Abstract
Background
Lymphoedema is caused by dysfunction of the lymphatic system resulting in accumulation of high-protein content fluid in the interstitial space. To date, the bacteria associated with wound infections of patients with lower limb lymphoedema in Ethiopia have not been studied. This study identified pathogenic bacteria involved in wound infection and assessed antimicrobial susceptibility patterns in patients with lymphoedema in Ethiopia.
Methods
Swab samples were collected from the wounds of patients with lymphoedema and cultured using standard microbiological techniques. Micro-organisms were identified by colony morphology followed by identification and antimicrobial susceptibility testing using the automated VITEK 2 COMPACT Microbial Detection System.
Results
Swabs were collected from 103 patients and 84 were culture positive: 44 (52.4%) culture-positive samples showed polymicrobial growth and 40 (47.6%) grew single bacterial isolates. In total, 134 isolates were obtained, of which 26 gram-negative and 12 gram-positive bacterial species were identified. A total of 28/63 (44.4%) gram-negative isolates and 3/57 (5.3%) gram-positive isolates were multiple drug resistant. There was no resistance to ciprofloxacin, moxifloxacin or gentamycin among gram-negative or gram-positive bacteria.
Conclusion
In this study, many infections were polymicrobial and showed multiple drug resistance. Fluoroquinolones and gentamycin, however, seemed to be effective against bacterial wound infection in this setting.
Keywords: antimicrobial drug resistance, bacteria, Ethiopia, lymphoedema, wound infection
Introduction
Lymphoedema is caused by failure of lymphatic drainage leading to the accumulation of protein-rich fluid in the interstitial space. It is classified into primary and secondary lymphoedema. The causes of primary lymphoedema are poorly described, but may arise from genetic disorders, while secondary lymphoedema is attributed to damage to the lymphatic system, resulting from lymphatic vessel infestation, lymphadenectomy or radiotherapy in cancer patients.1 The two main causes of lymphoedema in the tropics are lymphatic filariasis followed bypodoconiosis.2 In Africa, lymphatic filariasis is caused by Wuchereria bancrofti species transmitted by blood-feeding mosquitoes.3 Podoconiosis is a form of lymphoedema arising among barefoot subsistence farmers who have contact with irritant red clay soil of volcanic origins over long periods of time.4
Lymphoedema has a marked physical and psychological impact in affected patients and significantly reduces their quality of life.2 Wound ulcer development is one of the most serious complications and often makes it impossible for patients to work.5 Patients with lymphoedema have a high risk of wound formation resulting from infection, including fungal infection in skin folds, moisture build-up and trauma.5
Skin lesions, including wounds, fissures, paronychia and eczema, allow the penetration of bacteria and fungus into the underlying tissues. Secondary infection along with inflammation also seems to play a major role in the skin changes seen in the limbs of individuals affected by lymphoedema and the development of elephantiasis.3
The lower limbs are more prone to infection than other parts of the skin because of exposure to contaminated environments, which predisposes the skin to bacterial colonisation. In patients with lymphoedema, the lymphatic system is partially or completely halted.6 As a result, the lymph transport is restricted and the patient predisposed to infection and chronic dermatolymphangioadenitis.7
Patients with secondary lymphoedema are predisposed to the development of cellulitis, which is likely caused by group A, C or G streptococci, or Staphylococcus aureus if associated with an abscess or boil.6 Cellulitis is an acute, diffuse, spreading, oedematous, suppurative infection of deeper subcutaneous tissues and is associated with abscess formation.6
The most frequent micro-organisms reported to cause wound infection are Sta. aureus, Streptococcus pyogenes, Enterococci, Escherichia coli, Klebsiella pneumonia, Proteus species and Pseudomonas aeruginosa.8 Continued use of systemic and topical antimicrobial agents has provided selective pressure that has led to the emergence of antibacterial-resistant strains that, in turn, has driven the continued search for new agents.9 Furthermore, lymphoedema and its associated wounds are a major concern in terms of increased disability, stigma and financial impact in regions where podoconiosis and lymphatic filariasis are prevalent.10
To date, the organisms found in the wounds of patients with lower limb lymphoedema in Ethiopia have not been identified. Therefore, this study aims to investigate the bacterial profile and assess antimicrobial susceptibility patterns among patients with lower limbs lymphoedema-associated wounds in East Wollega, Oromia Regional State, Ethiopia.
Methods
Study design and area
This cross-sectional study was conducted from 15 August to 5 September 2019 in the East Wollega zone, Oromia Regional State, Ethiopia. Patients who visited the Konchi, Sire, Boneyya and Bata Beseka clinics were screened for lymphoedema with associated wounds and enrolled in the study for swab collection. There is a high burden of podoconiosis and lymphatic filariasis with lymphoedema in these areas.11
Written informed consent was obtained from each participant. Ethical clearance was obtained from the Brighton and Sussex Medical School, Research Governance and Ethics Committee (ref. ER/BSMS9DY2/1), and Institutional Review Board (ref. 004/19/CDT), College of Health Sciences, Addis Ababa University.
Demographics and patient characteristics
Demographics and socioeconomic characteristics (such as age, gender, educational background, occupation, residence) and clinical characteristics (stage of lymphoedema, type of wound, treatment received) were collected using a structured questionnaire.
Swab collection and processing
Wounds were cleaned with sterile normal saline then wound swabs and discharge were obtained from all study participants aseptically using a sterile moistened cotton swab. Swabs were then immersed in a container of Amies transport medium with charcoal (Biomark Laboratories, Pune, India).
All samples were transported on ice to the Ethiopian Public Health Institute, National Referral Bacteriology and Mycology Laboratory (Ethiopian National Accreditation Office-accredited and ranked as five star by the American Society for Microbiology), where all laboratory tests were conducted. Swabs were used to inoculate MacConkey agar (Becton Dickinson and Company, Cockeysville, MD, USA), blood agar and mannitol salt agar (both from HiMedia Laboratories, Mumbai, India) and incubated aerobically at 37°C and 5% CO2 for 24 h. After 24 h, plates without growth were incubated further for up to 48 h.
Identification and antimicrobial susceptibility tests
Growth of micro-organisms was identified by examining colony morphology followed by biochemical identification using the automated VITEK 2 COMPACT Microbial Detection System (bioMerieux, Marcyl'Etoile, France). The antibiotic susceptibility tests for each bacterial species were completed using the VITEK 2 system susceptibility testing cards for gram-positive (antimicrobial susceptibility tests-gram-positive [AST-GP67] panel) and gram-negative (antimicrobial susceptibility tests-gram-negative [AST- GN71] panel) bacteria according to the manufacturer's instructions.
