, 22]. This process, called “gating”, slows the elongation of FH2-bound filaments relative to the rate at which free filaments elongate.
Although FH2 dimers are necessary and sufficient to maintain processive formin association with barbed ends, engagement of the FH1 domain dramatically increases filament elongation rates [13]. The FH1 domain contains a number of polyproline tract sequences, which are separated by regions of intrinsic disorder that confer flexibility and diffusivity to the domain [23]. The majority of actin monomers in cells are bound by profilin, a small protein that also binds polyproline sequences [24–28]. Upon association of a profilin-actin complex with a polyproline tract, the FH1 domain delivers the actin subunit to the FH2-bound barbed end in a diffusion-limited reaction (Figure 1A) [22, 29, 30]. FH1-mediated actin delivery occurs via the assembly of an intermediate state called the “ring complex”, in which the actin monomer is simultaneously bound to the FH1-associated profilin and to the barbed end [22, 31, 32]. Dissociation of profilin from the newly delivered actin subunit and the FH1 domain disassembles the ring complex, enabling incorporation of the actin into the filament via FH2 domain stepping.
Figure 1. Formin-mediated delivery of profilin-actin to the barbed end.
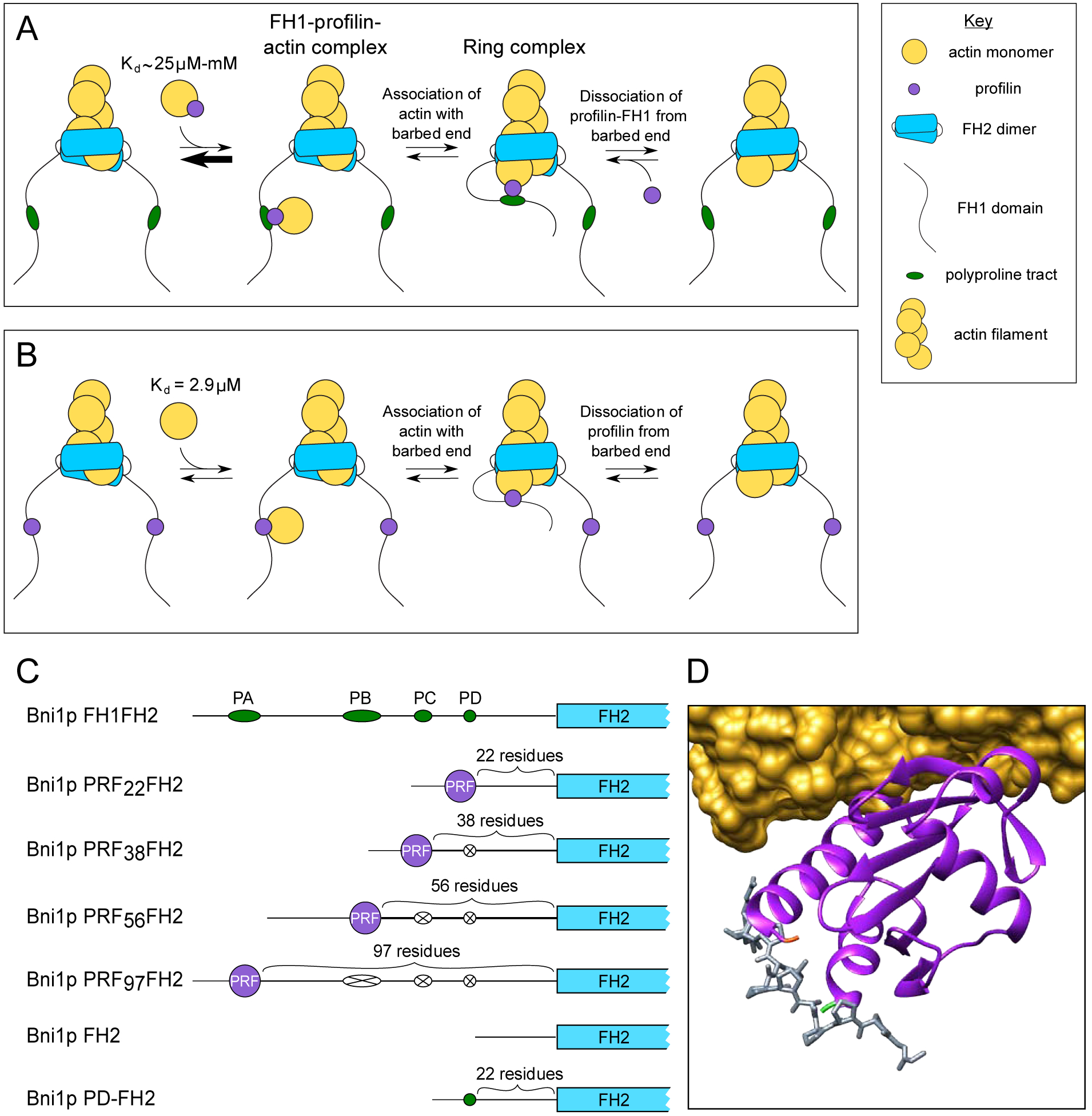
(A) Schematic representation of the steps involved in the delivery of a profilin-actin (purple and yellow circles) complex to the barbed end of a formin-bound actin filament. For simplicity, a single polyproline tract (green oval) is depicted in the formin’s FH1 domain. Profilin-actin binds polyproline with an affinity that depends on the length of the tract. Barbed end delivery proceeds through the association of the “ring complex” intermediate state, in which the profilin-actin complex is simultaneously bound to the barbed end and the FH1 domain. Dissociation of the profilin and subsequent stepping of one FH2 domain (not shown) complete the cycle of subunit addition. (B) Barbed end delivery of an actin monomer (yellow circle) by a formin whose FH1 domain encodes profilin. Actin monomers bind S. cerevisiae profilin with an affinity of 2.9 μM, and delivery proceeds through the assembly of the ring complex intermediate state. Dissociation of profilin from the barbed end and subsequent FH2 domain stepping (not shown) complete the cycle of subunit addition. (C) Domain architectures of formin constructs used in this study. FH1 domains and polyproline tracts are shown to-scale, whereas FH2 domains and profilin are not. Polyproline tract sequences that have been substituted with poly-Gly/Ser sequences are each represented by a white oval with an X. (D) Molecular model depicting simultaneous binding of profilin to an actin monomer and a polyproline sequence. A ribbon diagram of S. cerevisiae profilin (purple, PDB entry 1YPR) is shown bound to rabbit skeletal muscle alpha-actin (shown in yellow as a solvent-excluded surface model) and a peptide corresponding to the terminal polyproline repeat of human VASP (shown as a stick model in gray) from PDB entry 2PAV. The N- and C-termini of profilin are shown in green and orange.
In this study, we investigated the energetic determinants of profilin’s role as a dynamic adaptor protein in formin-mediated elongation. By covalently tethering profilin to the FH1 domain of the S. cerevisiae formin Bni1p, we synthetically promoted the assembly of the FH1-profilin-actin ternary complex that facilitates actin delivery. This chimeric formin mediates slow filament elongation despite robust delivery of actin monomers by FH1-tethered profilin. Using kinetic modeling, we determined that the slow rate of subunit addition results from an increase in the probability of profilin binding at formin-bound barbed ends. A decrease in the mobility of tethered profilin also likely slows the rate of actin monomer binding. Varying the position at which profilin is tethered to the FH1 domain modulates the likelihood of profilin occupancy at the barbed end and alters the filament elongation rate. By regulating the availability of the barbed end, we propose that profilin binding establishes a secondary point of control over the rate of formin-mediated filament elongation independent of FH2 domain gating. Profilin’s differential affinities for actin monomers, barbed ends and polyproline thus enable formins to frequently and transiently populate the ring complex state and mediate an optimal rate of actin polymerization.
Results
Profilin stimulates formin-mediated actin polymerization through its interactions with actin monomers, filament ends and the polyproline tracts encoded in formin FH1 domains. Profilin’s affinity for each of its binding partners has been shown to directly influence the rate of filament elongation [22, 30, 33]. To investigate the contributions of profilin’s dynamic interactions with FH1 domains to the kinetics of filament elongation, we sought to restrict profilin’s ability to dissociate from formin. We designed a chimeric variant of the S. cerevisiae formin Bni1p in which we covalently tethered profilin to the FH1 domain (Figure 1B). In this variant, the amino acid sequence of S. cerevisiae profilin replaces the sequence of polyproline tract PD, which is the tract located closest to the FH2 domain in Bni1p’s FH1 domain (Figure 1C). By circumventing profilin-polyproline binding, this strategy ensures that assembly of the FH1-profilin-actin ternary complex requires only the binding of an actin monomer to profilin. This interaction occurs with a 10-fold tighter affinity than the interaction of profilin with the longest polyproline tract in Bni1p’s FH1 domain [30, 34] (Figure 1A and B).
Profilin is particularly amenable to insertion into an FH1 domain because of the locations of profilin’s N- and C-termini on its polyproline-binding surface [35] (Figure 1D). As a result, covalent attachment of profilin’s termini to the FH1 domain positions profilin in an orientation that mimics the position it adopts when bound to a polyproline tract. This orientation exposes profilin’s actin-binding surface [36], making it available for binding and delivery of actin subunits to the barbed end.
To simplify the interpretation of polymerization data, we did not include any polyproline tracts in the FH1 domain sequence of our chimeric formin construct. We named this variant “Bni1p PRF22FH2”, where “22” corresponds to the number of residues separating profilin from the FH2 domain (Figure 1C).
