Abstract
Background
Porcine circovirus type 2 (PCV2) is recognized as virulent porcine pathogen and has been linked to porcine circovirus diseases (PCVD). However, there remain many unknowns regarding the spread and epidemic growth of PCV2.
Methods
To assess the genetic diversity of PCV2 in the southern China, a total of 92 sequences of PCV2 strains from this region were retrieved from GenBank and were subjected to amino acid variation and phylogenetic analyses together with 28 representative sequences, based on the sequence of the ORF2 gene, from different swine‐producing countries.
Results
All 92 PCV2 strains shared between 93.7% and 100% sequence similarity and could be divided into four genotypes (PCV2a, PCV2b, PCV2d and PCV2h), of which PCV2d had surpassed PCV2b and became the most prevalent PCV2 genotype in this region. Alignment of the deduced amino acid sequences of the capsid protein revealed that the obtained PCV2 strains possess two major heterogenic regions/hypervariable regions (positions 52–68 and 185–191), which were within or close to the epitopic regions in the capsid (Cap) protein. Meanwhile, the 92 PCV2 sequences also show evidence of at least five unique recombination events.
Conclusion
The data in this study indicate that the PCV2 strains in the southern China are undergoing constant genetic variation and that the predominant strain and its antigenic epitopes in this area have been gradually changing in recent years.
Keywords: capsid protein, genetic variation, phylogenetic analysis, porcine circovirus type 2, recombination
PCV2 strains from the Beibu Gulf of China could be divided into four genotypes (PCV2a, PCV2b, PCV2d and PCV2h), of which PCV2d had surpassed PCV2b and became the most prevalent PCV2 genotype in this area. In addition, the Beibu Gulf PCV2 strains possess two major heterogenic regions/hypervariable regions (positions 52–68 and 185–191), which were within or close to the epitopic regions in the capsid (Cap) protein. Meanwhile, these PCV2 sequences also show evidence of at least five unique recombination events.
![]()
1. INTRODUCTION
Porcine circovirus type 2 (PCV2), a member of the genus Circovirus of the Circoviridae family, is the smallest virus known to infect mammals. Increasing evidences indicate that PCV2 is the primary causative agent of PCV2‐systemic disease (PCV2‐SD) and PCV2‐subclinical infection (PCV2‐SI), and is associated with other pathological conditions designated by porcine circovirus diseases (PCVD), which cause a relevant economic impact in the global swine industry (Segalés, 2015). As a non‐enveloped, single‐stranded DNA virus, the PCV2 virion is icosahedral and 17 nm in diameter (Lv, Guo, & Zhang, 2014). The PCV2 has an ambisense, closed, circular genome with a size of 1,766–1,768 and 1,777 nucleotides that is computationally predicted to possess 11 overlapping open reading frames (ORFs) (Nguyen et al., 2012). To date, six ORFs have been characterized in detail: ORF1 codes for two replication‐associated proteins (Rep and Rep'), ORF2 codes for the capsid protein (Cap)involved in the host immune response, ORF3 codes for the apoptotic protein, ORF4 codes for the anti‐apoptotic protein (Lv, Guo, Zhang, & Zhang, 2016), ORF5 codes for a novel potentially endoplasmic reticulum stress‐inductive protein (Lv, Guo, Xu, Wang, & Zhang, 2015) and ORF6 codes for a newly discovered protein that may be involved in caspases regulation and the expression of multiple cytokines in PCV2‐infected cells (Li, Wang, et al., 2018; Li, He, et al., 2018). Particularly, ORF2 is a common target gene used for epidemiological and phylogenetic analyses on PCV2 strains as the analysis has been shown to be representative of full genome analysis. Previous studies have substantiated that the Cap protein encoded by ORF2 possesses three specific antigenic sites (aa 69–83, aa 117–131 and aa 169–183) and three spatial overlapping antigenic epitopes (aa 47–63, aa 165–200 and aa 230–233) (Lv et al., 2014).
PCV2 can be divided into eight genotypes (PCV2a to PCV2h), which were compiled based on the updated phylogeny‐grounded genotype definition for PCV2 strains that have been described by Franzo and Segalés (2018). PCV2a and PCV2b are the most common strains. In approximately 2003, there was a global‐scale shift in PCV2 from genotype PCV2a to PCV2b, which is highly prevalent in many countries. PCV2c is a genotype that was first identified in Denmark (Dupont, Nielsen, Baekbo, & Larsen, 2008), while PCV2d and PCV2e are two later discovered genotypes in China and other countries (Guo, Lu, Wei, Huang, & Liu, 2010; Wang et al., 2009). PCV2f is a novel genotype that was first identified by Bao et al. (2018), while PCV2g and PCV2h are two more genotypes that are recently proposed by Franzo and Segalés (2018). Additionally, PCV2 might be divided into group 1 (PCV2b) and group 2 (PCV2a) with eight clusters (1A–1C and 2A–2E) in another PCV2 genotype definition (Olvera, Cortey, & Segalés, 2007). The link between PCV2 group and disease status has been investigated many times but the results are not clear‐cut. Of these results, it is generally accepted that PCV2a, PCV2b and PCV2d are able to experimentally reproduce PCV2‐SD under appropriate circumstances, such as co‐infection with other swine pathogens or immunostimulation by vaccines or adjuvants (Gillespie, Opriessnig, Meng, Pelzer, & Buechner‐Maxwell, 2009; Opriessnig et al., 2014; Segalés, 2015).
In China, PCV2 infection was first recognized in 1996, and was then subsequently identified in most pig farms in different regions (Bao et al., 2018). Currently, several studies has revealed that Chinese strains share a high‐nucleotide sequence identity and mainly belong to genotypes PCV2b andPCV2d, with PCV2d becoming more dominant (Liu et al., 2018). There are known recombinant events of PCV2 within the ORF2 gene that are considered to be type‐specific and closely related to the pathogenesis (Cai et al., 2012; Wang et al., 2009). However, there remain many unknowns regarding the spread and epidemic growth of PCV2. In the present study, we obtained the complete genomes of 92 PCV2 strains from the Guangxi Beibu Gulf economic zone in China from GenBank and subjected them to amino acid variation and phylogenetic analyses, along with 28 PCV2 strains from different geographic regions throughout the world, based on the sequence of the ORF2. This information may provide valuable insights in to the genetic variation and phylogenetic characteristics of PCV2 populations circulating in this area and shed new light on the choice of vaccines that are ultimately used.
2. MATERIALS AND METHODS
2.1. Collection of PCV2 genome sequence
A total of 92 PCV2 full‐genome sequences were collected from different regions of the Guangxi Beibu Gulf economic zone in China from 2006 to 2016. Meanwhile, an additional 28 PCV2 strains derived from other countries or regions were retrieved from GenBank. Specifically, a significant amount of PCV2 strains were isolated from sick pigs with various clinical syndromes, such as postweaning multisystemic wasting syndrome (PMWS), respiratory signs, wasting, death etc, although the small number of strains are unpublished or their clinical status of the host have not been specifed. The information on geographical origin, year of strain isolation and genotype/subgroup are summarized in Tables 1 and 2. The geographical distribution of the 92 samples is shown in Figure 1. To facilitate the analysis, all of these obtained sequences were linearized at the same point and aligned with the Clustal W component of the MegAlign program (DNASTAR, version 7.10) as described previously (Mu et al., 2012).
TABLE 1.