After the VITEK 2 COMPACT automated ID/AST instrument was validated, according to the manufacturer's instructions, using the standard strains, the 24-h bacterial cultures were tested. Aseptically, 3.0 ml of sterile saline (0.45–0.5% NaCl, pH 4.5–7.0) was transferred into a clear glass test tube (12 mm x 75 mm) and morphologically similar colonies were transferred to the saline using a sterile plastic loop. A homogenous suspension was prepared with a density equivalent to the appropriate McFarland standard (0.5 to 0.63) using the VITEK 2DensiCHEK Plus instrument. A second tube of 3.0 ml saline contained 145 μl of the suspension for AST-GN or 280 μl of the suspension for AST-GP susceptibility testing cards. The time between preparation of inoculum and filling of the card was always <30 min. The tubes were then placed in the cassette with a susceptibility card. A barcode reader was used to scan the order number of identification and susceptibility cards and information from the cassette worksheet was entered into the Maintain Virtual Cassette window on the workstation. The cassette was loaded into the filler station and transferred to the VITEK 2COMPACT cassette loading station within 10 min. Results were obtained after 8–10 h. For Shewanella algae, the Kirby Bauer disk diffusion method was used for the antimicrobial susceptibility tests.12
Statistical analysis
Data were entered and analysed using Statistical Package for Social Science version 20 (SPSS Inc., Chicago, IL, USA). Descriptive analyses such as frequencies and mean SD were used and the results are presented using tables and charts.
Results
Characteristics of study participants
A total of 103 participants with lymphoedema were screened and had swab samples collected. Of these, 33 were male (32.0%) and 70 were female (68.0%). Patients included in the study had a mean±SD age of 44.86±14.23 (range 19–75) y. The majority (74.8%) of study participants were farmers and 81.6% could neither read nor write (Table 1).
Table 1.
Demographics and socioeconomic and clinical characteristics of patients with lymphoedema included in the study
| Characteristics | Number (%) patients |
|---|---|
| Gender | |
| Male | 33 (32.0) |
| Female | 70 (68.0) |
| Age group (y) | |
| 19–25 | 7 (6.8) |
| 26–35 | 28 (27.2) |
| 36–45 | 20 (19.4) |
| 46–55 | 23 (22.3) |
| ≥55 | 25 (24.3) |
| Occupational status | |
| Farmers | 77 (74.8) |
| Daily labourers | 10 (9.7) |
| Government employee | 1 (1.0) |
| Housewives | 8 (7.8) |
| Merchants | 7 (6.8) |
| Stage of lymphoedemaa | |
| Mild | 5 (4.9) |
| Moderate | 34 (33.0) |
| Severe | 64 (62.1) |
| Time lived with lymphoedema (y) | |
| 1–10 | 46 (44.7) |
| 11–20 | 36 (35.0) |
| 21–30 | 10 (9.7) |
| 31–40 | 9 (8.7) |
| 41–50 | 2 (1.9) |
| Education level | |
| Could not read or write | 84 (81.6) |
| Could read and write | 2 (1.9) |
| Completed grades 2–5 | 8 (7.8) |
| Completed grades 6–9 | 6 (5.8) |
| Completed grades ≥10 | 3 (2.9) |
Lymphoedema was categorized into three stages (mild, moderate and severe) based on the International Society of Lymphology classification.9
According to the International Society of Lymphology classification,13 64 participants (62.1%) had advanced disease, 34 (33.0%) had moderate disease and 5 (4.9%) had mild disease (Table 1). In all participants, lymphoedema was confined to below the knee and no patient had hydrocele. All study participants had open, contaminated and chronic wounds. In total, 93 participants (90.3%) had bilateral lymphoedema and the others had just one affected leg. Patients had lived with lymphoedema for a mean±SD of 15.8±11.53 (range 1– 50) y.
The majority (83 [80.6%]) of patients had visited the clinic for lymphoedema treatment at least once previously while 20 (19.4%) were new cases. Among those who came to the clinic and took medications and herbal ointments, symptoms had partially resolved for 63 (75.9%) of the participants, relapsed for 16 (19.3%), not changed for 2 (2.4%) and worsened for 1 (1.2%).
Bacterial profile of study participants
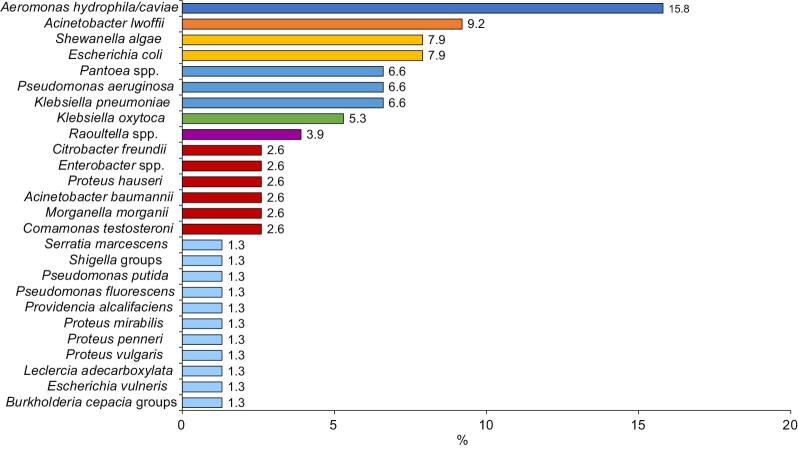
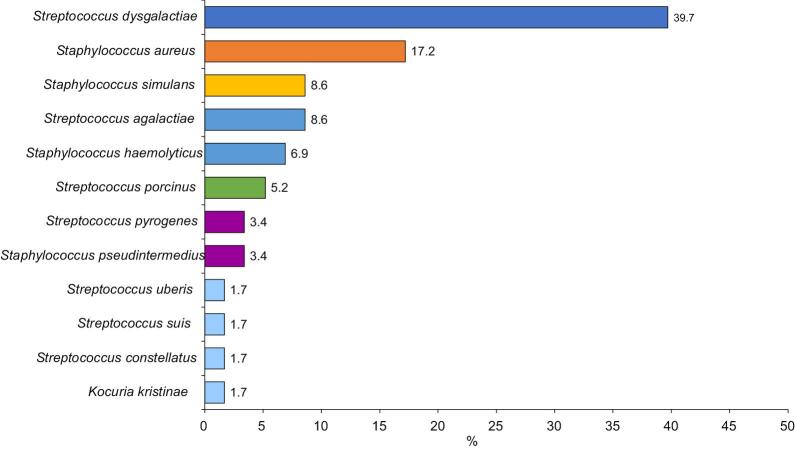
Most (84 [81.6%]) samples were culture positive, indicating wound infection; 44 (52.4%) samples showed polymicrobial growth while 40 (47.6%) grew single bacterial isolates. Among the 134 isolates, 26 gram-negative and 12 gram-positive bacterial species were isolated (Figures 1 and 2). Two species of bacterial infection were observed most frequently (37 [44.0%]), followed by three (4 [4.8%]) and four species (1 [1.2%]).