Bni1p PRF22FH2 mediates slow actin filament elongation
To assess the polymerization activity of Bni1p PRF22FH2, we introduced it into reactions containing 0.75 μM actin monomers (33% Oregon Green-labeled). We used total internal reflection fluorescence (TIRF) microscopy to visualize elongating actin filaments (Figure 2A). In the presence of PRF22FH2, we observed two populations of polymerizing filaments. One population elongated at an average rate of 10.4 subunits/s, which matches the elongation of control filaments (measured in the absence of formin) and indicates that these filaments are not formin-bound (Figures 2B and 2C). The second population of filaments elongated continuously at a rate of 2.6 subunits/s, suggesting that Bni1p PRF22FH2 competently binds barbed ends and slows the rate of actin subunit addition.
Figure 2. PRF22FH2 mediates slow actin filament elongation.
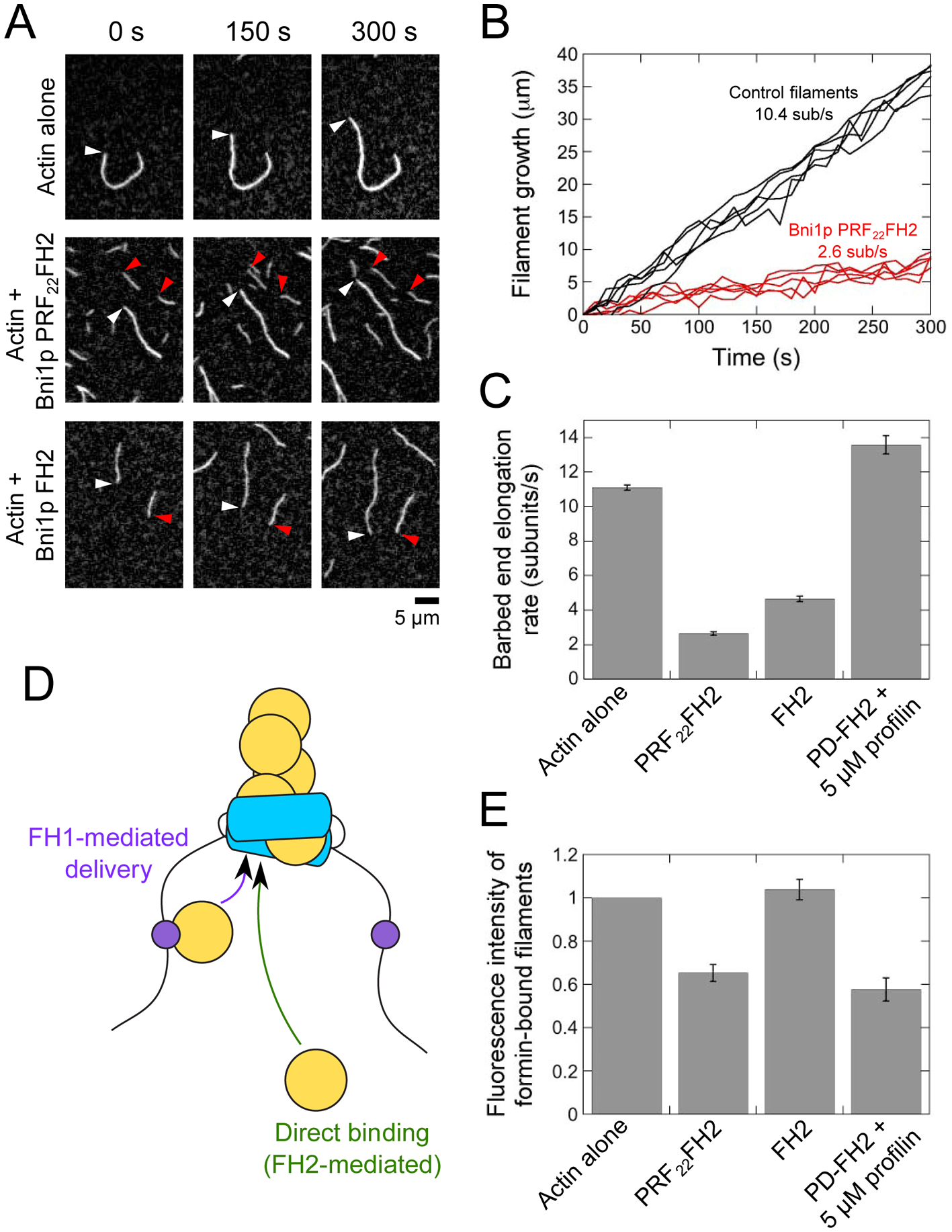
The experimental conditions were as follows: 0.75 μM chicken skeletal muscle actin monomers (33% Oregon Green-labeled) in microscopy buffer. The data were collected by TIRF microscopy. (A) Time series of TIRF micrographs of actin filament elongation in the absence and presence of PRF22FH2. White and red arrowheads indicate the barbed ends of free and formin-bound actin filaments, respectively. (B) Time courses of the elongation of five representative free barbed ends (black) and five barbed ends associated with Bni1p PRF22FH2 (red). (C) Barbed end elongation rates of filaments polymerizing in the absence or presence of Bni1p PRF22FH2, Bni1p FH2, or Bni1p PD-FH2 with 5 μM profilin. Error bars are standard errors of the mean elongation rate of at least 15 filaments. (D) Schematic representation of a formin-bound filament. Subunit addition can occur via FH1-mediated delivery (purple arrow) or direct binding of an actin monomer to the barbed end (green arrow; FH2-mediated). (E) Fluorescence intensities of filaments polymerized in the absence or presence of formin. Fluorescence intensities of formin-bound filaments were normalized to the intensities of filaments that are not formin-bound. Error bars are standard errors of the mean intensities of at least 15 filaments.
FH2 gating regulates the availability of barbed ends for monomer binding and slows filament elongation. To determine whether FH1-tethered profilin contributes to subunit addition mediated by PRF22FH2, or if the slow rate of polymerization can be attributed wholly to FH2 domain gating, we performed analogous polymerization experiments using a construct of Bni1p that contains only its FH2 domain (Figure 2A; Bni1p FH2). Filaments bound by Bni1p FH2 elongated at 4.7 subunits/s (Figure 2C), which is ~80% faster than the rate measured for Bni1p PRF22FH2-bound filaments. A Student’s t test confirmed that the difference in the elongation rates mediated by Bni1p PRF22FH2 and Bni1p FH2 is significant (p < 0.0001). Thus, Bni1p PRF22FH2 slows the rate of subunit addition to a greater extent than is produced by FH2 gating alone.
FH1-tethered profilin delivers actin to barbed ends
To elucidate the role played by profilin during elongation mediated by PRF22FH2, we characterized its mechanism of action. Formins polymerize actin filaments both by incorporating monomers that bind directly to FH2-bound barbed ends (i.e. FH2-mediated elongation) and by delivering FH1-bound profilin-actin complexes to barbed ends (i.e. FH1-mediated elongation) (Figure 2D). As a result, filament elongation rates reflect the sum of the contributions of both modes of subunit addition to polymerization.
To quantify the contribution of each mode of subunit addition to filament elongation mediated by Bni1p PRF22FH2, we took advantage of profilin’s 10-fold tighter affinity for unlabeled actin monomers than for actin that is fluorescently labeled at Cysteine 374 [37]. As a result of this difference, FH1-mediated polymerization, which requires the interaction of profilin with actin, gives rise to the incorporation of a larger fraction of unlabeled actin subunits than is observed for filaments that assemble in a profilin-independent manner [13, 38]. Quantification of the fluorescence intensities of filaments polymerized by formins therefore provides a direct measurement of the fraction of actin subunits that are incorporated via FH1-mediated elongation [38] (See Materials and Methods).
We measured the fluorescence intensities of formin-bound and free filaments in polymerization reactions containing Bni1p PRF22FH2 and Bni1p FH2 (Figure 2E). Consistent with published studies, filaments polymerized by Bni1p FH2 were as bright as control filaments [13, 29, 38]. In contrast, filaments assembled by Bni1p PRF22FH2 were 35% dimmer than control filaments, revealing the incorporation of a larger fraction of unlabeled subunits by this formin. This difference suggests that FH1-tethered profilin competently delivers actin subunits to barbed ends.
Using the fluorescence intensities of our formin-bound filaments and the published affinities of profilin for unlabeled and fluorescently labeled actin monomers, we quantified the fractions of the actin subunits polymerized by Bni1p PRF22FH2 that were incorporated through either FH1-mediated delivery or direct binding to the barbed end (see Materials and Methods) [33, 38]. We found that 38% of actin subunits are delivered by FH1-tethered profilin, whereas 62% of subunits are incorporated following direct binding to the barbed end.
To guide our interpretation of these results, we compared the rate of actin delivery mediated by FH1-tethered profilin to the rate generated by the wild-type PD polyproline tract. To do so, we measured the fluorescence intensities of filaments polymerized by Bni1p PD-FH2, which contains the native PD polyproline tract in Bni1p’s FH1 domain, followed by the FH2 domain (Figure 1C). In the presence of 5 μM profilin, the PD tract efficiently binds and delivers profilin-actin complexes to filament barbed ends [29, 33] (Figure 2C). As a result, the PD tract incorporates a large fraction of unlabeled subunits into filaments, which appear 42% dimmer than control filaments (Figure 2E). Quantification of the fraction of actin subunits incorporated via the FH1 domain reveals that the PD tract delivers ~50% of actin subunits to the barbed end in these conditions [38]. Thus, covalent attachment of profilin at the position normally occupied by the PD polyproline tract decreases the relative contribution of FH1-mediated actin delivery to filament elongation. As a result, a larger fraction of actin subunits is incorporated into Bni1p PRF22FH2-bound filaments via direct binding to the barbed end.