PCV2 strains isolated from the southern China used in this study
| Isolate's name | Year of isolation | Genome size (nt) | Genotype | Accession number | References |
|---|---|---|---|---|---|
| Huanan‐10 | 2006 | 1,767 | PCV2b | EF493839 | unpublished |
| Huanan‐9 | 2006 | 1,767 | PCV2b | EF493838 | unpublished |
| GX0601 | 2006 | 1,768 | PCV2a | EF524532 | Wang et al., 2009 |
| GX0602 | 2006 | 1,768 | PCV2a | EF524533 | Wang et al., 2009 |
| GXNN0603 | 2006 | 1,767 | PCV2b | MH465418 | Yao et al., 2019 |
| GXYL0601 | 2006 | 1,767 | PCV2b | MH465433 | Yao et al., 2019 |
| GXWM | 2007 | 1,767 | PCV2d | EF675241 | Huang et al., 2006 |
| GXLC | 2007 | 1,767 | PCV2b | EF675240 | Yin et al., 2007 |
| GXHP | 2007 | 1,767 | PCV2b | EF675239 | Huang et al., 2006 |
| GXHK | 2007 | 1,767 | PCV2b | EF675238 | Huang et al., 2006 |
| GXGW | 2007 | 1,767 | PCV2b | EF675237 | Huang et al., 2006 |
| GXNN0804 | 2008 | 1,767 | PCV2d | MH465458 | Yao et al., 2019 |
| GXNN0806 | 2008 | 1,767 | PCV2b | MH465420 | Yao et al., 2019 |
| GXNN0803 | 2008 | 1,767 | PCV2b | MH465419 | Yao et al., 2019 |
| GXBH0801 | 2008 | 1,767 | PCV2b | MH465398 | unpublished |
| GXCZ0805 | 2008 | 1,767 | PCV2b | MH465402 | Yao et al., 2019 |
| GXNN0904 | 2009 | 1,767 | PCV2d | MH465460 | Yao et al., 2019 |
| GXNN0901b | 2009 | 1,767 | PCV2d | MH465459 | Yao et al., 2019 |
| GXNN0902 | 2009 | 1,767 | PCV2b | MH465422 | Yao et al., 2019 |
| GXNN0901a | 2009 | 1,767 | PCV2b | MH465421 | Yao et al., 2019 |
| GXBH1008 | 2010 | 1,767 | PCV2b | MH465399 | Yao et al., 2019 |
| GXFC11 | 2011 | 1,767 | PCV2d | KJ680370 | unpublished |
| BL12 | 2012 | 1,767 | PCV2d | KJ680369 | unpublished |
| BLFC12 | 2012 | 1,767 | PCV2d | KJ680368 | unpublished |
| GXYQ12 | 2012 | 1,767 | PCV2d | KJ680367 | unpublished |
| GXNN1209b | 2012 | 1,767 | PCV2b | MH465424 | Yao et al., 2019 |
| GXNN1209a | 2012 | 1,767 | PCV2b | MH465423 | Yao et al., 2019 |
| GXWM121203 | 2012 | 1,767 | PCV2d | MH756618 | unpublished |
| GXSL121231 | 2012 | 1,767 | PCV2d | MH756617 | unpublished |
| GXYL1208 | 2012 | 1,767 | PCV2h | MH465473 | Yao et al., 2019 |
| GXNN1304a | 2013 | 1,767 | PCV2d | MH465461 | Yao et al., 2019 |
| GXNN1312 | 2013 | 1,767 | PCV2b | MH465426 | Yao et al., 2019 |
| GXNN1304b | 2013 | 1,767 | PCV2b | MH465425 | Yao et al., 2019 |
| GXNN130121 | 2013 | 1,767 | PCV2b | MH756615 | unpublished |
| GXLA130701 | 2013 | 1,767 | PCV2d | MH756613 | unpublished |
| GXBB130624 | 2013 | 1,767 | PCV2d | MH756608 | unpublished |
| GXBL130228 | 2013 | 1,767 | PCV2d | MH756610 | unpublished |
| GXBL130313 | 2013 | 1,767 | PCV2d | MH756611 | unpublished |
| GXYL1307c | 2013 | 1,767 | PCV2d | MH465475 | Yao et al., 2019 |
| GXYL1307a | 2013 | 1,767 | PCV2d | MH465474 | Yao et al., 2019 |
| GXYL1310 | 2013 | 1,767 | PCV2b | MH465437 | Yao et al., 2019 |
| GXYL1307d | 2013 | 1,767 | PCV2b | MH465436 | Yao et al., 2019 |
| GXYL1305 | 2013 | 1,767 | PCV2b | MH465435 | Yao et al., 2019 |
| GXYL1304 | 2013 | 1,767 | PCV2b | MH465434 | Yao et al., 2019 |
| GXYL1307b | 2013 | 1,768 | PCV2a | MH465490 | Yao et al., 2019 |
| BH5 | 2014 | 1,767 | PCV2d | KM245558 | Zhang et al., 2017 |
| NN2 | 2014 | 1,767 | PCV2b | KJ956692 | Zhang et al., 2017 |
| HX02 | 2014 | 1,767 | PCV2b | KJ956691 | Zhang et al., 2017 |
| HX1 | 2014 | 1,767 | PCV2b | KJ956690 | Zhang et al., 2017 |
| BH6 | 2014 | 1,767 | PCV2b | KJ956689 | Zhang et al., 2017 |
| GXNN1410c | 2014 | 1,767 | PCV2d | MH465464 | Yao et al., 2019 |
| GXNN1410a | 2014 | 1,767 | PCV2d | MH465463 | Yao et al., 2019 |
| GXNN1409a | 2014 | 1,767 | PCV2d | MH465462 | Yao et al., 2019 |
| GXNN1406 | 2014 | 1,767 | PCV2b | MH465427 | Yao et al., 2019 |
| BH7 | 2014 | 1,767 | PCV2d | KY305198 | unpublished |
| GXNN141225 | 2014 | 1,767 | PCV2d | MH756616 | unpublished |
| GXLA140815 | 2014 | 1,767 | PCV2d | MH756614 | unpublished |
| GXYL1410 | 2014 | 1,767 | PCV2d | MH465480 | Yao et al., 2019 |
| GXYL1405 | 2014 | 1,767 | PCV2d | MH465479 | Yao et al., 2019 |
| GXYL1403b | 2014 | 1,767 | PCV2d | MH465478 | Yao et al., 2019 |
| GXYL1403a | 2014 | 1,767 | PCV2d | MH465477 | Yao et al., 2019 |
| GX140420 | 2014 | 1,767 | PCV2d | MH756607 | unpublished |
| GXYL1401 | 2014 | 1,767 | PCV2d | MH465476 | Yao et al., 2019 |
| GXYL1409 | 2014 | 1,767 | PCV2b | MH465438 | Yao et al., 2019 |
| GXCZ1410 | 2014 | 1,767 | PCV2b | MH465403 | Yao et al., 2019 |
| GXNN1410b | 2014 | 1,768 | PCV2a | MH465488 | Yao et al., 2019 |
| GXNN1409b | 2014 | 1,768 | PCV2a | MH465487 | Yao et al., 2019 |
| GXYL1408 | 2014 | 1,767 | PCV2a | MH465491 | Yao et al., 2019 |
| GXNN1504 | 2015 | 1,767 | PCV2d | MH465467 | Yao et al., 2019 |
| GXNN1503 | 2015 | 1,767 | PCV2d | MH465466 | Yao et al., 2019 |
| GXNN1501 | 2015 | 1,767 | PCV2d | MH465465 | Yao et al., 2019 |
| GXNN1511 | 2015 | 1,767 | PCV2b | MH465428 | Yao et al., 2019 |
| GXNN5 | 2015 | 1,767 | PCV2b | KY305202 | unpublished |
| GXNN3 | 2015 | 1,767 | PCV2b | KY305201 | unpublished |
| GXQZ2 | 2015 | 1,767 | PCV2d | KY305200 | unpublished |
| GXQZ1 | 2015 | 1,767 | PCV2d | KY305199 | unpublished |
| GXYL1512 | 2015 | 1,767 | PCV2d | MH465481 | Yao et al., 2019 |
| GXBB1501211 | 2015 | 1,767 | PCV2d | MH756609 | unpublished |
| GXFC1501 | 2015 | 1,767 | PCV2d | MH465443 | Yao et al., 2019 |
| GXCZ1510b | 2015 | 1,767 | PCV2d | MH465442 | Yao et al., 2019 |
| GXNN1612b | 2016 | 1,767 | PCV2d | MH465471 | Yao et al., 2019 |
| GXNN1612a | 2016 | 1,767 | PCV2d | MH465470 | Yao et al., 2019 |
| GXNN1603a | 2016 | 1,767 | PCV2d | MH465469 | Yao et al., 2019 |
| GXNN1602 | 2016 | 1,767 | PCV2d | MH465468 | Yao et al., 2019 |
| GXNN1612c | 2016 | 1,767 | PCV2b | MH465431 | Yao et al., 2019 |
| GXNN1604b | 2016 | 1,767 | PCV2b | MH465430 | Yao et al., 2019 |
| GXNN1603b | 2016 | 1,767 | PCV2b | MH465429 | Yao et al., 2019 |
| GXNN2 | 2016 | 1,767 | PCV2d | KY305204 | unpublished |
| GXNN1 | 2016 | 1,767 | PCV2d | KY305203 | unpublished |
| GXYL1607 | 2016 | 1,767 | PCV2d | MH465482 | Yao et al., 2019 |
| GXQZ1601 | 2016 | 1,767 | PCV2d | MH465472 | Yao et al., 2019 |
| GXNN1604a | 2016 | 1,768 | PCV2a | MH465489 | Yao et al., 2019 |
TABLE 2.