Figure 1.
Percentage of swab samples taken from the infected wounds of patients with lymphoedema that contained gram-negative bacteria
Figure 2.
Percentage swab samples taken from the infected wounds of patients with lymphoedema that contained gram-positive bacteria
Antimicrobial susceptibility pattern of isolated gram-negative bacteria
Gram-negative bacteria were tested against 18 selected antibacterial drugs and susceptibility varied with the type of organism and antibacterial drug employed. In a total of 76 gram-negative isolates, levels of resistance were highest against ampicillin (43 [56.6%]) followed by cefazolin (28 [36.8%]), ampicillin/sulbactam (21 [27.6%]), trimethoprim/sulfamethoxazole (10 [13.2%]) and tigecycline (7 [9.2%]). All isolates were susceptible to ertapenem and ciprofloxacin and showed low levels of resistance to amikacin (1 [1.3%]), gentamicin (1 [1.3%]), moxifloxacin (1 [1.3%]), cefepime (1 [1.3%]), tobramycin (2 [2.6%]), aztreonam (4 [5.3%]), ceftriaxone (6 [7.9%]), meropenem (7 [9.2%]) and imipenem (8 [10.5%]) (Table 2).
Table 2.
Antibiotic resistance profiles of gram-negative bacteria isolated from the infected wounds of patients with lymphoedema
| Penicillins | Cephalosporin | Carbapenem | Aminoglycosides | Fluoroquinolones | Glycylcycline | Sulfonamides | Penicillins | Cephalosporin | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microbial species isolated (n) | ESBL n (%) | Resistance pattern | A | AS | CFZ | CTR | CP | AZM | E | IPM | M | AK | G | TM | CX | MXF | TGC | TS | PB | CZ |
| Total gram-negative isolates (n=76) | 1 (1.3) | 43 (56.6) | 21 (27.6) | 28 (36.8) | 6 (7.9) | 2 (1.3) | 4 (5.3) | 0 (0) | 8 (10.5) | 7 (9.2) | 1 (1.3) | 1 (1.3) | 2 (2.6) | 0 (0) | 1 (1.3) | 7 (9.2) | 10 (13.2) | 0 (0) | 0 (0) | |
| Acinetobacter baumannii (n=2) | NA | S | 0 (0) | 1 (50) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | ND | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 2 (100) | 1 (50) | 2 (100) | 0 (0) | 0 (0) | 2 (100) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Acinetobacter lwoffii (n=7) | NA | S | 5 (71.4) | 6 (85.7) | 6 (85.7) | 6 (85.7) | 7 (100) | 7 (100) | ND | 7 (100) | 6 (85.7) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 6 (85.7) | 6 (85.7) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 2 (28.6) | 1 (14.3) | 1 (14.3) | 1 (14.3) | 0 (0) | 0 (0) | ND | 0 (0) | 1 (14.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (14.3) | ND | ND | ||
| Aeromonas hydrophila/caviae (n=12) | NA | S | 1 (8.3) | 2 (16.7) | 4 (33.3) | 12 (100) | 12 (100) | 12 (100) | ND | 10 (833) | 7 (58.3) | 12 (100) | 11 (91.7) | 11 (91.7) | 12 (100) | 12 (100) | 12 (100) | 11 (91.7) | ND | ND |
| I | 1 (8.3) | 1 (8.3) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | ND | 1 (8.3) | 0 (0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 10 (83.3) | 9 (75) | 7 (58.3) | 0 (0) | 0 (0) | 0 (0) | ND | 1 (8.3) | 5 (41.7) | 0 (0) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 1 (8.3) | ND | ND | ||
| Burkholderia cepacia groups (n=1) | NA | S | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 0 (0) | 1 (100) | 0(0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Citrobacter freundii (n=2) | NEG | S | ND | ND | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | ND | ND | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Enterobacter species (n=2) | NEG | S | ND | ND | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | ND | ND | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Morganella morganii (n=2) | NA | S | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Escherichia vulneris (n=1) | NEG | S | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Escherichia coli (n=6) | NEG | S | 2 (33.3) | 2 (33.3) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | 5 (83.3) | 5 (83.3) | 0 (0) | 4 (66.7) | ND | ND |
| I | 0 (0) | 2 (33.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (16.7) | 0 (0) | 6 (100) | 0 (0) | ND | ND | ||
| R | 4 (66.7) | 2 (33.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (16.7) | 0 (0) | 2 (33.3) | ND | ND | ||
| Klebsiella oxytoca (n=4) | NEG | S | 0 (0) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Klebsiella pneumoniae (n=5) | 1 (20%) | S | 0 (0) | 4 (80) | 4 (80) | 4 (80) | 4 (80) | 4 (80) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 4 (80) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 5 (100) | 1 (20) | 1 (20) | 1 (20) | 1 (20) | 1 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | ND | ND | ||
| Leclercia adecarboxylata (n=1) | NA | S | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Morganella morganii (n=2) | NA | S | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Pantoea species (n=5) | NA | S | ND | ND | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | ND | ND |
| I | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Proteus vulgaris (n=1) | NA | S | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Proteus hauseri (n=2) | NA | S | 0 (0) | 0 (0) | 2 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Proteus penneri (n=1) | NA | S | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | ND | ND | ||
| Proteus mirabilis (n=1) | NA | S | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | ND | ND | ||
| Providencia alcalifaciens (n=1) | NA | S | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | ND | ND | ||
| Pseudomonas fluorescens (n=1) | NA | S | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Pseudomonas putida (n=1) | NA | S | 0 (0) | 0 (0) | 0 (0) | 1 (100) | ND | ND | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 1 (100) | 1 (100) | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | ND | ND | ||
| Pseudomonas aeruginosa (n=5) | NA | S | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (100) | 5 (100) | 5 (100) | 4 (80) | 4 (80) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 2 (40) | 2 (40) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 2 (40) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 5 (100) | 5 (100) | 5 (100) | 3 (60) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (60) | 3 (60) | ND | ND | ||
| Raoultella species (n=3) | NA | S | 0 (0) | 1 (33.3) | 2 (66.7) | 2 (66.7) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | ND | ND |
| I | 0 (0) | 1 (33.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 3 (100) | 1 (33.4) | 1 (33.3) | 1 (33.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Shewanella algae (n=6) | NA | S | ND | ND | ND | ND | 6 (100) | ND | ND | 6 (100) | ND | 6 (100) | 6 (100) | 6 (100) | 6 (100) | ND | ND | ND | 6 (100) | 6 (100) |
| I | ND | ND | ND | ND | 0 (0) | ND | ND | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ND | 0 (0) | 0 (0) | ||
| R | ND | ND | ND | ND | 0 (0) | ND | ND | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ND | 0 (0) | 0 (0) | ||
| Shigella groups (n=1) | NA | S | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| Serratia marcescens (n=1) | NA | S | ND | ND | 0 (0) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ND | ND |
| I | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
| R | ND | ND | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ND | ND | ||
A: ampicillin; AK: amikacin; AS: ampicillin/sulbactam; AZM: aztreonam; CFZ: aefazolin; CP: cefepime; CT, cefotaxime; CTR: ceftrixaone; CX: ciprofloxacin; CZ: ceftazidime; E: ertapenem; ESBL: extended spectrum ß-lactamase; FA, fosfomycin; G: gentamicin; I: intermediate; IPM: imipenem; M: meropenem; MXF: moxifloxacin; ND: not determined; NEG: negative; PB: piperacillin/tazobactam; R: resistant; S: susceptible; TGC: tigecycline; TM: tobramycin; TS: trimethoprim/sulfamethoxazole.