Free profilin attenuates FH1-mediated actin delivery by Bni1p PRF22FH2
To confirm the contribution of FH1-tethered profilin to filament elongation mediated by Bni1p PRF22FH2, we introduced free profilin into our polymerization reactions. Profilin-actin complexes can bind barbed ends directly, so inclusion of low micromolar concentrations of profilin did not affect the rates of elongation of control or Bni1p FH2-bound filaments (Figure 3A, open squares and open circles). In contrast, binding of free profilin precludes interactions between actin monomers and FH1-tethered profilin, effectively lowering the concentration of actin monomers available for FH1-mediated delivery. As a result, inclusion of profilin slowed the rate of elongation of PRF22FH2-bound filaments (Figure 3A, filled circles).
Figure 3. Free profilin attenuates actin delivery by FH1-tethered profilin.
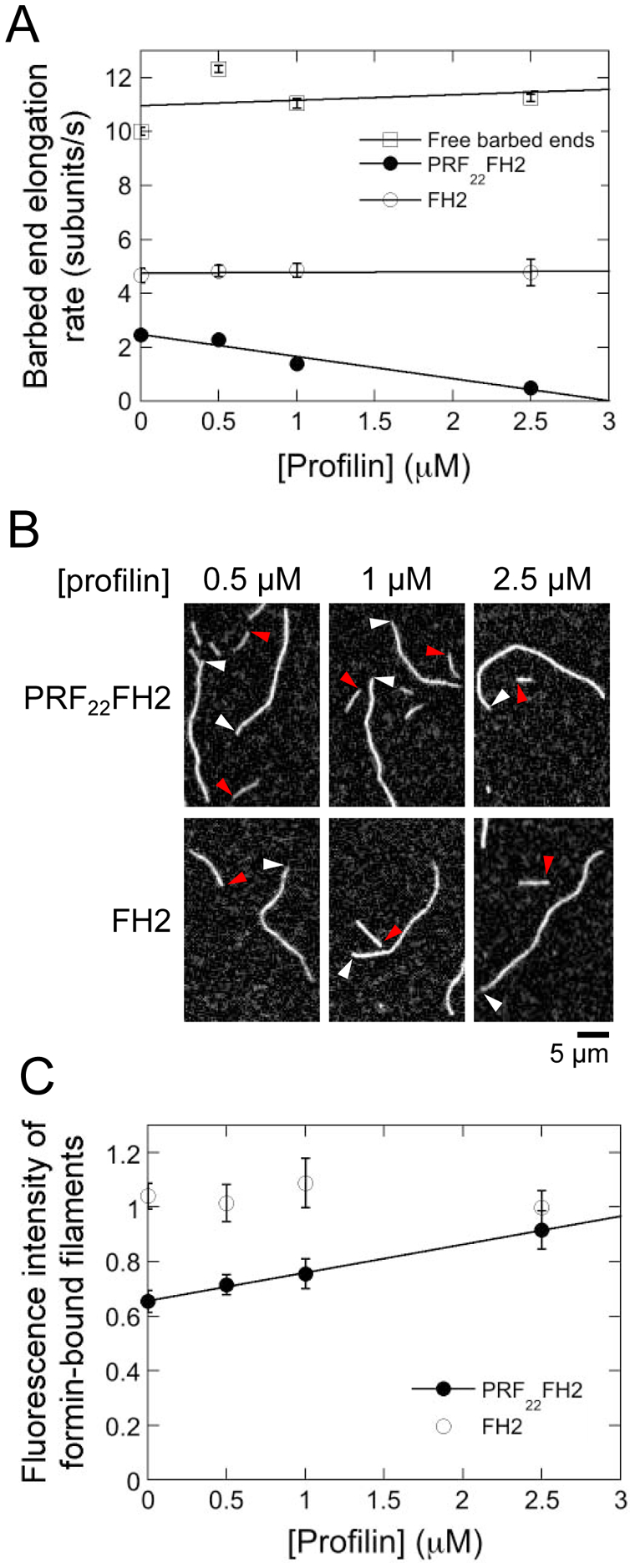
The experimental conditions were as follows: 0.75 μM actin monomers (33% Oregon Green-labeled) were allowed to polymerize in the presence of a range of S. cerevisiae profilin in microscopy buffer. The data were collected by TIRF microscopy. (A) Dependence of the rates of elongation of barbed ends bound by Bni1p PRF22FH2 (filled circles) or Bni1p FH2 (open circles) on the concentration of S. cerevisiae profilin. Error bars are standard errors of the mean elongation rate of at least 15 filaments. Lines are linear fits to the data. (B) Representative TIRF micrographs of actin filaments elongating in the presence of Bni1p PRF22FH2 or Bni1p FH2 and a range of S. cerevisiae profilin concentrations. White and red arrowheads indicate the barbed ends of free and formin-bound actin filaments, respectively. (C) Dependence of the fluorescence intensities of actin filaments bound by Bni1p PRF22FH2 and Bni1p FH2 on the concentration of S. cerevisiae profilin. Fluorescence intensities of formin-bound filaments were normalized to the intensities of filaments that are not formin-bound. Error bars are standard errors of the mean intensities of at least 15 filaments. Line is a linear fit to the data for Bni1p PRF22FH2.
Because of profilin’s tighter affinity for unlabeled actin monomers, the majority of profilin-actin complexes formed in reactions that include free profilin are unlabeled, whereas a large fraction of the fluorescent actin monomers remains free. Inclusion of profilin did not alter the fluorescence of filaments assembled by Bni1p FH2 (Figures 3B and 3C). In contrast, filaments assembled by Bni1p PRF22FH2 became brighter as the profilin concentration increased (Figure 3B and 3C), consistent with a role for FH1-mediated delivery in filament elongation mediated by this formin.
FH1-tethered profilin does not remove terminal subunits from FH2-bound barbed ends
Despite successful FH1-mediated actin delivery, filaments bound by Bni1p PRF22FH2 elongate more slowly than filaments bound by the FH2 domain alone. One possible explanation for this is the removal of actin subunits from barbed ends by FH1-tethered profilin (i.e. depolymerization), which might attenuate an otherwise rapid rate of subunit addition. In the absence of formin, introduction of 100-fold excess concentrations of profilin promotes dramatic filament shortening even in the presence of ATP-bound actin monomers, indicating that profilin binding at the barbed end stimulates the dissociation of the terminal actin subunit [39, 40]. Association of FH2 dimers with barbed ends has been shown to protect filaments from depolymerization in the absence of profilin [41, 42]. However, binding of FH1-tethered profilin to the barbed end might induce a conformational change in the terminal actin subunit that would promote its dissociation.
To test for profilin-stimulated depolymerization, we assembled actin filaments bound by Bni1p PRF22FH2 and measured changes in filament length following removal of actin monomers. We identified control and formin-bound filaments based on their elongation rates and fluorescence intensities during the assembly phase of our experiments. In the absence of actin monomers, filaments with free barbed ends shortened at an average rate of 3.7 ± 0.7 subunits/s, consistent with published studies using Oregon Green-labeled actin (Figure 4A) [39, 42]. Filaments bound by Bni1p PRF22FH2 depolymerized significantly more slowly (0.5 ± 0.2 subunits/s), matching the published rate for filaments bound by Bni1p FH2 (0.6 subunits/s [42]). This suggests that binding of FH1-tethered profilin does not stimulate removal of the terminal actin subunit from FH2-bound barbed ends (Figure 4A). Thus, the slow rate of filament elongation mediated by Bni1p PRF22FH2 arises from infrequent actin subunit addition rather than from the removal of subunits from the barbed end.
Figure 4. Actin monomers associate infrequently with barbed ends bound by Bni1p PRF22FH2.