Representative PCV2 strains used in the genetic variation and phylogenetic analyses
| Isolate's name | Year of isolation | Geographic origin | Genome size (nt) | Group/Cluster | Accession number | References |
|---|---|---|---|---|---|---|
| NAVET_vietnam3 | 2004 | Vietnam | 1,767 | PCV2h | JX506730 | Franzo & Segalés, 2018 |
| FJ | 2004 | Fujian‐China | 1,768 | PCV2a | AY556474 | Nguyen et al., 2012 |
| BJW | 2004 | Beijing‐China | 1,767 | PCV2b | AY847748 | Olvera et al., 2007 |
| SCNB/CHA/05 | 2005 | Sichuan‐China | 1,767 | PCV2g | FJ998185 | Franzo & Segalés, 2018 |
| USA/MN‐088/2006 | 2006 | USA | 717 ORF2 gene | PCV2e | KT867799 | Franzo & Segalés, 2018 |
| JX0602 | 2006 | Jiangxi‐China | 1,768 | PCV2a | EF524541 | Wang et al., 2009 |
| DK1980PMWSfree | 2007 | Denmark | 1,767 | PCV2c | EU148503 | Dupont et al., 2008 |
| DK1987PMWSfree | 2007 | Denmark | 1,767 | PCV2c | EU148504 | Dupont et al., 2008 |
| MDJ | 2007 | Heilongjiang‐China | 1,766 | PCV2d | HM038031 | Guo et al., 2010 |
| LG | 2008 | China | 1,768 | PCV2a | HM038034 | Guo et al., 2010 |
| C6 | 2008 | South Korea | 702 ORF2 gene | PCV2b | EU450638 | Franzo & Segalés, 2018 |
| 762,454 | 2008 | England | 702 ORF2 gene | PCV2b | KY806003 | Grierson, Werling, Bidewell, & Williamson, 2018 |
| P2425NT | 2008 | Vietnam | 1,767 | PCV2g | JX099786 | Huynh et al., 2014 |
| 549‐QNa | 2009 | Vietnam | 1,767 | PCV2h | KM042398 | Franzo & Segalés, 2018 |
| TY1 | 2010 | Taiwan‐China | 1,768 | PCV2a | HQ202949 | Franzo & Segalés, 2018 |
| PM163 | 2010 | Brazil | 1,767 | PCV2c | KJ094599 | Franzo et al., 2015 |
| ZrBd_wb UKR | 2010 | Ukraine | 1,767 | PCV2g | KP420197 | Franzo & Segalés, 2018 |
| 201207JS | 2011 | Jiangsu‐China | 1,767 | PCV2d | KX960929 | Franzo & Segalés, 2018 |
| BG0‐1 | 2011 | Vietnam | 1,767 | PCV2h | JQ181592 | Huynh et al., 2014 |
| 3196‐LE | 2012 | Slovakia | 702 ORF2 gene | PCV2b | KP768478 | Sliz, Vlasakova, Jackova, & Vilcek, 2015 |
| PCV‐Y22 | 2012 | China | 1,767 | PCV2d | KC515014 | Franzo & Segalés, 2018 |
| MZ‐9 | 2012 | India | 1,767 | PCV2f | LC008135 | Franzo & Segalés, 2018 |
| CBI090 | 2013 | Thailand | 705 ORF2 gene | PCV2d | MF314285 | Thangthamniyom et al., 2017 |
| MZ‐5 | 2013 | India | 1,767 | PCV2f | LC004750 | Franzo & Segalés, 2018 |
| AS‐2 | 2013 | India | 1,767 | PCV2f | LC008137 | Franzo & Segalés, 2018 |
| USA/NE‐002/2015 | 2015 | USA | 1,777 | PCV2e | KT870147 | Franzo & Segalés, 2018 |
| USA/43520/2015 | 2015 | USA | 1,777 | PCV2e | KT795289 | Harmon et al., 2015 |
| KU‐1609 | 2016 | South Korea | 1,768 | PCV2a | KX828215 | Franzo & Segalés, 2018 |
FIGURE 1.
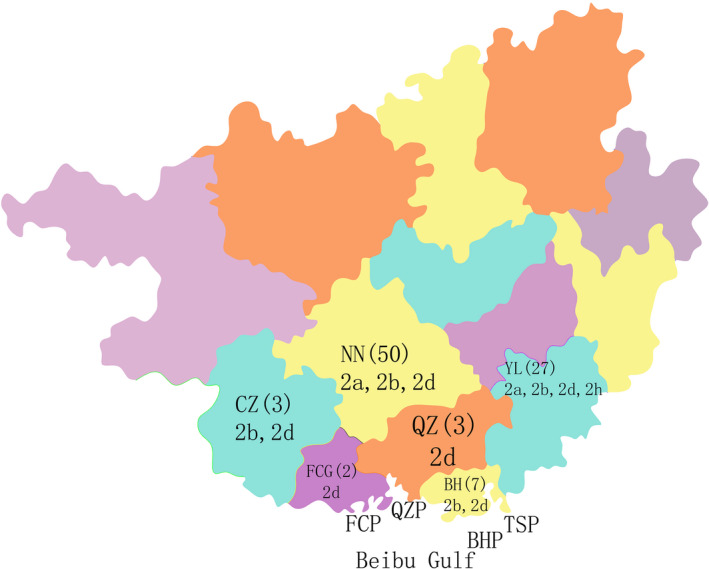
Geographical distribution of archived PCV2 strains. The abbreviations represent names of regions, the numbers represent the number of PCV2 strains and the genotypes detected in each region are also indicated under the numbers
2.2. Phylogenetic analyses of PCV2sequences
To provide better insight into the extent of genetic heterogeneity among PCV2 strains in southern China, a phylogenetic tree of the ORF2 gene was constructed with MEGA v.10.0.5 software using the neighbour‐joining (NJ) method (Kumar, Stecher, & Tamura, 2016). A bootstrap value was calculated using 1,000 replicates. The newly proposed method for genotyping by Franzo and Segalés (2018) was applied.
2.3. Analysis of antigenic structure of PCV2 ORF2 gene
It is widely accepted that the variation in the antigenic structure of the deduced capsid protein encoded by the ORF2 gene is important evidence for viral adaptability. Therefore, we performed an epitope cluster analysis via the newly developed cluster‐breaking algorithm with an identity threshold of 80% on ORF2 genes used in this study (http://tools.iedb.org/cluster2/). In addition, predictions of B‐cell epitopes, secondary structures and surface locations were also performed on this target gene by using the BepiPred method (Minin, Bloomquist, & Suchard, 2008). These synthetic datasets displayed the variation in this gene sequence in different PCV2 strains.
2.4. Nucleotide and amino acid substitutions analysis of PCV2 ORF2 sequences
Substitution rates of the nucleotide sequences and the deduced amino acid sequences of ORF2 gene products were analysed by the Tamura–Nei method using MEGA v.10.0.5. Tajima's neutrality test was performed to calculate the Θ, π and D values, which are the common indicators within the neutrality test that are used for evaluating the selection pressure on the group being tested, following the procedures described elsewhere (Nielsen, 2001). In addition, we gathered the frequencies of the amino acids used by PCV2 strains and the average frequencies and their standard deviations (SD) of each amino acid among different PCV2 genome sequences.
2.5. Recombination analysis of PCV2 sequences
To investigate the recombination rates, putative breakpoints and potential parental sequences of the PCV2 genomes, Recombination Detection Program (RDP v.4.97) was utilized according to the recommendations of previous studies (Mu et al., 2012). To further comprehensively confirm the identified recombinant events, the seven recombination detection methods, namely RDP, MaxChi, GeneConv, BootScan, SiScan, 3Seq and Chimaera, which are abbreviated to R, M, G, B, S, T and C, respectively, that had been implemented in the RDP4 software were employed again to ensure an acceptably low rate of false positives. The correlation parameters were identical to those reported by Li, He, et al. (2018)
3. RESULTS
3.1. Nucleotide sequences analysis
The sequence analysis showed that the complete genomes of these 92 PCV2 Chinese strains were 1,767 or 1,768 bp in length, while the ORF2 nucleotide sequences were 702 or 705 bp. The pairwise comparison analysis revealed that the nucleotide homologies of the complete genomes among these 92 PCV2 Chinese strains ranged from 93.7% and 100% (data not shown), while the pairwise similarities within 92 Chinese strains and 8 representative strains ranged between 91.1% and 100% (Table 3).
TABLE 3.