Data show the type of antibiotic tested and the number (%) of bacterial isolates that are resistant, susceptible or intermediate to that antibiotic.
The multiple drug resistance (MDR) of 63 gram-negative samples to different antibacterial drugs is summarised in Table 3. Among the identified gram-negative isolates, 28 (44.4%) were multiple drug resistant, while a low level of extensive drug resistance (XDR) (1 [1.6%]) was observed (Table 3).
Table 3.
Patterns of multidrug resistance of gram-negative bacteria isolated from the infected wounds of patients with lymphoedema
| Antimicrobial patterns, n (%) | Multiple drug resistance, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial isolates | N (%) | R0 | R1 | R2 | R3 | R4 | ≥R5 | MDR | XDR | PDR | ESBL |
| Gram negatives | 63 (100) | 9 (14.3) | 19 (30.2) | 9 (14.3) | 7 (11.1) | 11 (17.5) | 8 (12.7) | 28 (44.4) | 1 (1.6) | 0 (0) | 1 (20) |
| Acinetobacter baumannii | 2 (3.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 1 (50) | 0 (0) | NA |
| Acinetobacter lwoffii | 7 (11.1) | 5 (71.4) | 1 (14.3) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 1 (14.3) | 0 (0) | 0 (0) | NA |
| Aeromonas hydrophila | 12 (19.0) | 1 (8.3) | 1 (8.3) | 4 (33.3) | 4 (33.3) | 1 (8.3) | 1 (8.3) | 8 (66.7) | 0 (0) | 0 (0) | NA |
| Burkholderiacepacia groups | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | NA |
| Citrobacter freundii | 2 (3.2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Enterobacter species | 2 (3.2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Comamonas testosteroni | 2 (3.2) | 1 (50) | 0 (0) | 1 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | N/A |
| Escherichia coli | 6 (9.5) | 2 (33.3) | 2 (33.3) | 1 (16.7) | 1 (16.7) | 0 (0) | 0 (0) | 2 (33.3) | 0 (0) | 0 (0) | 0 (0) |
| Klebsiella oxytoca | 4 (6.3) | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Klebsiella pneumoniae | 5 (7.9) | 0 (0) | 4 (80) | 0 (0) | 0 (0) | 1 (20) | 0 (0) | 1 (20) | 0 (0) | 0 (0) | 1 (20) |
| Morganella morganii | 2 (3.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | NA |
| Proteus vulgaris | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | NA |
| Proteus hauseri | 2 (3.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 0 (0) | NA |
| Proteus penneri | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | NA |
| Proteus mirabilis | 1 (1.6) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Providencia alcalifaciens | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | NA |
| Pseudomonas fluorescens | 1 (1.6) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Pseudomonas putida | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | NA |
| Pseudomonas aeruginosa | 5 (7.9) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 1 (20) | 3 (60) | 4 (80) | 0 (0) | 0 (0) | NA |
| Raoultella species | 3 (4.8) | 0 (0) | 1 (33.3) | 2 (66.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
| Shigella groups | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | NA |
| Serratia marcescens | 1 (1.6) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA |
ESBL: extended spectrum ß-lactamase; MDR: multidrug resistant—resistant to at least one agent in ≥3 antimicrobial classes; NA: not applicable; PDR: pan-drug resistant—resistant to all antimicrobial agents in all antimicrobial classes; R0: no antibiotic resistant; R1: resistant to one antimicrobial category; R2: resistant to two antimicrobial categories; R3: resistant to three antimicrobial categories; R4: resistant to four antimicrobial categories; ≥R5: resistant to five or more antimicrobial categories; XDR: extensive drug resistant—resistant to at least one agent in all but ≤2 antimicrobial categories
Antimicrobial susceptibility pattern of isolated gram-positive bacteria
Gram-positive isolates were tested against 20 selected antibacterial drugs and most of the isolates were susceptible. Across 58 gram-positive isolates, the level of resistance was highest to erythromycin (21 [36.2%]) and clindamycin (18 [31.0%]), followed by tetracycline (11 [19.0%]) and penicillin (10 [17.2%]) (Table 4). Low levels of resistance were observed against oxacillin (2 [3.4%]), trimethoprim/sulfamethoxazole (5 [8.6%]), tigecycline (2 [3.4%]) and ciprofloxacin (1 [1.7%]). All isolates of gram-positives were susceptible to gentamicin, levofloxacin, moxifloxacin, quinupristin/dalfopristin, linezolid and vancomycin.
Table 4.