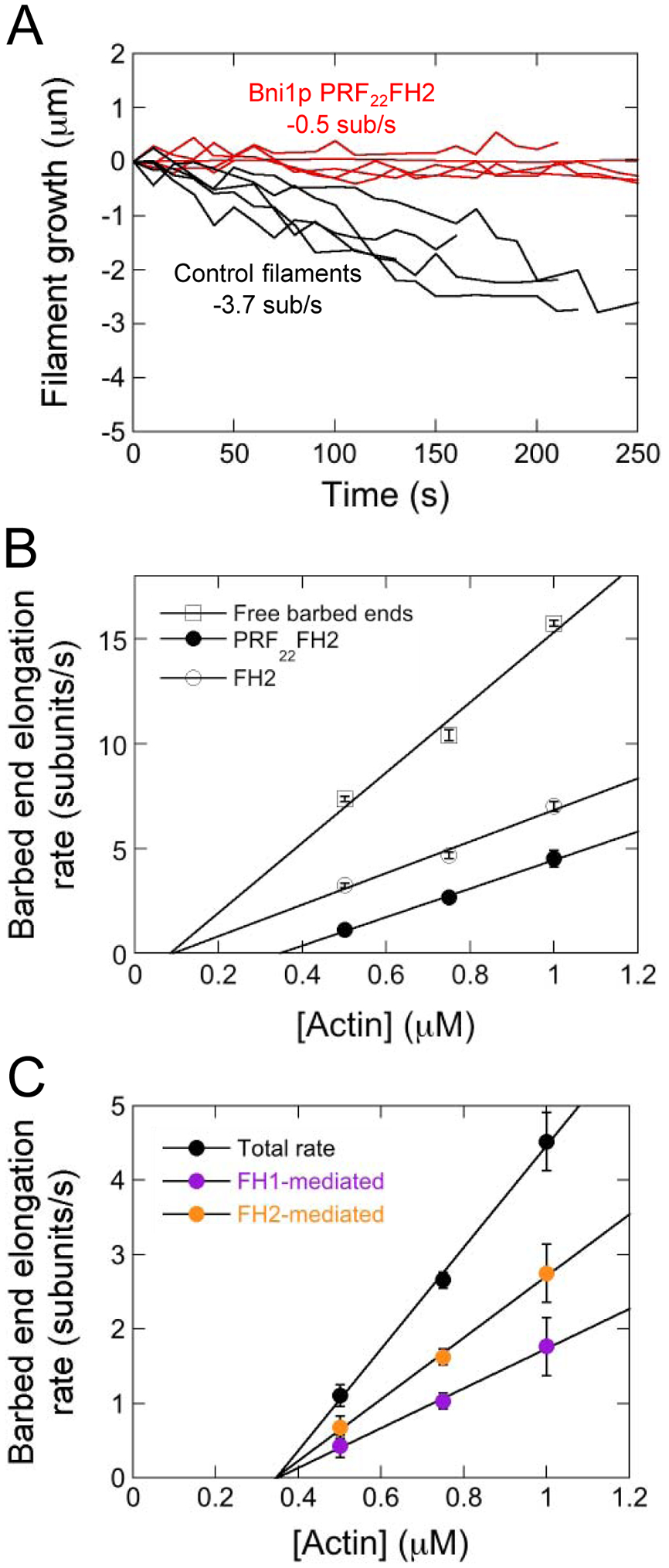
The experimental conditions were as follows: A range of concentrations of actin monomers (33% Oregon Green-labeled) were allowed to polymerize in microscopy buffer. The data were collected by TIRF microscopy. (A) Time courses of depolymerization following washout of actin monomers for five representative free barbed ends (black) and five barbed ends associated with Bni1p PRF22FH2 (red). (B) Dependence of the rates of barbed end elongation of free (open squares), Bni1p PRF22FH2-bound (filled circles) and Bni1p FH2-bound (open circles) filaments on the concentration of actin monomers. Error bars are standard errors of the mean elongation rate of at least 15 filaments. Lines are linear fits to the data. (C) Dependence of the total (black data), FH1-mediated (purple data) and FH2-mediated (orange data) barbed end elongation rates mediated by PRF22FH2 on the concentration of actin monomers. Error bars are standard errors of the mean elongation rates measured for at least 15 formin-bound filaments. Lines are linear fits to the data.
Binding of actin monomers to FH1-tethered profilin regulates the rate of filament elongation
Our finding that FH1-tethered profilin competently delivers actin to barbed ends indicates that Bni1p PRF22FH2 populates the ring complex intermediate state following the association of an actin monomer with profilin (Figure 1A). To quantify the relationship between FH1-profilinactin complex assembly and the rate of filament elongation, we varied the concentration of actin monomers in our polymerization experiments (Figure 4B). We found that the rate of elongation of PRF22FH2-bound filaments is proportional to the concentration of actin monomers, as has been observed for control filaments (Figure 4B, filled circles and open squares) [43]. Application of linear fits to the elongation data yields a 2.5-fold slower rate of actin subunit association for filaments bound by Bni1p PRF22FH2 compared to filaments with free barbed ends (kon = 16.8 subunits*μM−1s−1 for control filaments and kon = 6.8 subunits*μM−1s− for filaments bound by PRF22FH2). In comparison, the rate of actin association with barbed ends bound by Bni1p FH2 is 7.5 subunits*μM−1s−1 (Figure 4B, open circles). Bni1p PRF22FH2 also increases actin’s critical concentration whereas Bni1p FH2 does not, suggesting that FH1-tethered profilin partially inhibits the polymerization activity of Bni1p’s FH2 domain.
Using measurements of filament fluorescence, we calculated the FH1- and FH2-mediated elongation rates generated by Bni1p PRF22FH2 and plotted them as a function of the concentration of actin monomers (Figure 4C, purple and orange circles). Applying linear fits to these data reveals that direct binding of actin to barbed ends depends more steeply on the actin concentration than does FH1-mediated delivery (4.1 subunits*μM−1s−1 and 2.7 subunits*μM−1s−1, respectively).
FH1-tethered profilin regulates filament elongation in a position-specific manner
The slow elongation of filaments bound by Bni1p PRF22FH2 suggests the existence of a rate-limiting step in the polymerization reaction that regulates the frequency of subunit binding at the barbed end. In addition to promoting the assembly of the FH1-profilin-actin complex, covalent attachment of profilin to the FH1 domain increases the local concentration of profilin relative to the barbed end [44]. This increases the likelihood of profilin binding at the barbed end, which can occur via either FH1-mediated actin delivery or a direct, actin-independent binding event (Figure 5A). Structural modeling of the barbed end has shown that binding of profilin to the terminal subunit occludes the actin monomer binding site at the penultimate position, thus preventing actin subunit addition [40]. Thus, we hypothesized that the rate-limiting step might involve the dissociation of profilin from the barbed end.
Figure 5. The rate of actin delivery depends on the distance between profilin and the barbed end.
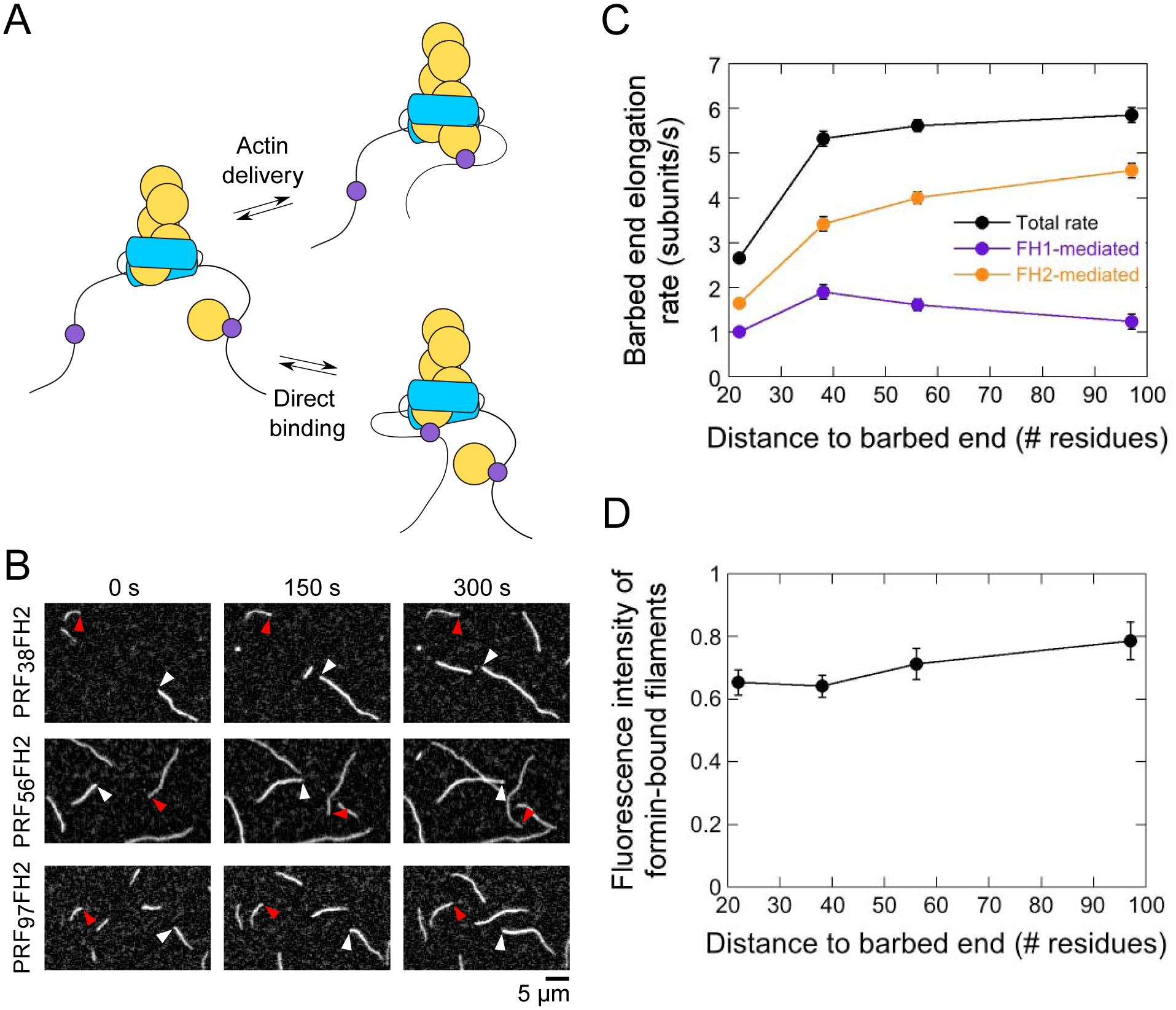
The experimental conditions were as follows: 0.75 μM actin (33% Oregon Green-labeled) in microscopy buffer. The data were collected by TIRF microscopy. (A) FH1-tethered profilin can bind the barbed end either via FH1-mediated actin delivery (upper reaction scheme) or via a direct binding event (lower reaction scheme). (B) Time series of TIRF micrographs of actin filament elongation in the absence and presence of PRF38FH2, PRF56FH2 and PRF97FH2. White and red arrowheads indicate the barbed ends of free and formin-bound actin filaments, respectively. (C) Dependence of the total (black data), FH1-mediated (purple data) and FH2-mediated (orange data) barbed end elongation rates mediated by the PRFxxFH2 constructs on the number of residues separating the profilin and Bni1p’s FH2 domain. Error bars are standard errors of the mean elongation rates measured for at least 15 formin-bound filaments. (D) Dependence of the fluorescence intensities of actin filaments polymerized by the Bni1p PRFxxFH2 constructs on the number of residues separating the profilin and Bni1p’s FH2 domain. Fluorescence intensities of formin-bound filaments were normalized to the intensities of filaments that are not formin-bound. Error bars are standard errors of the mean intensities of at least 15 filaments.