Homology comparison of the ORF2 nucleotide sequence of PCV2 strains
| Isolates | Strains for nucleotide sequence comparison | |||||||
|---|---|---|---|---|---|---|---|---|
| PCV2a | PCV2b | PCV2c | PCV2d | PCV2e | PCV2f | PCV2g | PCV2h | |
| AY556474 | AY847748 | EU148503 | KX960929 | KT870147 | LC008135 | FJ998185 | KM042398 | |
| EF493839 | 95.2 | 98.2 | 94.7 | 95.6 | 90.2 | 95.6 | 95.2 | 96.2 |
| EF493838 | 95.3 | 98.1 | 94.8 | 95.9 | 90.2 | 95.7 | 94.8 | 96.4 |
| EF524532 | 99.1 | 95.3 | 93.9 | 94.5 | 89.8 | 95.7 | 92.5 | 95.6 |
| EF524533 | 99.0 | 95.2 | 93.9 | 94.4 | 89.9 | 95.6 | 93.1 | 95.5 |
| MH465418 | 95.1 | 99.1 | 94.5 | 96.2 | 91.0 | 95.8 | 95.3 | 96.1 |
| MH465433 | 95.2 | 98.8 | 94.7 | 96.2 | 91.0 | 95.8 | 95.5 | 96.2 |
| EF675241 | 94.8 | 95.9 | 94.3 | 98.7 | 91.8 | 95.3 | 97.3 | 96.1 |
| EF675240 | 95.2 | 98.8 | 94.7 | 96.1 | 91.0 | 95.8 | 95.5 | 96.1 |
| EF675239 | 95.5 | 98.5 | 95.1 | 96.0 | 90.5 | 96.0 | 95.1 | 96.6 |
| EF675238 | 94.9 | 98.2 | 95.3 | 95.7 | 91.0 | 95.5 | 97.3 | 96.2 |
| EF675237 | 95.4 | 98.4 | 95.1 | 96.0 | 90.5 | 95.9 | 96.0 | 96.5 |
| MH465458 | 94.7 | 95.7 | 94.3 | 98.7 | 91.6 | 95.2 | 97.1 | 96.1 |
| MH465420 | 95.2 | 97.8 | 94.5 | 96.9 | 90.7 | 95.5 | 95.7 | 96.3 |
| MH465419 | 95.0 | 98.0 | 94.5 | 95.7 | 90.3 | 95.4 | 94.4 | 96.1 |
| MH465398 | 95.4 | 98.7 | 94.7 | 96.3 | 91.0 | 95.9 | 95.5 | 96.3 |
| MH465402 | 95.2 | 98.7 | 94.8 | 96.1 | 91.0 | 95.9 | 94.9 | 96.1 |
| MH465460 | 94.4 | 95.7 | 93.9 | 99.2 | 91.0 | 95.1 | 95.7 | 96.5 |
| MH465459 | 94.7 | 96.0 | 94.2 | 99.5 | 91.4 | 95.3 | 96.7 | 96.8 |
| MH465422 | 95.2 | 98.1 | 94.5 | 96.9 | 90.8 | 95.5 | 95.8 | 96.4 |
| MH465421 | 95.2 | 98.1 | 94.5 | 96.9 | 90.8 | 95.6 | 95.9 | 96.4 |
| MH465399 | 95.3 | 98.9 | 94.7 | 96.1 | 90.9 | 95.8 | 95.6 | 96.1 |
| KJ680370 | 94.6 | 95.7 | 94.1 | 99.4 | 91.4 | 95.2 | 96.6 | 96.6 |
| KJ680369 | 94.7 | 96.0 | 94.3 | 99.6 | 91.5 | 95.4 | 96.8 | 96.8 |
| KJ680368 | 94.7 | 96.0 | 94.3 | 99.6 | 91.5 | 95.4 | 96.8 | 96.8 |
| KJ680367 | 94.7 | 95.9 | 94.2 | 99.5 | 91.4 | 95.3 | 96.8 | 96.8 |
| MH465424 | 95.5 | 99.0 | 94.9 | 96.4 | 91.0 | 96.1 | 95.8 | 96.4 |
| MH465423 | 95.3 | 98.8 | 94.7 | 96.2 | 90.9 | 95.9 | 94.8 | 96.2 |
| MH756618 | 94.6 | 96.0 | 94.2 | 99.4 | 91.4 | 95.3 | 96.7 | 96.6 |
| MH756617 | 94.6 | 96.4 | 94.2 | 98.8 | 91.4 | 95.3 | 95.5 | 96.5 |
| MH465473 | 95.4 | 96.8 | 94.4 | 97.0 | 91.3 | 95.9 | 95.9 | 98.2 |
| MH465461 | 94.7 | 95.9 | 94.3 | 99.5 | 91.5 | 95.3 | 96.6 | 96.8 |
| MH465426 | 95.5 | 98.9 | 94.8 | 96.5 | 91.1 | 96.0 | 95.5 | 96.5 |
| MH465425 | 95.5 | 98.8 | 94.8 | 96.5 | 91.1 | 96.0 | 95.6 | 96.5 |
| MH756615 | 95.4 | 98.9 | 94.7 | 96.5 | 91.0 | 96.0 | 95.5 | 96.5 |
| MH756613 | 94.7 | 96.1 | 94.2 | 99.7 | 91.5 | 95.5 | 96.7 | 96.9 |
| MH756608 | 94.7 | 96.0 | 94.3 | 99.5 | 91.4 | 95.4 | 96.7 | 96.8 |
| MH756610 | 94.7 | 96.0 | 94.3 | 99.6 | 91.5 | 95.4 | 96.8 | 96.8 |
| MH756611 | 94.6 | 95.9 | 94.2 | 99.4 | 91.4 | 95.3 | 96.7 | 96.7 |
| MH465475 | 94.7 | 96.0 | 94.3 | 99.5 | 91.3 | 95.3 | 96.7 | 96.8 |
| MH465474 | 94.7 | 96.0 | 94.3 | 99.6 | 91.5 | 95.4 | 96.8 | 96.8 |
| MH465437 | 95.6 | 98.9 | 94.9 | 96.6 | 91.1 | 96.1 | 95.7 | 96.6 |
| MH465436 | 95.3 | 98.6 | 94.8 | 96.4 | 90.9 | 95.9 | 95.3 | 96.4 |
| MH465435 | 95.3 | 98.7 | 94.5 | 96.1 | 90.8 | 95.7 | 94.6 | 96.1 |
| MH465434 | 95.2 | 98.5 | 94.7 | 96.3 | 90.8 | 95.7 | 94.8 | 96.2 |
| MH465490 | 99.0 | 95.1 | 93.9 | 94.6 | 89.9 | 95.6 | 96.4 | 95.6 |
| KM245558 | 94.7 | 96.0 | 94.3 | 99.4 | 91.5 | 95.5 | 96.5 | 96.8 |
| KJ956692 | 95.6 | 99.0 | 94.9 | 96.7 | 91.2 | 96.2 | 96.1 | 96.6 |
| KJ956691 | 95.6 | 99.0 | 94.9 | 96.7 | 91.2 | 96.2 | 96.1 | 96.6 |
| KJ956690 | 95.6 | 99.0 | 94.9 | 96.7 | 91.2 | 96.2 | 96.1 | 96.6 |
| KJ956689 | 95.6 | 98.9 | 95.1 | 96.6 | 91.1 | 96.1 | 96.2 | 96.7 |
| MH465464 | 94.7 | 96.0 | 94.4 | 99.4 | 91.4 | 95.3 | 96.5 | 96.8 |
| MH465463 | 94.6 | 95.9 | 94.2 | 99.5 | 91.3 | 95.3 | 96.6 | 96.7 |
| MH465462 | 94.6 | 95.9 | 94.2 | 99.4 | 91.4 | 95.3 | 96.6 | 96.7 |
| MH465427 | 95.6 | 99.1 | 94.8 | 96.4 | 91.1 | 96.0 | 95.7 | 96.4 |
| KY305198 | 94.6 | 96.6 | 94.3 | 99.3 | 91.7 | 95.5 | 96.6 | 96.5 |
| MH756616 | 94.8 | 96.7 | 94.4 | 98.9 | 91.5 | 95.5 | 95.6 | 96.7 |
| MH756614 | 94.8 | 96.3 | 94.4 | 99.4 | 91.2 | 95.4 | 95.9 | 96.9 |
| MH465480 | 94.8 | 96.1 | 94.3 | 99.3 | 91.5 | 95.6 | 96.6 | 96.8 |
| MH465479 | 94.6 | 95.9 | 94.2 | 99.4 | 91.3 | 95.3 | 96.6 | 96.6 |
| MH465478 | 94.8 | 96.1 | 94.4 | 99.5 | 91.5 | 95.5 | 96.6 | 96.9 |
| MH465477 | 94.7 | 96.1 | 94.3 | 99.5 | 91.5 | 95.4 | 96.7 | 96.8 |
| MH756607 | 94.4 | 95.6 | 94.2 | 99.2 | 91.2 | 95.1 | 96.7 | 96.4 |
| MH465476 | 94.8 | 96.8 | 94.4 | 99.0 | 91.6 | 95.6 | 95.7 | 96.7 |
| MH465438 | 95.5 | 98.7 | 94.7 | 96.2 | 90.8 | 96.1 | 95.6 | 96.2 |
| MH465403 | 95.5 | 98.8 | 94.8 | 96.5 | 91.1 | 96.0 | 95.5 | 96.5 |
| MH465488 | 99.4 | 95.6 | 94.2 | 94.9 | 90.0 | 96.0 | 92.9 | 96.0 |
| MH465487 | 97.8 | 95.0 | 93.8 | 94.6 | 90.2 | 96.4 | 92.2 | 95.5 |
| MH465491 | 97.8 | 95.2 | 93.9 | 94.8 | 90.4 | 96.5 | 92.2 | 95.6 |
| MH465467 | 94.6 | 95.8 | 94.2 | 99.4 | 91.2 | 95.2 | 96.6 | 96.6 |
| MH465466 | 94.6 | 95.9 | 94.2 | 99.4 | 91.2 | 95.3 | 96.5 | 96.7 |
| MH465465 | 94.6 | 95.8 | 94.1 | 99.4 | 91.6 | 95.3 | 96.6 | 96.6 |
| MH465428 | 95.4 | 98.9 | 94.9 | 96.5 | 91.0 | 96.0 | 95.6 | 96.5 |
| KY305202 | 95.2 | 98.8 | 94.7 | 96.1 | 90.8 | 95.7 | 94.8 | 96.2 |
| KY305201 | 95.5 | 98.8 | 94.8 | 96.6 | 91.1 | 96.0 | 96.0 | 96.5 |
| KY305200 | 94.5 | 95.9 | 94.2 | 99.3 | 91.3 | 95.2 | 96.3 | 96.6 |
| KY305199 | 94.6 | 95.9 | 94.3 | 99.3 | 91.4 | 95.2 | 96.5 | 96.6 |
| MH465481 | 94.3 | 96.1 | 93.9 | 98.2 | 91.2 | 94.9 | 94.6 | 96.2 |
| MH756609 | 94.9 | 96.2 | 94.5 | 98.9 | 91.7 | 95.5 | 96.5 | 96.5 |
| MH465443 | 94.8 | 95.9 | 94.3 | 99.5 | 91.4 | 95.3 | 96.7 | 96.8 |
| MH465442 | 94.5 | 95.7 | 94.1 | 99.2 | 91.2 | 95.1 | 96.3 | 96.5 |
| MH465471 | 94.4 | 95.7 | 93.8 | 98.9 | 91.0 | 95.1 | 96.2 | 96.4 |
| MH465470 | 94.9 | 96.1 | 94.5 | 99.4 | 91.4 | 95.6 | 96.8 | 96.8 |
| MH465469 | 94.8 | 96.6 | 94.3 | 98.8 | 92.0 | 95.5 | 95.5 | 96.6 |
| MH465468 | 94.6 | 96.3 | 94.2 | 98.6 | 91.5 | 95.2 | 95.2 | 96.4 |
| MH465431 | 95.1 | 98.3 | 94.5 | 96.3 | 91.2 | 95.6 | 95.0 | 96.2 |
| MH465430 | 95.1 | 98.4 | 94.5 | 96.1 | 90.9 | 95.6 | 95.0 | 96.0 |
| MH465429 | 95.4 | 98.9 | 94.7 | 96.3 | 91.2 | 95.9 | 95.6 | 96.4 |
| KY305204 | 94.7 | 96.0 | 94.3 | 99.4 | 91.4 | 95.4 | 96.5 | 96.8 |
| KY305203 | 94.9 | 96.1 | 94.3 | 99.5 | 91.5 | 95.5 | 96.6 | 96.9 |
| MH465482 | 94.8 | 96.1 | 94.2 | 99.2 | 91.3 | 95.6 | 96.5 | 96.8 |
| MH465472 | 95.2 | 96.5 | 94.8 | 99.3 | 91.9 | 95.9 | 97.0 | 96.9 |
| MH465489 | 96.5 | 94.5 | 93.1 | 94.1 | 89.9 | 95.1 | 91.7 | 95.1 |
3.2. Phylogenetic relationships of PCV2 isolated in southern China
Based on the subgroup terminology in a previous report [13], the phylogenetic analysis indicated that 92 strains could be divided into four genotypes: PCV2a (7/92), PCV2b (37/92), PCV2d (47/92) and PCV2h (1/92), with PCV2d being the currently prevalent genotype (Figure 2; Table 1) [18]. Further research on the genotyping of these PCV2 strains found that PCV2 strains coming from the same or different regions may fall into the different or the same subgroups (e.g. MH465458 and MH465420; EF675240 and MH465419) (Table 1; Figure 1) (Wang et al., 2009; Zhang et al., 2014), and this discovery is similar to findings of a previous study in which the hypothesis was put forward that the prevalence of different PCV2 subgroups does not have a significant geographical characteristic (Li et al., 2010). Consistently, the genetic distances between the Chinese PCV2 strains were relatively far away from one another (data not shown) and the strains of each genotype were interspersed with the same genotypic PCV2 strains worldwide.