Antibiotic resistance profiles of gram-positive bacteria isolated from the infected wounds of patients with lymphoedema
| Penicillin | Aminoglycoside | Quinolone | Macrolides | lincosamide | Streptogramins | Glycopeptide | Tetracyclines | Glycylcycline | Sulfonamides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microbial species isolated (n) | Cefoxitin screening (MRSA) | ICM | Resistance pattern | PEN | A | OX | GMHL | STRH | GM | CX | LVX | MXF | ERY | CM | QDA | LNZ | VA | TE | TGC | TS |
| Total gram positive isolates (n=58) | 2 (3.4) | 8 (13.8) | 10 (17.2) | 0 (0) | 2 (3.4) | ND | ND | 0 (0) | 1 (1.7) | 0 (0) | 0 (0) | 21 (36.2) | 18 (31.0) | 0 (0) | 0 (0) | 0 (0) | 11 (19.0) | 2 (3.4) | 5 (8.6) | |
| Kocuria kristinae (n=1) | 1 (100) | 1 (100) | S | 0 (0) | ND | 0 (0) | ND | ND | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| R | 1 (100) | ND | 1 (100) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Staphylococcus aureus (n=10) | 0 (0) | 3 (30) | S | 3 (30) | ND | 10 (100) | ND | ND | 10 (100) | 10 (100) | 10 (100) | 10 (100) | 6 (60) | 7 (70) | 10 (100) | 10 (100) | 10 (100) | 9 (90) | 10 (100) | 8 (80) |
| R | 7 (70) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (40) | 3 (30) | 0 (0) | 0 (0) | 0 (0) | 1 (10) | 0 (0) | 2 (20) | |||
| Staphylococcus haemolyticus (n=4) | 1 (25) | 1 (25) | S | 3 (75) | ND | 3 (75) | ND | ND | 4 (100) | 3 (75) | 3 (75) | 4 (100) | 2 (50) | 3 (75) | 4 (100) | 4 (100) | 4 (100) | 3 (75) | 4 (100) | 1 (25) |
| I | 0 (0) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| R | 1 (25) | ND | 1 (25) | ND | ND | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 2 (50) | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 3 (75) | |||
| Staphylococcus pseudintermedius (n=2) | 0 (0) | NG | S | 1 (50) | ND | 2 (100) | ND | ND | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| R | 1 (50) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Staphylococcus simulans (n=5) | 0 (0) | 2 (40) | S | 5 (100) | ND | 5 (100) | ND | ND | 5(100) | 5(100) | 5 (100) | 5 (100) | 4 (60) | 4 (60) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) |
| R | 0 (0) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (40) | 2 (40) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| Streptococcus agalactiae (n=5) | 0 (0) | ND | S | 5 (100) | 5 (100) | ND | ND | ND | ND | 5 (100) | 5 (100) | 5 (100) | 2 (40) | 2 (40) | 5 (100) | 5 (100) | 5 (100) | 3 (60) | 3 (60) | 5 (100) |
| R | 0 (0) | 0 (0) | ND | ND | ND | ND | 0 (0) | 0 (0) | 0 (0) | 3 (60) | 3 (60) | 0 (0) | 0 (0) | 0 (0) | 2 (40) | 2 (40) | 0 (0) | |||
| Streptococcus constellatus (n=1) | 0 (0) | ND | S | 1 (100) | 1 (0) | ND | ND | ND | ND | ND | 1 (100) | ND | 1 (100) | 1 (100) | ND | 1 (100) | 1 (100) | 1 (100) | ND | 1 (100) |
| R | 0 (0) | 2 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 0 (0) | 0 (0) | ND | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | |||
| Streptococcus dysgalactiae (n=23) | 0 (0) | ND | S | 23 (100) | 3 (0) | ND | ND | ND | 23 (100) | 23 (100) | 23 (100) | 23 (100) | 18 (78.3) | 19 (82.6) | ND | 23 (100) | 23 (100) | 17 (73.9) | ND | 23 (100) |
| R | 0 (0) | 4 (0) | ND | ND | ND | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (21.7) | 4 (17.4) | ND | 0 (0) | 0 (0) | 6 (26.1) | ND | 0 (0) | |||
| Streptococcus porcinus (n=3) | 0 (0) | ND | S | 3 (100) | 5 (0) | ND | ND | ND | ND | ND | 3 (100) | ND | 0 (0) | 0 (0) | ND | 3 (100) | 3 (100) | 3 (100) | ND | 3 (100) |
| R | 0 (0) | 6 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 3 (100) | 3 (100) | ND | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | |||
| Streptococcus pyrogenes (n=2) | 0 (0) | 1 (50) | S | 2 (100) | 7 (0) | ND | ND | ND | ND | ND | 2 (100) | ND | 2 (100) | ND | ND | 2 (100) | 2 (100) | 0 (0) | ND | 2 (100) |
| I | 0 (0) | 8 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 1 (50) | ND | 0 (0) | |||
| R | 0 (0) | 9 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 0 (0) | ND | ND | 0 (0) | 0 (0) | 1 (50) | ND | 0 (0) | |||
| Streptococcus suis (n=1) | 0 (0) | 0 (0) | S | 1 (100) | 10 (0) | ND | ND | ND | ND | ND | 1 (100) | ND | 0 (0) | 0 (0) | ND | 1 (100) | 1 (100) | 1 (100) | ND | 1 (100) |
| R | 0 (0) | 11 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 1 (100) | 1 (100) | ND | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | |||
| Streptococcus uberis (n=1) | 0 (0) | 0 (0) | S | 1 (100) | 12 (0) | ND | ND | ND | ND | ND | 1 (100) | ND | 0 (0) | 0 (0) | ND | 1 (100) | 1 (100) | 1 (100) | ND | 1 (100) |
| R | 0 (0) | 13 (0) | ND | ND | ND | ND | ND | 0 (0) | ND | 1 (100) | 1 (100) | ND | 0 (0) | 0 (0) | 0 (0) | ND | 0 (0) | |||
A: ampicillin; CM: clindamycin; CX: ciprofloxacin; ERY: erythromycin; FD: nitrofurantoin; GM: gentamicin; GMHL: gentamicin high level; ICM: inducible clindamycin; I: intermediate; LNZ: linezolid; LVX: levofloxacin; MXF: moxifloxacin; ND: not determined; NEG: negative; OX: oxacillin; PEN: penicillin; POS: positive QDA: quinupristin/dalfopristin; R: resistance; RA: rifampin; S: susceptible; STRHL: streptomycin high level; TE: tetracycline; TGC: tigecycline; TS: trimethoprim/sulfamethoxazole; VA: vancomycin.
Data show the type of antibiotic tested and the number (%) of bacterial isolates that are resistant, susceptible or intermediate to that antibiotic.
MDR patterns of 57 gram-positive bacterial isolates from wounds of lymphoedema patients are summarised in Table 5. Much lower levels of MDR (3 [5.3%]) were identified among the gram-positive compared with gram-negative isolates. Overall, two gram-positive isolates (3.5%) were cefoxitin-screening (methicillin-resistant Sta. aureus [MRSA]) positive and seven (12.3%) showed inducible clindamycin resistance.
Table 5.