To test this hypothesis, we varied the position at which profilin was tethered to Bni1p’s FH1 domain. We reasoned that increasing the distance between profilin and the barbed end would decrease the likelihood of their interaction by reducing the local concentration of profilin. We constructed three additional chimeric variants by inserting profilin into the FH1 domain at sites corresponding to the positions of the PA, PB or PC polyproline tracts (Figure 1C). These positions are located 97, 56 and 38 residues away from the FH2 domain. To maintain the overall flexibility of the FH1 domain and simplify data interpretation, we replaced all other polyproline tracts in these variants with repeated Gly-Gly-Gly-Ser sequences that span the length of each replaced tract.
When included in polymerization reactions, each Bni1p variant assembled actin filaments that elongated more slowly than control filaments (Figures 5B and 5C). However, as the separation between profilin and the FH2 domain increased, the formin-mediated elongation rate increased non-linearly and reached an apparent plateau at ~6 subunits/s. The fluorescence intensities of the formin-bound filaments also increased by 30% over the range of distances sampled by our variants (Figure 5D). Thus, constructs in which profilin is located distal to the barbed end incorporate a smaller fraction of unlabeled subunits into filaments, suggesting a smaller contribution of FH1-mediated actin delivery to the overall rate of filament elongation.
We calculated the elongation rates mediated via direct binding of actin monomers to the barbed end (i.e. FH2-mediated) and delivery by FH1-tethered profilin (Figure 5C). As the separation between profilin and the FH2 domain increases from 22 to 97 residues, the rate of FH2-mediated subunit addition increases approximately 3-fold. Thus, a reduction in the local concentration of profilin enables more frequent direct binding of actin subunits to the barbed end. In contrast, the rate of FH1-mediated elongation peaks when profilin is separated by 38 residues from the barbed end and decreases at larger distances (Figure 5C). This non-linear relationship suggests that, as the separation between profilin and the barbed end increases, a slower rate of delivery offsets the increase in the availability of the barbed end for subunit binding.
Discussion
To characterize profilin’s dynamic role in formin-mediated actin polymerization, we covalently tethered profilin to the FH1 domain of the S. cerevisiae formin Bni1p. By physically linking profilin to its binding site in the FH1 domain, we effectively lowered the binding constant for profilin-FH1 binding to zero. As a result, assembly of the FH1-profilin-actin complex requires only the interaction of an actin monomer with FH1-tethered profilin, which occurs with low micromolar affinity [34]. We found that FH1-tethered profilin successfully delivers actin subunits to barbed ends (Figure 2). However, filaments bound by our chimeric formins elongate slowly (Figures 2 and 5).
Slow profilin-actin binding attenuates the contribution of the FH1 domain to elongation
To explore the effects of the covalent attachment of profilin on the dynamics of FH1-mediated delivery, we simulated formin-mediated actin polymerization with a kinetic model that accounts for interactions among actin, profilin, formin and filament ends [22, 29, 30]. We used published values for the on- and off-rates that govern each set of intermolecular associations [22]. To capture the properties of FH1-tethered profilin, we created a single molecular species that binds actin monomers and filament ends with affinities matching those of profilin and mediates ring complex assembly at rates dictated by the diffusive properties of the FH1 domain (Supplemental Figure S1 and Supplemental Table S1).
Initial simulations performed using published kinetic parameters [22] produced a relatively rapid rate of filament elongation and showed FH1-mediated delivery as the dominant mode of subunit addition (Figure 6A, rates intercepted by the green dashed line). In contrast, our experimental data indicate that delivery of actin by FH1-tethered profilin occurs more slowly than FH2-mediated subunit incorporation (Figure 4B). This suggests that actin monomers associate less frequently with FH1-tethered profilin than with barbed ends. To capture the relative contributions of FH1- and FH2-mediated subunit addition to the overall rate of elongation mediated by our chimeric formin, we therefore considered the effects of tethering profilin to the FH1 domain on profilin’s diffusive properties. Whereas profilin can ordinarily diffuse in three dimensions and rotate freely, physically linking profilin to a formin’s FH1 domain significantly decreases its lateral and rotational freedom. As a result, FH1-tethered profilin likely collides less frequently with actin monomers than would be observed for free profilin. This reduction in the collision frequency decreases the rate of association of actin monomers with profilin. A similar effect on rotational freedom is predicted to slow the diffusion of free profilin following its association with a wild-type FH1 domain. Published simulations of filament elongation mediated by wild-type formins account for this by including an association rate for actin monomers and FH1-bound profilin that is slower by a factor of two than the rate measured for free profilin [22, 27, 37].
Figure 6. FH1-tethered profilin associates slowly with actin monomers and frequently with barbed ends.
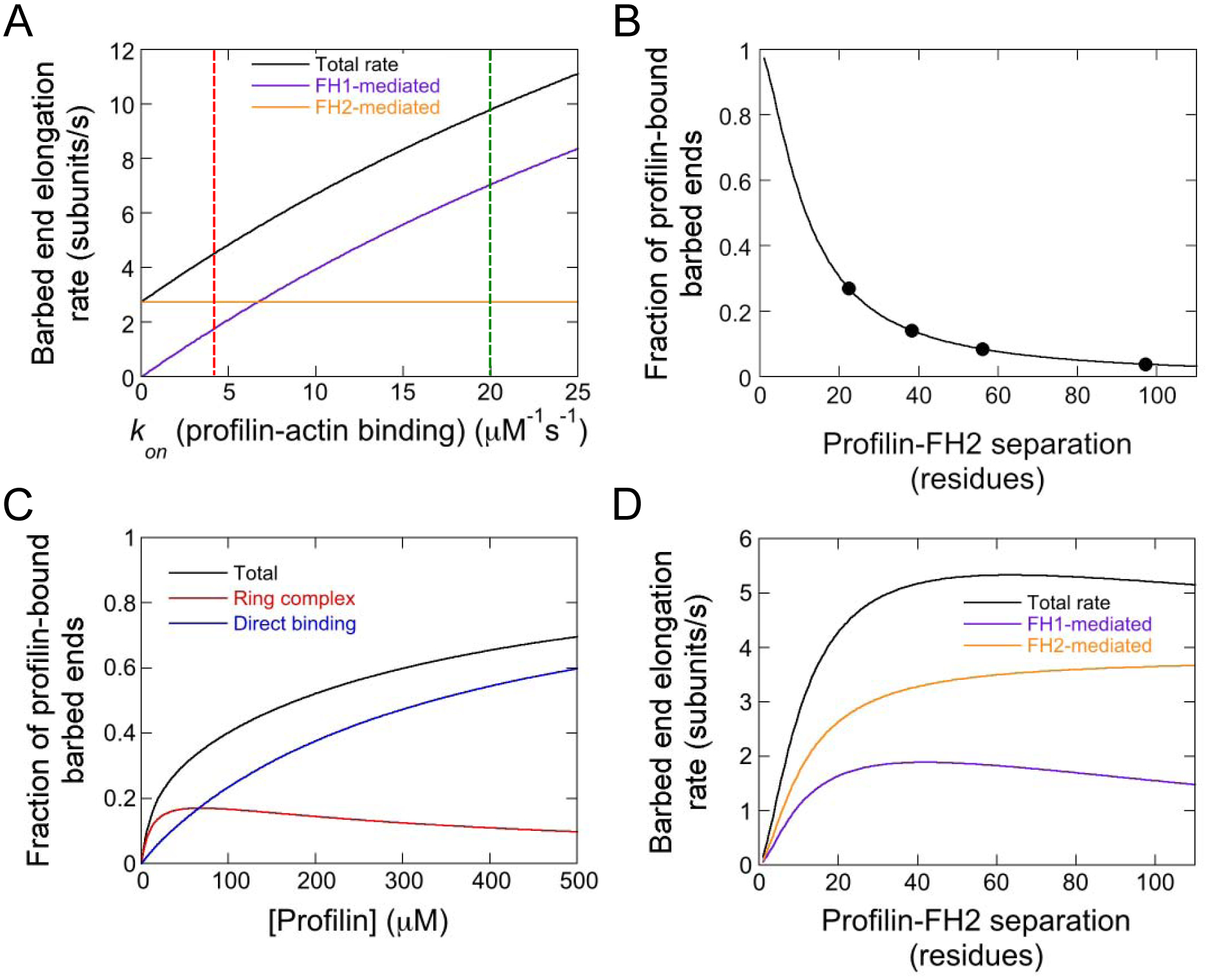
(A) Dependence of the simulated rate of barbed end elongation mediated by Bni1p PRF22FH2 on the on-rate for the association of profilin with actin monomers. Total (black line), FH1-mediated (purple line) and FH2-mediated (orange line) elongation rates are shown. The green dashed line intersects the elongation rates obtained when using the published association rate (i.e. kon = 20 μM−1s−1 [22, 37]). The red dashed line indicates the association rate that most closely reproduces our experimental elongation rates (i.e. kon = 4 μM−1s−1). Simulations were performed with 0.75 μM actin monomers. (B) Dependence of the fraction of formin-bound filament barbed ends that are bound by profilin in our simulations on the number of residues separating FH1-tethered profilin from the FH2 domain. (C) Dependence of the fraction of barbed ends that are bound by profilin on the concentration of free profilin in simulations of wild-type formin. Simulations were carried out in the presence of 0.75 μM actin monomers. Profilin can bind the barbed end via assembly of the ring complex (red line) or through direct, FH1-independent association of profilin or a profilin-actin complex with the filament end (blue line). The total fraction of profilin-bound barbed ends (black line) was calculated by summing the fractions of barbed ends bound via both modes of profilin association. (D) Dependence of the simulated total (black line), FH1-mediated (purple line) and FH2-mediated (orange line) barbed end elongation rates on the number of residues separating FH1-tethered profilin from the FH2 domain.