FIGURE 2.
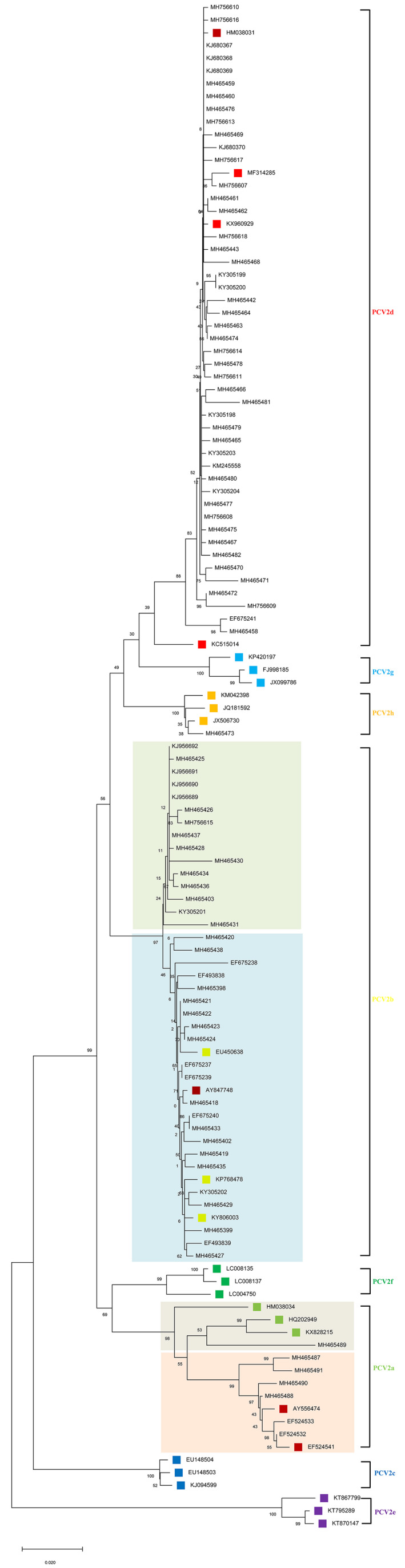
The phylogenetic tree was constructed by the neighbour‐joining method for 120 ORF2 sequences of PCV2 using MEGA v.10.0.5 software. Bootstrap values were calculated on 1,000 replicates. The 28 reference sequences are labelled by filled square
3.3. Amino acid analysis
To investigate variation in the amino acid sequences of the putative capsid protein, the deduced amino acid sequences of the ORF2 gene of 100 PCV2 strains, including the 92 Chinese strains and 8 selected representative strains, were subjected to pairwise alignment. As shown in Figure 3, the divergence at the amino acid level was greater than that of the nucleotide sequence (Table 3), displaying lower similarity that ranged from 78.6% to 100% (data not shown). Analysis of amino acid substitutions in ORF2 genes indicated that there were several major regions, located at positions 52–68 and 185–191 within the Chinese PCV2 ORF2 sequences, of high heterogeneity in this study, in spite of ubiquitous variations in the whole capsid protein sequence. In addition, the ORF2 amino acid variations have also been found at several other positions, such as at 8 (tyrosine to phenylalanine or leucine), 13 (histidine to leucine, arginine or threonine), 57 (isoleucine to valine), and 63 (lysine to threonine, serine or arginine). The nucleotide replacement rate analysis showed that the amino acids with greatly changed operating frequencies during the evolutionary process of PCV2 were asparagine, serine and lysine, suggesting that these amino acids may be involved in PCV2 evolution, although many important questions remain. The results of Tajima's neutrality test demonstrated that the indicator D value was significantly lower than zero, implying that the group size of PCV2 Chinese strains was potentially enlarging or these strains were possibly suffering some kind of directional selection. Epitope cluster analysis with the BepiPred method disclosed that there may be also several other new short epitopes, located at positions 5–40, 60–70, 80–98, 113–123, 210–213 and 220–227, which distributed dispersively across the whole capsidprotein sequence (Figure 4). This information may provide new evidence for PCV2 phenotyping.
FIGURE 3.
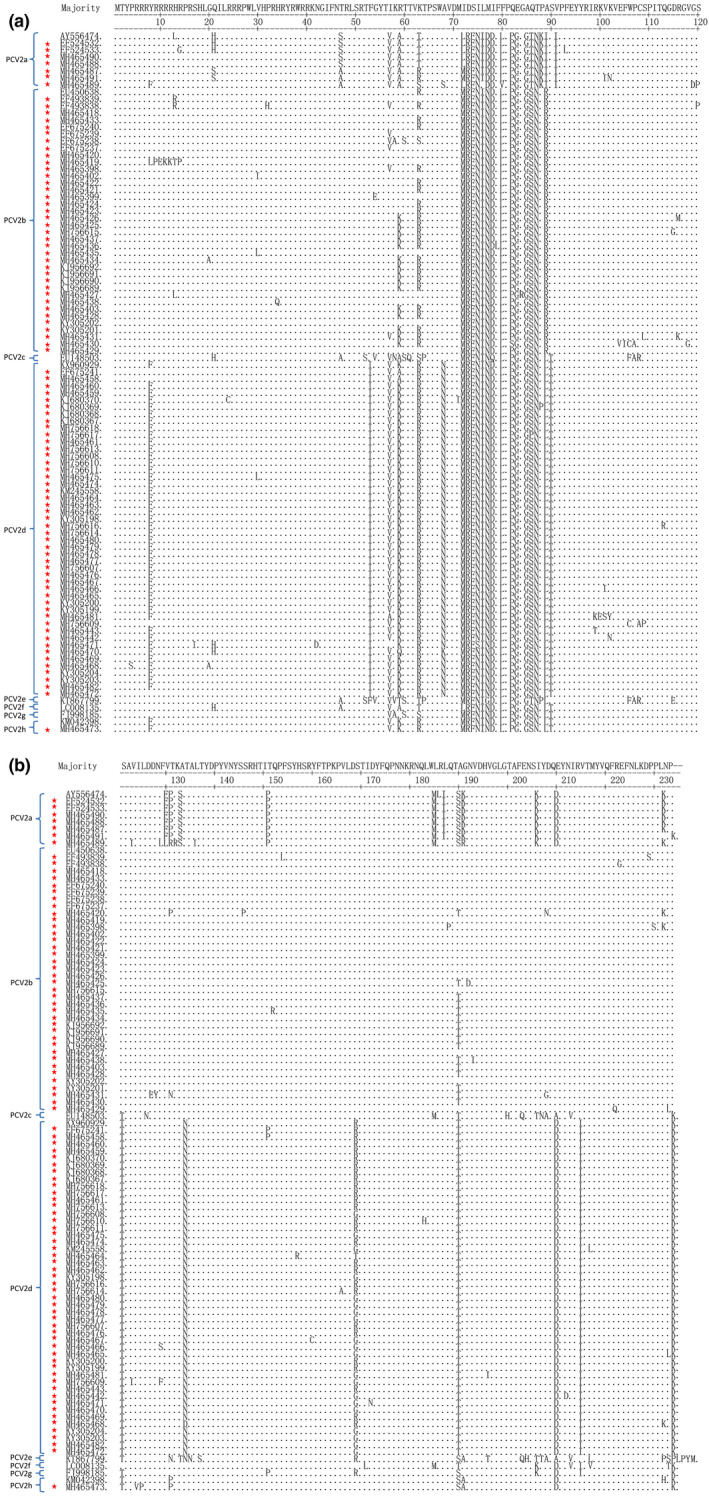
Pairwise alignment of amino acid sequences for the capsid protein of 100 PCV2 strains including 92 Guangxi strains as well as 8 selected representative sequences in the study. Residues that are exactly consistent with the consensus are indicated with dots. BJW is used as the majority sequence for this alignment (AY847748). The 92 Guangxi Beibu Gulf strains are marked with pentagram
FIGURE 4.