Patterns of multidrug resistance of gram-positive bacteria isolated from the infected wounds of patients with lymphoedema
| Antimicrobial resistance patterns, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial isolates | N (%) | R0 | R1 | R2 | R3 | R4 | ≥R5 | MDR, n (%) | Cefoxitin screening (MRSA), n (%) | Inducible clindamycin, n (%) |
| Gram-positives | 57 (100) | 17 (29.8) | 31 (54.4) | 8 (14.0) | 2 (3.5) | 1 (1.8) | 0 (0) | 3 (5.3) | 2 (3.5) | 7 (12.3) |
| Kocuriakristinae | 1 (1.8) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| Staphylococcus aureus | 10 (17.5) | 2 (20) | 4 (40) | 3 (30) | 0 (0) | 1 (10) | 0 (0) | 1 (10) | 0 (0) | 3 (30) |
| Staphylococcus haemolyticus | 4 (7.0) | 0 (0) | 2 (50) | 1 (25) | 0 (0) | 1 (25) | 0 (0) | 1 (25) | 1 (25) | 1 (25) |
| Staphylococcus pseudintermedius | 2 (3.5) | 1 (50) | 1 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Staphylococcus simulans | 5 (8.8) | 3 (60) | 2 (40) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (40) |
| Streptococcus agalactiae | 5 (8.8) | 1 (20) | 2 (40) | 1 (20) | 1 (20) | 0 (0) | 0 (0) | 1 (20) | 0 (0) | 0 (0) |
| Streptococcus dysgalactiae | 23 (40.4) | 9 (39.1) | 11 (47.8) | 3 (13.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Streptococcus porcinus | 3 (5.3) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Streptococcus pyogenes | 2 (3.5) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (50) |
| Streptococcus suis | 1 (1.8) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Streptococcus uberis | 1 (1.8) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
ESBL: extended spectrum ß-lactamase; MDR: multidrug resistant—resistant to at least one agent in ≥3 antimicrobial classes; ND: not determined; R0: no antibiotic resistant; PDR: resistant to all antimicrobial agents in all antimicrobial classes; R1: resistant to one antimicrobial category; R2: resistant to two antimicrobial categories; R3: resistant to three antimicrobial categories; R4: resistant to four antimicrobial categories; ≥R5: resistant to five or more antimicrobial categories; XDR: extensive drug resistant—resistant to at least one agent in all but ≤2 antimicrobial categories.
Discussion
Bacterial contamination of the wounds of patients with lympho-edema is a serious problem in regions where tropical lymphoedema is common. Proper identification of pathogenic micro-organisms and knowledge of their susceptibility to commonly used antibacterial drugs will help clinicians in the management of wounds in these patients. Our study characterised gram-negative and gram-positive bacteria, including the proportion of samples with MDR and XDR to antibacterial agents, from the wounds of Ethiopian patients with lymphoedema. Most participants (74.8%) in the current study were farmers. Since most farmers in rural communities do not use footwear,14 there is a high probability of injury with subsequent infection of the wound with micro-organisms from the environment. Furthermore, delayed healthcare seeking creates opportunities for bacterial contamination and multiplication.14
The development of bacterial biofilm by normal flora on human skin is also an important factor for persistent wound infections. Staphylococcus aureus and Ps. aeruginosa are widely known to cause chronic biofilm-based wound infections, which maintain chronic infection and impair wound healing.15
The types of wound pathogens identified in our study, and their relative prevalence, were consistent with earlier studies.16 As reported previously,17 the most common gram-negative bacteria isolated from wound infections in our study was Aeromonas hydrophila/caviae (15.9%). Aeromonas hydrophila/caviae causes mild to severe wound infection and typically occurs on the extremities upon exposure of skin lesions to contaminated mud and river water.17
In agreement with previous studies,18 the next most common gram-negative species isolated from the wounds of patients with lymphoedema in our study was Acinetobacter lwoffii (9.2%), which is known to colonise wounds and cause infections, including cellulitis, followed by Es. coli (7.9%), Kl. pneumoniae (6.6%) and Ps. aeruginosa (6.6%). In another study from India, Es. coli was isolated from local lesions of patients with filarial lymphoedema along with other gram-negative and gram-positive bacteria, potentially due to contamination of the wound with faeces.19
Shewanella algae, another frequent gram-negative isolate in our study (7.9%), is common in skin and soft tissue infections, especially in immunocompromised patients with pre-existing cutaneous ulcers and is associated with exposure to aquatic environments, as shown by Goyal et al.,20 who reported wound infection by She. algae among people in Iran with a history of swimming.
Streptococcus dysgalactiae represented 39.7% of gram-positive isolates in the current study. Our data are supported by findings showing that Str. dysgalactiae is known to cause soft tissue infection and cellulitis.21
Similar to previous findings from Ethiopia,9 Sta. aureus made up 17.8% of all gram-positive isolates in our study. Acute inflammation of the skin and tissue (cellulitis) of the lower limbs of lymphoedema patients is commonly caused by Sta. aureus and Str. pyogenes.16
Staphylococcus simulans, which was isolated from 8.9% of wounds in the current study, is a coagulase-negative staphylococcus (CoNS) species. Infection in humans predominantly occurs among patients who have contact with animals.22 Another CoNS member of the staphylococcus genus, Staphylococcus haemolyticus, was isolated from 5.4% of patients, in line with Czekaj et al., who isolated Sta. haemolyticus from toe-web swabs and lymph nodes of lymphoedematous legs of European patients.23
Streptococcus pyogenes made up 3.6% of gram-positive bacterial isolates in the current study, supporting previously published data from Ethiopia.24 Streptococcus pyogenes is a member of the β-haemolytic group A streptococci, and its coinfection with other micro-organisms is the most common cause of infection in lymphoedematous limbs, causing erysipelas, particularly in the lower limbs.6
In the current study, bacterial isolates were tested for their susceptibility to the antibacterial drugs most commonly used for treatment in Ethiopia. Higher rates of MDR were noted among the gram-negative (44.4%) than the gram-positive (5.3%) bacteria.
Among the gram-negative bacteria, all isolates of Kl. pneumonia were resistant to ampicillin whereas 20% were resistant to ampicillin/sulbactam, cefazolin, ceftriaxone, cefepime, aztreonam and trimethoprim/sulfamethoxazole. A similar finding on Kl. pneumonia resistance to ampicillin has been reported previously in Ethiopia.9 However, all isolates of Kl. pneumonia were susceptible to carbapenems (ertapenem, imipenem and meropenem), aminoglycosides (amikacin, gentamicin and tobramycin), fluoroquinolones (ciprofloxacin and moxifloxacin) and tigecycline, in agreement with Lin et al.25
All the isolates of Ps. aeruginosa (n=5) were resistant to ampicillin, ampicillin/sulbactam and first-generation cephalosporins (cefazolin), whereas three of the isolates were not susceptible to a third-generation cephalosporin (ceftriaxone) or trimethoprim/sulfamethoxazole. Resistance to ampicillin and ceftriaxone was reported previously by Dessie et al.26 All isolates of Ps. aeruginosa in this study were susceptible to aminoglycosides (amikacin, gentamycin and tobramycin), fluoroquinolones (ciprofloxacin and moxifloxacin), carbapenems (aztreonam, ertapenem and imipenem) and fourth-generation cephalosporin (cefepime).
In a previous study in Ethiopia, Ae. hydrophila was resistant to ampicillin (100%), trimethoprim-sulfamethoxazole (100%) and ceftriaxone (75%).27 However, in the current study, none of the isolates of Ae. hydrophila showed resistance to the aminoglycosides (amikacin and gentamicin), fluoroquinolones (ciprofloxacin and moxifloxacin), carbapenems (aztreonam and ertapenem) or cephalosporins (ceftriaxone and cefepime).