We tested the effects of varying the rate at which FH1-tethered profilin binds actin monomers on the simulated elongation rate mediated by our chimeric formin (Figure 6A). We found that filament elongation slows as the rate of profilin-actin association decreases. Further, the contribution of FH1-mediated delivery to elongation decreases when binding of actin to profilin is slow, whereas the rate of FH2-mediated elongation is unaffected. Using these simulation results, we determined that decreasing the profilin-actin association rate fivefold relative to the published value (i.e. 20 μM−1s−1 [22, 27, 37]; Supplemental Table S1) reproduces our experimental results (Figure 6A, rates intercepted by red dashed line). This association rate is slower than the rate of spontaneous subunit addition at barbed ends, consistent with the emergence of FH2-mediated elongation as the dominant mode of subunit addition.
FH1-tethered profilin binds barbed ends with high probability
Barbed ends bound by Bni1p PRF22FH2 elongate more slowly than filaments bound by Bni1p FH2. Thus, in addition to mediating slow actin subunit delivery, FH1-tethered profilin inhibits actin binding at the barbed end. Profilin binds actin monomers and barbed ends with differing affinities, and it is important to consider how tethering profilin to the FH1 domain might influence both sets of interactions. S. cerevisiae profilin binds actin monomers with an affinity of 2.9 μM [34], so the nanomolar concentrations of formin included in our polymerization experiments ensure dynamic and non-saturating interactions between FH1-tethered profilin and the actin monomers in solution. Profilin’s affinity for barbed ends is nearly 100-fold weaker than for monomers (Kd = 238 μM for human profilin 1 [40]), so nanomolar concentrations of profilin would not normally be expected to measurably bind barbed ends. However, tethering profilin to the FH1 domain significantly increases its local concentration relative to the barbed end. This in turn increases the likelihood of profilin binding to the barbed end.
Because of the physical linkage between profilin and the FH1 domain in Bni1p PRF22FH2, binding of profilin to the barbed end occurs via population of the ring complex. This complex can form either following actin monomer delivery or via direct binding of profilin to the terminal subunit at the barbed end (Figure 5A). To quantify the effects of increasing the local concentration of profilin on its interactions with the barbed end, we calculated the fractional population of the ring complex state (which corresponds to the fraction of barbed ends bound by FH1-tethered profilin) in our simulations. We found that approximately 30% of Bni1p PRF22FH2-bound barbed ends are bound by profilin via formation of the ring complex state at any given time (Figure 6B, data point at a profilin-FH2 separation of 22 residues). This fraction of profilin-bound barbed ends corresponds to an effective local profilin concentration of approximately 100 μM (calculated based on a Kd of 238 μM for profilin-barbed end binding [40]).
In simulations of wild-type formin, rapid dissociation of profilin from the polyproline tracts in the FH1 domain disfavors a long-lived ring complex state. In these reactions, barbed end binding can also occur independently of the FH1 domain via direct association of profilin or a profilin-actin complex with the filament end. Despite this additional binding mode, the fraction of barbed ends occupied by profilin is negligible in the presence of low micromolar concentrations of free profilin. The barbed end occupancy increases in proportion to the concentration of profilin and reaches 30% in the presence of 45 μM profilin (Figure 6C).
By regulating the availability of the barbed end, we propose that profilin binding establishes a secondary point of control over the rate of formin-mediated actin polymerization independent of FH2 domain gating. A similar mode of profilin-mediated barbed end regulation has been observed for actin filaments with free barbed ends elongating in physiological conditions [25]. In mammalian cells, profilin-actin concentrations range from 50–200 μM, depending on cell type. Although profilin-actin complexes can readily associate with filament ends, dissociation of profilin from the barbed end becomes rate-limiting and slows filament elongation under these conditions [25]. At these cellular concentration levels, 18–45% of barbed ends are predicted to be bound by profilin at any given time [40], which is similar to the fraction of barbed ends bound by FH1-tethered profilin in our experiments with Bni1p PRF22FH2.
Notably, barbed end regulation by profilin has been observed to slow the elongation of filaments bound by wild-type formins less dramatically than filaments with free barbed ends [25]. Our results with Bni1p PRF22FH2 suggest that the presence of an FH2 dimer does not automatically promote the dissociation of profilin from formin-bound filament ends. Rather, it is likely that diffusion-limited delivery of FH1-bound profilin-actin complexes enables formins to maintain rapid elongation rates despite a high probability of binding of profilin at the barbed end. The high local concentrations of the polyproline tracts encoded by FH1 domains may also facilitate the removal of profilin from the barbed end. This has been demonstrated to be an effective mechanism for rapid elongation even for formins with very small FH2 gating factors [13].
The rate of ring complex formation influences both FH1- and FH2-mediated elongation
To vary the local concentration of profilin, we generated a series of chimeric formins that sample a physiologically-relevant range of positions spanning the length of Bni1p’s FH1 domain (Figure 1C). We found that the rate of filament elongation increases as the separation between profilin and the barbed end increases (Figure 5C). This rise in the elongation rate stems largely from an increase in the rate of FH2-mediated subunit addition (Figure 5C). In contrast, FH1-mediated subunit delivery is less sensitive to the profilin positions sampled in our experiments.
The volume sampled by profilin via diffusion of the FH1 domain depends on the location at which profilin is attached (or bound) to the FH1 domain. This relationship in turn dictates the frequency of collisions between profilin and the barbed end, and thus the rate of ring complex assembly. We simulated the dependence of the filament elongation rate on the position at which profilin is tethered to the FH1 domain by varying the rate of ring complex assembly in proportion to its distance from the barbed end (Figure 6D). Our simulations reproduced the general trend in the elongation rates that we observed in our experiments (Figure 5C).
To quantify the relative availability of the barbed end for actin binding, we calculated the fractional population of the ring complex state in our simulations (Figure 6B). We found a sharp dependence on the separation between profilin and the barbed end. Whereas ~30% of barbed ends are bound by profilin when it is positioned 22 residues away from the FH2 domain, this fraction decreases at larger separations. Profilin occupies approximately 15%, 8% and 4% of barbed ends at profilin-barbed end separations of 38, 56 and 97 residues, respectively. These fractions of profilin-bound barbed ends correspond to effective local profilin concentrations of 45, 20 and 10 μM (calculated based on a Kd of 238 μM).
These results reveal two ways in which formin-mediated elongation depends on the position at which profilin is attached to the FH1 domain. First, modulating the profilin occupancy at the barbed end tunes the frequency of actin monomer binding. As profilin binding decreases, the rate of FH2-mediated elongation increases, as does the rate of FH1-mediated elongation at short profilin-FH2 separations. Second, the rate of collisions between the FH1-profilin-actin complex and the barbed end (i.e. the ring complex assembly rate) decreases as the separation between profilin and the FH2-bound barbed end increases. Thus, whereas the rate of FH2-mediated elongation increases robustly, FH1-mediated delivery depends non-linearly on profilin’s position in the FH1 domain. The modest peak we observe in FH1-mediated elongation at intermediate distances in both our experimental and simulated data reflects the position at which the probability of barbed end binding by FH1-tethered profilin and the efficiency of FH1-mediated actin delivery are optimized.
Profilin’s affinities for actin monomers, barbed ends and polyproline are tuned for rapid formin-mediated polymerization
Whereas most actin nucleation-promoting proteins bind directly to actin monomers via WASP homology 2 (WH2) or similar binding motifs [2–4, 45–47], formins utilize profilin as an adaptor protein to enable efficient delivery of actin subunits to filament ends. Profilin’s relatively tight affinity for actin monomers ensures that profilin-actin complexes are plentiful and available for frequent association with FH1 domains under most physiological conditions. Following incorporation into a filament, actin monomers undergo a conformational change that weakens their affinity for profilin by two orders of magnitude [40, 48]. This dramatic reduction in affinity promotes rapid dissociation of profilin following profilin-actin binding at the barbed end. Our results demonstrate that this combination of disparate affinities is crucial for efficient formin-mediated filament elongation, because it promotes both the frequent assembly and rapid disassembly of the ring complex intermediate state.
The requirement for differing affinities for monomers and barbed ends complements the tuning of profilin binding affinities along formin FH1 domains [30]. Whereas a tight profilin-polyproline interaction is advantageous in assembling a long-lived FH1-profilin-actin complex, profilin must also efficiently dissociate from the FH1 domain to enable disassembly of the ring complex following profilin-actin delivery (Figure 7A). Profilin’s dynamic interactions with polyproline thus enable it to bridge actin monomers and barbed ends via transient associations with formin FH1 domains. Its rapid dissociation from both actin and formin frees the barbed end for the next cycle of subunit addition. In sum, a discrete range of affinities for actin monomers and polyproline enables profilin to maximize the rate of formin-mediated filament elongation (Figure 7B).
Figure 7. Profilin’s differential affinities for actin monomers, filament ends and polyproline modulate population of the ring complex intermediate state and filament elongation.