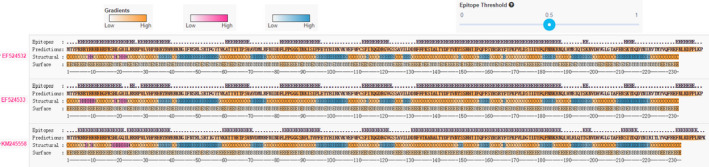
Epitopes and predictions in genes of part of the species. Epitopes: Positions above epitope threshold. Predictions: The protein sequence is displayed with an orange gradient, illustrating BepiPred‐2.0 predictions. Structural: Helix (H ‐ pink probability gradient), Sheet (E ‐ blue probability gradient) and Coil (C‐ Orange probability gradient) predicted. Surface: Buried(B)/Exposed(E) and orange gradient illustrating predicted relative surface accessibility. The 3 representative Guangxi Beibu Gulf strains are shown here
3.4. Recombination analysis
Based on the analysis performed using the RDP v.4.97 software, a total of 18 potential recombination events were detected within all 92 of the analysed PCV2 strains from southern China, and only the five recombination events that had been confirmed by at least three of the seven methods with p value cut‐off of 0.05 were reported here (Table 4). The only recombination event that was confirmed separately using the seven methods implemented in RDP4 was shown in Figure 5. Three of the recombination events likely involved direct exchanges of genetic material contained in the PCV2b and PCV2d sequences, although no evidence of the exchange of PCV2b and PCV2a sequences was detected. It is quite clear that the PCV2h sequences (MH465473) contained evidence of having acquired sequences from PCV2b sequences (EF675238 and MH465430). All the five reported recombination events involved transfers of large fragments of the ORF2 gene, a different group of the five recombination events involved transfers of small fragments of the ORF1 gene (see Figure 5 as an example). Specifically, it was observed that the recombination events primarily targeted the region between nucleotides 1,215–1,389 within the ORF2 gene (Figure 6).
TABLE 4.
Results of the identification of recombination events among 92 PCV2 genomes isolated from the southern China using the RDP, MaxChi, GeneConv, BootScan, SiScan, 3Seq and Chimaera algorithms, respectively
| Event number | Recombinant/genotype | Major parent/genotype | Minor parent/genotype | Detection methods | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | G | B | M | C | S | T | ||||
| 1 | MH465421/PCV2b | MH465460/PCV2d | MH465423/PCV2b | − | + | + | + | + | + | + |
| 2 | MH465420/PCV2b | MH465458/PCV2d | MH465418/PCV2b | + | − | − | + | − | + | + |
| 3 | MH465468/PCV2d | MH465398/PCV2b | MH465460/PCV2d | + | + | + | + | + | + | + |
| 4 | EF493839/PCV2b | MH465473/PCV2h | MH465399/PCV2b | − | − | + | + | + | + | + |
| 5 | MH465473/PCV2h | EF675238/PCV2b | MH465430/PCV2b | − | − | − | + | − | + | + |
FIGURE 5.
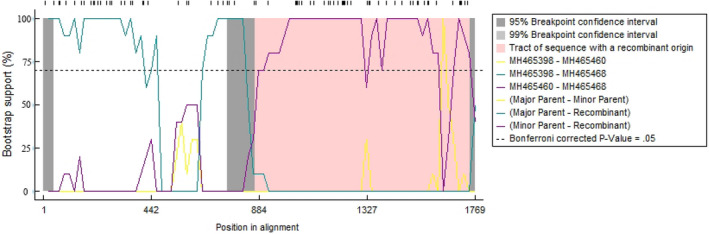
Identification of the recombination events. The most representative recombinant (MH465468) derived from recombination between MH465398 and MH465460 was shown by the means of the BootScan method with a window size 30. The y‐axis refers to bootstrap support, x‐axis represents the positions of informative sites, and the left and right bounds of the pink tract region indicate breakpoint positions
FIGURE 6.
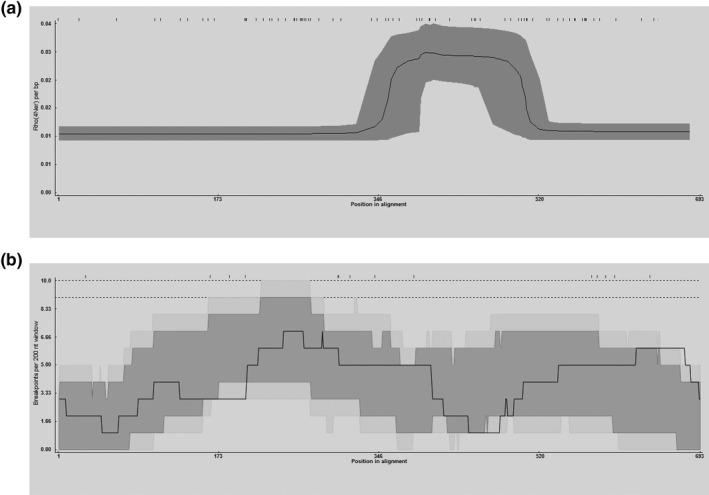
Recombination rates and distribution of breakpoints. The x‐axis refers to the positions of informative sites. The y‐axis in A represents Rho (4Ner) per base pair and the y‐axis in B represents breakpoints per 200‐nucleotide window
4. DISCUSSION
Homogeneous analysis demonstrated that the overall diversity of PCV2 strains in southern China is still low, as the lowest sequence similarity observed between any two Chinese strains was 93.7%, which is similar to the sequence similarity of 94.6% found in a previous study in 2009 (Harmon et al., 2015) and the sequence similarity of 92.7% described in another report in 2012 (Table 3) (Mu et al., 2012). However, phylogenetic and other genetic variation analyses that have aimed to elucidate the evolution and spread of PCV2 are still compelling, because the ORF2 gene of PCV2 is relatively free of recombination, which is a prominent feature of PCV2 evolution (Olvera et al., 2007).
Based on the ORF2 gene, a phylogenetic tree was generated using the neighbour‐joining method. The results indicated that the 92 PCV2 isolates from the southern China could be grouped based on two major genotypes (PCV2b and PCV2d), with PCV2d being the more predominant PCV2 genotype in this region (Figure 2 and Table 1). Meanwhile, we found that the ORF1 region in three Chinese strains was sharing high homology with that of a strain of PCV2c that was first recognized in Denmark and reported in a recent study, although no evidence of the presence of PCV2c was found in this region (Liu, Wang, Zhu, Sun, & Wu, 2016). Currently, however, whether the three Chinese strains derived from Denmark by international trade transportation or genovariation are still unclear. Notably, the size of the PCV2d group is significantly larger than those reported by Wang et al. (2009) and Mu et al. (2012), and is significantly larger than the PCV2b group, which indicates that PCV2d is undergoing an increase in population size due to some directional selection and is becoming the predominant genotype in China. This conclusion was further confirmed by a nucleotide replacement rate analysis that indicated that the D value derived from Tajima's neutrality test was significantly less than zero. Nevertheless, all of the 92 PCV2 strains analysed in this study were collected after 2006 (Table 1) (Wang et al., 2009; Zhang et al., 2014), which coincided with an increase in the severity of PCVD cases in China (Li et al., 2010). Moreover the findings are consistent with a previous report that indicated that the genotypic shift from PCV2b to PCV2d likely occurred in approximately 2010 (Franzo & Segalés, 2018; Yang et al., 2018). Coincidentally, global genetic analysis indicated that the PCV2 evolution trace was PCV2a to PCV2b to PCV2d and has occurred in many countries (Franzo & Segalés, 2018). In the USA, a variant PCV2 mutant strain designated as mPCV2b, and now grouped in PCV2d, was detected in several PCVD cases in 2012 and the prevalence rate of it appears to have increased in recent years, indicating that there is an ongoing genotype shift occurring from PCV2b to PCV2d on a global scale in these subsequent years (Franzo & Segalés, 2018; Jiang et al., 2017; Xiao, Halbur, & Opriessnig, 2012). Currently, there are series commercial vaccines that are PCV‐2b or PCV‐2a based are extensively used in this region, which phenomenon may also contribute to the evolution of PCV2 from PCV2b to PCV2d genetic subtype, as PCV2 virus is able to evolve by way of mutation and recombination in response to the wide‐spread application of these vaccines. Further phylogenetic analysis indicated that the PCV2b strains obtained from different parts of the region were closely related to each other but were more distantly related to other Chinese and foreign PCV2b strains. This finding suggests that these distinct PCV2b genotypic strains originated in and are endemic to the southern China. However, it is worth pondering whether there are more genotypes should be proposed as two different clusters that are marked by shadows with pale yellow and wathet blue, respectively are found within PCV2b genotype, and another two different clusters that are marked by shadows with grey and pink separately were also discovered within PCV2a genotype (Figure 2).