A high proportion of Es. coli isolates showed resistance to ampicillin (66.7%), ampicillin/sulbactam (33.3%), moxifloxacin (16.7%) and trimethoprim-sulfamethoxazole (33.3%). However, all isolates were susceptible to cephalosporins (cefazolin, ceftriaxone and cefepime), carbapenems (aztreonam, ertapenem, imipenem and meropenem) and aminoglycosides (amikacin, gentamycin and tobramycin). These data suggest lower resistance than a study from Gondar, Ethiopia.24
More than 44% of gram-negative isolates were found to be multiple drug-resistant bacteria, of which 1.6% were bacteria with XDR. Acinetobacter baumannii (100%), Ac. lwoffii (14.3%), Ae. hydrophila (66.7%), Es. coli (33.3%), Ps. aeruginosa (80%) and Proteus species (100%) were among the multiple drug-resistant bacteria. One isolate of Ac. baumannii showed XDR. MDR of each of these organisms has been reported previously.9,22
In the current study, 20% of the isolates of Kl. pneumonia were extended spectrum ß-lactamase (ESBL)-producing isolates. ESBL enzymes confer resistance to most ß-lactam antibacterial drugs, including penicillin, cephalosporins and the monobactam, aztreonam. The emergence of ESBL-producing isolates has important clinical and therapeutic implications.28
In the current study, gram-positive isolates were most commonly resistant to erythromycin (36.2%) and clindamycin (31.0%) followed by tetracycline (19.0%) and penicillin G (17.2%). However, all gram-positive isolates were susceptible to glycopeptides (linezolid and vancomycin), fluoroquinolones (levofloxacin and moxifloxacin), gentamycin and quinupristin-dalfopristin. Among the gram-positive isolates, 10% of Sta. aureus and 25% of Sta. haemoliticus and Streptococcus agalactiae (group B streptococcus) isolates demonstrated MDR.
Staphylococcus aureus was resistant to penicillin (70%), erythromycin (40%), clindamycin (30%), trimethoprim-sulfamethoxazole (20%) and tetracycline (10%). All isolates of Sta. aureus were susceptible to oxacillin, gentamycin, ciprofloxacin, levofloxacin, moxifloxacin, linezolid, vancomycin, quinupristin-dalfopristin and rifampicin, in agreement with studies from other parts of Ethiopia9 and elsewhere.28
Similar to the findings of Czekaj et al.,23 70% of Sta. haemolyticus isolates were resistant to trimethoprim-sulfamethoxazole, 50% to erythromycin and 25% to penicillin, oxacillin, ciprofloxacin, clindamycin and tetracycline. However, all isolates were susceptible to glycopeptides (linezolid and vancomycin), gentamycin, quinolones (moxifloxacin and levofloxacin), quinupristin/dalfopristin and tigecycline.
In the current study, Sta. haemolyticus and Kocuria kristinae were positive for cefoxitin-screening MRSA. A similar study from Brazil reported that 91% of Sta. haemolyticus isolates were positive for MRSA screening.29
In line with a report from Pakistan,30 13.8% of gram-positive isolates were positive for inducible clindamycin resistance. Among these, 50% were Str. pyogenes, 40% Sta. simulans, 30% Sta. aureus and 25% Sta. haemolyticus. A positive test indicates the presence of macrolide-induced resistance to clindamycin, which induced production of methylase that alters the common ribosomal binding site for macrolides, clindamycin and quinupristin.
There were limitations to the current study. No attempt was made to determine the source of infection (environment or hospital acquired). The number of samples used was small and the study did not recruit patients from different parts of the country to compare the national distribution of wound infection in lymphoedematous limbs. In addition, bacteria that present as normal flora on the skin of patients (biofilm) were not identified. Anaerobic micro-organisms were not included since anaerobic transport systems and culture facilities were not available, and advanced molecular techniques were not used to characterise and identify the species.
Conclusions
This study identifies current bacterial species involved in colonising wounds of lymphoedematous limbs in patients from Ethiopia. More than 40% of wound infections were polymicrobial. The most predominant bacteria contributing to wound infections were Ae. hydrophila/caviae, Ac. lwoffii, Es. coli, Kl. pneumoniae, Ps. aeruginosa, She. algae, Sta. aureus, Str. pyogenes, Str. dysgalactiae, Sta. haemolyticus, Str. agalactiae and Sta. simulans. A higher rate of antimicrobial resistance was detected among the gram-negative than the gram-positive isolates and MDR was also identified. We observed a high percentage of isolate resistance to ampicillin, cefazoline, clindamycin, erythromycin and tetracycline, which are the most commonly used antibacterial drugs for the management of bacterial infections in the study area.
Microbiological tests of wound infection and bacterial antibiotic susceptibility testing are recommended to guide treatment and reduce the emergence of resistant bacteria. Given the antimicrobial resistance documented, we recommend use of the fluoroquinolones (ciprofloxacin or moxifloxacin) or gentamycin for the management of both gram-negative and gram-positive wound infections in patients with lower limb lymphoedema. These antibacterial drugs are available and affordable in these areas. In addition, carbapenems are recommended for the management of resistant gram-positive bacteria while glycopeptides (linezolid or vancomycin) or quinupristin/dalfopristin are recommended for resistant gram-negative bacteria.
Extensive drug-resistant Ac. baumannii requires special attention and we recommend further research to establish its wider prevalence and to monitor its resistance. Similar research should be replicated to establish the microbial profile and antimicrobial susceptibility patterns associated with wounds in other areas where tropical lymphoedema is prevalent. In addition, further study of the biofilm of lymphoedematous limbs in these environmental settings is recommended.
Acknowledgements
We would like to thank CDT-Africa, Addis Ababa University for the field support, the Ethiopian Public Health Institute for allowing us to use their laboratory and Konchi Catholic Church Clinic for supporting sample collection. Our special gratitude to Clare Callow, Grit Gansch and Manuela McDermid for supporting the procurement of laboratory supplies. Special thanks to Dr. Abrham Tesfaye, Dr. Yimitubezenash Wolde-Amanuel, Mr Tesfa Addis, Mr Zeleke Ayenew, Sister Cicily Joseph Cherriakara, Mr Abdi Samuel and Mr Estifanos Tsige for the support during study design, sample collection and laboratory work. Catriona McKay, PhD, provided help with editing the manuscript.
Contributor Information
Dereje Nigussie, Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University, PO Box 9086, Addis Ababa, Ethiopia; Centre for Global Health Research, Brighton and Sussex Medical School, University of Sussex, Brighton, BN1 9PX, UK.
Eyasu Makonnen, Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University, PO Box 9086, Addis Ababa, Ethiopia; Department of Pharmacology and Clinical Pharmacy, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia.
Belete Adefris Legesse, Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University, PO Box 9086, Addis Ababa, Ethiopia.