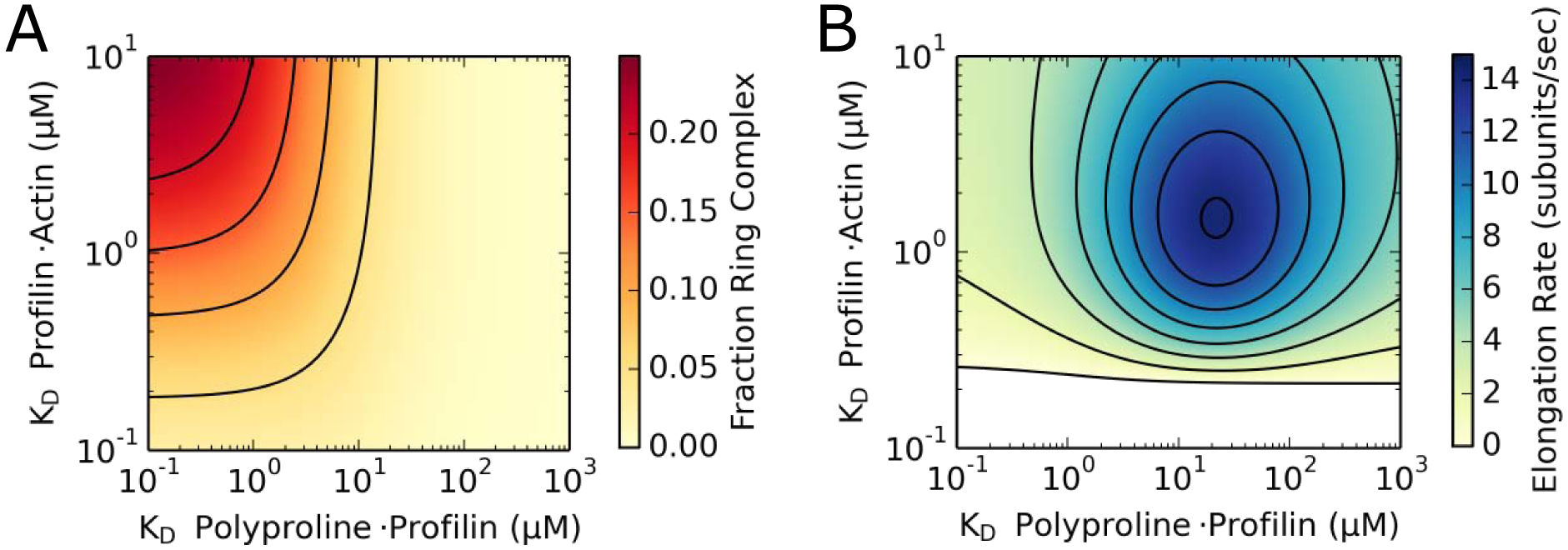
(A) Contour plot depicting the dependence of the simulated fraction of formin-bound barbed ends that populate the ring complex state at any given time on profilin’s affinities for actin monomers and polyproline. Simulations were carried out in the presence of 1 μM actin monomers and 5 μM profilin. Axes are shown in log scale. Contour lines are shown at intervals of 0.05. (B) Contour plot depicting the dependence of the simulated formin-mediated actin filament elongation rate on profilin’s affinities for actin monomers and polyproline. Simulations were carried out in the presence of 1 μM actin monomers and 5 μM profilin. For simplicity, simulations were performed using a formin with a single polyproline tract. Maximal rates of filament elongation (dark blue region) occur at affinities that promote low levels of ring complex assembly (light yellow region in plot A). Axes are shown in log scale. Contour lines are shown at intervals of 2 subunits/s. The white portion of the plot corresponds to a region of depolymerization.
Materials and Methods
Design of chimeric profilin-formin variants
Design and construction of chimeric profilin-formin variants occurred in two steps. First, a series of variants of a construct encoding the FH1 and FH2 domains of the S. cerevisiae formin Bni1p were designed and cloned into a pGEX-4T-3 plasmid (GE Healthcare Life Sciences). In each variant, all but one of the polyproline tracts were replaced with a repeated Gly-Gly-Gly-Ser sequence corresponding to the length of each replaced tract. In addition to the N-terminal GST tag that is included in pGEX vectors, each construct encoded an N-terminal TEV protease recognition sequence and a C-terminal 6-His tag. To maximize protein solubility, each variant was N-terminally truncated to include only 10–28 residues of FH1 domain sequence immediately preceding the polyproline tract. The residue boundaries for the single-tract variants are as follows: 1228–1766 (Bni1p PA-FH2), 1251–1766 (Bni1p PB-FH2), 1292–1766 (Bni1p PC-FH2) and 1310–1766 (Bni1p PD-FH2).
Second, the polyproline tract sequence in the FH1 domain of each single-tract variant was replaced with the nucleotide sequence encoding S. cerevisiae profilin. An additional Gly-Gly-Ser sequence was included following the profilin sequence to promote flexibility in the linkage between the profilin and the FH1 domain sequence. These hybrid profilin-formin constructs were named “PRFxxFH2”, where “xx” corresponds to the number of residues in the FH1 domain that separate the profilin from the FH2 domain.
Protein purification
All Bni1p PRFxxFH2 proteins were expressed at 16 °C overnight in 1 L cultures of BL21 DE3 RP Codon Plus cells (Agilent Technologies). Cell pellets were resuspended and lysed by sonication in 50 mM Tris (pH 8.0), 500 mM NaCl, 1 mM DTT. Following centrifugation at 16,000 rpm for 40 minutes, lysate supernatants were incubated with 2 mL glutathione-sepharose resin (Gold Biotechnology) with rotation for 1 hour at 4 °C. Resin and protein solutions were transferred to empty chromatography columns and washed with 20 mL of lysis buffer, followed by 20 mL of low-salt wash buffer (50 mM Tris (pH 8.0), 100 mM NaCl, 1 mM DTT). GST-tagged proteins were eluted with 6 mL of 100 mM glutathione (pH 8.0) in low-salt wash buffer, and glycerol was added to a final concentration of 10% (v/v). Each protein was incubated with 2–5 μM TEV protease overnight at 4 °C to remove the GST tag. Formins were separated from the TEV protease and cleaved GST via nickel affinity chromatography, concentrated using 30,000 MWCO spin columns (Millipore Sigma) and dialyzed into KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole (pH 7.0)) with 10% (v/v) glycerol and 1 mM DTT. Proteins were assayed for purity by SDS-PAGE (Supplemental Figure S2). Purified proteins were flash-frozen in liquid nitrogen and stored at −80 °C. We used ProtParam (www.web.expasy.org/protparam) [49] to calculate extinction coefficients for all constructs.
Skeletal muscle actin was purified from an acetone powder prepared from frozen chicken breasts (Trader Joe’s) via one cycle of polymerization and depolymerization [50]. Actin was labeled on cysteine 374 with Oregon Green 488 iodoacetamide (Thermo Fisher Scientific) [51]. Monomers were gel-filtered on S-300 resin (GE Healthcare) in G-Buffer (2 mM Tris (pH 8.0), 0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2) and stored at 4 °C. We used extinction coefficients of 26,000 M−1cm−1 at λ = 290 nm for unlabeled actin and 78,000 M−1cm−1 at λ = 491 nm for Oregon Green, and the following equation to calculate the concentration of Oregon Green-labeled actin: [Oregon Green-labeled actin] = (A290 – (A491*0.171))/26,000 M−1 cm−1.
Microscopy and data analysis
Glass coverslips (22 mm × 50 mm; Fisher Scientific) and slides were sonicated in 1% Hellmanex III and rinsed with water. Chambers were prepared as previously described [29] and stored at room temperature for up to 1 week.
The following solutions were introduced sequentially into each chamber and allowed to incubate for 1 minute: (1) 0.5% Tween 20 in HS-TBS (50 mM Tris (pH 7.5), 600 mM NaCl), (2) ~1 μM NEM-inactivated chicken skeletal muscle myosin [51] in HS-TBS, and (3) 100 mg/mL BSA in HS-TBS. Chambers were washed with HS-TBS following each incubation step and with KMEI prior to the introduction of a polymerization reaction.
Mixtures of unlabeled and Oregon Green-labeled Ca2+-ATP actin were converted to Mg2+-ATP actin via the addition of 50 μM MgCl2 and 0.2 mM EGTA, followed by a 5-minute incubation. Polymerization of actin monomers with or without formins was initiated by addition of 2x microscopy buffer (1x microscopy buffer: 10 mM imidazole (pH 7.0), 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 15 mM glucose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, 0.5% (w/v) methylcellulose (4,000 cP at 2%)). Reactions were immediately introduced into the chamber upon the initiation of polymerization.
Time-lapse images of elongating actin filaments were collected via through-objective total internal reflection fluorescence (TIRF) microscopy on an Olympus Ti83 motorized microscope equipped with a cellTIRF system using a 60x, 1.49 N.A. objective and a 488-nm laser. Images were collected every 10 s using a Hamamatsu C9100–23B ImagEM X2 EMCCD camera and cellSens Dimension software (Olympus).
Movies were processed with ImageJ software (National Institutes of Health) [52]. For each reaction, changes in length for at least 15 formin-bound and 15 control filaments were measured, typically over a span of at least 20 frames or 200 seconds. Linear fits were applied to plots of filament length over time to determine elongation rates. We measured the fluorescence intensities of control and formin-bound filaments by performing line scans along 5–10 μm stretches at filament barbed ends, as well as neighboring background regions using ImageJ.