As reported previously, the Cap protein is considered the most variable structural protein of PCV2, and the amino acid variation in this region might be associated with pathogenicity and/or immunogenicity (Fenaux, Opriessnig, Halbur, Elvinger, & Meng, 2004; Mu et al., 2012). Therefore, an amino acid alignment of the Cap protein, which is encoded by the ORF2 gene, was also conducted. Our results show that there are two major regions of variation, residues 52–68 and 185–191, that correspond to two of the three dominant immunoreactive areas identified by Lekcharoensuk et al. (2004). In addition, these data show that patterns specific to each group exist, such as Chinese PCV2 strains clustered within PCV2h had one amino acid marker region located at positions 57–63 and two specific amino acid variations found at positions 124–125. However, it is strange that less variation or no significant differences were observed within the newly reported antigenic recognition regions (residues 117–131, 132–146, 156–162, 195–202 and 230–233) (Li et al., 2010; Shang et al., 2009). All of the ORF2 genes of the 47 PCV2d strains from the southern China encoded 234 aa, while the vast majority of the remaining strains encoded 233 aa, which is the same number that was encoded by ORF2 in the PCV2 strains isolated from other parts of the world (Figure 2). The extra amino acid encoded by Chinese PCV2d strains is lysine and is the penultimate residue of the Cap protein; this extra residue was identical in other PCV2d strains identified by Wang et al. (2009). Further observation indicated that some amino acid mutations of the Cap protein could be used for PCV2 genotyping, especially for differentiating the novel genotype PCV2d from other genotypes, as many amino acid variations found at specific positions were unique to the PCV2d genotype. For example, the ORF2 amino acid variations at positions F53I and A68N were unique to the PCV2d genotype. Thus, whether the above‐mentioned amino acid variations at specific positions contribute to the pathogenicity and virulence of PCV2 strains requires further investigation. In addition, some previous reports have demonstrated that there is a highly conserved putative N‐glycosylation site within the PCV2 Cap protein at amino acid residues 143–145 (NYS), which may have a tight link with the host immune responses due to its potential impact on the immunogenicity of Cap‐based DNA vaccines (Gu et al., 2012). A similar phenomenon occurred in the antigenic epitope 26‐RPWLVHPRHRY‐36 in the nuclear localization signal region of the PCV2 Cap protein (Guo, Lu, Huang, Wei, & Liu, 2011). Interestingly, the N‐terminus of the Cap protein encompassing the nuclear localization signals in all the 92 PCV2 sequences was also found to be fairly well conserved (Figure 3), further confirming the inferred importance of this site. Epitope cluster analysis revealed that there were eight potential epitopes within the Cap protein in the 92 PCV2 strains (Figure 4), of which two epitopes (residues 5–40 and 130–190) were mainly related to antigen recognition and were highly conserved in all of the 92 analysed PCV2 sequences. We noted, however, that the remaining short epitopes do not correspond to the six dominant immunoreactive areas identified in other studies, and this finding may therefore present new evidence for PCV2 adaptability.
Genetic recombination is an important evolutionary mechanism that generates diversity in PCV2. For instance, a novel PCV genotype that had apparently arisen through recombination between the ORF2 genes in the PCV2a and PCV2b strains was discovered by Cai et al. (2012). In this study, we have identified five recombinant events that occurred primarily within the ORF2 and/or ORF1 gene; four out of the five recombination events resulted in inter‐genotype recombination, whilst only one recombination event resulted in intra‐genotype recombination between PCV2b sequences (Table 4), which implies that recombination has likely played an important role in the genetic diversity of PCV2 in the southern China. Coincidentally, the finding was consistent with the previously determined result that PCV2 strains have been involved in both inter‐ and intra‐genotype recombination events (Mu et al., 2012). Due to the obvious evidence of recombination between Chinese PCV2a and PCV2b genotypes detected in other studies, we propose that recombination between different PCV2 strains in China should not be restricted to PCV2b lineages and that recombination that occurred in the ORF2 genes between different PCV2 genotypes has likely contributed to the difference in viral pathogenicity. In addition, the southern China may have acquired PCV2h strain from Vietnam by international trade, as three PCV2h representative strains were all found in Vietnam and the China borders Vietnam on the southwest; however, the specific details regarding how this occurred are still unclear, but at least further demonstrating clearly that the recombination patterns between different PCV2 strains in China are complicated and more careful research is needed in the future.
5. ETHICS STATEMENT
All the 120 PCV2 full‐genome sequences were retrieved from GenBank and thereby no ethical approval was required.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTION
Qi‐zhuang Lv: Conceptualization; Funding acquisition; Writing‐original draft. Tao Wang: Data curation; Methodology. Jia‐hua Deng: Data curation; Methodology. Yan Chen: Formal analysis; Software. Qiu Yan: Formal analysis; Software. Dao‐bo Wang: Conceptualization; Funding acquisition; Supervision; Validation; Writing‐review & editing. Yu‐lin Zhu: Conceptualization; Supervision; Validation; Writing‐review & editing.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 31860708), the Guangxi Natural Science Foundation (No. 2017GXNSFBA198025) and the Scientific Research and Technology Development Program of Guangxi (No. GK‐AA17202037), and we would like to thank PhD Xiao‐Hu Hu for technical assistance.
Lv Q, Wang T, Deng J, et al. Genomic analysis of porcine circovirus type 2 from southern China. Vet Med Sci. 2020;6:875–889. 10.1002/vms3.288
The peer review history for this article is available at https://publons.com/publon/10.1002/VMS3.288
[Correction added on 3 August 2020, after first online publication: URL for peer review history has been corrected.]
Contributor Information
Daobo Wang, Email: juanliu012@163.com.
Yulin Zhu, Email: qizhuanglv-16@ylu.edu.cn.
REFERENCES
- Bao, F. , Mi, S. , Luo, Q. , Guo, H. , Tu, C. , Zhu, G. , & Gong, W. (2018). Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transboundary & Emerging Diseases, 65(2), 432–440. 10.1111/tbed.12721 [DOI] [PubMed] [Google Scholar]
- Cai, L. , Ni, J. , Xia, Y. , Zi, Z. , Ning, K. , Qiu, P. , … Tian, K. (2012). Identification of an emerging recombinant cluster in porcine circovirus type 2. Virus Research, 165(1), 95–102. 10.1016/j.virusres.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Dupont, K. , Nielsen, E. O. , Baekbo, P. , & Larsen, L. E. (2008). Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case–control study supportsa shift in genotypes with time. Veterinary Microbiology, 128(1–2), 56–64. 10.1016/j.vetmic.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Fenaux, M. , Opriessnig, T. , Halbur, P. G. , Elvinger, F. , & Meng, X. J. (2004). Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. Journal of Virology, 78(24), 13440–13446. 10.1128/JVI.78.24.13440-13446.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo, G. , Cortey, M. , Castro, A. M. M. G. D. , Piovezan, U. , Szabo, M. P. J. , Drigo, M. , … Richtzenhain, L. J. (2015). Genetic characterisation of porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Veterinary Microbiology, 178(1–2), 158–162. 10.1016/j.vetmic.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Franzo, G. , & Segalés, J. (2018). Porcine circovirus 2 (PCV‐2) genotype updateand proposal of a new genotypingmethodology. PLoS One, 13(12), e0208585 10.1371/journal.pone.0208585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie, J. , Opriessnig, T. , Meng, X. J. , Pelzer, K. , & Buechner‐Maxwell, V. (2009). Porcine circovirus type 2 and porcine circovirus‐associated disease. Journal of Veterinary Internal Medicine, 23(6), 1151–1163. 10.1111/j.1939-1676.2009.0389.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson, S. S. , Werling, D. , Bidewell, C. , & Williamson, S. (2018). Characterisation of porcine circovirus type 2 in porcine circovirus disease cases in England and Wales. Veterinary Record, 182(1), 22–26. 10.1136/vr.104450 [DOI] [PubMed] [Google Scholar]