Abebaw Fekadu, Center for Innovative Drug Development and Therapeutic Trials for Africa (CDT-Africa), College of Health Sciences, Addis Ababa University, PO Box 9086, Addis Ababa, Ethiopia; Centre for Global Health Research, Brighton and Sussex Medical School, University of Sussex, Brighton, BN1 9PX, UK.
Gail Davey, Centre for Global Health Research, Brighton and Sussex Medical School, University of Sussex, Brighton, BN1 9PX, UK; School of Public Health, Addis Ababa University, Addis Ababa, Ethiopia.
Authors’ contributions
DN, GD, EM, BAL and AF were involved in conceptualisation and design of the study and in collection, analysis and interpretation of the data. DN wrote the first draft of the manuscript and DN, GD, EM, BAL and AF critically reviewed the manuscript for intellectual content. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Institute for Health Research (NIHR) Global Health Research Unit on NTDs at Brighton and Sussex Medical School using Official Development Assistance (ODA) funding. The views expressed here are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Competing interests
None declared.
Ethical approval
Ethical approval was obtained from the Brighton and Sussex Medical School, Research Governance and Ethics Committee (ER/BSMS9DY2/1) and Institutional Review Board (004/19/CDT), Addis Ababa University, College of Health Sciences, Addis Ababa, Ethiopia.
Data availability
None.
References
- 1. Geroulakos G, Peter R, Jane L. Lymphoedema. Surgery. 2007;26(1):2–5. [Google Scholar]
- 2. Ayman A, Grada TJP. Lymphedema pathophysiology and clinical manifestations. Am Acad Dermatol. 2017;77(6):12. [DOI] [PubMed] [Google Scholar]
- 3. Tekola-Ayele F, Yeshanehe WE. Podoconiosis: Tropical lymphedema of the lower legs. In: Dermatology and Allergology - Principles and Practice. 1st ed Hong Kong: iConcept Press Ltd, 2014:14. [Google Scholar]
- 4. Davey G. Podoconosis, non-filariasis elepheantiasis and lymphology. Lymphology. 2010;43:168–77. [PubMed] [Google Scholar]
- 5. Karnasula VM. Management of ulcers in lymphoedematous limbs. Indian J Plast Surg. 2012;45(2):261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper R, White R. Cutaneous infections in lymphoedema. J Lymphoedema. 2009;4(1):44–8. [Google Scholar]
- 7. Khanna AK, Tiwary SK. Ulcers of the Lower Extremity. London, UK: Springer India, 2016. [Google Scholar]
- 8. Mahat P, Manandhar S, Baidya R. Bacteriological profile of wound infection and antibiotic susceptibility pattern of the isolates. Microbiol Exp. 2017;4(5):1–8. [Google Scholar]
- 9. Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Negussie H, Molla M, Ngari M et al. Lymphoedema management to prevent acute dermatolymphangioadenitis in podoconiosis in northern Ethiopia (GoLBeT): a pragmatic randomised controlled trial. Lancet Glob Heal. 2018;6(7):e795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bekele K, Deribe K, Amberbir T et al. Burden assessment of podoconiosis in Wayu Tuka woreda , east Wollega zone, western Ethiopia: A community-based cross-sectional study. BMJ Open. 2016;6(9):2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CLSI Performance Standards for Antimicrobial Suceptiability testing. 2019 ed CLSI. supplimented M100 Wayen, PA: Clinical and Standards Institute, 2019. [Google Scholar]
- 13. Executive Committee The diagnosis and treatment of peripheral lymphedema: 2016 consensus document of the International Society of Lymphology. Lymphology. 2016;49(4):170–84. [PubMed] [Google Scholar]
- 14. Alemu G, Ayele FT, Daniel T et al. Burden of podoconiosis in poor rural communities in Gulliso woreda, West Ethiopia. PLoS Negl Trop Dis. 2011;5(6):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sisay M, Worku T, Edessa D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: A meta-analysis of laboratory-based cross-sectional studies. BMC Pharmacol Toxicol. 2019;20(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Niaimi F, Cox N. Cellulitis and lymphoedema: a vicious cycle. J Lymphoedema. 2009;4(2):38–42. [Google Scholar]
- 17. Aravena-Román M, Inglis TJJ, Henderson B et al. Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob Agents Chemother. 2012;56(2):1110–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barghouthi S, Hammad G, Kurdi M. Acinetobacter lwoffii induced cellulitis with allergy-like symptoms. Internet J Microbiol. 2012;10(1):13–5. [Google Scholar]
- 19. Pal B, Mohanty S, Plant R et al. Incidence of different bacterial pathogens associated with filaria patients from coastal areas of Odisha. J Pure Appl Microbiol. 2014;8(6):2–5. [Google Scholar]
- 20. Goyal R, Kaur N, Thakur R. Case report of human soft tissue infection by the emerging pathogen Shewanella algae. J Infect Dev Ctriese. 2011;5(4):310–2. [DOI] [PubMed] [Google Scholar]
- 21. Kittang BR, Langeland N, Skrede S et al. Two unusual cases of severe soft tissue infection caused by streptococcus dysgalactiae subsp. equisimilis. J Clin Microbiol. 2010;48(4):1484–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romero FT, García-Rodrigo CG, Tamariz VV et al. Acute infection by Staphylococcus simulans in the hand of a man focal photodamage on the occipital Scalp. JAMA Dermatol. 2016;152(9):2020. [DOI] [PubMed] [Google Scholar]
- 23. Czekaj T, Ciszewski M, Szewczyk EM. Staphylococcus haemolyticus - an emerging threat in the twilight of the antibiotics age. Microbiology. 2015;161:2061–8. [DOI] [PubMed] [Google Scholar]
- 24. Mohammed A, Seid ME, Gebrecherkos T et al. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017;2017:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin W, Wang J, Chang S et al. The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in Taiwan, a trend analysis. Sci Rep. 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dessie W, Mulugeta G, Fentaw S et al. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. Int J Microbiol. 2016;2016:2418902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arega B, Wolde-amanuel Y, Adane K et al. Rare bacterial isolates causing bloodstream infections in Ethiopian patients with cancer. Infect Agent Cancer. 2017;12(40):4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hope D, Ampaire L, Oyet C et al. Antimicrobial resistance in pathogenic aerobic bacteria causing surgical site infections in Mbarara regional referral hospital, Southwestern Uganda. Sci Rep. 2019;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Secchi C, Lúcia A, Antunes S et al. Identification and detection of methicillin resistance in non - epidermidis coagulase-negative Staphylococci. Brazilian J Infect Dis. 2008;12:316–20. [DOI] [PubMed] [Google Scholar]
- 30. Afridi FI, Zeb M, Hussain A et al. Inducible clindamycin resistance in Staphylococcus Species. J Coll Physicians Surg Pakistan. 2014;24(7):481–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
None.