Calculation of FH1-mediated elongation rates
Rates of filament elongation arising from FH1-mediated subunit delivery were calculated from total elongation rates as described previously [38]. Briefly, the fluorescence intensities of formin-bound filaments were normalized to those of control filaments. The fraction of unlabeled actin subunits incorporated via FH1-mediated delivery (FH1Black) was then calculated as follows:
| (1) |
where FluorescenceFormin-bound is the normalized fluorescence intensity of the formin-bound filaments in the reaction. The fraction of labeled actin subunits incorporated by the FH1 domain (FH1Green) was then calculated using the following equation:
| (2) |
where Green PA is the labeled fraction of the actin that binds profilin and Black PA is the unlabeled fraction of the actin that binds profilin. Green PA and Black PA were calculated using the published affinities of S. cerevisiae profilin for unlabeled actin and actin that is labeled at cysteine 374 (2.9 and 29 μM, respectively) [34, 37] and the following relation:
| (3) |
where [actin] is the concentration of either labeled or unlabeled actin subunits. The total fraction of actin subunits incorporated via FH1-mediated delivery was calculated by summing FH1Black and FH1Green, and the polymerization rate mediated by the FH1 domain was obtained by multiplying this sum by the total elongation rate mediated by the formin. The polymerization rate mediated via direct binding of subunits to barbed ends (i.e. FH2-mediated elongation) was obtained by subtracting the rate of FH1-mediated elongation from the total elongation rate.
Kinetic modeling
The kinetic scheme described by Vavylonis and coworkers [22] was modified to include equilibria incorporating binding, ring complex assembly and delivery of actin monomers via a profilin moiety within the FH1 domain of the formin (Figure 1B). Mathematical modeling of actin polymerization time courses was performed using COPASI [53] using previously published rate constants. Time courses were calculated in deterministic mode using the LSODA algorithm [54, 55] at fixed bulk actin and profilin concentrations.
Molecular model of binding of profilin to an actin monomer and a polyproline sequence
The structure of S. cerevisiae profilin (Protein Data Bank (PDB) entry 1YPR) [34] was superimposed onto that of human profilin complexed with alpha-actin from rabbit skeletal muscle and a peptide corresponding to the terminal polyproline tract from human VASP (PDB entry 2PAV) [56]. The resulting molecular structure was further adjusted through rigid body refinement and energy minimization using MODELLER [57] and AutoDock Vina [58]. Molecular models were constructed using the UCSF Chimera [59]. The solvent-excluded molecular surface was constructed using the program MSMS [60] with a probe radius of 1.4 Å. Figures were rendered using POV-Ray (http://www.povray.org/).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (NIH) research grant GM-122787. MEZ was supported by NIH Training Grant AR007612.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Pollard TD (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct 36, 451–77. [DOI] [PubMed] [Google Scholar]
- 2.Quinlan ME, Heuser JE, Kerkhoff E & Mullins RD (2005) Drosophila Spire is an actin nucleation factor. Nature 433, 382–8. [DOI] [PubMed] [Google Scholar]
- 3.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB & Mullins RD (2009) p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell. Biol 11, 451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG & Goode BL (2010) Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J. Cell. Biol 189, 1087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode BL & Eck MJ (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem 76, 593–627. [DOI] [PubMed] [Google Scholar]
- 6.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A & Boone C (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–5. [DOI] [PubMed] [Google Scholar]
- 7.Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J & Pring M (2003) Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol 13, 1820–3. [DOI] [PubMed] [Google Scholar]
- 8.Paul AS & Pollard TD (2009) Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J. Biol. Chem 284, 12533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campellone KG & Welch MD (2010) A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell. Biol 11, 237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faix J & Grosse R (2006) Staying in shape with formins. Dev. Cell 10, 693–706. [DOI] [PubMed] [Google Scholar]
- 11.Higgs HN (2005) Formin proteins: a domain-based approach. Trends. Biochem. Sci 30, 342–53. [DOI] [PubMed] [Google Scholar]
- 12.Pruyne D (2016) Revisiting the Phylogeny of the Animal Formins: Two New Subtypes, Relationships with Multiple Wing Hairs Proteins, and a Lost Human Formin. PLoS One 11, e0164067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovar DR, Harris ES, Mahaffy R, Higgs HN & Pollard TD (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–35. [DOI] [PubMed] [Google Scholar]
- 14.Neidt EM, Skau CT & Kovar DR (2008) The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem 283, 23872–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott BJ, Neidt EM & Kovar DR (2011) The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell 22, 3826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang F, Drubin D & Nurse P (1997) cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell. Biol 137, 169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feierbach B & Chang F (2001) Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol 11, 1656–65. [DOI] [PubMed] [Google Scholar]
- 18.Petersen J, Nielsen O, Egel R & Hagan IM (1998) FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell. Biol 141, 1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M & Rosen MK (2005) Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–94. [DOI] [PubMed] [Google Scholar]
- 20.Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D & Goode BL (2004) A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL & Eck MJ (2004) Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–23. [DOI] [PubMed] [Google Scholar]
- 22.Vavylonis D, Kovar DR, O’Shaughnessy B & Pollard TD (2006) Model of formin-associated actin filament elongation. Mol. Cell 21, 455–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horan BG, Zerze GH, Kim YC, Vavylonis D & Mittal J (2018) Computational modeling highlights the role of the disordered Formin Homology 1 domain in profilin-actin transfer. FEBS Lett. 592, 1804–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V & Kovar DR (2015) Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 32, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funk J, Merino F, Venkova L, Heydenreich L, Kierfeld J, Vargas P, Raunser S, Piel M & Bieling P (2019) Profilin and formin constitute a pacemaker system for robust actin filament growth. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrella EC, Machesky LM, Kaiser DA & Pollard TD (1996) Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 35, 16535–43. [DOI] [PubMed] [Google Scholar]
- 27.Perelroizen I, Marchand JB, Blanchoin L, Didry D & Carlier MF (1994) Interaction of profilin with G-actin and poly(L-proline). Biochemistry 33, 8472–8. [DOI] [PubMed] [Google Scholar]
- 28.Pimm ML, Hotaling J, Henty-Ridilla JL (2020) Profilin choreographs actin and microtubules in cells and cancer. Int. Rev. Cell. Mol. Biol 355, 155–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul AS & Pollard TD (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol 18, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtemanche N & Pollard TD (2012) Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem 287, 7812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Courtemanche N (2018) Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev 10, 1553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao L, Kerleau M, Suzuki EL, Wioland H, Jouet S, Guichard B, Lenz M Romet-Lemonne G & Jegou A (2018) Modulation of formin processivity by profilin and mechanical tension. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zweifel ME & Courtemanche N (2020) Competition for delivery of profilin-actin to barbed ends limits the rate of formin-mediated actin filament elongation. J Biol Chem 295, 4513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK & Almo SC (1998) Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry 37, 11171–81. [DOI] [PubMed] [Google Scholar]
- 35.Mahoney NM, Janmey PA & Almo SC (1997) Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol 4, 953–60. [DOI] [PubMed] [Google Scholar]
- 36.Chik JK, Lindberg U & Schutt CE (1996) The structure of an open state of beta-actin at 2.65 A resolution. J. Mol. Biol 263, 607–23. [DOI] [PubMed] [Google Scholar]
- 37.Vinson VK, De La Cruz EM, Higgs HN & Pollard TD (1998) Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 37, 10871–80. [DOI] [PubMed] [Google Scholar]
- 38.Sherer LA, Zweifel ME & Courtemanche N (2018) Dissection of two parallel pathways for formin-mediated actin filament elongation. J. Biol. Chem 293, 17917–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jégou A, Niedermayer T, Orbán J, Didry D, Lipowsky R, Carlier MF & Romet-Lemonne G (2011) Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS. Biol 9, e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courtemanche N & Pollard TD (2013) Interaction of profilin with the barbed end of actin filaments. Biochemistry 52, 6456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovar DR, Kuhn JR, Tichy AL & Pollard TD (2003) The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell. Biol 161, 875–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiwara I, Zweifel ME, Courtemanche N & Pollard TD (2018) Latrunculin A Accelerates Actin Filament Depolymerization in Addition to Sequestering Actin Monomers. Curr. Biol 28, 3183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard TD (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell. Biol 103, 2747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erickson HP (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause M, Dent EW, Bear JE, Loureiro JJ & Gertler FB (2003) Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell. Dev. Biol 19, 541–64. [DOI] [PubMed] [Google Scholar]
- 46.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM & Qualmann B (2007) Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 131, 337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan Q, Zelter A, MacCoss MJ, Davis TN & Nolen BJ (2018) Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc. Natl. Acad. Sci. USA 115, E1409–E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oda T, Iwasa M, Aihara T, Maéda Y & Narita A (2009) The nature of the globular- to fibrous-actin transition. Nature. 457, 441–5. [DOI] [PubMed] [Google Scholar]
- 49.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD & Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. (Walker JM, ed) Humana Press, Totowa, NY. [Google Scholar]
- 50.Spudich JA, Watt S (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem 246, 4866–71. [PubMed] [Google Scholar]
- 51.Kuhn JR & Pollard TD (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J 88, 1387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS & Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P & Kummer U (2006) COPASI--a COmplex PAthway SImulator. Bioinformatics 22, 3067–74. [DOI] [PubMed] [Google Scholar]
- 54.Hindmarsh AC (1983) ODEPACK, A Systematized Collection of ODE Solvers (Stepleman RS, ed) pp. 55–64, Scientific Computing, Elsevier, Amsterdam. [Google Scholar]
- 55.Petzold L (1983) Automatic selection of methods for solving stiff and nonstiff systems of ordinary differential equations. S.I.A.M. J. Sci. Stat. Comput 4, 136–48. [Google Scholar]
- 56.Ferron F, Rebowski G, Lee SH & Dominguez R (2007) Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 26, 4597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb B & Sali A (2016) Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trott O & Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem 31, 455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC & Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–12. [DOI] [PubMed] [Google Scholar]
- 60.Sanner MF, Olson AJ & Spehner JC(1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