- Gu, J. Y. , Cao, R. B. , Zhang, Y. , Lian, X. , Ishag, H. , & Chen, P. Y. (2012). Deletion of the single putative N‐glycosylation site of the porcine circovirus type 2 Cap protein enhances specific immune responses by DNA immunisation in mice. The Veterinary Journal, 192(3), 385–389. 10.1016/j.tvjl.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Guo, L. J. , Lu, Y. H. , Huang, L. P. , Wei, Y. W. , & Liu, C. M. (2011). Identification of a new antigen epitope in the nuclear localization signal region of porcine circovirus type 2 capsid protein. Intervirology, 54(3), 156–163. 10.1159/000319838 [DOI] [PubMed] [Google Scholar]
- Guo, L. J. , Lu, Y. H. , Wei, Y. W. , Huang, L. P. , & Liu, C. M. (2010). Porcine circovirus type 2 (PCV2): Genetic variation and newly emerging genotypes in China. Virology Journal, 7, 273–284. 10.1186/1743-422X-7-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, K. M. , Gauger, P. C. , Zhang, J. Q. , Piñeyro, P. E. , Dunn, D. D. , & Chriswell, A. J. (2015). Whole‐genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announcements, 3(6), e01315–15. 10.1128/genomeA.01315-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. J. , Lian, H. X. , Yin, Y. S. , Qu, S. Z. , Li, J. , Yu, K. L. , … Liu, F. (2006). Sequencing and analysis of genome of local epidemic PCV‐ 2 strains. Progress in Veterinary Medicine, 27(7), 66–73. http://en.cnki.com.cn/Article_en/CJFDTOTAL‐DYJZ200607016.htm [Google Scholar]
- Huynh, T. M. L. , Nguyen, B. H. , Nguyen, V. G. , Dang, H. A. , Mai, T. N. , Tran, T. H. G. , … Park, B. K. (2014). Phylogenetic and phylogeographic analyses of porcine circovirus type 2 among pig farms in Vietnam. Transboundary & Emerging Diseases, 61(6), e25–e34. 10.1111/tbed.12066 [DOI] [PubMed] [Google Scholar]
- Jiang, C. G. , Wang, G. , Tu, Y. B. , Liu, Y. G. , Wang, S. J. , Cai, X. H. , & An, T. Q. (2017). Genetic analysis of porcine circovirus type 2 in China. Archives of Virology, 162(9), 2715–2726. 10.1007/s00705-017-3414-1 [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekcharoensuk, P. , Morozov, I. , Paul, P. S. , Thangthumniyom, N. , Wajjawalku, W. , & Meng, X. J. (2004). Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. Journal of Virology, 78(15), 8135–8145. 10.1128/JVI.78.15.8135-8145.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Wang, J. , Xu, S. , Cai, S. , Ao, C. , Fang, L. , … Jiang, Y. (2018). Identification and functional analysis of the novel ORF6 protein of porcine circovirus type 2 in vitro. Veterinary Research Communications, 42(1), 1–10. 10.1007/s11259-017-9702-0 [DOI] [PubMed] [Google Scholar]
- Li, G. , He, W. , Zhu, H. , Bi, Y. , Wang, R. , Xing, G. , … Su, S. (2018). Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. Advanced Science, 5(9), 1800275 10.1002/advs.201800275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. L. , Wang, X. W. , Ma, T. , Feng, Z. X. , Li, Y. F. , & Jiang, P. (2010). Genetic analysis of porcine circovirus type 2 (PCV2) strainsisolated between 2001 and 2009: Genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes, 40(2), 244–251. 10.1007/s11262-009-0438-y [DOI] [PubMed] [Google Scholar]
- Liu, J. , Wei, C. , Dai, A. , Lin, Z. , Fan, K. , Fan, J. , … Yang, X. (2018). Detection of PCV2e strains in Southeast China. Peer Journal, 6, e4476 10.7717/peerj.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Wang, F. X. , Zhu, H. W. , Sun, N. , & Wu, H. (2016). Phylogenetic analysis of porcine circovirus type 2 (PCV2) isolates from China with high homology to PCV2c. Archives of Virology, 161(6), 1591–1599. 10.1007/s00705-016-2823-x [DOI] [PubMed] [Google Scholar]
- Lv, Q. Z. , Guo, K. K. , Xu, H. , Wang, T. , & Zhang, Y. M. (2015). Identification of putative ORF5 protein of porcine circovirus type 2 and functional analysis of GFP‐fused ORF5 protein. PLoS One, 10(6), e0127859 10.1371/journal.pone.0127859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, Q. Z. , Guo, K. K. , Zhang, G. F. , & Zhang, Y. M. (2016). The ORF4 protein of porcine circovirus type 2 (PCV2) antagonizes apoptosis by stabilizing the concentration of ferritin heavy chain (FHC) through physical interaction. Journal of General Virology, 97(7), 1636–1646. 10.1099/jgv.0.000472 [DOI] [PubMed] [Google Scholar]
- Lv, Q. Z. , Guo, K. K. , & Zhang, Y. M. (2014). Current understanding of genomic DNA of porcine circovirus type 2. Virus Genes, 49(1), 1–10. 10.1007/s11262-014-1099-z [DOI] [PubMed] [Google Scholar]
- Minin, V. N. , Bloomquist, E. W. , & Suchard, M. A. (2008). Smooth skyride through a rough skyline: Bayesian coalescent‐brased inference of population dynamics. Molecular Biology and Evolution, 25(7), 1459–1471. 10.1093/molbev/msn090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, C. , Yang, Q. , Zhang, Y. , Zhou, Y. , Zhang, J. , Martin, D. P. , … Cui, B. (2012). Genetic variation and phylogenetic analysis of porcine circovirustype 2 infections in central China. Virus Genes, 45(3), 463–473. 10.1007/s11262-012-0789-7 [DOI] [PubMed] [Google Scholar]
- Nguyen, V. G. , Kim, H. K. , Moon, H. J. , Park, S. J. , Keum, H. O. , Rho, S. , … Park, B. K. (2012). Population dynamics and ORF3 gene evolution of porcine circovirus type 2 circulating in Korea. Archives of Virology, 157(5), 799–810. 10.1007/s00705-012-1234-x [DOI] [PubMed] [Google Scholar]
- Nielsen, R. (2001). Statistical tests of selective neutrality in the age of genomics. Heredity, 86(Pt 6), 641–647. 10.1046/j.1365-2540.2001.00895.x [DOI] [PubMed] [Google Scholar]
- Olvera, A. , Cortey, M. , & Segalés, J. (2007). Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology, 357(2), 175–185. 10.1016/j.virol.2006.07.047 [DOI] [PubMed] [Google Scholar]
- Opriessnig, T. , Xiao, C. T. , Gerber, P. F. , Halbur, P. G. , Matzinger, S. R. , & Meng, X. J. (2014). Mutant USA strain of porcine circovirus type 2 (mPCV2) exhibits similar virulence to the classical PCV2a and PCV2b strains in caesarean‐derived, colostrum‐deprived pigs. Journal of General Virology, 95(Pt 11), 2495–2503. 10.1099/vir.0.066423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés, J. (2015). Best practice and future challenges for vaccination against porcine circovirus type 2. Expert Review of Vaccines, 14(3), 473–487. 10.1586/14760584.2015.983084 [DOI] [PubMed] [Google Scholar]
- Shang, S.‐B. , Jin, Y.‐L. , Jiang, X.‐T. , Zhou, J.‐Y. , Zhang, X. , Xing, G. , … Yan, Y. (2009). Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Molecular Immunology, 46(3), 327–334. 10.1016/j.molimm.2008.10.028 [DOI] [PubMed] [Google Scholar]
- Sliz, I. , Vlasakova, M. , Jackova, A. , & Vilcek, S. (2015). Characterisation of porcine circovirus type 3 and porcine circovirus type 2 in wild boars (sus scrofa) in Slovakia. Journal of Wildlife Diseases, 51(3), 703–711. 10.7589/2015-01-005 [DOI] [PubMed] [Google Scholar]
- Thangthamniyom, N. , Sangthong, P. , Poolperm, P. , Thanantong, N. , Boonsoongnern, A. , Hansoongnern, P. , … Lekcharoensuk, P. (2017). Genetic diversity of porcine circovirus type 2 (PCV2) in Thailand during 2009–2015. Veterinary Microbiology, 208, 239–246. 10.1016/j.vetmic.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Wang, F. , Guo, X. , Ge, X. N. , Wang, Z. T. , Chen, Y. H. , Cha, Z. L. , & Yang, H. C. (2009). Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Research, 145(1), 151–156. 10.1016/j.virusres.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Xiao, C. T. , Halbur, P. G. , & Opriessnig, T. (2012). Complete genome sequence of a novel porcine circovirus type 2b variant present in cases of vaccine failures in the United States. Journal of Virology, 86(22), 12469 10.1128/JVI.02345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Yin, S. , Shang, Y. , Liu, B. , Yuan, L. , Zafar Khan, M. U. , … Cai, J. (2018). Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transboundary & Emerging Diseases, 65(2), e383–e392. 10.1111/tbed.12768 [DOI] [PubMed] [Google Scholar]
- Yao, J. , Qin, Y. R. , Zeng, Y. , Ouyang, K. , Chen, Y. , Huang, W. J. , & Wei, Z. Z. (2019). Genetic analysis of porcine circovirus type2 (PCV2) strains between 2002 and 2016 reveals PCV2 mutant predominating in porcine population in Guangxi, China. BMC Veterinary Research, 15, 118–128. 10.1186/s12917-019-1859-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y. S. , Huang, W. J. , Qu, S. Z. , Xie, L. H. , Liu, H. P. , Luo, T. R. , … Liao, S. H. (2007). Sequencing and analysis of genome of 9 local epidemic PCV2 strains. Journal of Guangxi Agricultural and Biological Science, 26(2), 101–105. http://www.cqvip.com/QK/97024B/200702/24835756.html [Google Scholar]
- Zhang, M. , Xie, Z. , Xie, L. , Deng, X. , Xie, Z. , Luo, S. , … Zeng, T. (2014). Two subgroups of porcine circovirus 2 appearing among pigs in southern China. Genome Announcements, 2(5), e00942–14. 10.1128/genomeA.00942-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. X. , Xie, Z. X. , Xie, L. J. , Huang, L. , Xie, Z. Q. , Liu, J. B. , … Huang, J. L. (2017). Complete genomic sequence analysis of porcine circovirus type 2 from Guangxi. Chinese Veterinary Science, 47(4), 481–488. http://en.cnki.com.cn/Article_en/CJFDTOTAL‐ZGSY201704014.htm [Google Scholar]


